Abstract
The ribosomal RNA (rrn) operon is a key suite of genes related to the production of protein synthesis machinery and thus to bacterial growth physiology. Experimental evidence has suggested an intrinsic relationship between the number of copies of this operon and environmental resource availability, especially the availability of phosphorus (P), because bacteria that live in oligotrophic ecosystems usually have few rrn operons and a slow growth rate. The Cuatro Ciénegas Basin (CCB) is a complex aquatic ecosystem that contains an unusually high microbial diversity that is able to persist under highly oligotrophic conditions. These environmental conditions impose a variety of strong selective pressures that shape the genome dynamics of their inhabitants. The genus Bacillus is one of the most abundant cultivable bacterial groups in the CCB and usually possesses a relatively large number of rrn operon copies (6–15 copies). The main goal of this study was to analyze the variation in the number of rrn operon copies of Bacillus in the CCB and to assess their growth-related properties as well as their stoichiometric balance (N and P content). We defined 18 phylogenetic groups within the Bacilli clade and documented a range of from six to 14 copies of the rrn operon. The growth dynamic of these Bacilli was heterogeneous and did not show a direct relation to the number of operon copies. Physiologically, our results were not consistent with the Growth Rate Hypothesis, since the copies of the rrn operon were decoupled from growth rate. However, we speculate that the diversity of the growth properties of these Bacilli as well as the low P content of their cells in an ample range of rrn copy number is an adaptive response to oligotrophy of the CCB and could represent an ecological mechanism that allows these taxa to coexist. These findings increase the knowledge of the variability in the number of copies of the rrn operon in the genus Bacillus and give insights about the physiology of this bacterial group under extreme oligotrophic conditions.
Keywords: rRNA operon copies, oligotrophy, bacterial growth, Bacillus, Cuatro Ciénegas
Introduction
Population genetics is the most direct tool for use in understanding adaptation to the ecological challenges imposed upon microbial communities by the environment (Whitaker et al., 2003; Xu, 2006; Spencer et al., 2008). Functional traits can aid in the study of population genetics, because they help to define species in terms of their ecological roles, such as how they use environmental resources or how they interact with other species (McGill et al., 2006; Hughes et al., 2008). These functional traits are often considered to be ecological strategies because they are useful in understanding why certain bacteria live in a particular environment and how they respond to environmental challenges (Green et al., 2008).
The ribosomal RNA operon (rrn hereafter) is the key genetic structure for protein synthesis and thus a functional trait related to bacterial life history (Stevenson and Schmidt, 2004). Ecologically, the rrn operon has been related with the bacterial capacity to respond to changes in environmental conditions (Codon et al., 1995; Prüβ et al., 1999; Green et al., 2008). In particular, the variation in the number of copies of the rrn operon has been considered an ecological strategy related to resource availability, with physiological implications associated with bacterial growth rate and fitness (Klappenbach et al., 2000; Shrestha et al., 2007). The rrn operon is comprised of three genes (5S, 16S, and 23S rDNA) and its copy number varies from 1 to 15 among bacterial genomes (Klappenbach et al., 2001; Acinas et al., 2004; Stoddard et al., 2015) and even more dramatically among eukaryotes (Elser et al., 2000). Experimentally, it has been shown that deletions of one or more copies of the rrn operon have a considerable impact on growth rate, affecting various stress-response mechanisms (Nanamiya et al., 2010; Yano et al., 2013). Hence, it has been suggested that the multiplicity of the rrn operon is a potential mechanism for adaptation to different environmental conditions (Elser et al., 2000; Green et al., 2008). In general terms, bacteria that possess more rrn operon copies may cope better with fluctuating nutrient inputs than bacteria with fewer rrn operon copies, which tend to live in environments where nutrients are scarce (Klappenbach et al., 2001; Elser, 2003; Jeyasingh and Weider, 2007). Moreover, the relationship between rrn operon copy number and the bacterial biotic potential for the cellular allocation of key resources could be analogous to the ecological strategies described in other macro-biota (r- and K-strategies), (Pianka, 1970; Elser et al., 2000; Dethlefsen and Schmidt, 2007; Shrestha et al., 2007; Lipowsky et al., 2012).
Bacillus is a genus that is well-known because of its ecological versatility (Feldgarden et al., 2003). The fact that it can sporulate increases its long-range dispersal and allows it to explore diverse environments (Nanamiya et al., 2010; Yano et al., 2013). Coincidentally, Bacilli have a relatively high number of rrn operon copies per genome, ranging from six to 15 (rrnDB, Stoddard et al., 2015), a degree of variation that may aid in this lifestyle strategy of colonizing new environments and provide great adaptability in response to stress, as well as being able to uptake a wide variety of resources (Feldgarden et al., 2003; Connor et al., 2010). If the new environment is rich in phosphorus (P), high rrn operon copy number may be favored because the rich P supply could then support the rapid production of P-rich rrn to meet the protein demands of rapid growth, as stated by the “Growth Rate Hypothesis” (GRH), a core idea within the theory of biological stoichiometry (Elser et al., 2000; Elser, 2006). However, in environments with low phosphorus availability, multiple rrn operon copies could represent a competitive cost if a high rrn operon copy number leads to the over-production of P-rich rrn (Sterner and Elser, 2002; Maciá, 2005; Jeyasingh and Weider, 2007). Indeed, aquatic bacteria isolated from oligotrophic environments usually have low rrn operon copy numbers (Fegatella et al., 1998; Strehl et al., 1999; Lauro et al., 2009), as well as various other adaptations to decrease cellular phosphorus demand (Cavicchioli et al., 2003; Alcaraz et al., 2008; Martiny et al., 2009; Van Mooy et al., 2009). Thus, it has been proposed that there is a connection between the number of rrn operon copies and environmental P availability (Elser et al., 2000; Weider et al., 2005; Jeyasingh and Weider, 2007). However, to our knowledge, we lack extensive studies that document this variation in rrn operon copy number and other associated ecological strategies employed by bacteria coexisting in oligotrophic environments, especially those characterized by severe P limitation.
The aims of this study were to describe rrn operon copy number variation in different lineages of Bacillus strains isolated from an extremely oligotrophic ecosystem and to analyze the possible association between the copy number and its physiological implications for growth rate and chemical composition (P content and N:P stoichiometry). Severe P limitation is considered a primary selective pressure that drives bacterial evolution in this environment (Souza et al., 2008, 2012). For example, we have previously reported on an endemic and moderately halophilic Bacillus (type strain of B. coahuilensis: m4-4 = NRRL B-41737T), (Cerritos et al., 2008) that has a typical number of rrn copies (nine) but also clear adaptations to the extreme oligotrophic conditions, including a small genome (3.5 Mb), a diversity of phosphate acquisition genes (Moreno-Letelier et al., 2011), and a cellular membrane composed of sulfolipids (Alcaraz et al., 2008). Similar adaptations to low P levels have been reported only from oligotrophic marine cyanobacteria with low rrn copy numbers (Cavicchioli et al., 2003; Lauro et al., 2009; Martiny et al., 2009; Van Mooy et al., 2009). Hence, the present study represents the first attempt to link biological stoichiometry to Bacillus diversity and rrn operon copy number, as well as the first report in which the numbers of rrn copies are analyzed in several members of this genus that coexist in the same habitats.
Materials and Methods
Site Description and Selection of Bacillus Strains
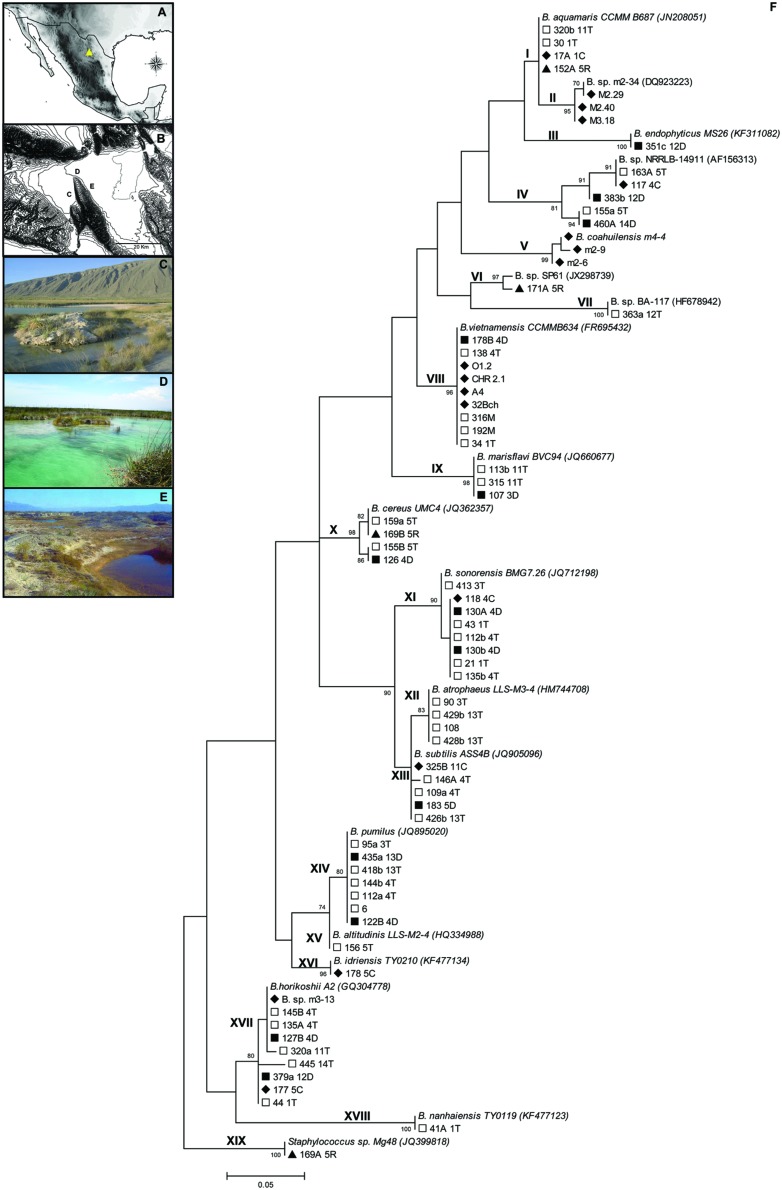
The Cuatro Ciénegas Basin (CCB hereafter) is a hydrologic system in the Chihuahuan Desert in northeastern México (Souza et al., 2006), (Figure 1A, yellow triangle). This basin represents an oasis with a high microbial diversity in extremely oligotrophic conditions (<1 μmol PO43-; Peimbert et al., 2012; Souza et al., 2012). Interestingly, ca. 50% of the bacterial communities in the CCB are most closely related to marine relatives (Souza et al., 2006). Isolates related to the genus Bacillus were identified from samples collected at various sites in the CCB during 15 years of field work (Souza et al., 2006; Alcaraz et al., 2008, 2010; Cerritos et al., 2010; Pérez-Gutiérrez et al., 2013), (Figure 1B).
FIGURE 1.
The Cuatro Ciénegas Basin (CCB) in the Chihuahuan Desert, in northeastern México (A,B) and the sites where Bacillus strains were previously isolated.
(C) The Churince system, (D) Pozas Rojas (Los Hundidos), and (E) Río Mesquites. (F) Maximum-Likelihood (ML) tree of the 19 phylogenetic groups identified using the 5′ HV region of the 16S rDNA. Bootstrap values higher than 70% are shown. Symbols represent the sample type of isolation:  = Top section of sediment,
= Top section of sediment,  = Bottom section of sediment,
= Bottom section of sediment,  = Water sediment adjacent to a plant, and
= Water sediment adjacent to a plant, and  = Water.
= Water.
Our isolates are from three primary sampling sites within the basin: (i) the Churince site consists of a freshwater spring that connects to an intermediate shallow pond via a small stream and eventually terminates in a shallow desiccated lagoon (Figure 1C); (ii) the Río Mesquites is a stable system composed of a river and some lateral desiccated ponds (Figure 1D) with low nutrient concentrations and highly imbalanced C:N:P ratios [C:N:P, 900:150:1 (molar); Souza et al., 2012]; and (iii) the Pozas Rojas site (Figure 1E) is located in a system called Los Hundidos and consists of a shallow lake and nine to 12 small semi-permanent ponds with strongly fluctuating conditions (high salinity and temperature in summer, both decrease in winter).
The Bacillus strains isolated from the CCB are part of a larger collection (several thousands of isolates) of microbes that is maintained at the Molecular Evolution and Experimental Laboratory at the Instituto de Ecología, UNAM (Valeria Souza) and at the Molecular Bacteriology Laboratory in the CINVESTAV, Irapuato (Gabriela Olmedo); cultures are available upon request. We selected 71 of these isolates and classified them according to the site of isolation and sample type (plant root, sediment, or water). Sixty-seven Bacillus isolates were sampled from Churince, one was from Río Mesquites and three were from Pozas Rojas.
Phylogenetic Analysis
To obtain biomass for DNA extraction, Bacillus isolates were grown in the standard medium used for their isolation in the field (Marine agar, DifcoTM 2216). Genomic DNA extractions were performed using the QIAmp® DNA Mini Kit (USA), according to the manufacturer’s instructions. The 5′ hypervariant (HV) region of the 16S rDNA was amplified (275 bp; 70–344 position), following Goto et al., (2000). This region has a high level of conservation and is useful for the clustering of Bacillus species. The polymerase chain reaction (PCR) products were confirmed via 1.5% agarose gel electrophoresis. The sequencing of the HV region was performed by the High Throughput Genomics Center (htSEQ), University of Washington (USA), and compared with the GenBank database using BLAST (NCBI). Sequences were aligned using CLUSTAL W (Thompson et al., 2002), and a maximum-likelihood tree was constructed using MEGA5, with a bootstrap of 1000 replicates (Tamura et al., 2011). The sequences of the 5′ HV region of the 16S rDNA were submitted to GenBank with the following accession numbers: KT781592–KT781661.
I-CeuI Cleavage Map of the Bacillus Strains
The I-CeuI (CeuI hereafter) restriction endonuclease recognizes a 26-bp sequence from position 1911–1936 of the 23S rRNA gene in rrn operons, with the number of CeuI (New England Biolabs) fragments usually representing the number of rrn operons. Pulsed-field gel electrophoresis (PFGE) was used to construct the rrn profile of the chromosome from the Bacillus isolates. Bacterial genomic DNA from Salmonella enterica serovar Typhimurium LT2 cleaved with CeuI and the 0.1–200 kb Sigma Plus Marker were used as molecular weight markers. Because the size of S. enterica Typhimurium LT2 identifying fragments had been determined previously (Liu et al., 1993), their inclusion improved the precision of the band-size estimation.
Preparation and Digestion of Genomic DNA in Agarose Blocks
Bacillus isolates were cultured aerobically in DifcoTM Marine Broth 2216 (BD & Co.) and incubated overnight at 35°C. The genomic DNA of each strain was prepared in agarose blocks using a previously described method, with some modifications (Nakasone et al., 2000; Delgado et al., 2013). Two processes of incubation in proteinase K solution (12 h at 37°C) were carried out to increase the purity of the DNA. Agarose blocks were pre-incubated in 1X NEBuffer 4 (New England Biolabs) for 30 min at 4°C. Finally, the digestion of the genomic DNA was achieved with 100 μl fresh 1X NEBuffer 4 containing 15 U of I-CeuI restriction enzyme, and it was incubated overnight at 37°C.
Pulsed-Field Gel Electrophoresis (PFGE) and DNA Fragments Transfer
The CeuI fragments were separated by a CHEF-DR II electrophoresis system (Bio-Rad). Electrophoresis was performed on a 1% agarose (Seakem Gold agarose, BioWhittaker Molecular Applications) gel and 0.5X TBE buffer (Bio-Rad) at 11°C. The electrophoresis conditions were divided into two stages to separate the DNA fragments of various sizes: First stage, pulse time ramped from 6.75 s to 2 min for 20 h at 4 V cm-1 and in a second stage, pulse time ramped from 0.22 to 5.10 s for 15 h at 6 V cm-1.
The agarose gels were radiated with UV light for 1 min in a UV Crosslinker (UVP) to fix the DNA. The gels were washed in 250 mM HCl solution for 15 min with moderate shaking. Next, the gels were washed in denaturing buffer (1.5 M NaCl, 0.5 M NaOH) for 2 h and later washed in a neutralization buffer (0.5 M Tris/HCl, 1.5 M NaCl; pH 8.0) for 2 h. The DNA fragments were then transferred onto N+nylon membrane (Amersham Biosciences) via Southern blotting as described previously (Sambrook et al., 1989). Finally, the membrane was radiated with UV light to fix the DNA (1 min; UV Crosslinker, UVP).
Preparation of DNA Probes and Hybridization
The rrn profiles of Bacillus isolates were validated by probing the Southern blotting membranes with PCR products of the 16S and 23S rrn operon from the Bacillus horikoshii ATCC 700161 strain. The primer sets used to amplify the rrs gene were designed using the 5′ HV region (described above), and an internal region of the rrl gene was designed from the 2283 to 2696 position (23S3) of the rrn. Then, the 23S3 region (413 bp) was amplified using the forward primer F23S3 5′-ACG GAG GCG CCC AAA GGT T-3′ and the reverse primer R23S3 5′-CCA GCG GTG CGT CCA TCC-3′. The primer set used to amplify the 23S3, was designed based on previously sequenced genomes using the Primer Select program of the DNASTAR Lasergene 7 package (DNASTAR, Inc., Madison, WI, USA).
The PCR amplification conditions were as follows: 95°C for 5 min for the initial denaturation, 30 cycles of denaturation at 95°C for 40 s, annealing at 60°C for 40 s, an extension of 1 min at 72°C and a final extension of 5 min at 72°C (Gene Amp, PCR System 9700). The presence and size of PCR products were subsequently confirmed via 1.5% agarose gel electrophoresis. The PCR products were purified with the PCR Clean-up Gel Extraction Kit (Macherey–Nagel products) and then DIG-labeled using the random primer method of the DIG High Prime DNA Labeling system (Roche). The membrane was incubated in 10 ml hybridization solution (DIG Easy Hyb buffer). Incubation was carried out at 58°C with constant, gentle shaking for 1 h. The labeled probe was then added to fresh hybridization solution and hybridization was carried out overnight at 58°C with constant and gentle shaking. The membrane was exposed to X-ray film after being washed at high astringency (64°C).
Growth Parameter Estimations
Genotypes of Bacillus with different numbers of copies of the rrn operon were chosen from among the groups described in the phylogenetic analysis. Prior to growth parameter estimation, all the strains were pre-cultured in fresh marine broth for 24 h to homogenize their metabolic condition. All cultures were incubated at 35°C, the maximum water temperature during summer at the CCB (Pérez-Gutiérrez et al., 2013), with shaking at 150 rpm. Additionally, experiments were carried out for nutritional conditions similar to CCB; for this, we inoculated the strains into sterile water collected from the Churince field site but supplemented with tryptone (5 g per liter; BactoTM Tryptone, BD and Company; hereafter, CCBwt) and incubated for 12 h (overnight), (Supplementary Figure S1A).
The growth parameters were then determined using the previously described overnight culture. Three new 50 ml flask of fresh CCBwt medium were inoculated to reach an optical density of 0.05 (600 nm wavelength; BioPhotometer Plus, Eppendorf), which corresponded to ∼107 colony-forming units (CFU) ml-1. CFU counts were made taking at least seven samples distributed through a period of 12 h to cover all the phases of the growth curve, the samples were diluted appropriately in 0.85% NaCl to perform a plate count analysis.
We estimated the lag phase period (λ; units: hours), the tangential growth rate (Gtan; units: cells/h), and maximal biomass reached [A; units: Ln(CFU/CFU(t = 0))] from our data using a non-linear regression (CurveExpert Professional 2.0.3 software) to fit a Gompertz equation according to Zwietering et al. (1990), (Supplementary Figure S1B). We obtained the final parameters from the predicted curve, defining Gtan as the tangent of the inflection point of the curve, λ as the X intercept of the tangential line through the inflection point (where the X-axis is time), and A as the Y-value of the asymptote [where the Y-axis is Ln(CFU/CFU(t = 0))] for plate-count assays. In addition, we estimated the maximum specific growth rate (μmax; units: hours-1) as follows:
and the bacterial doubling (generation) time as follows:
N0 and Ne are the cell densities reached at the beginning and at the end of the exponential phase, respectively, while t0 and te are the times (h) at which the exponential phase started and ended, respectively.
Cell Contents of Carbon (C), Nitrogen (N), and Phosphorus (P) During the Exponential Phase
The samples of bacterial biomass were harvested in the exponential phase during the determination of growth dynamics. Biomass samples were spin in a centrifuge for the removal of the growth medium. To avoid the influence of remains of the growth medium in the elemental composition analysis, the biomass samples were washed three times with 250 μl water (Mili-Q)® and after of each washing, supernatant was removed by centrifuge. Finally, the biomass samples were vacuum dried to be shipped frozen to Arizona State University for analysis.
At ASU total phosphorus content was measured using a modified ascorbic acid method with persulfate digestion (APHA, 2005). The dried biomass samples were weighed and treated with a potassium persulfate and sulfuric acid solution and then autoclaved for 30 min at 121°C and 15–20 psi. The samples were allowed to cool and then neutralized before the addition of the color reagent. After 30 min, the absorbance was read on a spectrophotometer at 880 nm. The samples were analyzed with a triplicate standard curve and triplicate NIST reference material.
The total carbon and nitrogen content was measured via combustion in a Perkin Elmer model 2400 elemental analyzer. The samples were combusted at 1760°C. Elemental detection was conducted via a thermal conductivity detector.
The C, N, and P data were expressed as percentages of dry mass and referred to as “C content,” “N content,” and “P content,” respectively (Supplementary Table S2).
Statistical Analysis
All statistical analyses, including the estimation of Pearson correlation coefficients (r) and principal component analysis (PCA) were performed with the R Statistic program Version 3.3.1 (24-07-10).
Results
Identification and Clustering of Bacillus Isolates from the CCB
The 71 isolates used for this study were clustered into 19 phylogenetic groups, forming a large “marine” cluster (30 isolates; i.e., formed by strains with marine affinities). Most of the lineages include representatives from a variety of habitats (e.g., soil, water, sediment); however, some “marine” groups included strains only sampled from water, while the B. atrophaeus lineage was only found in the top layers of sediment. Strains related to B. pumilus were also only found in sediment (Figure 1F).
The “marine” cluster is composed of nine phylogenetic groups (arbitrarily numbered I to IX) composed of Bacillus strains isolated mostly from CCB aquatic samples, such as B. sp. m2-34 (group II) and B. coahuilensis (group V). In addition, three CCB phylogenetic groups of Bacillus were related to type strains isolated from water: B. endophyticus (group III), related to a pollutant-degrading strain isolated from industrial effluent, as well as B. sp. NRRLB-14911 (group IV) and B. marisflavi (group IX), both isolated from seawater. The B. aquamaris (group I), B. sp. SP61 (group VI), and B. vietnamensis (group VIII) groups are related to strains isolated from hypersaline environments (salterns and a microbial mat). Finally, group VII is related to a strain isolated from a biofilm of a lake (B. sp. BA-117). The small B. cereus cluster is related to type strains isolated from marine sediments (group X). The B. subtilis cluster (group XIII), was similar to soil type Bacillus. Group XI is most closely related to B. sonorensis isolated from the soil of the Sonoran desert. Organisms in group XII are related to a B. atrophaeus strain isolated from soil and water samples, and group XIII is related to a B. subtilis strain from marine samples, although this is a well-known cosmopolitan species. Strains related to a B. altitudinis strain (group XV), were present in soil and water samples; B. idriensis isolates (group XVI), were related to strains from soil samples with halotolerance. Finally, the B. horikoshii cluster (group XVII) has as a representative a strain isolated from a fish pond as well as a lineage related to a halotolerant B. nanhaiensis (group XVIII), isolated from a non-saline soil sample. Unexpectedly, the 169A 5R isolate was closely related to the strain Staphylococcus sp. Mg48 (JQ399818), isolated from a saline lake (group XIX), but it is not unusual to isolate a Staphylococcus strain when aiming to select Bacillus.
Phylogenetic Variability of the rrn Operon Copy Number
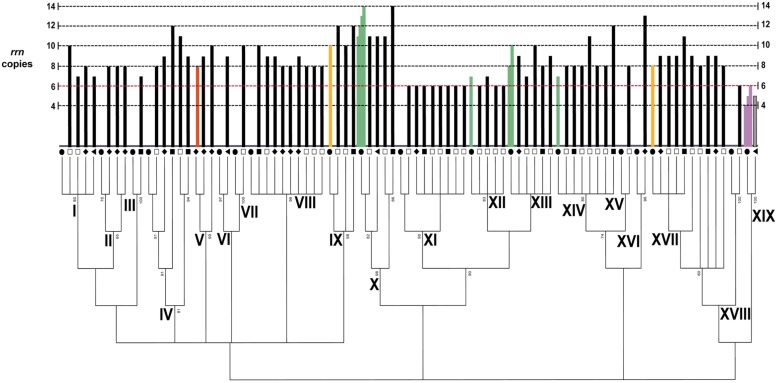
The rrn operon copy number was determined for every isolate described above in the maximum-likelihood tree via PFGE and hybridization analyses (Figure 2 and Supplementary Figure S2). To obtain a benchmark for the rrn copy number in the Bacillus diversity from the CCB, we analyzed the type strain of B. coahuilensis (m4-4 = NRRL B-41737T) that was isolated from the Churince site (Cerritos et al., 2008) and that has already been sequenced (Alcaraz et al., 2008). After genomic digestion and hybridization analysis, eight rrn operons were quantified (Figure 2 and Supplementary Figure S2; group V). The 70 strains of Bacillus from the CCB showed a range of between six and 14 rrn operon copies (Figure 2 and Supplementary Figure S2). Some groups showed intraspecific variation from one to four copies. Interestingly, we quantified only six copies of this operon in some phylogenetic groups, such as B. sonorensis (XI), B. atrophaeus (XII), and B. nanhaiensis (XVIII); the lowest number of copies quantified in other strains of the genus (rrnDB, Stoddard et al., 2015). The highest number of copies was observed in the B. cereus group (X) (14 copies).
FIGURE 2.
Variability of the rRNA operon copy number in the Bacillus diversity from the CCB. ML tree of the 5′ HV region of the 16S rDNA with a 50% of cut-off value. The black circles represent the type strain of Bacillus for every phylogenetic group. The squares, triangles and rhomboids symbols correspond to the environment as described in Figure 1. The dotted red line represents the low number of the rrn operon in Bacillus according to the rrnDB. The black bars represent the number of operon copies in the isolates from the CCB, and the green bars represent the number of copies in Bacillus species reported in the rrnDB. Gray bar represents the number of copies in the isolate related to the genus Staphylococcus. The light purple bars represent the number of operon copies in different Staphylococcus species analyzed by the rrnDB. The orange bar represents the number of copies quantified in the type strain of B. coahuilensis (m4-4 = NRRL B-41737T), group V. The yellow bars represent the number of copies in the type strains of B. marisflavi (JCM 11544 = KCCM 41588) and B. horikoshii (ATTCC 700161), groups IX and XVII, respectively.
To further increase knowledge about the number of copies of the rrn operon in the genus Bacillus, we analyzed two type strains similar to those observed in the CCB, B. marisflavi (JCM 11544 = KCCM 41588), and B. horikoshii (ATTCC 700161). The genomes of these species have not yet been sequenced and the number of copies of this functional gene is unknown. The B. marisflavi and B. horikoshii type strains showed ten and eight copies of the rrn, respectively (Figure 2 and Supplementary Figure S2, groups IX and XVII). Thus, the number of copies quantified in these Bacillus type species was similar to the number described in the strains isolated from the CCB (Figure 2).
Homogeneity in rrn operon copy number was observed in the B. sp. m2-34 (II; eight copies) and B. sonorensis (XI; six copies) groups. However, considerable heterogeneity and intraspecific variation were observed in several other groups: B. aquamaris (I; seven to nine copies), B. sp. NRRLB-14911 (IV; eight, nine, eleven and twelve copies), B. vietnamiensis (VIII; eight to ten copies), B subtilis (XIII; seven to eleven copies), B. pumilus (XIV; eight, eleven, and twelve copies) and B. horikoshii (XVII; eight, nine and eleven copies). In addition, the phylogenetic groups composed of only one isolate showed different numbers of rrn operon copies: B. endophyticus (III; seven copies), B. sp. SP61 (VI; ten copies), B. sp. BA-117 (VII; nine copies), B. altitudinis (XV; eight copies), B. idriensis (XVI; 13 copies) and B. nanhaiensis (XVIII; six copies; Figure 2). The Staphylococcus isolate had five copies of the rrn.
Growth Parameters
Growth parameters were estimated for a subsample of 15 Bacillus isolates from the CCB representative of the phylogenetic diversity present and the range of rrn operon copy numbers observed. We also characterized the isolate related to the Staphylococcus genus (Table 1). Not surprisingly, given the large diversity in this genus within the CCB, the results show a high heterogeneity in the growth parameters estimated. In agreement with these results, the lag phase period of these Bacilli is variable and it is not related with their growth rate (μmax; Supplementary Figure S3A). In addition, the maximum biomass reached was correlated with the maximum specific growth rate (μmax; Supplementary Figure S3B).
Table 1.
Growth parameters estimated in the Bacillus isolates in the CCB.
| Growth parameters | |||||||
|---|---|---|---|---|---|---|---|
| rrn copies | Isolate | Phylogenetic group | A | BBB (h) | μmax (h-1) | td (h) | |
| A | 14 | 155B_5T | X. B. cereus | 9.98 | 0.92 | 0.26 | 2.66 |
| B | 6 | 118_4C | XI. B. sonorensis | 9.89 | 1.13 | 0.24 | 2.88 |
| C | 8 | m3-18 | II. B. sp. m2-34 | 9.78 | 2.89 | 0.24 | 2.88 |
| D | 9 | m2-9 | V. B. coahuilensis | 7.95 | 1.08 | 0.22 | 3.15 |
| E | 10 | m2-6 | V. B. coahuilensis | 7.68 | 1.45 | 0.21 | 3.30 |
| F | 12 | 112B_4D | XIV. B. pumilus | 7.35 | 1.70 | 0.31 | 2.23 |
| G | 5 | 169A_5R | XIX. Staphylococcus | 7.30 | 0.92 | 0.26 | 2.66 |
| H | 12 | 107_3D | IX. B. marisflavi | 7.28 | 0.25 | 0.19 | 3.64 |
| I | 13 | 178_5C | XVI. B. idriensis | 7.02 | 2.56 | 0.57 | 1.21 |
| J | 11 | 126_4D | X. B. cereus | 6.69 | 2.78 | 0.23 | 3.01 |
| K | 7 | 152A_5R | I. B. aquamaris | 6.48 | 0.22 | 0.16 | 4.33 |
| L | 8 | 44_1T | XVII. B. horikoshii | 5.20 | 4.12 | 0.19 | 3.64 |
| M | 10 | 315_11T | IX. B. marisflavi | 4.44 | 1.44 | 0.12 | 5.77 |
| N | 11 | 144b_14T | XIV. B. pumilus | 3.67 | 2.79 | 0.11 | 6.30 |
| O | 6 | 108 | XII. B. atrophaeus | 3.38 | 2.06 | 0.08 | 8.66 |
| P | 11 | 127B_4D | XVII. B. horikoshii | 3.22 | 1.70 | 0.09 | 7.70 |
A = maximum biomass reached [Ln(CFU/CFU(t =0))]; BBB = lag phase; μmax = maximum specific growth rate; td = doubling (generation) time.
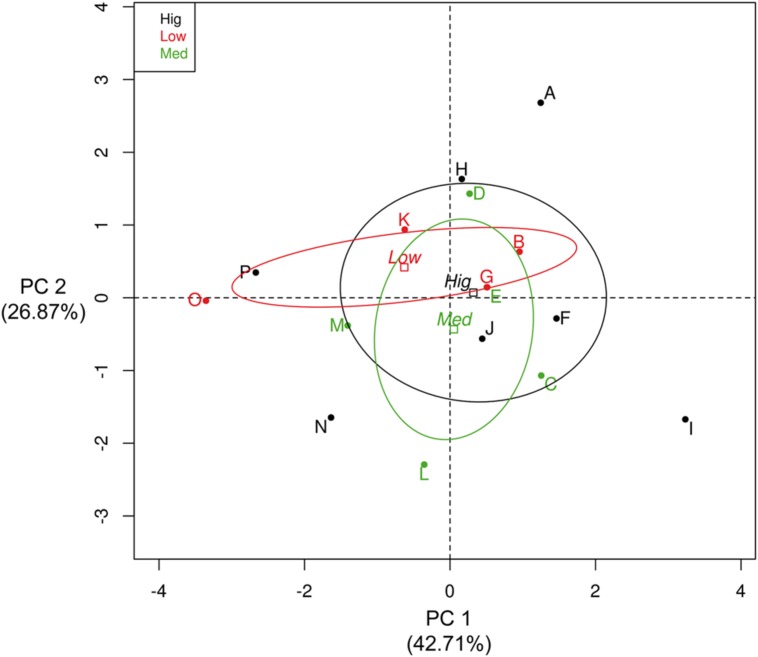
Interestingly, an exploratory analysis showed no overall correlation between the number of copies of the rrn operon and the growth parameters estimated (Supplementary Table S1). However, an arbitrary categorization of this copy number (where “low” was from five to seven copies, “mid” was from eight to ten copies and “high” was from 11 to 14 copies) showed that the isolates with the fewest copies had lower levels of dispersion in their growth parameters (Figure 3). Principal component analysis (PCA) was performed to describe the influence of the various growth parameters in these categories in the isolates from the CCB (Figure 3). It seems that 42.71% of the variance was explained by Component 1, which was defined by doubling time (td), maximum growth rate (μmax) and the maximum biomass reached (A). Component 2 explained 26.87% of the variance, and was principally defined by adaptation time (λ) and the rrn operon copies (Supplementary Figure S4). Meanwhile, isolates with the highest copy numbers showed greater dispersion, having the most extreme parameter values (Table 1). These results may indicate that the rrn operon copy number in the Bacillus from the CCB may be related to the integrated suite of growth dynamics parameters, but not exclusively to the growth rate. Then, the heterogeneity in the growth dynamics of the isolates of Bacillus from the CCB could be a response to the low availability of nutrients and the competitive cost that represents the high number of copies of the rrn operon.
FIGURE 3.
Principal component analysis (PCA) of the growth parameters estimated in the Bacillus isolates from the CCB. The names of isolates (letters, from A to P) were in agreement with the maximum biomass reached (A) in Table 1. Color labels represent the isolates of the arbitrary categorization of the number of copies of the rrn operon (upper left box), where red labels are the isolates with “low copy numbers” (five to seven), green labels are the isolates with “intermediate copy numbers” (8 to 10) and black labels are the isolates with “high copy numbers” (from 11 to 14).
P and N Contents and N:P Ratios in the Exponential Phase
To assess potential eco-physiological implications associated with the oligotrophic conditions of the CCB regarding the genus Bacillus, biomass C, N, and P contents were estimated during the exponential phase of growth for all strains (Supplementary Table S2). All the isolates showed a relatively low but variable P (%) content (Mean = 0.496; SD = 0.616; Median = 0.258). While C (%) and N (%) content also showed high variability among the isolates [N (%): Mean = 7.14; SD = 5.61; Median = 5.49; C (%): Mean = 64.64; SD = 26.63; Median = 59.63]. Both the C:N ratio (Mean = 13.18; SD = 7.18; Median = 12.32) and the N:P ratio (Mean = 126.7; SD = 235.1; Median = 52.1) showed a considerable range (Table 2). Finally, the N:P ratios estimated in the Bacillus from the CCB were substantially higher than the ratios reported for other Bacilli (B. subtilis 10.6; Loladze and Elser, 2011).
Table 2.
C:N and N:P ratio during the exponential phase of growth in the isolates of Bacillus from the CCB.
| rrn copies | Isolate | Phylogenetic group | C:N | N:P | |
|---|---|---|---|---|---|
| A | 14 | 155B_5T | X. B. cereus | 4.94 | 965.38 |
| B | 6 | 118_4C | XI. B. sonorensis | 19.28 | 10 |
| C | 8 | m3-18 | II. B. sp. m2-34 | 8.35 | 286.47 |
| D | 9 | m2-9 | V. B. coahuilensis | 24.82 | 79.09 |
| E | 10 | m2-6 | V. B. coahuilensis | 18.18 | 52.91 |
| F | 12 | 112B_4D | XIV. B. pumilus | 18.77 | 194.61 |
| G | 5 | 169A_5R | XIX. Staphylococcus | 5.75 | 25.83 |
| H | 12 | 107_3D | IX. B. marisflavi | 8.87 | 19.55 |
| I | 13 | 178_5C | XVI. B. idriensis | 17.85 | 40.67 |
| J | 11 | 126_4D | X. B. cereus | 24.73 | 67.41 |
| K | 7 | 152A_5R | I. B. aquamaris | 0.62 | 72.58 |
| L | 8 | 44_1T | XVII. B. horikoshii | 13.79 | 4.61 |
| M | 10 | 315_11T | IX. B. marisflavi | 16.7 | 82.72 |
| N | 11 | 144b_14T | XIV. B. pumilus | 6.59 | 50.75 |
| O | 6 | 108 | XII. B. atrophaeus | 10.77 | 23.6 |
| P | 11 | 127B_4D | XVII. B. horikoshii | 10.85 | 51.32 |
| Mean ± SD | 13.18 ± 7.18 | 126.72 ± 235.13 | |||
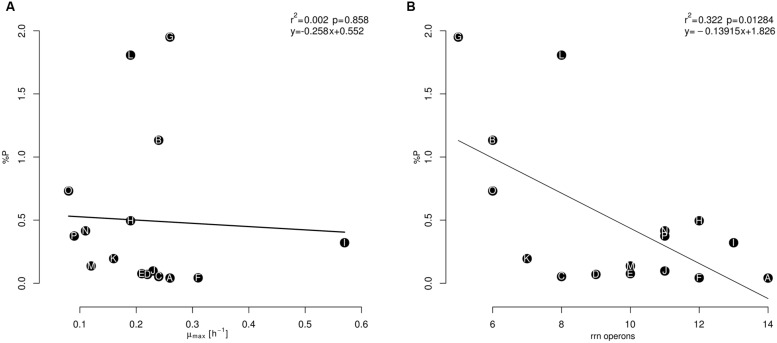
Despite the growth rate not being related to cellular P content (Figure 4A), the correlation between rrn operon copy number and P (%) content is negative and significant (Figure 4B). Isolate G, with a low copy number (five copies; Staphylococcus), had the highest cellular content of both elements. In addition, isolates B and O, with six rrn operon copies, had high P-content levels (B. sonorensis and B. atrophaeus, respectively). Meanwhile, isolate A (14 rrn operon copies; B. cereus) had the lowest P content of all the isolates analyzed; this isolate also had the second-highest N content. The isolates D and E (B. coahuilensis; nine and ten rrn operon copies, respectively) showed low P contents and had the lowest N content. Moreover, the previous isolates with low rrn operon copies (B, G, and O) showed low N:P ratios, while isolate A, with a high number of rrn operon copies had the highest estimated N:P ratio. In addition, the two isolates related to B. coahuilensis showed intermediate values of this elemental ratio (Table 2). These results were not consistent with the GRH and it seems that the isolates with low number of copies of the rrn operon may cope better in this oligotrophy of the CCB; although the number of operon copies quantified is high in comparison with other bacterial groups that live in other oligotrophic environments (e.g., cyanobacteria; Fegatella et al., 1998).
FIGURE 4.
Relationship between the maximum specific growth rate (μmax) (A) and the number of copies of the rrn operon (B), along P (%) content, during the exponential phase. The isolates names (letters, from A to P) are within the circles in black and were in agreement with the maximum biomass reached (A) in Table 1.
Discussion
The general objectives of this work were to assess the variability of a particular ecological trait, the rrn operon copy number in Bacillus strains isolated from extremely oligotrophic aquatic ecosystems, and to evaluate whether there is any association between the variation of this trait and strain physiology. We analyzed 71 isolates of this ecosystem and found considerable variation in the ribosomal operon copy number with a tendency toward an intermediate number of rrn operon copies. We also documented variation in growth rate dynamics and elemental composition. While we did observe physiological associations consistent with the GRH (e.g., the isolate with the slower-growth showed the lowest P content, and a high ratio of N:P). However, there were no consistent associations between copy number and growth parameters. Instead, it is likely that a variety of genomic strategies beyond variation in rrn copy number are employed to modulate growth in this clade of Bacillus, potentially allowing for their coexistence in different niches.
Phylogenetic Clustering of Bacillus Isolates
Our phylogenetic reconstruction showed a large diversity of species with isolates obtained from different habitats generally widespread in the tree. This suggests that each sampling site, as well as the overall system, contains several coexisting taxa. The microbial diversity in the Churince ecosystem has been deeply documented, particularly its Bacillus population, yielding 55 thermo-resistant strains and several extremely halotolerant strains (Cerritos et al., 2010). Moreover, endemic Bacillus strains have been described and genotyped. For instance, the genomes of B. coahuilensis and Bacillus m3-13 show several interesting low-nutrient adaptations (Alcaraz et al., 2008, 2010), as well as an ancient ancestry (Moreno-Letelier et al., 2011). Further sampling of pond sediments in Churince led to a demonstration of how antagonistic interactions between the Bacilli contribute to the large observed diversity, while maintaining a large local differentiation via either resistance or avoidance as in a paper-rock-scissors model (Pérez-Gutiérrez et al., 2013; Aguirre-von-Wobeser et al., 2014).
Most of our isolates came from Churince, whose large terminal lake is now mostly dry due to water overexploitation (Souza et al., 2006, 2012). Interestingly, several times during this sampling period, we recovered the same phylogroups in the sediments in the same sampling sites. It has been argued that sediment is the “native” habitat of Bacillus in the CCB because many phylogenetic groups coexist there and can be recovered consistently at different sampling times (Pérez-Gutiérrez et al., 2013). Our results support what has been observed previously: the considerable levels of bacterial diversity in this basin are the outcome of complex biotic interactions within the community, CCB’s ancient geological history, low nutrient availability, and considerable spatial and seasonal variability in environmental conditions (Souza et al., 2012).
Variation in rrn Operon Numbers
We found no simple answer regarding variations in rrn operon copy number as a response on the part of bacteria in a low-nutrient environment. This was somewhat unexpected because rrn operon copy number is a well-studied functional trait that has been reported to be associated with bacterial lifestyle and represents an ecological strategy for nutrient use (Klappenbach et al., 2000; Stevenson and Schmidt, 2004; Green et al., 2008). A variety of studies have quantified the copy number of this operon in strains isolated from environmental samples, including some for which some Bacillus strains were analyzed (Klappenbach et al., 2000; Shrestha et al., 2007; Vieria-Silva and Rocha, 2015). However, no previous studies have focused on rrn operon copy number in such a diverse group of coexisting species within the genus Bacillus, much less in a shared environment with extremely low nutrient availability.
Previous work has described the genomic properties of some Bacillus isolated from the CCB (Alcaraz et al., 2008, 2010). For example, the genome analysis of B. coahuilensis documented nine rrn operon copies (Alcaraz et al., 2010; unpublished results). However, in our analysis, we quantified eight copies, perhaps because the differences in size among operons were too small to be detected via the pulse field method. Nevertheless, such discrepancies are common when these types of data are compared (Vishnivetskaya et al., 2009). Bacillus groups isolated from the CCB showed from six to 14 rrn operon copies, which is consistent with previous quantifications of the rrn operon (from six to 15 copies reported in rrnDB, Stoddard et al., 2015). This range of operon copies is not what would be expected for isolates from the CCB because the idea is that they should match a slower (i.e., “K-selected”) life history in an oligotrophic environment such as the CCB. This could be the case for all of the CCB isolates related to B. sonorensis, a soil strain that was first described in a desert sample with similar environmental conditions to those of the CCB (Palmisano et al., 2001; Souza et al., 2006, 2012). To evaluate this further, we compared our results with data from the rrnDB database. B. atrophaeus strain 1942 has seven rrn operon copies, but as mentioned above, in our analysis, we found a related strain with six copies. The rrnDB B. subtilis strains from the database showed from eight to ten copies; in our analysis, we observed a wider range (seven to eleven copies). On the other hand, seven copies of the rrn operon were observed in the B. pumilus genome, while B. pumilus relatives isolated in the CCB have eight, eleven, and twelve copies. Such intraspecific variability was evaluated by Acinas et al., (2004) for different bacterial genomes. They documented three species of Bacillus that had normal variation from one to three copies. Subsequent analysis with a larger number of Bacillus genomes showed similar variability (Rastogi et al., 2009). Accordingly, the CCB’s closely related, pumilus-like Bacillus showed a similar level of intraspecific variation, from one to four copies. Overall, the number of copies of the rrn operon in the genus Bacillus at the CCB shows considerable variability, but it does not show evidence of any clear directional change from previously published values for various taxa. This could be due to the fact that very large ranges of taxa within the Bacilli are being selected for a wide variety of responses to cope with the oligotrophic environment. For example, while some save P in their ribosomes with a slow growth rate (“K strategists”), others may maintain a high growth capacity (“r strategists”) that is compensated by other tactics, such as phospholipid to sulfolipid substitution, the presence of high-affinity P-uptake systems, and small genomes (Alcaraz et al., 2008; Moreno-Letelier et al., 2011).
Growth Parameters and rrn Operon Copy Number
The analysis of bacterial growth is considered to be an important tool in understanding and characterizing an organism because it describes potential bacterial response to changes in environmental conditions, as well as ecological responses to other microorganisms (Monod, 1949; Neidhardt, 1999). As mentioned in the results section, the growth dynamics of Bacillus inhabitants of the CCB showed a high degree of variability in various parameters. This heterogeneity in growth rate has been previously observed for environmental strains with different numbers of rrn operons (Dethlefsen and Schmidt, 2007). In addition, several of the estimated parameters were similar to those described in other Bacillus strains under extreme experimental growth conditions (Supplementary Table S3); however, many of these previous studies were performed with model or economically important species. Previous work has considered the ecological importance of the rrn operon copy numbers in bacterial adaptation to different environmental conditions (Elser et al., 2000; Klappenbach et al., 2000; Shrestha et al., 2007; Green et al., 2008). Variation in the number of rrn operon copies is potentially related to the bacterial growth rate because of the need to sustain high levels of rRNA synthesis (Codon et al., 1995; Nanamiya et al., 2010; Yano et al., 2013). However, in our analysis, rrn operon copy number was not correlated with the estimated growth rate parameters. Instead, the growth dynamics of the Bacillus from the CCB may be dictated by a combination of different physiological responses that are uncoupled from the rrn operon copies, such as differences in transcription rates or intra-cellular allocation processes.
Among the various growth dynamics of the strains from the CCB, we observed not only that many parameters were out of range when compared with those estimated for other previously studied Bacillus but also that they presented high variability that at first inspection, does not seem to relate either to evolutionary history, isolation site, or rrn operon copy number. We hypothesize that the oligotrophic condition in the CCB may have had a significant effect on the Bacillus growth dynamics that contribute to this variability. For example, long adaptation times are related to stressful conditions affecting the speed of bacterial growth, like limited nutrient availability (Chorin et al., 1997; Schaechter, 2006; Antolinos et al., 2011, 2012; Bren et al., 2013). Furthermore, estimates of maximum biomass reached can be quite variable because of the influence of overall nutrient availability and sensitivity to the waste material that accumulates during the exponential phase (Buchanan et al., 1997; Chorin et al., 1997). In addition, long doubling times are characteristic of bacteria in oligotrophic environments (Vieria-Silva and Rocha, 2015). Indeed, bacteria from other ecosystems that are extremely limited in terms of nutrients can achieve generation times of thousands of years (Jørgensen and Boetius, 2007; Labonté et al., 2015). Thus, the bacterial growth properties of Bacillus that live in the extremely oligotrophic ecosystems of the CCB likely involve a complex response to environmental and nutritional conditions acting in concert with genomic potential.
rrn Operon Copy Number and Phosphorus Availability in the CCB
As previously mentioned, ecosystems in the CCB are characterized by very low P availability in water, soil, and sediments (Elser et al., 2005; Peimbert et al., 2012), a condition that makes its high variation in rrn operon copy number somewhat surprising because we expected that such habitats would be dominated by taxa with low operon copy numbers. Previous work has suggested that rrn operon copy number is related to nutrient availability and especially with P because the copy number is linked to the growth rate, which is associated with the production of P-rich rrn (Elser, 2003; Jeyasingh and Weider, 2007). Indeed, it has been shown that bacteria that live in oligotrophic environments do tend to have very low rrn operon copy numbers (< two copies; Fegatella et al., 1998; Strehl et al., 1999; Lauro et al., 2009). In theory, multiple copies could allow a growth rate advantage when resources are abundant but would impose a competitive cost when resources are limited due to the costs of the over-production of rrn (Weider et al., 2005; Dethlefsen and Schmidt, 2007; Jeyasingh and Weider, 2007). While we detected many Bacillus isolates with six rrn operon copies, even this number is relatively high in comparison with other bacterial lineages that live under low nutrient conditions (Fegatella et al., 1998; Strehl et al., 1999; Lauro et al., 2009). Thus, it seems that the number of copies of the rrn operon is a functional trait related with the evolutionary history of the genus Bacillus, and defines its ecological versatility and adaptability to different environmental conditions (Klappenbach et al., 2000; Feldgarden et al., 2003; Stevenson and Schmidt, 2004; Connor et al., 2010).
A simple explanation of this incongruity is that the genus Bacillus does not pay for the full cost of high copy number under nutrient limitations due to its ability to escape from scarcity by forming spores and then germinating under better conditions. Indeed, stress response capacity has been shown to be related to rrn operon multiplicity (Nanamiya et al., 2010; Yano et al., 2013). However, spore formation in Bacillus of the CCB is not a given, due to the loss of many of the genes of the spore-forming complex in the sequenced genomes (Alcaraz et al., 2010) and the difficulty of obtaining spores experimentally in all the isolated strains. Nevertheless, B. coahuilensis m4-4 and B. sp. m3-13 have eight and nine rrn operon copies, respectively, like other spore-forming Bacilli (B. halodurans C-125, eight rrn operon copies; B. amyloliquefaciens CC178, 9 rrn operon copies). Thus, the variability in the rrn operon copy number could reflect broader life history strategies in which each taxa uses different sets of resources or inhabits distinct microhabitats in structured sediments, potentially decreasing competition. In agreement with this view, various bacterial communities have been shown to have similar functional heterogeneity regarding other important ecological traits (Martínez-Alonso et al., 2004; Giovannoni and Stingl, 2005; Martiny et al., 2006). Additional work is needed to understand the importance of conserving high rrn operon copy numbers in CCB Bacillus.
Our overall findings about diversity in Bacillus rrn operon number in the CCB do not seem to conform to the broader context of the GRH (Elser et al., 2000; Elser, 2006). For example, isolate A (B. cereus) had the highest number of copies of the rrn operon (14) and also had the slowest growth rate and highest N:P ratio (965) when growing on media produced using the CCB’s natural waters. Furthermore, several isolates with low rrn operon numbers showed some of the highest P-content values, contrary to the GRH. However, the high N:P ratio and slow growth rate of isolate A is consistent with the development of severe P limitations for this high-copy-number strain, resulting in its high biomass N:P ratio. Then, it seems that the isolates with lower number of copies of the rrn operon may cope better in this oligotrophy of the CCB. To more effectively test the genetic dimension of the GRH, each strain would need to be raised under optimal conditions at its genetically constrained maximal growth rate. Indeed, this inference is supported by the higher dispersion of growth parameters seen for high-copy-number strains. That is, low-copy-number strains may have a limited range of growth variation, regardless of media, while high-copy-number strains may have a considerable range of growth, depending on whether or not the environment is well-matched to their needs. Another possible explanation for the decoupling of rrn operon number from growth rate and stoichiometric properties is that CCB Bacillus are selected for fine-tuned signaling with resource supplies or for variation in the rates of rRNA genes expression, thus disconnecting copy number from RNA production and growth rate (i.e., low-copy-strains may have high levels of transcription for each copy, while high-copy strains may more stringently express each copy). In any case, our data do not provide a clear resolution regarding the validity of the genetic components of the GRH within the Bacillus of the CCB. It is possible that the consideration of a broader range of bacterial taxa, as well as the more extensive testing of growth conditions, are needed in order to more rigorously test the GRH in the microbial realm, using approaches that can overcome the possible impacts of physiological conditions, phylogenetic inertia, and taxon-specific lifecycle strategies (e.g., sporulation) in terms of confounding the interpretations.
We suggest that the growth patterns described in the Bacillus isolates from the CCB, as well as the high variability in rrn operon copy number, represent ecological strategies that allow them to persist in this oligotrophic ecosystem. Analogous strategies have been described in other organisms whose growth parameters are also affected by various environmental and biotic factors (Pianka, 1970; Page, 2002; Lipowsky et al., 2012). Variable lag phases may help in adapting to changing environmental conditions to reach optimum growth with long generation times (Crooks, 2005; Wangen and Webster, 2006; Daehler, 2009). We also note that the retardation of growth is a common result of intense interaction with other organisms, as well as stressful conditions (Gao et al., 2013; Tsugama et al., 2014). To date, these ecological strategies in bacteria have been largely related only to nutrient availability (Klappenbach et al., 2000; Fierer et al., 2007; Shrestha et al., 2007). However, the intensity of direct inter-specific interactions (such as chemical antagonisms) can also establish coexistence, and these interactions are known to be particularly intense in the CCB (Souza et al., 2012; Pérez-Gutiérrez et al., 2013; Aguirre-von-Wobeser et al., 2014). These inferences suggest a bet-hedging strategy on the part of CCB bacteria in which expression of RNA genes is tightly controlled due to low P-conditions (reflected in their overall low P content and high N:P ratio), but when chemical antagonism is successful, resources suddenly arrive, and the rrn operons are activated to grow rapidly under the nutrient bounty.
The main results of our work indicate that the rrn operon copy number exhibits considerable variation among field-isolated Bacilli and that considerable variation also exist in their growth properties and chemical composition. However, rrn operon copy number appears to be largely uncoupled from growth and chemical properties in this clade. Further investigation is needed to understand the ecological and physiological importance of this rrn operon variability, as mediated by gene transcriptional regulation, and its influence on ribosome and protein content and thus N:P stoichiometry (Gourse et al., 1996; Fegatella and Cavicchioli, 2000; Dethlefsen and Schmidt, 2007; Scott et al., 2010; Piir et al., 2011). It may be that the extreme oligotrophic conditions in the CCB have imposed important physiological constraints on resource allocation and growth rate, as well as the expression of the rRNA genes, and thus, the rate of production of ribosomes per rrn operon copy differs considerably among strains. More detailed studies, including competition experiments involving Bacillus strains isolated from different CCB environments and subject to various environmental limitations (such as differences in nutrient supply concentrations, ratios, and supply schedules), may be needed to identify the ecological and evolutionary significance of rrn operon copy number variation among microbes in the habitats of Cuatro Ciénegas and similar nutrient-deficient habitats.
Author Contributions
JV-A: Primary author, experimental design, amplification and analysis of genetic material, PFGE, analysis of data. LE-F: Experimental design, analysis of data. GD-S: PFGE standardization, analysis of data. PM-Z: Growth curves standardization, parameter estimations. JG-P: Statistical analysis. JL: Cell contents of phosphorus (P) and nitrogen (N). JE: Experimental design, analysis of data. GO-A: Experimental design, analysis of data. VS: Group leader, experimental design, analysis of data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks are due to Posgrado en Ciencias Biológicas of the Universidad Nacional Autónoma de México (UNAM). This work is the doctoral research of JV-A in the Doctorado en Ciencias Biológicas program (CVU: 216032; CONACyT fellow number: 207187). We would like to thank Ariadnna del C. Cruz-Córdoba and Juan Xicohtencatl-Cortés of the Laboratorio de Bacteriología Intestinal (Hospital Infantil de México “Federico Gómez”), and José Luis Méndez and Rosario Morales of the Laboratorio de Genómica Bacteriana (UNAM), because of their technical assistance in PFGE standardization, Africa Islas of the Laboratorio de Bacteriología Molecular (CINVESTAV-Unidad Irapuato) for providing the strains needed to perform this work, Felipe García-Oliva (Biogeoquímica de Suelos; CIEco, UNAM) and Mario Soberón-Chávez (Departamento de Microbiología Molecular; IBT, UNAM) for their advice and input of ideas throughout the development of this work, Ana Gutiérrez-Preciado (Institut Cavanilles de Biodiversitat i Biologia Evolutiva, Universitat de València) for her comments and observations that contributed to the improvement of the manuscript. Grants from Alianza WWF-Fundación Carlos Slim to VS and LE-F and U.S. NSF (DEB-0950179) to JE supported this study. Laura Espinosa-Asuar and Erika Aguirre-Planter helped in the general technical logistics during the entire project.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01486
FIGURE S1 | Growth conditions assays and Gompertz growth curves of the Bacillus isolates from the CCB. (A) Different nutritional conditions were analyzed to define the optimum conditions and thus describe the growth dynamics of the Bacillus: A = Marine broth (DifcoTM 2216); B = CCB water; C = CCB water + P (KH2PO4, 5 g per liter); and D = CCB water + N (tryptone, 5 g per liter). Growth was quantified via culture turbidity (OD600; BioPhotometer Plus, Eppendorf) after 12 h of growth, and the data were normalized to define the optimal conditions for growth. (B) Growth curves were described after linear regression analysis according to the Gompertz equation. Isolate names were in agreement with the maximum growth reached (A), followed by the number of copies of the rrn operon for every isolate. Growth curves were plotted with the data of the plate count assays [Ln(CFU/CFU(t = 0))].
FIGURE S2 | rRNA operon copy number in the genus Bacillus isolated in the CCB. For every isolate, the separation of fragments with PFGE is shown on the left and Southern-blot confirmation is shown on the right. At the bottom of the figure is the total number of rRNA operon copies quantified. The red asterisk represents the fragments identified with hybridization analysis.
FIGURE S3 | Relationship between the growth parameters estimated in the Bacillus isolates from the CCB. (A) λ vs μmax and, (B) μmax vs A. Isolate names (letters, from A to P) are within the circles in black and were in agreement with the maximum biomass reached (A) in Table 1. A = maximum biomass reached [Ln(CFU/CFU(t=0))]; λ = lag phase; μmax = maximum specific growth rate.
FIGURE S4 | Relative contribution (%) per variable in the PCA of the growth parameters estimated in the Bacillus isolates from the CCB. A = maximum growth reached; λ = lag phase; td = doubling time; and μmax = maximum specific growth rate.
References
- Acinas S. G., Marcelino L. A., Klepac-Ceraj V., Polz M. F. (2004). Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186 2629–2635. 10.1128/JB.186.9.2629-2635.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-von-Wobeser E., Soberón-Chávez G., Eguiarte L. E., Ponce-Soto G. Y., Vázquez-Rosas-Landa M., Souza V. (2014). Two-role model of an interaction network of free-living γ-proteobacteria from an oligotrophic environment. Environ. Microbiol. 16 1366–1377. 10.1111/1462-2920.12305 [DOI] [PubMed] [Google Scholar]
- Alcaraz L. D., Moreno-Hagelsieb G., Eguiarte L. E., Souza V., Herrera-Estrella L., Olmedo G. (2010). Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics. BMC Genomics 11:332 10.1186/1471-2164-11-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaraz L. D., Olmedo G., Bonilla G., Cerritos R., Hernández G., Cruz A., et al. (2008). The genome of Bacillus coahuilensis reveals adaptations essential for survival in the relic of an ancient marine environment. Proc. Natl. Acad. Sci. U.S.A. 105 5803–5808. 10.1073/pnas.0800981105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolinos V., Muñoz M., Ros-Chumillas M., Aznar A., Periago P. M., Fernández P. S. (2011). Combined effect of lysozyme and nisin at different incubation temperature and mild heat treatment on the probability of time growth of Bacillus cereus. Food Microbiol. 28 305–310. 10.1016/j.fm.2010.07.021 [DOI] [PubMed] [Google Scholar]
- Antolinos V., Muñoz-Cuevas M., Ros-Chumillas M., Periago P. M., Fernández P. S., Marc Y. L. (2012). Modelling the effects of temperature and osmotic shifts on the growth kinetics of Bacillus weihenstephanensis in broth and food products. Int. J. Food Microbiol. 158 36–44. 10.1016/j.ijfoodmicro.2012.06.017 [DOI] [PubMed] [Google Scholar]
- APHA (2005). Standard Methods for the Examination of Water and Wastewater. Washington, DC: American Publish Health Association. [Google Scholar]
- Bren A., Hart Y., Dekel E., Koster D., Alon U. (2013). The last generation of bacterial growth in limiting nutrient. BMC Syst. Biol. 7:27 10.1186/1752-0509-7-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan R. L., Whiting R. C., Damert W. C. (1997). When in simple good enough: a comparison of the Gompertz, Baranyi, and three-phase linear models for fitting bacterial growth curves. Food Microbiol. 14 313–326. 10.1006/fmic.1997.0125 [DOI] [Google Scholar]
- Burdett I. D. J., Kirkwood T. B. L., Whalley J. B. (1986). Growth kinetics of individual Bacillus subtilis cells and correlation with nucleoid extension. J. Bacteriol. 167 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavicchioli R., Ostrowski M., Fegatella F., Goodchild A., Guixa-Boixereu N. (2003). Life under nutrient limitation in oligotrophic marine environments: an eco/physiological perspective of Sphingopyxis alaskensis (formerly Sphingophyxis alaskensis). Microb. Ecol. 45 203–217. [DOI] [PubMed] [Google Scholar]
- Cerritos R., Eguiarte L. E., Avitia M., Siefert J., Travisano M., Rodríguez-Verdugo A., et al. (2010). Diversity of culturable thermo-resistant aquatic bacteria along an environmental gradient in Cuatro Ciénegas, Coahuila, México. Antonie Van Leeuwenhoek 98 1–16. 10.1007/s10482-010-9490-9 [DOI] [PubMed] [Google Scholar]
- Cerritos R., Vinuesa P., Eguiarte L. E., Herrera-Estrella L., Alcaraz-Peraza L. D., Arvizu-Gómez J. L., et al. (2008). Bacillus coahuilensis sp. nov., a moderately halophilic species from a desiccated lagoon in the Cuatro Cienegas valley in Coahuila, Mexico. Int. J. Syst. Evol. Microbiol. 58 919–923. 10.1099/ijs.0.64959-0 [DOI] [PubMed] [Google Scholar]
- Chorin E., Thuault D., Cléret J. J., Bourgeois C. M. (1997). Modeling Bacillus cereus growth. Int. J. Food Microbiol. 38 229–234. 10.1016/S0168-1605(97)00110-4 [DOI] [PubMed] [Google Scholar]
- Chubukov V., Sauer U. (2014). Environmental dependence of stationary-phase metabolism in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 80 2901–2909. 10.1128/AEM.00061-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codon C., Liveris D., Squires C., Schwartz I., Squires C. L. (1995). rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177 4152–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. F., Richmound M. H. (1962). Rate of increase in length of bacteria between divisions. J. Gen. Microbiol. 28 15–33. 10.1099/00221287-28-1-15 [DOI] [PubMed] [Google Scholar]
- Connor N., Sikorski J., Rooney A. P., Kopac S., Koeppel A. F., Burger A., et al. (2010). Ecology of speciation in the genus Bacillus. Appl. Environ. Microbiol. 76 1349–1358. 10.1128/AEM.01988-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks J. A. (2005). Lag times and exotic species: the ecology and management of biological invasions in slow-motion. Ecoscience 12 316–329. 10.2980/i1195-6860-12-3-316.1 [DOI] [Google Scholar]
- Daehler C. C. (2009). Short lag times for invasive tropical plants: evidence from experimental planting in Hawaii. PLoS ONE 4:e4462 10.1371/journal.pone.0004462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado G., Souza V., Morales R., Cerritos R., González-González A., Méndez J. L., et al. (2013). Genetic characterization of atypical Citrobacter freundii. PLoS ONE 8:e74120 10.1371/journal.pone.0074120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., Schmidt T. M. (2007). Performance of the translational apparatus varies with the ecological strategies of bacteria. J. Bacteriol. 189 3237–3245. 10.1128/JB.01686-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser J. J. (2003). Biological stoichiometry: a theoretical framework connecting ecosystem ecology, evolution, and biochemistry for application in astrobiology. Int. J. Astrobiol. 2 185–193. 10.1017/S1473550403001563 [DOI] [Google Scholar]
- Elser J. J. (2006). Biological stoichiometry: a chemical bridge between ecosystem ecology and evolutionary biology. Am. Nat. 168 S25–S35. 10.1086/509048 [DOI] [PubMed] [Google Scholar]
- Elser J. J., Sterner R. W., Gorokhova E., Fagan W. F., Markow T. A., Cotner J. B., et al. (2000). Biological stoichiometry from genes to ecosystems. Ecol. Lett. 3 540–550. 10.1046/j.1461-0248.2000.00185.x [DOI] [Google Scholar]
- Elser J. J., Watts J., Schampel J. H., Farmer J. (2005). Early Cambrian food webs on a trophic knife-edge? A hypothesis and preliminary data from a modern stromatolite-based ecosystem. Ecol. Lett. 9 295–303. 10.1111/j.1461-0248.2005.00873.x [DOI] [PubMed] [Google Scholar]
- Fegatella F., Cavicchioli R. (2000). Physiological responses to starvation in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 66 2037–2044. 10.1128/AEM.66.5.2037-2044.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fegatella F., Lim J., Kjelleberg S., Cavicchioli R. (1998). Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64 4433–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldgarden M., Byrd N., Cohan F. M. (2003). Gradual evolution in bacteria: evidence from Bacillus systematics. Microbiology 149(Pt 12), 3565–3573. 10.1099/mic.0.26457-0 [DOI] [PubMed] [Google Scholar]
- Fierer N., Brandford M. A., Jackson R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88 1354–1364. 10.1890/05-1839 [DOI] [PubMed] [Google Scholar]
- Gao R., Wan Z. Y., Wong S. M. (2013). Plant growth retardation and conserved miRNAs are correlated to hibiscus chlorotic ringspot virus infection. PLoS ONE 8:e85476 10.1371/journal.pone.0085476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., Stingl U. (2005). Molecular diversity and ecology of microbial plankton. Nature 437 343–348. 10.1038/nature04158 [DOI] [PubMed] [Google Scholar]
- Goto K., Omura T., Hara Y., Sadaie Y. (2000). Application of the partial 16S rDNA sequence as an index for rapid identification of species in the genus Bacillus. J. Gen. Appl. Microbiol. 46 1–8. 10.2323/jgam.46.1 [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Gaal T., Bartlett M. S., Appleman J. A., Ross W. (1996). rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50 645–677. 10.1146/annurev.micro.50.1.645 [DOI] [PubMed] [Google Scholar]
- Green J. L., Bohannan B. J. M., Whitaker R. J. (2008). Microbial biogeography: from taxonomy to traits. Science 320 1039–1042. 10.1126/science.1153475 [DOI] [PubMed] [Google Scholar]
- Hughes A. R., Inouye B. D., Johnson M. T. J., Underwood N., Vellend M. (2008). Ecological consequences of genetic diversity. Ecol. Lett. 11 609–623. 10.1111/j.1461-0248.2008.01179.x [DOI] [PubMed] [Google Scholar]
- Jeyasingh P. D., Weider L. J. (2007). Fundamental links between genes and elements: evolutionary implications of ecological stoichiometry. Mol. Ecol. 16 4649–4661. 10.1111/j.1365-294X.2007.03558.x [DOI] [PubMed] [Google Scholar]
- Jørgensen B. B., Boetius A. (2007). Feast and famine-microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5 770–781. 10.1038/nrmicro1745 [DOI] [PubMed] [Google Scholar]
- Kacena M. A., Merrell G. A., Manfredi B., Smith E. E., Klaus D. M., Todd P. (1999). Bacterial growth in space flight: logistic growth curve parameters for Escherichia coli and Bacillus subtilis. Appl. Microbiol. Biotechnol. 51 229–234. 10.1007/s002530051386 [DOI] [PubMed] [Google Scholar]
- Klappenbach J. A., Dunbar J. M., Schmidt T. M. (2000). rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66 1328–1333. 10.1128/AEM.66.4.1328-1333.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klappenbach J. A., Saxman P. R., Cole J. R., Schmidt T. M. (2001). rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29 181–184. 10.1093/nar/29.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté J. M., Field E. K., Lau M., Chivian D., Van Heerden E., Wommack K. E., et al. (2015). Single cell genomics indicates horizontal gene transfer and viral infections in a deep subsurface Firmicutes population. Front. Microbiol. 22:349 10.3389/fmicb.2015.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro F. M., McDougald D., Thomas T., Williams T. J., Egan S., Rice S., et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 106 15527–15533. 10.1073/pnas.0903507106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky A., Roscher C., Schumacher J., Schmid B. (2012). Density-independent mortality and increasing plant diversity are associated with differentiation of Taraxacum officinale into r-and K-strategists. PLoS ONE 7:e28121 10.1371/journal.pone.0028121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Hessel A., Sanderson K. E. (1993). Genomic mapping with I-CeuI, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U.S.A. 90 6874–6878. 10.1073/pnas.90.14.6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loladze I., Elser J. J. (2011). The origins of the Redfield nitrogen-to-phosphorus ratio are in a homoeostatic protein-to-rRNA ratio. Ecol. Lett. 14 244–250. 10.1111/j.1461-0248.2010.01577.x [DOI] [PubMed] [Google Scholar]
- Maciá E. (2005). The role of phosphorus in chemical evolution. Chem. Soc. Rev. 34 691–701. 10.1039/b416855k [DOI] [PubMed] [Google Scholar]
- Martínez-Alonso M., Mir J., Caumette P., Gaju N., Guerrero R., Esteve I. (2004). Distribution of phototrophic populations and primary production in a microbial mat from the Ebro Delta, Spain. Int. Microbiol. 7 19–25. [PubMed] [Google Scholar]
- Martiny A. C., Coleman M. L., Chisholm S. W. (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc. Natl. Acad. Sci. U.S.A. 103 12552–12557. 10.1073/pnas.0601301103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny A. C., Huang Y., Li W. (2009). Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ. Microbiol. 11 1340–1347. 10.1111/j.1462-2920.2009.01860.x [DOI] [PubMed] [Google Scholar]
- McGill B. J., Enquist B. J., Weiher E., Westoby M. (2006). Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21 178–185. 10.1016/j.tree.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Membré J. M., Leporq B., Vialette M., Mettler E., Perrier L., Thuault D., et al. (2005). Temperature effect on bacterial growth rate: quantitative approach including cardinal values and variability estimates to perform growth simulations on/in food. Int. J. Food Microbiol. 100 179–186. 10.1016/j.ijfoodmicro.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Monod J. (1949). The growth of bacterial cultures. Annu. Rev. Microbiol. 3 371–394. 10.1146/annurev.mi.03.100149.002103 [DOI] [Google Scholar]
- Moreno-Letelier A., Olemdo G., Eguiarte L. E., Martinez-Castilla L., Souza V. (2011). Parallel evolution and horizontal gene transfer of the pst operon in Firmicutes from oligotrophic environments. Int. J. Evol. Biol. 2011:781642 10.4061/2011/781642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasone K., Noriaki M., Yoshihiro T., Rumie S., Go M., Tokuki S., et al. (2000). Characterization and comparative study of the rrn operons of alkaliphilic Bacillus halodurans C-125. Extremophiles 4 209–214. 10.1007/PL00010713 [DOI] [PubMed] [Google Scholar]
- Nanamiya H., Sato M., Masuda K., Sato M., Wada T., Suzuki S., et al. (2010). Bacillus subtilis mutants harbouring a single copy of the rRNA operon exhibit severe defects in growth and sporulation. Microbiology 156 2944–2952. 10.1099/mic.0.035295-0 [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C. (1999). Bacterial growth: constant obsession with dN/dt. J. Bacteriol. 181 7405–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. M., Schaffner D. W. (1997). Mathematical models for the effects of pH, temperature, and sodium chloride on the growth of Bacillus stearothermophilus in salty carrots. Appl. Environ. Microbiol. 63 1237–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page C. N. (2002). Ecological strategies in fern evolution: a neopteridological overview. Rev. Palaeobot. Palynol. 119 1–33. 10.1016/S0034-6667(01)00127-0 [DOI] [Google Scholar]
- Palmisano M. M., Nakamura L. K., Duncan K. E., Istock C. A., Cohan F. M. (2001). Bacillus sonorensis sp. nov., a close relative of Bacillus licheniformis, isolated from soil in the Sonoran Desert, Arizona. Int. J. Syst. Evol. Microbiol. 51 1671–1679. 10.1099/00207713-51-5-1671 [DOI] [PubMed] [Google Scholar]
- Peimbert M., Alcaraz L. D., Bonilla-Rosso G., Olmedo-Alvarez G., García-Oliva F., Segovia L., et al. (2012). Comparative metagenomics of two microbial mats at Cuatro Ciénegas Basin I: ancient lessons on how to cope with an environment under severe nutrient stress. Astrobiology 12 648–658. 10.1089/ast.2011.0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Gutiérrez R. A., López-Ramírez V., Islas A., Alcaraz L. D., Hernández-González I., Luna Oliviera B. C., et al. (2013). Antagonism influences assembly of a Bacillus guild in a local community and is depicted as a food-chain network. ISME J. 7 487–497. 10.1038/ismej.2012.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianka E. R. (1970). On r- and K-Selection. Am. Nat. 104 592–597. 10.1086/282697 [DOI] [Google Scholar]
- Piir K., Paier A., Liiv A., Tenson T., Maiväli Ü. (2011). Ribosome degradation in growing bacteria. EMBO Rep. 12 458–462. 10.1038/embor.2011.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell E. O. (1956). Growth rate and generation time of bacteria, with special reference to continuous culture. J. Gen. Microbiol. 15 492–511. 10.1099/00221287-15-3-492 [DOI] [PubMed] [Google Scholar]
- Prüβ B. M., Francis K. P., von Stetten F., Scherer S. (1999). Correlation of 16S ribosomal DNA signature with temperature-dependent growth rates of mesophilic and psychrotolerant strains of the Bacillus cereus group. J. Bacteriol. 181 2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi R., Wu M., Dasgupta I., Fox G. E. (2009). Visualization of ribosomal RNA operon copy number distribution. BMC Microbiol. 9:208 10.1186/1471-2180-9-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratkowsky D. A., Lowry R. K., McMeekin T. A., Stokes A. N., Chandler R. E. (1983). Model for bacterial culture growth rate troughout the entire biokinetic temperature range. J. Bacteriol. 154 1222–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press, 31–39. [Google Scholar]
- Schaechter M. (2006). From growth physiology to system biology. Int. Microbiol. 9 157–161. [PubMed] [Google Scholar]
- Scott M., Gunderson C. W., Mateescu E. M., Zhang Z., Hwa T. (2010). Interdependence of cell growth and gene expression: origins and consequences. Science 330 1099–1102. 10.1126/science.1192588 [DOI] [PubMed] [Google Scholar]
- Shrestha P. M., Noll M., Liesack W. (2007). Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ. Microbiol. 9 2464–2474. 10.1111/j.1462-2920.2007.01364.x [DOI] [PubMed] [Google Scholar]
- Souza V., Eguiarte L. E., Siefert J. S., Elser J. J. (2008). Microbial endemism: does phosphorous limitation enhance speciation? Nat. Rev. Microbiol. 6 559–564. 10.1038/nrmicro1917 [DOI] [PubMed] [Google Scholar]
- Souza V., Espinosa-Asuar L., Escalante A. E., Eguiarte L. E., Farmer L. E., Forney L., et al. (2006). An endangered oasis of aquatic microbial biodiversity in the Chihuahuan desert. Proc. Natl. Acad. Sci. U.S.A. 103 6565–6570. 10.1073/pnas.0601434103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza V., Siefert J. L., Escalante A. E., Elser J. J., Eguiarte L. E. (2012). The Cuatro Ciénegas Basin in Coahuila, Mexico: an astrobiological Precambriam park. Astrobiology 12 641–647. 10.1089/ast.2011.0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. C., Tyerman J., Bertrand M., Doebeli M. (2008). Adaptation increases the likelihood of diversification in an experimental bacterial linage. Proc. Natl. Acad. Sci. U.S.A. 105 1585–1589. 10.1073/pnas.0708504105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner R. W., Elser J. J. (2002). Ecological Stoichiometry: The Biology of Elements from Molecules to the Biosphere. Princeton, NJ: Princeton University Press. [Google Scholar]
- Stevenson B. S., Schmidt T. M. (2004). Life history implications of rRNA gene copy number in Escherichia coli. Appl. Environ. Microbiol. 70 6670–6677. 10.1128/AEM.70.11.6670-6677.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard S. F., Smith B. J., Hein R., Roller B. R. K., Schmidt T. M. (2015). rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43 D593–D598. 10.1093/nar/gku1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehl B., Holtzendorff J., Partensky F., Hess W. R. (1999). A small and compact genome in the marine cyanobacterium Prochlorococcus marinus CCMP 1375: lack of an intron in the gene for tRNAr (Leu)UAA and a single copy of the rRNAop. FEMS Microbiol. Lett. 181 261–266. 10.1111/j.1574-6968.1999.tb08853.x [DOI] [PubMed] [Google Scholar]
- Sutherland J. P., Aherne A., Beaumont A. L. (1996). Preparation and validation of growth model for Bacillus cereus: the effects of temperature, pH, sodium chloride and carbon dioxide. Int. J. Food Microbiol. 30 359–372. 10.1016/0168-1605(96)00962-2 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Higgins D. G. (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics Chapter 2 Unit2.3 10.1002/0471250953.bi0203s00 [DOI] [PubMed] [Google Scholar]
- Tsugama D., Liu S., Takano T. (2014). Analysis of functions of VIP1 and its close homologs in osmosensory responses of Arabidopsis thaliana. PLoS ONE 9:e103930 10.1371/journal.pone.0103930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valík L., Görner F., Lauková D. (2003). Growth dynamics of Bacillus cereus and self-life of pasteurised milk. Czech J. Food Sci. 21 195–202. [Google Scholar]
- Van Mooy B. A. S., Fredricks H. F., Pedler B. E., Dyhrman S. T., Karl D. M., Koblízek M., et al. (2009). Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458 69–72. 10.1038/nature07659 [DOI] [PubMed] [Google Scholar]
- Vieria-Silva S., Rocha E. P. C. (2015). The systemic imprint of growth and its uses in ecological (Meta) genomics. PLoS Genet. 6:e1000808 10.1371/journal.pgen.1000808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskaya T. A., Kathariou S., Tiedje J. M. (2009). The Exiguobacterium genus: biodiversity and biogeography. Extremophiles 13 541–555. 10.1007/s00792-009-0243-5 [DOI] [PubMed] [Google Scholar]
- Wangen S. R., Webster C. R. (2006). Potential for multiple lag phases during biotic invasions: reconstructing an invasion of the exotic tree Acer platanoides. J. Appl. Ecol. 43 258–268. 10.1111/j.1365-2664.2006.01138.x [DOI] [Google Scholar]
- Warth A. D. (1978). Relationship between the heat resistance of spores and the optimum and maximum growth temperatures of Bacillus species. J. Bacteriol. 134 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider L. J., Elser J. J., Crease T. J., Mateos M., Cotner J. B., Markow T. A. (2005). The functional significance of ribosomal (r) DNA variation: impacts on the evolutionary ecology of organisms. Annu. Rev. Ecol. Evol. Syst. 36 219–242. 10.1146/annurev.ecolsys.36.102003.152620 [DOI] [Google Scholar]
- Whitaker R. J., Grogan D. W., Taylor J. W. (2003). Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301 976–978. 10.1126/science.1086909 [DOI] [PubMed] [Google Scholar]
- Xu J. (2006). Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Mol. Ecol. 15 1713–1731. 10.1111/j.1365-294X.2006.02882.x [DOI] [PubMed] [Google Scholar]
- Yano K., Wada T., Suzuki S., Tagami K., Matsumoto T., Shiwa Y., et al. (2013). Multiple rRNAops are essential for efficient cell growth and sporulation as well as outgrowth in Bacillus subtilis. Microbiology 159 2225–2236. 10.1099/mic.0.067025-0 [DOI] [PubMed] [Google Scholar]
- Zwietering M. H., Jongenburger I., Rombouts F. M., van’t Riet K. (1990). Modeling of the bacterial growth cure. Appl. Environ. Microbiol. 56 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | Growth conditions assays and Gompertz growth curves of the Bacillus isolates from the CCB. (A) Different nutritional conditions were analyzed to define the optimum conditions and thus describe the growth dynamics of the Bacillus: A = Marine broth (DifcoTM 2216); B = CCB water; C = CCB water + P (KH2PO4, 5 g per liter); and D = CCB water + N (tryptone, 5 g per liter). Growth was quantified via culture turbidity (OD600; BioPhotometer Plus, Eppendorf) after 12 h of growth, and the data were normalized to define the optimal conditions for growth. (B) Growth curves were described after linear regression analysis according to the Gompertz equation. Isolate names were in agreement with the maximum growth reached (A), followed by the number of copies of the rrn operon for every isolate. Growth curves were plotted with the data of the plate count assays [Ln(CFU/CFU(t = 0))].
FIGURE S2 | rRNA operon copy number in the genus Bacillus isolated in the CCB. For every isolate, the separation of fragments with PFGE is shown on the left and Southern-blot confirmation is shown on the right. At the bottom of the figure is the total number of rRNA operon copies quantified. The red asterisk represents the fragments identified with hybridization analysis.
FIGURE S3 | Relationship between the growth parameters estimated in the Bacillus isolates from the CCB. (A) λ vs μmax and, (B) μmax vs A. Isolate names (letters, from A to P) are within the circles in black and were in agreement with the maximum biomass reached (A) in Table 1. A = maximum biomass reached [Ln(CFU/CFU(t=0))]; λ = lag phase; μmax = maximum specific growth rate.
FIGURE S4 | Relative contribution (%) per variable in the PCA of the growth parameters estimated in the Bacillus isolates from the CCB. A = maximum growth reached; λ = lag phase; td = doubling time; and μmax = maximum specific growth rate.