Abstract
Viral glycoproteins, such as influenza hemagglutinin (HA) and human immunodeficiency virus gp41, are anchored by a single helical segment transmembrane domain (TMD) on the viral envelope membrane. The fusion peptides (FP) of the glycoproteins insert into the host membrane and initiate membrane fusion. Our previous study showed that the FP or TMD alone perturbs membrane structure. Interaction between the influenza HA FP and TMD has previously been shown, but its role is unclear. We used PC spin labels dipalmitoylphospatidyl-tempo-choline (on the headgroup), 5PC and 14PC (5-C and 14-C positions on the acyl chain) to detect the combined effect of FP-TMD interaction by titrating HA FP to TMD-reconstituted 1,2-dimyristoyl-sn-glycero-3-phosphocholine/1,2-dimyristoyl-sn-glycero-3-phospho-(1’-rac-glycerol)/cholesterol lipid bilayers using electron spin resonance. We found that the FP-TMD increases the lipid order at all positions, which has a greater lipid ordering effect than the sum of the FP or TMD alone, and this effect reaches deeper into the membranes. Although HA-mediated membrane fusion is pH dependent, this combined effect is observed at both pH 5 and pH 7. In addition to increasing lipid order, multiple components are found for 5PC at increased concentration of FP-TMD, indicating that distinct domains are induced. However, the mutation of Gly1 in the FP and L187 in the TMD eliminates the perturbations, consistent with their fusogenic phenotypes. Electron spin resonance on spin-labeled peptides confirms these observations. We suggest that this interaction may provide a driving force in different stages of membrane fusion: initialization, transition from hemifusion stalk to transmembrane contact, and fusion pore formation.
Introduction
Influenza hemagglutinin (HA)-mediated membrane fusion is the most extensively studied viral membrane fusion. HA consists of two subunits HA1 and HA2. HA1 binds to target cell receptors, and HA2 catalyses membrane fusion. At low pH, HA trimers expose their fusion peptide, which inserts into the target membrane, whereas their transmembrane domain is anchored in the viral membrane. Further conformational changes in the HA trimer result in the formation of a HA-trimer-hairpin that brings the cellular and viral membranes into close proximity, allowing them to fuse. Currently, the most popular model for membrane fusion is the stalk model, or hemifusion hypothesis. It was proposed that membrane fusion starts with formation of an intermediate membrane structure called stalk, in which the outer leaflets of the two interacting membranes are fused, forming a stalk; whereas the inner leaflets are intimately apposed, forming a diaphragm. A lateral expansion of the stalk opens a fusion pore in the diaphragm. Enlargement of the fusion pore will lead to complete membrane fusion. The hemifusion diaphragm was visualized in a model membrane fusion system recently (1), suggesting the correctness of the stalk model. However, the details of the stalk model, especially the mechanism of pore opening is still unclear.
The glycoproteins on a viral membrane, such as influenza HA2 and human immunodeficiency virus (HIV) gp41, are anchored by a single-helical-segment transmembrane domain (TMD) on the viral membranes. The TMDs share a highly conserved sequence with various envelope viruses (2). The function of the TMDs is still unclear, but it is speculated to be related to the finalization of membrane fusion, which is much less understood than the initialization of membrane fusion (2). The wild-type (WT) influenza TMD increases lipid order (3) and it associates with membrane rafts (4). However, lack of the TMDs does not have any effect on lipid mixing (i.e., the fusion between the outer layers of the opposing membranes) (5). Instead, a minimum length of TMD is required for full fusion; otherwise, the membrane fusion stops at the hemifusion stage (6). A point mutation in the TMD (G520L) induces the hemifusion phenotype (7). These results strongly suggest that the TMD is related to membrane fusion in its final steps. The interaction between influenza HA fusion peptide (FP) and TMD (8) and between HIV gp41 FP and TMD (9) has previously been shown experimentally, and for parainfluenza virus FP and TMD (10) it has been suggested theoretically. Thus, we hypothesize that the interaction between the FP and TMD is a driving force for pore opening in the final steps in membrane fusion.
Previously, we studied the effect of influenza FP (11) and TMD (3) and HIV FP (12) alone on the membranes using the electron spin resonance (ESR) method. We found that both peptides induce an ordering effect in lipid bilayers, and this effect is correlated to the fusion activity of the peptides. The effect is attributed to a result of dehydration. Therefore, we hypothesize that the interaction between FP and TMD changes the membrane structure to a greater extent than the FP or TMD does alone. The stronger perturbation is required to disrupt the relatively stable hemifusion stage as suggested in the stalk model. The formation of the extension of the transmonolayer contact (TMC) or hemifusion diaphragms (HD) requires a transition from negative curvature to zero curvature. We expect the interaction between FP and TMD will induce a perturbation to the lipid structure to facilitate this transition.
Materials and Methods
Materials
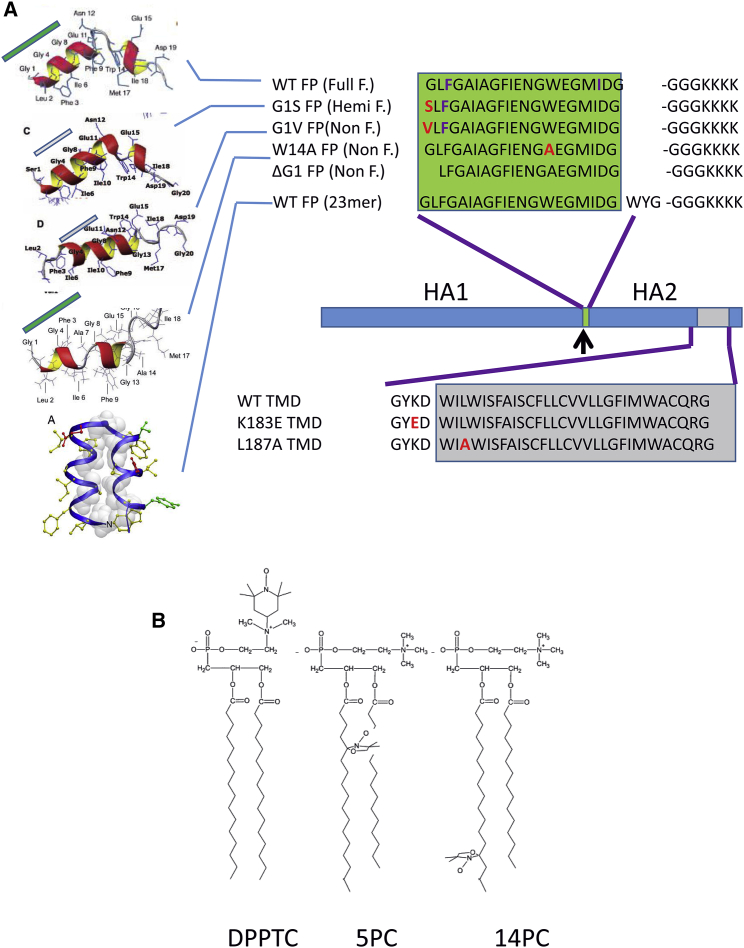
The lipids 1-palmitoyl-2-oleoyl-sn-gycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-gycero-3-phosphoglycerol (POPG), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DMPG), and the chain spin labels 5PC- and 14PC- and a headgroup spin label dipalmitoylphospatidyl-tempo-choline (DPPTC) were purchased from Avanti (Alabaster, AL). Cholesterol was purchased from Sigma (St. Louis, MO). The peptide that corresponds to the first 20 and 23 residues of the N-terminal sequence of X-31 strain influenza hemagglutinin HA2 and its mutants, and the TMD corresponding to the 188–213 residues of HA2 were synthesized by SynBioSci Co. (Livermore, CA). The structure of the spin-labeled lipids and the sequences of the peptides are shown in Fig. 1.
Figure 1.
(A) Sequences and structures of influenza hemagglutinin FP and TMD. The structures of WT (13), G1S (14), G1V (14), W14A (15), and 23 mer (16) are adapted from references. (B) Structures of spin-labeled lipids DPPTC, 5PC, and 14PC.
Vesicle preparation
The desired amount of DMPC, DMPG, cholesterol, and 0.5% (mol:mol) spin-labeled lipids in chloroform were mixed well and dried by N2 flow. The mixture was evacuated in a vacuum drier overnight to remove any trace of chloroform. For the TMD reconstituted membrane, the TMD dissolved in 1,1,1,3,3,3-hexafluoro-2-propanol (Sigma) were well mixed with the lipid mixture and dried and evacuated together. To prepare multilamellar vesicles (MLVs), the lipids were resuspended and fully hydrated using 1 ml of pH 7 or pH 5 buffer (50 mM Tris, 150 mM NaCl and 0.1 mM EDTA, pH 7, or pH 5) at room temperature (RT) for 2 h. To prepare small unilamellar vesicles (SUVs), the lipids were resuspended in pH 7 buffer and sonicated in ice bath for 20 min. To prepare large unilamellar vesicles (LUVs), the lipids were frozen and thawed five times before they were extruded in an Avanti extruder through a membrane with 100 nm pore size.
Isothermal titration calorimetry (ITC)
ITC experiments were performed in an N-ITC III calorimeter (TA Instrument, New Castle, DE). FP at 20 μM was titrated into 1 ml 5 mM SUVs at 37°C. Each addition was 10 μl and the injection time was 15 s for each injection and the interval time was 5 min. Each experiment comprised ∼25 to 30 injections. The data were analyzed with Origin (OriginLab, Northampton, MA).
ESR spectroscopy and nonlinear least squares fit of ESR spectra
To prepare the samples for lipid ESR study, the desired amounts of stock solution of the FP (1 mg/ml) was added into the lipid MLV dispersion. After 20 min of incubation, the dispersion was spun at 13,000 rpm for 10 min. The pellet was transferred to a quartz capillary tube for ESR measurement. ESR spectra were collected on an ELEXSYS ESR spectrometer (Bruker Instruments, Billerica, MA) at X-band (9.5 GHz) at 37°C using a N2 Temperature Controller (Bruker Instruments).
The ESR spectra from the labeled lipids were analyzed using the nonlinear least squares (NLLS) fitting program based on the stochastic Liouville equation (17, 18) using the microscopic order macroscopic disorder model as in previous studies (3, 11, 19, 20, 21). The fitting strategy is described below. We employed the Budil et al. NLLS fitting program (17) to obtain convergence to optimum parameters. We required a good fit with a small value of χ2 and in addition good agreement between the details of the final simulation and the experimental spectrum. All our fits in this work and our previous work meet these criteria. Because there could be more than one local minimum to which the least squares minimization converges, the process was repeated several times with different starting or seed values for the parameters. Most of the time, they converged to a common set of final parameters, but there were occasional outliers (typically unphysical), which were discarded. In addition, each experiment (and subsequent fit) was repeated two or three times to check reproducibility and estimate experimental uncertainty. Specifically, we initially varied N, Rbar, C20, C22, gib0, and gib2 in the fitting, as has been our common procedure in works (3, 11, 12, 19). We find quite generally that N, gib0, and gib2 hardly changed across a set of spectral fittings for a given spin label in a set of related experiments (e.g., the same membrane with typically six different amounts of FP), so we would then fix these parameters at the average over all six values. The NLLS fitting program was then rerun for the parameters Rbar, C20, and C22. This procedure reduced the uncertainty in these fitting parameters. Also, the very small variations in N, gib0, and gib2 across a set of related experiments hardly affected the fits. The additional features for fitting the two component spectra are discussed in the Supporting Materials and Methods.
Two sets of parameters that characterize the rotational diffusion of the nitroxide radical moiety in spin labels are generated. The first set is the rotational diffusion constants. As in the previous work (3, 11, 12) we report on the R⊥ and R||, which are derived from Rbar and N. R⊥ and R|| are respectively the rates of rotation of the nitroxide moiety around a molecular axis perpendicular and parallel to the preferential orienting axis of the acyl chain. The second set consists of the ordering tensor parameters, S0 and S2, which are defined as follows: S0 = 〈D2,00〉 = 〈1/2(3cos2θ-1)〉, and S2 = 〈D2,02 + D2,0–2〉 = 〈√(3/2)sin2θcos2φ〉, where D2,00, D2,02, and D2,0–2 are the Wigner rotation matrix elements and θ and φ are the polar and azimuthal angles for the orientation of the rotating axes of the nitroxide bonded to the lipid relative to the director of the bilayer, i.e., the preferential orientation of lipid molecules (11, 18), and the angular brackets imply ensemble averaging. S0 and its uncertainty were then calculated in well-known fashion (19) from its definition and the dimensionless ordering potentials C20 and C22 and their uncertainties found in the fitting. The typical uncertainties we find for S0 range from 1–5 × 10−3, whereas the uncertainties from repeated experiments are 4–8 × 10−3 or <±0.01 (cf. Tables S1–S12). S0 indicates how strongly the chain segment to which the nitroxide is attached is aligned along the normal to the lipid bilayer, which is strongly correlated with hydration/dehydration of the lipid bilayers (19). As previously described, S0 is the more important parameter for this study (3, 11, 12).
Spin labeling and ESR on peptides
The desired amounts of WT, G1S, G1V, and W14A FPs with a F3C or I18C mutation and a −GGGKKKK sequence in their C-termini were dissolved in 50:50 acetonitrile/buffer and mixed with 10-fold excess (S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) dissolved in the same solution for overnight in dark at RT as described previously (22, 23). The free spins were removed by dialysis against pH 7 buffer in a membrane with 2 kD cut-off size. The spin-labeled peptides were confirmed by ESR and mass spectrometry. The spin-labeled peptides were lyophilized and kept at −80°C before the experiments. The HA2 FPs were incubated with the SUVs with or without 1:200 TMD reconstituted in an 1:400 peptide/lipid ratio for 20 min before the samples were measured in the ELEXSYS ESR spectrometer at X-band (9.5 GHz) at RT. Power saturation experiments were performed on RT samples in 1) O2, 2) deoxygenated and then argon filled, and 3) deoxygenated and argon filled containing 20 mM Ni(II)EDDA as previously described (24). The insertion depth parameter Φ, based upon the saturation behavior of the samples containing oxygen and Ni(II)EDDA, was calculated as previously described (13, 24, 25). For low-temperature ESR, each sample (∼1 mg lipids with 0.5% spin-labeled FP) was rapidly frozen in thin capillaries by quickly submerging in liquid nitrogen before measurement.
Results
The binding of FP increases the ordering of membranes
We have previously shown that the binding of FP alone increases the ordering of membranes composed of POPC or DMPC in a pH-dependent fashion. We have also shown that the TMD alone increases the ordering of membranes composed of DMPC/DMPG/Chol = 40:30:30 (3). We used this lipid composition in the current study on the effects of FP-TMD on a membrane to compare it with our previous study (3). To investigate the synergistic effect of both FP and TMD, we prepared the DMPC/DMPG/Chol MLVs with different spin-labeled lipids and with or without TMD reconstitution, and determined the S0 from the ESR spectra both before and after the binding of FP. Three spin labels were used: DPPTC has a tempo-choline headgroup and the spin is sensitive to changes of environment at the headgroup region; 5PC and 14PC have a doxyl group in the C5 or C14 position of the acyl chain, respectively (cf. Fig. 1 B), and they are sensitive to the changes of environment in the hydrophobic acyl chain region at different depths. These three spin-labeled lipids have been used in previous studies and their usage in detecting the change in membrane structure has been validated (3, 11, 12).
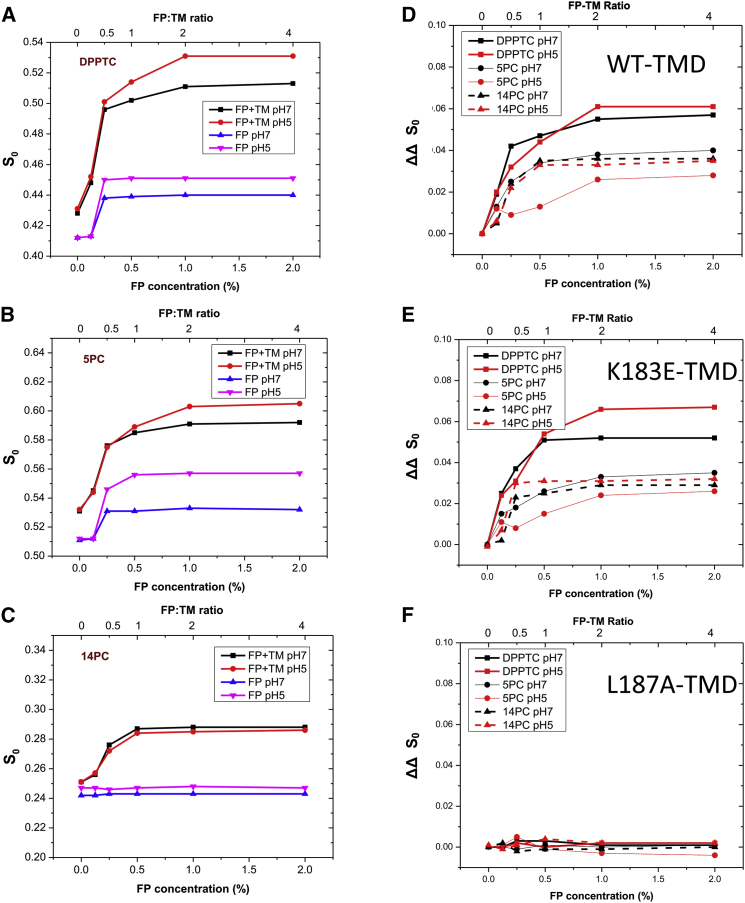
As shown in Fig. 2, A–C, and Tables S1–S12, in the pure lipid vesicles, when the peptide/lipid ratio (P/L ratio) increases from 0% to 0.25%, the S0 of DPPTC and 5PC spin labels increases significantly at both pH 5 and pH 7 conditions. But at the pH 5 condition, the increase of S0 is more significant: for DPPTC, the ΔS0,0.25% (defined as S0 with 0.25% FP binding minus S0 without FP binding) are 0.038 at pH 5 and 0.026 at pH 7; for 5PC, the ΔS0,0.5% is 0.034 and 0.021 at pH 5 and pH 7 conditions, respectively. The binding of FP has no effect on the S0 of 14PC. The results are consistent with our previous work on POPC and DMPC membranes, which confirms that the ordering effect of FP is applicable to membranes with different compositions.
Figure 2.
The plot of order parameters of DPPTC (A), 5PC (B), and 14PC (C) versus HA2 FP concentration in DMPC/DMPG/Chol = 40:30:30 MLV with (black, red) or without (blue, pink) 0.5% TMD (mol:mol) peptide in pH 7 (black, blue) and pH 5 (red, pink) buffer with 150 mM NaCl at 37°C. (D–F) ΔΔS0 of DPPTC (thick line), 5PC (thin line), and 14PC (dashed line) versus FP concentration in DMPC/DMPG/:Chol = 40:30:30 MLV with WT TMD (D) K183E TMD (E) and L187A TMD (F) reconstitution. Black, pH 7, red, pH 5.
The binding of FP further increases the ordering of TMD reconstituted membranes
It has been shown that the FP interacts with the TMD by fluorescence (8). We then tested the effect of FP binding onto the TMD reconstituted membranes. As also shown in Fig. 2, A–C, and Tables S1–S12, the binding of FP increases the ordering of DPPTC, 5PC, and 14PC at both pH 5 and pH 7 conditions. The S0 (in 0% FP binding) is different in the presence and the absence of TMD, showing that the TMD itself increases the membrane ordering. We define ΔS0 = S0 with FP binding minus S0 without FP binding, and ΔΔS0 as the ΔS0 (TMD) minus ΔS0 (pure lipid) at the corresponding FP concentrations. Fig. S4 shows the differences in the ESR spectra that we have observed, which we associate with an increase in S0 (i.e., ΔS0) only seen when WT FP is used and ΔΔS0 when both FP and TMD are used. We compare in (A), (B), and (C) the spectra of DPPTC, 5PC, and 14PC for 0.5% versus 0.125% WT FP in the 0.5% TMD reconstituted MLVs at pH 5. In Fig. S4, D–F, we show the equivalent spectra of DPPTC, 5PC, and 14PC where the WT FP has been replaced by the nonfusogenic G1V FP, for which no discernible differences were observed. In Fig. S4, G–I, we show the spectra of DPPTC, 5PC, and 14PC, for 0.5% WT FP in MLVs with 0.5% TMD versus without (both at pH 5), where key differences were observed.
As shown in Fig. 2 D, the binding of FP increases ΔΔS0 of DPPTC when the FP concentration increases. This effect is saturated when the FP/TMD ratio equals to two. The effect on ΔΔS0 of 5PC has the same pattern. To our surprise, the ΔΔS0 of 14PC is also affected by the binding of FP. Because FP alone has no effect on the ordering of 14PC, the ordering of 14PC in the TMD reconstituted membrane must be due solely from the FP-TMD synergy. The c50 (i.e., the FP/TMD ratio to induce 50% of the ΔΔS0) for DPPTC, 5PC, and 14PC are 0.38, 0.37, and 0.38, respectively. Although the FP/TMD ratio is not supposed to exceed 1:1 in the biological scenario, the ordering effects occur well before the 1:1 ratio.
Although the effect of FP alone is affected by pH, its effect from the FP-TMD interaction is much less pH-dependent. On DPPTC and 14PC, the pH basically has no effect at all; on 5PC, although there are only slight differences between two acidity conditions, the difference is not significant.
It was reported that the 23-mer FP containing WYG in the C-terminal adopts a different structure to the 20-mer WT FP (16). Our ESR shows that it exhibits a similar membrane ordering effect as the 20-mer (Fig. S5).
The binding of FP in high TMD concentration membranes induces distinct microdomains
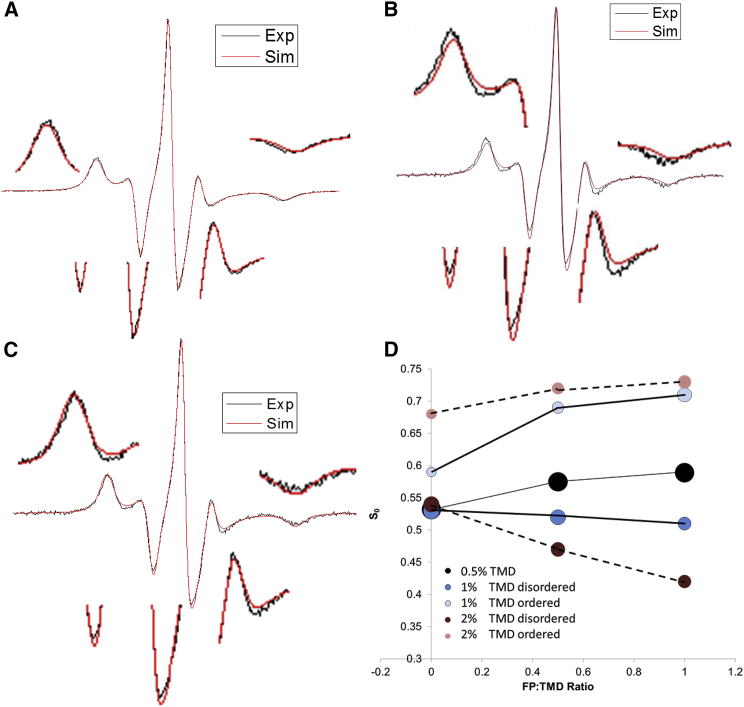
It was proposed that multiple copies of HA protein aggregate near the site of the fusion pore (26). Thus, we wanted to see whether the concentration of FP-TMD plays a role in membrane structure. We increased the TMD concentration to 1% mol:mol, and repeated the titration experiments. Previous studies showed that the 1% TMD induces two components (75% of S0 = 0.53 and 25% of S0 = 0.59 in the repeated experiments) in the bilayer, which indicates that microdomains have been induced by the TMD (3). We found that FP-TMD also induces microdomains in membranes with >1% TMD. We compare in Fig. 3, B and C, the single and double component fit of the same spectrum (1% FP + 1% TMD in MLVs at 37°C, pH 5). The reduced χ2 for the double component fit is smaller than that of the single component fit (2.23 vs. 6.41), and it is similar to the typical χ2 for the single component fit of those spectra that actually contain only one component (see Fig. 3 A). The differences between the experimental and simulated spectra are magnified in the figure by 2× in insets, and show clearly that the two-component fit is better.
Figure 3.
(A) Experimental (black) and simulated (red) 5PC ESR spectra of DMPC/DMPG/Chol = 40:30:30 MLVs with 1% FP at 37°C, pH 5. (B and C) Single (B, red) and double (C, red) component fit of the experimental (black) 5PC ESR spectra of DMPC/DMPG/Chol MLVs with 1% FP and 1% TMD at 37°C, indicate a poorer fit (B) and a good fit (C) for the same spectrum. The differences are magnified by 2× in the insets. (D) The plot of order parameters (S0) of 5PC versus HA2 FP/TMD ratio in DMPC/DMPG/Chol MLV with 0.5% (black circles, thin line), 1% (blue and shallow blue circles, thick line), and 2% (brown and pink circles, dashed line) TMD in pH 5 buffer at 37°C. In the 1% and 2% TMD MLV, two components with different S0s coexist. The sizes of the circles represent the relative proportion of each component.
We repeated the experiments on these two component samples at two temperatures (25 and 30°C) in addition to the original temperature (37°C). As shown in Table S39, fitting these spectra yields the trend in Rbar and S0, which we expected based on our previous work (19, 21) and little change in fractions of the components (also expected).
As shown in Fig. 3 D and Tables S37 and S38, the binding of 0.5% FP increased the S0 as well as the proportion of the more ordered components (63% of S0 = 0.52 and 37% of S0 = 0.69). The binding of 1% FP has a greater effect (48% of S0 = 0.52 and 52% of S0 = 0.71). The S0 of the more ordered component is higher than the ones with a 0.5% TMD and 2% FP as shown in Fig. 3 D. Control experiments (shown in Fig. S2 and Table S38) indicate that membrane with 2% TMD alone does not have such a large effect (70% of S0 = 0.54 and 30% of S0 = 0.68). Thus, it can be concluded that the FP-TMD complex has greater ability to induce discrete domains than the TMD alone. Titration of the FP into 2% TMD containing membrane (shown in Fig. 3 D and Table S38) induces two components with a larger difference of S0 between the two components. More significantly, the S0 of the less ordered domain is even lower than that of the membrane without TMD (S0 = 0.42 for 1% FP in 2% TMD containing membrane).
The more ordered domain may represent lipid associated with the FP-TMD. The spectral distinction of the domains indicates a slow exchange rate between the less ordered (e.g., the bulk lipids) and the more ordered lipids (e.g., the boundary lipids), which suggests that the FP-TMD localizes in patches in membranes with 1% and 2% TMD instead of being evenly distributed as in membrane with 0.5% TMD. The membrane with lower FP-TMD concentration may also consist of two types of lipids. However, they cannot be detected from the ESR simulations, either because of the limits of the spectral resolution, or because of fast exchange between them or both. The localization of HA near the fusion pore is observed during viral entry (27). The higher proportion of the more ordered component indicates that the area of the localized patches is larger, which may correspond to the expansion of the TMC during the real membrane fusion scenario. In the 1% TMD + 1% FP situation, ∼50% of the lipid is in the boundary state, corresponding to each peptide having ∼25 bound lipids, which is similar to the peptide/lipid ratio of gramicidin (28). The unbound lipid, constituting less ordered domains, is usually the sparser and is distributed between more ordered domains, resulting in heterogeneity of the membrane and a higher probability of water penetration through the less ordered domains and/or the boundaries between the more ordered and less ordered domains. It may facilitate a pore opening.
G1S and G1V cannot induce an FP-TMD interaction type of membrane ordering
The Gly1 mutations of influenza HA FP G1S and G1V have been studied both functionally and structurally. G1S is a hemifusion phenotype, meaning that it can mediate lipid mixing, i.e., the mixture between the two outer layers of opposite membranes, but not the inner layers. Thus, the fusion may stop at an intermediate structure (29). G1S FP has a similar structure as WT, adopting a boomerang structure (14). G1V is a nonfusion phenotype (29), adopting a linear structure instead (14).
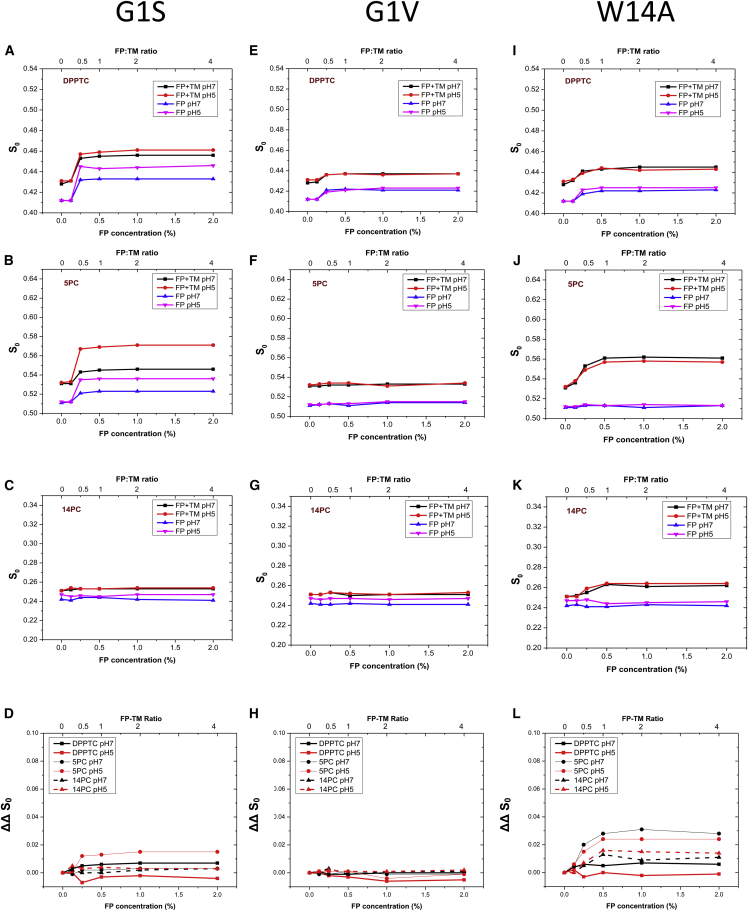
As shown in Fig. 4, A–D, and Tables S13–S18, G1S alone induces membrane ordering of DPPTC and 5PC as does WT. However, the effect is slightly smaller than that of WT. G1S has no effect on 14PC ordering. The effects on TMD reconstituted membrane are significantly different. Although G1S induces lipid ordering of DPPTC and 5PC in TMD reconstituted membrane, the effect largely results from G1S alone, not from the G1S-TMD interaction. As shown in Fig. 4 D, the ΔΔS0 for all spin labels are small compared to those of WT. The results suggest that G1S has no effect on a membrane involving FP-TMD interactions. G1V (Fig. 4, E–H, and Tables S19–S24), however, has no effect on the ordering of all three spins in both pure lipid and reconstituted membranes. The results are consistent with the fact that G1S induces lipid mixing at a similar rate as does WT, whereas G1V mediates lipid mixing at a much lower rate.
Figure 4.
The plot of order parameters of DPPTC (A, E, and I), 5PC, (B, F, and J), and 14PC (C, G, and K) versus FP concentration in DMPC/DMPG/Chol = 40:30:30 MLV with (black, red) or without (blue, pink) 0.5% TMD (mol:mol) peptide in pH 7 (black, blue) and pH 5 (red, pink) buffer with 150 mM NaCl at 37°C. (A–C), G1S FP; (E–G), G1V FP mutant; (I–K), W14A FP. (D, H, and L) ΔΔS0 of G1S FP (D) and G1V (H) and W14A (L), thick line, DPPTC; thin line, 5PC; dashed line, 14PC; black, pH 7, red, pH 5.
W14A cannot increase membrane ordering alone but can induce a FP-TMD type of membrane ordering
We next examined the effect of W14A on membrane ordering. W14A is a nonfusion phenotype. It adopts a superficially boomerang structure, but its kink region is highly flexible. Therefore, it cannot position itself in the membrane the same way as WT does. Instead, its N-terminal arm lies in the hydrophobic-hydrophilic interface and its C-terminal arm points outward from the membrane (15). As shown in Fig. 4, I–L, and Tables S25–S30, W14A alone cannot increase the ordering of spins at all three positions, which is consistent with its nonfusogenicity. However, it has a significant effect on TMD reconstituted membranes. Although W14A shows no effect on DPPTC, its effect on 5PC is similar to that of WT. Surprisingly, W14A even has ordering effects on 14PC, although it is about half of that of the WT. These results suggest that the effect of W14A is due solely to the FP-TMD interaction.
Mutation at TMD changes the membrane ordering effect
It has been shown that the TMD itself increases the membrane ordering, and two conserved mutations eliminate this effect (3). K183E and L187A are located in the hydrophilic and hydrophobic region, respectively. The insertion depth of L187 is approximately at the depth of 5PC. We studied whether these two mutations have any effect on the FP-TMD interaction-induced membrane ordering. We reconstituted these mutants in the MLV and repeated the ESR experiments using WT FP. The results (see Fig. 2, E and F, and Tables S31–S36) suggest that the FP induces similar membrane ordering effects in the K183E reconstituted membrane as in the WT TMD reconstituted membrane, but none in the L187A reconstituted membrane, which suggests that L187 is a critical residue for the FP-TMD interaction.
The N-terminal arm of the FP is the interaction site
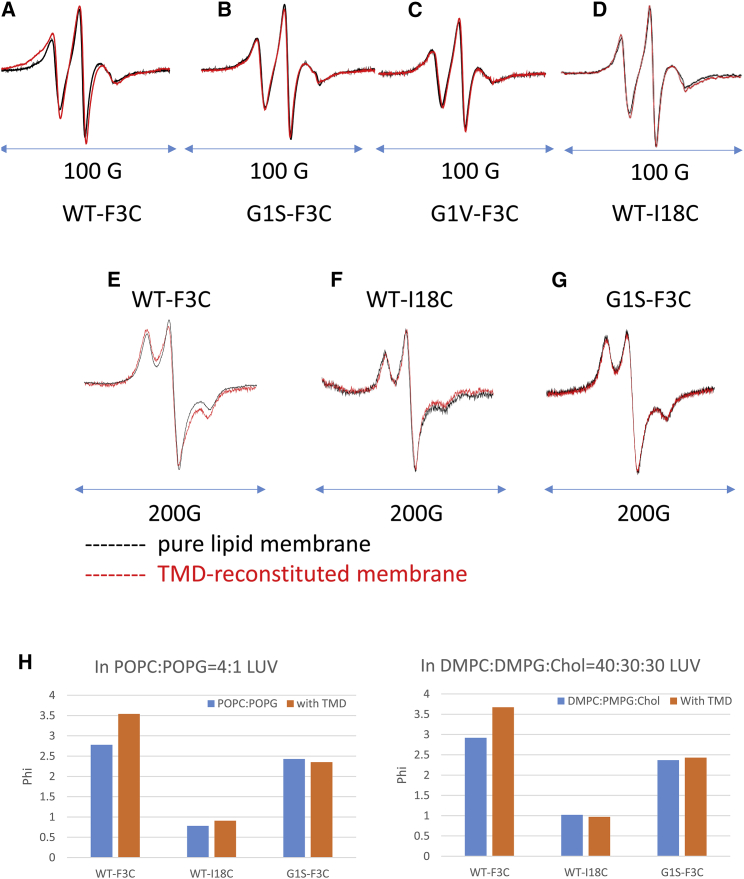
To directly investigate the interaction site of FP-TMD, we synthesized mutants with cysteine substitution, F3C and I18C and spin labeled them with MTSL (S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) (designated as F3R1 and I18R1, respectively). Those mutants were used in determining the insertion depth of the FP, and it was shown that they do not change the structure and membrane insertion of the FP (13, 23). To compare our results with the previous literature using POPC/POPG LUVs (13, 23, 30), we collected the ESR spectra in both POPC/POPG = 4:1 and DMPC/DMPG/Chol LUVs. The spin-labeled FP was bound to the LUVs with or without 0.5% TMD reconstituted, and the spectra were compared. The broadening of the spectra indicates contact between the FP-TMD in the sites that are close to the spin-labeled site. As shown in Fig. 5, A–D, in which the spectra are scaled to the same peak-to-peak amplitude, when the WT-F3R1 was bound to the TMD reconstituted membrane, there is a broadening in the central peak and a more immobile component to the low field is observed, suggesting a state of the FP that binds to the TMD (Fig. 5 A), whereas there is no significant change for the WT-I18R1 spectrum (Fig. 5 D). These results indicate that the interaction is in the N-terminal arm instead of the C-terminal arm. The G1S-F3R1 (Fig. 5 B) and G1V-F3R1(Fig. 5 C) peptides, however, do not exhibit broadening effects in the TMD reconstituted membranes. The experiments in DMPC/DMPG/Chol SUVs exhibit similar results (Fig. S3, A–D).
Figure 5.
(A–D) ESR Spectra of WT (A), G1S (B), and G1V (C) with spin labeled at F3R1 and WT with spin label at I18R1 (D), in 0.25% mol:mol ratio in POPC:POPG = 4:1 LUV membranes without (black) and with 0.5% TMD (red) at RT. (E–G), ESR spectra recorded at 90 K of WT F3R1 (E), WT I18R1 (F), and G1S F3R1 (G) mutant in POPC:POPG = 4:1 LUV membranes without (black) and with (red) 0.5% TMD. (H) Φ of WT F3R1, WT I18R1, and G1S F3R1 in POPC/POPG = 4:1 (left) and DMPC/DMPG/Chol = 40:30:30 (right) LUVs without (blue) or with 0.5% TMD (orange).
Thermodynamics of FP-TMD interaction
We determined the thermodynamics of the FP-TMD interaction by the ITC technique. The lipid-peptide interaction is characterized by the thermodynamics of partitioning instead of a classical ligand binding (31), and the interaction between FP and TMD may involve complex binding thermodynamics. To avoid such complications, we studied the FP-TMD interaction in a qualitative spirit without also measuring the binding constant of FP-TMD interaction. We used POPC/POPG = 4:1 SUVs to directly compare our results to previous literature (15, 32). In these experiments, small aliquots of FP were injected into a reaction cell containing SUVs, either composed of pure lipids or with 1% TMD reconstituted. The concentration of the lipid and TMD is much higher than the final concentration of the FP. Therefore, through the titration, the membranes and TMD can be approximately considered unsaturated, and each injection releases the same amount of heat, reflecting the enthalpy ΔH of the mixing. In the reaction of FP and TMD reconstituted SUVs, the reaction heat is the sum of heat of fusion peptide interaction with lipid, and the heat of FP-TMD interaction. The first part can be measured by injecting the peptide into pure lipid SUVs.
As shown in Table S41 and Fig. S1, the ΔH of FP-lipid interaction is around −16.08 kCal/mol, and ΔH of FP-TMD-lipid is around −21.75 kCal/mol. Thus, the net enthalpy generated by FP-TMD (ΔH (FP-TMD)) is −5.67 kCal/mol. The relatively small value of the net enthalpy indicates that the FP-TMD interaction is weak. The net enthalpy for G1S-TMD is −1.45 kCal/mol, significantly lower than that of WT. G1V and ΔG1 have little enthalpy change, which is consistent with the fact that the mutants cannot interact with TMD. These results suggest that G1S and G1V have much lower affinities to TMD, which is consistent with the fact that they have very little synergetic effects with TMD. W14A-TMD, on the other hand, has −3.19 kCal/mol reaction enthalpy, which is smaller than that of WT but significantly larger than that of G1S, indicating there is substantial binding.
The interaction energy between WT FP and the two TMD mutants was also measured. The FP-K183E has a similar enthalpy change compared to the FP-WT TMD (−4.63 kCal/mol), whereas the L187A has a significant smaller enthalpy change (−1.93 kCal/mol). Thus, this mutation in the hydrophobic region hampers the interaction between FP-TMD. The results are again consistent with the ESR experiments.
The energy barriers for the formation of an opening pore was estimated around 11–13 kbT (33), which corresponds to 7 kCal/mol. The enthalpy generated by the FP-TMD interaction is smaller than this requirement. However, multiple FP-TMD interactions could possibly provide enough energy to overcome the barrier.
The FP inserts deeper into the TMD-reconstituted membrane
We used low-temperature ESR to determine the relative insertion depth of the FP in the LUV membranes (12). When the temperature decreases to 90 K, the spectra of frozen samples of labeled FP reach the rigid limit. The 2Azz is directly measurable as the separation between the minimum of the high field and the maximum of the low field parts of the spectrum. With the exception of polarity, all factors that affect the value of outer splitting, 2Azz, are frozen in the rigid limit (24). A larger 2Azz indicates a more hydrophilic environment and a smaller 2Azz indicates a more hydrophobic environment. The hydrophobic environment indicates a deeper region in the bilayer. Thus, we correlated the 2Azz values with the insertion depth. We rapidly froze the samples to minimize any structural change during freezing. This method has been successfully exploited to study the insertion depth of HIV FP in various lipid compositions (12).
As shown in Fig. 5, E–G, when WT F3R1 binds to a membrane the 2Azz is 71.0 G. The 2Azz value in membranes containing 0.5% TMD decreases to 68.3 G. However, the 2Azz values of G1S F3R1 in the TMD reconstituted membrane and the pure lipid membranes do not differ significantly and are similar to that of WT F3R1 in pure membrane (71.5 G and 71.3 G, respectively). The WT I18R1 value changes slightly in TMD reconstituted membrane but not as significantly as the F3R1 mutant (71.6 G in TMD membrane, and 72.5 G in pure membrane). Our results suggest that N-terminus of WT FP inserts deeper into the TMD reconstituted membrane than in the pure membrane, whereas the C-terminus inserts into a similar depth in both membranes.
We also performed power saturation ESR on F3R1 and I18R1 in POPC/POPG = 4:1 LUVs with/without 0.5% TMD in the presence of Argon, O2, and Ni(II)ethylenediaminediacetic acid. As shown in Fig. 5 H, the F3R1 has a greater Φ value than I18R1 (2.7 vs. 0.8) in pure lipid membranes, indicating that F3R1 inserts deeper into the membranes than I18R1. The Φ values we obtained are similar to those previously reported (13). In the membranes with TMD, F3R1 has an even greater Φ (3.5), whereas the Φ of I18R1 is basically unchanged (0.9), indicating F3R1 inserts deeper into the membrane with TMD than into the pure lipid membrane. The Φ values of G1S-F3R1 are 2.4 and 2.5 in membranes without and with TMD, respectively, suggesting that the insertion depth is basically unchanged. Equivalent measurements carried out in DMPC/DMPG/Chol LUVs show a similar trend. The results are consistent with our low-temperature ESR results.
Discussion
The mechanism of membrane fusion is still only partially understood. Our previous work showed that the fusion peptide and TMD individually induce increased membrane ordering in a collective fashion, and we suggested that this ordering increase is associated with membrane dehydration (19). We further suggested that this dehydration due to peptide insertion is an important step to remove repulsive forces between the opposite membranes and thereby facilitate the initialization of membrane fusion (3, 11, 12). Our current study provides several further insights into this process. First of all, the combined FP-TMD interaction can induce membrane ordering to a greater extent than either alone and it extends deeper into the hydrophobic core of the membrane; it can also induce microdomains in the membrane. The nonfusogenic mutants do not have such effects. Second, the interacting site between the two is on the N-terminal arm of the FP and on the hydrophobic segment of the TMD. Third, the interaction lets the FP insert deeper into the membrane. Fourth, the hemifusion phenotype G1S induces membrane ordering, whereas the nonfusogenic G1V has no effect.
To summarize this and our previous studies on the jump in S0 as a function of FP concentration: on HA2 FP/PC interactions at pH 5 there were seven such examples with ΔS0 ∼0.02 to 0.05 (11); on HIV gp41 FP/PC interactions there were 10 examples with ΔS0 ∼0.05 to 0.11 (12); and when the concentration of TMD peptide from HA virus was varied, there were six examples with ΔS0 ∼0.04 to 0.08 (3). In this research, there are 24 examples with ΔS0 ∼0.03 to 0.10. In all 47 examples these results were compared with the appropriate nonfusogenic mutant peptides that do not show this jump. (Note that each result noted here is the average over three or two independent experiments.) We regard this as overwhelming evidence for this collective phenomenon. Similar arguments apply to the ΔΔS0 studied in this work, although with somewhat greater uncertainty. Here, we consider the individual experiments, with results that range from 0.02 to 0.08, and we have 46 such results with a ΔΔS0 in this range.
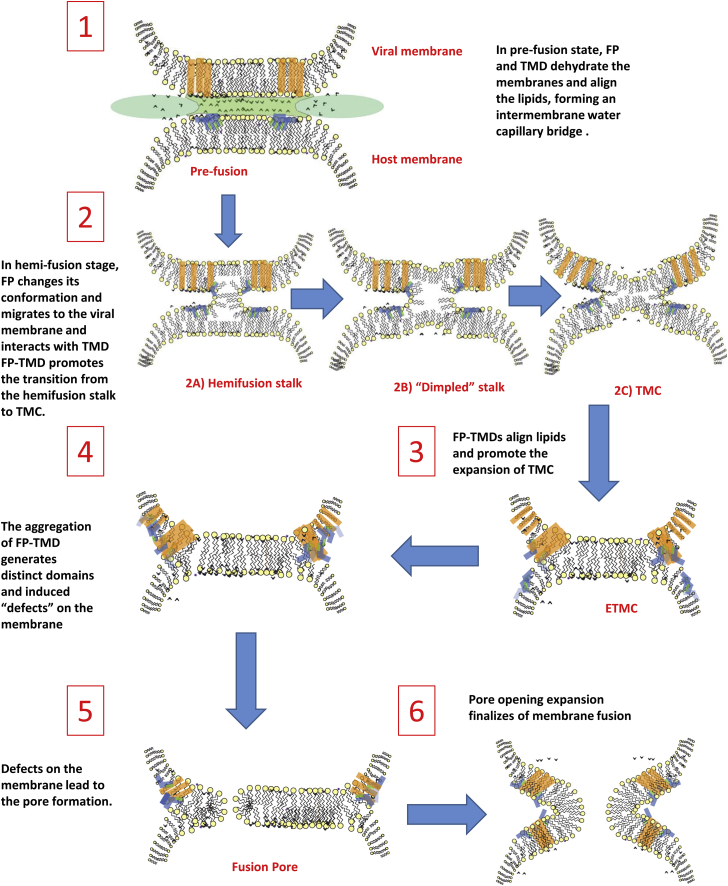
However, the implications of our studies need to be addressed. Based on our current and previous results, we considered a model of fusion, adapted from the models proposed by Tamm (34), Lentz (35) and Grubmuller (36). We emphasize the roles of the FP and TMD in the viral membrane fusion in light of our results, as discussed below (illustrated in Fig. 6).
Figure 6.
Schematic representation of the model of HA fusion peptide induced membrane fusion, adapted from (34, 35, 36). Blue, FP; orange, TMD.
Membrane bending moment induced by HA FP and highly ordered membrane domains induced by HA TMD are a prerequisite for initialization of membrane fusion: step 1
The ability for membrane ordering of the FP is correlated with the structure of the peptide as we have previously shown (11, 12). In this work, we found that the linear G1V and the flexible-kink-boomerang W14A, which cannot induce hemifusion (14, 15, 29), also cannot induce an ordering effect. On the other hand, the G1S, sharing an overall similar structure as WT and inducing hemifusion, shows the ability to induce membrane ordering. These results suggest that the ordering effect is strongly correlated with the initialization of membrane fusion (11, 19). The reason for the inability of the HA nonfusogenic peptides to induce membrane order may be their shallower insertion into the membrane because of the lack of fixed kinked structure.
It was shown that multiple HA trimers are required for fusion and these trimers interact cooperatively such that the initial fusion rate is positively correlated to the density of the trimer (37) The result may suggest that the bending moment induced by the FP must be large enough for fusion to start, which requires the cooperation of the FP.
Previously, we showed that the major effect of the HA TMD on the model membrane structure is to induce highly ordered (3), thus presumably strongly dehydrated (19), membrane domains in which the negatively charged lipids and the peptide of HA TMD are enriched. These domains form discrete hydrophobic islands on the membrane. Because HA TMD forms membrane spanning coiled-coils in both viral and model membranes (38), it is likely that the orderings in both leaflets of the bilayer are increased. This implies that incorporation of the TMD condenses both leaflets of the bilayers, thus it is not likely to generate a significant bending moment such as induced by the FP.
In the biological scenario, the FPs insert into the host membrane and the TMDs remain in the viral membrane in a close position (34). The two membranes are then more ordered than the pure lipid membrane. The effect of FP on membrane ordering is different from that of the TMD in three respects: 1) the ΔS0 is smaller; 2) the perturbation affects a shallower region, e.g., the ordering of 14PC is only affected by the TMD but not by the FP; and 3) only the outer leaflet is affected by the FP. An increase in ordering in the acyl chain region or in the headgroup region indicates that the lateral packing density in that region is increased, and/or that the local region becomes more condensed and more solid-like. Due to a coupling (39) between the different mechanical responses of two leaflets of the bilayer to the FP binding, a nonuniform distribution of stress across the bilayer, extensile in the outer leaflet and compressive in the inner leaflet, is created (11). Thus, a negative bending moment in the bilayer would be generated, which tends to bend the bilayer toward the outer surface of the vesicle (39). For a vesicle that is not closed, such as a vesicle with a fusion pore, the coupling no longer exists, but a negative bending moment in the bilayer would still be generated due to a larger cohesive (extensile) force in the more condensed outer leaflet relative to that in the fluid inner leaflet (3).
Given these results, we suggest further details in the first step of viral membrane fusion (34, 35) (Fig. 6, step 1) in the model, which is the transition from the prefusion state to the hemifusion intermediate. We suggest that when an influenza virion attacks a host membrane in a low pH medium, due to hydrophobic interaction, a liquid-liquid capillary bridge (40) is formed around the HA trimers, which link the dehydrated host membrane and the highly ordered membrane domain of the virion. Two different water phases are separated by the capillary meniscus. The capillary bridge is made up of the ordered water, which has the same phase structure as that of the ordered water on the two membrane surfaces around the HA trimers, whereas the bulk water surrounding the capillary is nonordered. The orientations of the water molecules and hydrogen bonding structure in the two water phases are significantly different (41). An attractive force is generated between the two membranes by an electric field inside the bridge as a result of polarization of water molecules, i.e., due to the preferential orientation of electric dipoles of water molecule along the axis of the bridge (42). This capillary force is responsible for dragging a PC vesicle and an influenza particle close together as observed by Lee in an electron cryo-tomography study of viral fusion (27). Thus, the removal of hydrorepulsion water and the attraction of a highly ordered water capillary pushes the membranes together and leads to the fusion of outer layers of opposite membranes, which is the hemifusion state. The mutant FPs that cannot dehydrate the membranes cannot induce the formation of the water capillary bridge, and thus they are not able to induce lipid mixing.
The FP-TMD interaction may play a role in stalk-TMC transition: steps 2 and 3
Our experiments show the FP-TMD interaction in model membranes, and indicate the structural factors for this interaction, as well as the effect of this interaction on the structure of the model membranes. This interaction requires the glycine 1 in the N-terminus of the FP and L187 of the TMD, which may be the interaction site of this complex. Our results show that the FP-TMD complex increases the lipid order to a larger extent than does the TMD or FP alone. The ordering effect also reaches deeper into the membrane than for the TMD or FP alone, which is due to the deeper insertion of the FP helped by the FP-TMD interaction as shown in our low-temperature ESR and room temperature power saturation results.
The FP-TMD interaction could have two roles in the early steps of viral membrane fusion in the biological scenario. First of all, as Bentz and Mittal suggested, in the initialization step, the fusion peptide can locate in either the host membrane or the viral membrane after its exposure (43). When inserted into the viral membrane, it allows for the FP-TMD interaction. Following the discussion in the previous sections, the larger ordering effect, interpreted as implying a greater degree of dehydration, will promote membrane fusion more efficiently. The FP-TMD interaction may not be a prerequisite, but it should facilitate membrane fusion in its first step. This could be a reason why G1S exhibits a lower lipid mixing rate in a cell-cell fusion experiment (29), because it cannot interact with the TMD.
Second, the FP-TMD induced membrane ordering effect can play a role in the stalk-TMC transition (step 2). In the biological scenario, the FP-TMD interaction will also occur in the hemifusion stage, in which the outer leaflets of the opposing membranes merge together, initially form a stalk, and then the stalk expands to a TMC intermediate (44, 45). There is no widely accepted model for the transition from the stalk to the TMC in viral membrane fusion, but the stalk may experience a transition from hemifusion-stalk to a dimpled stalk, and then to a TMC and then to an extended TMC (ETMC) or HD (35, 45, 46) (Figs. 6, steps 2A–2C and 3). It is still unclear how the FP/TMD/FP-TMD fit in the stalk structure. It is also difficult to separate the stalk intermediates for experimental studies. Therefore, the results from our model system cannot indicate directly whether FP-TMD has a role in this transition. However, we suggest that the FP-TMD-induced membrane ordering effect may play a role in pushing the stalk toward the ETMC. In the initial hemifusion stalk, the TMD most likely remains in the viral membrane because there is an energy barrier for it to overcome the void region of the stalk as it spans the bilayer. The FP, however, can migrate from the host membrane to the viral membrane via the stalk, because it only penetrates into the outer leaflet of the host membrane (Fig. 6, step 2A). Once the FP interacts with the TMD, it can align the lipids around it. This alignment would drive the balance from the hemifusion stalk to the dimpled stalk via a zipper-like mechanism. It could further drive the dimpled stalk to the TMC and to the ETMC. More studies would be required to validate this.
The FP-TMD interaction is important for the pore opening: steps 4 and 5
Our results indicate that the high FP-TMD concentration is able to induce distinct microdomains in model membranes: the more ordered domain surrounding the FP-TMD and the less ordered domain in the remaining areas. In the biological scenario, the formation of FP-TMD and the aggregation of hemagglutinin during the membrane fusion (43) increases the concentration of FP-TMD in the pore-forming site. Because of the geometric constraint on the ectodomain remaining outside of the cells the FP-TMDs are distributed in the ring area around the HD. Thus, the more ordered domains are arranged around the ring and the less ordered domain are in the core area of HD, and the ordered domains also generate a tension to the core area by the tendency to recruit more lipids in the ordered ring. The membrane consists of less ordered domains indicating that the membrane is easier for water penetration, thus making a pore opening event more likely. In the models for membrane fusion that require a stable hemifusion intermediate, the formation of fusion pores has been suggested to be a flickering expansion process (47). A large membrane tension in the HD facilitates the transition from hemifusion stage to full fusion when the HD tension is greater than the membrane tension required for membrane rupture (48). It has been suggested that the membrane rupture may arise from some molecular scale defect and the fusion-mediated protein may produce such defects (49). Our results showing the formation of ordered and disorder domains produced by FP-TMD could be just such a defect on the membrane (Fig. 6, step 4) that leads to fusion pore formation (Fig. 6, step 5). In the molecular dynamics study, when an external force was applied, the pore formation in a single bilayer is associated with disordered lipids (50, 51). The formation of the pore during viral entry possibly has a similar mechanism, i.e., the disordered domains are defects on the membrane. Our assumption that the disordered domains are away from the fusion proteins may seem somehow to disagree with the molecular dynamics study on SNARE-mediated fusion, which suggests that the leakage site is close to the rim of the HD (i.e., the protein-binding sites) (36). However, because the FP-TMDs only locate in the rim of the HD, the disordered domains are likely also near the rim of the HD.
The importance of FP-TMD for the finalization of membrane fusion is suggested by our study. Those mutations that cannot induce full fusion or pore opening (G1S and G1V FP and L187A TMD) have a lower FP-TMD interaction and a lower ability to perturb the membrane, as shown by ESR (and ITC). Interestingly, W14A, which cannot even induce lipid mixing, exhibits a certain degree of FP-TMD interaction, indicating that the ability to induce lipid mixing and the ability to interact with TMD depends on different structural factors.
Conclusions
Based on our results and discussion, we have suggested the roles of FP and TMD for the overall process of viral membrane fusion (Fig. 6). In the first step, the fusion peptide inserts into the host membrane, squeezes water molecules out of the headgroup region, and induces membrane ordering (as shown by our extensive results). The membrane ordering effect reduces the repulsion force between the membranes, and establishes a highly ordered water capillary bridge between the opposite membranes, and generates an attractive force by strong polarization of the water. These two effects drag the membranes to a proximate position and initiate the fusion of the outer layers (step 2A). The fusion of the outer layer makes the formation of FP-TMD possible. This interaction has a greater ordering effect than just the FP (as shown in our results) and will promote the alignment of lipid molecules and may promote the transition from a hemifusion stalk (step 2A) to the dimpled stalk (step 2B), and to the TMC (step 2C) and expand the TMC (step 3). When the FP-TMD aggregates in the fusion site, the highly concentrated FP-TMD then induces microdomains (as shown in our results), the more ordered domains are associated with the FP-TMD around the fusion site (step 4). The less ordered domains around the more ordered domain are perhaps also located around the edge of the TMC, which may facilitate the formation of fusion pores in the edge of the TMC (step 5). The expansion of the fusion pores then finalizes the membrane fusion (step 6).
Author Contributions
A.L.L. and J.H.F. designed research; A.L.L. performed research; A.L.L. and J.H.F. analyzed data; and A.L.L. and J.H.F. wrote the article.
Acknowledgments
We thank the Protein Facility in Department of Chemistry and Chemical Biology for allowing us to use the ITC instrument in the research. We thank Dr. Mingtao Ge, Dr. Peter Borbat, and Dr. Boris Dzikovski for helpful discussions and suggestions, and B.D. as well for his critical reading of the manuscript.
This research was supported by National Institute of Biomedical Bioengineering/National Institutes of Health (NIBIB/NIH) grant No. R01EB003150 and National Institute of General Medical Sciences/NIH (NIHGMS/NIH) grant No. P41GM103521.
Editor: David Cafiso.
Footnotes
Supporting Materials and Methods, five figures, and forty-two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)01121-2.
Supporting Material
References
- 1.Nikolaus J., Stöckl M., Herrmann A. Direct visualization of large and protein-free hemifusion diaphragms. Biophys. J. 2010;98:1192–1199. doi: 10.1016/j.bpj.2009.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroth-Diez B., Ludwig K., Herrmann A. The role of the transmembrane and of the intraviral domain of glycoproteins in membrane fusion of enveloped viruses. Biosci. Rep. 2000;20:571–595. doi: 10.1023/a:1010415122234. [DOI] [PubMed] [Google Scholar]
- 3.Ge M., Freed J.H. Two conserved residues are important for inducing highly ordered membrane domains by the transmembrane domain of influenza hemagglutinin. Biophys. J. 2011;100:90–97. doi: 10.1016/j.bpj.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 5.Leikina E., LeDuc D.L., Chernomordik L.V. The 1-127 HA2 construct of influenza virus hemagglutinin induces cell-cell hemifusion. Biochemistry. 2001;40:8378–8386. doi: 10.1021/bi010466+. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong R.T., Kushnir A.S., White J.M. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melikyan G.B., Markosyan R.M., Cohen F.S. A point mutation in the transmembrane domain of the hemagglutinin of influenza virus stabilizes a hemifusion intermediate that can transit to fusion. Mol. Biol. Cell. 2000;11:3765–3775. doi: 10.1091/mbc.11.11.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang D.K., Cheng S.F., Liu Y.T. Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 2008;6:2. doi: 10.1186/1741-7007-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reuven E.M., Dadon Y., Shai Y. HIV-1 gp41 transmembrane domain interacts with the fusion peptide: implication in lipid mixing and inhibition of virus-cell fusion. Biochemistry. 2012;51:2867–2878. doi: 10.1021/bi201721r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donald J.E., Zhang Y., DeGrado W.F. Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc. Natl. Acad. Sci. USA. 2011;108:3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge M., Freed J.H. Fusion peptide from influenza hemagglutinin increases membrane surface order: an electron-spin resonance study. Biophys. J. 2009;96:4925–4934. doi: 10.1016/j.bpj.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai A.L., Freed J.H. HIV gp41 fusion peptide increases membrane ordering in a cholesterol-dependent fashion. Biophys. J. 2014;106:172–181. doi: 10.1016/j.bpj.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han X., Bushweller J.H., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 14.Li Y., Han X., Tamm L.K. Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion. J. Virol. 2005;79:12065–12076. doi: 10.1128/JVI.79.18.12065-12076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai A.L., Park H., Tamm L.K. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J. Biol. Chem. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- 16.Lorieau J.L., Louis J.M., Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc. Natl. Acad. Sci. USA. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budil D.E., Lee S., Freed J.H. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J. Magn. Reson. A. 1996;120:155–189. [Google Scholar]
- 18.Liang Z.C., Freed J.H. An assessment of the applicability of multifrequency ESR to study the complex dynamics of biomolecules. J. Phys. Chem. B. 1999;103:6384–6396. [Google Scholar]
- 19.Ge M., Freed J.H. Hydration, structure, and molecular interactions in the headgroup region of dioleoylphosphatidylcholine bilayers: an electron spin resonance study. Biophys. J. 2003;85:4023–4040. doi: 10.1016/S0006-3495(03)74816-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge M.T., Costa-Filho A., Freed J. The structure of bleb membranes of RBL-2H3 cell is heterogenous: an ESR study. Biophys. J. 2001;80:332a. [Google Scholar]
- 21.Smith A.K., Freed J.H. Determination of tie-line fields for coexisting lipid phases: an ESR study. J. Phys. Chem. B. 2009;113:3957–3971. doi: 10.1021/jp808412x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai A.L., Moorthy A.E., Tamm L.K. Fusion activity of HIV gp41 fusion domain is related to its secondary structure and depth of membrane insertion in a cholesterol-dependent fashion. J. Mol. Biol. 2012;418:3–15. doi: 10.1016/j.jmb.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai A.L., Tamm L.K. Locking the kink in the influenza hemagglutinin fusion domain structure. J. Biol. Chem. 2007;282:23946–23956. doi: 10.1074/jbc.M704008200. [DOI] [PubMed] [Google Scholar]
- 24.Georgieva E.R., Xiao S., Eliezer D. Tau binds to lipid membrane surfaces via short amphipathic helices located in its microtubule-binding repeats. Biophys. J. 2014;107:1441–1452. doi: 10.1016/j.bpj.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altenbach C., Greenhalgh D.A., Hubbell W.L. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang R., Prorok M., Weliky D.P. A trimeric HIV-1 fusion peptide construct which does not self-associate in aqueous solution and which has 15-fold higher membrane fusion rate. J. Am. Chem. Soc. 2004;126:14722–14723. doi: 10.1021/ja045612o. [DOI] [PubMed] [Google Scholar]
- 27.Lee K.K. Architecture of a nascent viral fusion pore. EMBO J. 2010;29:1299–1311. doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dzikovski B.G., Borbat P.P., Freed J.H. Spin-labeled gramicidin a: channel formation and dissociation. Biophys. J. 2004;87:3504–3517. doi: 10.1529/biophysj.104.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao H., Armstrong R.T., White J.M. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamm L.K., Han X., Lai A.L. Structure and function of membrane fusion peptides. Biopolymers. 2002;66:249–260. doi: 10.1002/bip.10261. [DOI] [PubMed] [Google Scholar]
- 31.Seelig J. Titration calorimetry of lipid-peptide interactions. Biochim. Biophys. Acta. 1997;1331:103–116. doi: 10.1016/s0304-4157(97)00002-6. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Han X., Tamm L.K. Thermodynamics of fusion peptide-membrane interactions. Biochemistry. 2003;42:7245–7251. doi: 10.1021/bi0341760. [DOI] [PubMed] [Google Scholar]
- 33.Gao L.H., Lipowsky R., Shillcock J. Tension-induced vesicle fusion: pathways and pore dynamics. Soft Matter. 2008;4:1208–1214. doi: 10.1039/b801407h. [DOI] [PubMed] [Google Scholar]
- 34.Tamm L.K. Hypothesis: spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. Biochim. Biophys. Acta. 2003;1614:14–23. doi: 10.1016/s0005-2736(03)00159-7. [DOI] [PubMed] [Google Scholar]
- 35.Chakraborty H., Sengupta T., Lentz B.R. pH Alters PEG-mediated fusion of phosphatidylethanolamine-containing vesicles. Biophys. J. 2014;107:1327–1338. doi: 10.1016/j.bpj.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Risselada H.J., Smirnova Y., Grubmüller H. Free energy landscape of rim-pore expansion in membrane fusion. Biophys. J. 2014;107:2287–2295. doi: 10.1016/j.bpj.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danieli T., Pelletier S.L., White J.M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatulian S.A., Tamm L.K. Secondary structure, orientation, oligomerization, and lipid interactions of the transmembrane domain of influenza hemagglutinin. Biochemistry. 2000;39:496–507. doi: 10.1021/bi991594p. [DOI] [PubMed] [Google Scholar]
- 39.Evans E.A., Skalak R. CRC Press; Boca Raton, FL: 1980. Mechanics and Thermodynamics of Biomembranes. [Google Scholar]
- 40.Gögelein C., Brinkmann M., Herminghaus S. Controlling the formation of capillary bridges in binary liquid mixtures. Langmuir. 2010;26:17184–17189. doi: 10.1021/la103062s. [DOI] [PubMed] [Google Scholar]
- 41.Binder H. Water near lipid membranes as seen by infrared spectroscopy. Eur. Biophys. J. 2007;36:265–279. doi: 10.1007/s00249-006-0110-6. [DOI] [PubMed] [Google Scholar]
- 42.van Honschoten J.W., Brunets N., Tas N.R. Capillarity at the nanoscale. Chem. Soc. Rev. 2010;39:1096–1114. doi: 10.1039/b909101g. [DOI] [PubMed] [Google Scholar]
- 43.Bentz J., Mittal A. Architecture of the influenza hemagglutinin membrane fusion site. Biochim. Biophys. Acta. 2003;1614:24–35. doi: 10.1016/s0005-2736(03)00160-3. [DOI] [PubMed] [Google Scholar]
- 44.Chernomordik L.V., Kozlov M.M. Membrane hemifusion: crossing a chasm in two leaps. Cell. 2005;123:375–382. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 45.Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knecht V., Marrink S.J. Molecular dynamics simulations of lipid vesicle fusion in atomic detail. Biophys. J. 2007;92:4254–4261. doi: 10.1529/biophysj.106.103572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chanturiya A., Chernomordik L.V., Zimmerberg J. Flickering fusion pores comparable with initial exocytotic pores occur in protein-free phospholipid bilayers. Proc. Natl. Acad. Sci. USA. 1997;94:14423–14428. doi: 10.1073/pnas.94.26.14423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warner J.M., O’Shaughnessy B. The hemifused state on the pathway to membrane fusion. Phys. Rev. Lett. 2012;108:178101. doi: 10.1103/PhysRevLett.108.178101. [DOI] [PubMed] [Google Scholar]
- 49.Siegel D.P. Lipid Membrane Fusion. In: Yeagle P.L., editor. The Structure of Biological Membranes. 2nd ed. CRC Press; Boca Raton, FL: 2005. [Google Scholar]
- 50.Shigematsu T., Koshiyama K., Wada S. Molecular dynamics simulations of pore formation in stretched phospholipid/cholesterol bilayers. Chem. Phys. Lipids. 2014;183:43–49. doi: 10.1016/j.chemphyslip.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Koshiyama K., Wada S. Molecular dynamics simulations of pore formation dynamics during the rupture process of a phospholipid bilayer caused by high-speed equibiaxial stretching. J. Biomech. 2011;44:2053–2058. doi: 10.1016/j.jbiomech.2011.05.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.