Abstract
Dissociation encompasses changes in a series of phenotypes: colony and cell morphology, inmunological and biochemical reactions and virulence. The concept is generally associated to the in vitro transition between smooth (S) and rough (R) colonies, a phenotypic observation in Gram-negative bacteria commonly made since the beginning of microbiology as a science. It is also well known that the loss of the O-polysaccharide, the most external lipopolysaccharide (LPS) moiety, triggers the change in the colony phenotype. Although dissociation is related to one of the most basic features used to distinguish between species, i.e., colony morphology, and, in the case of pathogens, predict their virulence behavior, it has been considered a laboratory artifact and thus did not gain further attention. However, recent insights into genetics and pathogenesis of members of Brucella, causative agents of brucellosis, have brought a new outlook on this experimental fact, suggesting that it plays a role beyond the laboratory observations. In this perspective article, the current knowledge on Brucella LPS genetics and its connection with dissociation in the frame of evolution is discussed. Latest reports support the notion that, by means of a better understanding of genetic pathways linked to R phenotype and the biological impact of this intriguing “old” phenomenon, unexpected applications can be achieved.
Keywords: Brucella, lipopolysaccharide, O-polysaccharide, rough, smooth
The Brucella genus includes Gram-negative microorganisms that cause brucellosis, a major worldwide zoonosis. The taxonomical criteria used to divide the genus into several species include host preference, physiological differences, phage susceptibility and cell envelope structural features. Based on the aspect of colonies on agar plates, which is in accordance with the cell surface and lipopolysaccharide (LPS) structure, Brucella may occur either as smooth (S) or rough (R) species. The zoonotically more relevant S species B. melitensis, B. suis and B. abortus express a full LPS molecule (S-LPS) that is anchored in the outer membrane (OM) (Whatmore, 2009). This group furthermore comprises species that have been isolated from rodents (B. neotomae) and marine mammals (B. ceti and B. pinnipedialis; Foster et al., 2007). More recently, B. microti, primarily isolated from voles and red foxes, was isolated directly from soil, a fact that has not been reported for any other S species (Scholz et al., 2008). In contrast, the naturally occurring R species B. ovis and B. canis express R-LPS that lacks the O-antigen, a trait linked to their reduced virulence.
Brucella LPS: Overall structure and function
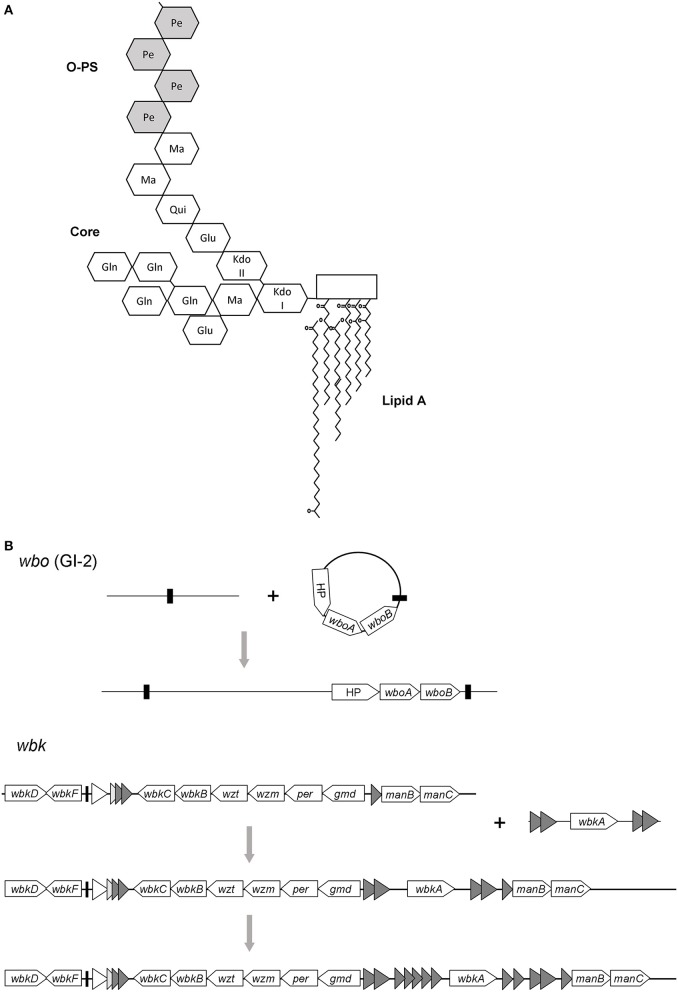
Similar to many LPS of Gram-negative pathogens, the Brucella LPS plays a fundamental role in the interaction with the corresponding host. In contrast to the well-known endotoxic properties manifested by enterobacterial LPS, the Brucella LPS is a paradigm of a poorly endotoxic, barely proinflammatory molecule unable to activate the innate immunity to a great extent (Moreno et al., 1981; Lapaque et al., 2005). Its structure consists of three covalently bound canonical motifs anchored in the OM: lipid A, the most hydrophobic part that is embedded in the OM; a core oligosaccharide that creates a bridge to the O-antigen or O-polysaccharide (O-PS), the third and most external surface moiety. Brucella lipid A contains a diaminoglucose backbone with reduced quantities of phosphate. The backbone is substituted with saturated C16 and C18 fatty acids, but also with unusual long-chain hydroxilated C28 and other very long acyl chains, a feature that Brucella share with members of the plant-symbiont genus Rhizobium (Moreno et al., 1990; Iriarte et al., 2004). The lipid A is linked to an oligosaccharide core chemically composed of 3-deoxy-D-manno-2-octulosonic acid (KDO), glucosamine, glucose, mannose and quinovosamine (Iriarte et al., 2004). Even though the core structure has not yet been definitively solved, recent insights suggest that it holds a mannose-containing lateral branch that hampers the recognition by complement, antimicrobial peptides and pathogen recognition receptor complex TLR4-MD2 (Conde-Álvarez et al., 2012). In addition, genetic and structural analyses demonstrated that some core sugars are not connecting the O-PS with the lipid A, which confirms such a branched array (Conde-Álvarez et al., 2012; Kubler-Kielb and Vinogradov, 2013; Gil-Ramírez et al., 2014). The O-PS is a linear homopolymer of N-formylperosamine, an unusual chemical composition among members of α-2 Proteobacteria (Moreno and Moriyón, 2006). The O-PS confers resistance to the innate bactericidal response by preventing deposition of complement factors at the cell surface (Eisenschenk et al., 1999), but also by impairing the binding of antimicrobial peptides to the membrane (Martínez de Tejada et al., 1995). The O-PS also hinders the production of proinflammatory cytokines (Barquero-Calvo et al., 2007), and along with the core provides receptor moieties for brucellaphages (Monreal et al., 2003). Hence, the interference with innate immunity mechanisms induced by Brucella LPS is critical to avoid an early host immune response, thus allowing a successful intracellular infection (Gorvel and Moreno, 2002). A scheme of LPS structure is given in Figure 1A.
Figure 1.
(A) A summary of Brucella LPS structure showing the sugar backbone of O-PS and core plus lipid A. Glu, glucose; Gln, glucosamine; Ma, mannose; Pe, N-formylperosamine; Qui, quinovosamine (based on Iriarte et al. (2004) and Gil-Ramírez et al. (2014). (B) Evolutionary scenario proposed for the O-polysaccharide in Brucella. The wbo locus was probably acquired by a single horizontal transfer event involving the unstable element GI-2 and its cognate integrase. In contrast, the wbk locus might evolve from a primary integration, followed by the addition of the wbkA gene by transposition. Thus, the wbkA region could serve as bait for transposition of related elements (shaded triangles). Black bars indicate direct repeats and a tRNA gene in wbo and wbk regions, respectively.
Horizontal transfer and O-PS acquisition
Contrary to the structure of lipid A and core of LPS, the O-PS is quite variable in bacteria. The diversity of repeat units within the O-PS and linkages between them is responsible for the O-PS variation and provides the basis for the O-serotyping classification in Enterobacteriaceae (Wang et al., 2010). The genes responsible for the O-PS synthesis in Gram-negative bacteria are mainly clustered in the chromosome and often form a single transcriptional unit (Reeves and Wang, 2002). They can be categorized into three groups: nucleotide sugar pathway genes; those encoding glycosyltransferases (GT), which can also be found scattered throughout the genome; and those for processing and transport. Brucella is no exception from this rule, its O-PS genes are encoded in two main loci wbk and wbo. The O-PS synthesis depends on two GT genes carried by wbo (wboA and wboB) (McQuiston et al., 1999; González et al., 2008) that are included in the Brucella genomic island GI-2 (Rajashekara et al., 2008), an unstable genetic element of 15.1 kb (Mancilla et al., 2010). This region carries an additional gene, located close to GT genes, that encodes a hypothetical protein (BMEI0999) that apparently is part of the O-PS synthetic machinery since attempts to complement ΔGI-2 mutants using a plasmid carrying only wboA-wboB have failed (Rajashekara et al., 2008). Major O-PS locus wbk is located between a ribose transport system (rbs) and an rnc gene (Moriyón et al., 2004). The low GC content of approximately 50%, a feature shared with many O-PS clusters, has been linked to its hypothetical acquisition via horizontal transfer (Cloeckaert et al., 2000; Godfroid et al., 2000). This region encodes the genes putatively necessary to synthesize perosamine (gmd, per), n-formylation of perosamine residues (wbkC), GT for polymerization (wbkE, wbkA), to prime bactoprenol (wbkD, wbkF), and ABC transporters that translocate the O-PS (wzm and wzt). Genes for mannose synthesis have also been identified in the same region (manAOAg, manBOAg, and manCOAg), but mutational analysis of manBOAg indicated that it is not essential (González et al., 2008), since independent homologs located in the chromosome II (manBAcore) are able to meet mannose demands (Monreal et al., 2003). To date, there is no confirmed function for wbkB (encodes a putative perosamine synthetase) because the corresponding mutant preserved the S phenotype and the hypothetical ligase that binds the amino sugar O-PS to the lipid A-core in the periplasmic interface has not yet been identified (Moriyón et al., 2004).
Concerning horizontal acquisitions outside from wbk and wbo, the role of a gene cluster encoding enzymes for LPS biosynthesis has been proposed (Vizcaíno et al., 2001). The cluster was named GI-8 and can be found in the majority of “classic” Brucella species although it is absent from B. abortus (Rajashekara et al., 2004). Consistent with their functionality, the expression of several genes carried by GI-8 has been detected in B. melitensis (Rossetti et al., 2009). Moreover, an exopolysaccharide consisting of glucosamine, glucose and mostly mannose has been described in B. melitensis 16M (Godefroid et al., 2010). The annotation matches with genes expected for the biosynthesis of constituents previously mentioned, therefore the data strongly support a role of GI-8 in the production of such an exopolysaccharide.
Since members of Brucella are facultative intracellular parasites that usually inhabit a constrained environment that precludes horizontal transfer with other bacteria, this ecological niche might explain the homogeneity of their O-PS structures or, in other terms, the restricted O-serotyping diversity found across Brucella species. This point of view does not contradict the acquisition of O-PS genes via HGT because it is thought that they were captured and integrated into the ancestral Brucella genome before the speciation (Figure 1B).
Mechanisms of dissociation
The phenotypic change from S to R colonies experienced by Brucella under cultivation is widely known (Braun, 1946). Nevertheless, the S-R dissociation is far from being a behavior exclusively manifested by the brucellae. Several reports (McCallum et al., 1989; Liu and Reeves, 1994; Walsh and Moran, 1997) and reviews (Reeves, 1995; Reeves and Wang, 2002; Wang et al., 2010) have reported on this phenomenon in major Enterobacteriaceae species and even in microorganisms phylogenetically remote such as Mycobacteriaceae (Eckstein et al., 2000). It has been speculated that the propensity to become rough under laboratory conditions reflects the fact that the O-PS is needed in natural niches (Reeves, 1995). Indeed, mutants defective in O-PS are generally serum sensitive, since their lipid A represents an exposed target for complement killing activity (Rautemaa and Meri, 1999). Despite its importance in virulence for Gram-negative pathogens, the mechanisms affecting the stability of O-PS, i.e., dissociation, have only been marginally explored. In Brucella, this phenomenon has been described very early (Henry, 1933; Braun, 1946) and is of great importance in basic research and vaccine production (Alton et al., 1988). Furthermore, the organization of its O-PS clusters and the location of the O-PS GT is representative of several Gram-negative bacteria and, therefore, clues from Brucella dissociation mechanisms may be extrapolated to other bacteria.
According to the stochasticity of the mutations implicated in dissociation, we can distinguish two main pathways. There are unpredictable mutations in which the LPS gene(s) affected originate from an apparently coincidental event. In contrast, mutations that arise upon a discrete and reproducible recombination event follow well-defined pathways. Typical examples for random mutations are those displayed by attenuated vaccine strains obtained through several culture passages (Alton et al., 1988; Moriyón et al., 2004). In some instances, the corresponding R-linked genetic defect could be identified as was the case with the B. abortus RB51 strain. Transposition of the IS711 element into the wboA caused the disruption of the gene, a fact that contributed to the roughness of the RB51 strain (Vemulapalli et al., 1999). In B. melitensis B115 R strain, a nonsense mutation in wzm was identified that interrupts the O-PS transport and thus causes accumulation of O-PS in the cytoplasm of this mutant (Adone et al., 2008). Further reports have shown that point mutations, indels and rearrangements of the manBAcore locus occurred in spontaneous B. melitensis 16M R mutants (Turse et al., 2011) and that R strains maintained in the laboratory over years can accumulate mutations on LPS genes (Adone et al., 2011).
It has become clear that Brucella and other organisms carry unstable, mobile elements implicated in chromosomal deletions that lead to the loss of LPS genes. These DNA fragments are often maintained under selective pressure from specific exposure to the host, which means that upon environmental changes they can be released from the chromosome. This statement especially applies for brucellae, whose O-PS loci are located in spots of recombination. For instance, the wbo locus can spontaneously excise from the chromosome due to being part of a genomic island (Mancilla et al., 2010). The excision is prompted by site-specific recombination between 41 bp flanking repeats catalyzed by the phage-related integrase of GI-2 (Mancilla et al., 2010). Interestingly, this mechanism, along with spontaneous mutations in wbk genes (Zygmunt et al., 2009), could be involved in the evolution of the naturally R species B. ovis that lacks GI-2 (Vizcaíno et al., 2004). The role of homologous recombination mediated by the RecA protein in dissociation has also been investigated (Mancilla et al., 2012). This recombination activity is responsible for the spontaneous excision of wbkA, which resides in a putative transposon remnant. wbkA- flanking ISBm1 elements are involved in a recombination event that leads to the excision and loss of a 5.5 kb fragment including the wbkA gene from a small fraction of the bacterial population. It must be pointed out that the excessive recombination affecting these loci has been observed under unfavorable culture conditions, but the biological significance remains unclear. On the other hand, a deletion of 351 bp comprising the wbkF-wbkD genes has given rise to the R phenotype of B. canis (Zygmunt et al., 2009). The mutation might have occurred due to a slipped mispairing mechanism involving short direct repeats. Although, it may follow a reproducible pathway, the deletion has not been detected in samples positive for GI-2 and wbkA deletions (unpublished results). The genetic events related to Brucella dissociation are summarized in Table 1.
Table 1.
Stochastic and non-stochastic events related to dissociation.
| S-R mutation | Mechanism | Species/strain | References |
|---|---|---|---|
| GI-2 deletion | Site-specific recombination | B. abortus, B. melitensis, and B. suis | Mancilla et al., 2010 |
| wbkA deletion | Homologous recombination | B. abortus, B. melitensis, and B. suis | Mancilla et al., 2012 |
| wbkFD deletion | Strand-slippage during replication | B. canis | Zygmunt et al., 2009 |
| manBAcore indels, large deletion | Strand-slippage, homologous recombination | B. abortus 2308, B. melitensis 16M | Turse et al., 2011 |
| wboA::IS711 | Gene disruption by IS transposition | B. abortus RB51 | Vemulapalli et al., 1999 |
| wzm mutation | Frameshift derived from a point mutation | B. melitensis B115 | Adone et al., 2011 |
In this context, the repeat units found in the wbk region deserve special attention. The presence of IS and related remnants may offer sequence substrates for homologous recombination which could result in extensive deletions. Indeed, the atypical Brucella spp. BO2 strain seems to have lost a large portion of the wbk locus. Instead of that, a cluster of rhamnose-based O-PS biosynthetic genes is responsible for the S-LPS phenotype depicted by this strain, which explains the untypeable character using conventional antibodies (Wattam et al., 2012; Zygmunt et al., 2012).
The biological significance of dissociation
It has been stated that Brucella O-PS is a critical virulence factor of classical S species. Spontaneous R variants undergo enhanced intracellular killing by macrophages (Fernandez-Prada et al., 2001), consistent with an increased activation of these cells in culture (Fernandez-Prada et al., 2003; Rittig et al., 2003). In the murine brucellosis model, R mutants are cleared faster than their S counterpart (Allen et al., 1998). We know that the O-PS is largely responsible for the stealthy behavior manifested by the S cells, which is why it does not seem likely that dissociation plays a role under field conditions, where the pressure imposed by the host limits growth and spread. However, in a recent study concerning the LPS expression of B. melitensis in infected cell cultures, it has been shown that it is possible to isolate R types from mice infected with S cells (Turse et al., 2011). The authors argued that dissociation is a natural process taking place during infection, a conclusion also supported by the enhanced growth shown by the manBAcore mutants recovered. Later, the same research team demonstrated that the cytotoxicity of R cells are necessary for in vitro egress and dissemination of B. melitensis S cells from infected host cells, suggesting for the first time a biological role for dissociation (Pei et al., 2014). Contrary to what would have been expected, this fact might change the current model of infection in which the S cells are able to induce cell lysis and spread by themselves, without relying on R mutants (Starr et al., 2012). As a consequence, we can speculate that this finding would also explain the presence of R strains originated from S species in collections of field strains (Dorneles et al., 2014; Bertu et al., 2015).
Examples of genomic changes that impact the infection outcome suggest that there are loci, mainly encoding virulence genes, that are preferentially mutated and even lost during the infection. These mutations may lead to lifelong host-pathogen relationship, a fact that has already been described for some uropathogenic Escherichia coli strains (Zdziarski et al., 2010). Interestingly, spontaneous mutations on LPS genes of Burkholderia pseudomallei, causative agent of melioidosis, have been proposed to be involved in the persistence of some R strains (Tuanyok et al., 2012). Moreover, B. melitensis R variants have been ocassionally isolated from goat milk samples, suggesting that R types can survive in the mammary gland (Mancilla et al., 2012). It might be inferred that only those mutations that originate R types with a balanced intracellular fitness that allows competition with S cells would prevail. Therefore, the abolition of dissociation mechanisms or pathways predicts a deleterious effect on the fitness of the resulting R-mutants that may impact the infection outcome. If S brucellae have evolved into dissociation-prone species able to change their major “pathogen credential” when necessary is yet a matter of speculation. But certainly, the emergence of R types in vitro as well as in vivo suggests that brucellae may not only experience phenotypic heterogeneity but also that this fact could be needed for displaying full virulence of S cells. The idea that dissociation may occur in the extracellular milieu is stressed by the recent finding of B. microti environmental isolates devoid of O-PS (Al Dahouk et al., 2012). Aditionally, reversion (from R to S) has been reported in vivo, a fact that accounts for the mentioned heterogeneity during host-pathogen interaction (Pérez-Sancho et al., 2014). But this phenomenon, that suggests reversibility of dissociation, still awaits for further genetic characterization.
Future directions
The acquisition of clusters of genes is recognized as the major driving force for prokaryotic evolution (Hacker and Carniel, 2001). These clusters are part of the flexible, not essential gene pool that is maintained due to the environmental pressure, the integrative machinery encoded by the foreign fragment and/or endogenous homologous recombination (Gal-Mor and Finlay, 2006). Once the selective pressure is removed, spontaneous gene or even cluster deletions are triggered by recombination often by the system involved in the chromosomal integration process. The result of such deletions is a dramatic loss of virulence of the pathogen and over-attenuation in the case of vaccine strains. An important example concerning the impact of excessive recombination on the stability of vaccine strains is M. bovis BCG, the strain used to prevent tuberculosis. In fact, the deletion of a single 9 kb fragment known as RD1 is responsible for the marked attenuation of BCG in comparison with M. tuberculosis and M. bovis pathogenic strains (Pym et al., 2002).
In the case of Brucella, the licensed B. abortus S19 and B. melitensis Rev 1 s strains are superior vaccines with a long history of successful utilization in the eradication of brucellosis but unfortunately tend to dissociate, a characteristic related to a low efficacy (Alton et al., 1988; Grilló et al., 2000). The discovery of genes and cis elements involved in LPS-loci deletion pathways prepare the ground to control undesirable excessive recombination events and thus to avoid over-attenuation of established S live vaccines. A proof of this concept is the recent development of an improved version of B. melitensis Rev 1. Interestingly, the newly engineered strain called Rev 2 exhibits an enhanced stability owing to the abolishment of two well-proven dissociation mechanisms (Mancilla et al., 2013). Noteworthy, similarities between Brucella LPS loci organization and other Gram-negative bacteria suggest that those may also have acquired their O-PS by horizontal transfer and might undergo the same dissociation mechanisms. Therefore, awareness of these processes could be generally useful to obtain more stable bacterial strains for antigen and vaccine production, regarding not only control and eradication of brucellosis: this knowledge may be applied to improve vaccines against a broader spectrum of bacterial infections. However, dissociation pathway(s) to be abrogated should be carefully studied in order to avoid disruption of mechanisms important in the context of infection and development of a specific immunity.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the FONDECYT 11130347 grant from the National Comission for Scientific and Technological Research, CONICYT, Chile. The author thanks Dr. Melanie Kaiser for her valuable commentaries on the manuscript.
References
- Adone R., Francia M., Ciuchini F. (2008). Evaluation of Brucella melitensis B115 as rough-phenotype vaccine against B. melitensis and B. ovis infections. Vaccine 26, 4913–4917. 10.1016/j.vaccine.2008.07.030 [DOI] [PubMed] [Google Scholar]
- Adone R., Muscillo M., La Rosa G., Francia M., Tarantino M. (2011). Antigenic, immunologic and genetic characterization of rough strains B. abortus RB51, B. melitensis B115 and B. melitensis B18. PLoS ONE 6:e24073. 10.1371/journal.pone.0024073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Dahouk S., Hofer E., Tomaso H., Vergnaud G., Le Flèche P., Cloeckaert A., et al. (2012). Intraspecies biodiversity of the genetically homologous species Brucella microti. Appl. Environ. Microbiol. 78, 1534–1543. 10.1128/AEM.06351-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alton G., Jones L., Angus R., Verger J. M. (eds.). (1988). The production of Brucella vaccines, in Techniques for the Brucellosis Laboratory (Paris: INRA; ), 143–156. [Google Scholar]
- Allen C. A., Adams L. G., Ficht T. A. (1998). Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect. Immun. 66, 1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquero-Calvo E., Chaves-Olarte E., Weiss D. S., Guzmàn-Verri C., Chacón-Díaz C., Rucavado A., et al. (2007). Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS ONE 2:e631. 10.1371/journal.pone.0000631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertu W. J., Ducrotoy M. J., Muñoz P. M., Mick V., Zúñiga-Ripa A., Bryssinckx W., et al. (2015). Phenotypic and genotypic characterization of Brucella strains isolated from autochthonous livestock reveals the dominance of B. abortus biovar 3a in Nigeria. Vet. Microbiol. 180, 103–108. 10.1016/j.vetmic.2015.08.014 [DOI] [PubMed] [Google Scholar]
- Braun W. (1946). Dissociation in Brucella abortus: a demonstration of the role of inherent and environmental factors in bacterial variation. J. Bacteriol. 51, 327–349. [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., Grayon M., Verger J. M., Letesson J. J., Godfroid F. (2000). Conservation of seven genes involved in the biosynthesis of the lipopolysaccharide O-side chain in Brucella spp. Res. Microbiol. 151, 209–216. 10.1016/S0923-2508(00)00141-8 [DOI] [PubMed] [Google Scholar]
- Conde-Alvarez R., Arce-Gorvel V., Iriarte M., Mancek-Keber M., Barquero-Calvo E., Palacios-Chaves L., et al. (2012). The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition. PLoS Pathog. 8:e1002675. 10.1371/journal.ppat.1002675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorneles E. M., Santana J. A., Alves T. M., Pauletti R. B., Mol J. P., Heinemann M. B., et al. (2014). Genetic stability of Brucella abortus isolates from an outbreak by multiple-locus variable-number tandem repeat analysis (MLVA16). BMC Microbiol. 14:186. 10.1186/1471-2180-14-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein T. M., Inamine J. M., Lambert M. L., Belisle J. T. (2000). A genetic mechanism for deletion of the ser2 gene cluster and formation of rough morphological variants of Mycobacterium avium. J. Bacteriol. 182, 6177–6182. 10.1128/JB.182.21.6177-6182.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenschenk F. C., Houle J. J., Hoffmann E. M. (1999). Mechanism of serum resistance among Brucella abortus isolates. Vet. Microbiol. 68, 235–244. 10.1016/S0378-1135(99)00075-9 [DOI] [PubMed] [Google Scholar]
- Fernandez-Prada C. M., Nikolich M., Vemulapalli R., Sriranganathan N., Boyle S. M., Schurig G. G., et al. (2001). Deletion of wboA enhances activation of the lectin pathway of complement in Brucella abortus and Brucella melitensis. Infect. Immun. 69, 4407–4416. 10.1128/IAI.69.7.4407-4416.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Prada C. M., Zelazowska E. B., Nikolich M., Hadfield T. L., Roop R. M., II, Robertson G. L., et al. (2003). Interactions between Brucella melitensis and human phagocytes: bacterial surface O-Polysaccharide inhibits phagocytosis, bacterial killing, and subsequent host cell apoptosis. Infect. Immun. 71, 2110–2119. 10.1128/IAI.71.4.2110-2119.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster G., Osterman B. S., Godfroid J., Jacques I., Cloeckaert A. (2007). Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int. J. Syst. Evol. Microbiol. 57, 2688–2693. 10.1099/ijs.0.65269-0 [DOI] [PubMed] [Google Scholar]
- Gal-Mor O., Finlay B. B. (2006). Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell. Microbiol. 8, 1707–1719. 10.1111/j.1462-5822.2006.00794.x [DOI] [PubMed] [Google Scholar]
- Gil-Ramírez Y., Conde-Álvarez R., Palacios-Chaves L., Zúñiga-Ripa A., Grilló M. J., Arce-Gorvel V., et al. (2014). The identification of wadB, a new glycosyltransferase gene, confirms the branched structure and the role in virulence of the lipopolysaccharide core of Brucella abortus. Microb. Pathog. 73, 53–59. 10.1016/j.micpath.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Godefroid M., Svensson M. V., Cambier P., Uzureau S., Mirabella A., De Bolle X., et al. (2010). Brucella melitensis 16M produces a mannan and other extracellular matrix components typical of a biofilm. FEMS Immunol. Med. Microbiol. 59, 364–377. 10.1111/j.1574-695x.2010.00689.x [DOI] [PubMed] [Google Scholar]
- Godfroid F., Cloeckaert A., Taminiau B., Danese I., Tibor A., de Bolle X., et al. (2000). Genetic organisation of the lipopolysaccharide O-antigen biosynthesis region of Brucella melitensis 16M (wbk). Res. Microbiol. 151, 655–668. 10.1016/S0923-2508(00)90130-X [DOI] [PubMed] [Google Scholar]
- González D., Grilló M. J., De Miguel M. J., Ali T., Arce-Gorvel V., Delrue R. M., et al. (2008). Brucellosis vaccines: assessment of Brucella melitensis lipopolysaccharide rough mutants defective in core and O-polysaccharide synthesis and export. PLoS ONE 3:e2760. 10.1371/journal.pone.0002760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvel J. P., Moreno E. (2002). Brucella intracellular life: from invasion to intracellular replication. Vet. Microbiol. 90, 281–297. 10.1016/S0378-1135(02)00214-6 [DOI] [PubMed] [Google Scholar]
- Grilló M. J., Bosseray N., Blasco J. M. (2000). In vitro markers and biological activity in mice of seed lot strains and commercial Brucella melitensis Rev 1 and Brucella abortus B19 vaccines. Biologicals 28, 119–127. 10.1006/biol.2000.0249 [DOI] [PubMed] [Google Scholar]
- Hacker J., Carniel E. (2001). Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2, 376–381. 10.1093/embo-reports/kve097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B. (1933). Dissociation in the genus Brucella J. Infect. Dis. 52, 374–402. 10.1093/infdis/52.3.374 [DOI] [Google Scholar]
- Iriarte M., Gonzalez D., Delrue R. M., Monreal D., Conde R., Lopez-Goni I., et al. (2004). Brucella lipopoyscharide: structure, biosynthesis and genetics, in Brucella: Molecular and Cellular Biology, eds Lopez-Goni I., Moriyon I. (Pamplona: Horizon Bioscience; ), 159–191. [Google Scholar]
- Kubler-Kielb J., Vinogradov E. (2013). The study of the core part and non-repeating elements of the O-antigen of Brucella lipopolysaccharide. Carbohydr. Res. 366, 33–37. 10.1016/j.carres.2012.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapaque N., Moriyon I., Moreno E., Gorvel J. P. (2005). Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol. 8, 60–66. 10.1016/j.mib.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Liu D., Reeves P. R. (1994). Escherichia coli K12 regains its O antigen. Microbiology 140 (Pt 1), 49–57. 10.1099/13500872-140-1-49 [DOI] [PubMed] [Google Scholar]
- Mancilla M., Grilló M. J., de Miguel M. J., López-Gońi I., San-Román B., Zabalza-Baranguá A., et al. (2013). Deletion of the GI-2 integrase and the wbkA flanking transposase improves the stability of Brucella melitensis Rev 1 vaccine. Vet. Res. 44:105. 10.1186/1297-9716-44-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancilla M., López-Goñi I., Moriyón I., Zárraga A. M. (2010). Genomic island 2 is an unstable genetic element contributing to Brucella lipopolysaccharide spontaneous smooth-to-rough dissociation. J. Bacteriol. 192, 6346–6351. 10.1128/JB.00838-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancilla M., Marín C. M., Blasco J. M., Zárraga A. M., López-Goñi I., Moriyon I. (2012). Spontaneous excision of the O-polysaccharide wbkA glycosyltranferase gene is a cause of dissociation of smooth to rough Brucella colonies. J. Bacteriol. 194, 1860–1867. 10.1128/JB.06561-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez de Tejada G., Pizarro-Cerdá J., Moreno E., Moriyón I. (1995). The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect. Immun. 63, 3054–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum K. L., Schoenhals G., Laakso D., Clarke B., Whitfield C. (1989). A high-molecular-weight fraction of smooth lipopolysaccharide in Klebsiella serotype O1:K20 contains a unique O-antigen epitope and determines resistance to nonspecific serum killing. Infect. Immun. 57, 3816–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuiston J. R., Vemulapalli R., Inzana T. J., Schurig G. G., Sriranganathan N., Fritzinger D., et al. (1999). Genetic characterization of a Tn5-disrupted glycosyltransferase gene homolog in Brucella abortus and its effect on lipopolysaccharide composition and virulence. Infect. Immun. 67, 3830–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monreal D., Grilló M. J., González D., Marín C. M., De Miguel M. J., López-Goñi I., et al. (2003). Characterization of Brucella abortus O-polysaccharide and core lipopolysaccharide mutants and demonstration that a complete core is required for rough vaccines to be efficient against Brucella abortus and Brucella ovis in the mouse model. Infect. Immun. 71, 3261–3271. 10.1128/IAI.71.6.3261-3271.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E., Berman D. T., Boettcher L. A. (1981). Biological activities of Brucella abortus lipopolysaccharides. Infect. Immun. 31, 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E., Moriyón I. (2006). The genus Brucella, in The Prokaryotes, Vol. 5, Part 1, Section 31, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrant E. (New York, NY: Springer-Verlag; ), 315–456. [Google Scholar]
- Moreno E., Stackebrandt E., Dorsch M., Wolters J., Busch M., Mayer H. (1990). Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 172, 3569–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyón I., Grilló M. J., Monreal D., González D., Marín C., López-Goñi I., et al. (2004). Rough vaccines in animal brucellosis: structural and genetic basis and present status. Vet. Res. 35, 1–38. 10.1051/vetres:2003037 [DOI] [PubMed] [Google Scholar]
- Pei J., Kahl-McDonagh M., Ficht T. A. (2014). Brucella dissociation is essential for macrophage egress and bacterial dissemination. Front. Cell. Infect. Microbiol. 4:23. 10.3389/fcimb.2014.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Sancho M., Adone R., García-Seco T., Tarantino M., Diez-Guerrier A., Drumo R., et al. (2014). Evaluation of the immunogenicity and safety of Brucella melitensis B115 vaccination in pregnant sheep. Vaccine, 32, 1877–1881. 10.1016/j.vaccine.2014.01.070 [DOI] [PubMed] [Google Scholar]
- Pym A. S., Brodin P., Brosch R., Huerre M., Cole S. T. (2002). Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46, 709–717. 10.1046/j.1365-2958.2002.03237.x [DOI] [PubMed] [Google Scholar]
- Rajashekara G., Covert J., Petersen E., Eskra L., Splitter G. (2008). Genomic island 2 of Brucella melitensis is a major virulence determinant: functional analyses of genomic islands. J. Bacteriol. 190, 6243–6252. 10.1128/JB.00520-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajashekara G., Glasner J. D., Glover D. A., Splitter G. A. (2004). Comparative whole-genome hybridization reveals genomic islands in Brucella species. J. Bacteriol. 186, 5040–5051. 10.1128/JB.186.15.5040-5051.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautemaa R., Meri S. (1999). Complement-resistance mechanisms of bacteria. Microbes Infect. 1, 785–794. 10.1016/S1286-4579(99)80081-1 [DOI] [PubMed] [Google Scholar]
- Reeves P. (1995). Role of O-antigen variation in the immune response. Trends Microbiol. 3, 381–386. 10.1016/S0966-842X(00)88983-0 [DOI] [PubMed] [Google Scholar]
- Reeves P. P., Wang L. (2002). Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264, 109–135. 10.1007/978-3-642-56031-6_7 [DOI] [PubMed] [Google Scholar]
- Rittig M. G., Kaufmann A., Robins A., Shaw B., Sprenger H., Gemsa D., et al. (2003). Smooth and rough lipopolysaccharide phenotypes of Brucella induce different intracellular trafficking and cytokine/chemokine release in human monocytes. J. Leukoc. Biol. 74, 1045–1055. 10.1189/jlb.0103015 [DOI] [PubMed] [Google Scholar]
- Rossetti C. A., Galindo C. L., Lawhon S. D., Garner H. R., Adams L. G. (2009). Brucella melitensis global gene expression study provides novel information on growth phase-specific gene regulation with potential insights for understanding Brucella: host initial interactions. BMC Microbiol. 9:81. 10.1186/1471-2180-9-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz H. C., Hubalek Z., Nesvadbova J., Tomaso H., Vergnaud G., Le Flèche P., et al. (2008). Isolation of Brucella microti from soil. Emerging Infect. Dis. 14, 1316–1317. 10.3201/eid1408.080286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T., Child R., Wehrly T. D., Hansen B., Hwang S., López-Otin C., et al. (2012). Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 11, 33–45. 10.1016/j.chom.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuanyok A., Stone J. K., Mayo M., Kaestli M., Gruendike J., Georgia S., et al. (2012). The genetic and molecular basis of O-antigenic diversity in Burkholderia pseudomallei lipopolysaccharide. PLoS Negl. Trop. Dis. 6:e1453. 10.1371/journal.pntd.0001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turse J. E., Pei J., Ficht T. A. (2011). Lipopolysaccharide-deficient Brucella variants arise spontaneously during infection. Front. Microbiol. 2:54. 10.3389/fmicb.2011.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemulapalli R., McQuiston J. R., Schurig G. G., Sriranganathan N., Halling S. M., Boyle S. M. (1999). Identification of an IS711 element interrupting the wboA gene of Brucella abortus vaccine strain RB51 and a PCR assay to distinguish strain RB51 from other Brucella species and strains. Clin. Diagn. Lab. Immunol. 6, 760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno N., Caro-Hernández P., Cloeckaert A., Fernández-Lago L. (2004). DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6, 821–834. 10.1016/j.micinf.2004.04.009 [DOI] [PubMed] [Google Scholar]
- Vizcaíno N., Cloeckaert A., Zygmunt M. S., Fernández-Lago L. (2001). Characterization of a Brucella species 25-kilobase DNA fragment deleted from Brucella abortus reveals a large gene cluster related to the synthesis of a polysaccharide. Infect. Immun. 69, 6738–6748. 10.1128/IAI.69.11.6738-6748.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E. J., Moran A. P. (1997). Influence of medium composition on the growth and antigen expression of Helicobacter pylori. J. Appl. Microbiol. 83, 67–75. 10.1046/j.1365-2672.1997.00164.x [DOI] [PubMed] [Google Scholar]
- Wang L., Wang Q., Reeves P. R. (2010). The variation of O antigens in gram-negative bacteria. Subcell. Biochem. 53, 123–152. 10.1007/978-90-481-9078-2_6 [DOI] [PubMed] [Google Scholar]
- Wattam A. R., Inzana T. J., Williams K. P., Mane S. P., Shukla M., Almeida N. F., et al. (2012). Comparative genomics of early-diverging Brucella strains reveals a novel lipopolysaccharide biosynthesis pathway. MBio 3, e00246–e00211. 10.1128/mBio.00388-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore A. M. (2009). Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 9, 1168–1184. 10.1016/j.meegid.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Zdziarski J., Brzuszkiewicz E., Wullt B., Liesegang H., Biran D., Voigt B., et al. (2010). Host imprints on bacterial genomes–rapid, divergent evolution in individual patients. PLoS Pathog. 6:e1001078. 10.1371/journal.ppat.1001078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygmunt M. S., Blasco J. M., Letesson J. J., Cloeckaert A., Moriyón I. (2009). DNA polymorphism analysis of Brucella lipopolysaccharide genes reveals marked differences in O-polysaccharide biosynthetic genes between smooth and rough Brucella species and novel species-specific markers. BMC Microbiol. 9:92. 10.1186/1471-2180-9-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygmunt M. S., Jacques I., Bernardet N., Cloeckaert A. (2012). Lipopolysaccharide heterogeneity in the atypical group of novel emerging Brucella species. Clin. Vaccine Immunol. 19, 1370–1373. 10.1128/CVI.00300-12 [DOI] [PMC free article] [PubMed] [Google Scholar]