Abstract
Long noncoding RNA cancer upregulated drug resistant (CUDR) is overexpressed in many tumors and promotes tumorigenesis. Herein, we demonstrate CUDR could enhance the human embryonic stem cells (ESC) differentiation into hepatocyte-like cells by reducing trimethylation on histone H3 twenty-seventh lysine (H3K27me3). On the other hand, excessive CUDR triggers hepatocyte-like cells malignant transformation. Mechanistically, we identify CUDR causes highly upregulated in liver cancer (HULC) and β-catenin abnormal expression by inhibiting HULC promoter methylation and promoting β-catenin promoter-enhancer chromatin looping formation mediated by CUDR-ccctc-binding factor (CTCF) complex, which recruits more RNA polII and P300. Strikingly, HULC and β-catenin activity are crucial for CUDR oncogenic function. These findings provide the first demonstration that CUDR plays a positive potential role in liver cancer stem cell through the cascade of CUDR-HULC/CUDR-β-catenin signaling, and offer insights into a novel link between long noncoding RNA (lncRNA) and the epigenetic modification in cancer stem cells.
Introduction
The liver consists of endodermal components, hepatocytes and cholangiocytes, and various types of nonparenchymal cells such as sinusoidal endothelial cells, satellite cells, kupffer cells, and blood cells. Hepatocytes originate from a common progenitor, the hepatoblasts, which are derived from the ventral foregut endoderm and the main component of the liver primordium.1,2 Hepatoblasts give rise to mature hepatocytes in the liver parenchyma. Recently, several monoclonal antibodies against cell surface molecules were used to isolate hepatoblasts from fetal livers and the isolated hepatoblasts were shown to proliferate clonally and differentiate into hepatocyte lineages.3,4 Thus, it became possible to characterize the growth and differentiation potential of hepatoblasts and to investigate the mechanism by which they give rise to hepatocytes. Several transcription factors known as liver-enriched transcription factors play key roles in liver organogenesis and metabolic functions of the liver.5,6 Among them, hepatic nuclear factors (HNF) HNF1β (TCF2) and HNF6 (Onecut1) are highly expressed in cholangiocytes and have been implicated in the formation of bile ducts.7 By contrast, HNF1α, HNF4 are strongly expressed in mature hepatocytes and play essential roles in the differentiation and metabolic functions of hepatocytes.8 Hepatocellular carcinoma (HCC) was thought historically to arise from hepatocytes, as well as studies have suggested that it can also arise from fetal progenitor cells or their adult progenitor progeny.
It is well known that HCC is a leading cause of cancer death in the world. Lack of early diagnosis tools is one of the clinical obstacles for effective treatment of HCC. Thus, enhanced understanding of the molecular changes associated with HCC is urgently needed to develop novel strategies for the diagnosis and treatment of this dismal disease. While aberrant expression of long noncoding RNAs (lncRNAs) has been functionally associated with certain cancers, the expression profiles and biological relevance of lncRNAs in HCC remain unclear. lncRNAs played important roles in proliferation, apoptosis, and invasiveness of tumor cells, and participated in metastatic capacity of cancers. In addition to regulating transcription, lncRNAs also control various aspects of post-transcriptional mRNA processing. LncRNAs can regulate gene expression in many ways, including chromosome remodeling, transcription, and post-transcriptional processing. Moreover, the dysregulation of lncRNAs has increasingly been linked to many human diseases, especially in cancers.9 A handful of studies have implicated lncRNAs in a variety of disease states and support an involvement and cooperation in oncogenesis.10
Cancer upregulated drug resistant (CUDR), or called urothelial cancer-associated 1 (UCA1) was spliced and polyadenylated with 3 exons and form multiple isoforms (1.4, 2.2, and 2.7 kb). CUDR is upregulated in various human tumors, including colon, cervical, lung, and bladder cancer. CUDR may play an important role in the growth and tumorigenesis of human bladder cancer.11 CUDR is also a very sensitive and specific unique marker for bladder cancer. Thus, it could have important implications in postoperative noninvasive follow-up.12 CUDR expression in bladder cancer cells was upregulated by transcription factor CCAAT/enhancer binding protein α (C/EBPα). Reversing the upregulation of CUDR can prevent bladder cancer progression.13,14 Moreover, CUDR increases the cisplatin resistance of bladder cancer cells by enhancing the expression of Wnt6, and thus represents a potential target to overcome chemoresistance in bladder cancer.15,16 Intriguingly, CUDR may be involved in the activation of Akt signaling pathway by Ets-217 and regulate cell cycle through CREB.11,18 Importantly, stable transfection with the CUDR gene was found to induce resistance to doxorubicin as well as drug-induced apoptosis by downregulations of caspase3.19 Moreover, studies suggest knockdown of CUDR could attenuate the migrational ability of melanoma cells, and increased expression of CUDR might have a correlation with melanoma metastasis.20 Of significance, patients with high CUDR expression had a significantly poorer prognosis than those with low CUDR expression. Strikingly, CUDR also promotes glycolysis through inducing hexokinase 2 (HK2) functions21 and is a direct target of CAPERα/TBX3 repression whose overexpression is sufficient to induce senescence, and CUDR sequesters hnRNPA1 and thus stabilizes CDKN2A-p16INK.22
To address whether CUDR influences on the human embryonic stem cells (ESC) differentiation into hepatocyte-like cells and plays a potential role in liver stem malignant transformation, we induce the ESC differentiation and explore the cascade of CUDR-HULC/CUDR-β-catenin signaling during liver stem cell malignant transformation.
Results
CUDR expression in hepatoblasts, hepatocytes, and liver cancer cells
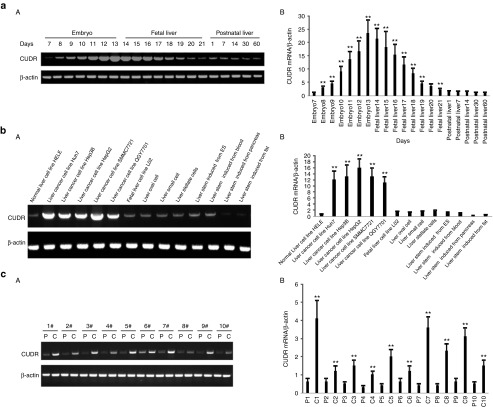
To explore CUDR existence in hepatoblasts, hepatocytes, and liver cancer cells, we examined the CUDR expression by reverse-transcription polymerase chain reaction (RT-PCR). As shown in Figure 1a (A,B), CUDR expression was gradually increased in Balb/c mouse embryo from the 7th to the 13th days, as well as CUDR expression was gradually decreased in Balb/c mouse fetal liver from the 14th to the 21th days. However, CUDR expression was lower in Balb/c mouse postnatal liver from the 1st to 60th days. As shown in Figure 1b (A,B), CUDR expression was lower in normal liver cell line HELE. On the contrary, CUDR expression was higher in liver cancer cell lines Huh7, Hep3B, HepG2, SMMC7721, and QGY7701. Furthermore, CUDR expression was lower in fetal liver cell line L02, human liver oval cells, liver small cells, liver satellite cells, the ESC-derived hepatocyte-like cells, and the blood stem cell-derived hepatocyte-like cells. However, CUDR was rarely expressed in the pancreas stem cell-derived hepatocyte-like cells and the fat stem cell-derived hepatocyte-like cells. In 10 cases of human liver cancer tissues, CUDR was significantly higher in liver cancer tissues than in its para-cancerous liver tissues (the upregulated expression rate 100%, P < 0.01) (Figure 1c (A,B). Together, these observations suggest that CUDR expression was altered during the benign to malignant differentiation of liver stem cells.
Figure 1.
Cancer upregulated drug-resistant (CUDR) expression analysis in hepatoblast, hepatocytes, and liver cancer cells. (a) CUDR expression in Balb/c mouse embryo (7th to 13th days), Balb/c mouse fetal liver (14th to 21th days), and Balb/c mouse postnatal liver (1st, 7th, 14th, 30th, and 60th after birth). β-actin as internal control. (A) Regular reverse-transcription polymerase chain reaction (RT-PCR). (B) Real-time RT-PCR. (b) CUDR expression in normal liver cell line HELE, liver cancer cell lines Huh7, Hep3B, HepG2, SMMC7721, QGY7701, fetal liver cell line L02, human liver oval cell, liver small cell, liver stellate cells, induced hepatocyte-like cell from embryonic stem cells, pancreas stem cells, blood stem cells, and fat stem cells (indicated in upper). β-actin as internal control. (A) Regular RT-PCR. (B) Real-time RT-PCR. (c) RT-PCR CUDR expression anlysis in liver cancer tissue (C) and its paracancerous liver tissues (P) respectively (indicated in upper). β-actin as internal control. (A) Regular RT-PCR. (B) Real-time PCR.
CUDR favors to induce hepatocyte-like cells from ESC
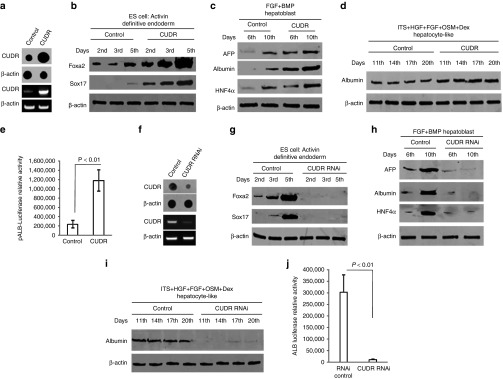
ESC is capable of unlimited self-renewal and can differentiate into any cell type in the adult. We selected the human ESC MEL-2 and these cells were stably transfected with pCMV6-A-GFP and pCMV6-A-GFP-CUDR. As shown in Figure 2a, CUDR was overexpressed in MEL-2 transfected with pCMV-A-GFP-CUDR compared to the control. Next, we induced the embryic stem cells to diffentiate into the hepatocyte-like cells. Foxa2, anti-Sox17 expression were gradually increased in these induced cells transfected with pCMV6-A-GFP-CUDR from 2nd to 5th day compared to the control respectively (P < 0.01) (Figure 2b). AFP, Albumin, and HNF4α expression were gradually increased in these induced cells transfected with pCMV6-A-GFP-CUDR from 6th to 10th day compared to the control respectively (Figure 2c). Albumin expression were higher in these induced cells transfected with pCMV6-A-GFP-CUDR from 11th to 20th day compared to the control respectively (Figure 2d). Furthermore, albumin promoter luciferase activity was higher in induced hepatocyte-like cells transfected with pCMV6-A-GFP-CUDR than in control cells (P < 0.01) (Figure 2e).
Figure 2.
Cancer upregulated drug-resistant (CUDR) enhances the embryonic stem cells (ESC) diffentiation into the hepatocyte-like cells. (a) Nuclear run-on (upper) and reverse-transcription polymerase chain reaction (RT-PCR) (lower) analysis of CUDR in human ESC line MEL-2 transfected with pCMV-A-GFP-CUDR and pCMA6-A-GFP respectively. β-actin as internal control. (b) Human ESC line MEL-2 transfected with pCMV-A-GFP-CUDR or pCMA6-A-GFP could efficiently generate definitive endoderm (DE) tissue by treating the the modified cultures with high concentrations of the TGFβ family ligand activin A (100 ng/ml) for 5 days. Western blot analysis with anti-Foxa2, anti-Sox17 in these induced cells (the 2nd, 3rd, and 5th day). β-actin as internal control. (c) A number of groups have generated hepatoblasts using this DE tissue as a starting material, plating the DE on matrix to mimic the hepatic ECM and then added FGF4 (100 ng/ml) and BMP (100 ng/ml) to mimic hepatic induction for 5 days. Western blotting with anti-AFP, anti-Albumin, and anti-HNF4α in the induced cells (the 6th and 10th day). β-actin as internal control. (d) Some combination of insulin-transferrinselenite (ITS, 5 μg/ml), hepatocyte growth factor (HGF) (20 ng/ml), oncostatin M (OSM) (10 ng/ml), acid fibroblast growth factor (aFGF) (50 ng/ml), and dexamethasone (10−7M) to expand the hepatoblast population and to promote hepatic maturation for 10 days. Western blotting with anti-Albumin in the induced cells (the 11th, 14th, 17th, and 20th day). β-actin as internal control. (e) Albumin promoter luciferase activity assay in derived hepatocyte-like cells. Each value was presented as mean ± standard error of the mean (SEM). **P < 0.01. (f) Nuclear run-on (upper) and RT-PCR (lower) analysis of CUDR in human ESC line MEL-2 transfected with pGFP-V-RS-CUDR and pGFP-V-RS, respectively, β-actin as internal control. (g) Western blot analysis with anti-Foxa2, anti-Sox17 in these derived cells (the 2nd, 3rd, and 5th day). β-actin as internal control. (h) Western blotting with anti-alpha fetoprotein (AFP), anti-Albumin, and anti-HNF4α in the derived cells (the 6th and 10th day), β-actin as internal control. (i) Western blotting with anti-Albumin in the induced cells (the 11th, 14th, 17th, and 20th day), β-actin as internal control. (j) Albumin promoter luciferase activity assay in derived hepatocyte-like cells (CUDR knockdown or control). Each value was presented as mean ± SEM. **P < 0.01).
Further on, as shown Figure 2f, CUDR was knocked down in MEL-2 transfected with pGFP-V-RS-CUDR compared to the control. Next, we induced the embryic stem cells to differentiate into the hepatocyte-like cells. Foxa2, anti-Sox17 was rarely expressed in these induced cells transfected with pGFP-V-RS-CUDR from 2nd to 5th day compared to the control respectively (Figure 2g), as well as AFP, Albumin, and HNF4α expression were rarely expressed in these induced cells transfected with pGFP-V-RS-CUDR from 6th to 10th day compared to the control respectively (Figure 2h). Albumin expression were not expressed in these induced cells transfected with pGFP-V-RS-CUDR from 11th to 20th day compared to the control respectively (Figure 2i). Moreover, albumin promoter luciferase activity was not activated in the induced cells transfected with pGFP-V-RS-CUDR than in control cells (P < 0.01) (Figure 2j). Taken together, the results suggest that CUDR promotes the differentiation of ESC into the hepatocyte-like cells.
CUDR promotes hepatocyte-like cells malignant growth
To investigate the CUDR oncogenic function, we constructed the human ESC MEL-2 stable cell line transfected with pCMV6-A-GFP-CUDR, pCMV6-A-GFP, pGFP-V-RS-CUDR, and pGFP-V-RS, respectively. Next, the MEL-2 cell lines were differentiated into the hepatocyte-like cells. As shown Figure 3a, CUDR was overexpressed in MEL-2-derived hepatocyte-like cells transfected with pCMV-A-GFP-CUDR compared to the pCMV6-A-GFP control and CUDR was knocked down in MEL-2-derived hepatocyte-like cells transfected with pGFP-V-RS-CUDR compared to pGFP-V-RS control. As shown in Figure 3b, CUDR overexpression enhanced and CUDR knockdown inhibited hepatocyte-like cells proliferation capacity at the 1st, 2nd, 3rd, 4th, and 5th day, respectively (for these time points, P < 0.01). The soft-Agar colony-formation was 7.8 ± 2.0% in MEL-2-derived hepatocyte-like cells with excessive CUDR, as well as the other groups did not form soft-agar colony (P < 0.01) (Figure 3c). Cell spheres were only formed in MEL-2-derived hepatocyte-like cells with excessive CUDR (Figure 3d). Further on, we performed tumorigenesis test in vivo. As shown in Figure 3e, xenograft tumors were only produced in group of MEL-2-derived hepatocyte-like cells with excessive CUDR (average wet weight 0.91 ± 0.23 g, P < 0.01). Furthermore, these xenograft tumor all displayed poor differentiation malignant grade (Figure 3f). Taken together, these observations suggest that excessive CUDR promotes malignant transformation of ESC-derived hepatocyte-like cells.
Figure 3.
Cancer upregulated drug-resistant (CUDR) promotes malignant transformation of derived hepatocyte-like cells. (a) Nuclear run-on (upper) and reverse-transcription polymerase chain reaction (RT-PCR) (lower) analysis of CUDR in derived hepatocyte-like cells (CUDR overexpression or knockdown), β-actin as internal control. (b) Cell proliferation assay in vitro. Cell growth curve was based on the corresponding relative values of OD450 and each point represents the mean of three independent samples. Data are means of value from three independent experiments, bar ± standard error of the mean (SEM); **P < 0.01; *P < 0.05. (c) Soft-Agar colony-formation efficiency assay. Imaging the colonies (left) and count colonies using a dissecting microscope. Data are means of value from three independent experiments, bar ± SEM; **P < 0.01 (right). (d) Cells sphere formation ability. (e) Tumorigenesis test in vivo in derived hepatocyte-like cells (left). Mice were stratified and the tumors were recovered. The photography of xerograft tumor in the four groups (right). The wet weight of each xenograft tumor was determined for each mouse. Each value was presented as mean ± SEM; **P < 0.01. (f) A portion of each xenograft tumor was fixed in 4% paraformaldehyde, embedded in paraffin and micrometers of sections (4 µm) for histological hematoxylin-eosin (HE) staining (original magnification ×100).
CUDR alters genes expression through inhibiting three methylation on twenty-seventh lysine of histone H3
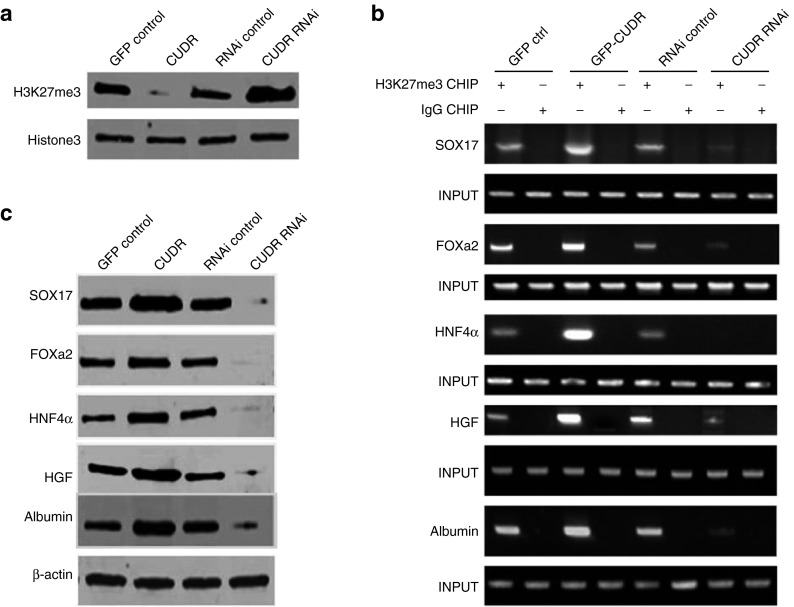
To explore CUDR involved molecular signaling pathway from human ESC to hepatocyte-like cells, we constructed the human ESC MEL-2 stable cell lines transfected with pCMV6-A-GFP-CUDR, pCMV6-A-GFP, pGFP-V-RS-CUDR, and pGFP-V-RS respectively. These MEL-2 cells with excessive or CUDR overexpression or knockdown were differentiated into the hepatocyte-like cells. As shown Figure 4a, three methylation on twenty-seventh lysine of histone H3 (H3K27me3) was decreased in hepatocyte-like cells with CUDR overexpression compared to control cells and was increased in hepatocyte-like cells CUDR knockdown compared to control cells. Furthermore, CUDR overexpression increased and CUDR knockdown decreased the H3K27me3 binding to the promoter region of Sox17, FOXa2, HNF4α, HGF, and Albumin in hepatocyte-like cells, respectively (Figure 4b). Therfore, Sox17, FOXa2, HNF4α, HGF, and Albumin were increased in hepatocyte-like cells with CUDR overexpression compared to control cells and were decreased in hepatocyte-like cells with CUDR knockdown compared to control cells (Figure 4c). Together, these observations suggest that CUDR altered genes expression in human ES-derived hepatocyte-like cells through inhibiting H3K27me3 epigentic modification.
Figure 4.
Cancer upregulated drug-resistant (CUDR) altered genes expression in derived hepatocyte-like cells. (a) Western blotting analysis with anti-H3K27me3 in derived hepatocyte-like cells with CUDR overexpression or knockdown (indicated in upper). Histone3 as internal control. (b) Anti-H3K27me3 CHIP in hepatocyte-like cells with CUDR overexpression or control (indicated in upper). PCR with Sox17, FOXa2, HNF4α, HGF, and Albumin promoter respectively. INPUT as internal control (indicated in the left). (c) Western blotting analysis with anti-Sox17, anti-FOXa2, anti-HNF4α, anti-HGF, anti-Albumin (indicated in the left) in derived hepatocytes-like cells with CUDR overexpression or depletion (indicated in upper). β-actin as internal control.
CUDR induces HULC expression by inhibiting HULC promoter methylation
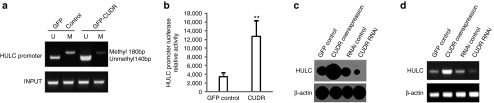
Given that long noncoding RNA highly upregulated in liver cancer (HULC) was highly upregulated in human liver cancer, we considered whether CUDR could impact on HULC expression. At the first time, we analyzed the promoter region methylation by methylation-specific polymerase chain reaction analysis. As shown in Figure 5a, the promoter region methylation was lower in hepatocyte-like cells with CUDR overexpression compared to control cells. Next, we performed HULC promoter luciferase activity assay in these hepatocyte-like cells. HULC promoter luciferase activity was decreased in hepatocyte-like cells with CUDR overexpression compared to control cells (P < 0.01) (Figure 5b). Further on, we performed nuclear run-on analysis for lncRNA HULC expression. HULC was increased in hepatocyte-like cells with CUDR overexpression compared to control cells and was decreased in hepatocyte-like cells with CUDR knockdown compared to control cells (Figure 5c). Furthermore, we also performed RT-PCR to detect HULC in these hepatocyte-like cells. As shown in Figure 5d, HULC was increased in hepatocyte-like cells with CUDR overexpression compared to control cells and was decreased in hepatocyte-like cells with CUDR knockdown compared to control cells. Taken together, these observations suggest that CUDR increases the long noncoding RNA HULC expression by blocking HULC promoter region methylation.
Figure 5.
Cancer upregulated drug-resistant (CUDR) promotes highly upregulated in liver cancer (HULC) expression. (a) Methylation-specific polymerase chain reaction analysis on promoter region of HULC in hepatocyte-like cells with CUDR overexpression or control (indicated in upper). INPUT as internal control. Methyl DNA fragment is 180 bp. Unmethyl DNA fragment is 140 bp. (b) HULC promoter luciferase activity assay in hepatocyte-like cells with CUDR overexpresion or control. Data are means of values from three independent experiments, bar ± standard error of the mean. ** means P < 0.01. (c) Nuclear run-on analysis of HULC expression in induced hepatocyte-like cells with CUDR overexpression or knockdown (indicated in upper). β-actin as internal control. (d) Reverse-transcription polymerase chain reaction analysis of HULC in derived hepatocyte-like cells with CUDR overexpression or knockdown (indicated in upper). β-actin as internal control.
CUDR promotes β-catenin promoter-enhancer chromatin looping formation
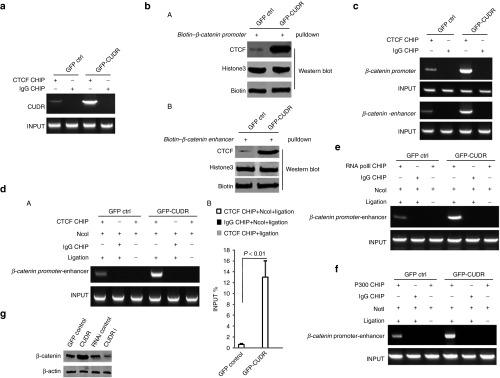
Seeing that malignant transformation to hepatocellular carcinoma was shown to be strongly associated with activating β-catenin, we infer CUDR may promote β-catenin expression. To address this regulation and control mechanism based on CUDR, we first preformed RNA immunoprecipitation, DNA pulldown, and CHIP experiments. Our results indicated that CUDR increased the interaplay between insulator ccctc-binding factor (CTCF) and CUDR (Figure 6a). The interaction between insulator CTCF and β-catenin promoter DNA was enhanced in hepatocyte-like cells with CUDR overexpression compared to control cells (Figure 6b, A). The interplay between insulator CTCF and β-catenin enhancer DNA was enhanced in hepatocyte-like cells with CUDR overexpression compared to control cells (Figure 6b, B). More CTCT molecular was loaded onto the β-catenin promoter or enhancer region in hepatocyte-like cells with CUDR overexpression compared to control cells (Figure 6c). Further on, we performed 3C-CHIP with anti-CTCF experiment. The DNA looping linked-up β-catenin promoter and enhancer by CTCF was formed, as well as the DNA loopings were significantly increased in hepatocyte-like cells with CUDR overexpression than in control cells (Figure 6d, A,B). Next, we performed 3C-CHIP with anti-RNA polII or anti-P300. The DNA loopings recruit more RNApolII in hepatocyte-like cells with CUDR overexpression than in control cells (Figure 6e). Similarly, the DNA loopings recruit more P300 (A acetyltransferase) in hepatocyte-like cells with CUDR overexpression than in control cells (Figure 6f). Finally, β-catenin expression was greatly increased in hepatocyte-like cells with CUDR overexpression compared to control cells and was significantly decreased in hepatocyte-like cells with CUDR knockeddown compared to control cells (Figure 6g). Together, these observations suggest that excessive CUDR promotes β-catenin expression through forming β-catenin promoter-enhancer DNA looping mediated by insulator CTCF.
Figure 6.
Cancer upregulated drug-resistant (CUDR) promotes β-catenin expression mediated by insulator ccctc-binding factor (CTCF). (a) RNA immunoprecipitation (RIP) with anti-CTCF followed by reverse-transcription polymerase chain reaction (RT-PCR) with CUDR mRNA primers in hepatocyte-like cells with CUDR overexpression or control. IgG RIP as negative control. CUDR mRNA as INPUT. (b) (A) β-catenin promoter DNA pulldown in hepatocyte-like cells with CUDR overexpression or control (indicated in upper). (B) β-catenin enhancer DNA pulldown in hepatocyte-like cells with CUDR overexpression or control (indicated in upper).Western blot with anti-CTCF. Biotin as INPUT, Histone3 as loading control. (c) Anti-CTCF CHIP in hepatocyte-like cells with CUDR overexpression or control (indicated in upper). PCR with β-catenin promoter (the first and second row) and enhancer Primers (the third and fourth row) respectively. INPUT as internal control. (d) 3C-CHIP with anti-CTCF, Ncol digestion in hepatocyte-like cells with CUDR overexpression or control (indicated in upper). PCR primers using β-catenin promoter (F primer) and enhancer (R primer) respectively. INPUT (β-catenin promoter and enhancer DNA fragment) as internal control. (e) 3C-CHIP with anti-RNApolII, Ncol digestion in hepatocyte-like cells with CUDR overexpression or control (indicated in upper). PCR primers using β-catenin promoter (F primer) and enhancer (R primer) respectively. INPUT (β-catenin promoter and enhancer DNA fragment) as internal control. (f) 3C-CHIP with anti-P300, Ncol digestion in hepatocyte-like cells with CUDR overexpression or control (indicated in upper). PCR primers using β-catenin promoter (F primer) and enhancer (R primer) respectively. INPUT (β-catenin promoter and enhancer DNA fragment) as internal control. (g) Western blotting analysis with anti-β-catenin in derived hepatocyte-like cells with CUDR overexpression or knockdown (indicated in upper). β-actin as internal control.
HULC and β-catenin activity is crucial for CUDR oncogenic function
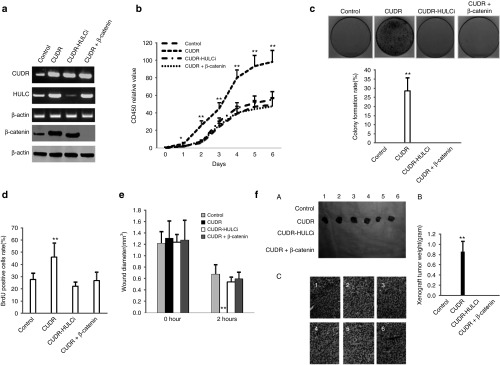
To explicit whether CUDR oncogenic function was controlled by HULC and β-catenin, we performed rescue test in the four groups of hepatocyte-like cells, including control, CUDR overexpression, CUDR overexpression plus HULC knockdown, and CUDR overexpression plus β-catenin knockdown. As shown in Figure 7a, CUDR was overexpressed in CUDR overexpression, CUDR overexpression plus HULC knockdown and CUDR overexpression plus β-catenin knockdown. HULC was overexpressed in CUDR overexpression and CUDR overexpression plus β-catenin knockdown, as well as HULC was knocked down in CUDR overexpression plus HULC knockdown. β-catenin was overexpressed in CUDR overexpression and CUDR overexpression plus HULC knockdown, as well as β-catenin was knocked down in CUDR overexpression plus β-catenin knockdown. Excessive CUDR overexpression promoted the hepatocyte-like cells proliferation at the 1st, 2nd, 3rd, 4th, 5th, and 6th day respectively (for each time points, P < 0.01), as well as HULC knockdown or β-catenin knockdown abrogated the excessive CUDR oncogenic function (P > 0.05) (Figure 7b). The soft-agar colony-formation rate was 28.5 ± 7.2% in group with CUDR overexpression, as well as the soft-agar colony-formation rate was 0% in control group (P < 0.01). Furthermore, soft-agar colony was also not produced in the three groups of CUDR overexpression plus HULC knockdown and CUDR overexpression plus β-catenin knockdown (Figure 7c). BrdU-positive rates in CUDR overexpression group (45.9 ± 11.6%) was significantly higher than that in control group (27.6 ± 5.3%) (P < 0.01), CUDR overexpression plus HULC knockdown group (22.2 ± 3.2%) (P < 0.01), and CUDR overexpression plus β-catenin knockdown group (26.7 ± 6.8%) (P < 0.01), as well as there were no significant differences among control group (27.6 ± 5.3%), CUDR overexpression plus HULC knockdown group (22.2 ± 3.2%), and CUDR overexpression plus β-catenin knockdown group (26.7 ± 6.8%) (P > 0.05) (Figure 7d). Then, we performed the wound test in these cells at 0 and 24 hours. As shown in Figure 7e, the wound diameters (mm) at 0 hour were 1.21 ± 0.21, 1.3 ± 0.31, 1.24 ± 0.13, and 1.27 ± 0.35 in CUDR overexpression group, CUDR overexpression plus HULC knockdown, and CUDR overexpression plus β-catenin knockdown, respectively (P > 0.01). The wound diameters (mm) at 24 hours in CUDR overexpression group (0 mm) was significantly shorter than in control group (0.67 ± 0.17 mm), CUDR overexpression plus HULC knockdown group (0.54 ± 0.08 mm), and CUDR overexpression plus β-catenin knockdown group (0.59 ± 0.12 mm) (P < 0.01), as well as there were no significant differences among control group (0.67 ± 0.17 mm), CUDR overexpression plus HULC knockdown group (0.54 ± 0.08 mm), and CUDR overexpression plus β-catenin knockdown group (0.59 ± 0.12 mm) (P > 0.05). Then, we preformed in vivo test in these derived hepatocyte-like cells. As shown in Figure 7f, A, CUDR overexpression promoted the xenograft tumor formation, as well as xenograft tumor was not produced in control group, CUDR overexpression plus HULC knockdown group, and CUDR overexpression plus β-catenin knockdown group. And the wet weight of xenograft tumor added up to 0.85 ± 0.21 g in CUDR overexpression group (P < 0.01) (Figure 7f, B). In addition, histological hematoxylin-eosin staining presented poor differentiation grade (Figure 7f, C). Taken together, these observations suggest that the excessive CUDR oncogenic function was abrogated by the knockdown of HULC or β-catenin in the hESC-derived hepatocyte-like cells.
Figure 7.
The excessive cancer upregulated drug-resistant (CUDR) oncogenic action was abrogated by the knockdown of highly upregulated in liver cancer (HULC) or β-catenin in the derived hepatocyte-like cells. (a) Reverse-transcription polymerase chain reaction (RT-PCR) analysis for CUDR and HULC, and western blotting analysis for β-catenin in derived hepatocyte-like cells with CUDR overexpression, CUDR overexpression plus HULC knockdown, or CUDR overexpression plus β-catenin knockdown. Loading based on β-actin. (b) Cells proliferation assay in vitro. Data are means of values from three independent experiments, bar ± standard error of the mean (SEM). **P < 0.01 or *P < 0.05. (c) Soft-agar colony-formation efficiency assay. Data are means of values from three independent experiments, bar ± SEM. **P < 0.01. (d) BrdU staining analysis. Data are means of values from three independent experiments, bar ± SEM. **P < 0.01. (e) Wound test analysis at 0 and 24 hours, respectively. Data are presented as mean ± SEM, **P < 0.01. (f) Tumorigenesis test in vivo. (A) Mice were stratified and the tumors were recovered. The photography of xenograft tumor in the four groups (indicated in left). (B) The wet weight of each xenograft tumor was determined for each mouse (n = 6). Each value was presented as mean ± standard error of the mean (SEM). **P < 0.01 shows a positive difference. (C) A portion of each tumor was fixed in 4% paraformaldehyde, embedded in paraffin and micrometers of sections (4 µm) for histological hematoxylin-eosin (HE) staining (original magnification ×100).
Discussion
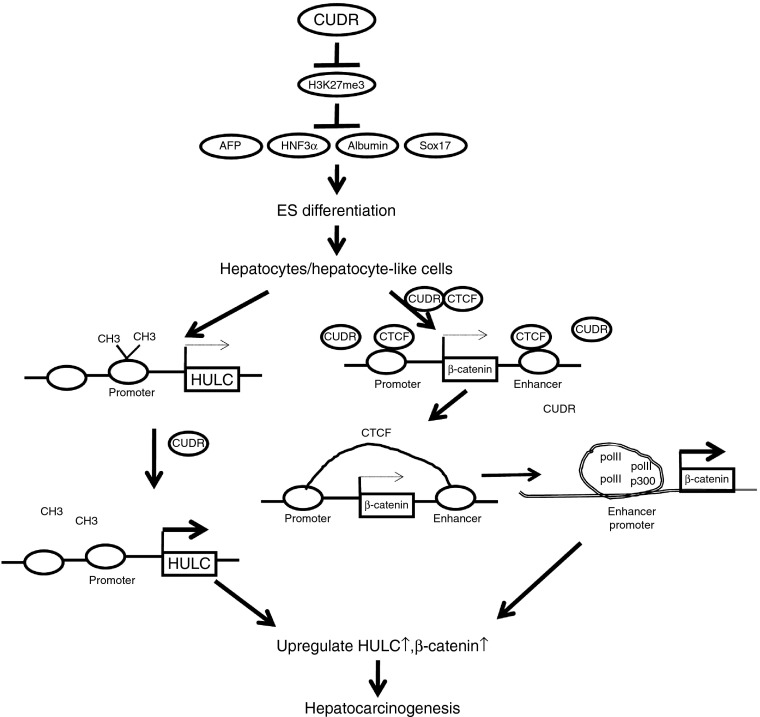
To our knowledge, this is the first report demonstrating CUDR function in liver development and malignant progression. Studies are now indicated to evaluate the effects in experimental models and illustrate the mechanisms for CUDR-promoting hepatocyte-like cells malignant transformation (Figure 8). Our findings identify excessive CUDR could promote the human ESC differentiation into hepatocyte-like cell, however, CUDR also promotes hepatocyte-like cells malignant transformation. Mechanistically, we reveal CUDR causes HULC and β-catenin abnormal expression by inhibiting HULC promoter methylation and promoting β-catenin promoter-enhancer chromatin looping formation, which recruits more RNA polII and P300, respectively. Evidently, HULC and β-catenin activity are crucial for CUDR oncogenic function, suggesting a new strategy for cancer gene therapy based on CUDR.
Figure 8.
Schematic diagram illustrating the mechanisms for excessive cancer upregulated drug-resistant (CUDR) promoting derived hepatocyte-like cells malignant transformation. CUDR could enhance the human embryonic stem cells differentiation into hepatocyte-like cell. On the other hand, excessive CUDR further triggers hepatocyte-like cells malignant transformation. Mechanistically, we reveal excessive CUDR causes highly upregulated in liver cancer (HULC) or β-catenin abnormal expression by inhibiting HULC promoter methylation, and promoting β-catenin promoter-enhancer chromatin DNA looping formation mediated by CUDR-CTCF complex, which recruits more RNA polII and P300. Thereby, HULC or β-catenin activity is crucial for CUDR oncogenic function.
For years, investigators attempted to promote hepatic differentiation in ESC. The critical first step was the demonstration that human ESC could efficiently generate definitive endoderm tissue by treating the cultures with high concentrations of the TGFβ family ligand activin, to mimic the role of Nodal in the gastrula embryo.23 Using these developmental protocols on mouse and human ESC, researchers have successfully generated “hepatocyte-like” phenotype. These cells exhibit many features of hepatocytes including: expression of hepatic enzymes, hepatocyte morphology, robust glycogen storage, uptake and metabolizism of drugs, and secretion of albumin.24 Our observations suggest CUDR may participate in the regulation of these processes. CUDR possess potential to promote hepatocyte-like cells transformation. The involvement of CUDR promotion of hepatoblast differentiation is supported by results from the following four parallel sets of experiments: (i) our results show that CUDR expression is associated with embryo liver development. CUDR favors to induce hepatocyte-like cells from ESC; (ii) we clearly indicated that CUDR triggers bipotential hepatoblast differentiate into hepatocyte-like cells, which express fetal liver genes (AFP) as well as Albumin. (iii) The demonstration that haploinsufficiency of CUDR may block hepatocyte-like cell formation provided an understanding of novel function of CUDR. (iv) CUDR controls hepatocyte-like cell-related gene expression by reducing trimethylation on histone H3 twenty-seventh lysine (H3K27me3). Actually, we will further underscore the need for new approaches to uncover the mechanisms underlying CUDR-mediated functions in vivo.
Our result shows that CUDR possesses strong oncogenic function. LncRNAs can regulate gene expression in many ways, including chromosome remodeling, transcription, and post-transcriptional processing. Moreover, the dysregulation of lncRNAs has increasingly been linked to many human diseases, especially in cancers.9 A report indicates that the knockdown of CUDR expression phenocopied the effects of upregulation of hsa-miR-1, which inhibited bladder cancer cell growth, induced apoptosis, and decreased cell motility.25 In this study, although CUDR possesses a capacity that promotes stem cell differentiation, on the other hand, it may also trigger liver stem cell malignant transformation under certain conditions. The involvement of CUDR promotion of liver stem cell malignant transformation is supported by results from the following three parallel sets of experiments: (i) we clearly reveal that CUDR is overexpressed in human liver cancer tissue and liver cancer cell lines; (ii) CUDR promotes human liver stem cell malignant growth in vitro and in vivo. (iii) CUDR may activate some vital tumor-signaling pathway. Obviously, our observations demonstrate that CUDR is crucial for cell growth and viability during liver stem cells malignant transformation.
Importantly, our observations suggest that CUDR alters genes expression involved in liver stem cells malignant transformation. Intriguingly, we clearly found that CUDR enhanced β-catenin expression by promoter-enhancer DNA looping formation through CTCF. Some reports indicate that CTCF and cohesin are integral components of most human subtelomeres.26 CTCF may also play a key role in the pluripotency of cells through the regulation of miR-290 cluster.27 The cohesin complex holds sister chromatids together and is essential for chromosome segregation. Cohesins have been implicated in transcriptional regulation and insulation through genome-wide colocalization with the insulator protein CTCF.28,29 It is identied that Wnt/β-catenin pathway is a highly complex and unique signaling pathway, which has ability to regulate gene expression, cell invasion, migration, proliferation, and differentiation for the initiation and progression of cancer.30 Recently, a genotype and phenotype classification system was suggested malignant transformation to HCC and was shown to be strongly associated with activating mutations in β-catenin.31 Of significance, Wnt/β-catenin signaling is of fundamental importance in the regulation of self-renewal, migration/invasion, and differentiation of human mesenchymal stem cells (hMSCs).32 Stabilization of β-catenin, through inhibition of glycogen synthase kinase 3 (GSK3) activity, in conjunction with inhibition of mitogen-activated protein kinase kinase 1/2 (MEK) promotes self-renewal of naive-type mouse ESC.33 It has been reported that the abnormal localization of β-catenin observed in HCC tissues and the truncated β-catenin proteins may represent an initiating or contributing factor in the development of HCC.34 According to our findings, CUDR may positively control β-catenin during hepatocarcinogenesis.
On the other hand, our data suggest that CUDR upregulates HULC expression by inhibiting its promoter methylation. HULC is the first ncRNA with highly specific upregulation in HCC. HULC has recently been revealed to be involved in HCC development and progression. Silencing of HULC effectively reversed the epithelial-to-mesenchymal transition phenotype.35,36 Strikingly, depletion of HULC resulted in a significant deregulation of several genes involved in liver cancer.37 Of significance, HULC lncRNA is detected with higher frequency in the plasma of HCC patients compared to healthy controls.38 Because HULC was detected in blood of HCC patients, a potential use as novel biomarker can be envisaged.39 Notably, Hepatitis B virus X protein (HBx) upregulated HULC in human hepatoma. HBx activated the HULC promoter via cAMP-responsive element-binding protein. In turn, the upregulated HULC by HBx promotes proliferation of hepatoma cells through suppressing p18 located near HULC in the same chromosome.40 We infer CUDR may also enhance the HBx activity that lead to the HULC overexpression. Thereby, CUDR-HULC axis may play an important role during liver stem cell malignant transformation.
Obviously, these findings are noteworthy that upregulation of HULC and β-catenin partly contribute to CUDR-medicated promotion of liver stem cell malignant transformation. Our findings in this study provide novel evidence for an active role of HULC and β-catenin in CUDR-mediated promotion of liver stem cell malignant growth. This assertion is based on the observation that the overexpressed CUDR oncogenic action was abrogated by the knockdown of HULC, β-catenin in the induced hepatocyte-like cells.
In conclusion, the present study depicts a novel evidence for CUDR to play tumorigenesis roles by HULC and β-catenin. Anyhow, we provide the demonstration that haploinsufficiency of lncRNA CUDR is very important in liver stem cell development and hepatocarcinogenesis. Our findings underscore the need for new approaches to uncover the mechanisms underlying lncRNA CUDR-mediated functions in vivo.
Materials and Methods
Cell lines and plasmids. Human ES cell line MEL-2 (Merck Millipore, Darmstadt, Germany) was maintained in HEScGRO medium (1,000 IU/ml LIF; Merck Millipore, Darmstadt, Germany) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Grand Island, NY) on matrigel (0.1% gelatin solution or human collagen IV-coating material, Gibco) in a humidified atmosphere of 5% CO2 incubator (Thermo Scientific, Waltham, MA) at 37 °C. Mitocally inactivated Detroit 551 or HS27 feeder cells (ATCC, Manassas, VA) were plated at 80,000 cells/cm2. Plasmid pGFP-V-RS and pCMV6-A-GFP were purchased from Origene (Rockville, MD). pGL3 was purchased from Addgene (Cambridge, MA). Recombinant plasmids pGFP-V-RS-CUDR, pCMV6-A-CUDR, and pGL3-ALB were constructed by the authors.
Isolation and induced differentiation. The isolation of hepatoblasts, liver oval cells, liver small cells, liver satellite cells, ESC, pancreas stem cells, blood stem cells, and adipose-deived stem cells differentiate into hepatocyte-like cell in vitro. Also, see the detail in Supplementary Materials and Methods.
Cell transfection and stable cell lines. Cells were transfected with DNA plasmids using transfast transfection reagent lipofectamineR 2000 (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. For screening stable cell lines, 48 hours after transfection, cells were plated in the selective medium containing Puromycin (1–2 μg/ml, Calbiochem) for the next 4 weeks, and the selective media were replaced every 3 days.
Reverse-transcriptase polymerase chain reaction. cDNA was prepared by a SuperScript First-Strand Synthesis System (Invitrogen). The PCR amplification kit (Takara, Japan) was adopted according to the manufacturer's instructions. PCR products were analyzed by 1.0% agarose gel electrophoresis and visualized by ethidium bromide staining. Also, see the detail in Supplementary Materials and Methods.
Western blotting. Cells lysate proteins were separated on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes. The blots were incubated with 0.2 µg/ml of antibody (appropriate dilution) overnight at 4 °C. Following three washes, membranes were then incubated with secondary antibody for 60 minutes at 37 °C. Signals were visualized by enhanced chemiluminescence plus kit or infrared imaging system. Also, see the detail in Supplementary Materials and Methods.
Chromosome conformation capture (3C)-chromatin immunoprecipitation (ChIP) (3C-ChIP). Chromatin bound to the antibody-Protein-A/G-Sepharose beads were resuspended in 500 μl of 1.2× restriction enzyme buffer at 37 °C for 1 hour. After digestion, 40 μl of 20% SDS was added to the digested Chromatin, and the mixture was incubated at 65 °C for 10 minutes. T4 DNA ligase was added at 16 °C for 4 hours incubation. Samples were then decrosslinked at 65 °C overnight. After purification, the ChIP-3C material was detected for long range interaction with specific primers. Also, see the detail in Supplementary Materials and Methods.
Cells proliferation CCK8 assay. Cells were grown in complete medium for assay using cell proliferation reagent CCK8 (Roche, Basel, Switzerland) according to the manufacturer instruction. In brief, cells at a concentration of 4 × 103 were seeded into 96-well culture plates in 100 μl culture medium containing 10% heat-inactivated fetal calf serum (Gibico). Before detection, 10 μg/well cell proliferation reagent CCK8 was added and incubated for 4 hours at 37 °C and 5% CO2. Cell growth curve was based on the corresponding normalized values of OD450.
Soft agar colony formation assay. 2 × 102 cells were plated on a six-well plate containing 0.5% (lower) and 0.35% (upper) double layer soft-agar. Then, the six-well plates were incubated at 37 °C in humidified incubator for 21 days. The cells were fed one to two times per week with cell culture media (Dulbecco's Modified Eagle Medium). The soft-agar colony formation ability was assessed by counting the number of colonies under a microscope after 0.05% crystal violet (Sigma-Aldrich, St. Louis, MO) staining for more than 1 hour. Representative views were photographed.
Cells sphere formation ability assay. Cells were collected and washed to remove serum; then suspended in serum-free Dulbecco's Modified Eagle Medium/F12 supplemented with 20 ng/ml human recombinant epidermal growth factor, 10 ng/ml human recombinant basic fibroblast growth factor, 2% B27 supplement without vitamin A, and 1% N2 supplement (Invitrogen). The cells were subsequently cultured in ultra low attachment six-well plates (Corning, Corning, NY) at a density of no more than 5,000 cells/well. The spheres were collected by gentle centrifugation, then dissociated with trypsin-ethylene diamine tetraacetic acid and mechanically disrupted with a pipette. The resulting single cells were then centrifuged to remove the enzyme and resuspended in serum-free medium allowed to reform spheres. The spheres should be passaged every 5–8 days before they reached a diameter of 100 μm. The sphere from 10 random chosen fields of at least three independent samples were counted.41
Xenograft transplantation in vivo. Four-week-male athymic Balb/C mice were purchased from Shi laike Company (Shanghi, China) and feed in the Tongji animal facilities approved by the China Association for accreditation of laboratory animal care. The athymic Balb/C mouse (totally 72 mice, maintained in specific pathogen-free environment) was injected at the armpit area subcutaneously with a suspension of 1 × 107 induced hepatocyte-like cells in 100 μl of phosphate-buffered saline. The mice were observed over 4 weeks, and then sacrificed to recover the tumors. The wet weight of each tumor was determined for each mouse. A portion of each tumor was fixed in 4% paraformaldehyde and embedded in paraffin for histological hematoxylin-eosin staining. The use of mice for this work was reviewed and approved by the institutional animal care and use committee in accordance with China National Institutes of Health guidelines.
SUPPLEMENTARY MATERIAL Materials and Methods
Acknowledgments
This study was supported by grants from National Natural Science Fundation of China (NCSF No. 81272291) and Science and Technology Commission of Shanghai Municipality (No. 13JC1405500-13JC1405501). The authors disclose no conflicts.
Supplementary Material
References
- 1Duncan, SA and Watt, AJ (2001). BMPs on the road to hepatogenesis. Genes Dev 15: 1879–1884. [DOI] [PubMed] [Google Scholar]
- 2Shiojiri, N (1997). Development and differentiation of bile ducts in the mammalian liver. Microsc Res Tech 39: 328–335. [DOI] [PubMed] [Google Scholar]
- 3Kubota, H and Reid, LM (2000). Clonogenic hepatoblasts, common precursors for hepatocytic and biliary lineages, are lacking classical major histocompatibility complex class I antigen. Proc Natl Acad Sci USA 97: 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Tanimizu, N, Nishikawa, M, Saito, H, Tsujimura, T and Miyajima, A (2003). Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J Cell Sci 116(Pt 9): 1775–1786. [DOI] [PubMed] [Google Scholar]
- 5Parviz, F, Matullo, C, Garrison, WD, Savatski, L, Adamson, JW, Ning, G et al. (2003). Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 34: 292–296. [DOI] [PubMed] [Google Scholar]
- 6Coffinier, C, Gresh, L, Fiette, L, Tronche, F, Schütz, G, Babinet, C et al. (2002). Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1beta. Development 129: 1829–1838. [DOI] [PubMed] [Google Scholar]
- 7Clotman, F, Lannoy, VJ, Reber, M, Cereghini, S, Cassiman, D, Jacquemin, P et al. (2002). The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development 129: 1819–1828. [DOI] [PubMed] [Google Scholar]
- 8Pontoglio, M, Barra, J, Hadchouel, M, Doyen, A, Kress, C, Bach, JP et al. (1996). Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell 84: 575–585. [DOI] [PubMed] [Google Scholar]
- 9Shi, X, Sun, M, Liu, H, Yao, Y and Song, Y (2013). Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 339: 159–66. [DOI] [PubMed] [Google Scholar]
- 10Pibouin, L, Villaudy, J, Ferbus, D, Muleris, M, Prospéri, MT, Remvikos, Y et al. (2002). Cloning of the mRNA of overexpression in colon carcinoma-1: a sequence overexpressed in a subset of colon carcinomas. Cancer Genet Cytogenet 133: 55–60. [DOI] [PubMed] [Google Scholar]
- 11Wang, Y, Chen, W, Yang, C, Wu, W, Wu, S, Qin, X et al. (2012). Long non-coding RNA UCA1a(CUDR) promotes proliferation and tumorigenesis of bladder cancer. Int J Oncol 41: 276–284. [DOI] [PubMed] [Google Scholar]
- 12Wang, XS, Zhang, Z, Wang, HC, Cai, JL, Xu, QW, Li, MQ et al. (2006). Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res 12: 4851–4858. [DOI] [PubMed] [Google Scholar]
- 13Xue, M, Li, X, Wu, W, Zhang, S, Wu, S, Li, Z et al. (2014). Upregulation of long non-coding RNA urothelial carcinoma associated 1 by CCAAT/enhancer binding protein α contributes to bladder cancer cell growth and reduced apoptosis. Oncol Rep 31: 1993–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Hiemer, SE, Szymaniak, AD and Varelas, X (2014). The transcriptional regulators TAZ and YAP direct transforming growth factor β-induced tumorigenic phenotypes in breast cancer cells. J Biol Chem 289: 13461–13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Fan, Y, Shen, B, Tan, M, Mu, X, Qin, Y, Zhang, F et al. (2014). Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J 281: 1750–1758. [DOI] [PubMed] [Google Scholar]
- 16Huang, J, Zhou, N, Watabe, K, Lu, Z, Wu, F, Xu, M et al. (2014). Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 5: e1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Wu, W, Zhang, S, Li, X, Xue, M, Cao, S and Chen, W (2013). Ets-2 regulates cell apoptosis via the Akt pathway, through the regulation of urothelial cancer associated 1, a long non-coding RNA, in bladder cancer cells. PLoS One 8: e73920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Kaneko, K, Ito, Y, Ono, Y, Tainaka, T, Tsuchiya, H, Shimoyama, Y et al. (2011). Gene expression profiling reveals upregulated UCA1 and BMF in gallbladder epithelia of children with pancreaticobiliary maljunction. J Pediatr Gastroenterol Nutr 52: 744–750. [DOI] [PubMed] [Google Scholar]
- 19Tsang, WP, Wong, TW, Cheung, AH, Co, CN and Kwok, TT (2007). Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA 13: 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Tian, Y, Zhang, X, Hao, Y, Fang, Z and He, Y (2014). Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res 24: 335–341. [DOI] [PubMed] [Google Scholar]
- 21Xue, M, Li, X, Li, Z and Chen, W (2014). Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biol 35: 6901–6912. [DOI] [PubMed] [Google Scholar]
- 22Kumar, PP, Emechebe, U, Smith, R, Franklin, S, Moore, B, Yandell, M et al. (2014). Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. Elife 2014: e02805. [DOI] [PMC free article] [PubMed]
- 23D'Amour, KA, Agulnick, AD, Eliazer, S, Kelly, OG, Kroon, E and Baetge, EE (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23: 1534–1541. [DOI] [PubMed] [Google Scholar]
- 24Gouon-Evans, V, Boussemart, L, Gadue, P, Nierhoff, D, Koehler, CI, Kubo, A et al. (2006). BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol 24: 1402–1411. [DOI] [PubMed] [Google Scholar]
- 25Wang, T, Yuan, J, Feng, N, Li, Y, Lin, Z, Jiang, Z et al. (2014). Hsa-miR-1 downregulates long non-coding RNA urothelial cancer associated 1 in bladder cancer. Tumour Biol 35: 10075–10084. [DOI] [PubMed] [Google Scholar]
- 26Deng, Z, Wang, Z, Stong, N, Plasschaert, R, Moczan, A, Chen, HS et al. (2012). A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J 31: 4165–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Saito, Y and Saito, H (2012). Role of CTCF in the regulation of microRNA expression. Front Genet 3: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Lin, S, Ferguson-Smith, AC, Schultz, RM and Bartolomei, MS (2011). Nonallelic transcriptional roles of CTCF and cohesins at imprinted loci. Mol Cell Biol 31: 3094–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Tiffen, JC, Bailey, CG, Marshall, AD, Metierre, C, Feng, Y, Wang, Q et al. (2013). The cancer-testis antigen BORIS phenocopies the tumor suppressor CTCF in normal and neoplastic cells. Int J Cancer 133: 1603–1613. [DOI] [PubMed] [Google Scholar]
- 30Petrov, NS and Popov, BV (2014). Study of Wnt2 secreted by A-549 cells in paracrine activation of β-catenin in co-cultured mesenchymal stem cells. Biochemistry (Mosc) 79: 524–530. [DOI] [PubMed] [Google Scholar]
- 31Kim, GJ, Seok, JY, Rhee, H, Choi, JY, Choi, JS, Kim, KS et al. (2014). β-Catenin activated hepatocellular adenoma: a report of three cases in Korea. Gut Liver 8: 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Kolben, T, Peröbner, I, Fernsebner, K, Lechner, F, Geissler, C, Ruiz-Heinrich, L et al. (2012). Dissecting the impact of Frizzled receptors in Wnt/β-catenin signaling of human mesenchymal stem cells. Biol Chem 393: 1433–1447. [DOI] [PubMed] [Google Scholar]
- 33Meek, S, Wei, J, Sutherland, L, Nilges, B, Buehr, M, Tomlinson, SR et al. (2013). Tuning of β-catenin activity is required to stabilize self-renewal of rat embryonic stem cells. Stem Cells 31: 2104–2115. [DOI] [PubMed] [Google Scholar]
- 34Li, P, Cao, Y, Li, Y, Zhou, L, Liu, X and Geng, M (2014). Expression of Wnt-5a and β-catenin in primary hepatocellular carcinoma. Int J Clin Exp Pathol 7: 3190–3195. [PMC free article] [PubMed] [Google Scholar]
- 35Zhao, Y, Guo, Q, Chen, J, Hu, J, Wang, S and Sun, Y (2014). Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep 31: 358–364. [DOI] [PubMed] [Google Scholar]
- 36Li, CH and Chen, Y (2013). Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol 45: 1895–1910. [DOI] [PubMed] [Google Scholar]
- 37Wang, J, Liu, X, Wu, H, Ni, P, Gu, Z, Qiao, Y et al. (2010). CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 38: 5366–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Xie, H, Ma, H and Zhou, D (2013). Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int 2013: 136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Panzitt, K, Tschernatsch, MM, Guelly, C, Moustafa, T, Stradner, M, Strohmaier, HM et al. (2007). Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132: 330–342. [DOI] [PubMed] [Google Scholar]
- 40Du, Y, Kong, G, You, X, Zhang, S, Zhang, T, Gao, Y et al. (2012). Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem 287: 26302–26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Cao, L, Zhou, Y, Zhai, B, Liao, J, Xu, W, Zhang, R et al. (2011). Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol 11:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.