Abstract Abstract
Background
Over the past 50 years, Southeast Asia has suffered the greatest losses of biodiversity of any tropical region in the world. Malaysia is a biodiversity hotspot in the heart of Southeast Asia with roughly the same number of mammal species, three times the number of butterfly species, but only 4% of the land area of Australia. Consequently, in Malaysia, there is an urgent need for biodiversity monitoring and also public engagement with wildlife to raise awareness of biodiversity loss. Citizen science is “on the rise” globally and can make valuable contributions to long-term biodiversity monitoring, but perhaps more importantly, involving the general public in science projects can raise public awareness and promote engagement. Butterflies are often the focus of citizen science projects due to their charisma and familiarity and are particularly valuable “ambassadors” of biodiversity conservation for public outreach.
New information
Here we present the data from our citizen science project, the first “Peninsular Malaysia Butterfly Count”. Participants were asked to go outdoors on June 6, 2015, and (non-lethally) sample butterfly legs for species identification through DNA barcoding. Fifty-seven citizens responded to our adverts and registered to take part in the butterfly count with many registering on behalf of groups. Collectively the participants sampled 220 butterfly legs from 26 mostly urban and suburban sampling localities. These included our university campus, a highschool, several public parks and private residences. On the basis of 192 usable DNA barcodes, 43 species were sampled by the participants. The most sampled species was Appias olferna, followed by Junonia orithya and Zizina otis. Twenty-two species were only sampled once, five were only sampled twice, and four were only sampled three times. Three DNA barcodes could not be assigned species names. The sampled butterflies revealed that widely distributed, cosmopolitan species, often those recently arrived to the peninsula or with documented "invasive" potential, dominated the habitat types sampled by the participants. Data from this first Butterfly Count helps establish a baseline from which we can monitor the patterns and changes in butterfly communities in Peninsular Malaysia.
Keywords: DNA barcoding, species identification, Barcode Index Numbers, butterflies, citizen science, Malaysia, invasive species
Background
Citizen Science
Over the past 50 years, Southeast Asia has suffered the greatest losses of biodiversity of any tropical region in the world (Gibson et al. 2011). Malaysia is a biodiversity hotspot in the heart of Southeast Asia with roughly the same number of mammal species, three times the number of butterfly species, but only 4% of the land area of Australia (Table 1). Consequently, in Malaysia, there is urgent need for biodiversity monitoring and also public engagement with wildlife to raise awareness of biodiversity loss.
Table 1.
Comparison of Malaysia and Australia in terms of demography, biogeography, and public engagement with biodiversity.
| Country | Malaysia | Australia |
| Human population | 30,608,552 (http://www.statistics.gov.my/) |
23,849,269 (http://www.abs.gov.au/) |
| Land area (km2)(http://www.data.un.org) | 330,803 | 7,692,024 |
| World bank status in 2015 (http://data.worldbank.org) | Upper-middle-income economy (Vision to be high-income economy by 2020; http://rmk11.epu.gov.my) |
High-income economy |
| Internet users in 2014 (http://www.InternetLiveStats.com) | 20,140,125 | 21,176,595 |
| Facebook users in 2012 (http://www.InternetLiveStats.com) | 13,589,520 | 11,808.360 |
| Butterfly species | 1,182 (http://malaysiabutterflies.myspecies.org) |
416 (Braby 2004) |
| Endangered mammal species in 2015 (http://data.worldbank.org) |
70 | 55 |
| National nature societymembers in 2014 | 4,000 (Malaysian Nature Society; http://www.mns.my) |
40,000 (Wilderness Society; http://www.wilderness.org.au) |
| Participants in national birdcount in 2014 | 222 (http://www.mygardenbirdwatch.com) |
9,000 (http://aussiebirdcount.org.au) |
Citizen science is “on the rise” globally and can make valuable contributions to long-term biodiversity monitoring (Tulloch et al. 2013), although data tends to remain underutilised (Theobald et al. 2015). Perhaps even more importantly, involving the general public in science projects can raise public awareness and promote civic engagement (Loos et al. 2015). Citizen science is well established in high-income economies where projects regularly attract thousands of participants (e.g., 52,000 people took part in the UK’s Big Butterfly Count in 2015 http://www.bigbutterflycount.org; Table 1). In transitioning economies, such as Malaysia, citizen science is less mainstream, and efforts to engage citizens face a different set of challenges. These include lack of money, time and taxonomic skills among potential participants, but also mental, cultural and socio-economic barriers (Loos et al. 2015). Participants from high-income economies often contribute their own financial resources to citizen science activities, whereas in transitioning countries, participants may require financial support to cover the cost of materials (e.g., field guides, butterfly nets) (Loos et al. 2015). More complex factors influencing participation include low levels of interpersonal trust, civic participation and social capital among the populations of transitioning economies, and the dominance of individualistic values (Loos et al. 2015). Furthermore, corruption in transitioning economies seeds mistrust of formal, and even informal, institutions (Loos et al. 2015), and feeds apathy towards all civic activities.
Against the backdrop of these challenges, here we present data and insights from our citizen science project, the first “Peninsular Malaysia Butterfly Count”.
Butterfly Counts
Butterflies are often the focus of citizen science projects (e.g., http://scistarter.com/blog/2012/07/summer-is-busy-season-for-butterflies-and-citizen-scientists; http://www.pierisproject.org) due to their charisma and familiarity and are particularly valuable “ambassadors” of biodiversity conservation for public outreach (http://www.thestar.com.my/News/Education/2014/10/26/The-butterfly-effect/). Butterflies are thought to react rapidly to environmental changes due to their short generation time and high mobility (McIntyre 2000), and patterns of butterfly diversity are reflected in other distantly related taxonomic groups (e.g., bats; Syaripuddin et al. 2015) making them useful indicators of environmental change and degradation. Data concerning butterfly diversity is valuable in itself, as populations of butterflies are dwindling globally (New 1997) with tropical butterflies disappearing at the fastest rates (Brook et al. 2003). The butterflies of Peninsular Malaysia have been the focus of a series of comprehensive field guides written by British naturalists, beginning with Distant in 1882–1886, and followed by four editions of Corbet and Pendlebury’s classic checklist, first published in 1934 and most recently revised by Eliot in 1992 (Wilson et al. 2013). This latest edition, and accounting for some minor taxonomic changes since publication, puts the number of butterfly species recorded in Peninsular Malaysia at 1,182 (Table 1).
Material and methods
Preparing for Count Day
This project builds on our experience with another ongoing citizen science project, "The School Butterfly Project", which is reported elsewhere (Jisming-See et al. 2015). When we first thought about holding a butterfly count day, we searched the internet (using Google) to find out about similar projects held in other countries and discovered "Butterfly Education and Awareness Day" (also know by the acronym BEAD). BEAD is promoted by the Association for Butterflies as the first Saturday in June of each year (http://afbeducation.org/bead/). By coincidence, in Malaysia, the first Saturday of June is the Yang di-Pertuan Agong's (King's) official birthday, a public holiday. On such occasions families often like to go out for picnics in local parks, so it seemed ideal to have our first Butterfly Count on BEAD - June 6, 2015. We restricted our project to Peninsular Malaysia (West Malaysia) to avoid complications due to the shipping overseas and because the peninsula and East Malaysia (part of the island of Borneo) are governed by a different set of wildlife laws.
As facebook is very popular in Malaysia (Table 1) the "main face" of the project was a facebook page (http://www.facebook.com/butterflycount). We created an advertisement for the project in facebook and purchased “boost post” across Malaysia to encourage maximum participation across the peninsula. According to facebook statistics the advert "reached" 27,392 people at a cost of RM73.00. We also contacted national newspapers to request coverage to encourage registration. This request was taken up by two national English-language newspapers (http://www.star2.com/living/2015/05/11/help-count- butterflies/; http://www.nst.com.my/node/83845) and one national Chinese-language newspaper (the Sin Chew Daily; Fig. 1).
Figure 1.

Advertisement for the first Peninsular Malaysia Butterfly Count in the Sin Chew Daily national newspaper.
Interested citizens could register online, using a Google form (in English and Malay language) linked to the facebook page, or by phone. Registration was closed on May 31, 2015.
Registered citizens were sent (via Pos Laju) a Butterfly Count Pack containing:
i) Butterfly Count Guide (Suppl. material 1), which includes details about our motivation for running the project, a brief explanation of DNA barcoding, the plan for the count day, how to make a butterfly net, how to distinguish butterfly families, how to collect butterfly legs (a video was also available on the facebook page; Fig. 2), and a form to use on the count day.
Figure 2.
How to collect butterfly legs for DNA barcoding. The video is also available here https://youtu.be/yebuyCYRZzs.
ii) Ten 1.5ml microcentifuge tubes.
iii) Pair of tweezers.
iv) Butterfly net.
v) Prepaid addressed envelope (Pos Laju).
vi) Souvenir button badges.
Following guidelines in the Butterfly Count Guide (Suppl. material 1), citizens were asked to go outdoors on June 6, 2015, to collect butterfly legs and then mail their butterfly legs to the Museum of Zoology, University of Malaya, using the prepaid addressed envelope. Participants were also encouraged to share photographs taken on the Butterfly Count day on the Peninsular Malaysia Butterfly Count facebook page (https://www.facebook.com/butterflycount/photos_stream).
Butterfly Identification
In September 2015, the national parks board of Singapore (NParks), also conducted an inaugural butterfly count in neighbouring Singapore (https://www.nparks.gov.sg/butterflycount). The NParks program involved a butterfly identification training workshop and assigned participants to a specified count location, requiring a significant commitment (time) and investment (travel costs) by the participants. In contrast, for the Peninsular Malaysia Butterfly Count, in order to reduce costs and encourage participation, we allowed the participants to choose their own count location, and did not provide identification training (although a simple guide to distinguish butterfly families was provided in the Butterfly Count Guide). The participants were asked to collect non-lethal tissue samples (butterfly legs) to enable accurate species identification through DNA barcoding (a DNA barcode reference library for local butterfly species has been generated previously from museum specimens; Wilson et al. 2013). Such methods have been shown to have no effect on survivorship or reproductive potential of sampled butterflies (Crawford et al. 2013, Koscinski et al. 2011, Marschalek et al. 2013) and have been used previously in Peninsular Malaysia for butterflies surveys (Syaripuddin et al. 2015, Sing et al. 2015). This method also has the advantage of providing a more personal interaction with the butterflies, matching the project objective, rather than providing dubious identifications of species "on the wing". The family-level identifications, when attempted by the participants, were compared to those obtained by DNA barcoding.
DNA Barcoding
Genomic DNA was extracted from butterfly legs using a modified alkaline lysis method whereby legs were digested in 17.5 µl alkaline buffer for 20 minutes before adding 32.5 µl of neutralization buffer (following Ivanova et al. 2009). The DNA extracts were diluted 1/10 in ddH2O prior to PCR. All the DNA extracts were used for COI DNA barcoding following standard methods with the primer pair LCO1490 and HCO2198 (see Wilson 2012) or mlCOIintF and HCO2198 (Leray et al. 2013, Brandon-Mong et al. 2015). PCR amplification was performed in a 12 µl volume containing 0.125 µl of Accura Taq (Lucigen, USA), 6.25 µl of Accura 2x buffer, 1.0 µl of dNTP, 1.625 µl of ddH2O, 1.25 µl of each primer and 0.5 µl of diluted DNA. The thermocycle profile was 120 s at 94 °C followed by 40 cycles of 60 s at 94 °C, 60 s at 40 °C, 90 s at 72 °C, and a final extension step for 7 minutes at 72 °C. PCR products were visualised on a 1.5% agarose gel. PCR products were sequenced by a local company (MYTACG Bioscience, Malaysia) and the resulting chromatograms edited with CodonCode Aligner (CodonCode Corp.) and BioEdit (following Wilson 2012). The COI DNA barcodes, together with collection metadata, were submitted to the Barcode of Life Data systems (BOLD; Ratnasingham and Hebert 2007). The longer COI DNA barcodes (around 500bp or longer) were assigned to species on the basis of their BIN allocations (Ratnasingham and Hebert 2013). Shorter DNA barcodes, not allocated to BINS, were assigned species names based on >97% similarity with named DNA barcodes on BOLD.
Results
Participation
Fifty-seven citizens responded to our adverts and registered to take part in the Butterfly Count with 19 registering on behalf of groups, usually families (Fig. 3 but also one highschool. Of the 57 Butterfly Count Packs dispatched, we received 32 batches of butterfly legs (56% return rate). Some participants reported being unable to find butterflies on the count day, others told us they were afraid to collect butterfly legs and sent us photographs instead. Many required prompting via phone calls and Whatsapp messages before sending the butterfly legs to our Museum. The returned packages amounted to 220 butterfly legs from 26 mostly urban and suburban sampling localities These included our university campus, a highschool, several public parks and private residences (Fig. 4).
Figure 3.

Participants on the first Peninsular Malaysia Butterfly Count day.
Figure 4.
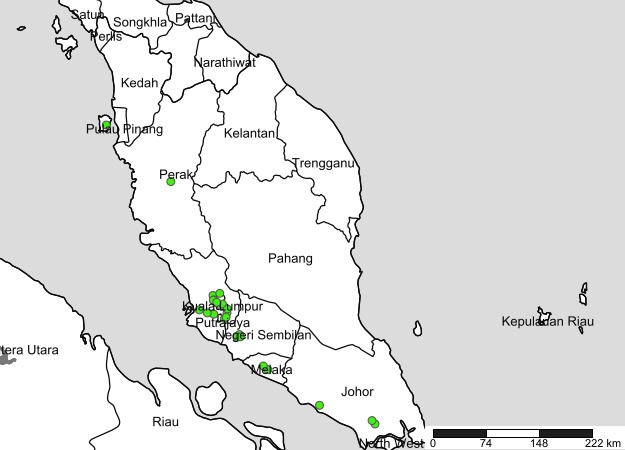
Sampling localities for the first Peninsular Malaysia Butterfly Count.
DNA Barcodes
Of the 220 legs received, 192 (87%) generated 'usable' DNA barcodes of varying quality and length. The DNA barcodes and associated collection data are available on BOLD in the publicly accessible datatset - Peninsular Malaysia Butterfly Count [PMBC] (http://www.boldsystems.org/index.php/Public_SearchTerms?query=<PMBC> and dx.doi.org/10.5883/DS-PMBC).
Butterfly Species Counted
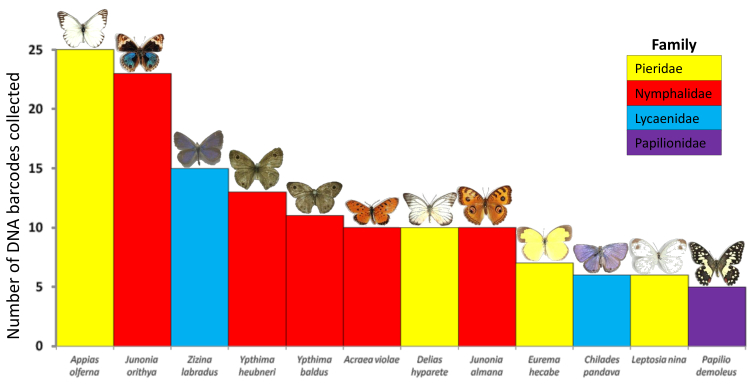
On the basis of 192 usable DNA barcodes, 43 species were sampled by the participants. The most sampled species was Appias olferna (BOLD:AAZ4640), followed by Junonia orithya (BOLD:ABZ6191) (Fig. 5). Twenty-one species were only sampled once, four were only sampled twice, and five were only sampled three times. Three DNA barcodes could not be assigned species names. One could only be assigned to the family Hesperiidae, the two others, representing two distinct BINs (BOLD:ACW8027 and BOLD:ACX2349) were assigned to the genus Ypthima (Nymphalidae) using the strict tree-based criterion for DNA barcode-based higher-taxon assignment (Wilson et al. 2011).
Figure 5.
Top sampled species during the first Peninsular Malaysia Butterfly Count (Suppl. material 3).
Family-Level Identification Success by Participants
Of the 192 usable DNA barcodes, the participants had attempted a family-level identification for 108 (56%) of these butterflies. Based on the DNA barcode assigments, 60% of these family-level identifications were correct (Fig. 6).
Figure 6.
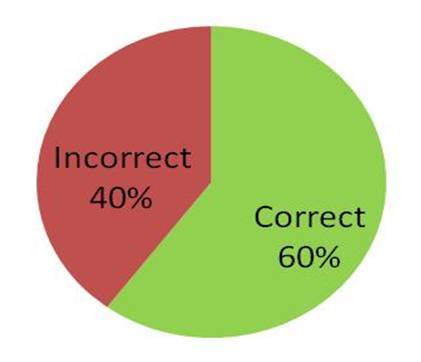
Family-level (field) identifications (108) by the first Peninsular Malaysia Butterfly Count participants compared with DNA barcode identifications (Suppl. material 3).
Feedback to Participants
The major findings of the Butterfly Count were collated into a newsletter (Suppl. material 2) which was posted to the Peninsular Malaysia Butterfly Count facebook page and also mailed (via Pos Malaysia) to the participants.
Discussion
Participation
In our initial proposal for the Peninsular Malaysia Butterfly Count, our target was to dispatch 100 Butterfly Count Packs. After registration closed we were able to send 57 packs, falling short on this target. Considering that 222 people/groups took part in the sixth annual "Malaysian Garden Bird Watch", the only comparable citizen science project in Malaysia which we are aware of (Table 1, 57 registrations in the first year of the Peninsular Malaysia Butterfly Count suggests to us a very promising start. We predicted that of the Butterfly Count Packs dispatched, roughly half would result in butterfly legs being sent to our Museum. The return rate closely matched our expectation. Several participants needed prompting before sending the collected butterfly legs, even though prepaid addressed envelopes were provided for this purpose. This was somewhat unexpected and consequently the importance of promptly returning samples will be given more prominence in any further count materials.
All of those who registered to take part in the Butterfly Count live on the west coast of Peninsular Malaysia. Furthermore, most of the participants live in the Klang Valley, the large urban agglomeration surrounding Kuala Lumpur (a similar pattern was seen with the Malaysian Garden Bird Watch; http://www.mygardenbirdwatch.com/?cur=bird/search). We need to review how to attract participation from the east coast of Peninsular Malaysia and rural areas, as it was clear we failed to reach out to those communities. Based on the registered names of participants it was also clear we were more successful in attracting participants from Malaysia's ethnic Chinese community than the other ethnic groups represented in Peninsular Malaysia. We were unable to obtain any coverage in Malay language newspapers, which may partially account for this trend, but it also likely reflects the rural-urban divide.
DNA Barcodes and Taxonomic Identifications
Of the 220 legs received, 192 (87%) generated 'usable' DNA barcodes (i.e., with significant hits on BOLD) of varying quality and length. There are several explanations for the relatively low success rate of PCR and sequencing. As noted above, it took some butterfly legs quite a while to reach us after the count day. Depending on the storage conditions, this could have boosted DNA degradation, which is particularly a problem in hot and humid Malaysia (Wilson et al. 2013). Secondly, in order to keep the costs of the project low, and provide a sustainable model for project continuation, we used the "quick, cheap and dirty" alkaline lysis method for DNA extraction. Although, in our experience, alkaline lysis usually provides ample DNA for successful PCR, when coupled with the prolonged pre-extraction storage conditions, this could have affected the quality of the DNA extracts. Thirdly, our labwork coincided with a period of difficulty for staff at our external DNA sequencing company who are currently revising their protocols.
A comparison of the DNA barcode identifications and the family-level identifications provided by the participants revealed that the participants were able to correctly identify the family of butterfly specimens 60% of the time. The relatively low success rate suggests that our butterfly family identification guide could undergo some improvement. Preparation of an identification guide represents a trade-off between being "user-friendly" and technical, and it is important not to discourage participants from attempting identifications by providing overly-complex guidelines. The relatively low success of family identifications suggests that asking participants for species identifications would not be very useful, either as a learning experience, or for contributing data on species occurrences. This finding further validates the continued use of the non-lethal DNA barcoding model used for this project.
The Butterflies
Although the primary purpose of the Peninsular Malaysia Butterfly Count was to promote awareness and engage the public with biodiversity, the Butterfly Count did produce some ecologically interesting findings.
The most sampled butterfly species was Appias olferna, commonly known as the Striped Albatross. Although most often treated as a distinct species (http://www.nic.funet.fi/pub/sci/bio/life/insecta/lepidoptera/ditrysia/papilionoidea/pieridae/pierinae/appias/index.html), Appias olferna is sometimes considered a subspecies of Appias libythea and DNA barcodes currently named as A. libythea and A. olferna share the same BIN in BOLD (BOLD:AAZ4640). A. olferna shows extreme sexual dimorphism with the male being predominantly white, but the female having broad black stripes on the upperside of the wings. This could potentially explain some of the family-level misidentifications by the participants, as our guide relied heavily on wing colour. According to Corbet and Pendlebury (Corbet et al. 1992), A. olferna was rare in Peninsular Malaysia until 60 years ago but by the early 1980s it had become one of the most common butterflies in gardens and along roadsides (Corbet et al. 1992). In Singapore, the host plant of A. olferna is reported to be Cleome rutidosperma, the Fringed Spider Flower (http://butterflycircle.blogspot.my/2010/06/life-history-of-striped-albatross.html). C. rutidosperma is a common weed found growing in disturbed habitats such as roadsides, gardens, and abandoned land. C. rutidosperma is native to Africa and is an invasive species in Peninsular Malaysia as well as other parts of Asia, Australia and the Domican Republic (http://www.cabi.org/isc/datasheet/14044#20087204071).
The second most sampled species was Junonia orithya, the Blue Pansy (BOLD:ABZ6191), which has a distribution covering Central Asia, India, Southeast Asia, southern China, Taiwan, Australia, and Africa (Corbet et al. 1992), and utilises a broad range of host plants. The top two most sampled species were relatively large, showy butterflies, which could be easier to spot and net by the participants. In contrast, the third most sampled species, the Common Grass Blue, Zizina otis (BOLD:AAB2267, also known as Zizina labradus; Yago et al. 2008), is a small, inconspicuous, lycaenid. Z. otis was the most sampled species during a survey of city parks in Kuala Lumpur (Sing et al. 2015), is one of the most abundant and widespread butterflies in Australia (New 2011), and has been considered invasive in New Zealand (Dugdale 1989). The Ypthima species (Y. heubneri BOLD:AAZ4966 and Y. baldus BOLD:AAN9479), at positions four and five amongst the most collected species are, together with Eurema hecabe (BOLD:AAA6082) at position nine, known to be paticularly common in gardens and along roadsides (Corbet et al. 1992). This is reflective of the localities where the participants conducted their sampling. The Ypthima group, known as the "rings", is taxonomically difficult and a review of the species in Peninsular Malaysia, including a study of the classic "ring" wing characters, is presently underway by our research group. Interestingly, two DNA barcodes collected in Penang Island (BOLD:ACW8027 and BOLD:ACX2349), fell within the Ypthima group of DNA barcodes on BOLD but without species-level matches. This suggests new species records for Peninsular Malaysia. At position six-equal amongst the most sampled species was Acraea violae, the Tawny Coster (BOLD:ABY2739, also known as Acraea terpsicore). The species, native to India, was first recorded in Peninsular Malaysia in 1992. The recent range expansion, reaching Australia in 2012, has been reviewed by Braby et al. 2013 who speculate on possible explanations for the expansion, including a response to climate change or habitat loss. Braby et al. 2014) also discuss the invasive potential of this species. Chilades pandava (BOLD:AAJ4577, also known as Luthrodes pandava), the tenth-equal collected species, is of concern in Taiwan and China as an "invasive" species with a history of populations "outbreaks" (Wu et al. 2010).
Summary
1) The level of participation in the first Peninsular Malaysia Butterfly Count was encouraging, but reaching and engaging rural communities remains a challenge.
2) The non-lethal DNA barcoding approach for species identification worked effectively, however, protocols could be improved to limit the number of returned samples which could not be identified. The family-level identification guide could use some improvement but provides an important educational tool for the participants.
3) The sampled butterflies revealed that widely distributed, cosmopolitan species, often recently arrived to the peninsula or with documented "invasive" potential, dominate the habitats sampled by the participants. Data from the first Butterfly Count helps establish a baseline from which we can monitor changes in butterfly communities in Peninsular Malaysia.
Supplementary Material
Butterfly Count Guide
Shi-Wei Jisming-See, Guo-Jie Brandon-Mong, Kong-Wah Sing, John-James Wilson
Data type: Butterfly Count Guide
File: oo_48731.pdf
Bulletin of the Museum of Zoology (3)(4)
John-James Wilson, Shi-Wei Jisming-See, Guo-Jie Brandon-Mong, Aik-Hean Lim, Voon-Ching Lim, Ping-Shin Lee, Kong-Wah Sing
Data type: Newsletter
File: oo_60225.pdf
Butterfly Count Raw Data
John-James Wilson, Shi-Wei Jisming-See, Guo-Jie Brandon-Mong, Aik-Hean Lim, Voon-Ching Lim, Ping-Shin Lee, Kong-Wah Sing
Data type: Data spreadsheet
File: oo_63256.xlsx
Acknowledgements
The citizen scientists - Huwa Yean Yin, Chia Chue Hon, Lai Sook Ling, Lee Soon Teck, Lee Sze Ying, Lee Chia Hao, Lee Chia Hsien, Lee Chia Sheng, Lee Shin Yee, Nor Azni Abdullah, Kerk Kim Lian, Julie Khoo, Alexandra Zieritz, Koo Si Hui, Noraina binti Jamal Rashid, Chong Yen Chin, Sean Vooi, Leong Sum Yee, Yu Cheng Swan, Sofwan bin Badruddin, Koo Si Hui, See Li Lian, Lilianne Yu, Soo Said Chyn, Chai Shut Kee, Chin Ah Yim, Ting Yun Yee, Lee See Mun, Muna Najihah binti Hj. Mokhtarruddin, Ng Yean Wah, Lee Qian Hui, Lee Jin Ming, Ma Bao Yi, Zaira Jane, Hanna, Azam, Joyce Lean, Ryan Teoh, Dylan Teoh, Leong Kai Jie, Lee Wai Yee, Lee Wai Kuan, Sei Hwui Tan, Chen Siew Ping, Nurzuhaila binti Jamaludin, Salman, Salahuddin, Saad, Mdm. Goh, Gerald Lee Zheng Yang, Innes Lee Jia Hui, Teo Yew Guan, Students from Senior Three Science – Chinese High School Batu Pahat Johor, Ng Mei Eng, Pang Chee Yee, Pang Chee Kok, Hew Lee Yoon, Gan Hui Hui, Ng Say Chong and three friends, Ting Mei Ying, Mr. Ong, David Foon, Phoebe Wong Sau May and family, See Sow Pang, Ernest Tang Onn Sheng, Eddie Tang Gwo Huei, Lai Yik Chuen, Virgo Tjan, Ng Zhi Yu with Mother, and Elder Brother, lam Panny with Bapa, Ibu, Kakak and Adik, Hannah Muhammad Akram, Maryam, Hasan, Narong Jaturas, Emily Lim, "Reptilia Kingdom", Chu Bah Boh, Crystal Sophillea Alexander, Clarence Gabriel Alexander.
The team at BOLD for their database support.
The journalists for promoting our project.
The helpful and supportive staff at UMCares.
Author contributions
JJW and KWS devised the project. All authors were involved in the implementation and administration of the project. SWJS, GJBM and KWS performed the labwork. JJW, SWJS, GJBM and AHL analysed the data. JJW wrote the manuscript with input from all the authors.
References
- Braby M. F. The complete field guide to butterflies of Australia. CSIRO PUBLISHING; 2004. 340 [Google Scholar]
- Braby Michael F, Thistleton Brian M, Neal Michael J. Host plants, biology and distribution of Acraea terpsicore (Linnaeus, 1758) (Lepidoptera: Nymphalidae): a new butterfly for northern Australia with potential invasive status. http://dx.doi.org/10.1111/aen.12078. Austral Entomology. 2014;53(3):288–297. doi: 10.1111/aen.12078. [DOI] [Google Scholar]
- Braby M. F., Bertelsmeier C., Sanderson C., Thistleton B. M. Spatial distribution and range expansion of the Tawny Coster butterfly, Acraea terpsicore (Linnaeus, 1758) (Lepidoptera: Nymphalidae), in South-East Asia and Australia. http://dx.doi.org/10.1111/icad.12038. Insect Conservation and Diversity. 2013;7(2):132–143. doi: 10.1111/icad.12038. [DOI] [Google Scholar]
- Brandon-Mong G. J., Gan H. M., Sing K. W., Lee P. S., Lim P. E., Wilson J. J. DNA metabarcoding of insects and allies: an evaluation of primers and pipelines. http://dx.doi.org/10.1017/s0007485315000681. Bulletin of Entomological Research. 2015:First View. doi: 10.1017/s0007485315000681. [DOI] [PubMed]
- Brook B. W., Sodhi N. S., Ng P. K. L. Catastrophic extinctions follow deforestation in Singapore. http://dx.doi.org/10.1038/nature01795. Nature. 2003;424(6947):420–426. doi: 10.1038/nature01795. [DOI] [PubMed] [Google Scholar]
- Corbet A. S., Pendlebury H. M., Eliot J. N. The butterflies of the Malay Peninsula. 4th edition. Malayan Nature Society; Kuala Lumpur: 1992. 664 [Google Scholar]
- Crawford Lindsay A., Koscinski Daria, Watt Kaitlin M., McNeil Jeremy N., Keyghobadi Nusha. Mating success and oviposition of a butterfly are not affected by non-lethal tissue sampling. http://dx.doi.org/10.1007/s10841-013-9566-8. Journal of Insect Conservation. 2013;17(5):859–864. doi: 10.1007/s10841-013-9566-8. [DOI] [Google Scholar]
- Dugdale J. S. New Zealand Lepidoptera: basic biogeography. New Zealand Journal of Zoology. 1989;16(4):679–687. doi: 10.1080/03014223.1989.10422926. [DOI] [Google Scholar]
- Gibson L., Lee T. M., Koh L. P., Brook B. W., Gardner T. A., Barlow J., Peres C. A., Bradshaw C. J. A., Laurance William F., Lovejoy T. E., Sodhi N. S. Primary forests are irreplaceable for sustaining tropical biodiversity. http://dx.doi.org/10.1038/nature10425. Nature. 2011;478(7369):378–381. doi: 10.1038/nature10425. [DOI] [PubMed] [Google Scholar]
- Ivanova N. V., Borisenko A. V., Hebert P. D. N. Express barcodes: racing from specimen to identification. http://dx.doi.org/10.1111/j.1755-0998.2009.02630.x. Molecular Ecology Resources. 2009;9:35–41. doi: 10.1111/j.1755-0998.2009.02630.x. [DOI] [PubMed] [Google Scholar]
- Jisming-See S. W., Sing K. W., Wilson J. J. Monitoring of butterflies by schoolchildren to increase environmental awareness and assess effects of climate change in peninsular Malaysia. Genome. 2015;58(5):233–233. [Google Scholar]
- Koscinski Daria, Crawford Lindsay A., Keller Heidi A., Keyghobadi Nusha. Effects of different methods of non-lethal tissue sampling on butterflies. http://dx.doi.org/10.1111/j.1365-2311.2011.01272.x. Ecological Entomology. 2011;36(3):301–308. doi: 10.1111/j.1365-2311.2011.01272.x. [DOI] [Google Scholar]
- Leray M., Yang J. Y., Meyer C. P., Mills S. C., Agudelo N., Vincent Ranwez, Boehm J. T., Machida R. J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. http://www.frontiersinzoology.com/content/10/1/34. Frontiers in Zoology. 2013;10(1):34. doi: 10.1186/1742-9994-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos J., Horcea-Milcu A. I., Kirkland P., Hartel T., Osváth-Ferencz M., Fischer J. Challenges for biodiversity monitoring using citizen science in transitioning social–ecological systems. http://dx.doi.org/10.1016/j.jnc.2015.05.001. Journal for Nature Conservation. 2015;26:45–48. doi: 10.1016/j.jnc.2015.05.001. [DOI] [Google Scholar]
- Marschalek Daniel A., Jesu Julia A., Berres Mark E. Impact of non-lethal genetic sampling on the survival, longevity and behaviour of the Hermes copper (Lycaena hermes) butterfly. http://dx.doi.org/10.1111/icad.12024. Insect Conservation and Diversity. 2013;6(6):658–662. doi: 10.1111/icad.12024. [DOI] [Google Scholar]
- McIntyre N. E. Ecology of Urban Arthropods: A Review and a Call to Action. http://dx.doi.org/10.1603/0013-8746(2000)093[0825:eouaar]2.0.co;2. Annals of the Entomological Society of America. 2000;93(4):825–835. doi: 10.1603/0013-8746(2000)093[0825:eouaar]2.0.co;2. [DOI] [Google Scholar]
- New T. R. Are Lepidoptera an effective ‘umbrella group ‘for biodiversity conservation? Journal of Insect Conservation. 1997;1(1):5–12. doi: 10.1023/A:1018433406701. [DOI] [Google Scholar]
- New T. R. ‘In Considerable Variety’: Introducing the Diversity of Australia’s Insects. Springer Netherlands; 2011. Origins, Distributions and Diversity.37-54. [DOI] [Google Scholar]
- Ratnasingham S., Hebert P. D. N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org) http://dx.doi.org/10.1111/j.1471-8286.2007.01678.x. Molecular Ecology Notes. 2007;7(3):355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasingham Sujeevan, Hebert P. D. N. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. http://dx.doi.org/10.1371/journal.pone.0066213. PLoS ONE. 2013;8(7):e66213. doi: 10.1371/journal.pone.0066213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing K. W., Jusoh W. F. A., Hashim N. R., Wilson J. J. Urban parks: refuges for tropical butterflies? Genome. 2015;58(5):281–281. [Google Scholar]
- Syaripuddin K., Sing K. W., Wilson J. J. Comparison of butterflies, bats and beetles as bioindicators based on four key criteria and DNA barcodes. http://tropicalconservationscience.mongabay.com/content/v8/tcs_v8i1_138-149_Syaripuddin.pdf Tropical Conservation Science. 2015;8(1):138–149. [Google Scholar]
- Theobald E. J., Ettinger A. K., Burgess H. K., DeBey L. B., Schmidt N. R., Froehlich H. E., Wagner C., HilleRisLambers J., Tewksbury J., Harsch M. A., Parrish J. K. Global change and local solutions: Tapping the unrealized potential of citizen science for biodiversity research. http://dx.doi.org/10.1016/j.biocon.2014.10.021. Biological Conservation. 2015;181:236–244. doi: 10.1016/j.biocon.2014.10.021. [DOI] [Google Scholar]
- Tulloch Ayesha I. T., Possingham Hugh P., Joseph Liana N., Szabo Judit, Martin Tara G. Realising the full potential of citizen science monitoring programs. http://dx.doi.org/10.1016/j.biocon.2013.05.025. Biological Conservation. 2013;165:128–138. doi: 10.1016/j.biocon.2013.05.025. [DOI] [Google Scholar]
- Wilson J. J. DNA barcodes for insects. In: Kress W. John, Erikson David L., editors. DNA Barcodes. Vol. 858. Humana Press; 2012. [DOI] [Google Scholar]
- Wilson J. J., Sing K. W., Sofian-Azirun M. Building a DNA Barcode Reference Library for the True Butterflies (Lepidoptera) of Peninsula Malaysia: What about the Subspecies? http://dx.doi.org/10.1371/journal.pone.0079969. PLoS ONE. 2013;8(11):e79969. doi: 10.1371/journal.pone.0079969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. J., Rougerie R., Schonfeld J., Janzen D. H., Hallwachs W., Hajibabaei M., Kitching I. J., Haxaire J., Hebert P. D. N. When species matches are unavailable are DNA barcodes correctly assigned to higher taxa? An assessment using sphingid moths. http://dx.doi.org/10.1186/1472-6785-11-18. BMC Ecology. 2011;11(1):18. doi: 10.1186/1472-6785-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. W., Yen S. H., Lees D. C., Hsu Y. F. Elucidating genetic signatures of native and introduced populations of the Cycad Blue, Chilades pandava to Taiwan: a threat both to Sago Palm and to native Cycas populations worldwide. http://dx.doi.org/10.1007/s10530-009-9672-4. Biological Invasions. 2010;12(8):2649–2669. doi: 10.1007/s10530-009-9672-4. [DOI] [Google Scholar]
- Yago M., Hirai N., Kondo M., Tanikawa T., Ishii M., Wang M., Williams M., Ueshima R. Molecular systematics and biogeography of the genus Zizina (Lepidoptera: Lycaenidae) Zootaxa. 2008;1746:15–38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Butterfly Count Guide
Shi-Wei Jisming-See, Guo-Jie Brandon-Mong, Kong-Wah Sing, John-James Wilson
Data type: Butterfly Count Guide
File: oo_48731.pdf
Bulletin of the Museum of Zoology (3)(4)
John-James Wilson, Shi-Wei Jisming-See, Guo-Jie Brandon-Mong, Aik-Hean Lim, Voon-Ching Lim, Ping-Shin Lee, Kong-Wah Sing
Data type: Newsletter
File: oo_60225.pdf
Butterfly Count Raw Data
John-James Wilson, Shi-Wei Jisming-See, Guo-Jie Brandon-Mong, Aik-Hean Lim, Voon-Ching Lim, Ping-Shin Lee, Kong-Wah Sing
Data type: Data spreadsheet
File: oo_63256.xlsx