Abstract
The water channel aquaporin-4 (AQP4) forms supramolecular clusters whose size is determined by the ratio of M1- and M23-AQP4 isoforms. In cultured astrocytes, differences in the subcellular localization and macromolecular interactions of small and large AQP4 clusters results in distinct physiological roles for M1- and M23-AQP4. Here, we developed quantitative superresolution optical imaging methodology to measure AQP4 cluster size in antibody-stained paraffin sections of mouse cerebral cortex and spinal cord, human postmortem brain, and glioma biopsy specimens. This methodology was used to demonstrate that large AQP4 clusters are formed in AQP4−/− astrocytes transfected with only M23-AQP4, but not in those expressing only M1-AQP4, both in vitro and in vivo. Native AQP4 in mouse cortex, where both isoforms are expressed, was enriched in astrocyte foot-processes adjacent to microcapillaries; clusters in perivascular regions of the cortex were larger than in parenchymal regions, demonstrating size-dependent subcellular segregation of AQP4 clusters. Two-color superresolution imaging demonstrated colocalization of Kir4.1 with AQP4 clusters in perivascular areas but not in parenchyma. Surprisingly, the subcellular distribution of AQP4 clusters was different between gray and white matter astrocytes in spinal cord, demonstrating regional specificity in cluster polarization. Changes in AQP4 subcellular distribution are associated with several neurological diseases and we demonstrate that AQP4 clustering was preserved in a postmortem human cortical brain tissue specimen, but that AQP4 was not substantially clustered in a human glioblastoma specimen despite high-level expression. Our results demonstrate the utility of superresolution optical imaging for measuring the size of AQP4 supramolecular clusters in paraffin sections of brain tissue and support AQP4 cluster size as a primary determinant of its subcellular distribution.
Introduction
Aquaporin-4 (AQP4) transports water across the astrocyte plasma membrane in response to osmotic gradients and contributes to physiological regulation of water homeostasis in the central nervous system (CNS), extracellular space volume and K+ dynamics after neuronal excitation and lamellipodial extension during cell migration (1). AQP4 is expressed as two major isoforms: a long isoform (M1) with translation initiation at Met-1, and a short isoform (M23) with translation initiation at Met-23 (2, 3). M1- and M23-AQP4 form both homo- and hetero-tetramers with similar water permeability. Supramolecular clustering of AQP4 was initially deduced from the observation that orthogonal arrays of particles (OAPs), a prominent feature of astrocyte end-foot membranes observed by freeze-fracture electron microscopy (FFEM) (4, 5), are absent in AQP4 null mice and that overexpression of AQP4 is sufficient to induce the formation of OAPs in heterologous systems (6, 7, 8). Subsequent work demonstrated that, when expressed alone, M1-AQP4 is found mainly as isolated tetramers, whereas M23-AQP4 forms large arrays; when both isoforms are coexpressed, array size and cell surface diffusional mobility are determined by the relative M1:M23 expression level (9, 10, 11).
Populations of AQP4 localized to specific subcellular sites are associated with distinct physiological functions. Enrichment of AQP4 at the leading edge of migrating astrocytes supports lamellipodial extension (12). Extracellular space volume and K+ dynamics are regulated by AQP4-mediated volume change in local astrocytic processes in the parenchyma (13, 14). Enrichment of AQP4 at the glia limitans and in perivascular end-feet (15) confers high water permeability to astrocyte membranes adjacent to the blood-brain barrier. Recently, we demonstrated that large AQP4 clusters preferentially localize to adhesion complexes in cultured cells and to foot-processes of cortical astrocytes in vivo, but are excluded from lamellipodial regions in migrating cells (16). These findings demonstrate that supramolecular clustering of AQP4 is an important determinant of subcellular localization and hence the water permeability of specific regions of the astrocyte plasma membrane.
Further characterization of the relationship between AQP4 cluster size and subcellular localization in situ has been hindered by the inability of conventional optical microscopy to resolve these small, dense structures in brain tissue sections. Point localization-based superresolution optical imaging methods can localize molecules with ∼20 nm precision (17, 18), and can be combined with spatial correlation analysis to determine the average size of protein clusters over defined areas of cell membranes (19, 20). Direct stochastic optical reconstruction microscopy (dSTORM) and photoactivation localization microscopy have been used previously to characterize the size of AQP4 clusters, which are believed to correspond to OAPs, on the adherent surface of cultured cells (16, 21). Point spread function (PSF) engineering with cylindrical optics allows three-dimensional (3D) localization of single molecules with high precision (22), which has been used for 3D superresolution imaging in cultured cells (23) and brain tissue slices (24). Here, we extend these methods to image the distribution of AQP4 clusters in antibody-stained paraffin sections of mouse and human CNS and in brain tumor. The methodology was validated theoretically and experimentally, and used to demonstrate the involvement of AQP4 clusters in subcellular localization to specific plasma membrane areas of astrocytes and macromolecular complexation with a K+ channel.
Materials and Methods
Materials
Immunofluorescence staining was done with rabbit or goat polyclonal AQP4 antibody (Cat. No. sc-20812/sc-9888; Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal GFAP antibody (Cat. No. MAB360; Millipore, Billerica, MA), rabbit polyclonal Kir4.1 antibody (Cat. No. APC-035; Alomone Labs, Jerusalem, Israel), goat polyclonal Myelin Basic Protein antibody (Cat. No. sc-13914, MBP; Santa Cruz Biotechnology), and AlexaFluor 488-, 546-, or 647-labeled secondary antibodies (Life Technologies, Thermo Fisher Scientific, Waltham, MA). Other reagents were from Sigma-Aldrich (St. Louis, MO).
Tissue sections
C57/Bl6 mice (age 10–12 weeks) were perfusion-fixed, and the brain or spinal cord was then removed and postfixed in 4% paraformaldehyde overnight, dehydrated and embedded in paraffin blocks. Specimens of control human brain cortex were from autopsy, and glioblastoma from a surgically removed right-brain tumor from a 63-year-old patient. A number of 5-μm-thick microtome sections were mounted directly on clean, poly-D-lysine-coated coverslips, deparaffinized and rehydrated. Antigen recovery with 10 mM sodium citrate/0.05% TWEEN (pH 6.0) at 95°C was used to facilitate staining with Kir4.1 antibody. Sections were blocked in PBS (phosphate-buffered saline) with 1% BSA (bovine serum albumin) and 0.1% Triton X-100 for 30 min, stained sequentially with primary (1 μg/mL) and secondary antibodies (0.5 μg/mL) in the same buffer, and stored in PBS before imaging.
Cell culture and transfections
Adenoviral transduction of primary AQP4−/− astrocyte cultures and injection of adenoviruses encoding M1- or M23-AQP4 into the cortex of AQP4−/− mice was done as described in Smith et al. (16) using protocols approved by the University of California at San Francisco Institutional Animal Care and Use Committee. Primary cultures of mouse astrocytes were generated from the cortex of AQP4+/+ and AQP4−/− mice and used for experiments after 14 days. Cell monolayers were infected with adenovirus encoding M1-AQP4 or M23-AQP4 (25) at 4 × 107 colony forming units/mL for 24 h, after which time the media was changed and cells were allowed to recover for a further 24 h before fixation and staining. Cells transfected with these AQP4-expression cassettes produce only a single band at 32 kDa (M1-AQP4) or 30 kDa (M23-AQP4) after lysis and immunoblotting (26). For intracerebral injection of adenovirus, AQP4−/− mice (C57/Bl6 background, 10–12 weeks) were anesthetized and mounted on a stereotactic frame. A midline scalp incision was made and a burr hole of 1 mm diameter was drilled in the skull 2 mm to the right of the bregma. A 30-gauge needle attached to a 50 μL gas-tight glass syringe (Hamilton Instrument, Cinnaminson, NJ) was used to infuse adenovirus solution containing 105 CFU/μL (∼1 μL/min). Mice were sacrificed after Day 7 and brains were processed for sectioning. Sparsely labeled individual astrocytes in the cortex were imaged after staining for AQP4. U87-MG cells were cultured and transiently transfected with M1-AQP4 or M23-AQP4 in pcDNA3.1 as described previously in Smith et al. (16), and cotransfection was done with Kir4.1 in pCMV-XL4 (OriGene Technologies, Rockville, MD).
Methods
Confocal microscopy
Fluorescently stained tissue sections prepared as above were mounted in ProLong Gold antifade (Life Technologies) and imaged using a laser-scanning confocal microscope (Eclipse C1; Nikon, Melville, NY) using 20× N.A. 0.5 or 100× N.A. 1.4 objective lenses.
Superresolution microscopy
Imaging was done on a TE-2000E inverted microscope (Nikon) equipped with a 100×, 1.49-NA total-internal-reflection-fluorescence objective, autofocus unit, 405, 488, 561, and 642 nm diode lasers, acousto-optic tunable filter (AA Opto-Electronic, Orsay, France), quad-band filter cube with fluorophore-specific emission filters (Semrock, Rochester, NY) mounted on a Lambda 10-3 optical filter changer (Sutter Instrument, Novato, CA), and an electron-multiplying charge-coupled device camera (QuantEM:512SC; Photometrics, Tucson, AZ) under control of Micro-Manager 1.4 software (https://www.micro-manager.org). Experiments were performed using highly inclined wide-field illumination (27) resulting in an ∼500-nm excitation-field depth. A 5 m focal-length cylindrical lens (CVI Laser Optics, Albuquerque, NM) was used to introduce astigmatism in the image plane. Imaging was done in PBS containing an enzymatic oxygen-depleting system (0.5 mg/mL glucose oxidase, 40 μg/mL catalase, 10% glucose) and 100 mM MEA (β-mercaptoethylamine); depletion of oxygen was allowed to proceed for at least 20 min before addition of MEA. Single images showing total fluorophore distribution in the inclined illumination mode were captured at low laser intensity, then fluorophores were switched to dark-state by increase of laser power; spontaneous reactivation of individual fluorophores (blinking) was then recorded for 20,000 frames and 20 Hz as described by van de Linde et al. (28). For two-color imaging with Alexa 647 and Alexa 546, the same buffer system was used, and the Alexa647 images were captured first, in the manner described above. Alexa 546 blinking was then captured for 50,000 frames at 50 Hz.
Image processing
Initial image processing and analysis were performed using the SMLM plug-ins for the software Image J (http://www.sussex.ac.uk/gdsc/intranet/microscopy/imagej/smlm_plugins). The 3D localization and image generation were done using the software MATLAB (The MathWorks, Natick, MA). Candidate peaks were detected based on local signal at least 50 photons above background and then fit to a 2D Gaussian function with the Levenberg-Marquardt algorithm. Blinking events that spanned multiple frames and fast flickering events that occurred within 10 frames (0.5 s) and 1 pixel (107 nm) of the preceding blink were extracted and refit from combined images. Drift correction was performed using either added fluorescent spheres or 6–10 heavily labeled endogenous structures as reference. Localization precision was calculated from the number of detected photons in each blink using the definition of Mortensen et al. (29), with the precision adjusted by a factor of to account for the increased shot noise associated with the use of an electron-multiplying charge-coupled device camera. Blinks with localization precision >50 nm in the x,y plane were discarded from further analysis. Astigmatism was calibrated from stack images of 100-nm-diameter fluorescent microspheres (Tetraspeck Microspheres; Life Technologies) imaged at 25 nm steps along the z axis. The width in x (wx cal) and y (wy cal) at each z position were separately fitted to a function, . In experimental samples, 2D Gaussian fits of individual blinks were then localized in the z axis by minimization of D in the function, (23). Localizations were rejected if the minimum value of D was >100 nm. The z axis coordinates were multiplied by 0.8 to correct for distortion due to refractive index mismatch. Blink localization coordinates were rendered as image stacks with 10-nm voxels and filtered for display with a 3D-Gaussian blur of width 20 nm in the x- and y dimensions and 50 nm in the z dimension.
dSTORM simulations
Simulations used the create data function of SMLM plugins. Label blinking characteristics were experimentally measured from the blinking intensity and duration of single, immobilized Alexa647-conjugated secondary antibodies in standard dSTORM buffer. Blinking parameters, together with the measured 3D microscope PSF, were used to generate simulated image sequences over a 100 × 100 pixel region. Analysis and processing of these image sequences was done as described above for experimental data sets.
Statistical analysis of AQP4 clustering
Segmentation analysis was performed with a watershed algorithm followed by binary thresholding and particle counting. Pair autocorrelation was calculated by fast Fourier transform (FFT) of binary dSTORM images using the method of Veatch et al. (20), , where FFT and FFT−1 are forward and reverse Fourier transforms, Im is the image, ρ is the density of blinks, and N(r) is a normalization factor to account for the size of the region of interest (ROI) being analyzed. The pair-correlation function was calculated for small, contiguous ROIs of at least 2 μm2 within planar membrane regions where the Euclidean distance between points is a reasonable approximation of geodesic distance along the 3D surface. Analysis was limited to 2D projections of areas that were flat in the x,y plane (<100-nm variation in z coordinate over a 2 μm2 area) due to the greater uncertainty of localization precision in the z axis. ROIs were drawn to exclude contributions from endogenous autofluorescence. Correlations were corrected for repetitive fluorophore blinking by subtracting the effective PSF, , where d is the average number of blinks/molecule, σ is the average localization precision of blinks, and ρ is the density of blinks. The pair correlation function was then fit to the exponential , where A is related to the ratio of the density of localizations in clusters to the total density, and ξ is the domain radius. The number of molecules per cluster (N) was calculated as described by Sengupta et al. (19), where N = 2Aπξ2ρ/d. Calculation of the cross-correlation function of Alexa-647 and Alexa-546 localizations in two-color experiments was performed using Eq. 9 of Sengupta et al. (30).
Results
Quantitative validation of AQP4 superresolution imaging in astrocyte end-feet
Astrocyte end-feet contain AQP4 in orthogonal arrays of particles composed of 10–100 tetramers at densities as high as 100–300 μm−2 (8). System resolution in superresolution microscopy depends on both the localization precision of individual blinking events and the density of detected molecules with typical resolution estimates of 30–50 nm (31); localization of molecules at high density is facilitated by the use of fluorophores with low blinking frequency (32). To determine whether point localization-based superresolution microscopy can resolve the small, dense AQP4 clusters in astrocyte end-feet, we measured the blinking characteristics of individual fluorescent labels and used these in computational simulations of the effects of cluster size and density in dSTORM imaging. Blinking parameters of individual Alexa-647-labeled antibodies immobilized on glass coverslips at low density were measured in buffer containing 100 mM MEA as a reducing agent over the course of 20,000 frames acquired at 20 Hz (Fig. 1 A).
Figure 1.
Influence of cluster size and density on interpretation of dSTORM experiments. (A) Fluorescence of a single, immobilized Alexa647-labeled secondary antibody in buffer containing 100 mM MEA for 20,000 frames at 20 Hz. Fluorophore blinking seen as upward ticks in fluorescence intensity. (B) Distribution of on-time per blink (ton), off-time per blink (toff), number of blinks/antibody, and emission rate measured for 297 individual blink events from 99 individual labeled antibodies. (C) Superresolution images generated from simulations for AQP4 under disperse conditions, in small clusters (30 tetramers) and in large clusters (90 tetramers), at low (300 tetramers/μm2) and high (3000 tetramers/μm2) AQP4 density. (D). Blinking-corrected pair-correlation function, g(r), calculated at 10-nm intervals from the data sets in (C) (solid circles) and fitted to a single exponential (solid line). (E) Cluster size calculated from fitted amplitude and exponential time constant of g(r) as a function of AQP4 density, for small and large clusters (dotted lines). To see this figure in color, go online.
Images of 99 individual antibodies, with 297 detected blinks, were used to generate histograms of blink duration, interblink interval, number of blinks/antibody, and photon emission rate during each blink (Fig. 1 B). Blink duration was fitted to a single exponential with a time constant of 120 ms; interblink interval was fitted to a double exponential with a fast time constant of 250 ms and a slow time constant of 250 s; the mean emission rate was 4000 photons/s; and, on average, there were 1.5 fast and 1.5 slow blinks per antibody over the experimental course of 20,000 frames. AQP4 labeling density was measured in CHO cells transfected with M23-AQP4 (data not shown), which forms large and spatially well-segregated clusters of >100 individual tetramers (9). Similar labeling efficiency is expected to occur in brain sections. The deduced labeling density was 10,000 μm−2 within clusters, equivalent to 1 label for every 2.8 AQP4 tetramers given the unit spacing of ∼6 nm for AQP4 in orthogonal arrays (4). The staining conditions used are therefore unlikely to generate spurious clustering due to multiple labeling of AQP4 tetramers and the labeling density and localization precision predict a system resolution of ∼31 nm (from Eq. 3 in Gould et al. (31)).
We performed experimental simulations to determine whether fluorophore-labeled antibodies, with the experimentally determined blinking characteristics and labeling densities, would be able to distinguish clustered antigens with varied cluster size and density. Fig. 1 C shows simulated superresolution images for a 2 × 2 μm region containing randomly distributed AQP4 tetramers, small clusters (average size 30 molecules) and large clusters (average size 90 molecules), at low (300 tetramers/μm2) and high (3000 tetramers/μm2) density. Individual small and larger clusters can be resolved visually at low and high densities. Pair-correlation analysis has been used to correct for multiple appearances of individual molecules and to quantify cluster size (19), but the effect of cluster size and molecular density on the accuracy of this approach has not been systematically explored. Fig. 1 D shows calculated pair-correlation functions, g(r), and fitted single-exponential functions for each of the conditions in Fig. 1 C, demonstrating that disperse labeling, small clusters, and large clusters can be readily distinguished at both high and low densities. Fig. 1 E shows the estimated cluster size deduced from the amplitude and interaction distance of the pair correlation function for large and small clusters at AQP4 tetramer densities between 30 and 10,000 molecules/μm2. These results support the feasibility of measuring AQP4 cluster size at the densities observed in astrocyte end-feet; however, at very high density, the accuracy of cluster size determination is reduced.
To apply this methodology in brain and spinal cord tissue sections, where astrocyte end-feet are sectioned at arbitrary angles within complex 3D geometry, we localized each molecule in 3D space by deforming the PSF with a cylindrical lens. Calibration of the degree of deformation in the x- and y dimensions as a function of distance from the focal plane was performed with subdiffractive fluorescent microspheres imaged along the axial plane (Fig. 2 A, top and left) and fitted to an elliptical Gaussian function (Fig. 2 A, bottom right). Paraffin sections, mounted directly on glass coverslips, were processed using standard immunostaining techniques and illuminated in a highly inclined mode (27) (Fig. 2 B, top left). The Alexa-647 label was induced to blink by intense laser illumination in reducing buffer; localizations were captured for 20,000 frames and plotted as image stacks, where intensity represents localization density, with 50-nm axial resolution (Fig. 2 B, bottom and right).
Figure 2.
Quantification of AQP4 cluster size in planar regions of tissue sections. (A) Axial position of 0.1-μm-diameter fluorescent beads measured using a cylindrical lens in the emission path. Images show the average of 10 beads imaged at the indicated axial position. (Graph, lower-right) Average width in x and y (wx, wy) as a function of z position. (B) Wide-field image of a paraffin section from mouse brain cortex immunolabeled for AQP4 (top left), and reconstructed superresolution images (see solid boxed region, pseudocolored for localization density), showing localizations from three axial planes through the indicated capillary. (C) Magnified view of dashed-boxed region from (B) with individual AQP4 clusters visible (left) and blinking-corrected pair correlation function, g(r) (right). (D) Detection of clusters from the same region as in (C) by watershed-based segmentation and thresholding (left) and quantification of cluster size distribution by particle counting (right). To see this figure in color, go online.
Two separate approaches were used to quantify AQP4 cluster size in 2D optical sections of 50 nm depth. First, pair-correlation analysis (with correction for multiple blinking of secondary labels) was applied to raw localizations within contiguous, planar regions in a single axial section to determine the pair correlation function for localizations within that 2D region (Fig. 2 C). The pair correlation function was then used to estimate the mean cluster size within that region, calibrated for labeling efficiency as determined in vitro. Second, to estimate the size distribution of clusters within the same regions, segmentation and thresholding of localization density maps (Fig. 2 D, left) was used to identify individual clusters, and the cluster size was measured from the resulting binary image (Fig. 2 D, right). The cluster size, as estimated by fitting an exponential to the pair correlation function in Fig. 2 C, was 39 AQP4 molecules; the mean cluster size in Fig. 2 D was 1290 nm2, corresponding to 35 AQP4 molecules.
M23-AQP4 is required for formation of clusters in vivo
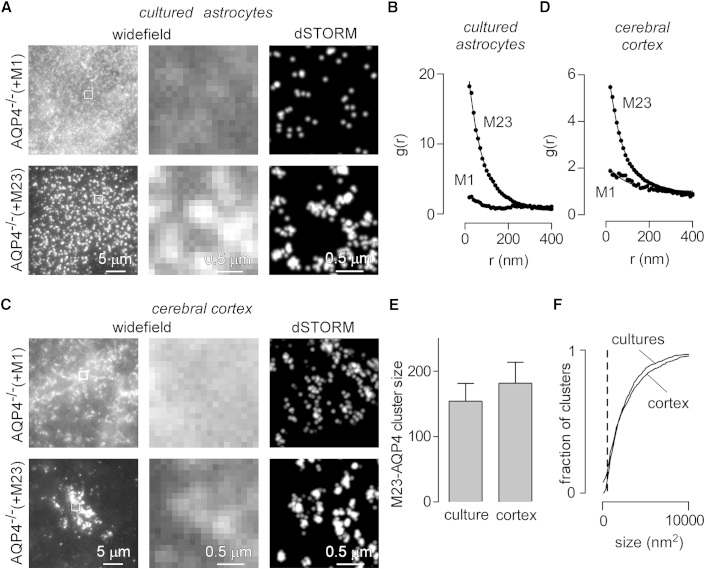
Cultured cells transfected with only M23-AQP4 form large clusters whereas those expressing only M1-AQP4 do not (9). M23-AQP4 clusters do not polarize in cultured astrocytes, but they are polarized to perivascular regions in cortical sections of AQP4−/− mice virally transduced with M23-AQP4 (16). To determine whether the interactions involved in polarization change cluster size, we imaged astrocyte cultures and cortical sections from AQP4−/− mice after viral transduction of M1- or M23-AQP4. Fig. 3 A (left panels) shows the diffuse localization of M1-AQP4 and the clustered localization of M23-AQP4 at the basal plasma membrane of primary astrocyte cultures from neonatal brain cortex of AQP4−/− mice expressing M1- or M23-AQP4 isoforms individually. Discrete AQP4 clusters could be resolved by superresolution imaging (Fig. 3 A, right panels). Label density was similar in M1- and M23-AQP4-transduced cells; differences in the pair correlation function of AQP4 localizations in the two cell types are attributable to the marked clustering of M23-AQP4 but not of M1-AQP4 (Fig. 3 B).
Figure 3.
M23-AQP4 clusters in astrocyte cultures and cortical astrocytes in mouse brain. (A) Wide-field (left panel, with center panel showing magnified view of the boxed region) and dSTORM (right panel) images of astrocyte cultures from AQP4−/− mice that were transduced with M1- or M23-AQP4 and immunostained for AQP4. (B) Averaged g(r) measured from 10 separate regions of astrocytes expressing M1- or M23-AQP4. (C) Wide-field and superresolution images of AQP4 in paraffin sections of cerebral cortex of AQP4−/− mice at 3 days after injection of adenovirus encoding M1- or M23-AQP4. (D) Averaged g(r) of AQP4 localizations in cortex, showing average from 10 separate regions of at least 1 μm2 for each condition. (E) Average M23-AQP4 cluster size in astrocyte cultures and cortical sections determined from g(r) in (B) and (D). (F) Size distribution of AQP4 clusters determined by segmentation analysis and particle counting of the regions analyzed in (B) and (D); (dashed line) minimum resolvable size of 400 nm2.
Confocal imaging of brain sections from AQP4−/− mice virally transduced with individual AQP4 isoforms in vivo showed diffuse localization of M1-AQP4 throughout the plasma membrane of astrocytes, whereas M23-AQP4 is enriched around blood vessels (16). A similar difference in distribution was seen in wide field images of paraffin sections under highly inclined illumination (Fig. 3 C, left panels). dSTORM imaging, under conditions where localizations were restricted to a 100-nm-thick section in the axial plane, showed well-demarcated individual clusters in the M23-AQP4-transduced astrocytes (Fig. 3 C, right). As in the astrocyte cultures, pair-correlation analysis, applied to areas with similar labeling density for M1- and M23-AQP4, demonstrated marked clustering of M23-AQP4 but not of M1-AQP4 (Fig. 3 D).
The M23-AQP4 cluster size calculated from the pair correlation function, after correction for difference in the amount of label in vivo and in vitro, demonstrated that the M23-AQP4 cluster size was similar in astrocyte cultures and astrocytes in brain tissue (Fig. 3 E). This finding was confirmed when the size distribution of M23-AQP4 clusters was measured by segmentation analysis (Fig. 3 F). These results demonstrate that differential clustering of M1- and M23-AQP4 can be detected by superresolution imaging in tissue sections, extending prior in vitro results (21) and showing that M23-AQP4 forms similar-sized clusters in vivo to those observed in cell culture models.
Subcellular distribution of AQP4 clusters in cerebral cortex and spinal cord
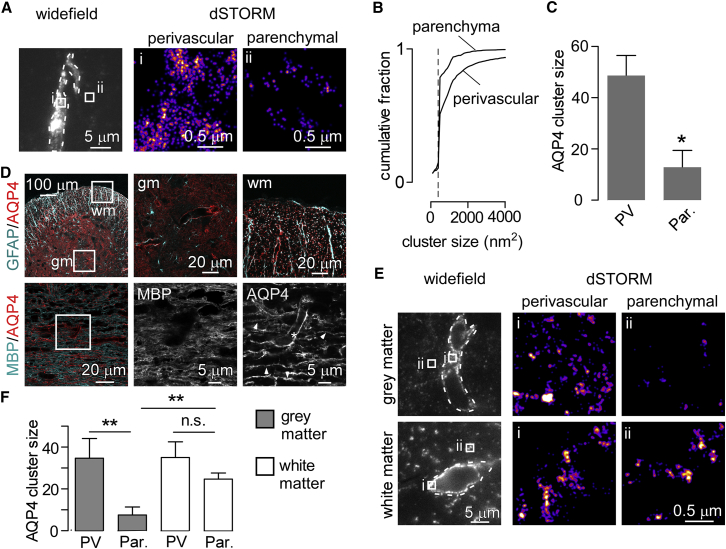
Coexpressed M1- and M23-AQP4 form heterotetramers that further assemble into clusters with a broad distribution of size and cell surface diffusional mobility (10). M23-AQP4-mediated cluster formation is required for AQP4 polarization (16) suggesting that, within a heterogeneous population of cluster sizes, larger clusters may preferentially polarize to end-feet. AQP4 polarization along the perivascular wall was clearly seen in wide-field images (Fig. 4 A, left panel), and individual clusters could be resolved in superresolution images where the vessel wall is sectioned in the x,y plane (Fig. 4 A, center panel). In parenchymal areas, both small clusters and disperse AQP4 labeling were seen (Fig. 4 A, right panel). Segmentation-based quantification of cluster size in these regions demonstrated a difference in the distribution of cluster sizes between end-feet and the rest of the astrocyte, with a higher fraction of large clusters in perivascular regions (Fig. 4 B). Mean cluster size, deduced from pair-correlation analysis in perivascular and parenchymal regions, also showed large differences in average cluster size (Fig. 4 C). These data support the conclusion that larger AQP4 clusters are found in end foot than in parenchymal areas of the astrocyte plasma membrane.
Figure 4.
Subcellular distribution of AQP4 clusters in cortical and spinal cord astrocytes. (A) Distribution of AQP4 clusters in perivascular and parenchymal regions of astrocytes in cerebral cortex. (Left panel) Wide-field images of AQP4 staining obtained with highly inclined illumination. Superresolution images (center and right panels) of AQP4 clusters in perivascular and parenchymal regions of cortex from the indicated regions. (Images are pseudocolored for localization density.) (B) Distribution of cluster sizes by segmentation analysis and counting in perivascular or parenchymal regions; (dashed line) minimum resolvable size of 400 nm2. (C) Average cluster size calculated from fitting g(r) in perivascular and neuropil regions (9 perivascular and 12 neuropil regions from three sections, ∗∗p < 0.01 by t-test). (D) Distribution of AQP4 in gray and white matter astrocytes of spinal cord. (Top panels) Transverse spinal cord section with AQP4 labeling in GFAP-positive astrocytes in gray matter (gm) and white matter (wm). (Lower panels) Longitudinal sections of white matter demonstrating enrichment of AQP4 in both perivascular regions (asterisk) and in processes adjacent to myelinated fibers (arrowheads). (E) Wide-field and superresolution images of AQP4 clusters in perivascular and parenchymal regions of spinal-cord gray and white matter. (F) Mean AQP4 cluster size determined from g(r) in paravascular and parenchymal region of gray and white matter (9–12 regions from three separate experiments, ∗∗p < 0.01 by t-test; n.s., not significant). To see this figure in color, go online.
Substantial variation in the subcellular distribution of AQP4 has been observed in different regions of the CNS and between different subpopulations of glial cells (33, 34, 35). White matter astrocytes of the spinal cord display AQP4 enrichment throughout GFAP-positive processes, and AQP4 is less polarized than in gray matter (36). AQP4 and orthogonal arrays have been detected in perinodal processes of white matter astrocytes in optic nerve where they may participate in K+ clearance from the perinodal space (37, 38). We used superresolution imaging to investigate possible differences in the subcellular distribution of AQP4 clusters in gray and white matter astrocytes of the thoracic spinal cord in mice. Confocal imaging confirmed that AQP4 was concentrated in end-feet of gray matter astrocytes, but showed a more diffuse distribution throughout GFAP-labeled processes in white matter astrocytes (Fig. 4 D, top panels). Confocal images of longitudinal sections immunostained for AQP4 and MBP demonstrated dense AQP4 staining in astrocytic processes surrounding myelinated fibers in white matter (Fig. 4 D, bottom panels). Superresolution imaging of paraffin sections revealed AQP4 clusters in perivascular regions of gray matter astrocytes in the ventral spinal horn with a similar distribution to that observed in cortical gray matter (Fig. 4 E, top). In contrast, large AQP4 clusters were found in both perivascular and parenchymal regions of white matter tracts (Fig. 4 E, bottom). Quantification of AQP4 cluster size demonstrated a significant difference in average cluster size between perivascular and parenchymal regions of gray matter but not of white matter (Fig. 4 F).
AQP4 clusters colocalize with K+ channel Kir4.1 in astrocyte end-feet
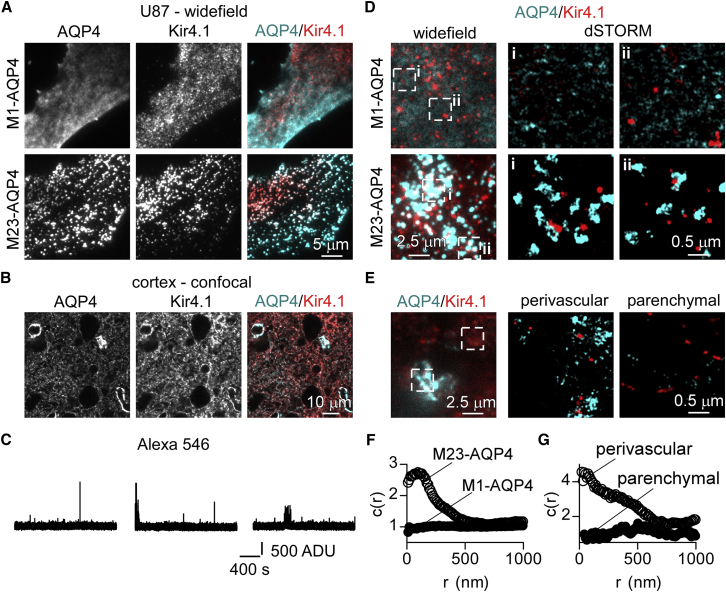
We previously reported that large, M23-AQP4-enriched clusters preferentially remain in detergent-insoluble complexes on the basal surface of cultured cells (16), suggesting that these clusters interact with macromolecular anchoring complexes. AQP4 has been reported to bind anchoring complexes containing the inwardly rectifying K+ channel, Kir4.1 (39). Here, we found partial colocalization of M23-AQP4, but not M1-AQP4, with Kir4.1 in U87-MG glioma cells that were cotransfected with individual AQP4 isoforms and Kir4.1 (Fig. 5 A). In brain tissue sections, Kir4.1 colocalized with AQP4 in both end foot and parenchymal regions, but did not show polarization to end-feet (Fig. 5 B).
Figure 5.
Two-color superresolution imaging of AQP4 and Kir4.1 labeled with Alexa 647 and 546, respectively. (A) Wide-field fluorescence microscopy showing AQP4 and Kir4.1 immunofluorescence in U87-MG cells cotransfected with Kir4.1 and M1-AQP4 or M23-AQP4. (B) Confocal microscopy of AQP4 and Kir4.1 immunofluorescence in brain cortical sections of wild-type mouse. (C) Blinking of single Alexa 546-labeled secondary antibodies in buffer containing 100 mM MEA. (D) Two-color wide-field (left panels) and superresolution images (right panels show the indicated boxed regions) of Kir4.1 and AQP4 in cultured U87-MG cells transfected with M1- or M23-AQP4. (E) Wide-field images of AQP4 and Kir4.1 (left panel) and superresolution images (right panels) showing distribution of AQP4 and Kir4.1 in parenchymal and perivascular regions of cortical sections. (F) Cross-correlation function, c(r), calculated for Kir4.1 versus AQP4 localizations in U87-MG cells transfected with M1- or M23-AQP4. The cross-correlation was averaged from four-to-six regions of the same size as those in (D). (G) The cross-correlation calculated for Kir4.1 versus AQP4 localizations in perivascular and parenchymal regions of cortical sections. To see this figure in color, go online.
Two-color dSTORM experiments have generally been limited from the lack of a second fluorophore with blinking characteristics that were as favorable as those of Alexa 647 and with appropriate buffer compatibility (40). We found that single, Alexa-546-labeled secondary antibodies displayed isolated blinking (Fig. 5 C, top traces) or intermittent fast-flickering (Fig. 5 C, bottom) in imaging buffer containing 100 mM MEA. The average on-time per blink was 30 ms, off-time was 1.53 s, average number of blinks per antibody was 17.7, and the emission rate in the on-state was 6000 photons/s (n = 12,390 blinks from 70 molecules). In two-color dSTORM measurements on cultured cells, Kir4.1 was visible as distinct puncta adjacent to large M23-AQP4 clusters in cells transfected with both isoforms, whereas M1-AQP4 was not closely associated with Kir4.1 (Fig. 5 D). Kir4.1 was closely colocalized with AQP4 clusters in end foot regions but was not in parenchymal regions of cortical slices (Fig. 5 E). To quantify these observations, we performed spatial cross-correlation analysis of AQP4 and Kir4.1 localizations as described by Sengupta et al. (30). This analysis demonstrated significant close-range correlation of M23-AQP4 and Kir4.1 localizations at distances <500 nm in transfected cells in culture. In contrast, no correlation between Kir4.1 and M1-AQP4 localizations was observed (Fig. 5 F). In tissue sections from the cerebral cortex, AQP4 and Kir4.1 showed significant short-range correlation in perivascular but not in parenchymal regions (Fig. 5 G), demonstrating that macromolecular complexes containing AQP4 clusters and Kir4.1 are confined to astrocyte end-feet.
AQP4 cluster disruption in brain tumor
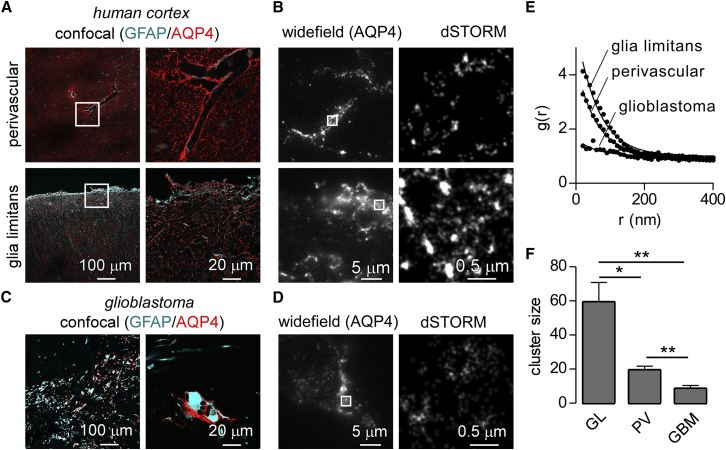
Loss of AQP4 polarization is a common feature of many neurological diseases, but it is unclear whether the loss of polarization occurs as a result of changes in AQP4 clusters, mislocalization of anchoring proteins from end-feet, and/or damage to the basement-membrane surrounding capillaries. We performed experiments to determine whether the AQP4 cluster size can be measured in histopathological human specimens. Confocal images of an autopsy specimen of human brain cortex showed AQP4 enrichment at the glia limitans and in end-feet around cerebral microcapillaries (Fig. 5 A). Superresolution imaging showed AQP4 clusters in both areas (Fig. 6 B). Perivascular clusters in human specimens were smaller than those observed in mouse, perhaps because of intrinsic differences between mouse and human astrocytes (41) or changes in the distribution of AQP4 during aging (42).
Figure 6.
Superresolution imaging of AQP4 clusters in normal human brain and glioblastoma. (A) AQP4 and GFAP immunofluorescence (by confocal microscopy) in perivascular areas and at the glia limitans in postmortem human brain cortex. (B) Wide-field images of AQP4 staining in perivascular areas and at the glia limitans (left panel) and dSTORM images of AQP4 clusters in the indicated areas (right panels). (C) Confocal images of AQP4 and GFAP expressing glioblastoma cells from a surgically resected specimen. (D) Wide-field and dSTORM images of AQP4 localizations in a region from the same section as in (C). (E) Average g(r) for AQP4-enriched areas of the glia limitans and perivascular end-feet in human brain and from membrane protrusions of glioblastoma cells. (F) Average AQP4 cluster size calculated from g(r) in (E); ∗p < 0.05, 10–15 regions from three separate experiments; ∗∗p < 0.01 (by t-test). To see this figure in color, go online.
AQP4 is expressed at high levels in glioblastomas but is mostly unpolarized (43). We studied a surgically resected glioblastoma that showed high levels of AQP4 and GFAP expression (Fig. 6 C). Remarkably, superresolution imaging of AQP4-enriched membrane areas of the glioblastoma cells did not show large AQP4 clusters (Fig. 6 D). Pair-correlation analysis of AQP4-enriched regions showed significant clustering in the glia limitans and around microcapillaries in normal brain, whereas short-range clustering of AQP4 was not seen in glioblastoma (Fig. 6 E). Average cluster size deduced from pair-correlation data revealed significantly reduced cluster size in glioblastoma when compared to either the glia limitans or perivascular regions; significant differences were also observed between the glia limitans and perivascular areas (Fig. 6 F). These results demonstrate the utility of superresolution imaging to determine AQP4 aggregation state in human histopathological specimens, and suggest that the disperse distribution of AQP4 in glioblastoma cells is associated with loss of aggregation.
Discussion
The mechanisms responsible for AQP4 aggregation into orthogonal arrays have been studied extensively (6, 9, 11, 44); however, the cellular function of these structures and the significance of their localization at high density in astrocyte end-feet remain poorly understood. Superresolution optical imaging methods allow characterization of subdiffractive biological structures such as orthogonal arrays. Here, we have demonstrated that these methods can be used to address questions relating to the structure and function of AQP4 clusters in intact CNS tissues, and find that: 1) M23-AQP4 mediates AQP4 clustering both in vitro and in vivo; 2) distinct subcellular distributions of AQP4 clusters exist in different classes of astrocytes; 3) AQP4 clusters colocalize with Kir4.1 in astrocyte end-feet but not in parenchymal areas; and 4) AQP4 clusters are largely absent in a human glioblastoma specimen despite strong AQP4 expression.
AQP4 clustering has been studied extensively using FFEM to visualize orthogonal arrays in astrocyte end-feet (4, 5). Here, we show that superresolution optical microscopy (dSTORM) can be combined with astigmatic optics and statistical clustering analysis to quantify AQP4 cluster size in membranes of 3D specimens. This approach complements and extends existing methodology; FFEM offers unparalleled spatial resolution but cannot detect AQP4 that is not aggregated into orthogonal arrays, requires fracture along the plane of the topologically complex astrocyte membrane, and is not well suited to colocalization analysis with other proteins. Superresolution optical imaging can be applied in paraffin tissue sections using standard staining methodology, allowing for high-resolution structural analysis in banked histopathological specimens. We note, however, that the lower resolution of dSTORM compared with FFEM, and the stochastic blinking of fluorophores, could limit the unambiguous detection of individual clusters at very high densities and we cannot definitely differentiate AQP4 in the orthogonal array from other hypothetical clustering mechanisms. Superresolution optical imaging and other approaches such as native gel electrophoresis would complement FFEM in further studies of AQP4-containing macromolecular protein complexes.
When expressed individually in AQP4−/− astrocytes, M23-AQP4 clusters polarize to end-feet, whereas nonclustered M1-AQP4 remains diffusely distributed in the plasma membrane (16). In cultured astrocytes from wild-type mice, where M1- and M23-AQP4 are coexpressed, there is a heterogeneous population of AQP4 cluster sizes (10, 11); here, we found evidence for size-based segregation of these clusters in cortical gray matter astrocytes. Larger clusters are preferentially polarized to end-feet for high-capacity water flux, whereas a less clustered population of AQP4 is dispersed throughout the astrocyte plasma membrane to facilitate rapid astrocyte volume change in response to neuronal activity. Fine astrocytic processes are structurally dynamic, and extend lamellipodia-like structures to perisynaptic regions (45, 46). Diffusionally mobile AQP4 tetramers and small clusters support lamellipodial extension (16), and may participate in this process. The mechanism for enrichment of larger AQP4 clusters at end-feet remains unknown but could be due to preferential anchoring of large clusters to cell-surface adhesion complexes or preferential vesicular trafficking to end-feet. An unexpected finding of our study was the difference in the subcellular distribution of AQP4 clusters between gray and white matter astrocytes of the spinal cord. Orthogonal arrays have previously been observed by FFEM in close proximity to gap junctions of juxtaparanodal astrocyte processes in optic nerve (37) and AQP4 may participate in K+ spatial redistribution within the panglial syncytium of astrocytes and oligodendrocytes surrounding myelinated axons (38). The significance of the distribution of AQP4 clusters in white matter is unclear but might be involved in anchoring to specific sites adjacent to myelinated fibers. The distribution of AQP4 clusters in gray and white matter astrocytes may also play a role in the pathogenesis of neuromyelitis optica, where autoantibodies that preferentially bind clustered AQP4 on astrocytes cause demyelination and axonal degeneration (47).
AQP4 transports water in response to osmotic gradients generated by ion channels and transporters. Biochemical measurements and immunogold electron microscopy have suggested AQP4 complexation with the inwardly rectifying K+ channel Kir4.1 (48) in dystroglycan-anchored complexes at astrocyte end-feet (49). M23-AQP4 forms large clusters that remain bound to adhesion complexes after detergent lysis of transfected U87 cells (16), and we found colocalization of Kir4.1 in these structures in cotransfected cells and with AQP4 at astrocyte end-feet. These results support the conclusion that M23-AQP4 and orthogonal arrays participate in macromolecular complex formation and polarization, whereas isolated M1-AQP4 tetramers do not interact strongly with other proteins. It remains unknown whether this is due to an increased local density of C-terminal PDZ binding domains when AQP4 is clustered or if exposed hydrophobic N-terminal residues in M23-AQP4 that mediate supramolecular clustering can also bind additional proteins. The functional significance of the close spatial colocalization between AQP4 and Kir4.1 in end-feet remains unclear. Patch-clamp studies have failed to find evidence for direct modulation of Kir4.1 currents by AQP4 under isosmotic conditions (50), but indirect regulation of the K+ reversal potential may occur in response to osmotic swelling (51).
AQP4 is expressed at high levels in glioblastoma where it facilitates cell migration (12). We found previously that only small AQP4 clusters and isolated tetramers can translocate to the leading edge of a glioma cell line to facilitate edge extension in vitro (16). We showed here that superresolution imaging can be applied to human specimens to quantify the AQP4 aggregation state. AQP4 was found to be diffusely localized in human glioblastoma, in agreement with the results of Noell et al. (43), who demonstrated that upregulation of AQP4 in glioblastoma was not associated with an increase in size or density of orthogonal arrays measured by FFEM. Changes in AQP4 expression and polarization occur in response to numerous neuropathological conditions including stroke (52) and Alzheimer’s disease (53). It is unclear whether changes in AQP4 aggregation state, due to changes in the proportion of M1- and M23-AQP4, or other factors such as degradation of the basement membrane and/or anchoring complexes cause the loss of AQP4 polarization. Application of the imaging approach here to mouse disease models and human specimens should facilitate investigation of the mechanistic relationship between AQP4 aggregation state and loss of polarization.
We have established methodology for optical imaging of AQP4 clusters in paraffin sections of brain and spinal cord tissue, and demonstrate that the major observations on AQP4 cluster size and cellular distribution made in astrocyte cultures translate to brain in vivo. Our work provides observations that the distribution of AQP4 cluster size is dependent on subcellular localization and astrocyte subtype and that formation of macromolecular clusters between AQP4 and Kir4.1 occurs in specific subcellular locations. The glioblastoma data here are consistent with previous FFEM studies demonstrating that increases in AQP4 expression in glioblastoma are not correlated with increases in orthogonal array density (43). The relative ease of this approach compared to FFEM should facilitate further studies of AQP4 clusters and their interactions under normal physiological conditions and in disease. The experimental data on the subcellular distribution of AQP4 clusters in normal brain and in glioblastoma support a model where the formation of supramolecular clusters, and by extension the M1:M23-AQP4 ratio, plays a pivotal role in targeting AQP4 to specific functional complexes and subcellular regions in the normal brain, and disruption of clusters results in distribution of AQP4 throughout the plasma membrane in astrocytes.
Acknowledgments
The authors thank Alex Herbert (University of Sussex, UK) for assistance with analysis and simulation software, Byung-Ju Jin for assistance with MATLAB code, Hua Zhang and Julien Ratelade for paraffin-embedded mouse brain specimens, and the University of California at San Francisco Brain Tumor Research Center Tissue Bank for glioblastoma specimens.
This work was funded by grant Nos. EB00415, DK35124, DK101373, DK72517, and EY13574 from the National Institutes of Health, Bethesda, MD.
Editor: Leonid Brown.
Contributor Information
Alex J. Smith, Email: alex.smith@ucsf.edu.
Alan S. Verkman, Email: alan.verkman@ucsf.edu.
References
- 1.Papadopoulos M.C., Verkman A.S. Aquaporin water channels in the nervous system. Nat. Rev. Neurosci. 2013;14:265–277. doi: 10.1038/nrn3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung J.S., Bhat R.V., Agre P. Molecular characterization of an aquaporin cDNA from brain: candidate osmoreceptor and regulator of water balance. Proc. Natl. Acad. Sci. USA. 1994;91:13052–13056. doi: 10.1073/pnas.91.26.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang B., Ma T., Verkman A.S. cDNA cloning, gene organization, and chromosomal localization of a human mercurial insensitive water channel. Evidence for distinct transcriptional units. J. Biol. Chem. 1995;270:22907–22913. doi: 10.1074/jbc.270.39.22907. [DOI] [PubMed] [Google Scholar]
- 4.Landis D.M., Reese T.S. Membrane structure in mammalian astrocytes: a review of freeze-fracture studies on adult, developing, reactive and cultured astrocytes. J. Exp. Biol. 1981;95:35–48. doi: 10.1242/jeb.95.1.35. [DOI] [PubMed] [Google Scholar]
- 5.Wolburg H., Wolburg-Buchholz K., Mack A.F. Structure and functions of aquaporin-4-based orthogonal arrays of particles. Int. Rev. Cell Mol. Biol. 2011;287:1–41. doi: 10.1016/B978-0-12-386043-9.00001-3. [DOI] [PubMed] [Google Scholar]
- 6.Yang B., Brown D., Verkman A.S. The mercurial insensitive water channel (AQP-4) forms orthogonal arrays in stably transfected Chinese hamster ovary cells. J. Biol. Chem. 1996;271:4577–4580. [PubMed] [Google Scholar]
- 7.Verbavatz J.M., Ma T., Verkman A.S. Absence of orthogonal arrays in kidney, brain and muscle from transgenic knockout mice lacking water channel aquaporin-4. J. Cell Sci. 1997;110:2855–2860. doi: 10.1242/jcs.110.22.2855. [DOI] [PubMed] [Google Scholar]
- 8.Rash J.E., Yasumura T., Nielsen S. Direct immunogold labeling of aquaporin-4 in square arrays of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Proc. Natl. Acad. Sci. USA. 1998;95:11981–11986. doi: 10.1073/pnas.95.20.11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furman C.S., Gorelick-Feldman D.A., Rash J.E. Aquaporin-4 square array assembly: opposing actions of M1 and M23 isoforms. Proc. Natl. Acad. Sci. USA. 2003;100:13609–13614. doi: 10.1073/pnas.2235843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crane J.M., Bennett J.L., Verkman A.S. Live cell analysis of aquaporin-4 M1/M23 interactions and regulated orthogonal array assembly in glial cells. J. Biol. Chem. 2009;284:35850–35860. doi: 10.1074/jbc.M109.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin B.J., Rossi A., Verkman A.S. Model of aquaporin-4 supramolecular assembly in orthogonal arrays based on heterotetrameric association of M1-M23 isoforms. Biophys. J. 2011;100:2936–2945. doi: 10.1016/j.bpj.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saadoun S., Papadopoulos M.C., Verkman A.S. Involvement of aquaporin-4 in astroglial cell migration and glial scar formation. J. Cell Sci. 2005;118:5691–5698. doi: 10.1242/jcs.02680. [DOI] [PubMed] [Google Scholar]
- 13.Strohschein S., Hüttmann K., Steinhäuser C. Impact of aquaporin-4 channels on K+ buffering and gap junction coupling in the hippocampus. Glia. 2011;59:973–980. doi: 10.1002/glia.21169. [DOI] [PubMed] [Google Scholar]
- 14.Jin B.J., Zhang H., Verkman A.S. Aquaporin-4-dependent K+ and water transport modeled in brain extracellular space following neuroexcitation. J. Gen. Physiol. 2013;141:119–132. doi: 10.1085/jgp.201210883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen S., Nagelhus E.A., Ottersen O.P. Specialized membrane domains for water transport in glial cells: high-resolution immunogold cytochemistry of aquaporin-4 in rat brain. J. Neurosci. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith A.J., Jin B.J., Verkman A.S. Aggregation state determines the localization and function of M1- and M23-aquaporin-4 in astrocytes. J. Cell Biol. 2014;204:559–573. doi: 10.1083/jcb.201308118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 19.Sengupta P., Jovanovic-Talisman T., Lippincott-Schwartz J. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat. Methods. 2011;8:969–975. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veatch S.L., Machta B.B., Baird B.A. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS One. 2012;7:e31457. doi: 10.1371/journal.pone.0031457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossi A., Moritz T.J., Verkman A.S. Super-resolution imaging of aquaporin-4 orthogonal arrays of particles in cell membranes. J. Cell Sci. 2012;125:4405–4412. doi: 10.1242/jcs.109603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao H.P., Verkman A.S. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys. J. 1994;67:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang B., Wang W., Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dani A., Huang B., Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L., Zhang H., Verkman A.S. Proinflammatory role of aquaporin-4 in autoimmune neuroinflammation. FASEB J. 2011;25:1556–1566. doi: 10.1096/fj.10-177279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crane J.M., Lam C., Verkman A.S. Binding affinity and specificity of neuromyelitis optica autoantibodies to aquaporin-4 M1/M23 isoforms and orthogonal arrays. J. Biol. Chem. 2011;286:16516–16524. doi: 10.1074/jbc.M111.227298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokunaga M., Imamoto N., Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 28.van de Linde S., Löschberger A., Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 29.Mortensen K.I., Churchman L.S., Flyvbjerg H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Methods. 2010;7:377–381. doi: 10.1038/nmeth.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengupta P., Jovanovic-Talisman T., Lippincott-Schwartz J. Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nat. Protoc. 2013;8:345–354. doi: 10.1038/nprot.2013.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gould T.J., Verkhusha V.V., Hess S.T. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat. Protoc. 2009;4:291–308. doi: 10.1038/nprot.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dempsey G.T., Vaughan J.C., Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobik M., Ellisman M.H., Martone M.E. Potassium channel subunit Kv3.2 and the water channel aquaporin-4 are selectively localized to cerebellar pinceau. Brain Res. 2004;1026:168–178. doi: 10.1016/j.brainres.2004.07.088. [DOI] [PubMed] [Google Scholar]
- 34.Nicchia G.P., Rossi A., Frigeri A. Dystrophin-dependent and -independent AQP4 pools are expressed in the mouse brain. Glia. 2008;56:869–876. doi: 10.1002/glia.20661. [DOI] [PubMed] [Google Scholar]
- 35.Sosunov A.A., Wu X., Goldman J.E. Phenotypic heterogeneity and plasticity of isocortical and hippocampal astrocytes in the human brain. J. Neurosci. 2014;34:2285–2298. doi: 10.1523/JNEUROSCI.4037-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oklinski M.K., Lim J.S., Kwon T.H. Immunolocalization of water channel proteins AQP1 and AQP4 in rat spinal cord. J. Histochem. Cytochem. 2014;62:598–611. doi: 10.1369/0022155414537495. [DOI] [PubMed] [Google Scholar]
- 37.Waxman S.G., Black J.A. Freeze-fracture ultrastructure of the perinodal astrocyte and associated glial junctions. Brain Res. 1984;308:77–87. doi: 10.1016/0006-8993(84)90919-3. [DOI] [PubMed] [Google Scholar]
- 38.Rash J.E. Molecular disruptions of the panglial syncytium block potassium siphoning and axonal saltatory conduction: pertinence to neuromyelitis optica and other demyelinating diseases of the central nervous system. Neuroscience. 2010;168:982–1008. doi: 10.1016/j.neuroscience.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amiry-Moghaddam M., Frydenlund D.S., Ottersen O.P. Anchoring of aquaporin-4 in brain: molecular mechanisms and implications for the physiology and pathophysiology of water transport. Neuroscience. 2004;129:999–1010. doi: 10.1016/j.neuroscience.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 40.Lampe A., Haucke V., Schmoranzer J. Multi-colour direct STORM with red emitting carbocyanines. Biol. Cell. 2012;104:229–237. doi: 10.1111/boc.201100011. [DOI] [PubMed] [Google Scholar]
- 41.Oberheim N.A., Takano T., Nedergaard M. Uniquely hominid features of adult human astrocytes. J. Neurosci. 2009;29:3276–3287. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kress B.T., Iliff J.J., Nedergaard M. Impairment of paravascular clearance pathways in the aging brain. Ann. Neurol. 2014;76:845–861. doi: 10.1002/ana.24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noell S., Wolburg-Buchholz K., Fallier-Becker P. Dynamics of expression patterns of AQP4, dystroglycan, agrin and matrix metalloproteinases in human glioblastoma. Cell Tissue Res. 2012;347:429–441. doi: 10.1007/s00441-011-1321-4. [DOI] [PubMed] [Google Scholar]
- 44.Crane J.M., Verkman A.S. Determinants of aquaporin-4 assembly in orthogonal arrays revealed by live-cell single-molecule fluorescence imaging. J. Cell Sci. 2009;122:813–821. doi: 10.1242/jcs.042341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirrlinger J., Hülsmann S., Kirchhoff F. Astroglial processes show spontaneous motility at active synaptic terminals in situ. Eur. J. Neurosci. 2004;20:2235–2239. doi: 10.1111/j.1460-9568.2004.03689.x. [DOI] [PubMed] [Google Scholar]
- 46.Heller J.P., Rusakov D.A. Morphological plasticity of astroglia: understanding synaptic microenvironment. Glia. 2015;63:2133–2151. doi: 10.1002/glia.22821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papadopoulos M.C., Verkman A.S. Aquaporin 4 and neuromyelitis optica. Lancet Neurol. 2012;11:535–544. doi: 10.1016/S1474-4422(12)70133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagelhus E.A., Horio Y., Ottersen O.P. Immunogold evidence suggests that coupling of K+ siphoning and water transport in rat retinal Müller cells is mediated by a coenrichment of Kir4.1 and AQP4 in specific membrane domains. Glia. 1999;26:47–54. doi: 10.1002/(sici)1098-1136(199903)26:1<47::aid-glia5>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 49.Connors N.C., Adams M.E., Kofuji P. The potassium channel Kir4.1 associates with the dystrophin-glycoprotein complex via α-syntrophin in glia. J. Biol. Chem. 2004;279:28387–28392. doi: 10.1074/jbc.M402604200. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H., Verkman A.S. Aquaporin-4 independent Kir4.1 K+ channel function in brain glial cells. Mol. Cell. Neurosci. 2008;37:1–10. doi: 10.1016/j.mcn.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soe R., Macaulay N., Klaerke D.A. Modulation of Kir4.1 and Kir4.1-Kir5.1 channels by small changes in cell volume. Neurosci. Lett. 2009;457:80–84. doi: 10.1016/j.neulet.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 52.Steiner E., Enzmann G.U., Engelhardt B. Loss of astrocyte polarization upon transient focal brain ischemia as a possible mechanism to counteract early edema formation. Glia. 2012;60:1646–1659. doi: 10.1002/glia.22383. [DOI] [PubMed] [Google Scholar]
- 53.Wilcock D.M., Vitek M.P., Colton C.A. Vascular amyloid alters astrocytic water and potassium channels in mouse models and humans with Alzheimer’s disease. Neuroscience. 2009;159:1055–1069. doi: 10.1016/j.neuroscience.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]