Abstract
Voltage-sensitive phosphatases (VSPs) are proteins that directly couple changes in membrane electrical potential to inositol lipid phosphatase activity. VSPs thus couple two signaling pathways that are critical for cellular functioning. Although a number of nonmammalian VSPs have been characterized biophysically, mammalian VSPs are less well understood at both the physiological and biophysical levels. In this study, we aimed to address this gap in knowledge by determining whether the VSP from mouse, Mm-VSP, is expressed in the brain and contains a functional voltage-sensing domain (VSD) and a phosphatase domain. We report that Mm-VSP is expressed in neurons and is developmentally regulated. To address whether the functions of the VSD and phosphatase domain are retained in Mm-VSP, we took advantage of the modular nature of these domains and expressed each independently as a chimeric protein in a heterologous expression system. We found that the Mm-VSP VSD, fused to a viral potassium channel, was able to drive voltage-dependent gating of the channel pore. The Mm-VSP phosphatase domain, fused to the VSD of a nonmammalian VSP, was also functional: activation resulted in PI(4,5)P2 depletion that was sufficient to inhibit the PI(4,5)P2-regulated KCNQ2/3 channels. While testing the functionality of the VSD and phosphatase domain, we observed slight differences between the activities of Mm-VSP-based chimeras and those of nonmammalian VSPs. Although the properties of VSP chimeras may not completely reflect the properties of native VSPs, the differences we observed in voltage-sensing and phosphatase activity provide a starting point for future experiments to investigate the function of Mm-VSP and other mammalian VSPs. In conclusion, our data reveal that both the VSD and the lipid phosphatase domain of Mm-VSP are functional, indicating that Mm-VSP likely plays an important role in mouse neurophysiology.
Introduction
The phospholipid composition of cellular membranes and the electrical potential across those membranes are two important signaling mechanisms that are involved in a wide variety of cellular functions. Accordingly, many diseases are known to involve improper regulation of lipid signaling (1) or the proteins that regulate the membrane potential (2). Voltage-sensitive phosphatases (VSPs) are a family of proteins comprising two domains that directly couple these two important signaling mechanisms (3). The first domain in VSPs is homologous to the voltage-sensing domains (VSDs) in voltage-gated ion channels (4, 5), whereas the second domain is homologous to the lipid phosphatase and tumor suppressor PTEN (6, 7, 8, 9, 10).
The VSP from the sea squirt Ciona intestinalis (Ci-VSP) is the best-characterized VSP so far. In Ci-VSP, depolarization increases the protein’s lipid phosphatase activity (3, 11), resulting in dephosphorylation of the 5′ position of phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) and phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3) (12, 13, 14). Although VSPs have been used as tools to study the role of phosphoinositides in regulating cellular processes (15, 16, 17), and as a structural model for studying VSD activation (5, 18, 19, 20, 21), little is known about the biological roles that VSPs play in vivo. Furthermore, much less is known about the properties of the mammalian VSP family members than is known about their counterparts in other species.
To better understand the functionality of VSPs, we studied the mouse VSP as a model mammalian VSP. We investigated its expression in the central nervous system, where both electrical signaling and lipid signaling play important roles, and found that neural expression of the protein is developmentally regulated. To test the functionality of Mm-VSP’s VSD and lipid phosphatase domain, we generated chimeric proteins containing each of these domains separately. These studies revealed that both the VSD and phosphatase domain of Mm-VSP are functional independently of one another. Analyses of the VSD also suggested a potential sensitivity to intracellular pH that may serve as a mechanism to regulate Mm-VSP’s activity.
As a note regarding nomenclature, although the mouse protein was originally termed transmembrane phosphatase with tensin homology (TPTE), our results indicate that the protein is not only voltage sensitive but also displays lipid phosphatase activity. Therefore, by analogy to Ci-VSP, we refer to the protein as the VSP from Mus musculus (Mm-VSP) from here on.
Materials and Methods
Protein sequence alignment
For amino acid sequence comparisons, the COBALT algorithm (22) was used to align the sequences of Mm-VSP (NP_954866.2), Ci-VSP (BAD98733.1), Dr-VSP (BAG50379.1), Hs-VSP2 (previously named TPTE)(NP_954868.1), Hs-VSP1 (previously named TPIP) (AAP45146.1), Gg-VSP (XP_417079), and Drosophila Shaker (CAA29917.1).
RT-PCR
RNA was purified from mouse whole-brain homogenate using a spin column system (NucleoSpin; Machery-Nagel). RT-PCR primers against the Mm-VSP mRNA sequence (NM_199257.2) were designed using Primer-BLAST software (23) (Table S2 in the Supporting Material). Reverse transcription and PCR were carried out using SuperScript III with the Platinum Taq kit (Invitrogen). PCR products were separated by electrophoresis on a 1.5% agarose gel and visualized with ethidium bromide. For sequencing of selected amplicons, bands were cut from the agarose gel, purified using a spin column (QIAquick Gel Extraction Kit; Qiagen), and sequenced in the forward and reverse directions using the associated RT-PCR primers.
DNA construct generation
GalT-oxBFP was a gift of Dr. Erik L. Snapp (Albert Einstein College of Medicine). KvSynth1 was given to us by the Moroni lab at the University of Milan. KCNQ2, KCNQ3, and Dr-VSP were provided by the Hille lab at the University of Washington. A BluntII-TOPO plasmid containing the DNA for Mm-VSP was purchased from Open Biosystems (clone ID 40054415). Additional constructs were engineered and subcloned based on these plasmids using standard molecular cloning techniques. Details regarding the primers and restriction sites used are listed in Table S3.
Cell culture
HEK293T/17 cells (ATCC) were grown at 37°C with 5% CO2 saturation in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. For live-cell imaging and electrophysiology, cells were transfected with DNA using calcium phosphate, and experiments were conducted 24–48 h later. Primary cultures of mouse cortical neurons and astrocytes were obtained on postnatal day 0–1 (P0–1). Cells were dissociated using papain trituration. Astrocytes were grown under the same conditions as HEK293T/17 cells until confluency was reached. Neurons were cultured on top of a confluent layer of astrocytes in minimum essential medium supplemented with 1× GlutaMAX, 10% fetal horse serum, 25 mM HEPES, 20 mM glucose, 1% penicillin/streptomycin, 1% N2 supplement, 1% B27 supplement, and 1 mM sodium pyruvate.
Microscopy
Immunolabeled samples were imaged on a Nikon Eclipse 90i with a 40× objective, and standard filter sets for green fluorescent protein (GFP) (excitation: 450–490 nm; dichroic: 495 nm; emission: 500–550 nm), RFP (excitation: 530–560 nm; dichroic: 570 nm; emission: 590–650 nm), and DAPI (excitation: 325–375 nm; dichroic: 400 nm; emission: 435–485 nm). Images were acquired with a CoolSNAP HQ2 cooled CCD camera (Photometrics) controlled through NIS-Elements software (Nikon). Live-cell imaging was conducted on a Zeiss inverted laser scanning confocal microscope with a 40× objective. The oxBFP protein was excited with 405 nm light, and emitted light was collected from 438–484 nm. GFP was excited with 488 nm light, and emitted light was collected from 504–552 nm. Images were collected using ZEN software (Zeiss).
Antibody development
DNA encoding the first 113 amino acids of Mm-VSP was amplified and inserted into the bacterial expression vector pRSF (Table S3), generating a construct encoding a fusion protein with 6X-His and maltose-binding protein (His-MBP) at the amino terminus. Recombinant peptide containing Mm-VSP amino acids 1–113 was purified from BL21 E. coli using the His-MBP dual-affinity tag system and then cleaved from the tags using an engineered tobacco etched virus cleavage site. The identity of the protein was confirmed by mass spectrometry and the protein was then injected as an antigen into two different rabbits (h5535 and h5536) by New England Peptide. Antibody-containing serum from each rabbit had enzyme-linked immunosorbent assay titers of 63,100 and 13,000, and both rabbits’ sera were validated for use in immunocytochemistry experiments (Fig. S1 C). Serum h5536 was chosen for affinity purification to generate pure polyclonal antibodies for use in western blotting. The immunizing peptide was immobilized on NHS-activated agarose beads (Pierce). Serum diluted 1:1 in binding buffer (25 mM Tris, 150 mM NaCl, pH 7.2) was passed over the peptide beads, allowing the antibodies to bind the peptide. The antibodies were eluted using high-salt buffer (2.5 M KCl, pH 7.7). Eight fractions were collected and tested for immunoreactivity via western blot against mouse whole-brain homogenate. The fraction with the highest immunoreactivity was verified by incubating the antibody with the immunizing peptide before probing the western blot, which blocked the majority of the band intensity at the expected size of Mm-VSP (Fig. S1 A).
Western blotting
Mouse brains were homogenized immediately after dissection in 1 mL buffer (0.32 M sucrose, 5 mM HEPES, pH 7.4, 1× Roche EDTA-Free cOmplete protease inhibitor) per 100 mg tissue. Whole-brain homogenate samples were run out on polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were probed first with the affinity-purified Mm-VSP rabbit polyclonal antibody and then with a microtubule-associated protein 2 (MAP2) monoclonal antibody raised in mouse (Sigma). Both antibodies were diluted 1:1000 in phosphate-buffered saline with Tween 20 (PBST) with 1% bovine serum albumin. Secondary goat anti-rabbit or goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Invitrogen) was diluted 1:1000 in PBST and incubated with the blot. It was then exposed using Clarity ECL substrate (BioRad) and imaged using a Kodak IS440CF digital imager.
Immunolabeling
Cells were fixed with 4% paraformaldehyde, permeabilized, and blocked in blocking buffer (2% normal goat serum, 0.4% saponin, 1% bovine serum albumin, in PBS). The sera containing rabbit polyclonal antibodies generated against Mm-VSP were used at 1:200 in blocking buffer. Both sera had high immunoreactivity and specificity in immunolabeling (Fig. S1 C), and were used interchangeably. Mouse monoclonal antibodies against FLAG (Sigma), MAP2 (Sigma), GFAP (Sigma), and SV2 (24) were all used at 1:1000. Alexa 488 and 568 conjugated to goat anti-rabbit or goat anti-mouse secondary antibodies were used at dilutions between 1:1000 and 1:2500. Hoechst dye was used at 1:10,000 in blocking buffer to label cell nuclei. For all samples, a corresponding control sample in which only secondary antibodies were added was used to assess background fluorescence.
Electrophysiology
HEK293T/17 cells were plated on a poly-lysine-coated coverslip at low density 4–24 h before experiments began. Cells were continuously perfused with an extracellular solution of (in mM) 160 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES, and 8 glucose, pH 7.4 with NaOH. Cells were recorded in whole-cell voltage-clamp mode after a gigaohm seal was achieved through a borosilicate patch pipette, with an intracellular solution of (in mM) 175 KCl, 5 MgCl2, 5 HEPES, 0.1 K4BAPTA, 3 Na2ATP, and 0.1 Na3GTP, pH 7.4 with KOH (or pH 6.6 where noted). Data were collected at sample intervals of 0.1 ms (KCNQ2/3 tail currents), 0.4 ms (KvSynth currents), or 5 ms (current decay at +100 mV). For KvSynth recordings, cells were held at −70 mV. Channels were activated as shown in Fig. 3 B. Recordings were obtained once every 10 s to allow the channels to completely deactivate between pulses. For KCNQ2/3 (M-current) recordings, cells were held at −60 mV. Channels were activated as shown in Fig. 5 C and tail currents were measured instantly after the cell was repolarized. Traces were analyzed using IGOR Pro version 6.3.4.1 (WaveMetrics) after they were imported using the PPT library (Dr. Francisco Mendez and Frank Würriehausen, Max-Planck Institute, Göttingen, Germany).
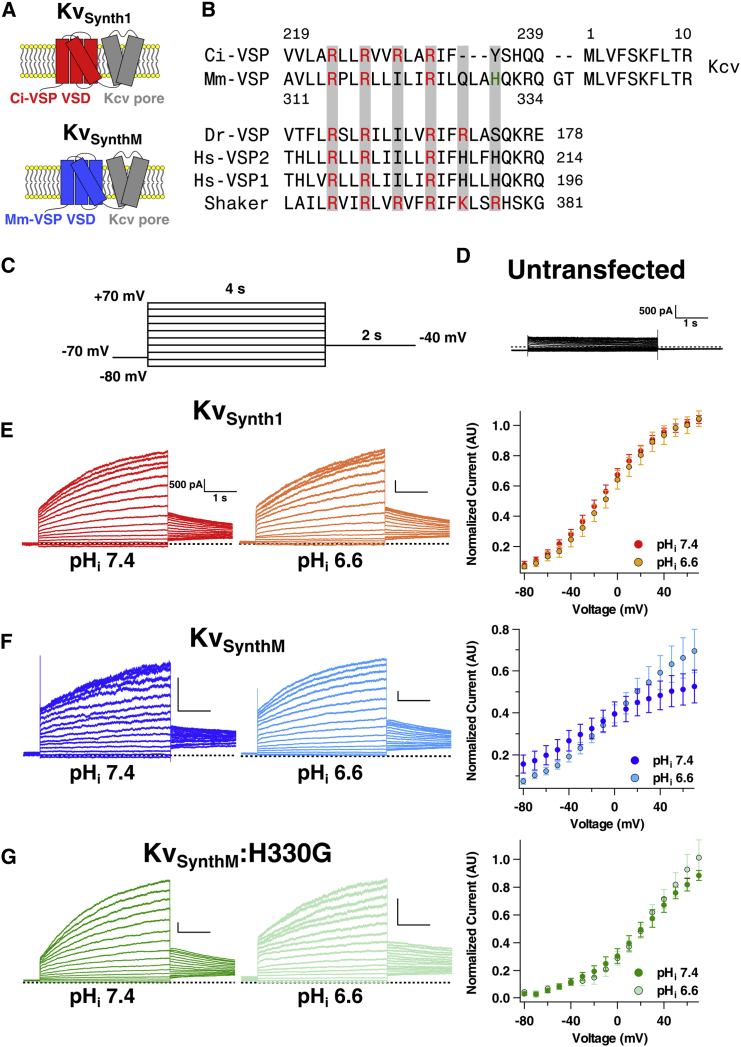
Figure 3.
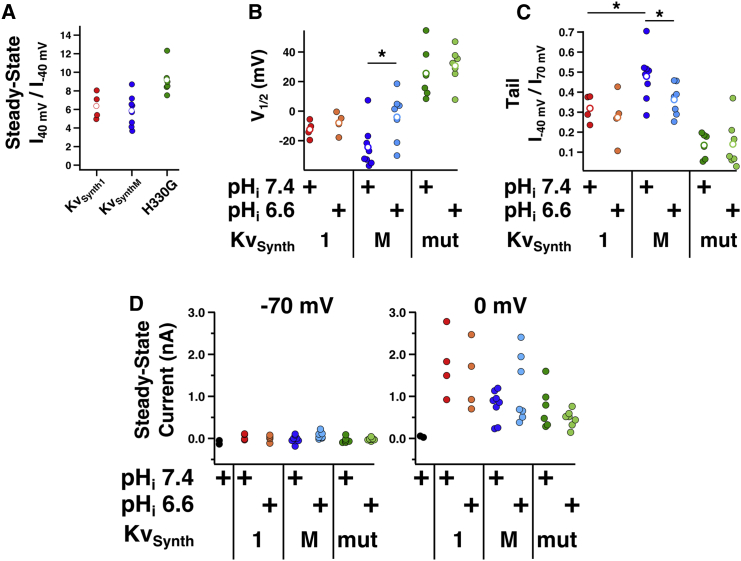
The transmembrane domain of Mm-VSP senses voltage and pH. (A) Schematics depicting chimeric KvSynth channels. KvSynth1 (top) contains the voltage sensor from Ci-VSP (red) and the Kcv potassium pore from the PBCV-1 virus (gray). KvSynthM (bottom) contains the voltage sensor from Mm-VSP (blue) and the Kcv potassium pore (gray). (B) S4 sequences of five VSP family members and the Drosophila Shaker potassium channel. The amino acid numbers of the first and last positions shown are noted. Positive-gating-charge residues are highlighted in red and the histidine residue that was mutated to produce the KvSynthM:H330G channel is highlighted in green. Shown with Ci-VSP and Mm-VSP are the first 10 residues of the Kcv pore, illustrating the juncture in the KvSynth1 and KvSynthM chimeras. Note that two residues were introduced into KvSynthM as part of the cloning strategy that were not present in KvSynth1. The presence of these residues does not appear to significantly impact coupling of the VSD to the Kcv pore (see Fig. 4A). (C) Voltage protocol used to generate voltage-dependent activation curves for the KvSynth channels. Cells were held at −70 mV in whole-cell mode. Four-second pulses from −80 mV to +70 mV were applied, increasing in 10 mV increments. Instantaneous tail currents were measured upon repolarization to −40 mV and used as an indication of voltage-sensor activation. Cells were kept at their holding potential for 10 s between sweeps. (D) Untransfected cells do not express KvSynth-like currents. Shown is a representative family of current traces from a HEK293T/17 cell that was not transfected with any KvSynth channel. Recordings were performed with an intracellular pH of 7.4. (E–G) Voltage-dependent gating of the KvSynth channels at different intracellular pH values. Representative current traces from HEK293T/17 cells transfected with the indicated KvSynth constructs are shown. Recordings were performed with intracellular solutions at pH 7.4 (darker traces) or pH 6.6 (lighter traces). Vertical scale bars indicate 500 pA and horizontal scale bars indicate 1 s. Dotted lines indicate zero current. To the right of the representative traces are average activation curves. Individual cells’ tail-current/voltage relationships were fitted using a two-state Boltzmann curve and were subsequently normalized to the extrapolated maximum and minimum values of these fits. The average of these normalized curves is shown, with error bars indicating standard error of the mean. Each color-coded data set corresponds to a given KvSynth channel: (E) KvSynth1 (pHi 7.4 n = 4, pHi 6.6 n = 4), (F) KvSynthM (pHi 7.4 n = 8, pHi 6.6 n = 7), and (G) KvSynthM:H330G (pHi 7.4 n = 6, pHi 6.6 n = 7). To see this figure in color, go online.
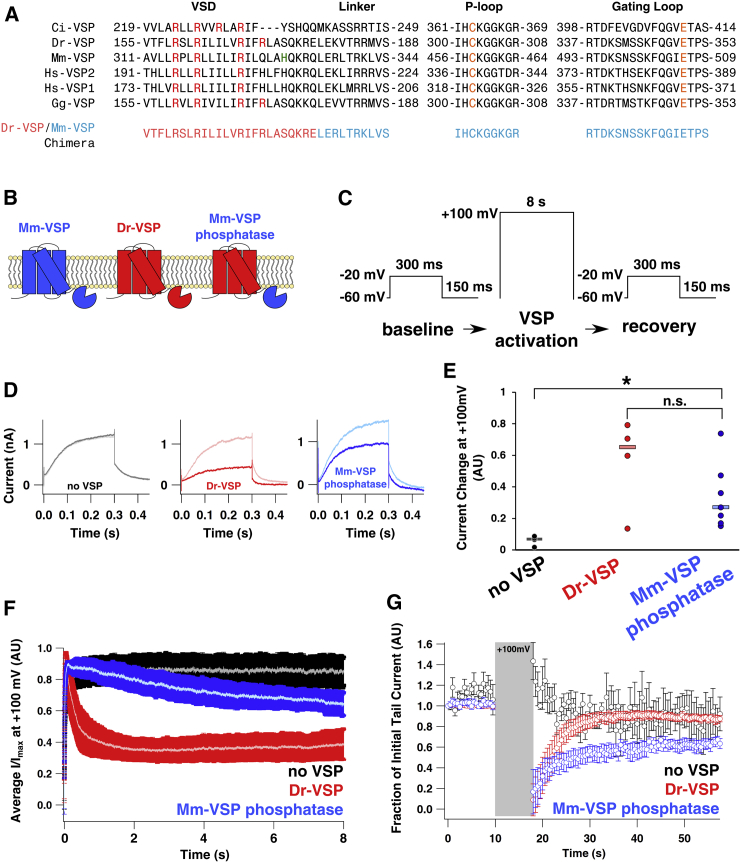
Figure 5.
A VSP chimera containing the Mm-VSP phosphatase inhibits M currents in HEK cells. (A) Sequence alignment of various VSP family members and an engineered Dr-VSP/Mm-VSP chimera. Amino acid positions are given by the numbers before and after each region of the sequence. Key regions implicated in the voltage-dependent phosphatase activity of VSPs are highlighted. In particular, the gating-charge residues of the VSD are shown in red and histidine 330 in Mm-VSP is shown in green, whereas the catalytic cysteine and gating-loop aspartate of the phosphatase active site are shown in orange. Below is the sequence of the Mm-VSP phosphatase-containing chimera. The voltage-sensor sequence of the chimera was taken from Dr-VSP and is depicted in light red, and the phosphatase domain, taken from Mm-VSP, is depicted in light blue. (B) Schematic of the topology of Mm-VSP (blue, left), Dr-VSP (red, center), and the chimera containing the Mm-VSP phosphatase fused to the Dr-VSP voltage sensor (right). (C) Voltage protocol used to probe VSP-based inhibition of PI(4,5)P2-dependent M currents. Baseline tail current amplitude was established by activating M currents at −20 mV for 300 ms and then measuring instantaneous currents when the cell was repolarized to −60 mV. Measurements were taken once every 500 ms for 10 s. Cells were then depolarized to +100 mV for 8 s to activate VSPs. After VSP activation, tail currents were again recorded once every 500 ms for 40 s. (D) Dr-VSP and the Mm-VSP phosphatase inhibit M currents. Shown are representative currents recorded from HEK293T/17 cells cotransfected with KCNQ2, KCNQ3, and a vector containing Dr-VSP (red), the Mm-VSP phosphatase chimera (blue), or no VSP (black). Traces recorded immediately before cells were depolarized to +100 mV are shown in lighter colors, and traces recorded immediately after depolarization are shown in darker colors. (E) Changes in normalized M current (Imax − Ifinal) at +100 mV. Changes in normalized M current observed in individual cells are plotted with circular markers, and medians for each condition are shown as rectangles. The resulting data were compared using a Wilcoxon rank-sum test (∗p < 0.05; n.s., p > 0.05). (F) Average normalized M current (I/Imax) of cells held at +100 mV. The lighter center of each trace represents the average I/Imax, and darker bars represent the per-time-point standard error of the mean. No VSP, n = 4 cells; Dr-VSP, n = 4 cells; Mm-VSP phosphatase, n = 7 cells. (G) Normalized tail current amplitudes plotted over time. The tail current amplitude was recorded at discrete time points and repeatedly measured at −60 mV after short pulses to −20 mV. Amplitudes were normalized to the initial tail current in each cell. Error bars are standard error of the mean for the same four to seven cells analyzed in (E). To see this figure in color, go online
Statistical analysis
Statistical tests were performed using IGOR Pro version 6.3.4.1 (WaveMetrics) and the R statistical programming environment (25). For all tests, α (the threshold for significance) was set to 0.05. Groups of data points were tested for approximate normality using the Jarque-Bera test. When the hypothesis of normality was not rejected, pairwise comparisons between families of data points were performed using Student’s t-test. When the data were found to be not normally distributed, the Wilcoxon rank-sum test was applied for pairwise comparisons. In instances involving multiple testing across families of data points, the Kruskal-Wallis test was performed to test the hypothesis that all data sets came from the same distribution. If this hypothesis was rejected, post hoc pairwise testing was performed using the Wilcoxon rank-sum test followed by Holm’s correction for multiple testing.
Results
Mm-VSP expression is developmentally regulated in the brain
Mm-VSP expression has been reported in the testis (26, 27), similarly to other VSP family members (3, 28, 29, 30). In most species, VSP expression has been detected in the central nervous system as well. A large-scale genomic study in mouse suggested that Mm-VSP might be expressed in the brain (31); however, these results have not yet been confirmed. More recent reports on VSP activity still refer to Mm-VSP as a testis-specific protein (29, 32), leaving unanswered the question of whether Mm-VSP is expressed in the brain. Because both electrical signaling and phospholipid signaling play important roles in the brain, neural expression of Mm-VSP would likely have important physiological consequences.
To determine whether Mm-VSP is expressed in the mouse brain, we first looked for mRNA transcript using RT-PCR. Primers were designed to amplify three different regions of Mm-VSP mRNA, to decrease the possibility of false negatives due to expression of different isoforms. RT-PCR targeting MAP2 was used as a positive control. Because expression of Mm-VSP has already been reported in the testis, RNA extracted from P37 mouse testis was used as a positive control for Mm-VSP expression. When RT-PCR was performed on RNA extracted from mouse brain, Mm-VSP transcript was detected using each of the primer pairs in both male and female mice at both P0 and P37 (Fig. 1 A).
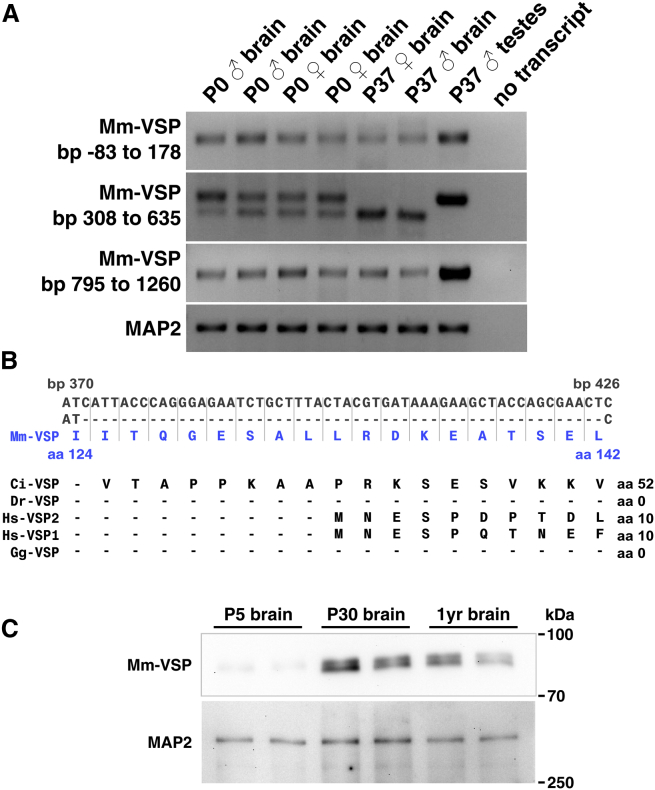
Figure 1.
Mm-VSP mRNA and protein are expressed and developmentally regulated in mouse brain. (A) Mm-VSP mRNA is expressed and developmentally regulated in the brain. RT-PCR amplicons were visualized in an agarose gel using ethidium bromide. Left-hand labels indicate the basepairs of Mm-VSP’s mRNA sequence amplified by each primer pair, where 0 is the beginning of the open reading frame. Primers against MAP2 mRNA were used as a positive control. Top labels indicate the postnatal age (in days), gender, and tissue of the mice from which RNA was collected. Each gel is representative of three replicates. (B) A novel Mm-VSP variant lacks exon 9. In dark gray (top row) is the cDNA sequence for Mm-VSP spanning exon 9 and flanking bases. In the second row, the sequence of the splice variant is shown with exon 9 indicated by dashes. The amino acids encoded by the cDNA are indicated below in blue. The alternate splice variant identified in (A) was found to be lacking exon 9 after sequencing of the amplicon. Shown below in black are the amino acid sequences of other VSP family members, aligned with Mm-VSP using COBALT. Note that the Mm-VSP amino terminus is much longer than that of other VSPs and lacks obvious sequence similarity to other orthologs in the region encoded by exon 9. (C) Mm-VSP protein is expressed and developmentally regulated in the brain. Whole-brain homogenates were probed in a western blot to detect the presence of Mm-VSP protein. MAP2, probed for on the same blot, served as a loading control. Tissue samples from two separate mice are shown for each age. The blot is representative of three technical replicates. The predicted mass of Mm-VSP is 77 kDa. Two bands near this size are seen to be present in whole-brain homogenate, and their intensity increases with the age of the mice. To see this figure in color, go online.
Surprisingly, a primer pair that targeted the mRNA region encoding part of the amino terminus amplified two differently sized products in the brains of P0 mice (Fig. 1 A, second row). By P37, only the shorter product could be detected in the brain, whereas only the longer product could be detected in the testis. Sequencing the DNA corresponding to the two bands revealed that the higher-molecular-weight band contained the expected 328 basepair sequence, whereas the lower-molecular-weight band contained a sequence lacking the 54 bases of exon 9 (Fig. 1 B). Alternate splicing of Mm-VSP has been predicted (26), although the exclusion of exon 9 has not been previously reported. We term this novel (to our knowledge) splice variant Mm-VSP-Ex9. Exon 9 encodes a region of Mm-VSP’s cytoplasmic amino terminus, and therefore this splice variant is not expected to affect the voltage-dependent properties or phosphatase activity of Mm-VSP. Furthermore, this region is one of the most evolutionarily divergent among VSPs, and exon 9 demonstrates very little homology to any other proteins, even among other VSP family members (Fig. 1 B).
To track Mm-VSP protein, we developed and validated a polyclonal antibody against a sequence in the amino terminus of Mm-VSP that is unique to the mouse protein (Fig. S1). To determine whether Mm-VSP protein is present in the brain, we used this antibody in Western blots of whole-brain homogenate (Fig. 1 C). Very little Mm-VSP was identified in mouse brain at P5, but by P30 a strong band that ran close to the expected molecular mass of Mm-VSP (76 kDa) was observed. This band was detected in the brains of 1-year-old mice as well. These data indicate that Mm-VSP is expressed in the adult mouse brain and is developmentally regulated at both the mRNA and protein levels.
The two predominant cell types within the brain are neurons and glia, and the most abundant glial cells are astrocytes (33). To address whether neurons, astrocytes, or both cell types express Mm-VSP, we investigated protein expression in primary cocultures of mouse cortical neurons and astrocytes. Cells were fixed after the neurons had grown for 14 days in vitro, and the cultures were then labeled with anti-Mm-VSP antisera. To distinguish between the cell types in the cultures, cells were colabeled with antibodies against protein markers of neurons (MAP2), synapses (SV2), or astrocytes (GFAP). MAP2-positive neurons consistently also showed high anti-Mm-VSP labeling (Fig. 2 A), whereas GFAP-positive astrocytes were consistently seen to lack anti-Mm-VSP labeling. These data suggest that within the brain, Mm-VSP is expressed exclusively in neurons.
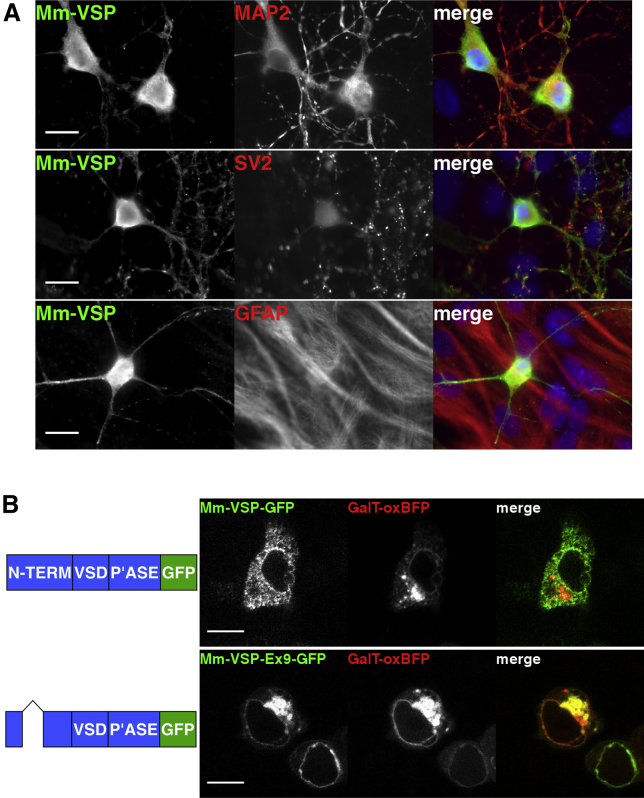
Figure 2.
Mm-VSP is expressed in neurons and localizes to intracellular compartments. (A) Mm-VSP is expressed in neurons, but not astrocytes. Mouse cortical neurons were cultured for 14 days in vitro on a confluent layer of astrocytes. Cells were double labeled with anti-Mm-VSP antibodies (left panels, green in the merged image) and an antibody against a protein marker of neurons (MAP2), synapses (SV2), or glia (GFAP) (center panels, red in the merged image). Cell nuclei were labeled with Hoechst dye (blue). Scale bars indicate 15 μm. (B) GFP-tagged Mm-VSP localizes to intracellular membranes in HEK293T/17 cells. Schematics depicting Mm-VSP-GFP variants (left) are shown alongside live-cell confocal fluorescence images of HEK293T/17 cells (right). N-term, amino terminal domain; VSD, voltage-sensing domain; P’ase, phosphatase domain. Removal of exon 9 is depicted as a carat connecting exons 8 and 10. Cells were cotransfected with the indicated Mm-VSP constructs (first panel, green in the merged image) and the GalT-oxBFP construct (center panel, red in the merged image). The fluorescent GalT-oxBFP protein localizes to the Golgi apparatus (34). Images were taken 24 h after transfection. Scale bars indicate 10 μm. To see this figure in color, go online.
Consistent with previous reports on Mm-VSP, we found that the protein localized to intracellular compartments when expressed in heterologous systems (26, 27). Indeed, when we expressed Mm-VSP-GFP fusion proteins in HEK293T/17 cells, we observed fluorescence in intracellular compartments (Fig. 2 B). Because previous studies indicated that Mm-VSP trafficked to the Golgi, we coexpressed our Mm-VSP fusion proteins with GalT-oxBFP, a fluorescent marker of the Golgi (34). We did not observe colocalization between fluorescence from full-length Mm-VSP-GFP and GalT-oxBFP. Rather, we saw Mm-VSP-GFP fluorescence in a compartment morphologically consistent with the endoplasmic reticulum (Fig. 2 B, top panel). This localization was not due to the presence of the fluorescent tag, because we observed a similar intracellular localization when a FLAG-Mm-VSP fusion protein was expressed in HEK293T/17 cells (Fig. S1 B). In contrast, we saw that fluorescence from the GFP-fusion of the exon 9-lacking splice variant, Mm-VSP-Ex9-GFP, did colocalize with GalT-oxBFP fluorescence, indicating localization to the Golgi (Fig. 2 C, bottom panel). This finding is comparable to previous reports that different splice variants of human VSPs localize to different subcellular compartments in heterologous expression systems (28, 35).
The VSD of Mm-VSP is voltage and pH sensitive
Whereas many VSPs generate sensing currents that can be used to characterize their voltage sensitivity (3, 29, 30, 36, 37), no such currents have yet been detected from mammalian VSPs, even those that have been observed to traffic to the plasma membrane (14). We thus sought another method to assess the voltage sensitivity of Mm-VSP. Recently, Arrigoni et al. (38) showed that the VSD from Ci-VSP could be expressed as a fusion protein with the pore of the viral potassium channel Kcv. The resulting chimera, KvSynth1, trafficked to the plasma membrane in Xenopus oocytes, and the voltage-dependent conformation change in the VSD could be used to drive gating of the pore. Hence, ionic currents generated by the KvSynth channel provide a readout of VSP voltage-sensor activation, a property that we utilized to investigate the voltage dependence of Mm-VSP.
To generate a KvSynth channel that would enable us to study Mm-VSP’s voltage sensor, we replaced the VSD of Ci-VSP in KvSynth1 with the VSD (amino acids 143–334) of Mm-VSP (Fig. 3, A and B), resulting in a chimera we termed KvSynthM. The KvSynth1 and KvSynthM channels were each transiently expressed in HEK293T/17 cells, and the cells were voltage clamped in whole-cell mode. To measure the voltage sensitivity of the Ci-VSP and Mm-VSP voltage sensors in the context of the KvSynth channels, we applied a series of depolarizing pulses and measured instantaneous tail currents at −40 mV (Fig. 3 C). The tail current amplitude was plotted against the activating voltage, and the data from each individual cell were fitted with a two-state Boltzmann function to derive the voltage of half-maximal activation (V1/2). The maximal and base values of the Boltzmann function were allowed to be fit to the data as free parameters, and the data from each cell were normalized based on these parameters. These curves were then averaged together for visual comparison (Fig. 3, D–F) and statistical analysis was performed on the families of values from the individual cells. Cells that produced values that could not be fit with a Boltzmann function were excluded from analysis.
HEK293T/17 cells expressing KvSynth1 produced voltage-dependent currents that were qualitatively similar to those reported when KvSynth1 was expressed in Xenopus oocytes (Fig. 3 E). These voltage-dependent currents comprised an instantaneous component and a slowly activating component, as previously reported (38). Analogous currents were not seen in cells that were not transfected with KvSynth channels (Fig. 3 D). There was, however, a large difference in the V1/2 we observed (−12 mV) and the V1/2 reported for KvSynth1 expressed in oocytes (+56 mV). This discrepancy may be due to the different cell types used and the different holding potentials from which test pulses were applied. For experiments in HEK293T/17 cells, we held the cells at −70 mV to model the neuronal resting membrane potential, whereas Arrigoni et al. held oocyte membranes at −20 mV (38). Although this difference is notable and warrants further investigation, it does not prevent us from using the KvSynth channels to address the question of whether the VSD in mammalian VSPs is functional. Nevertheless, because of this difference, and the fact that in this work our assessment of Mm-VSP voltage-sensor function was limited to the KvSynthM chimera, we did not seek to draw strong conclusions about the exact properties of voltage activation. Rather, we sought to determine the relative differences, if any, between the behavior of the Ci-VSP voltage sensor and that of the Mm-VSP voltage sensor.
Cells expressing the KvSynthM chimera produced large voltage-dependent currents, confirming that the transmembrane region of Mm-VSP acts as a VSD (Fig. 3 F). In the original report characterizing KvSynth1, Arrigoni and colleagues (38) demonstrated that the strong rectification observed in KvSynth1 was due to coupling between the VSD and the pore. Thus, we compared the degree of rectification of KvSynthM currents with that observed in cells expressing KvSynth1 by calculating the ratio of normalized current amplitude at 40 mV to current amplitude at −40 mV (Fig. 4 A). Values were normalized to the current at 70 mV to control for differences in expression levels. We observed similar degrees of rectification in KvSynthM and KvSynth1, leading us to conclude that our KvSynthM chimera demonstrates similar coupling between the VSD and pore domains.
Figure 4.
Electrophysiological properties of cells expressing KvSynth channels at neutral and low pHi, as determined by further analysis of recordings depicted in Fig. 3. Cells expressing KvSynth1 (1), KvSynthM (M), or the KvSynthM:H330G mutant (mut) at the indicated pH are depicted in different colors. Black circles represent values obtained from untransfected cells. For a given channel, data from cells with intracellular solutions of pHi 7.4 (darker colors) are shown to the left of data from cells with intracellular solutions of pHi 6.6 (lighter colors). Averages are shown as open circles. (A) KvSynthM couples voltage to pore opening comparably to KvSynth1. Current-voltage relationships for cells expressing KvSynth channels were normalized to the current observed at 70 mV to allow for comparison between different constructs and expression levels. To measure the degree of rectification, the ratio of the currents at 40 mV and −40 mV was compared between the KvSynth constructs. Individual cells’ current ratios are shown as solid circles and the average current ratios for each KvSynth construct are shown as open circles. Similar rectification was observed between constructs. All recordings were obtained at pHi 7.4. (B) V1/2 as a function of pHi. Solid circles represent the V1/2 of the tail currents measured from an individual cell expressing the indicated KvSynth channel at the indicated pHi. The open circles represent the average V1/2. Values were compared between pHi conditions for a given KvSynth channel using Student’s t-test (∗p < 0.05). Only KvSynthM demonstrated a statistically significant difference in V1/2 between pHi of 6.6 and 7.4. (C) Tail current ratios as a function of pHi. Each solid datum represents the tail current of an individual cell after pulsing to −40 mV divided by the tail current of that same cell after pulsing to 70 mV. Open circles represent the average. Values were compared between pHi conditions for a given KvSynth channel using Student’s t-test (∗p < 0.05). There was a statistically significant difference between the tail current ratios at pHi of 7.4 between KvSynth1 and KvSynthM. Only KvSynthM demonstrated a statistically significant difference in this current ratio between pHi 6.6 and pHi 7.4. (D) KvSynth current amplitudes are not noticeably different between pHi 7.4 and 6.6. Each datum represents the average whole-cell current in a HEK293T/17 cell at either −70 mV (left) or 0 mV (right). Note that none of the constructs produces appreciable current at −70 mV, indicating that expression of KvSynth channels does not produce significant leak current at resting membrane potentials. To see this figure in color, go online.
We assessed the voltage dependence of channel opening at negative potentials by calculating the ratio of current at −40 mV/−70 mV. This revealed that Mm-VSP-based KvSynthM was more active at hyperpolarized potentials than Ci-VSP-based KvSynth1 (Fig. 4 C). To attempt to understand the basis of this difference, we compared the residues of the fourth transmembrane segment (S4) between Mm-VSP and Ci-VSP. S4 is known to contain positively charged amino acid residues, termed gating charge residues, that confer sensitivity to voltage in voltage-gated ion channels and VSPs (4, 39). Mm-VSP appeared to lack one of the gating charge residues present in Ci-VSP (Fig. 3 B), which likely contributes to the weaker voltage sensitivity of the Mm-VSP VSD (Table 1). This comparison also revealed that the Mm-VSP VSD contains a histidine residue, H330, that is conserved in human VSPs but absent in Ci-VSP.
Table 1.
Values of Boltzmann Fits to KvSynth Chimera Currents
| KvSynth1, pHi 7.4 | KvSynth1, pHi 6.6 | KvSynthM, pHi 7.4 | KvSynthM, pHi 6.6 | KvSynthM:H330G, pHi 7.4 | KvSynthM:H330G, pHi 6.6 | |
|---|---|---|---|---|---|---|
| V1/2 (mV) | −12.43 ± 2.99 | −7.96 ± 3.70 | −24.39 ± 5.14 | −4.07 ± 6.28 | 25.60 ± 7.26 | 30.69 ± 4.62 |
| z | 1.00 ± 0.08 | 1.02 ± 0.04 | 0.84 ± 0.09 | 0.77 ± 0.05 | 0.93 ± 0.07 | 1.00 ± 0.15 |
Tail currents from individual HEK293T/17 cells expressing the indicated channels were plotted versus activating voltage and fitted with the two-state Boltzmann function:
where
and k is the Boltzmann constant, T is the absolute temperature in Kelvin, and e is the elementary charge. kT/e was taken to be a constant (25.693 mV) at room temperature to estimate values of z. Mean values for the recordings depicted in Fig. 3 are reported in the above table ± standard error of the mean.
The pKa of a histidine side chain (typically near 6.0) is such that histidine can be protonated at low physiological pH, which adds positive charge. Consequently, when gating charge residues of voltage-gated channels (40, 41) or Ci-VSP (39) are replaced with histidines, the mutant proteins demonstrate a pH-dependent influence on voltage-dependent properties. Furthermore, Sutton et al. (42) reported that a histidine in the same region (three residues before the analog of H330 in human VSPs) conferred proton flux in voltage-sensor chimeras. H330 in Mm-VSP is six residues away from the final gating charge of the VSD, and thus is likely near the same face of the α-helical S4 as the gating charges. We therefore hypothesized that this histidine, which is conserved in human VSPs, might result in pH-titratable gating effects for the VSD of Mm-VSP. Based on the known structure of Ci-VSP’s voltage sensor (5), we expected this histidine to be near the cytoplasmic side of Mm-VSP’s S4. This reasoning led us to predict that changing the intracellular pH (pHi) might affect the voltage-dependent properties of KvSynthM. To test this possibility, we repeated the KvSynth experiments using a pHi of 6.6 instead of 7.4.
Decreasing the pHi from 7.4 to 6.6 had very little effect on either the V1/2 (Fig. 4 B) or the strength of voltage dependence observed for the Ci-VSP-based KvSynth1 (Fig. 3 E). In contrast, the Mm-VSP-based KvSynthM demonstrated moderately different voltage-dependent properties at different pHi values (Fig. 3 F). In particular, the V1/2 of KvSynthM increased from −24 mV at pHi 7.4 to −4 mV at pHi 6.6 (Fig. 4 B; Table 1). Although the difference in the average V1/2 between pHi conditions was statistically significant, there was a large degree of variability in the measured V1/2 for individual cells expressing KvSynthM. To provide an additional test of pH effects, we employed an alternate measure of voltage-dependent gating that did not depend on the goodness of fit of any particular model. By measuring the ratio of the tail current amplitude after pulsing to −40 mV over the tail current amplitude after pulsing to 70 mV, we calculated the percentage of total channel activation observed at a moderate voltage. When we performed this analysis, we saw no effect of pHi on the current ratio of KvSynth1 (Fig. 4 C). KvSynthM, however, demonstrated a statistically significant (albeit slight) decrease in the current ratio at pHi 6.6. This result indicates less channel activity at −40 mV at pH 6.6 and is consistent with the difference in V1/2.
The properties of the pHi-dependent effects we observed in KvSynthM are distinct from the proton permeation in VSD chimeras reported by Sutton and colleagues (42). The proton conductance they reported is on the order of 1–4 pS, compared with the >100 pS conductance of KvSynth1 (38). A proton conductance on this scale would make up a small fraction of our observed current, and therefore is unlikely to account for the observed effect of pHi on the voltage dependence of KvSynthM. Conversely, if KvSynthM were to conduct a significant outward proton current in addition to the expected potassium current, cells with pHi 6.6 would be expected to demonstrate larger current amplitudes than cells with pHi 7.4, due to the increased proton driving force. Contrary to this, the KvSynthM current amplitudes did not differ significantly between cells with pHi 7.4 and cells with pHi 6.6 (Fig. 4 D). Furthermore, H330 is conserved in the human VSPs (Fig. 3 B), and Sutton and colleagues (42) found that this residue did not produce proton currents in their experiments.
To attempt to identify the mechanism underlying the apparent pH sensitivity in KvSynthM, we mutated the candidate pH-sensing residue H330 to a glycine to prevent protonation at this amino acid. The histidine-to-glycine mutation (H330G) did abolish the pH-dependent shift in V1/2 and the difference in the tail current ratio (Figs. 3 G and 4, B and C; Table 1). However, we were not able to conclusively determine that this residue behaves as a pH sensor in KvSynthM because the H330G mutant had a positively shifted activation curve and stronger voltage dependence compared with the wild-type KvSynthM (Table 1). Therefore, H330 appears to have a pH-independent influence on KvSynthM gating that complicates the interpretation of results obtained by changing the pHi in the context of this mutation.
Although these results suggest that both H330 and pH could play functional roles in the activity of Mm-VSP, it should be noted that the effect size of changing the pHi from 7.4 to 6.6 is small in KvSynthM, corresponding to a change in activation of ∼10%. While this is an intriguing finding, we acknowledge that further experiments will be required to fully evaluate the functional roles of intracellular pH and H330 in native Mm-VSP.
The phosphatase domain of Mm-VSP inhibits PI(4,5)P2-sensitive M currents
It has been shown that the enzymatic activity of the phosphatase domains of the tumor suppressor PTEN and the human Hs-VSP1 (previously named TPIP) can be controlled by the VSD from Ci-VSP (14, 43). VSP chimeras can thus be a useful tool for investigating the activity of phosphatase domains. Because we found no indication of Mm-VSP on the plasma membrane (Fig. 2 B), we turned to a chimeric approach to study Mm-VSP’s phosphatase activity. To this end, we generated a chimera in which the phosphatase from Mm-VSP is fused to the VSD of the zebrafish VSP (Dr-VSP) (Fig. 5, A and B). The VSD of Dr-VSP traffics readily to the plasma membrane and requires membrane potentials above 0 mV to be activated (36), making it ideally suited for studies in HEK293T/17 cells.
To assess the phosphatase activity of Dr-VSP and the Dr-VSP/Mm-VSP chimera, we measured their effect on M currents. The amplitudes of M currents, generated by potassium channels comprising the KCNQ2 and KCNQ3 subunits, are highly sensitive to the concentration of PI(4,5)P2 in the plasma membrane (44). Consequently, the amplitudes of M currents serve as an electrophysiological readout of the PI(4,5)P2 concentration in the plasma membrane. Because dephosphorylation of PI(4,5)P2 is the best-characterized enzymatic activity of VSPs examined to date, we coexpressed VSPs with KCNQ2/3 channels to assess lipid phosphatase activity. To test whether the Mm-VSP phosphatase produced a voltage-dependent decrease in PI(4,5)P2, cells were cotransfected with KCNQ2, KCNQ3, and the chimera containing the Mm-VSP phosphatase fused to the Dr-VSP VSD. Transfected cells were subjected to a stimulation protocol in which the membrane was depolarized to +100 mV (Fig. 5 C). In cells expressing the Dr-Mm-VSP chimera, the M current was suppressed (Fig. 5, D–F) with a time constant of τinhibition = 4.5 s (Table S1). We did not observe inhibition of M currents in cells that were coexpressing full-length Mm-VSP (Fig. S2), which is consistent with Mm-VSP being absent from the plasma membrane.
To verify that the observed suppression of M currents was due to the Dr-VSP/Mm-VSP chimera and not to an activity endogenous to HEK293T/17 cells, we repeated the experiment in cells cotransfected with KCNQ2, KCNQ3, and the pIRES2-EGFP vector (containing no VSP construct). Cells expressing these constructs generated appreciable M currents when they were held at −60 mV and depolarized to −20 mV (Fig. 5, D–F). Repeated measurements of tail currents in individual cells demonstrated that the M-current amplitudes did not show much variability (Fig. 5 G), indicating that the concentration of PI(4,5)P2 at the plasma membrane was stable over time. Additionally, the M-current amplitudes were stable when currents were continuously recorded for 8 s at +100 mV (Fig. 5, E and F).
To compare the effects of activation of the Mm-VSP phosphatase and a well-studied VSP, we cotransfected cells with KCNQ2, KCNQ3, and Dr-VSP. As in the absence of Dr-VSP, robust M currents were observed at −20 mV (Fig. 5 D). Repeated measurements of tail currents at discrete time points again showed very little variability, confirming that Dr-VSP was not active at this potential (Fig. 5 G). When cells expressing Dr-VSP were depolarized to +100 mV, M-current amplitudes rapidly decreased (Fig. 5, D–F), indicating depletion of PI(4,5)P2. Repolarizing cells and continuing to measure tail currents repeatedly after the +100 mV depolarization revealed tail current-amplitude recovery over time, indicating the resynthesis of PI(4,5)P2 by endogenous lipid kinases (Fig. 5 G). When M-current inhibition and tail current recovery after Dr-VSP activation were fitted with single exponentials (Table S1), the time constants from the fits (τinhibition = 0.3 s τrecovery = 4.4 s) were in good agreement with those previously reported under these conditions (16).
Suppression of KCNQ2/3 currents at +100 mV and recovery of tail currents after suppression both occurred more slowly in cells coexpressing the Mm-VSP phosphatase chimera than in cells coexpressing Dr-VSP (Fig. 5, F and G). Furthermore, fitting the time course of tail current recovery after Mm-VSP phosphatase activation required a double exponential, whereas recovery of the tail currents after Dr-VSP activation could be fit with a single exponential (Table S1). Interestingly, the fast component of tail-current recovery after activation of the Mm-VSP phosphatase-containing chimera (τfast recovery = 2.5 s) was comparable to that observed after Dr-VSP activation (τrecovery = 4.4 s); however, the second time constant was about seven times slower (τslow recovery = 18.6 s). Taken together, our data indicate that the phosphatase domain of Mm-VSP can be activated by the Dr-VSP voltage sensor, and that its activation can reduce the concentration of PI(4,5)P2 in the plasma membrane, though with different kinetics compared with Dr-VSP.
Discussion
The biological functions of VSPs are not yet understood, particularly in mammals. However, both electrical signaling and phospholipid signaling have been well studied with regard to a wide variety of cellular processes. In the mammalian brain, for example, these signaling mechanisms have been implicated in diseases as diverse as cancer (45, 46), epilepsy (47, 48), and autism (49, 50). This suggests that VSPs serve important physiological roles in the cells where they are expressed, and may provide useful drug targets.
The nonmammalian Ci-VSP (3) and Dr-VSP (36) are the two best-characterized VSPs to date; however, previous work on Mm-VSP and the Hs-VSPs suggests that mammalian and nonmammalian proteins may have distinct functional properties. The first line of evidence for this comes from the different subcellular localization observed for mammalian and nonmammalian VSPs when expressed in heterologous cells. Whereas both Ci-VSP and Dr-VSP localize to the plasma membrane (3, 36), most isoforms of the human and mouse VSPs appear to localize to intracellular membranes (26, 35). In this study, we observed intracellular localization of Mm-VSP in heterologous expression systems (Fig. 2 B). This suggests that if Mm-VSP is present on the surface of neurons, it may require a trafficking partner or splice variant that is absent in cultured fibroblasts.
Another putative difference between mammalian and nonmammalian VSPs is the voltage sensitivity of the proteins’ VSDs. Whereas nonmammalian VSPs are characteristically activated by very large depolarizations (11, 29, 30, 36), our data indicate that Mm-VSP may be more active at negative potentials (Fig. 4 C). Furthermore, the weak voltage sensitivity of the KvSynthM chimera indicates that factors other than membrane voltage may act as modulators of Mm-VSP. One such possible modulator suggested by our data is intracellular pH, although we note that because we performed our studies with chimeras, it remains unknown whether the properties revealed here accurately represent the properties of full-length Mm-VSP in its native environment.
The phosphatase activity of some mammalian VSPs also appears to differ from that of nonmammalian VSPs. Previous reports have shown that whereas the phosphatase domain of human Hs-VSP1 dephosphorylates PI(4,5)P2 (14), two residue substitutions at positions 342 and 343 in the P-loop region of Hs-VSP2 prevent phosphatase activity against lipid substrates (51) (Fig. 5 A). In this study, we found that activating Mm-VSP’s phosphatase domain resulted in an apparent decrease in PI(4,5)P2, as measured by a decrease in PI(4,5)P2-dependent M current. However, the kinetics of M-current recovery after activation of the Dr-VSP/Mm-VSP chimera was distinct from that observed upon activation of Dr-VSP. Unlike the effects of Dr-VSP, which could be fit with a single exponential (Fig. 5, F and G; Table S1), the time course of PI(4,5)P2 recovery we observed after Mm-VSP activation was biphasic and required two exponential time constants to fit.
One potential scenario that could account for the biphasic recovery of the M current after activation of Mm-VSP phosphatase activity is that PI(4,5)P2 resynthesis takes place in two steps: phosphorylation of PI to produce PI(4)P, and subsequent phosphorylation of PI(4)P to produce PI(4,5)P2. It is known that PI(4)P and PI(4,5)P2 are in dynamic equilibrium in cells, mediated by the activity of lipid kinases and phosphatases (52). As a result, decreasing the concentration of PI(4)P could have the result of indirectly decreasing PI(4,5)P2. It is thus intriguing to speculate that Mm-VSP functions as a PI(4)P phosphatase, inhibiting M current by causing an indirect decrease in plasma membrane PI(4,5)P2. In this context, it is interesting to note that PI(4)P is the primary phosphoinositide in the Golgi body (53), raising the question of whether the subcellular localization of Mm-VSP that we and others have observed in heterologous systems (26, 27) (Fig. 2) reflects a functional role for Mm-VSP in this organelle.
If Mm-VSP were found to have PI(4)P phosphatase activity, it would be unique among VSPs, as only 3- and 5-phosphatase activities have been reported for other VSP family members. In particular, using a chimeric approach similar to ours, Halaszovich et al. (14) showed that the closely related Hs-VSP1 acts as a PI(4,5)P2 phosphatase. Although the sequence similarity of VSPs in regions such as the catalytic P-loop suggests that they would have similar substrate specificities, it is worth noting that the structural determinants of VSP substrate specificity are incompletely understood. For instance, in PTEN, the residues corresponding to Ci-VSP’s G365 and E411 are alanine and threonine, respectively. These differences have been shown to be responsible for Ci-VSP’s ability to dephosphorylate PI(4,5)P2, in contrast to PTEN’s strong specificity for PI(3,4,5)P3 (54). However, the glycine and glutamate at these positions are conserved among VSP family members that display varying substrate specificity. As an example, Gg-VSP from chicken displays almost no phosphatase activity against PI(4,5)P2, but has activity against PI(3,4,5)P3 and PI(3,5)P2 (30, 54), despite the presence of the glycine and glutamate in these two positions (Fig. 5 A).
Furthermore, evidence suggests that the phosphatase activity of Ci-VSP can change in a voltage-dependent manner, with 5-phosphatase activity dominating at moderate depolarizations and 3-phosphatase activity emerging at more severe depolarizations (54). Therefore, the residues that confer substrate specificity could be ones that move as a result of voltage-induced conformational changes. Based on crystal structures of the phosphatase domain of Ci-VSP, a gating loop was proposed to be the mobile structural element that couples movements from the VSD to regulate phosphatase activation (9). This region contains the conserved glutamate (E411 in Ci-VSP) that is known to be important for substrate specificity. Interestingly, the surrounding residues of the gating loop are somewhat less conserved than in other regions of the catalytic domain, such as the P-loop (Fig. 5 A). Mutagenesis experiments investigating this region may therefore provide insight into which residues are responsible for the varied substrate specificity across VSP proteins.
The fact that wild-type Mm-VSP did not appear to traffic to the plasma membrane presented a technical hurdle against studying the native protein. Therefore, we leveraged the modular nature of VSP VSDs and phosphatase domains to reveal the functionality of these domains in a mammalian VSP. When the VSDs and phosphatase domains were expressed as chimeras that allowed trafficking to the plasma membrane, we observed voltage-dependent gating of a viral pore or PI(4,5)P2-depletion-dependent inhibition of M currents, respectively. The ability of the VSDs and phosphatase domains to retain function in the context of a chimeric protein indicates some level of functional and structural conservation. However, we also observed subtle differences between the activities of these domains from VSPs of different species. Although more work is needed to determine whether the differences observed in the chimeras represent the respective activities of the native proteins, our findings provide the basis for future studies directed at determining how this intriguing class of proteins regulates cellular functioning in both health and disease.
Author Contributions
M.G.R. conceived the project. M.G.R., S.E.G., and S.M.B. designed the experiments. M.G.R. performed the experiments and analyzed the data. M.G.R., S.E.G., and S.M.B. wrote the manuscript.
Acknowledgments
The authors thank Eamonn J. Dickson, Bertil Hille, Anna Moroni, Erik Snapp, and Carlos Villalba-Galea for providing invaluable materials and support for this project.
This work was supported by grants from the National Institutes of Health (5F31MH099828 to M.G.R., and 5T32GM008268).
Editor: Miriam Goodman.
Footnotes
Two figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)01155-8.
Supporting Material
References
- 1.Wymann M.P., Schneiter R. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 2.Kass R.S. The channelopathies: novel insights into molecular and genetic mechanisms of human disease. J. Clin. Invest. 2005;115:1986–1989. doi: 10.1172/JCI26011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murata Y., Iwasaki H., Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 4.Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol. Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 5.Li Q., Wanderling S., Perozo E. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat. Struct. Mol. Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J., Yen C., Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 7.Steck P.A., Pershouse M.A., Tavtigian S.V. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 8.Maehama T., Dixon J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 9.Liu L., Kohout S.C., Minor D.L., Jr. A glutamate switch controls voltage-sensitive phosphatase function. Nat. Struct. Mol. Biol. 2012;19:633–641. doi: 10.1038/nsmb.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda M., Takeshita K., Nakagawa A. Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity. J. Biol. Chem. 2011;286:23368–23377. doi: 10.1074/jbc.M110.214361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murata Y., Okamura Y. Depolarization activates the phosphoinositide phosphatase Ci-VSP, as detected in Xenopus oocytes coexpressing sensors of PIP2. J. Physiol. 2007;583:875–889. doi: 10.1113/jphysiol.2007.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki H., Murata Y., Okamura Y. A voltage-sensing phosphatase, Ci-VSP, which shares sequence identity with PTEN, dephosphorylates phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 2008;105:7970–7975. doi: 10.1073/pnas.0803936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halaszovich C.R., Schreiber D.N., Oliver D. Ci-VSP is a depolarization-activated phosphatidylinositol-4,5-bisphosphate and phosphatidylinositol-3,4,5-trisphosphate 5′-phosphatase. J. Biol. Chem. 2009;284:2106–2113. doi: 10.1074/jbc.M803543200. [DOI] [PubMed] [Google Scholar]
- 14.Halaszovich C.R., Leitner M.G., Oliver D. A human phospholipid phosphatase activated by a transmembrane control module. J. Lipid Res. 2012;53:2266–2274. doi: 10.1194/jlr.M026021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein R.M., Ufret-Vincenty C.A., Gordon S.E. Determinants of molecular specificity in phosphoinositide regulation. Phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2) is the endogenous lipid regulating TRPV1. J. Biol. Chem. 2008;283:26208–26216. doi: 10.1074/jbc.M801912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falkenburger B.H., Jensen J.B., Hille B. Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol. 2010;135:99–114. doi: 10.1085/jgp.200910345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itsuki K., Imai Y., Mori M.X. Voltage-sensing phosphatase reveals temporal regulation of TRPC3/C6/C7 channels by membrane phosphoinositides. Channels (Austin) 2012;6:206–209. doi: 10.4161/chan.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villalba-Galea C.A., Sandtner W., Bezanilla F. S4-based voltage sensors have three major conformations. Proc. Natl. Acad. Sci. USA. 2008;105:17600–17607. doi: 10.1073/pnas.0807387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubota T., Lacroix J.J., Correa A.M. Probing α-3(10) transitions in a voltage-sensing S4 helix. Biophys. J. 2014;107:1117–1128. doi: 10.1016/j.bpj.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villalba-Galea C.A., Sandtner W., Bezanilla F. Charge movement of a voltage-sensitive fluorescent protein. Biophys. J. 2009;96:L19–L21. doi: 10.1016/j.bpj.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohout S.C., Ulbrich M.H., Isacoff E.Y. Subunit organization and functional transitions in Ci-VSP. Nat. Struct. Mol. Biol. 2008;15:106–108. doi: 10.1038/nsmb1320. [DOI] [PubMed] [Google Scholar]
- 22.Papadopoulos J.S., Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 23.Ye J., Coulouris G., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buckley K., Kelly R.B. Identification of a transmembrane glycoprotein specific for secretory vesicles of neural and endocrine cells. J. Cell Biol. 1985;100:1284–1294. doi: 10.1083/jcb.100.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2010. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 26.Guipponi M., Tapparel C., Antonarakis S.E. The murine orthologue of the Golgi-localized TPTE protein provides clues to the evolutionary history of the human TPTE gene family. Hum. Genet. 2001;109:569–575. doi: 10.1007/s004390100607. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y., Dowbenko D., Lasky L.A. PTEN 2, a Golgi-associated testis-specific homologue of the PTEN tumor suppressor lipid phosphatase. J. Biol. Chem. 2001;276:21745–21753. doi: 10.1074/jbc.M101480200. [DOI] [PubMed] [Google Scholar]
- 28.Walker S.M., Downes C.P., Leslie N.R. TPIP: a novel phosphoinositide 3-phosphatase. Biochem. J. 2001;360:277–283. doi: 10.1042/0264-6021:3600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratzan W.J., Evsikov A.V., Jaffe L.A. Voltage sensitive phosphoinositide phosphatases of Xenopus: their tissue distribution and voltage dependence. J. Cell. Physiol. 2011;226:2740–2746. doi: 10.1002/jcp.22854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi S., Kurokawa T., Homma K.J. Potential role of voltage-sensing phosphatases in regulation of cell structure through the production of PI(3,4)P2. J. Cell. Physiol. 2014;229:422–433. doi: 10.1002/jcp.24463. [DOI] [PubMed] [Google Scholar]
- 31.Reymond A., Marigo V., Ballabio A. Human chromosome 21 gene expression atlas in the mouse. Nature. 2002;420:582–586. doi: 10.1038/nature01178. [DOI] [PubMed] [Google Scholar]
- 32.Okamura Y., Murata Y., Iwasaki H. Voltage-sensing phosphatase: actions and potentials. J. Physiol. 2009;587:513–520. doi: 10.1113/jphysiol.2008.163097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kandel E.R., Schwartz J.H., Jessell T.M. McGraw-Hill, Health Professions Division; New York: 2000. Principles of Neural Science. [Google Scholar]
- 34.Costantini L.M., Baloban M., Snapp E.L. A palette of fluorescent proteins optimized for diverse cellular environments. Nat. Commun. 2015;6:7670. doi: 10.1038/ncomms8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tapparel C., Reymond A., Antonarakis S.E. The TPTE gene family: cellular expression, subcellular localization and alternative splicing. Gene. 2003;323:189–199. doi: 10.1016/j.gene.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 36.Hossain M.I., Iwasaki H., Okamura Y. Enzyme domain affects the movement of the voltage sensor in ascidian and zebrafish voltage-sensing phosphatases. J. Biol. Chem. 2008;283:18248–18259. doi: 10.1074/jbc.M706184200. [DOI] [PubMed] [Google Scholar]
- 37.Mutua J., Jinno Y., Okamura Y. Functional diversity of voltage-sensing phosphatases in two urodele amphibians. Physiol. Rep. 2014;2:2. doi: 10.14814/phy2.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arrigoni C., Schroeder I., Moroni A. The voltage-sensing domain of a phosphatase gates the pore of a potassium channel. J. Gen. Physiol. 2013;141:389–395. doi: 10.1085/jgp.201210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villalba-Galea C.A., Frezza L., Bezanilla F. Sensing charges of the Ciona intestinalis voltage-sensing phosphatase. J. Gen. Physiol. 2013;142:543–555. doi: 10.1085/jgp.201310993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starace D.M., Bezanilla F. Histidine scanning mutagenesis of basic residues of the S4 segment of the shaker k+ channel. J. Gen. Physiol. 2001;117:469–490. doi: 10.1085/jgp.117.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starace D.M., Stefani E., Bezanilla F. Voltage-dependent proton transport by the voltage sensor of the Shaker K+ channel. Neuron. 1997;19:1319–1327. doi: 10.1016/s0896-6273(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 42.Sutton K.A., Jungnickel M.K., Florman H.M. Evolution of the voltage sensor domain of the voltage-sensitive phosphoinositide phosphatase VSP/TPTE suggests a role as a proton channel in eutherian mammals. Mol. Biol. Evol. 2012;29:2147–2155. doi: 10.1093/molbev/mss083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacroix J., Halaszovich C.R., Villalba-Galea C.A. Controlling the activity of a phosphatase and tensin homolog (PTEN) by membrane potential. J. Biol. Chem. 2011;286:17945–17953. doi: 10.1074/jbc.M110.201749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H., Craciun L.C., Logothetis D.E. PIP(2) activates KCNQ channels, and its hydrolysis underlies receptor-mediated inhibition of M currents. Neuron. 2003;37:963–975. doi: 10.1016/s0896-6273(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 45.Litan A., Langhans S.A. Cancer as a channelopathy: ion channels and pumps in tumor development and progression. Front. Cell. Neurosci. 2015;9:86. doi: 10.3389/fncel.2015.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Endersby R., Baker S.J. PTEN signaling in brain: neuropathology and tumorigenesis. Oncogene. 2008;27:5416–5430. doi: 10.1038/onc.2008.239. [DOI] [PubMed] [Google Scholar]
- 47.Catterall W.A., Dib-Hajj S., Pietrobon D. Inherited neuronal ion channelopathies: new windows on complex neurological diseases. J. Neurosci. 2008;28:11768–11777. doi: 10.1523/JNEUROSCI.3901-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole-Edwards K.K., Bazan N.G. Lipid signaling in experimental epilepsy. Neurochem. Res. 2005;30:847–853. doi: 10.1007/s11064-005-6878-4. [DOI] [PubMed] [Google Scholar]
- 49.Brown C.M., Austin D.W. Autistic disorder and phospholipids: a review. Prostaglandins Leukot. Essent. Fatty Acids. 2011;84:25–30. doi: 10.1016/j.plefa.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 50.Schmunk G., Gargus J.J. Channelopathy pathogenesis in autism spectrum disorders. Front. Genet. 2013;4:222. doi: 10.3389/fgene.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leslie N.R., Yang X., Weijer C.J. PtdIns(3,4,5)P(3)-dependent and -independent roles for PTEN in the control of cell migration. Curr. Biol. 2007;17:115–125. doi: 10.1016/j.cub.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dickson E.J., Jensen J.B., Hille B. Golgi and plasma membrane pools of PI(4)P contribute to plasma membrane PI(4,5)P2 and maintenance of KCNQ2/3 ion channel current. Proc. Natl. Acad. Sci. USA. 2014;111:E2281–E2290. doi: 10.1073/pnas.1407133111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Overduin M., Cheever M.L., Kutateladze T.G. Signaling with phosphoinositides: better than binary. Mol. Interv. 2001;1:150–159. [PubMed] [Google Scholar]
- 54.Kurokawa T., Takasuga S., Okamura Y. 3′ Phosphatase activity toward phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2] by voltage-sensing phosphatase (VSP) Proc. Natl. Acad. Sci. USA. 2012;109:10089–10094. doi: 10.1073/pnas.1203799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.