Abstract
Barrett’s esophagus (BE) is a common and important precursor lesion of esophageal adenocarcinoma (EAC). A third of patients with BE are asymptomatic, and our ability to predict the risk of progression of metaplasia to dysplasia and EAC (and therefore guide management) is limited. There is an urgent need for clinically useful biomarkers of susceptibility to both BE and risk of subsequent progression. This study aims to systematically identify, review, and meta-analyze genetic biomarkers reported to predict both. A systematic review of the PubMed and EMBASE databases was performed in May 2014. Study and evidence quality were appraised using the revised American Society of Clinical Oncology guidelines, and modified Recommendations for Tumor Marker Scores. Meta-analysis was performed for all markers assessed by more than one study. A total of 251 full-text articles were reviewed; 52 were included. A total of 33 germline markers of susceptibility were identified (level of evidence II–III); 17 were included. Five somatic markers of progression were identified; meta-analysis demonstrated significant associations for chromosomal instability (level of evidence II). One somatic marker of progression/relapse following photodynamic therapy was identified. However, a number of failings of methodology and reporting were identified. This is the first systematic review and meta-analysis to evaluate genetic biomarkers of BE susceptibility and risk of progression. While a number of limitations of study quality temper the utility of those markers identified, some—in particular, those identified by genome-wide association studies, and chromosomal instability for progression—appear plausible, although robust validation is required.
Electronic supplementary material
The online version of this article (doi:10.1007/s10620-015-3884-5) contains supplementary material, which is available to authorized users.
Keywords: Barrett’s esophagus, Esophageal cancer, Esophageal dysplasia, Genetic, Biomarkers
Introduction
The importance of Barrett’s esophagus (BE) lies in its increasing prevalence and strong predisposition to esophageal adenocarcinoma (EAC) [1]. Western prevalence is estimated at 0.5–2.0 % [2] (a third of whom are asymptomatic [3]), conferring a 30- to 125-fold increased risk of developing EAC [4]. As EAC becomes more common [5], the benefits of predicting susceptibility to BE and detecting established metaplasia as soon as possible are numerous. Population screening overall is not presently feasible, but identifying both at-risk individuals and established metaplasia sooner provides windows for risk factor modification, chemoprevention, ablation, resection, and surveillance.
However, the natural history of BE is incompletely understood. Overall, the incidence of progression from metaplasia to high-grade dysplasia (HGD) or EAC is approximately 0.26–0.63 % per year [6], and that to EAC alone 0.12 % [7]. However, only a minority of patients progress to low-grade dysplasia (LGD), and of those that do similarly only a minority ultimately progress to HGD or EAC [8]. These progressive grades of dysplasia can help identify patients at particular risk (in particular, those with HGD in whom the risk is considerable) [9], but are insufficient in isolation.
There is therefore an immediate clinical need for biomarkers to predict both susceptibility to BE and progression. A major subtype is genetic variants, both germline and somatic. The potential of the latter, in particular, has been highlighted by a number of recent next-generation sequencing (NGS) studies of both EAC and BE, which have identified a number of candidate genes for further study [10]. However, no systematic reviews have been performed. The aims of this study were firstly to identify and evaluate all genetic markers tested in association with BE susceptibility and progression. Secondly, we aimed to identify markers with statistically significant associations and perform meta-analysis for those assessed by more than one study.
Methods
Inclusion Criteria
Studies testing associations between DNA markers [germline single nucleotide polymorphisms [SNP], somatic single nucleotide variants, insertions/deletions, copy number variants (CNV), loss of heterozygosity (LOH), microsatellite instability (MSI) or chromosomal instability (CIN)] and diagnosis/progression of BE were eligible; diagnosis was defined as endoscopic and histopathological evidence with or without intestinal metaplasia (IM) [13] and progression as histopathological progression from metaplasia to LGD/HGD/EAC or LGD to HGD/EAC, or HGD to EAC during surveillance endoscopies.
Exclusion Criteria
Studies comparing grades of metaplasia/dysplasia/malignancy within samples at one time point were excluded, as were studies comparing grades between patients. Studies using cell line, epigenetic, or expression data were excluded unless patient or DNA-specific data were available.
Literature Search
A search was performed in May 2014 of the PubMed and EMBASE databases, using the MOOSE and PRISMA guidelines [14] and the following term: [((((esophageal OR esophagus OR gastro esophageal)) AND (Barrett’s OR metaplasia OR columnar)) AND (genomic OR genetic OR genome OR pharmacogenetic OR pharmacogenomic OR amplification OR copy OR mutation OR polymorphism OR polymorphic OR variant OR deletion OR insertion OR locus OR loci OR allele OR ploidy OR instability OR biomarker))]. The bibliographies of retrieved articles were also searched.
Study Data
The following data were extracted: study methodology; variants and genes assessed; endpoints; population; and effect size [odds ratio (OR) or hazard ratio (HR)] and variance [standard error or confidence interval (CI)]. For studies not presenting OR, these were calculated using provided allele/variant frequencies. If reference SNP identification numbers were not provided (http://www.ncbi.nlm.nih.gov/dbSNP), these were mapped by searching cited methodology and performing in vitro polymerase chain reaction (http://genome.ucsc.edu), with nucleotide flank BLAST® (http://blast.ncbi.nlm.nih.gov).
Evidence Quality
Overall study and evidence quality were evaluated using the revised American Society of Clinical Oncology (ASCO) level of evidence (LOE) scale for biomarker research [15]. This stratifies study quality on the basis of trial design, patients and data, specimen collection, processing and archival, and statistical design and analysis from A to D (supplementary table 1), and uses this in conjunction with subsequent validation to stratify overall LOE for a marker from I to V (supplementary table 2). Methodological quality was appraised using the recommendations for tumor marker score (REMARK) guidelines modified by the authors, which scored study methodology in detail to generate a score from 0 to 17 (GWAS) and 0 to 18 (candidate studies; supplementary table 3). Reported associations were appraised for appropriate correction for multiple comparisons (Bonferroni method, false discovery rate, or multivariate analysis of all markers). If not undertaken, this was performed via post hoc Bonferroni correction. Significance was taken at p < 5×10−8 for genome-wide association studies (GWAS). Genomic quality criteria included reporting of genotyping call rate or providing data to allow its calculation, and assessment of Hardy–Weinberg equilibrium for germline variants.
Meta-Analysis
Meta-analysis was undertaken for markers assessed by more than one study, irrespective of correction for multiple comparisons (other than variants assessed by GWAS and non-GWAS due to major methodological differences) using RevMan v5.2 (Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration). Sensitivity analyses were performed on the basis of IM-only versus non-IM studies. In the case of studies reporting updated results using cohorts previously reported (i.e., population overlap), the most recent study was used.
Heterogeneity and Bias
Heterogeneity was estimated using I2 and Chi-square statistics. For moderate heterogeneity (I2 ≥ 50 %), a random-effects model was used. Funnel plots were reviewed for analyses of ≥5 studies [16]. Statistical assessment of these was not performed due to the low number of studies per variant. Publication bias was corrected using the “trim and fill” method [17].
Results
Literature Search
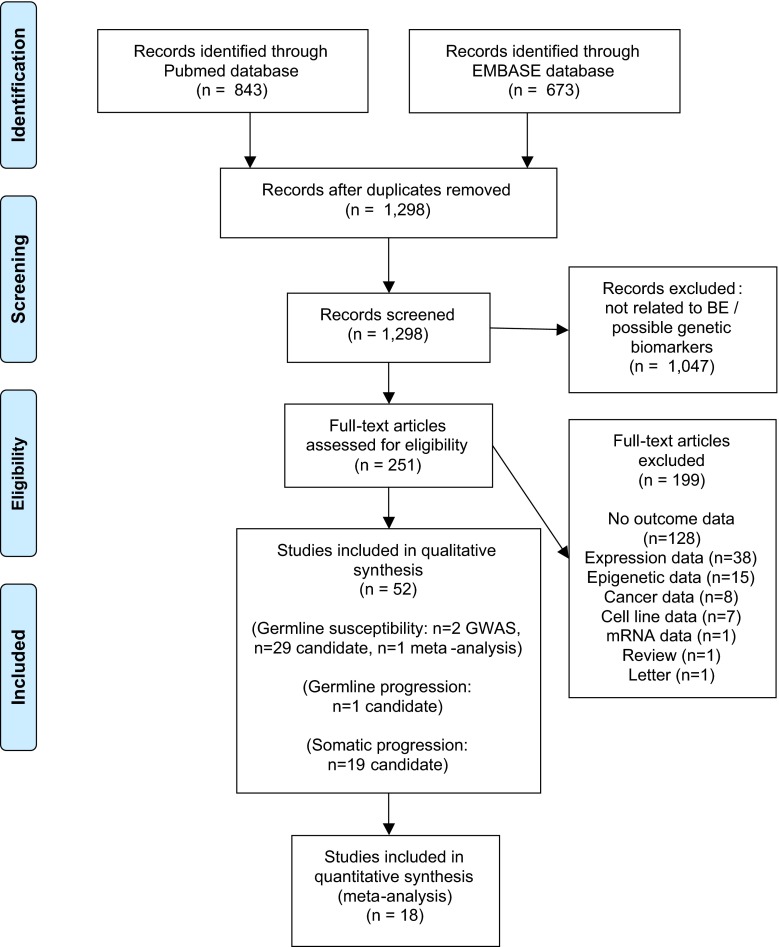
A total of 1516 articles were identified, of which 218 were duplicates. Therefore, 1298 articles were appraised on the basis of their title and abstract. Then, 251 full-text articles were retrieved, of which 52 met the inclusion criteria (Fig. 1).
Fig. 1.
PRISMA diagram
Studies Assessing Germline Markers of Susceptibility
A total of 32 studies were included: 2 GWAS, 29 candidate studies, and 1 meta-analysis of 4 candidate studies (supplementary tables 4 and 5). All original studies were LOE C (indicating prospective observational registries, without standardized treatment and follow-up), published between 1999 and 2014. Median modified REMARK scores were 15/17 (GWAS) and 13/18 (candidate studies; range 7–16; supplementary table 6). A number of frequent methodological issues were identified. The most common of these included failure to blind investigators (n= 18; 58.1 %), perform/report quality control procedures (19; 61.3 %), and match cases and controls (23; 74.2 %). Similarly, a number of recurrent reporting issues were identified. The most common of these were comparing markers with established risk factors (30; 96.8 %), adjusting for multiple comparisons (22; 71.0 %) and confounding factors (20; 64.5 %), reporting power calculations (15; 48.4 %) and reporting multivariate effect sizes (30; 96.8 %). Of the 27 studies assessing multiple variants, 24 did not perform multivariate analysis (88.9 %). For GWAS, there were a mean 5507 cases and 14,159 controls. For candidate studies, 134 cases and 196 controls. There were a number of cases of study population overlap, with 13 studies reported on subjects drawn from one of three populations.
Variants Associated with BE Susceptibility
A total of 187 candidate variants/haplotypes were tested. Twenty-eight significant associations were reported, of which 16 were excluded (Table 2). Twelve were therefore associated with BE (Table 1). However, for 2 of these (rs6785049 and rs9344) precise p values were not provided to allow for Bonferroni correction.
Table 2.
Reported germline markers of Barrett’s esophagus excluded on the basis of multiple comparisons
| Marker | Individual studies | Association (meta-analysis/individual study) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOE | Variant | Gene | Association | EV | LOE | Ethnicity | Methodology | ||||||
| Variant | Wild-type | Phenotype and OR (95 % CI) | I 2 | Chi | p | n | |||||||
| III | rs3776083 | CDX1 | Ren et al. [43] | AG | C | Caucasian | Candidate pro | BE-v-RE 1.91 (1.09–3.33) | NA | NA | 0.02 | 1 | |
| III | rs4769585 | CDX2 | Ren et al. [43] | CT | C | Caucasian | Candidate pro | BE-v-RE 2.68 (1.20–5.98) | NA | NA | 0.02 | 1 | |
| III | rs3812863 | CDX2 | Ren et al. [43] | AG | C | Caucasian | Candidate pro | BE-v-RE 2.53 (1.24–5.14) | NA | NA | 0.01 | 1 | |
| III | rs1143634 | IL1B | Izakovicoka-Holla et al. [27] | T | Caucasian (Czeck) | Candidate pro | BE -v-RE 0.56 (0.33–0.93) | NA | NA | 0.016 | 1 | ||
| III | VNTR polymorphism | IL1RN | Izakovicoka-Holla et al. [27] | 1/2 | Caucasian (Czeck) | Candidate pro | BE 0.62 (0.42–0.94) | NA | NA | 0.015 | 1 | ||
| III | rs1800587/rs16944/ rs1143634/IL1RN |
IL1B/ILRN | Izakovicoka-Holla et al. [27] | C/T/C/long | Caucasian (Czeck) | Candidate pro | BE 1.95 (1.09–1.30) | NA | NA | 0.03 | 1 | ||
| III | rs1800587/rs16944/r s1143634/IL1RN |
IL1B/ILRN | Izakovicoka-Holla et al. [27] | T/C/C/long | Caucasian (Czeck) | Candidate pro | BE 2.67 (1.15–6.24) | NA | NA | 0.02 | 1 | ||
| III | rs909253 | TNFB | Menke et al. [50] | AA | C | Caucasian (Netherlands) | Candidate retro archived | BEA 1.98 (1.03–3.81) | NA | NA | 0.04 | 1 | |
| III | rs2305764 | Myo9b | Menke et al. [51] | GG | C | Caucasian (Netherlands) | Candidate retro archived | BEA 2.96 (1.10–7.99) | NA | NA | 0.032 | 1 | |
| III | rs1946518 | IL18 | Babar et al. [29] | CC | Caucasian (Irish) | Candidate retro archived | BEA 1.45 (1.07–1.98) | NA | NA | 0.02 | 1 | ||
| III | rs1799750 | MMP1 | Bradbury et al. [52] | GG | Caucasian (USA) | Candidate retro archived | BEA 2.21 (1.08–4.53) | NA | NA | 0.03 | 1 | ||
| III | rs11209026 | IL23R | Gaj et al. [30] | A | Caucasians (Polish) | Candidate pro | BE 3.49 (1.37–8.90) | NA | NA | 0.011 | 1 | ||
| III | rs1800566 | NQO1 | di Martino et al. [53] | T | NP (UK) | Candidate pro | BEA-v-RE 0.22 (0.07–0.76) | NA | NA | 0.016 | 1 | ||
| III | B7 | HLA | Rajendra [54] | B7 | Asian (India) | Candidate pro | BEEx 21.4 (1.22–372 | NA | NA | 0.036 | 1 | ||
| III | rs415958/rs16944 | IL1RA/IL1B | Gough et al. [28] | CC/A | NS (UK) | Candidate pro | BE-v-RE 9.5 (1.19–75.9) | NA | NA | 0.010 | 1 | ||
| III | rs1800896 | IL10 | Gough et al. [28] | CC | NS (UK) | Candidate pro | BE-v-RE 1.84 (1.04–3.28) | NA | NA | 0.035 | 1 | ||
LOE level of evidence, EV effect variant, NP not presented; BE = Barrett’s esophagus; A adjusted OR, OR odds ratio, CI confidence interval, pro prospective, retro retrospective, NA not applicable, RE reflux esophagitis, Ex extrapolated
Table 1.
Reported germline markers of Barrett’s esophagus susceptibility
| Marker | Individual studies | Association (meta-analysis/individual study) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOE | Variant | Gene | Association | EV | LOE | Ethnicity | Methodology | |||||||
| Variant | Wild-type | None | Phenotype and OR (95 % CI) | I 2 | Chi | p | n | |||||||
| III | rs9257809 | MHC region | Su et al. [32], Palles et al. [33] | A | C | Caucasian | Multistage GWAS Pro/retro archived |
BEA 1.21 (1.13–1.28) | NA | NA | 4.09 × 10−9 | 1 | ||
| rs9936833 | FOXF1 | C | BEA 1.14 (1.10–1.19) | 2.74 × 10−10 | ||||||||||
| rs3072 | GDF7 | A | BEA 1.14 (1.09–1.18) | 1.75 × 10−11 | ||||||||||
| rs2701108 | TBX5 | A | BEA 0.90 (0.86–0.93) | 7.48 × 10−9 | ||||||||||
| rs2687201 | FOXP1 | T | BEA 1.16 (1.10–1.23) | 4.61 × 10−8 | ||||||||||
| III | rs3776082 | CDX1 | Ren et al. [43] | AA | C | Caucasian | Candidate Prospective |
BE-v-RE 4.02 (1.80–9.00) | NA | NA | <0.005 | 1 | ||
| III | rs2237091 | CDX1 | Ren et al. [43] | AA | C | Caucasian | Candidate Prospective |
BE-v-RE 0.28 (0.13–0.59) | NA | NA | <0.005 | 1 | ||
| III | rs717746 | CDX1 | Ren et al. [43] | GG | C | Caucasian | Candidate Prospective |
BE-v-RE 3.65 (1.73–7.69 | NA | NA | <0.005 | 1 | ||
| III | rs4444903 | EGFR | Menke et al. [20] | GG | C | Caucasian* | Candidate Retro archived |
BEA 3.00 (1.50–6.20) | NA | NA | 0.002 | 1 | ||
| III | rs917997 | IL18RAP | Babar et al. [29] | CC | C | Caucasian** | Candidate Retro archived |
BEA 0.59 (0.43–0.80) | NA | NA | 6.00 × 10−4 | 1 | ||
| III | rs6785049 | PXR | van de Winkel et al. [44] | G | C | Caucasian* | Candidate Retro archived |
BE 1.36 (1.03–1.79) | NA | NA | <0.05 (NS) | 1 | ||
| III | rs6214 | IGF1 | McElholm [21] | AA | C | Caucasian** | Candidate Retro archived |
BEA 0.43 (0.24–0.75) | NA | NA | 3.00 × 10−3 | 1 | ||
| III | rs2229765 + BMI > 30 | IGF1R | MacDonald et al. [22] | A | C | Caucasian | Candidate Retro archived |
BEA 3.11 (1.12–8.63) | NA | NA | <0.05 (NS) | 1 | ||
| III | rs3212227 | IL12B | Moons et al. [26] | C | C | Caucasian* | Candidate Retro archived |
BEA 1.82 (1.17–2.69) | NA | NA | 7.00 × 10−3 | 1 | ||
| II | rs1695 | GSTP | Bull et al. [18] | G | C | Caucasian | Meta-analysis | BE 1.50 (1.16–1.95) | NA | NA | <0.01 | 4 | ||
| Kala et al. [19] | G | Caucasian | Candidate (pro) | |||||||||||
| Murphy et al. [45] | G | Caucasian | Candidate (retro) | |||||||||||
| Casson et al. [46] | G | NP | Candidate (pro) | |||||||||||
| van Lieshout et al. [47] | G | Caucasian | Candidate (pro) | |||||||||||
| III | rs9344 | CCND1 | Casson et al. [31] | AA | C | NP | Candidate (pro) | BEA 3.69 (1.46–9.29) | NA | NA | <0.05 (NS) | 1 | ||
| III | rs25487 | XRCC1 | Casson et al. [48] | Ferguson et al. [49] | AA | C | NP | Candidate (pro) | BEA 0.65 (0.25–1.68) | 71 % | 3.42 | 0.370 | 2 | |
| AA | C | Caucasian | Candidate (retro) | |||||||||||
LOE level of evidence, EV effect variant, NS nonsignificant, NP not presented, BE Barrett’s esophagus, A adjusted OR, OR odds ratio, CI confidence interval, pro prospective, retro retrospective, RE reflux esophagitis, NA not applicable
*,** denote overlapping populations
The rs1695 (GSTP1) was assessed by 4 studies, which underwent meta-analysis by Bull et al. [18]. This calculated an OR of 1.50 (95 % CI 1.16–1.95 p = not presented; LOE II). While derived from a large total cohort (434 cases and 738 controls), none of the four studies adjusted for risk factors; indeed the sole study finding a significant association had only 22 cases [19]. The GSTM1 null genotype was also assessed by 4 studies and underwent updated meta-analysis in this study. Overall, no association was demonstrated. On sensitivity analysis, a significant negative association was apparent for the 2 studies not requiring IM [17, 20], although the relevance of this is unclear. Meta-analysis was performed for 5 other variants, none of which demonstrated associations (Table 3).
Table 3.
Meta-analyzed markers assessed in association with BE susceptibility
| Marker | Individual study | Meta-analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Association | ||||||||||
| Variant | Gene | EV | Variant | Wild-type | No association | OR (95 % CI) | I 2 | Chi | p | n |
| Null | GSTM1 | Null | Null | Kadioglu et al. [55] | BE 0.83 (0.61–1.12) | 27 % | 4.12 | 0.220 | 4 | |
| Kala et al. [19] | BEno IM 0.66 (0.44–0.99) | 0 % | 0.64 | 0.420 | 2 | |||||
| Casson et al. [46] | BEIM 1.12 (0.70–1.80) | 0 % | 0.64 | 0.630 | 2 | |||||
| van Lieshout et al. [56] | ||||||||||
| rs4880 | SOD2 | T | T | Kadioglu et al. [55], di Martino et al. [49] |
BEIM 0.90 (0.65–1.24) | 0.00 | 0.07 | 0.520 | 3 | |
| Murphy et al. [45] | ||||||||||
| rs16944 | IL1B | AA | AA | Izakovicova-Holla et al. [27], Gough et al. [28] |
BE 1.13 (0.63–2.03) | 0.00 | 0.00 | 0.680 | 2 | |
| rs1052133 | OGG1 | G | G | Kadioglu et al. [55], Ferguson et al. [49] |
BEA IM 1.34 (0.59–3.09) | 67 % | 3.01 | 0.480 | 2 | |
| rs25487 | XRCC1 | TT | TT | Casson et al. [48] | Ferguson et al. [49] | BEA IM 0.65 (0.25–1.68) | 71 % | 3.42 | 0.370 | 2 |
| rs13181 | ERCC2 | CC | CC | Casson et al. [48], Fergusons et al. [49] |
BEA IM 0.92 (0.57–1.48) | 31 % | 1.44 | 0.730 | 2 | |
EV effect variant, OR odds ratio, CI confidence interval, IM intestinal metaplasia, BE Barrett’s esophagus
Of the 12 significant candidate associations reported, only rs1695 (GSTP1) and rs25487 (XRCC1) were assessed by more than one study. This notwithstanding, 5 appear relatively robust on the basis of adjustment for clinical covariates. These include 3 growth factor variants: rs444903 (EGFR [20]; notably associated with reflux esophagitis and EAC), rs6214 (IGF1 [21]), and rs2229765 (IGF1R) [22]. Two interleukin variants also appear plausible: rs3212227 (IL12B) and rs917997 (IL18RAP) [23–25], with the former demonstrated to be independent of all other tested genotypes [26]. A number of other associations were reported in the IL1 [27, 28], IL10 [28], IL18 [29], and IL23 [30] clusters. Of these, however, only wild-type rs917997 (IL18RAP) persisted following correction for multiple comparisons [29].
The remaining 5 candidate variants included associations with 3 caudal homeobox 1 (CDX1) variants: rs3776082, rs2237091, and rs717767. The authors demonstrated these variants to be significantly associated with established risk factors for BE: age, gender, and the presence of hiatus hernia. However, multivariate analysis was not performed to demonstrate whether the association of these variants with BE was independent of these. Of the remaining 2, an association was demonstrated for the rs6785049 (NR1I2) variant; however, similarly this was not adjusted for risk factors. This was performed for the rs9344 (CCND1 [31]) variant, although the p value was not published.
Of the 5 GWAS variants identified, 4 were identified by the Wellcome Trust Centre Case Control Consortium (WTCCC). Two were identified by the initial report [32]: rs9257809 within the major histocompatibility complex (MHC; OR 1.21 [1.13–1.28]; p = 4.09 × 10−9) and rs9936833 (related to FOXF1; OR 1.14 [1.10–1.19]; p = 2.74 × 10−10). Subsequent replication identified rs3072 (related to GDF7: OR 1.14 [1.09–1.18]; p = 1.80 × 10−11) and rs2701108 (related to TBX5: OR 0.90 [0.86–0.93]; p = 7.50 × 10−9) [33]. All remained significant when meta-analyzed with data from the Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON) GWAS. The first 2 were also associated with EAC in the BEACON dataset and an independent candidate study [34]. Mechanistically, GWAS arrays rely on linkage disequilibrium between SNPs, by which SNPs act as proxies for others (i.e., genotyping one SNP allows the genotypes of others to be inferred with confidence). This “tagging”, however, means that variants identified may be functional or may in fact be bystanders tagging other SNPs [35]. However, the loci overall and related genes are consistent with the roles of immune-mediated inflammation (MHC) and thoracoembryogenesis (FOXF1, GDF7 and TBX5) in BE.
The BEACON GWAS identified 3 loci associated with either BE or EAC, although none reached the genome-wide threshold for BE alone. However, on subsequent meta-analysis of both GWAS, rs2687201 (FOXP1, similarly involved in developmental regulation) was significantly associated with BE alone, in addition to a further variant associated with either BE/EAC [33].
Studies Assessing Germline Variants Associated with Progression
One study was identified (supplementary table 7). LOE was C; modified REMARK score was 13.5/19. This assessed 4 variants in the IGF axis; none were associated with progression.
Studies Assessing Somatic Variants Associated with Progression
Sixteen studies were identified, published between 1989 and 2012 (supplementary table 8). LOE was C for 12 and D for 4. Mean modified REMARK score was 13.2 (range 10.5–18.5/19; supplementary table 9). Five classified progression as HGD/EAC, 10 as EAC, and 1 as EAC/CIN. Again, a number of recurrent methodological issues were identified. These included failure to: blind investigators (12; 75.0 %), perform appropriate quality control/reproducibility (9; 56.3 %), match controls (14; 87.5 %), and appropriate power calculations (14; 87.5 %). Recurrent reporting issues included failure to: report univariate association effects (9; 60.0 %), adjust for risk factors (particularly the presence of dysplasia at baseline; 14; 87.5 %), and fully report coefficients of multivariate models (13; 81.3 %).
Variants Associated with Progression
Of 7 variants assessed, 5 associations were identified: CIN, CNV (>70 Mbp), TP53 LOH, p16 LOH, and mutant TP53 (Table 4). Meta-analysis was possible for CIN, which was assessed by 11 studies. These defined CIN variably as aneuploidy (4), tetraploidy (1), and aneuploidy/tetraploidy (6). Six of these studies were derived from independent sample archives. However, this was not clear for 4. All studies reported associations of CIN with progression. However, only two adjusted for confounding variables, including the presence of dysplasia [36, 37]. Another two, while not adjusting for dysplasia, did adjust for length of Barrett’s segment [38, 39], with CIN remaining significant.
Table 4.
Reported somatic markers of Barrett’s esophagus progression to dysplasia/adenocarcinoma
| Marker | Individual study | Association (meta-analysis/individual study) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOE | Variant | Gene | Association | EV | LOE | Ethnicity | Phenotype | Adjusted for confounders? | ||||||
| Variant | None | Phenotype and OR/HR (95 % CI) | I 2 | Chi | p | n | ||||||||
| II | CIN | NA | Bird-Lieberman et al. [36] | CIN | C | NP | HGD/EAC | Yes (plus length, dysplasia) | HGD/EAC OR(A) IM PB 5.98 (2.10–17.1) | 57 % | 16.2 | 8.00 × 10−4 | 5 | |
| Sikkema et al. [37] | CIN | C | Cauc | HGD/EAC | Yes (plus dysplasia) | HGD/EAC HR(A) IM 1.36 (1.26–1.47) | 0 % | 0.98 | <1.00 × 10−5 | 2 | ||||
| Chao et al. [57]* | CIN | C | NP | EAC | No | |||||||||
| Galipeau et al. [38]* | CIN | C | NP | EAC | Yes (length, not dysplasia) | PB-adjusted for publication bias Fang 2004, Rabinovitch 2001, | ||||||||
| Fang et al. [58] | CIN | C | NP | EAC | No | |||||||||
| Maley et al. [39]* | CIN | C | NP | EAC | Yes (length, not dysplasia) | Teodori 1999 | ||||||||
| Rabinovitch et al. [59]* | CIN | C | NP | EAC | Yes (length, not dysplasia) | |||||||||
| Reid et al. [9]** | CIN | D | NP | EAC | No | Overlapping populations*** | ||||||||
| Teodori et al. [60] | CIN | C | NP | HGD/EAC | No | |||||||||
| Reid et al. [8]** | CIN | C | NP | EAC | No | |||||||||
| James et al. [61] | CIN | D | NP | EAC | No | |||||||||
| III | CNV | NA | Paulson et al. [62] | >70 Mbp | C | NP | EAC/CIN | No | EAC/CIN HR 4.90 (1.60–14.8) | NA | NA | 4.70 × 10−3 (NS) | 1 | |
| III | LOH | TP53 | Galipeau et al. [38]* | LOH | C | NP | EAC | Yes (length, not dysplasia) | EAC HRA 5.4 (2.5–12.0) | NA | NA | <1 × 10−3 | 1 | |
| Maley et al. [39]* | LOH | C | ||||||||||||
| Reid et al. [63]* | LOH | C | ||||||||||||
| III | LOH | P16 | Galipeau et al. [38]* | Maley et al. [39]* | LOH | C | NP | EAC | Yes (length, not dysplasia) | EACA HR 2.4 (1.0–5.5) | NA | NA | 0.045 | 1 |
| LOH | C | NP | EAC | Yes (length, not dysplasia) | ||||||||||
| III | Mutant | TP53 | Galipeau et al. [38]* | Mutant | C | NP | EAC | No | EAC HRA 1.27 (1.07–1.50) | NA | NA | <0.01 (NS) | 1 | |
| Maley et al. [39]* | Mutant | C | NP | EAC | Yes (length, not dysplasia) | |||||||||
| Dolan et al. [64] | Mutant | C | NP | EAC | No | No meta possible as study population overlap and differing effect measures | ||||||||
LOE level of evidence, EV effect variant, OR odds ratio, HR hazard ratio, CI confidence interval, CIN chromosomal instability, NA not applicable, NP not presented, NS nonsignificant, HGD high-grade dysplasia, EAC esophageal adenocarcinoma, A adjusted
*,**,*** denote overlapping populations
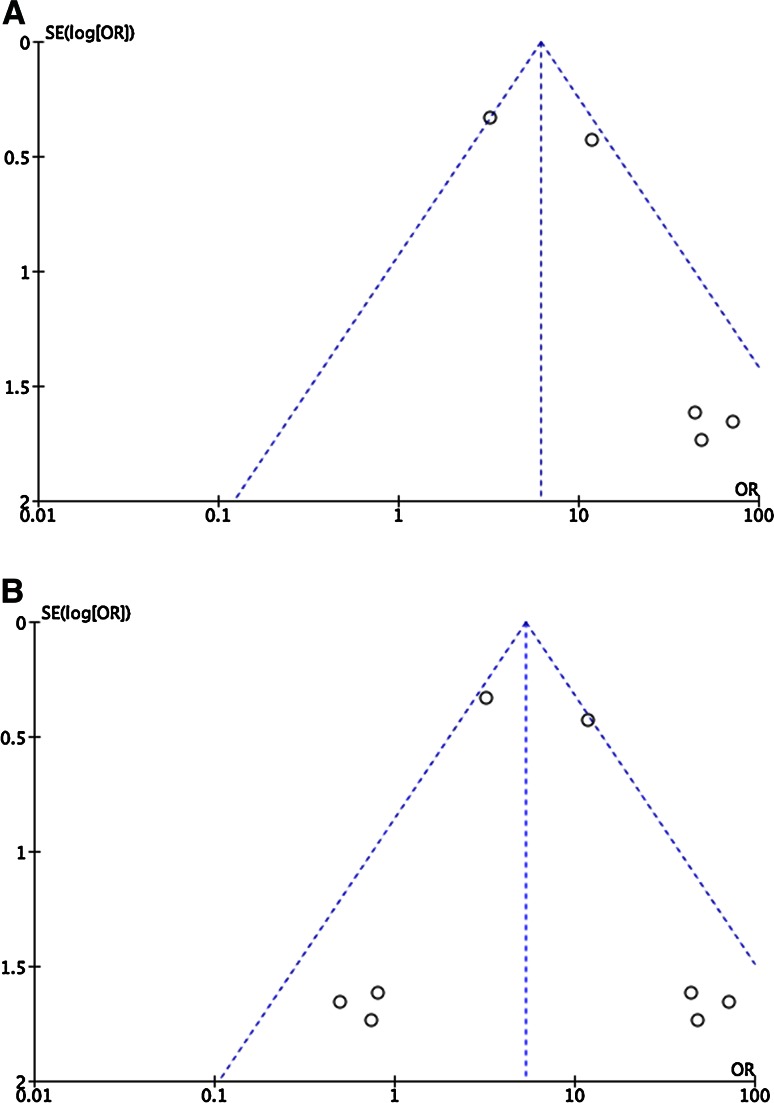
Meta-analysis was performed for both OR and HR of progression to HGD/EAC. Significant associations were demonstrated for both (Table 4; Fig. 2). All studies included patients with IM only. Meta-analyzed OR was 5.98 (2.10–17.1; p = 8.00 × 10−4; n = 5 studies; following exclusion of overlapping studies and correction for publication bias). However, only one of these studies adjusted for the presence of dysplasia. Meta-analyzed HR was 1.36 (1.26–1.47; n = 2 studies; p < 1.00 × 10−5; following exclusion of one overlapping study).Overall LOE for CIN was II. While more than one study assessed mutant TP53 (n = 3) and LOH TP53 (n = 2), meta-analysis was not possible for either. Two studies for each were derived from the same populations, and the third TP53 study used a different measure of effect size.
Fig. 2.
Uncorrected and corrected funnel plots for odds ratio of progression and chromosomal instability
Variants Associated with Risk of Progression Following Photodynamic Therapy
Three studies were identified (supplementary table 10). LOE was C for 2 and D for 1. Mean modified REMARK score was 16.2/20 (range 14.5–19; supplementary table 7). For 2 studies, while endoscopic mucosal resection was variably performed, this was only controlled for in 1 study. A total of 6 variants were assessed; while 5 were assessed by 2 studies, meta-analysis could not be performed due to population overlap. CIN at both 4 and 12 months was reported to be associated with risk of progression (Table 5).
Table 5.
Reported somatic biomarkers of Barrett’s esophagus progression to dysplasia/adenocarcinoma following photodynamic therapy
| Marker | Individual study | Association (meta-analysis/individual study) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOE | Variant | Gene | Association | EV | LOE | Ethnicity | Phenotype | Adjusted for confounders? | ||||||
| Variant | None | Phenotype HR (95 % CI) | I 2 | Chi | p | n | ||||||||
| IV | CIN | NA | Dunn et al. [65] | CIN 4 m post-PDT | D | NS | HGD | Yes | EAC/HGD HRA 4.10 (1.30–13.0) | NA | NA | 9.00 × 10−3 | 1 | |
| CIN 12 m post-PDT | EAC/HGD HRA 3.60 (1.05–12.3) | 2.90 × 10−3 | ||||||||||||
LOE level of evidence, CIN chromosomal instability, EV effect variant, HR hazard ratio, CI confidence interval, PDT photodynamic therapy, HGD high-grade dysplasia, EAC esophageal adenocarcinoma
Discussion
We believe this review to be the first to identify, synthesize, and evaluate the evidence for genetic markers of BE susceptibility and risk of progression. Thirty-three susceptibility markers were identified; however, just 17 remained significant after correcting for multiple comparisons. Five (rs9257809, rs9936833, and subsequently rs3072, rs2701108, and rs2687201) were derived from GWAS and are therefore most likely to be reproducible. Of the 6 candidate markers assessed by more than one study, meta-analysis was supportive for one (rs1695, GSTP1). Five non-meta-analyzed variants affecting either growth factors or inflammatory cytokines appear plausible and therefore represent priorities for validation: rs444903 (EGFR), rs6214 (IGF1), rs2229765 (IGF1R), rs3212227 (IL12B), and rs917997 (IL18RAP).
No germline markers of progression risk were identified. However, 5 somatic markers were reported, plus another of progression following photodynamic therapy. Meta-analysis was possible for CIN, demonstrating significant effects for both HR and OR after correction for publication bias. However, there was considerable heterogeneity regarding definition of CIN, duration, and frequency of follow-up, confounding risk factors (e.g., prevalence of HGD) in addition to minimum follow-up periods. Notably, however, both studies adjusting for HGD [36, 37] did demonstrated convincing effects.
The robustness of the associations between CIN and progression suggests it to be of immediate clinical utility. CIN is a constituent of genomic instability, a state of erroneous progression through the cell cycle. Inaccurate DNA replication, repair, and chromosomal segregation, results in accumulation of genomic errors and is a major factor driving tumorigenesis. CIN is associated with worse stage and prognosis in a range of tumors including esophageal [40] and has been demonstrated in a quarter of patients with BE [36]. Importantly, this subset appears to be at significantly higher risk of malignant progression, which can be readily demonstrated by flow cytometry. This does, however, remain somewhat imprecise and is unable to distinguish between stable and unstable, simple and complex abnormalities. Despite this, the incorporation of CIN into a biomarker panel such as that reported by Bird-Lieberman et al. [36] (comprising age, CIN, dysplasia, TP53 and Cyclin A expression, sialyl Lewis antigens, Aspergillus oryzae lectin, and binding of wheat germ agglutinin) may provide invaluable information with which to personalize management of BE.
By contrast, the immediate benefits of germline susceptibility biomarkers are less tangible. As predicted by the “common disease-common variant” hypothesis of complex traits, both GWAS suggested many common variants of small effect to contribute to development of BE. Consequently, germline associations may be weaker and more complex. Whilst this gives the potential to identify novel biology, variants may have little utility in isolation.
A number of recurrent methodological issues were identified, limiting the generalizability of reported variants. Ultimately, validation studies for prioritized variants should be designed with these in mind, with particular emphasis on the interaction between genomic and clinical factors. Other issues to be addressed include disparity as diagnostic criteria; confirmation of IM is a prerequisite in the USA, yet is not required in the UK [41], although this did not alter the findings of this review.
A number of exploratory NGS studies have recently been performed for both BE and EAC. These have served to highlight the mutational complexity of both conditions, providing biological context for markers and their genes (which are often considered in isolation) as well as suggesting new variants and genes for study. Recently, Streppel et al. [11] performed whole genome sequencing (WGS) of one patient, comparing normal squamous epithelium with metaplastic and neoplastic epithelium. This identified somatic nonsense mutations in genes including AT-rich interactive domain 1A (ARID1A), a member of the SWI/SNF family involved in gene expression via chromatin remodeling, which has been independently identified as a driver gene of EAC by of other NGS studies. The authors found ARID1A loss of expression to become progressively more common during the metaplasia-dysplasia-adenocarcinoma sequence, and to be associated with aberrant cellular proliferation and invasion in a knock-down model. NGS studies also provide valuable contextual information as to mutational spectra, as well as clonal and linear evolution to better understand to development of somatic mutations and genomic instability. Recently, Weaver et al. [12] performed WGS of 112 EACs, similarly identifying a number of significantly and recurrently mutated genes. One hundred and seven BE samples were then genotyped, with the notable finding that most such mutations were already present in non-dysplastic epithelium; just TP53 and SMAD4 mutations occurred later, in HGD and EAC. The advent of third-generation sequencing, for example from single cells, will undoubtedly shed yet further light on this process. Additionally, germline rather than somatic mutations in EAC driver genes have been shown to predispose to EAC, although this has yet to be demonstrated in BE [42].
This review has a number of limitations. While we searched two databases using a comprehensive search term, it is possible that relevant publications (including non-English articles) were not identified. As discussed, meta-analysis for CIN was performed within the context of considerably heterogeneity and must be interpreted with caution. Only one meta-analysis (CIN and OR of progression) involved more than 5 studies, with considerable funnel plot asymmetry, largely due to small studies reporting large effect sizes with significant variance. This was interpreted as publication bias, and was adjusted, without altering statistical significance. In addition, the “trim and fill” method used to correct for possible bias while widely used does make assumptions regarding the necessity for plot symmetry, while not incorporating study methodology. Unfortunately, the limited number of studies prevented useful meta-regression to assess this further. There are also limitations in using the revised ASCO guidelines, as these do not fully represent the complexity of methodological quality and also disagreement between studies. We therefore used modified REMARK guidelines to provide a further level of criticism. In particular, the ASCO guidelines do not allow for differences in methodology (for example, GWAS versus candidate studies). And more generically, while genomic biomarkers are typically considered in isolation, in reality their utility depends on innumerable variables (including transcriptional, translational and proteomic regulation, and clinical and environmental factors). Consequently, establishing their true utility will require parallel processing and consideration of these contexts.
In conclusion, this review has identified, evaluated, and synthesized the evidence for genomic biomarkers of BE susceptibility and dysplastic/malignant progression. Seventeen germline markers of susceptibility, 5 somatic markers of progression, and 1 marker of relapse following photodynamic therapy were identified. Meta-analysis demonstrated CIN to be a particularly plausible and clinically useful marker of progression and one which can be demonstrated readily. However, the overall evidence base is characterized by widespread methodological issues, which limit the immediate clinical utility of these markers. Consequently, larger studies with more robust design are required to validate these markers, identify novel variants, and incorporate them into clinical practice.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Core funding is provided to the Wellcome Trust Centre for Human Genetics from the Wellcome Trust (090532/Z/09/Z).
Compliance with ethical standards
Conflict of interest
Mark R. Middleton has received payment for advisory/consulting roles within the last 2 years from Amgen, Bristol-Myers Squibb, GlaxoSmithKline, Merck, Millennium, and Roche, and institutional funding from Amgen, AstraZeneca, Bristol-Myers Squibb, Clovis, Eisai, GlaxoSmithKline, Immunocore, Johnson & Johnson, Merck, Millennium, Novartis, Pfizer, Roche and Vertex. The authors declare no conflicts of interest.
References
- 1.Barrett NR. Chronic peptic ulcer of the oesophagus and ‘oesophagitis’. Br J Surg. 1950;38:175–182. doi: 10.1002/bjs.18003815005. [DOI] [PubMed] [Google Scholar]
- 2.Cameron AJ, Lomboy CT. Barrett’s esophagus: age, prevalence, and extent of columnar epithelium. Gastroenterology. 1992;103:1241–1245. doi: 10.1016/0016-5085(92)91510-b. [DOI] [PubMed] [Google Scholar]
- 3.Gerson LB, Edson R, Lavori PW, Triadafilopoulos G. Use of a simple symptom questionnaire to predict Barrett’s esophagus in patients with symptoms of gastroesophageal reflux. Am J Gastroenterol. 2001;96:2005–2012. doi: 10.1111/j.1572-0241.2001.03933.x. [DOI] [PubMed] [Google Scholar]
- 4.Wild CP, Hardie LJ. Reflux, Barrett’s oesophagus and adenocarcinoma: burning questions. Nat Rev Cancer. 2003;3:676–684. doi: 10.1038/nrc1166. [DOI] [PubMed] [Google Scholar]
- 5.Lepage C, Rachet B, Jooste V, Faivre J, Coleman MP. Continuing rapid increase in esophageal adenocarcinoma in England and Wales. Am J Gastroenterol. 2008;103:2694–2699. doi: 10.1111/j.1572-0241.2008.02191.x. [DOI] [PubMed] [Google Scholar]
- 6.Falk GW. Barrett’s oesophagus: frequency and prediction of dysplasia and cancer. Best Pract Res Clin Gastroenterol. 2015;29:125–138. doi: 10.1016/j.bpg.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett’s esophagus. N Engl J Med. 2011;365:1375–1383. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- 8.Reid BJ, Blount PL, Rubin CE, Levine DS, Haggitt RC, Rabinovitch PS. Flow-cytometric and histological progression to malignancy in Barrett’s esophagus: prospective endoscopic surveillance of a cohort. Gastroenterology. 1992;102:1212–1219. [PubMed] [Google Scholar]
- 9.Reid BJ, Levine DS, Longton G, Blount PL, Rabinovitch PS. Predictors of progression to cancer in Barrett’s esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95:1669–1676. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Streppel MM, Lata S, DelaBastide M, et al. Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett’s esophagus. Oncogene. 2014;33:347–357. doi: 10.1038/onc.2012.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weaver JM, Ross-Innes CS, Shannon N, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet. 2014;46:837–843. doi: 10.1038/ng.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellier MD, Shepherd NA. BSG Guidelines in Gastroenterology: Diagnosis of Columnar-Lined Oesophagus. British Society of Gastroenterology; 2005:13–17.
- 14.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 15.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sterne JA, Sutton AJ, Ioannidis JP, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343:d4002. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 17.Sutton AJ, Duval SJ, Tweedie RL, Abrams KR, Jones DR. Empirical assessment of effect of publication bias on meta-analyses. BMJ. 2000;320:1574–1577. doi: 10.1136/bmj.320.7249.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bull LM, White DL, Bray M, Nurgalieva Z, El-Serag HB. Phase I and II enzyme polymorphisms as risk factors for Barrett’s esophagus and esophageal adenocarcinoma: a systematic review and meta-analysis. Dis Esophagus. 2009;22:571–587. doi: 10.1111/j.1442-2050.2009.00947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kala Z, Dolina J, Marek F, Izakovicova Holla L. Polymorphisms of glutathione S-transferase M1, T1 and P1 in patients with reflux esophagitis and Barrett’s esophagus. J Hum Genet. 2007;52:527–534. doi: 10.1007/s10038-007-0148-z. [DOI] [PubMed] [Google Scholar]
- 20.Menke V, Pot RG, Moons LM, et al. Functional single-nucleotide polymorphism of epidermal growth factor is associated with the development of Barrett’s esophagus and esophageal adenocarcinoma. J Hum Genet. 2012;57:26–32. doi: 10.1038/jhg.2011.124. [DOI] [PubMed] [Google Scholar]
- 21.McElholm AR, McKnight AJ, Patterson CC, et al. A population-based study of IGF axis polymorphisms and the esophageal inflammation, metaplasia, adenocarcinoma sequence. Gastroenterology. 2010;139:204-e3–212-e3. doi: 10.1053/j.gastro.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 22.MacDonald K, Porter GA, Guernsey DL, Zhao R, Casson AG. A polymorphic variant of the insulin-like growth factor type I receptor gene modifies risk of obesity for esophageal adenocarcinoma. Cancer Epidemiology. 2009;33:37–40. doi: 10.1016/j.canep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 23.O’Riordan JM, Abdel-latif MM, Ravi N, et al. Proinflammatory cytokine and nuclear factor kappa-B expression along the inflammation–metaplasia–dysplasia–adenocarcinoma sequence in the esophagus. Am J Gastroenterol. 2005;100:1257–1264. doi: 10.1111/j.1572-0241.2005.41338.x. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins GJ, Harries K, Doak SH, et al. The bile acid deoxycholic acid (DCA) at neutral pH activates NF-kappaB and induces IL-8 expression in oesophageal cells in vitro. Carcinogenesis. 2004;25:317–323. doi: 10.1093/carcin/bgh032. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald RC, Abdalla S, Onwuegbusi BA, et al. Inflammatory gradient in Barrett’s oesophagus: implications for disease complications. Gut. 2002;51:316–322. doi: 10.1136/gut.51.3.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moons LM, Kusters JG, van Delft JH, et al. A pro-inflammatory genotype predisposes to Barrett’s esophagus. Carcinogenesis. 2008;29:926–931. doi: 10.1093/carcin/bgm241. [DOI] [PubMed] [Google Scholar]
- 27.Izakovicova Holla L, Borilova Linhartova P, Hrdlickova B, et al. Haplotypes of the IL-1 gene cluster are associated with gastroesophageal reflux disease and Barrett’s esophagus. Human Immunol. 2013;74:1161–1169. doi: 10.1016/j.humimm.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 28.Gough MD, Ackroyd R, Majeed AW, Bird NC. Prediction of malignant potential in reflux disease: are cytokine polymorphisms important? Am J Gastroenterology. 2005;100:1012–1018. doi: 10.1111/j.1572-0241.2005.40904.x. [DOI] [PubMed] [Google Scholar]
- 29.Babar M, Ryan AW, Anderson LA, et al. Genes of the interleukin-18 pathway are associated with susceptibility to Barrett’s esophagus and esophageal adenocarcinoma. Am J Gastroenterol. 2012;107:1331–1341. doi: 10.1038/ajg.2012.134. [DOI] [PubMed] [Google Scholar]
- 30.Gaj P, Mikula M, Wyrwicz LS, Regula J, Ostrowski J. Barrett’s esophagus associates with a variant of IL23R gene. Acta Biochim Pol. 2008;55:365–369. [PubMed] [Google Scholar]
- 31.Casson AG, Zheng Z, Evans SC, et al. Cyclin D1 polymorphism (G870A) and risk for esophageal adenocarcinoma. Cancer. 2005;104:730–739. doi: 10.1002/cncr.21229. [DOI] [PubMed] [Google Scholar]
- 32.Su Z, Gay LJ, Strange A, et al. Common variants at the MHC locus and at chromosome 16q24.1 predispose to Barrett’s esophagus. Nat Genet. 2012;44:1131–1136. doi: 10.1038/ng.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palles C, Chegwidden L, Li X, et al. Polymorphisms near TBX5 and GDF7 are associated with increased risk for Barrett’s esophagus. Gastroenterology. 2015;148:367–378. doi: 10.1053/j.gastro.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dura P, van Veen EM, Salomon J, et al. Barrett associated MHC and FOXF1 variants also increase esophageal carcinoma risk. Int J Cancer. 2013;133:1751–1755. doi: 10.1002/ijc.28160. [DOI] [PubMed] [Google Scholar]
- 35.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bird-Lieberman EL, Dunn JM, Coleman HG, et al. Population-based study reveals new risk-stratification biomarker panel for Barrett’s esophagus. Gastroenterology. 2012;143:927-e3–935-e3. doi: 10.1053/j.gastro.2012.06.041. [DOI] [PubMed] [Google Scholar]
- 37.Sikkema M, Kerkhof M, Steyerberg EW, et al. Aneuploidy and overexpression of Ki67 and p53 as markers for neoplastic progression in Barrett’s esophagus: a case–control study. Am J Gastroenterol. 2009;104:2673–2680. doi: 10.1038/ajg.2009.437. [DOI] [PubMed] [Google Scholar]
- 38.Galipeau PC, Li X, Blount PL, et al. NSAIDs modulate CDKN2A, TP53, and DNA content risk for progression to esophageal adenocarcinoma. PLoS Med. 2007;4:e67. doi: 10.1371/journal.pmed.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maley CC, Galipeau PC, Li X, et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 2004;64:7629–7633. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- 40.Yu JM, Yang LH, Guo Q, et al. Flow cytometric analysis DNA content in esophageal carcinoma. Correlation with histologic and clinical features. Cancer. 1989;64:80–82. doi: 10.1002/1097-0142(19890701)64:1<80::AID-CNCR2820640114>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 41.Watson A, Shepherd NA. The Definition of “Barrett’s” Columnar-Lined Oesophagus. British Society of Gastroenterology: Guidelines for the Diagnosis and Management of Barrett’s Columnar-Lined Oesophagus. 2005.
- 42.Buas MF, Levine DM, Makar KW, et al. Integrative post-genome-wide association analysis of CDKN2A and TP53 SNPs and risk of esophageal adenocarcinoma. Carcinogenesis. 2014;35:2740–2747. doi: 10.1093/carcin/bgu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren D, Zheng G, Bream S, Tevebaugh W, Shaheen NJ, Chen X. Single Nucleotide Polymorphisms of Caudal Type Homeobox 1 and 2 Are Associated with Barrett’s Esophagus. Dig Dis Sci. 2014;59:57–63. doi: 10.1007/s10620-013-2804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van de Winkel A, Menke V, Capello A, et al. Expression, localization and polymorphisms of the nuclear receptor PXR in Barrett’s esophagus and esophageal adenocarcinoma. BMC Gastroenterol. 2011;11:108. doi: 10.1186/1471-230X-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy SJ, Hughes AE, Patterson CC, et al. A population-based association study of SNPs of GSTP1, MnSOD, GPX2 and Barrett’s esophagus and esophageal adenocarcinoma. Carcinogenesis. 2007;28:1323–1328. doi: 10.1093/carcin/bgm007. [DOI] [PubMed] [Google Scholar]
- 46.Casson AG, Zheng Z, Porter GA, Guernsey DL. Genetic polymorphisms of microsomal epoxide hydroxylase and glutathione S-transferases M1, T1 and P1, interactions with smoking, and risk for esophageal (Barrett) adenocarcinoma. Cancer Detect Prev. 2006;30:423–431. doi: 10.1016/j.cdp.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 47.van Lieshout EM, Tiemessen DM, Witteman BJ, Jansen JB, Peters WH. Low glutathione and glutathione S-transferase levels in Barrett’s esophagus as compared to normal esophageal epithelium. Jpn J Cancer Res Gann. 1999;90:81–85. doi: 10.1111/j.1349-7006.1999.tb00669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casson AG, Zheng Z, Evans SC, Veugelers PJ, Porter GA, Guernsey DL. Polymorphisms in DNA repair genes in the molecular pathogenesis of esophageal (Barrett) adenocarcinoma. Carcinogenesis. 2005;26:1536–1541. doi: 10.1093/carcin/bgi115. [DOI] [PubMed] [Google Scholar]
- 49.Ferguson HR, Wild CP, Anderson LA, et al. No association between hOGG1, XRCC1, and XPD polymorphisms and risk of reflux esophagitis, Barrett’s esophagus, or esophageal adenocarcinoma: results from the factors influencing the Barrett’s adenocarcinoma relationship case-control study. Cancer Epidemiol Biomark Prev. 2008;17:736–739. doi: 10.1158/1055-9965.EPI-07-2832. [DOI] [PubMed] [Google Scholar]
- 50.Menke V, van Zoest KP, Moons LM, et al. NcoI TNF-beta gene polymorphism and TNF expression are associated with an increased risk of developing Barrett’s esophagus and esophageal adenocarcinoma. Scand J Gastroenterol. 2012;47:378–386. doi: 10.3109/00365521.2011.650192. [DOI] [PubMed] [Google Scholar]
- 51.Menke V, Van Zoest KP, Moons LM, et al. Myo9B is associated with an increased risk of Barrett’s esophagus and esophageal adenocarcinoma. Scand J Gastroenterol. 2012;47:1422–1428. doi: 10.3109/00365521.2012.722673. [DOI] [PubMed] [Google Scholar]
- 52.Bradbury PA, Zhai R, Hopkins J, et al. Matrix metalloproteinase 1, 3 and 12 polymorphisms and esophageal adenocarcinoma risk and prognosis. Carcinogenesis. 2009;30:793–798. doi: 10.1093/carcin/bgp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.di Martino E, Hardie LJ, Wild CP, et al. The NAD(P)H:quinone oxidoreductase I C609T polymorphism modifies the risk of Barrett esophagus and esophageal adenocarcinoma. Genetics Med. 2007;9:341–347. doi: 10.1097/GIM.0b013e3180654ccd. [DOI] [PubMed] [Google Scholar]
- 54.Rajendra S, Ackroyd R, Murad S, et al. Human leucocyte antigen determinants of susceptibility to Barrett’s oesophagus in Asians–a preliminary study. Aliment Pharmacol Ther. 2005;21:1377–1383. doi: 10.1111/j.1365-2036.2005.02496.x. [DOI] [PubMed] [Google Scholar]
- 55.Kadioglu E, Sardas S, Ergun M, Unal S, Karakaya AE. The role of oxidative DNA damage, DNA repair, GSTM1, SOD2 and OGG1 polymorphisms in individual susceptibility to Barrett’s esophagus. Toxicol Ind Health. 2010;26:67–79. doi: 10.1177/0748233709359278. [DOI] [PubMed] [Google Scholar]
- 56.van Lieshout EM, Roelofs HM, Dekker S, et al. Polymorphic expression of the glutathione S-transferase P1 gene and its susceptibility to Barrett’s esophagus and esophageal carcinoma. Cancer Res. 1999;59:586–589. [PubMed] [Google Scholar]
- 57.Chao DL, Sanchez CA, Galipeau PC, et al. Cell proliferation, cell cycle abnormalities, and cancer outcome in patients with Barrett’s esophagus: a long-term prospective study. Clin Cancer Res. 2008;14:6988–6995. doi: 10.1158/1078-0432.CCR-07-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang M, Lew E, Klein M, Sebo T, Su Y, Goyal R. DNA abnormalities as marker of risk for progression of Barrett’s esophagus to adenocarcinoma: image cytometric DNA analysis in formalin-fixed tissues. Am J Gastroenterol. 2004;99:1887–1894. doi: 10.1111/j.1572-0241.2004.30886.x. [DOI] [PubMed] [Google Scholar]
- 59.Rabinovitch PS, Longton G, Blount PL, Levine DS, Reid BJ. Predictors of progression in Barrett’s esophagus III: baseline flow cytometric variables. Am J Gastroenterol. 2001;96:3071–3083. doi: 10.1111/j.1572-0241.2001.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teodori L, Gohde W, Persiani M, et al. DNA/protein flow cytometry as a predictive marker of malignancy in dysplasia-free Barrett’s esophagus: thirteen-year follow-up study on a cohort of patients. Cytometry. 1998;34:257–263. doi: 10.1002/(SICI)1097-0320(19981215)34:6<257::AID-CYTO3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 61.James PD, Atkinson M. Value of DNA image cytometry in the prediction of malignant change in Barrett’s oesophagus. Gut. 1989;30:899–905. doi: 10.1136/gut.30.7.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paulson TG, Maley CC, Li X, et al. Chromosomal instability and copy number alterations in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:3305–3314. doi: 10.1158/1078-0432.CCR-08-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reid BJ, Prevo LJ, Galipeau PC, et al. Predictors of progression in Barrett’s esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–2848. doi: 10.1111/j.1572-0241.2001.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dolan K, Walker SJ, Gosney J, Field JK, Sutton R. TP53 mutations in malignant and premalignant Barrett’s esophagus. Dis Esophagus. 2003;16:83–89. doi: 10.1046/j.1442-2050.2003.00302.x. [DOI] [PubMed] [Google Scholar]
- 65.Dunn JM, Mackenzie GD, Oukrif D, et al. Image cytometry accurately detects DNA ploidy abnormalities and predicts late relapse to high-grade dysplasia and adenocarcinoma in Barrett’s oesophagus following photodynamic therapy. Br J Cancer. 2010;102:1608–1617. doi: 10.1038/sj.bjc.6605688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.