Abstract
The intestinal immune system maintains oral tolerance to harmless antigens or nutrients. One mechanism of oral tolerance is mediated by regulatory T cell (Treg)s, of which differentiation is regulated by a subset of dendritic cell (DC)s, primarily CD103+ DCs. The aryl hydrocarbon receptor (AhR), a ligand-activated transcription factor, plays an important role in regulating immunity. The intestines are exposed to various AhR ligands, including endogenous metabolites and phytochemicals. It was previously reported that AhR activation induced tolerogenic DCs in mice or in cultures of bone marrow-derived DCs. However, given the variety of tolerogenic DCs, which type of tolerogenic DCs is regulated by AhR remains unknown. In this study, we found that AhR ligand 3,3'-diindolylmethane (DIM) inhibited the development of CD103+ DCs from mouse bone marrow cells stimulated with Flt3L and GM-CSF. DIM interfered with phosphorylation of STAT3 and STAT5 inhibiting the expression of genes, including Id2, E2-2, IDO-1, and Aldh1a2, which are associated with DC differentiation and functions. Finally, DIM suppressed the ability of CD103+ DCs to induce Foxp3+ Tregs.
Keywords: AhR; Oral tolerance; 3,3'-diindolylmethane; CD103+ DC; Foxp3+ Tregs; STAT5
INTRODUCTION
The aryl hydrocarbon receptor (AhR) is a member of the basic region-helix-loop-helix (bHLH) superfamily of DNA binding proteins (1). Upon ligand binding, AhR translocates from the cytoplasm into the nucleus where it heterodimerizes with a second bHLH transcription factor, the Ah receptor nuclear translocator, and activates the transcription of target genes such as CYP1A1 and glutathione S-transferase (2,3). AhR is a cytosolic sensor of small synthetic compounds and natural chemicals, regulating cell growth and differentiation. Numerous studies of the AhR have been performed with halogenated aromatic hydrocarbons, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and benzo(a)pyrene (B(a)P) (4). However, a number of low-molecular weight, structurally diverse chemicals, including indoles, tetrapyroles, archidonic acid metabolites, and tryptophan metabolites, have been identified as naturally occurring exogenous and endogenous AhR ligands (5). Although AhR plays an important role in the detoxification of xenobiotics, physiological abnormalities as diverse as cardiac disorders, hepatocirrhosis, immunodeficiencies, and shortened lifespan have been reported in AhR-deficient mice, indicating that AhR is intimately involved in various physiological processes (6). AhR is widely expressed in a variety of animal species and humans (7). In mice and humans, AhR is expressed in several organs, including the lung, heart, liver, thymus, and kidney (8,9,10). One of most intensely studied systems associated with AhR is the immune system. AhR regulates the differentiation of Foxp3+ regulatory T cell (Treg)s and T helper 17 (Th17) cells (11).
The gastrointestinal tract is in constant contact with food proteins, commensals, and potentially pathogenic microorganisms. To maintain immune homeostasis in this environment, the intestinal immune system has evolved regulatory strategies, including Foxp3+ Tregs that play a central role in controlling intestinal homeostasis (12). CD103+ dendritic cell (DC)s, which make up 2~3% of the total leukocytes in the lamina propria of the small intestine, migrate to the mesenteric lymph nodes (13), and induce the development of Tregs by secreting retinoic acid (RA) and TGF-β (14). In addition, CD103+ DCs, which produce indoleamine 2,3-dioxygenase (IDO), catalyze the metabolism of tryptophan, depleting it from the microenvironment and stimulating Treg differentiation (15). Thus, CD103+ DCs in the intestinal mucosa play a central role in the tolerance to commensal bacteria and food antigens.
The intestines are exposed to various AhR ligands through foods contaminated with TCDD and other toxic chemicals, endogenous metabolites such as 6-formylindolo (3,2-b)carbazole and indoxyl 3-sulfate, phytochemicals including quercetin, kaempferol, and indole 3-carbinol (I3C), and metabolites produced by commensal bacteria such as I3C and indole-3-acetic acid (16,17,18,19,20). Thus, AhR may regulate intestinal homeostasis by directly acting on various types of cells in the intestine, including intraepithelial lymphocytes and innate lymphoid cells (21).
DCs are professional antigen-presenting cells that constitutively express AhR, responding to AhR ligands (22). AhR expression in bone marrow-derived DCs (BMDCs) was induced upon addition of lipopolysaccharide or CpG, which revealed that AhR was necessary for the induction of IDO expression and Foxp3+ Treg differentiation (23). Administration of VAF347 or 2-(1'H-indole-3'-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE), which are both AhR ligands, into mice suppressed allograft rejection or experimental autoimmune encephalomyelitis, respectively. The suppressive effect of the AhR ligands was mediated by Foxp3+ Tregs; the frequency of these cells in secondary lymphoid organs was increased upon AhR ligand addition (24,25). Additionally, AhR activation induced tolerogenic DCs in mice (24,25) or in cultures of BMDCs (25). However, given the variety of tolerogenic DCs (26) and the regulation of the differentiation of tolerogenic DCs by Foxp3+ Tregs (27), the type of tolerogenic DCs regulated by AhR remains unknown. In addition, AhR activation by TCDD was reported to impair oral tolerance, but underlying mechanisms have not been directly addressed (28,29).
In this study, we demonstrated that AhR ligand 3,3'-diindolylmethane (DIM), an acid-stimulated conversion product from I3C which is found in Cruciferous plants including broccoli and Brussels sprouts and produced by commensals in the intestine (20,30), inhibited the development of CD103+ DCs from bone marrow cells stimulated with Flt3L and GM-CSF. DIM interfered with the phosphorylation of STAT3 and STAT5 inhibiting expression of genes, including Id2, E2-2, IDO-1, and Aldh1a2, which are associated with DC differentiation and functions. Finally, DIM suppressed the ability of CD103+ DCs to induce Foxp3+ Tregs. These findings suggest that AhR interferes with the differentiation of tolerogenic CD103+ DCs, which may explain the breakdown of oral tolerance by AhR.
MATERIALS AND METHODS
Mice
C57BL/6 mice, 6~12 weeks of age, were purchased from the Korean Institute for Chemistry (Taejon, Korea). Five mice were housed per cage in a laminar airflow room maintained at 22±2℃ with a relative humidity of 55±5%. Mice were cared and treated in accordance with the guidelines established by the Changwon National University public health service policy on the use of laboratory animals. The animal study was performed in the immunology laboratory, Department of Biology, Changwon national University.
Chemicals and reagents
TCDD and DIM were purchased from Cambridge Isotope (Tewksbury, MA, USA) and Sigma-Aldrich (St. Louis, MO, USA), respectively. Human TGF-β and murine Flt3L and IL-2 were from eBioscience (San Diego, CA, USA). Mouse GM-CSF was from BioLegend (San Diego, CA, USA). Antibodies used in the present study included: anti-STAT3 and anti-phospho-STAT5 (Tyr 694), and anti-phospho-IκBα (Ser 32/36) from Santa Cruz Biotechnology (USA); anti-phospho-c-Src (Tyr416) and anti-STAT5 from Cell Signaling Technology (Danvers, MA, USA); anti-phospho-STAT3 (Tyr705) from BD Biosciences (Franklin Lakes, NJ, USA); FITC anti-mouse CD11c, Alexa Fluor 647 anti-mouse CD103, anti-CD3, anti-β actin, PE anti-mouse CD4, Alexa Fluor 488 anti-mouse FoxP3 from BioLegend.
Development of CD103+ DCs from bone marrow cells
The development of CD103+ DC from bone marrow cells was described by Jackson et al. (31). Briefly, bone marrow cells were extracted, and erythrocytes were removed by brief exposure to 0.168 M NH4Cl. Cells were cultured in a 24-well plate at a density of 1×106 cells/mL in RPMI 1640 containing 10 mM HEPES, 2 mM L-glutamine, 10% FBS, 50 µM β-mercaptoethanol, and Flt3L (200 ng/mL) for 7 days and subsequently for 2 days in the presence of GM-CSF (200 ng/mL). AhR ligands, DIM or TCDD, were added to the culture 1 h before the addition of Flt3L and GM-CSF. DMSO (0.1% v/v) was used as a vector control.
Flow cytometry and cell sorting
Cells were kept on ice or at 4℃ at all times. Immunofluorescence staining was performed in 96-well, round-bottomed cell culture plates. The cells were centrifuged and resuspended in 10 µL of primary antibody diluted with PBS at a previously determined optimal concentration. After 15 min of incubation, the cells were washed with washing buffer (PBS/0.5% BSA/0.09% sodium azide) three times and fixed with 0.9% buffered formalin. Cells were analyzed within 1 week on a FACSCalibur flow cytometer (BD Biosciences). Intracellular FoxP3 staining was performed by using BD Cytofix/Cytoperm Fixation/Permeabilization Kit (BD Biosciences). Briefly, the cells were first fixed and permeabilized using Fixation/Permeabilization solution and then stained with fluorochrome-conjugated anti-FoxP3 Ab. For cell sorting, FGBMCs were double stained with FITC-conjugated CD11c Ab and PE-conjugated with CD103 Ab, and sorted with FACS Aria III. The cut-off point was the co-expression of CD11c and CD103 at high levels. A small amount of cells were reanalyzed for CD11c and CD103 co-expression.
In vitro conversion assay
The conversion assay was performed as described previously (32). Briefly, 2×105 naïve CD4+CD62L+ helper T cells, which were prepared from murine spleen using the CD4+CD62L+ T cell isolation kit II (Milteyi Biotec, San Diego, CA, USA), were cultured with 4×104 Flt3/GM-CSF-stimulated bone marrow cultures in 1 mL of culture medium (RPMI 1640 containing 10 mM HEPES, 2 mM L-glutamine, 10% FBS, and 50 µM β-mercaptoethanol) in the presence of soluble anti-CD3 Ab (1 µg/mL), human TGF-β (3 ng/mL), and murine IL-2 (5 ng/mL) for 5 days.
RNA preparation, RT-PCR, and Real-Time PCR
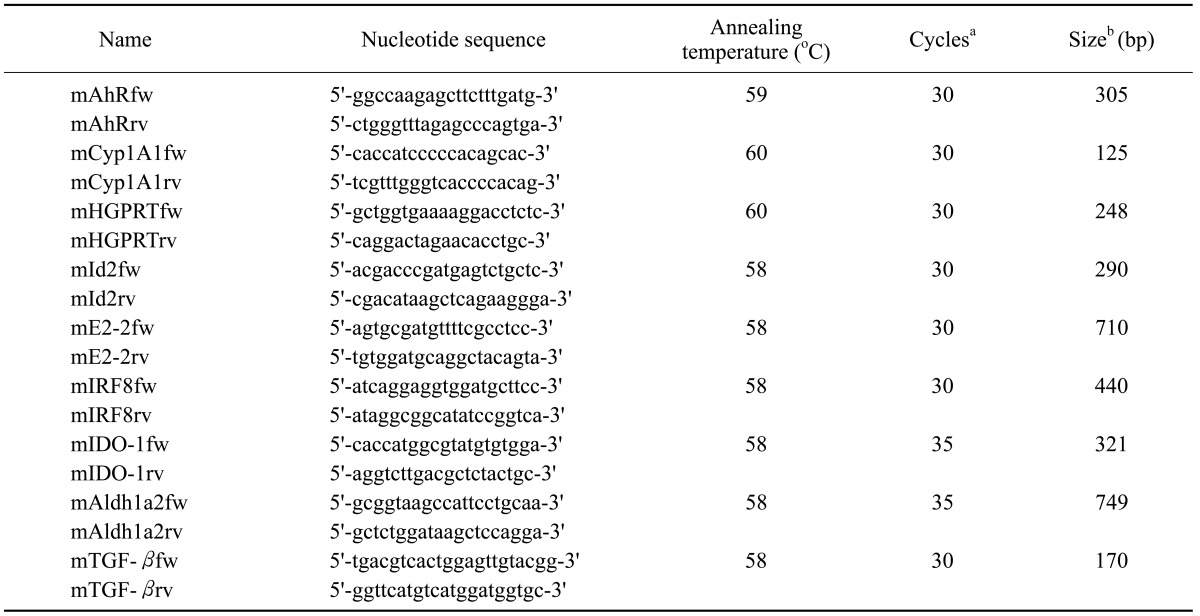
Total cellular RNA was extracted from cells using the RNAzol method (Tel-Test, Inc., Friendswood, TX, USA). For PCR analysis, RNA was used after contaminating DNA was completely removed by DNase I treatment. RT-PCR analysis was performed using pairs of oligonucleotide primers. The PCR products were confirmed to correspond to their original sequences by DNA sequencing. Gene-specific primers and the number of cycles of amplification, annealing temperature, and expected size of PCR products are listed in Table 1. Real-Time PCR was performed to quantitate PCR products. Power SYBR green PCR Master Mix and Real-time PCR system (7300, Applied Biosystems, Foster City, CA, USA) were used.
Table I. Primers used in RT-PCR.
aIndicates the number of cycles of amplification. bIndicates the expected size of PCR products.
Western blotting
Cells or tissues were homogenized in lysis buffer containing 20 mM Tris-HCl (pH 7.4), 1 mM EDTA (pH 8.0), 50 µM sodium vanadate, 20 mM p-nitrophenylphosphate, 50 mM sodium fluoride, leupeptin (0.5 µg/mL), aprotinin (10 µg/mL), and soybean trypsin inhibitor (10 µg/mL). Proteins size-fractionated on SDS/PAGE were transferred to PVDF membranes, and the blots were blocked with 3% BSA in TBS buffer (20 mM Tris-HCl pH 7.5/137 mM NaCl). The blots were sequentially treated with primary and secondary antibodies in TBST (20 mM Tris-HCl pH 7.5/137 mM NaCl/0.1% Tween 20) with intermittent washing with TBST. Immunodetection was performed with the ECL-plus kit (Amersham Phamacia Biotech, Amersham, UK). Densitometer analysis was performed using the Image Master 2-D Platinum software (Amersham Biosciences, Piscataway, NJ, USA) according to the protocols provided by the manufacturer.
Statistical analysis
Data were analyzed by the paired Student's t-test. A value of p<0.05 was considered as statistically significant.
RESULTS
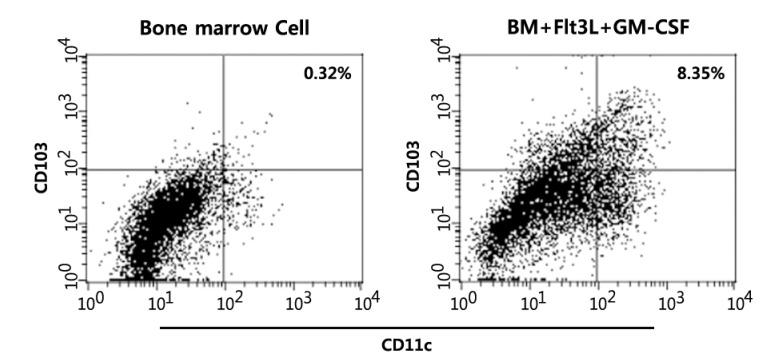
AhR is functionally expressed in Flt3/GM-CSF-stimulated bone marrow cultures and CD11+CD103+ dendritic cells
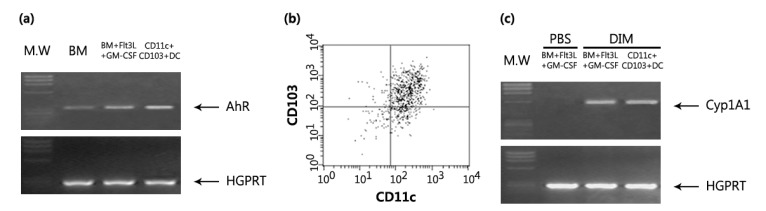
Although there are two main subsets of DCs, namely myeloid DCs and plasmacytoid DCs, various lineages of DCs have been defined by cell surface marker expression, anatomical location, and functional responses (33). It was previously reported that AhR was expressed in splenic DC, DC progenitors, and Langerhans cell (34,35). However, whether AhR is functionally expressed in tolerogenic DCs is unknown. Flt3L-induced differentiation of CD103+ DCs from BM cells was stimulated by GM-CSF (31). Thus, we examined the expression of AhR in Flt3L/GM-CSF-stimulated bone marrow cultures (FGBMCs). After a 9-day incubation of BM cells in the presence of Flt3L and GM-CSF, 8.35% of total cells expressed CD11c and CD103 (Fig. 1). AhR expression was increased in FGBMCs compared with that in BM cells (Fig. 2a). To examine whether CD11+CD103+ cells express AhR, FGBMCs were sorted by expression of CD11c/CD103 using FACS Aria III (Fig. 2b). AhR expression was also increased in CD11+CD103+ cells compared with that in BM cells (Fig. 2a). Next, we tested whether AhR is functional in FGBMCs by examining AhR ligand-induced expression of Cyp1A1, a target gene of AhR. Although TCDD is widely used as a surrogate ligand for AhR, the interpretation of the results related to TCDD should be considered with caution because TCDD is not quickly metabolized in the body (36). Thus, the physiological relevance of AhR signaling remains unclear. I3C, which is found in Cruciferous plants including broccoli and Brussels sprouts and produced by commensals in the intestine, is converted to higher-order AhR agonists in the acidic environment of the stomach (37). One of the condensation product of I3C is DIM, which induces AhR transformation and Cyp1A1 enzyme activity and thus has been widely used in AhR functional studies (5). Thus, in this study DIM was mainly used as the AhR ligand. When FGBMCs or CD11+CD103+ cells were incubated in the presence of DIM, Cyp1A1 expression was induced (Fig. 2c), suggesting that AhR is functional in FGBMCs and CD11+CD103+ cells.
Figure 1. In vitro differentiation of CD11c+CD103+ DCs from bone marrow cells. BM cells were induced to differentiate into CD11c+CD103+ DCs with Flt3L for the first 7 days and with Flt3L plus GM-CSF for the last 2 days. The cells were harvested, stained with fluorochrome conjugated-anti-CD11c and -anti-CD103 Abs, and analyzed by flow cytometry.
Figure 2. AhR is functionally expressed in Flt3L/GM-CSF-stimulated bone marrow cells and CD11c+CD103+ DCs. BM cells were induced to differentiate into CD11c+CD103+ DCs with Flt3L for the first 7 days and with Flt3L plus GM-CSF for the last 2 days. Then, the cells (FGBMCs) or CD11c+CD103+ sorted from FGBMCs were assayed for mRNA expression by PCR: AhR (a) and Cyp1A1 (c). The purity of the CD11c+CD103+ population was assessed post sorting (b). DIM (1 µM) was added to the culture 1 h before the addition of Flt3L/GM-CSF.
DIM inhibits the differentiation of BM cells into CD11c+CD103+ DCs in vitro
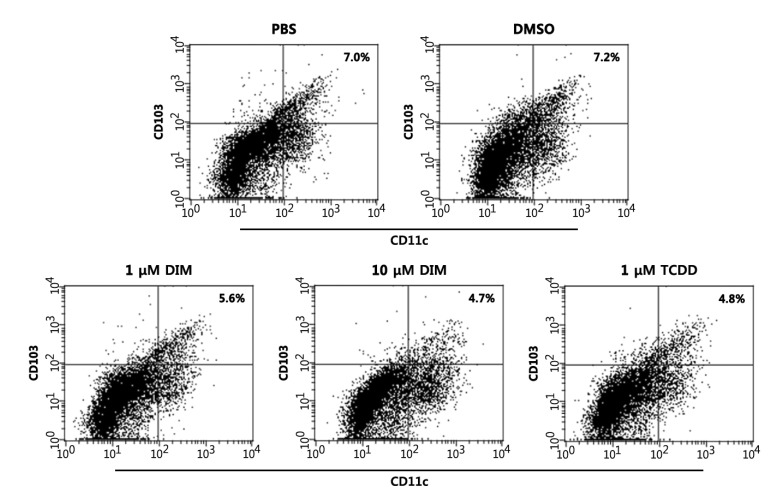
Next, we examined whether DIM regulates the differentiation of BM cells into CD11c+CD103+ DCs in vitro. On average, ~7% of BM cultures became CD11c+CD103+ by 9 days after culture (Fig. 3). DIM dose-dependently inhibited differentiation: a 1.25-fold reduction and a 1.48-fold reduction at 1 µM and 10 µM, respectively, compared with the PBS control. DMSO showed a negligible effect. This experiment was repeated twice, producing similar results (PBS, 7.0±0.8%; DMSO, 7.2±0.7%; 1 µM DIM, 5.6±0.6%; 10 µM DIM, 4.7±0.7%; TCDD, 4.8±0.9%), that were statistically significant: being compared with PBS, p=0.016 for 1 µM DIM and p=0.0003 for 10 µM DIM. CD11c+CD103+ DC differentiation was also reduced when TCDD, an AhR ligand, was added to the culture.
Figure 3. DIM and TCDD inhibit the development of CD11c+CD103+ DCs. BM cells were induced to differentiate into CD11c+CD103+ DCs with Flt3L for the first 7 days and with Flt3L plus GM-CSF for the last 2 days. The cells were harvested, stained with fluorochrome conjugated-anti-CD11c and -anti-CD103 Abs, and analyzed by flow cytometry. AhR ligands or DMSO (0.1% v/v) were added to the culture 1 h before the addition of Flt3L/GM-CSF. The results are representative of three separate experiments that showed similar results. Data were analyzed by the paired Student's t-test.
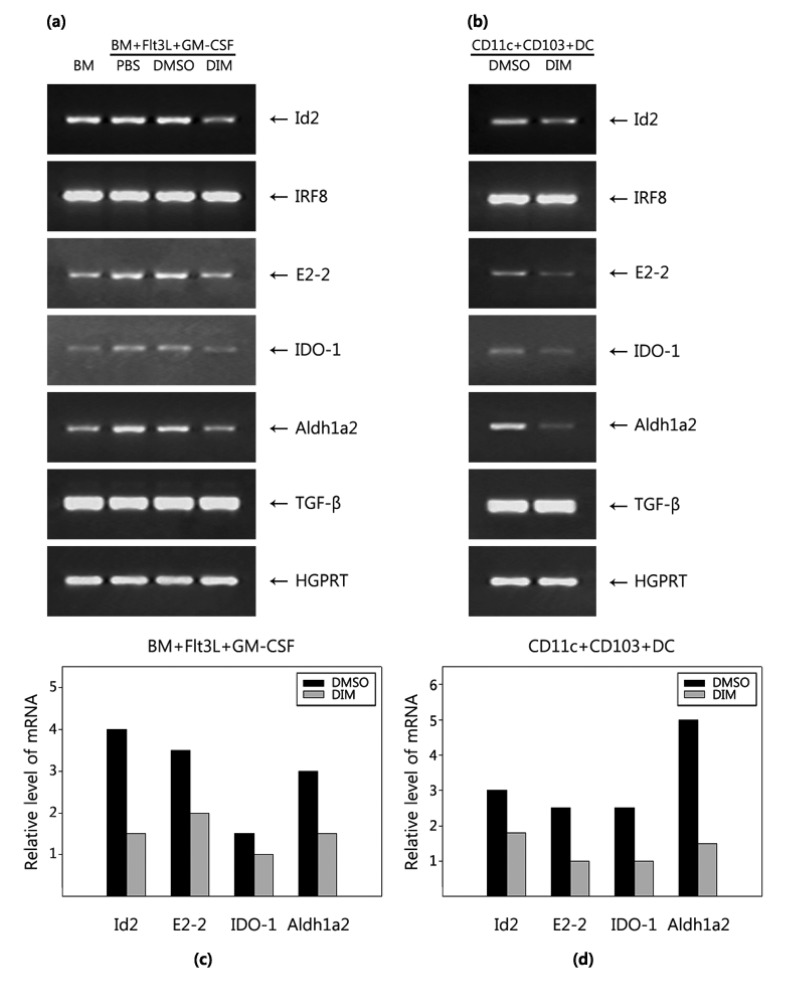
DIM differentially regulates genes associated with CD11c+CD103+ DC differentiation and functions
AhR functions either as a transcription factor or by interacting with effector molecules that mediate signal transduction pathways. DC differentiation is regulated by a combination of transcription factors including Id2, IRF8, and E2-2 (38). E2-2 expression was increased in FGBMCs compared with in BM cells, whereas the expression of Id2 and IRF8 was not modulated. Next, we examined whether DIM regulates the expression of genes associated with DC differentiation in FGBMCs (Fig. 4a). DIM inhibited the expression of the Id2 and E2-2 genes while exerting little effect on the IRF8 gene. Tolerologenic CD103+ DCs produce effector molecules such as retinoic acid, TGF-β, and tryptophan metabolites (39). CD103+ DCs produce IDO, which catalyzes the metabolism of tryptophan, generating toxic metabolites (kynurenines), and retinaldehyde dehydrogenase 2 (Aldh1a2), which metabolizes retinal to retinoic acid. The expression of IDO-1 and Aldh1a2 was increased in FGBMCs compared with in BM cells, whereas that of TGF-β was not modulated. DIM inhibited the expression of IDO-1 and Aldh1a2 genes while exerting little effect on TGF-β (Fig. 4a). The RT-PCR data were confirmed by real-time PCR (Fig. 4c). Next, we performed these experiments with purified CD11+CD103+ cells from FGBMCs. DIM inhibited the expression of the Id2, E2-2, IDO-1, and Aldh1a2 genes while showing little effect on the IRF8 and TGF-β genes compared with the vehicle control DMSO (Fig. 4b and d).
Figure 4. DIM regulates expression of genes associated with the development and functions of CD11c+CD103+ DCs. BM cells were induced to differentiate into CD11c+CD103+ DCs with Flt3L for the first 7 days and with Flt3L plus GM-CSF for the last 2 days. Next, the cells (FGBMCs) (a, c) or CD11c+CD103+ (b, d) sorted from FGBMCs were assayed for mRNA expression by PCR (a, b) or real-time PCR (c, d). DIM (1 µM) or DMSO (0.1% v/v) was added to the culture 1 h before the addition of Flt3L/GM-CSF. The results are representative of three separate experiments that showed similar results.
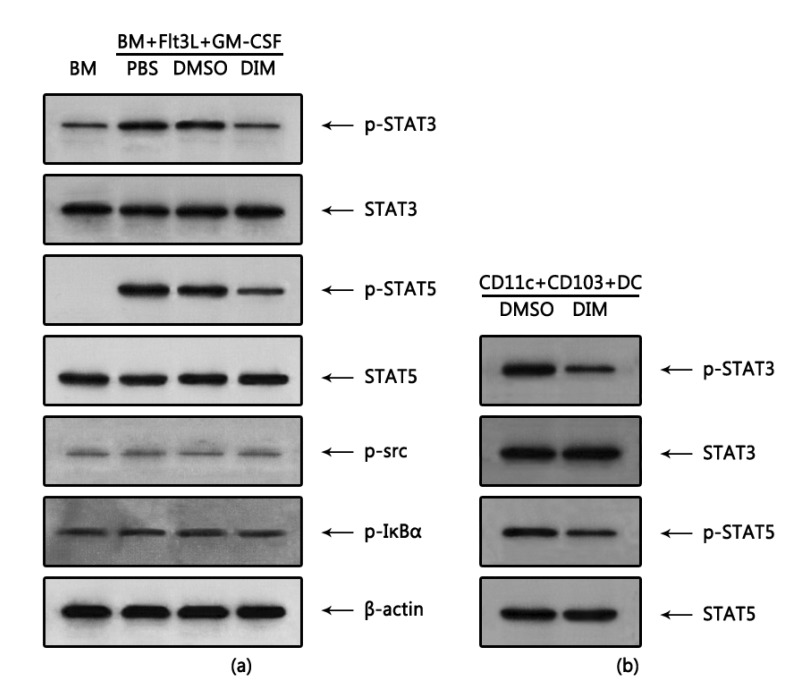
DIM suppresses phosphorylation of STAT3 and STAT5 but not of c-Src and IκBα
The differentiation of DCs is controlled mainly by STAT-dependent pathways (38), particularly by STAT3 and STAT5 in Flt3L- and GM-CSF-induced DCs (40). Thus, we examined whether DIM regulates signaling pathways in FGBMCs. Phosphorylation of STAT3 and STAT5, which was induced by Flt3L/GM-CSF treatment, was inhibited ~3-fold and ~2-fold by DIM, respectively (Fig. 5a). However, the levels of STAT3 and STAT5 were unchanged. In addition to STATs, the NF-κB and Src signaling pathways are also involved in GM-CSF-induced differentiation of DCs (41). However, the phosphorylation of c-Src and IκBα was neither induced by Flt3L/GM-CSF treatment nor modulated by DIM, suggesting that the modulating effects of DIM on signaling pathways are specific. When we performed these experiments with purified CD11+CD103+ cells from FGBMCs, it was observed that DIM inhibited phosphorylation of STAT3 and STAT5 ~4-fold and ~2-fold, respectively while exerting little effect on the levels of STAT3 and STAT5 compared with the vehicle control DMSO (Fig. 5b).
Figure 5. DIM inhibits phosphorylation of STAT3 and STAT5 associated with the development of CD11c+CD103+ DCs. BM cells were induced to differentiate into CD11c+CD103+ DCs with Flt3L for 7 days and then with Flt3L/GM-CSF for 1 h. Next, the cells (FGBMCs) were harvested for protein analysis by western blotting (a). For CD11c+ CD103+ DCs, BM cells were cultured with Flt3L for 7 days, harvested, and sorted for the expression of CD11c/CD103. Then, the CD11c+CD103+ DCs were cultured with Flt3L/GM-CSF for 1 h and harvested for protein analysis (b). DIM (1 µM) or DMSO (0.1% v/v) was added to the culture 1 h before the addition of Flt3L/GM-CSF. The results are representative of three separate experiments that showed similar results.
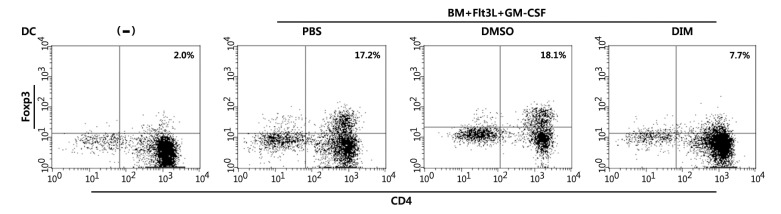
DIM suppresses the ability of tolerogenic DCs to induce Foxp3+ Tregs
One of the key functions of CD103+ DCs in tolerance induction is to induce Foxp3+ Tregs. We next examined whether FGBMCs could induce Treg differentiation. FGBMCs were co-cultured with naïve CD4 T cells in the presence of anti-CD3 Ab, TGF-β, and IL-2. For Treg differentiation assays, anti-CD3 Ab along with anti-CD28 is typically added to the culture supplemented with IL-2 (5 ng/mL) and higher concentration of TGF-β (10 ng/mL) (42). We co-cultured FGBMCs with naïve CD4 T cells in the presence of anti-CD3 Ab, TGF-β (3 ng/mL), and IL-2 for 5 days. Two percent of CD4 T cells became Foxp3+ when naïve CD4 T cells were cultured alone (Fig. 6). When FGBMCs were added to the culture, the frequencies of Foxp3+ CD4 T cells were remarkably increased to 17.2% (PBS) and 18.1% (DMSO). DIM reduced the frequency to 7.7%. This experiment was repeated twice, producing similar results (PBS, 17.2±2.5%; DMSO, 18.1±3.0%; 1 µM DIM, 7.7±2.1%), that were statistically significant: being compared with PBS, p=0.0002 for 1 µM DIM.
Figure 6. DIM inhibits the differentiation of Foxp3+ Tregs induced by tolerogenic DCs. Naïve CD4+CD62+ helper T cells (2×105) were cultured with 4×104 Flt3/GM-CSF-stimulated bone marrow cultures in 1 mL of culture medium (RPMI 1640 with 10 mM HEPES, 2 mM L-glutamine, 10% FBS, and 50 µM β-mercaptoethanol) in the presence of soluble anti-CD3 Ab (1 µg/mL), human TGF-β (3 ng/mL, and murine IL-2 (5 ng/mL) for 5 days. Then cells were harvested, stained with fluorochrome conjugated-anti-CD4 and -anti-Foxp3 Abs, and analyzed by flow cytometry. DIM (1 µM) or DMSO (0.1% v/v) was added to the Flt3/GM-CSF-stimulated bone marrow cultures 1 h before the addition of Flt3L/GM-CSF. The results are representative of three separate experiments that showed similar results. Data were analyzed by the paired Student's t-test.
DISCUSSION
In this study, we demonstrated that AhR was functionally expressed in FGBMCs and CD11+CD103+ DCs and that AhR activation by DIM inhibited the in vitro development of CD103+ DCs from bone marrow cells stimulated with Flt3L and GM-CSF. DIM interfered with the phosphorylation of STAT3 and STAT5 inhibiting the expression of Id2, E2-2, IDO-1, and Aldh1a2, which are associated with CD103+ DC differentiation and functions. Finally, DIM suppressed the ability of CD103+ DCs to induce Foxp3+ Tregs. This is the first direct evidence that AhR regulates the development of CD103+ DCs from bone marrow cells.
AhR regulates the proliferation and differentiation of various immune cells. In the intestines, AhR is indispensable for the maintenance and functions of intraepithelial lymphocytes and innate lymphoid cells (21). Treatment of mice with TCDD increased the number of bone marrow Lin-Sca-1+c-Kit+ hematopoietic stem cells (HSCs) (43,44). HSCs from AhR-/- knockout mice were hyperproliferative and had an altered cell cycle (45), suggesting that AhR has a significant role in the regulation of HSCs. The effects of AhR activation on the development of DCs appear to be dependent on DC lineages and culture conditions (24,25,46,47). AhR activation induced tolerogenic DCs in mice given AhR ligands 2-(1'-H-indole-3'-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) or VAF347 (24,25), but there is no direct evidence regarding which tolerogenic DCs were induced. The results of in vitro studies using BMDCs induced by GM-CSF/IL-4 are controversial. In one study using ITE, it was found that tolerogenic DCs were induced by ITE treatment based on increased production of TGF-β and IL-10 and a concomitant reduction in proinflammatory cytokines, including IL-1β, IL-6, and IL-12 (25). In other studies using TCDD or benzo(a)pyrene, TCDD inhibited the differentiation of BMDCs while benzo(a)pyrene interfering with the growth of BM cells (46,47). GM-CSF and Flt3L have different effects on DC lineage determination (41). GM-CSF-induced activation of STAT5 and canonical NF-κB transcription factors promotes the intrinsic immunogenicity of DCs, whereas DCs generated in response to Flt3L are tolerogenic. In this study, we found that DIM and TCDD inhibited the differentiation of CD103+ DCs from BM cells in the presence of GM-CSF and Flt3L. Additional studies using different lineages of DCs are necessary to understand the roles of AhR in DC differentiation.
GM-CSF and Flt3L mediate DC development by activating the signal transducers STAT5 and STAT3 (40). How the activation of STAT3 and STAT5 is regulated by AhR remains largely unknown. AhR regulates STAT signaling pathways either by modulating STAT expression or by modulating STAT phosphorylation. The levels of STAT proteins were reduced in BM-derived macrophages or antigen-stimulated mast cells derived from AhR-deficient mice (48), whereas the expression of AhR itself was under the control of STAT3 (49). Although little is known regarding the regulation of STAT activation by AhR ligands, the effects of AhR activation on the phosphorylation of STATs can be inhibitory or stimulatory. In a previous study, the activation of STAT1 and STAT3 under IL-17 producing-Th17 cell-polarizing conditions was compared in AhR WT and knock-out mice. In the study, activation of STAT1, but not of STAT3, was regulated by AhR activation (50). In other studies, indoxyl 3-sulfate and 6-formylindolo(3,2-b)carbazole (FICZ) inhibited the phosphorylation of STAT5 and STAT6 and of STAT6 during Th2 differentiation, respectively (51,52). In contrast, indoxyl 3-sulfate stimulated the phosphorylation of STAT3 during Th17 differentiation (42).
An increasing number of studies support a role for c-Src in the phosphorylation of STAT proteins. In A431 cells, a human epidermoid carcinoma cell line, that endogenously expresses high levels of epidermal growth factor receptor, it was shown that c-Src kinase activity was required for the epidermal growth factor-induced tyrosine phosphorylation of STAT1, STAT3, and STAT5 (53). In another study conducted in COS-1 cells, Src kinase activation resulted in the phosphorylation of both STAT5A and STAT5B (54).
c-Src, which is associated with hsp90 and AhR in the cytosol of adipose and liver cells was released from the AhR complex and activated upon TCDD exposure (55). c-Src was rapidly phosphorylated by the AhR ligands hexachlorobenzene and TCDD (42,56). AhR activity was also required for Src-dependent phosphorylation of IDO1 in DCs (57). Alternatively, c-Src was found to be activated by the inhibition of Csk kinase, which phosphorylates c-Src at a tyrosine residue (Tyr527 in mouse c-Src), inhibiting Src kinase activity (58). In AhR-null fibroblasts, the expression of C-terminal Src kinase-binding protein (Cbp), which is required for the recruitment of Csk to the plasma membrane to phosphorylate c-Src, was overexpressed, suggesting that AhR negatively regulates Cbp expression to result in c-Src activation (59). In the present study, the phosphorylation of STAT3 and STAT5 but not of c-Src and IκBα was greatly inhibited by DIM, suggesting that AhR has specific effects on signaling molecules. The Src kinase family consists of several members including c-Src, Fyn, and Yes (60). Thus, which Src kinase is regulated by AhR activation remains unclear.
The transcription factors inhibitor of DNA binding (Id)2, a member of the helix-loop-helix transcription family, and interferon regulatory factor (IRF)8 are required for the development of CD103+ DCs (61,62). Id, which is upregulated during development (31,61) and highly expressed in BM-derived CD103+ populations, antagonizes E-protein activity impairing pDC development (63). IRF8, which is expressed nearly exclusively in hematopoietic cells of both the myeloid and lymphoid lineages (64), is well known for its multiple effects on the development of various immune cells including macrophages, granulocytes, and subsets of DCs (62,65,66). It was previously reported that GM-CSF-mediated DC differentiation depends on IRF-4, whereas Flt3L-mediated differentiation depends mainly on IRF-8 (67). The E protein E2-2, a helix-loop-helix protein, that is highly expressed in plasmacytoid DCs (65), specifically regulates the generation and maintenance of plasmacytoid DCs (68). In this study, the expression of Id2 and E2-2, but not of IRF8, in FGBMCs and CD11+CD103+ DCs was inhibited by DIM. STAT5 stimulated expression of Id2 in response to GM-CSF in DC progenitors, while E2-2 mRNA was increased in DCs exposed to Flt3L (40). The relative importance of the regulated expression of Id2 and E2-2 by DIM in the differentiation of CD103+ DCs and the underlying regulatory mechanisms require further examination.
Tolerologenic CD103+ DCs produce effector molecules such as retinoic acid, TGF-β, and tryptophan metabolites (39). The expression of Aldh1a1 was reduced in melanoma cells exposed to AhR ligands (69). Exposure to benzo(a) pyrene, an AhR ligand, affected monocyte-derived DCs function without affecting IDO1 expression (70), whereas lipopolysaccharide stimulated IDO1 expression in splenic DCs (58). In the present study, DIM inhibited the expression of the IDO-1 and Aldh1a2 genes in FGBMCs CD11+CD103+ DCs while exerting little effect on TGF-β (Fig. 4). IDO expression in DCs was stimulated by activated STAT3 (71). It remains largely unknown how the expression of IDO-1 and Aldh1a2 is regulated by AhR.
In conclusion, our results indicate that AhR activation regulates oral tolerance to commensals and food antigens by regulating the development of CD103+ DCs via inhibiting the STAT3 and STAT5 signaling pathways and thus modulating expression of genes associated with the development and functions of CD103+ DCs. AhR activation by TCDD was reported to impair oral tolerance, but the underlying mechanisms have not been examined in detail (28,29). Thus, AhR-regulated CD103+ DCs differentiation may explain the breakdown of oral tolerance by AhR. Accumulating evidence indicates that AhR ligands can produce different, and nearly opposite effects in the immune system. Systemic administration of TCDD alleviated the symptoms of experimental autoimmune encephalomyelitis by inducing Treg cells, whereas FICZ worsened experimental autoimmune encephalomyelitis, interfering with Treg development while boosting Th17 differentiation (72). Further studies should be conducted to examine how AhR activation regulates the activation of STAT3 and STAT5, how the expression of Id2, E2-2, IDO-1, and Aldh1a2 is regulated by AhR activation, and whether the effects of DIM on CD103+ DC differentiation can be generalized for AhR activation by other ligands.
ACKNOWLEDGEMENTS
This work was supported by Changwon National University Grants (2015-2016).
Abbreviations
- DC
dendritic cell
- AhR
aryl hydrocarbon receptor
- DIM
3,3'-diindolylmethane
- Treg
regulatory T cell
- FGBMCs
Flt3L/GM-CSF-stimulated bone marrow cultures
Footnotes
CONFLICTS OF INTEREST: The authors have no financial conflict of interest.
References
- 1.Burbach KM, Poland A, Bradfield CA. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci U S A. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rushmore TH, Pickett CB. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J Biol Chem. 1990;265:14648–14653. [PubMed] [Google Scholar]
- 3.Swanson HI, Bradfield CA. The AH-receptor: genetics, structure and function. Pharmacogenetics. 1993;3:213–230. doi: 10.1097/00008571-199310000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Huff J, Lucier G, Tritscher A. Carcinogenicity of TCDD: experimental, mechanistic, and epidemiologic evidence. Annu Rev Pharmacol Toxicol. 1994;34:343–372. doi: 10.1146/annurev.pa.34.040194.002015. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barouki R, Coumoul X, Fernandez-Salguero PM. The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett. 2007;581:3608–3615. doi: 10.1016/j.febslet.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 7.Feng S, Cao Z, Wang X. Role of aryl hydrocarbon receptor in cancer. Biochim Biophys Acta. 2013;1836:197–210. doi: 10.1016/j.bbcan.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Abbott BD, Birnbaum LS, Perdew GH. Developmental expression of two members of a new class of transcription factors: I. Expression of aryl hydrocarbon receptor in the C57BL/6N mouse embryo. Dev Dyn. 1995;204:133–143. doi: 10.1002/aja.1002040204. [DOI] [PubMed] [Google Scholar]
- 9.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 10.Puga A, Ma C, Marlowe JL. The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem Pharmacol. 2009;77:713–722. doi: 10.1016/j.bcp.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quintana FJ, Sherr DH. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacol Rev. 2013;65:1148–1161. doi: 10.1124/pr.113.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powrie F, Read S, Mottet C, Uhlig H, Maloy K. Control of immune pathology by regulatory T cells. Novartis Found Symp. 2003;252:92–98. [PubMed] [Google Scholar]
- 13.Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL, Berg PL, Davidsson T, Powrie F, Johansson-Lindbom B, Agace WW. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med. 2008;205:2139–2149. doi: 10.1084/jem.20080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coombes JL, Siddiqui KR, rancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fallarino F, Grohmann U, You S, McGrath BC, Cavener DR, Vacca C, Orabona C, Bianchi R, Belladonna ML, Volpi C, Santamaria P, Fioretti MC, Puccetti P. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176:6752–6761. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- 16.Perdew GH, Babbs CF. Production of Ah receptor ligands in rat fecal suspensions containing tryptophan or indole-3-carbinol. Nutr Cancer. 1991;16:209–218. doi: 10.1080/01635589109514159. [DOI] [PubMed] [Google Scholar]
- 17.Heath-Pagliuso S, Rogers WJ, Tullis K, Seidel SD, Cenijn PH, Brouwer A, Denison MS. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry. 1998;37:11508–11515. doi: 10.1021/bi980087p. [DOI] [PubMed] [Google Scholar]
- 18.Yaktine AL, Harrison GG, Lawrence RS. Reducing exposure to dioxins and related compounds through foods in the next generation. Nutr Rev. 2006;64:403–409. doi: 10.1111/j.1753-4887.2006.tb00225.x. [DOI] [PubMed] [Google Scholar]
- 19.Aires A, Mota VR, Saavedra MJ, Rosa EA, Bennett RN. The antimicrobial effects of glucosinolates and their respective enzymatic hydrolysis products on bacteria isolated from the human intestinal tract. J Appl Microbiol. 2009;106:2086–2095. doi: 10.1111/j.1365-2672.2009.04180.x. [DOI] [PubMed] [Google Scholar]
- 20.Celloto VR, Oliveira AJ, Goncalves JE, Watanabe CS, Matioli G, Goncalves RA. Biosynthesis of indole-3-acetic acid by new Klebsiella oxytoca free and immobilized cells on inorganic matrices. ScientificWorldJournal. 2012:495970. doi: 10.1100/2012/495970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stange J, Veldhoen M. The aryl hydrocarbon receptor in innate T cell immunity. Semin Immunopathol. 2013;35:645–655. doi: 10.1007/s00281-013-0389-1. [DOI] [PubMed] [Google Scholar]
- 22.Simones T, Shepherd DM. Consequences of AhR activation in steady-state dendritic cells. Toxicol Sci. 2011;119:293–307. doi: 10.1093/toxsci/kfq354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen NT, Kimura A, Nakahama T, Chinen I, Masuda K, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauben E, Gregori S, Draghici E, Migliavacca B, Olivieri S, Woisetschlager M, Roncarolo MG. Activation of the aryl hydrocarbon receptor promotes allograft-specific tolerance through direct and dendritic cell-mediated effects on regulatory T cells. Blood. 2008;112:1214–1222. doi: 10.1182/blood-2007-08-109843. [DOI] [PubMed] [Google Scholar]
- 25.Quintana FJ, Murugaiyan G, Farez MF, Mitsdoerffer M, Tukpah AM, Burns EJ, Weiner HL. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2010;107:20768–20773. doi: 10.1073/pnas.1009201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Cao X. Regulatory dendritic cells in autoimmunity: A comprehensive review. J Autoimmun. 2015;63:1–12. doi: 10.1016/j.jaut.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Awasthi A, Carrier Y, Peron JP, Bettelli E, Kamanaka M, Flavell RA, Kuchroo VK, Oukka M, Weiner HL. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat Immunol. 2007;8:1380–1389. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- 28.Kinoshita H, Abe J, Akadegawa K, Yurino H, Uchida T, Ikeda S, Matsushima K, Ishikawa S. Breakdown of mucosal immunity in gut by 2,3,7,8-tetraclorodibenzo-p-dioxin (TCDD) Environ Health Prev Med. 2006;11:256–263. doi: 10.1007/BF02898015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chmill S, Kadow S, Winter M, Weighardt H, Esser C. 2,3,7,8-Tetrachlorodibenzo-p-dioxin impairs stable establishment of oral tolerance in mice. Toxicol Sci. 2010;118:98–107. doi: 10.1093/toxsci/kfq232. [DOI] [PubMed] [Google Scholar]
- 30.Bjeldanes LF, Kim JY, Grose KR, Bartholomew JC, Bradfield CA. Aromatic hydrocarbon responsiveness-receptor agonists generated from indole-3-carbinol in vitro and in vivo: comparisons with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Proc Natl Acad Sci U S A. 1991;88:9543–9547. doi: 10.1073/pnas.88.21.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson JT, Hu Y, Liu R, Masson F, D'Amico A, Carotta S, Xin A, Camilleri MJ, Mount AM, Kallies A, Wu L, Smyth GK, Nutt SL, Belz GT. Id2 expression delineates differential checkpoints in the genetic program of CD8alpha+ and CD103+ dendritic cell lineages. EMBO J. 2011;30:2690–2704. doi: 10.1038/emboj.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueno H, Klechevsky E, Morita R, Aspord C, Cao T, Matsui T, Di PT, Connolly J, Fay JW, Pascual V, Palucka AK, Banchereau J. Dendritic cell subsets in health and disease. Immunol Rev. 2007;219:118–142. doi: 10.1111/j.1600-065X.2007.00551.x. [DOI] [PubMed] [Google Scholar]
- 34.Frericks M, Meissner M, Esser C. Microarray analysis of the AHR system: tissue-specific flexibility in signal and target genes. Toxicol Appl Pharmacol. 2007;220:320–332. doi: 10.1016/j.taap.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Jux B, Kadow S, Esser C. Langerhans cell maturation and contact hypersensitivity are impaired in aryl hydrocarbon receptor-null mice. J Immunol. 2009;182:6709–6717. doi: 10.4049/jimmunol.0713344. [DOI] [PubMed] [Google Scholar]
- 36.Miniero R, De FE, Ferri F, di DA. An overview of TCDD half-life in mammals and its correlation to body weight. Chemosphere. 2001;43:839–844. doi: 10.1016/s0045-6535(00)00442-2. [DOI] [PubMed] [Google Scholar]
- 37.Bradfield CA, Bjeldanes LF. Structure-activity relationships of dietary indoles: a proposed mechanism of action as modifiers of xenobiotic metabolism. J Toxicol Environ Health. 1987;21:311–323. doi: 10.1080/15287398709531021. [DOI] [PubMed] [Google Scholar]
- 38.Watowich SS, Liu YJ. Mechanisms regulating dendritic cell specification and development. Immunol Rev. 2010;238:76–92. doi: 10.1111/j.1600-065X.2010.00949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott CL, Aumeunier AM, Mowat AM. Intestinal CD103+ dendritic cells: master regulators of tolerance? Trends Immunol. 2011;32:412–419. doi: 10.1016/j.it.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Li HS, Yang CY, Nallaparaju KC, Zhang H, Liu YJ, Goldrath AW, Watowich SS. The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development. Blood. 2012;120:4363–4373. doi: 10.1182/blood-2012-07-441311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van de Laar L, Coffer PJ, Woltman AM. Regulation of dendritic cell development by GM-CSF: molecular control and implications for immune homeostasis and therapy. Blood. 2012;119:3383–3393. doi: 10.1182/blood-2011-11-370130. [DOI] [PubMed] [Google Scholar]
- 42.Hwang SJ, Hwang YJ, Yun MO, Kim JH, Oh GS, Park JH. Indoxyl 3-sulfate stimulates Th17 differentiation enhancing phosphorylation of c-Src and STAT3 to worsen experimental autoimmune encephalomyelitis. Toxicol Lett. 2013;220:109–117. doi: 10.1016/j.toxlet.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 43.Murante FG, Gasiewicz TA. Hemopoietic progenitor cells are sensitive targets of 2,3,7,8-tetrachlorodibenzo-p-dioxin in C57BL/6J mice. Toxicol Sci. 2000;54:374–383. doi: 10.1093/toxsci/54.2.374. [DOI] [PubMed] [Google Scholar]
- 44.Sakai R, Kajiume T, Inoue H, Kanno R, Miyazaki M, Ninomiya Y, Kanno M. TCDD treatment eliminates the long-term reconstitution activity of hematopoietic stem cells. Toxicol Sci. 2003;72:84–91. doi: 10.1093/toxsci/kfg002. [DOI] [PubMed] [Google Scholar]
- 45.Gasiewicz TA, Singh KP, Bennett JA. The Ah receptor in stem cell cycling, regulation, and quiescence. Ann N Y Acad Sci. 2014;1310:44–50. doi: 10.1111/nyas.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JA, Hwang JA, Sung HN, Jeon CH, Gill BC, Youn HJ, Park JH. 2,3,7,8-Tetrachlorodibenzo-p-dioxin modulates functional differentiation of mouse bone marrow-derived dendritic cells Downregulation of RelB by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Lett. 2007;173:31–40. doi: 10.1016/j.toxlet.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 47.Hwang JA, Lee JA, Cheong SW, Youn HJ, Park JH. Benzo(a)pyrene inhibits growth and functional differentiation of mouse bone marrow-derived dendritic cells. Downregulation of RelB and eIF3 p170 by benzo(a)pyrene. Toxicol Lett. 2007;169:82–90. doi: 10.1016/j.toxlet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 48.Beamer CA, Seaver BP, Shepherd DM. Aryl hydrocarbon receptor (AhR) regulates silica-induced inflammation but not fibrosis. Toxicol Sci. 2012;126:554–568. doi: 10.1093/toxsci/kfs024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stobbe-Maicherski N, Wolff S, Wolff C, Abel J, Sydlik U, Frauenstein K, Haarmann-Stemmann T. The interleukin-6-type cytokine oncostatin M induces aryl hydrocarbon receptor expression in a STAT3-dependent manner in human HepG2 hepatoma cells. FEBS J. 2013;280:6681–6690. doi: 10.1111/febs.12571. [DOI] [PubMed] [Google Scholar]
- 50.Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U S A. 2008;105:9721–9726. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeong KT, Hwang SJ, Oh GS, Park JH. FICZ, a tryptophan photoproduct, suppresses pulmonary eosinophilia and Th2-type cytokine production in a mouse model of ovalbumin-induced allergic asthma. Int Immunopharmacol. 2012;13:377–385. doi: 10.1016/j.intimp.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 52.Hwang YJ, Yun MO, Jeong KT, Park JH. Uremic toxin indoxyl 3-sulfate regulates the differentiation of Th2 but not of Th1 cells to lessen allergic asthma. Toxicol Lett. 2014;225:130–138. doi: 10.1016/j.toxlet.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 53.Olayioye MA, Beuvink I, Horsch K, Daly JM, Hynes NE. ErbB receptor-induced activation of stat transcription factors is mediated by Src tyrosine kinases. J Biol Chem. 1999;274:17209–17218. doi: 10.1074/jbc.274.24.17209. [DOI] [PubMed] [Google Scholar]
- 54.Kazansky AV, Kabotyanski EB, Wyszomierski SL, Mancini MA, Rosen JM. Differential effects of prolactin and src/abl kinases on the nuclear translocation of STAT5B and STAT5A. J Biol Chem. 1999;274:22484–22492. doi: 10.1074/jbc.274.32.22484. [DOI] [PubMed] [Google Scholar]
- 55.Enan E, Matsumura F. Identification of c-Src as the integral component of the cytosolic Ah receptor complex, transducing the signal of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) through the protein phosphorylation pathway. Biochem Pharmacol. 1996;52:1599–1612. doi: 10.1016/s0006-2952(96)00566-7. [DOI] [PubMed] [Google Scholar]
- 56.Dong B, Cheng W, Li W, Zheng J, Wu D, Matsumura F, Vogel CF. FRET analysis of protein tyrosine kinase c-Src activation mediated via aryl hydrocarbon receptor. Biochim Biophys Acta. 2011;1810:427–431. doi: 10.1016/j.bbagen.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, Bicciato S, Mazza EM, Macchiarulo A, Vacca C, Iannitti R, Tissi L, Volpi C, Belladonna ML, Orabona C, Bianchi R, Lanz TV, Platten M, la Fazia MA, Piobbico D, Zelante T, Funakoshi H, Nakamura T, Gilot D, Denison MS, Guillemin GJ, DuHadaway JB, Prendergast GC, Metz R, Geffard M, Boon L, Pirro M, Iorio A, Veyret B, Romani L, Grohmann U, Fallarino F, Puccetti P. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Superti-Furga G, Fumagalli S, Koegl M, Courtneidge SA, Draetta G. Csk inhibition of c-Src activity requires both the SH2 and SH3 domains of Src. EMBO J. 1993;12:2625–2634. doi: 10.1002/j.1460-2075.1993.tb05923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rey-Barroso J, Colo GP, varez-Barrientos A, Redondo-Munoz J, Carvajal-Gonzalez JM, Mulero-Navarro S, Garcia-Pardo A, Teixido J, Fernandez-Salguero PM. The dioxin receptor controls beta1 integrin activation in fibroblasts through a Cbp-Csk-Src pathway. Cell Signal. 2013;25:848–859. doi: 10.1016/j.cellsig.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 60.Roskoski R., Jr Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun. 2004;324:1155–1164. doi: 10.1016/j.bbrc.2004.09.171. [DOI] [PubMed] [Google Scholar]
- 61.Hacker C, Kirsch RD, Ju XS, Hieronymus T, Gust TC, Kuhl C, Jorgas T, Kurz SM, Rose-John S, Yokota Y, Zenke M. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- 62.Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, Price J, Yin N, Bromberg J, Lira SA, Stanley ER, Nussenzweig M, Merad M. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagasawa M, Schmidlin H, Hazekamp MG, Schotte R, Blom B. Development of human plasmacytoid dendritic cells depends on the combined action of the basic helix-loop-helix factor E2-2 and the Ets factor Spi-B. Eur J Immunol. 2008;38:2389–2400. doi: 10.1002/eji.200838470. [DOI] [PubMed] [Google Scholar]
- 64.Driggers PH, Ennist DL, Gleason SL, Mak WH, Marks MS, Levi BZ, Flanagan JR, Appella E, Ozato K. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc Natl Acad Sci U S A. 1990;87:3743–3747. doi: 10.1073/pnas.87.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H, Nakabayashi J, Umehara M, Miyake N, Matsumoto N, Nakazawa M, Ozato K, Tamura T. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 2013;121:1839–1849. doi: 10.1182/blood-2012-06-437863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sasaki H, Kurotaki D, Osato N, Sato H, Sasaki I, Koizumi S, Wang H, Kaneda C, Nishiyama A, Kaisho T, Aburatani H, Morse HC, III, Ozato K, Tamura T. Transcription factor IRF8 plays a critical role in the development of murine basophils and mast cells. Blood. 2015;125:358–369. doi: 10.1182/blood-2014-02-557983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, O'Shea JJ, Singh H, Ozato K. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 68.Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier C, den Hollander NS, Kant SG, Holter W, Rauch A, Zhuang Y, Reizis B. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Contador-Troca M, varez-Barrientos A, Merino JM, Morales-Hernandez A, Rodriguez MI, Rey-Barroso J, Barrasa E, Cerezo-Guisado MI, Catalina-Fernandez I, Saenz-Santamaria J, Oliver FJ, Fernandez-Salguero PM. Dioxin receptor regulates aldehyde dehydrogenase to block melanoma tumorigenesis and metastasis. Mol Cancer. 2015;14:148. doi: 10.1186/s12943-015-0419-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kazantseva MG, Highton J, Stamp LK, Hessian PA. Dendritic cells provide a potential link between smoking and inflammation in rheumatoid arthritis. Arthritis Res Ther. 2012;14:R208. doi: 10.1186/ar4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noh KT, Son KH, Jung ID, Kang TH, Choi CH, Park YM. Glycogen Synthase Kinase-3beta (GSK-3beta) Inhibition Enhances Dendritic Cell-based Cancer Vaccine Potency via Suppression of Interferon-gamma-induced Indoleamine 2,3-Dioxygenase Expression. J Biol Chem. 2015;290:12394–12402. doi: 10.1074/jbc.M114.628578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]