Abstract
Abies alba (Mill.) is an important forest tree species, native to the mountainous regions of Europe but has been also widely introduced in the lowlands outside its native range. Like most forest tree species, A. alba forms obligate mutualisms with ectomycorrhizal (ECM) fungi. This investigation sought to examine ECM fungal communities of A. alba when the species grows 400 km north of its native range in the region of Pomerania in Poland. We surveyed for ECM fungi by sampling live roots from four mature forest stands where the A. alba component ranged from 20 to 100 %. Ectomycorrhizal fungal symbionts were identified based on morphotyping and sequencing of the internal transcribed spacer (ITS) of nuclear ribosomal DNA (rDNA). Thirty-five ECM fungal taxa were distinguished on root tips of A. alba from all tested stands with 22 to 27 ECM fungal taxa in the individual stand. The diversity and similarity metrics revealed a lack of statistical differences in the structure of the ECM fungal community between stands varying in overstory tree composition. Cenococcum geophilum was the most common fungal species at all investigated A. alba stands, with an abundance of 50 to 70 %. The ECM community was characterized by the lack of Abies-specific fungal symbionts and a rich and diverse suite of host-generalist mycobionts that seem to be sufficient for successful growth and development of A. alba outside of its native range.
Keywords: Silver fir, Symbiotic fungi, Ectomycorrhizal diversity, Host-generalist, Cenococcum geophilum
Introduction
Forest tree species may occur outside their historic natural range in response to climate change and global warming or because humans have deliberately introduced a tree species in a region where the species is very productive. The latter occurred with Abies alba Mill. (silver fir) in the lowlands of northern Poland (Pomerania) where this European mountain tree species found excellent conditions for growth and development 400 km north of its native range (Bijak 2010). The history of introduction of A. alba in the area of Polish Pomerania dates back to the end of the nineteenth century when experimental stands were established by German and Austrian foresters Schwappach, Wiedemann, and Cieślar (Bellon et al. 1977) and exist to the present time. These stands are still growing very well, and several productivity indicators (mean tree height, crown characteristics, tree-ring widths, health status, high abundance of natural regeneration) are similar to those obtained for A. alba grown in the native range of this tree species in southern Poland (Bijak 2010).
Within its native region, like most temperate and boreal tree species, A. alba develops obligate mutualisms with ectomycorrhizal (ECM) fungi, and this symbiosis plays a significant role in the survival and growth of trees (Smith and Read 2008). In general, A. alba has received little attention as an ECM host. Many studies on ECM fungi accompanying A. alba tend to focus on surveys of fungal fruiting bodies. In most other cases, ectomycorrhizas of A. alba were only characterized as “mycorrhizal types” without detailed identification of the fungal partner (Comandini et al. 2001 and references therein). A. alba ectomycorrhizas are also highly underrepresented in “An Information System for Characterization and DEtermination of EctoMYcorrhizae” (DEEMY), the largest online information system for morphological and anatomical determination of ectomycorrhizas (Agerer and Rambold 2004–2014). In this database comprising a total of 554 items, only four descriptions with taxonomic names of A. alba ectomycorrhizas are presented (Lactarius intermedius Krombh. ex Berk. & Broome, Lactarius salmonicolor R. Heim & Leclair, Lactarius subsericatus Kühner & Romagn., and Tricholoma bufonium (Pers.) Gillet). Still, less has been published about the ECM fungal symbionts that associate with A. alba based on molecular approaches (Eberhardt et al. 2000; Schirkonyer et al. 2013; Ważny 2014). As ECM symbiosis is required for the good growth and survival of many important forest tree species (Smith and Read 2008), the success of a tree species outside its natural range involves among other factors the availability of compatible symbiotic partners (Pringle et al. 2009; Vellinga et al. 2009; Dickie et al. 2010). Recently, several authors have intensively studied this issue in terms of different exotic tree species (e.g., Walbert et al. 2010; Bahram et al. 2013; O’Hanlon and Harrington 2012; Trocha et al. 2012; O’Hanlon et al. 2013; Lothamer et al. 2014). However, essentially, no data are available related to ECM fungal assemblages of A. alba when the tree species is growing outside its natural range. To fill gaps in this area of research, we surveyed for ECM fungi by sampling live roots from four mature forest stands of A. alba from the Polish Pomerania region where the fir component ranged from 20 to 100 %. A. alba was introduced ca. 100 years ago using seeds of unknown provenance; however, recent analyses of chloroplast microsatellites showed that the Sudety Mts. are the most probable region of origin of these fir trees (Dzialuk et al. 2013). Because suitable generalist ECM fungi are known to occur in this region (Stasińska 1999; Wojewoda 2003), and the introduced fir trees are exhibiting excellent growth rates, we hypothesized that the overall diversity of the ECM fungal community would be within the wide range of taxa reported from other coniferous forests. This assumption is supported by the finding that the non-native ECM trees often accept local mycorrhizal fungi as their symbiotic partners (Cullings et al. 2000; Tedersoo et al. 2007; Bahram et al. 2013; O’Hanlon et al. 2013; Kohout et al. 2011). Such overlap in compatibility with the local native ECM fungi (Watling 1995) likely appears in our studies, because A. alba and co-occurring trees from tested stands (Picea abies, Pinus sylvestris, Fagus sylvatica, Quercus sp.) come from temperate forest families that overlap in their native geographical ranges. A similar scenario was recently suggested and confirmed when non-native Pinus was introduced into forested areas of Iran (Bahram et al. 2013). Because A. alba trees have been introduced to the Pomerania region as seed without any fungal inoculum, and due to limitations of fungal dispersal (Galante et al. 2011; Horton et al. 2013) we further hypothesized that the ECM community of introduced trees will lack host-specific fungal symbionts. Results obtained will greatly enhance our understanding about the ECM fungal communities of A. alba when this tree species is growing outside its natural range.
Materials and methods
The study was conducted 400 km north of the natural range of A. alba in the central and eastern parts of Polish Pomerania (northern Poland) at four discreet forest stands, situated in the forest districts of Kartuzy (K), Lipusz (L), Osusznica (O), and Sławno (S) separated by at least 30 km. Selected stands were established after clear-cutting of managed mixed forests in the late nineteenth century and early twentieth century (Strzeliński, unpublished data). All stands had similar climatic and soil conditions with a mean annual temperature of +8 °C, ranging between −0.4 °C in January and 18.5 °C in July and a mean annual precipitation of about 745 mm. Details about stands characteristics are presented in Table 1.
Table 1.
Location, soil characteristics, plant association, and tree species composition of the study stands
| Stand | ||||
|---|---|---|---|---|
| K | L | O | S | |
| Geographic coordinates | 54° 13′ 27.2″ N, 17° 59′ 47.9″ E | 54° 14′ 25.0″ N, 17° 47′ 43.3″ E | 54° 06′ 58.8″ N, 17° 23′ 35.6″ E | 54° 29′ 02.8″ N, 16° 37′ 00.4″ E |
| Soil type | Brown acid | Brown acid | Luvisol | Brown acid |
| Humus form | Mull | Moder | Moder-mull | Moder-mor |
| Thickness of organic layer (cm) | 20 | 20 | 19 | 22 |
| pH H2O O-horizon | 3.9 | 3.9 | 3.9 | 4.0 |
| pH H2O A-horizon | 4.0 | 4.6 | 4.6 | 4.3 |
| Plant associationa | Fago-Quercetum | Abies alba-Deschampsia flexuosa/Luzulo pilosae-Fagetum | Luzulo pilosae-Fagetum | Luzulo pilosae-Fagetum |
| % Tree species composition and tree age (years) | 40 Abies (106) 40 Fagus (106) 10 Pinus (106) 10 Quercus (106) |
50 Picea (92) 40 Abies (92) 10 Fagus (162) |
100 Abies (115) | 60 Fagus (105) 20 Abies (105) 10 Picea (105) 10 Pinus (105) |
K Kartuzy, L Lipusz, O Osusznica, S Sławno, Abies Abies alba, Fagus Fagus sylvatica, Picea Picea abies Pinus Pinus sylvestris, Quercus Quercus spp.
aAccording to Matuszkiewicz 2001
Soil samples for mycorrhizal assessment were gathered in October 2012. A small spade was used to collect a 15 × 15 × 15 cm soil cube, starting from the organic layer after the removal of the litter layer. Sampling was done at least 30 m from the stand boundary to avoid an ecotone effect on the ECM fungal communities, and samples were separated by at least 6 m to avoid spatial autocorrelation. In each stand, 16 soil samples were randomly collected, from across each of four stands yielding a total of 64 samples. One non-random aspect of sampling was that samples were collected at about 50–80 cm from an A. alba tree trunk to ensure that we were sampling A. alba and no other tree sp. roots. Soil cores were secured by placing them into labeled individual ziplock plastic bags and transporting to the laboratory at the Institute of Dendrology, Kórnik, Poland. All samples were stored at 4 °C before being processed, but were not held longer than 2 weeks. Prior to analysis, soil samples were soaked in water at 4 °C overnight, and then, fine roots (<2 mm) were separated and cleaned from the soil in a sieve under cold running water. Fine roots with ectomycorrhizas were cut into 4–5-cm fragments. For each soil sample, three random root subsamples were counted until each subsample had approximately 250 root tips. Discrimination between ectomycorrhizas of Abies and ectomycorrhizas of other tree species, potentially present in the soil samples, was based on differences in morphology and the diameter of unramified mycorrhizal tips of Abies in relation to ectomycorrhizas of spruce, pine, or some deciduous trees. Diameter of Abies mycorrhizas is about 30 % larger than that of Pinus or Picea ectomycorrhizas and 50–60 % larger than that of ectomycorrhizas of broadleaf trees. Examination of the ectomycorrhizas was performed under a dissecting microscope at ×10 to ×100 magnification (SteREO Discovery V12, Zeiss). The ectomycorrhizas were described according to Agerer (1987–2008) based on macroscopic features (ramification system; color, shape, texture, and thickness of the mantle; presence and organization of the emanating hyphae; rhizomorphs; and other elements) and classified into separate morphotypes. The number of ectomycorrhizas of each morphotype and ECM colonization ratio were noted separately for each sample. For each ECM morphotype, five to 10 carefully cleaned ECM tips were placed in Eppendorf tubes in Milli-Q water and frozen at −20 °C until processing for molecular analysis. Throughout molecular identification, each morphotype sample was processed separately. Specimens were combined for abundance calculation only after the DNA analysis showed that morphotypes were the same. The relative abundance of each identified morphotype was calculated as a percentage of the total number of ectomycorrhizas of each subsample; then, data were averaged in each sample and in overall samples from each study stand. Ectomycorrhizal fungal symbionts were identified based on PCR amplification (wit primer pair ITS-1f and ITS-4) and sequencing of the internal transcribed spacer (ITS) of nuclear ribosomal DNA (rDNA). Total DNA was extracted from frozen ectomycorrhizas with the miniprep method developed by Gardes and Bruns (1996) or using a GeneMATRIX Plant & Fungi DNA Purification Kit (EURx Ltd., Gdańsk, Poland). Multiple PCR products (double DNA bands on the gel) were omitted from the analysis. Species-level identification of mycorrhizas was defined as the sharing of >98 % ITS region sequence identity with the reference sequence obtained from sporocarp vouchers, over a length of at least 450 bp. All fungal species names were updated to their current nomenclature using the online resources Index Fungorum (http://www.indexfungorum.org as of the date 07.04.2015). Selected sequences have been deposited in GenBank under the accession numbers KP230465-230499. Complete methods for molecular analysis and identification of ECM morphotypes are presented in our previous papers (Leski et al. 2010; Leski and Rudawska 2012).
The belowground ECM fungal diversity at each study stand was expressed as the number of recognized ECM taxa (taxa richness) and evaluated using Shannon–Wiener diversity and Simpson’s dominance indices. Jackknife 2 richness estimator was calculated with EstimateS 8.0 software (Colwell 2006), using 1000 randomized runs with sample replacement.
Multivariate analyses of the belowground community of ECM fungi were performed using PAST 2.13 software (Hammer et al. 2001), with the Bray–Curtis dissimilarity coefficient, based on square root-transformed data. The data matrix consisted of 64 samples with the relative abundance of each ECM fungal taxon within each sample. Nonmetric multidimensional scaling ordination (NMDS) was used to illustrate differences (based on the Bray–Curtis matrix) between ECM fungal communities from the investigated stands. Differences in relative abundance of ECM fungal taxa between study stands were tested with one-way analysis of similarity (ANOSIM). Analysis of variance with Tukey’s test was used to compare the mean taxa richness and ecological indices on a sample basis between tested A. alba stands.
Results
The examination of fine roots of A. alba from all stands revealed nearly 100 % ECM colonization. Morphological assessment was carried out on 35,825 ECM tips from 64 samples (16 samples per stand). Based on gross morphology, 38 mycorrhizal morphotypes were distinguished in all study stands. Overall, 221 out of the 248 ECM tips representing all morphotypes yielded PCR products and were subjected to ITS sequence analysis. The total successful amplification rate was 89 %. Based on sequencing of ITS fungal rDNA, 35 mycorrhizal fungal taxa were identified. Of these 35 ECM fungal taxa, 28 were assigned to a species level and seven to genus (Table 2). The taxa richness of mycorrhizal fungi ranged from 27 taxa (stand S) and 26 taxa (stand L) to 23 (stand O) and 22 taxa (stand K). Average taxa richness noted in soil samples ranged between 9.6 and 12.4, without significant differences between stands according to ANOVA (Table 2).
Table 2.
Molecular identification, relative abundance (±SE), observed total and mean species richness (±SE), estimated species richness, and ecological indices for ectomycorrhizal fungi on the roots of Abies alba trees from stands K, L, O, and S
| Identification | Accession | Closest match | Identity (%) | Relative abundance (%)—stand | |||
|---|---|---|---|---|---|---|---|
| K | L | O | S | ||||
| Cenococcum geophilum Fr. | KP230465 | Cenococcum geophilum (JN943885) | 99 | 68.0 ± 10.1 | 50.2 ± 6.2 | 69.8 ± 6.2 | 55.9 ± 6.0 |
| Tomentella stuposa (Link) Stalpers | KP230466 | Tomentella stuposa (UDB002429) | 100 | 7.4 ± 1.2 | 19.1 ± 1.6 | 7.5 ± 0.9 | 14.9 ± 2.1 |
| Lactarius camphoratus (Bull.) Fr. | KP230467 | Lactarius camphoratus (KF432971) | 100 | 3.5 ± 0.2 | 1.2 ± 0.1 | 2.0 ± 0.1 | 0.5 ± 0.1 |
| Russula fellea (Fr.) Fr. | KP230468 | Russula fellea (UDB000110) | 99 | 3.04 ± 0.3 | 1.0 ± 0.1 | 1.5 ± 0.1 | |
| Byssocorticium atrovirens (Fr.) Bondartsev & Singer | KP230469 | Byssocorticium atrovirens (UDB000075) | 99 | 2.8 ± 0.2 | 0.1 ± 0.1 | 0.1 ± 0.1 | |
| Tomentella albomarginata (Bourdot & Galzin) M.P. Christ. | KP230470 | Tomentella albomarginata (UDB018851) | 100 | 2.4 ± 0.3 | 3.4 ± 0.3 | 1.9 ± 0.2 | 4.2 ± 0.2 |
| Tomentella terrestris (Berk. & Broome) M.J. Larsen | KP230471 | Tomentella terrestris (UDB011638) | 100 | 2.4 ± 0.4 | 1.2 ± 0.2 | 1.2 ± 0.2 | 1.8 ± 0.1 |
| Cantharellus sp. | KP230472 | Cantharellus tubaeformis (UDB002389) | 96 | 2.0 ± 0.5 | 2.6 ± 0.3 | 1.2 ± 0.2 | 2.0 ± 0.2 |
| Xerocomellus pruinatus (Fr. & Hök) Šutara | KP230473 | Xerocomus pruinatus (UDB000481) | 99 | 1.5 ± 0.3 | 1.7 ± 0.3 | 0.9 ± 0.2 | 2.0 ± 0.1 |
| Genea sp. | KP230474 | Genea hispidula (AJ969623) | 96 | 1.3 ± 0.2 | 0.2 ± 0.1 | ||
| Cortinarius malachius (Fr.) Fr. | KP230475 | Cortinarius malachius (JQ888160) | 99 | 1.2 ± 0.4 | 2.6 ± 0.4 | 4.2 ± 0.9 | 1.9 ± 0.2 |
| Inocybe geophylla (Bull.) P. Kumm. | KP230476 | Inocybe geophylla (JQ888171) | 98 | 1.2 ± 0.5 | 1.7 ± 0.5 | 0.5 ± 0.1 | 1.5 ± 0.4 |
| Cortinarius sp. 2 | KP230477 | Cortinarius camphoratus (UDB011341) | 95 | 0.9 ± 0.1 | 1.1 ± 0.3 | 0.8 ± 0.3 | 0.3 ± 0.1 |
| Geopora cervina (Velen.) T. Schumach. | KP230478 | Geopora cervina (UDB016155) | 94 | 0.7 ± 0.2 | 1.4 ± 0.4 | 0.1 ± 0.1 | 0.6 ± 0.3 |
| Cortinarius fulvescens Fr. | KP230479 | Cortinarius fulvescens (HQ604731) | 99 | 0.6 ± 0.4 | 0.9 ± 0.2 | 0.3 ± 0.1 | 0.8 ± 0.4 |
| Meliniomyces variabilis Hambl. & Sigler | KP230480 | Meliniomyces variabilis (HQ157931) | 99 | 0.4 ± 0.2 | 1.0 ± 0.3 | 0.6 ± 0.3 | 0.4 ± 0.2 |
| Lactarius aurantiacus (Pers.) Gray | KP230481 | Lactarius aurantiacus (KF432974) | 99 | 0.4 ± 0.2 | 1.8 ± 0.6 | 0.5 ± 0.3 | |
| Lactarius rufus (Scop.) Fr. | KP230482 | Lactarius rufus (KF241543) | 99 | 0.1 ± 0.2 | >0.1 | 2.0 ± 1.1 | |
| Pseudotomentella tristis (P. Karst.) M.J. Larsen | KP230483 | Pseudotomentella tristis (UDB000032) | 100 | 0.1 ± 0.1 | 3.6 ± 1.1 | 0.5 ± 0.3 | 0.3 ± 0.1 |
| Tylopilus felleus (Bull.) P. Karst. | KP230484 | Tylopilus felleus (HM190016) | 100 | >0.1 | 0.2 ± 0.1 | 0.1 ± 0.2 | |
| Peziza sp. | KP230485 | Peziza succosa (UDB015317) | 97 | >0.1 | 0.9 ± 0.3 | 0.5 ± 0.2 | 1.6 ± 0.8 |
| Laccaria laccata (Scop.) Cooke | KP230486 | Laccaria laccata (UDB015789) | 96 | >0.1 | |||
| Piloderma fallax (Lib.) Stalpers | KP230487 | Piloderma fallax (UDB001614) | 99 | 2.6 ± 0.6 | 4.1 ± 0.9 | 3.1 ± 0.6 | |
| Tylospora asterophora (Bonord.) Donk | KP230488 | Tylospora asterophora (UDB002469) | 100 | 0.6 ± 0.2 | |||
| Tomentella botryoides (Schwein.) Bourdot & Galzin | KP230489 | Tomentella botryoides (UDB000258) | 100 | 0.6 ± 0.2 | |||
| Craterellus lutescens (Fr.) Fr. | KP230490 | Craterellus lutescens (UDB019796) | 96 | 0.3 ± 0.1 | 0.4 ± 0.2 | 0.8 ± 0.4 | |
| Amanita muscaria (L.) Lam. | KP230491 | Amanita muscaria (AB080984) | 99 | 0.1 ± 0.1 | 1.6 ± 0.3 | 0.2 ± 0.2 | |
| Imleria badia (Fr.) Vizzini | KP230492 | Xerocomus badius (HQ207696) | 100 | 0.1 ± 0.3 | 0.4 ± 0.2 | ||
| Tuber puberulum Berk. & Broome | KP230493 | Tuber puberulum (HM190013) | 99 | >0.1 | |||
| Cortinarius sp. 1 | KP230494 | Cortinarius traganus (UDB011350) | 97 | 0.2 ± 0.1 | |||
| Russula sp. 1 | KP230495 | Russula paludosa (JF908659) | 97 | 0.1 ± 0.1 | |||
| Russula sp. 2 | KP230496 | Russula xerampelina (UDB002533) | 95 | 1.1 ± 0.3 | |||
| Russula ochroleuca Fr. | KP230497 | Russula ochroleuca (JF908647) | 100 | 0.7 ± 0.2 | |||
| Cortinarius semisanguineus (Fr.) Gillet | KP230498 | Cortinarius semisanguineus (UDB001553) | 100 | 0.6 ± 0.3 | |||
| Laccaria amethystina Cooke | KP230499 | Laccaria amethystina (HM189776) | 100 | 0.1 ± 0.2 | |||
| Abundance of shared species | 92.4 | 92.6 | 91.9 | 88.6 | |||
| Abundance of stand characteristic species | >0.1 | 1.2 | 0.2 | 2.5 | |||
| Taxa richness | 22 | 26 | 23 | 27 | |||
| No. of stand characteristic taxa | 1 | 3 | 2 | 4 | |||
| Average taxa richness per sample | 9.62 ± 1.51 | 10.87 ± 2.64 | 10.62 ± 3.58 | 12.37 ± 1.41 | |||
| Jackknife 2 | 31.76 | 33.12 | 27.81 | 31.64 | |||
| Shannon diversity | 1.12 ± 0.51 | 1.31 ± 0.46 | 1.02 ± 0.42 | 1.40 ± 0.42 | |||
| Simpsons dominance | 0.51 ± 0.19 | 0.42 ± 0.15 | 0.55 ± 0.18 | 0.41 ± 0.17 | |||
K Kartuzy, L Lipusz, O Osusznica, S Sławno, abundance of shared species the abundance of species shared with at least one other site, abundance of the stand characteristic species abundance of species present in only one stand
The fungi identified were Basidiomycetes (orders: Agaricales, Atheliales, Boletales, Thelephorales, Russulales, and Cantharellales) and Ascomycetes (orders: Pezizales, Helotiales, and Mytilinidiales). In terms of taxa richness, the investigated ECM fungal communities were dominated by representatives of the orders Agaricales (nine taxa), Russulales (seven taxa), Thelephorales (five taxa), and Pezizales (four taxa). Fifteen fungal taxa were common in all study stands. One taxon was found exclusively in stand K, two in stand O, and three in stand L, and four taxa were restricted to stand S. The relative abundance of fungal taxa that were specific for the individual stand ranged from <0.1 % in stand K to 2.5 % in stand S (Table 2).
Irrespective of the stand, the ECM fungal communities were dominated by Cenococcum geophilum (relative abundance from 50.2 to 69.8 %). The second most abundant taxon was Tomentella sublilacina (relative abundance 7.4–19.1 %). Other species always occurred with a relative abundance of less than 5 % (Table 2).
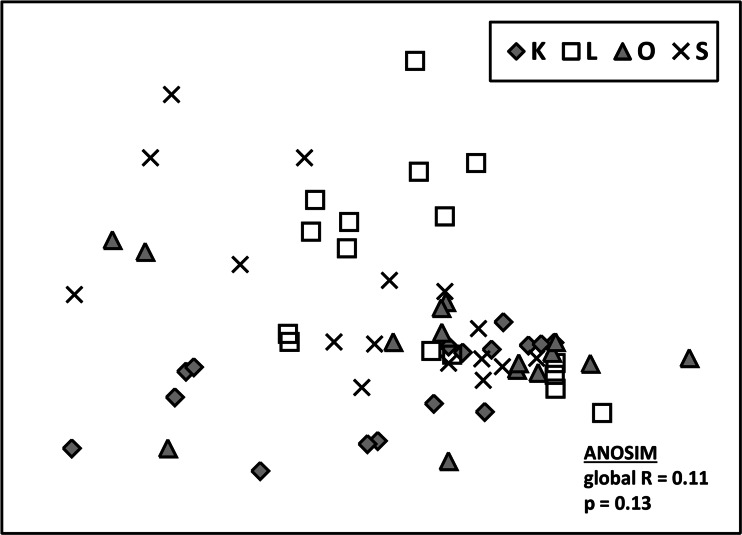
The Shannon’s diversity and Simpson dominance indices for the ECM assemblages of tested A. alba did not differ significantly between study stands (Table 2). ANOSIM and NMDS analyses were used to assess the influence of study stands on ECM fungal communities. Based on ANOSIM analysis, fungal communities were not significantly different between study stands (global R = 0.11; P = 0.13). Consistently, the NMDS ordinations of the ECM fungal communities (final stress = 0.16) showed no clear grouping of samples according to stand identity (Fig. 1).
Fig. 1.
Nonmetric multidimensional scaling (NMDS) ordination of ectomycorrhizal fungal communities of Abies alba trees from stands K, L, O, and S (NMDS parameters: R 2 of the axis 1 = 0.49, R 2 of the axis 2 = 0.37, final stress = 0.16)
Discussion
The overall species richness observed in our study support our assumption that the diversity of ECM fungal community of A. alba falls within the broad range of variability described in sequencing studies of other coniferous trees grown in their native temperate range. Examples include studies of Scots pine (29 taxa, Rudawska et al. 2011; 43 taxa, Pickles et al. 2012), Norway spruce (16 taxa, Kjøller et al. 2012), and Douglas fir (31–45 taxa depending on the site, Kranabetter et al. 2012). The 35 ECM fungal taxa characterized on root tips of A. alba from four mature forest stands correspond to 81 % of the potential estimated species richness of 44 taxa. Given the sampling strategy of our study, such level of discovered fungal species richness in relation to estimated richness may indicate that ECM symbionts compatible with A. alba outside of its native range have a rather homogenous spatial distribution. Moreover, low variation in relative abundances of the prevailing ECM fungal species further confirmed that certain fungal species were similarly abundant across the stands.
Comparison of our observed species richness with other data on ECM fungus community structure carried out directly in natural A. alba stands is difficult due to the scarcity of data that is mostly restricted to aboveground ECM fungal sporocarp diversity. Depending on the number of seasons and locations surveyed, Laganà et al. (2000, 2002) observed 43 to 61 ECM fungal species in A. alba monoculture forests in Tuscany, Italy. In contrast, aboveground ECM fungal diversity of A. alba grown outside its natural range in Mecklenburg-Western Pomerania, Germany, was much lower and amounted to only 22 species of ECM fungi (Unterseher et al. 2012). Belowground studies of the ECM fungal assemblages that used morphological characterization of ectomycorrhizas revealed 25 to 48 taxa on A. alba trees grown in the mountainous natural woods of the Abruzzo region, Italy (Comandini et al. 1998, 2001). However, resolution of ECM fungal diversity based on morphotyping is often low and only rarely allows for an unambiguous identification of mycorrhizas at the fungal species level (Sakakibara et al. 2002). Studies of the ECM fungal communities associated with A. alba using direct sequencing of the ECM root tip DNA are scarce compared with similar studies of other coniferous and deciduous trees (Comandini et al. 2012). Only 15 mycorrhizal taxa were identified with molecular methods in 100-year-old pure stands of A. alba in the Taunus Mountains in Central Germany (Schirkonyer et al. 2013) where A. alba is not native. The mycorrhizal species richness was much lower in that study than ours and was likely underestimated due to the low sampling effort of that research (only 46 mycorrhizal tips were analyzed). The only comprehensive sequencing data on the ECM fungal community of A. alba are from studies conducted on seedlings naturally regenerating under Silver fir and Scots pine canopy in the Polish Carpathians (Ważny 2014). Overall, ECM fungal richness in those stands was higher than the richness obtained in our studies, with a total of 49 ECM fungal taxa reported. Part of the discrepancy in overall species richness between the study in the Polish Carpathians and our studies in Polish Pomerania is likely due to sampling intensity (six vs four sites and 30 vs 16 samples per site, respectively). However, at the individual stand level, species richness was higher in our study ranging from 22 to 27 ECM fungal taxa, depending on the stand and canopy tree composition. Taken together, we believe that the ECM fungus suite of A. alba when grown outside the natural range is not restricted on a per stand basis, and A. alba trees are readily colonized by available and compatible ECM fungi.
It is generally accepted that the co-occurrence of different host tree species within a stand promotes ECM fungus diversity at the local scale (Ishida et al. 2007; Morris et al. 2008; Tedersoo et al. 2008). Therefore, we expected that that presence of closely related confamilials (P. abies and/or P. sylvestris) and more distantly related non-confamilials (Quercus spp. and/or F. sylvatica) in tested stands would affect ECM fungal species richness of A. alba trees. However, in our studies, the resulting diversity and similarity metrics revealed a lack of statistical differences in the structure of the ECM fungal community between stands that varied in overstory tree composition. This indicates that neighboring trees of different species had no influence on the ECM fungal community of A. alba. Interestingly, even the pure stand of A. alba had a similar ECM fungal community structure to stands with high overstory tree composition. This confirms previous findings that in the northern hemisphere where the range of many forest tree species overlap greatly, fine roots of non-native species can be colonized by indigenous ECM fungi (Trocha et al. 2012).
It has been well documented that moving different ECM tree species within/between the temperate and boreal regions (east–west movement) rarely presents problems for suitable indigenous ECM fungi to colonize the introduced tree species (Buée et al. 2011; O’Hanlon et al. 2013). In contrast, in north–south movements of ECM tree species, especially over thousands of kilometers (e.g., across the equator), situations can arise wherein the ECM fungal community is not suitable for ECM tree success (Dickie et al. 2010; Nuñez et al. 2009). Because both the direction and magnitude of A. alba movement in our study were small, we found suitable ECM fungi present at the destination site.
The ECM fungal taxa identified within all study stands are among the more common members of ECM fungal communities of temperate and boreal forests in Europe, including C. geophilum and members of the genera Tomentella, Lactarius, Russula, Cortinarius, and Laccaria. C. geophilum and many members of these genera exhibit broad host ranges and allow potential development of mycelial networks between co-occurring trees (Kennedy et al. 2003; Nara 2006). These taxa have been previously reported from mycocoenological studies from different regions of Poland (Wojewoda 2003).
Among the 35 ECM fungal species found in our study, only 10 species, including C. geophilum, Tomentella stuposa, Xerocomellus pruinatus, Lactarius aurantiacus, Piloderma fallax, Tylospora asterophora, Amanita muscaria, Imleria badia, Tuber puberulum, and Laccaria amethystina, were also common associates on A. alba seedlings grown in its native range (Ważny 2014). These 10 ECM fungal species colonized approximately 80 % of the root tips on mature A. alba trees outside of its native range, but only 25 % within its native range in the study of Ważny (2014). Contrary to the findings of Ważny (2014), we did not observe fungal symbionts belonging to the genera Amphinema, Clavulina, Elaphomyces, Hydnotrya, Hydnum, or Sebacina. Such discrepancy in relative abundance of shared species and species composition between ECM fungal communities of A. alba trees inside and outside its native range demonstrates the importance of site characteristics (pH, litter and soil quality, climate, etc.) in structuring ECM community (Jumpponen and Egerton-Warburton 2005).
Along with the occurrence of generalist fungal species as a component of ECM colonization of A. alba, we did not observe Abies-specific symbionts, such as L. salmonicolor, which is widespread in above- and belowground communities in the natural range of A. alba (Comandini et al. 1998, 2001; Laganà et al. 2000; Ważny 2014). Other fungal species that exhibited some level of Abies preference (Lactarius albocarneus Britzelm., L. intermedius, Russula cavipes Britzelm.) were also absent in our studies and in other surveys of A. alba grown outside its natural range (Unterseher et al. 2012; Schirkonyer et al. 2013). The absence of the fungal symbionts that show preference for Abies may be ascribed to several factors: (1) soil and environmental conditions are inappropriate for the growth and development of the mycelium of host-specific fungal symbionts; (2) geographic distance from the natural range of A. alba negatively influences the dispersal and competitive abilities of fungal propagules; and (3) sampling design of our studies was insufficient to detect the ECM host-specific symbionts, known to occur with low abundance (Ważny 2014). Our data suggest A. alba does not extensively rely on host-specific fungi, a feature characteristic of many late successional tree species; this supports the suggestion of Kropp and Trappe (1982) that was further confirmed by Horton et al. (2005), that selection pressure in later successional tree species pushes many species away from host specificity.
C. geophilum was the most common species at all investigated A. alba stands with an abundance of 50–70 %. Such abundance of C. geophilum might be somewhat overestimated due to the robust and melanized mantles of this fungus that allow the C. geophilum ectomycorrhizas to persist 4–10 times longer than ectomycorrhizas of other ECM fungal species (Fernandez et al. 2013). This unique feature makes morphological determination of this fungus possible even after the ectomycorrhizas are dead (Valentine et al. 2004). But even knowing that only 40 % of C. geophilum ectomycorrhizas are vital in autumn (Fernandez et al. 2013), this species remains the most abundant component of A. alba ECM fungal communities. An important factor that may contribute to the predominance of C. geophilum might be the thick organic layer on the forest floors of the tested stands. This layer is characterized by high fluctuations of soil temperature and moisture content, often to the point of drying. C. geophilum is well known as a very competitive fungus that grows very well in such conditions (Hasselquist et al. 2005; Matsuda et al. 2009; Dickie 2007).
It is generally recognized that tree species growing outside of their native range harbor relatively species-poor ECM communities (Tedersoo et al. 2007; Nuñez et al. 2009; Dickie et al. 2010; Walbert et al. 2010) because of the low number of compatible symbiotic fungi, dispersal limitations, and a frequent lack of host-specific symbionts. We did not observe evident impoverishment of ECM fungal assemblages during our study of A. alba stands in the Pomerania region when compared with the ECM constituents inside the native range (Ważny 2014). The ECM community was characterized by a lack of Abies-specific fungal symbionts and a rich and diverse suite of local host-generalist mycobionts. However, there are some limitations in our sampling strategy that are important to note. Due to time constraints, we only sampled at one point in time, so our findings cannot represent the year-round fungal community. In terms of vertical variation in the soil profile, sampling was limited to 15-cm depth; thus, some taxa with the preference to mineral soil horizons could be left undetected. Horizontal variation may also play a part in our study system, but in this case, we tried to avoid spatial autocorrelation between soil cores by sampling more than 6 m apart. Recently, the problem of horizontal, vertical, and temporal variations in ECM communities has been thoroughly reviewed (Bahram et al. 2014; O’Hanlon 2012) and should be carefully considered when planning future experiments.
Acknowledgments
We are grateful to Halina Narożna and Maria Wójkiewicz (Institute of Dendrology, Polish Academy of Sciences) for the technical assistance. We would also like to thank Randy Molina (the editor) and two anonymous reviewers for their constructive suggestions and comments on the manuscript. The present research was financially supported by the Polish Ministry of Science and Higher Education, project No. 2 P06L 017 30. Additional funding was provided by the Institute of Dendrology of the Polish Academy of Sciences.
Conflict of interest
The authors declare that they have no competing interests.
References
- Agerer R (1987–2008) Colour atlas of ectomycorrhizae. EinhornVerlag Eduard Dietenberger, Munich
- Agerer R, Rambold G (2004–2014) [first posted on 2004-06-01; most recent update: 2011-01-10]. DEEMY – An Information System for Characterization and Determination of Ectomycorrhizae. www.deemy.de – München, Germany
- Bahram M, Kõljalg U, Kohout P, Mirshahvaladi S, Tedersoo L. Ectomycorrhizal fungi of exotic pine plantations in relation to native host trees in Iran: evidence of host range expansion by local symbionts to distantly related host taxa. Mycorrhiza. 2013;23:11–19. doi: 10.1007/s00572-012-0445-z. [DOI] [PubMed] [Google Scholar]
- Bahram M, Peay KG, Tedersoo L. Local-scale biogeography and spatiotemporal variability in communities of mycorrhizal fungi. New Phytol. 2014;205:1454–1463. doi: 10.1111/nph.13206. [DOI] [PubMed] [Google Scholar]
- Bellon S, Tumiłowicz J, Król S. Obce gatunki drzew w gospodarstwie leśnym. Warsaw: PWRiL; 1977. [Google Scholar]
- Bijak S. Tree-ring chronology of silver fir and its dependence on climate of the Kaszubskie Lakeland (Northern Poland) Geochronometria. 2010;35:91–94. doi: 10.2478/v10003-010-0001-9. [DOI] [Google Scholar]
- Buée M, Maurice J-P, Zeller B, Andrianarisoa S, Ranger J, Courtecuisse R, Marçais B, Le Tacon F. Influence of tree species on richness and diversity of epigeous fungal communities in a French temperate forest stand. Fungal Ecol. 2011;4:22–31. doi: 10.1016/j.funeco.2010.07.003. [DOI] [Google Scholar]
- Colwell R (2006) EstimateS: Statistical estimation of species richness and shared species from samples, version 8.0. Available at http://purl.oclc.org/estimates
- Comandini O, Pacioni G, Rinaldi AC. Fungi in ectomycorrhizal associations of silver fir (Abies alba Miller) in Central Italy. Mycorrhiza. 1998;7:323–328. doi: 10.1007/s005720050200. [DOI] [Google Scholar]
- Comandini O, Pacioni G, Rinaldi AC. An assessment of below-ground ectomycorrhizal diversity of Abies alba Miller in central Italy. Plant Biosyst. 2001;135:337–350. doi: 10.1080/11263500112331350960. [DOI] [Google Scholar]
- Comandini O, Rinaldi AC, Kuyper TW. Measuring and estimating ectomycorrhizal fungal diversity: a continuous challenge. In: Pagano M, editor. Mycorrhiza Occur. Nat. Restored Environ. 1. New York: Nova; 2012. pp. 165–200. [Google Scholar]
- Cullings KW, Vogler DR, Parker VT, Finley SK. Ectomycorrhizal specificity patterns in a mixed Pinus contorta and Picea engelmannii forest in Yellowstone National Park. Appl Environ Microbiol. 2000;66:4988–4991. doi: 10.1128/AEM.66.11.4988-4991.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie IA. Host preference, niches and fungal diversity. New Phytol. 2007;174:230–233. doi: 10.1111/j.1469-8137.2007.02055.x. [DOI] [PubMed] [Google Scholar]
- Dickie IA, Bolstridge N, Cooper JA, Peltzer DA. Co-invasion by Pinus and its mycorrhizal fungi. New Phytol. 2010;187:475–484. doi: 10.1111/j.1469-8137.2010.03277.x. [DOI] [PubMed] [Google Scholar]
- Dzialuk A, Czarnecki J, Gout R, Filipiak M. Pochodzenie jodły pospolitej (Abies alba Mill.) z Nadleśnictwa Osusznica w świetle badań DNA cytoplazmatycznego-ostoja jodły sudeckiej na Pomorzu? (in Polish with English summary) Sylwan. 2013;157:139–148. [Google Scholar]
- Eberhardt U, Oberwinkler F, Verbeken A, Rinaldi AC, Pacioni G, Comandini O. Lactarius ectomycorrhizae on Abies alba: morphological description, molecular characterization, and taxonomic remarks. Mycologia. 2000;92:860–873. doi: 10.2307/3761582. [DOI] [Google Scholar]
- Fernandez CW, McCormack ML, Hill JM, et al. On the persistence of Cenococcum geophilum ectomycorrhizas and its implications for forest carbon and nutrient cycles. Soil Biol Biochem. 2013;65:141–143. doi: 10.1016/j.soilbio.2013.05.022. [DOI] [Google Scholar]
- Galante TE, Horton TR, Swaney DP. 95% of basidiospores fall within 1 m of the cap: a field-and modeling-based study. Mycologia. 2011;103:1175–1183. doi: 10.3852/10-388. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD (1996) Community structure of ectomycorrhizal fungi in a Pinus muricata forest: above- and below-ground views. Botany 74:1572–1583
- Hammer O, Harper DAT, Ryan PD (2001) PAST: Palaeontological statistics software package for education and data analysis. Palaeont Electron 4:9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm
- Hasselquist N, Germino MJ, Mcgonigle T, Smith WK. Variability of Cenococcum colonization and its ecophysiological significance for young conifers at alpine-treeline. New Phytol. 2005;165:867–873. doi: 10.1111/j.1469-8137.2005.01275.x. [DOI] [PubMed] [Google Scholar]
- Horton TR, Molina R, Hood K. Douglas-fir ectomycorrhizae in 40- and 400-year-old stands: mycobiont availability to late successional western hemlock. Mycorrhiza. 2005;15:393–403. doi: 10.1007/s00572-004-0339-9. [DOI] [PubMed] [Google Scholar]
- Horton TR, Swaney DP, Galante TE. Dispersal of ectomycorrhizal basidiospores: the long and short of it. Mycologia. 2013;105:1623–1626. doi: 10.3852/13-119. [DOI] [PubMed] [Google Scholar]
- Ishida TA, Nara K, Hogetsu T. Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer-broadleaf forests. New Phytol. 2007;174:430–440. doi: 10.1111/j.1469-8137.2007.02016.x. [DOI] [PubMed] [Google Scholar]
- Jumpponen A, Egerton-Warburton L. Mycorrhizal fungi in successional environments—a community assembly model incorporating host plant, environmental and biotic filters. In: Dighton J, White JF, Oudemans P, editors. The fungal community. New York: CRC Press; 2005. pp. 139–180. [Google Scholar]
- Kennedy PG, Izzo AD, Bruns TD. There is high potential for the formation of common mycorrhizal networks between understorey and canopy trees in a mixed evergreen forest. J Ecol. 2003;91:1071–1080. doi: 10.1046/j.1365-2745.2003.00829.x. [DOI] [Google Scholar]
- Kjøller R, Nilsson L-O, Hansen K, Schmidt IK, Vesterdal L, Gundersen P. Dramatic changes in ectomycorrhizal community composition, root tip abundance and mycelial production along a stand-scale nitrogen deposition gradient. New Phytol. 2012;194:278–286. doi: 10.1111/j.1469-8137.2011.04041.x. [DOI] [PubMed] [Google Scholar]
- Kohout P, Sýkorová Z, Bahram M, Hadincová V, Albrechtová J, Tedersoo L, Vohník M. Ericaceous dwarf shrubs affect ectomycorrhizal fungal community of the invasive Pinus strobus and native Pinus sylvestris in a pot experiment. Mycorrhiza. 2011;21:403–412. doi: 10.1007/s00572-010-0350-2. [DOI] [PubMed] [Google Scholar]
- Kranabetter J, Stoehr M, O’Neill G. Divergence in ectomycorrhizal communities with foreign Douglas-fir populations and implications for assisted migration. Ecol Appl. 2012;22:550–560. doi: 10.1890/11-1514.1. [DOI] [PubMed] [Google Scholar]
- Kropp BR, Trappe JM. Ectomycorrhizal fungi of Tsuga heterophylla. Mycologia. 1982;74:479–488. doi: 10.2307/3792970. [DOI] [Google Scholar]
- Laganà A, Salerni E, Barluzzi C, Perini C, De Dominicis V. Mycocoenology in Abies alba Miller woods of central-southern Tuscany (Italy) Acta Soc Bot Pol. 2000;69:293–298. doi: 10.5586/asbp.2000.039. [DOI] [Google Scholar]
- Laganà A, Angiolini C, Loppi S, Salerni E, Perini C, Barluzzi C, De Dominicis V. Periodicity, fluctuations and successions of macrofungi in fir forests (Abies alba Miller) in Tuscany, Italy. For Ecol Manag. 2002;169:187–202. doi: 10.1016/S0378-1127(01)00672-7. [DOI] [Google Scholar]
- Leski T, Rudawska M. Ectomycorrhizal fungal community of naturally regenerated European larch (Larix decidua) seedlings. Symbiosis. 2012;56:45–53. doi: 10.1007/s13199-012-0164-4. [DOI] [Google Scholar]
- Leski T, Aučina A, Skridaila A, Pietras M, Riepšas E, Rudawska M. Ectomycorrhizal community structure of different genotypes of Scots pine under forest nursery conditions. Mycorrhiza. 2010;20:473–481. doi: 10.1007/s00572-010-0298-2. [DOI] [PubMed] [Google Scholar]
- Lothamer K, Brown SP, Mattox JD, Jumpponen A. Comparison of root-associated communities of native and non-native ectomycorrhizal hosts in an urban landscape. Mycorrhiza. 2014;24:267–280. doi: 10.1007/s00572-013-0539-2. [DOI] [PubMed] [Google Scholar]
- Matsuda Y, Hayakawa N, Ito S. Local and microscale distributions of Cenococcum geophilum in soils of coastal pine forests. Fungal Ecol. 2009;2:31–35. doi: 10.1016/j.funeco.2008.10.002. [DOI] [Google Scholar]
- Matuszkiewicz W. Przewodnik do oznaczania zbiorowisk roślinnych Polski (in Polish) Warszawa: PWN; 2001. [Google Scholar]
- Morris MH, Smith ME, Rizzo DM, Rejmánek M, Bledsoe CS. Contrasting ectomycorrhizal fungal communities on the roots of co-occurring oaks (Quercus spp.) in a California woodland. New Phytol. 2008;178:167–176. doi: 10.1111/j.1469-8137.2007.02348.x. [DOI] [PubMed] [Google Scholar]
- Nara K. Ectomycorrhizal networks and seedling establishment during early primary succession. New Phytol. 2006;169:169–178. doi: 10.1111/j.1469-8137.2005.01545.x. [DOI] [PubMed] [Google Scholar]
- Nuñez MA, Horton TR, Simberloff D. Lack of belowground mutualisms hinders Pinaceae invasions. Ecology. 2009;90:2352–2359. doi: 10.1890/08-2139.1. [DOI] [PubMed] [Google Scholar]
- O’Hanlon R. Below-ground ectomycorrhizal communities: the effect of small scale spatial and short term temporal variation. Symbiosis. 2012;57:57–71. doi: 10.1007/s13199-012-0179-x. [DOI] [Google Scholar]
- O’Hanlon R, Harrington TJ. Similar taxonomic richness but different communities of ectomycorrhizas in native forests and non-native plantation forests. Mycorrhiza. 2012;22:371–382. doi: 10.1007/s00572-011-0412-0. [DOI] [PubMed] [Google Scholar]
- O’Hanlon R, Harrington TJ, Berch SM, Outerbridge RA. Comparisons of macrofungi in plantations of Sitka spruce (Picea sitchensis) in its native range (British Columbia, Canada) versus non-native range (Ireland and Britain) show similar richness but different species composition. Can J For Res. 2013;458:450–458. doi: 10.1139/cjfr-2012-0391. [DOI] [Google Scholar]
- Pickles BJ, Genney DR, Anderson IC, Alexander IJ. Spatial analysis of ectomycorrhizal fungi reveals that root tip communities are structured by competitive interactions. Mol Ecol. 2012;21:5110–5123. doi: 10.1111/j.1365-294X.2012.05739.x. [DOI] [PubMed] [Google Scholar]
- Pringle A, Bever JD, Gardes M, Parrent JL, Rillig MC, Klironomos JN. Mycorrhizal symbioses and plant invasions. Annu Rev Ecol Evol Syst. 2009;40:699–715. doi: 10.1146/annurev.ecolsys.39.110707.173454. [DOI] [Google Scholar]
- Rudawska M, Leski T, Stasińska M. Species and functional diversity of ectomycorrhizal fungal communities on Scots pine (Pinus sylvestris L.) trees on three different sites. Ann For Sci. 2011;68:5–15. doi: 10.1007/s13595-010-0002-x. [DOI] [Google Scholar]
- Sakakibara SM, Jones MD, Gillespie M, Hagerman SM, Forrest ME, Simard SW, Durall DM. A comparison of ectomycorrhiza identification based on morphotyping and PCR-RFLP analysis. Mycol Res. 2002;106:868–878. doi: 10.1017/S0953756202006263. [DOI] [Google Scholar]
- Schirkonyer U, Bauer C, Rothe GM. Ectomycorrhizal diversity at five different tree species in forests of the Taunus Mountains in Central Germany. Open J Ecol. 2013;3:66–81. doi: 10.4236/oje.2013.31009. [DOI] [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal symbiosis. 3. Great Britain: Academic Press in an Imprint of Elsevier; 2008. [Google Scholar]
- Stasińska M. Macromycetes in forest communities of the Ińsko Landscape Park (NW Poland) Acta Mycol. 1999;34:125–168. doi: 10.5586/am.1999.011. [DOI] [Google Scholar]
- Tedersoo L, Suvi T, Beaver K, Kõljalg U. Ectomycorrhizal fungi of the Seychelles: diversity patterns and host shifts from the native Vateriopsis seychellarum (Dipterocarpaceae) and Intsia bijuga (Caesalpiniaceae) to the introduced Eucalyptus robusta (Myrtaceae), but not Pinus caribea (Pinaceae) New Phytol. 2007;175:321–333. doi: 10.1111/j.1469-8137.2007.02104.x. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, Saar I, Kõljalg U. Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol. 2008;180:479–490. doi: 10.1111/j.1469-8137.2008.02561.x. [DOI] [PubMed] [Google Scholar]
- Trocha LK, Kałucka I, Stasińska M, Nowak W, Dabert M, Leski T, Rudawska M, Oleksyn J. Ectomycorrhizal fungal communities of native and non-native Pinus and Quercus species in a common garden of 35-year-old trees. Mycorrhiza. 2012;22:121–134. doi: 10.1007/s00572-011-0387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterseher M, Westphal B, Amelang N, Jansen F. 3,000 species and no end—species richness and community pattern of woodland macrofungi in Mecklenburg-Western Pomerania, Germany. Mycol Prog. 2012;11:543–554. doi: 10.1007/s11557-011-0769-7. [DOI] [Google Scholar]
- Valentine L, Fiedler T, Hart A, et al. Diversity of ectomycorrhizas associated with Quercus garryana in southern Oregon. Can J Bot. 2004;82:123–135. doi: 10.1139/b03-117. [DOI] [Google Scholar]
- Vellinga EC, Wolfe BE, Pringle A. Global patterns of ectomycorrhizal introductions. New Phytol. 2009;181:960–973. doi: 10.1111/j.1469-8137.2008.02728.x. [DOI] [PubMed] [Google Scholar]
- Walbert K, Ramsfield TD, Ridgway HJ, Jones EE. Ectomycorrhizal species associated with Pinus radiata in New Zealand including novel associations determined by molecular analysis. Mycorrhiza. 2010;20:209–215. doi: 10.1007/s00572-009-0277-7. [DOI] [PubMed] [Google Scholar]
- Watling R. Assessment of fungal diversity: macromycetes, the problems. Can J Bot. 1995;73:S15–S24. doi: 10.1139/b95-220. [DOI] [Google Scholar]
- Ważny R. Ectomycorrhizal communities associated with silver fir seedlings (Abies alba Mill.) differ largely in mature silver fir stands and in Scots pine forecrops. Ann For Sci. 2014;71:801–810. doi: 10.1007/s13595-014-0378-0. [DOI] [Google Scholar]
- Wojewoda W. Checklist of Polish Larger Basidiomycetes. Krytyczna Lista wielkoowocnikowych grzybów podstawkowych Polski. In: Mirek Z, editor. Biodiversity of Poland. Kraków: W. Szafer Institute of Botany, Polish Academy of Sciences; 2003. [Google Scholar]