NeuriteTracker is a computer vision approach used to analyze neuronal morphodynamics and to examine spatiotemporal Rho GTPase signaling networks regulating neurite outgrowth.
Abstract
Rho guanosine triphosphatases (GTPases) control the cytoskeletal dynamics that power neurite outgrowth. This process consists of dynamic neurite initiation, elongation, retraction, and branching cycles that are likely to be regulated by specific spatiotemporal signaling networks, which cannot be resolved with static, steady-state assays. We present NeuriteTracker, a computer-vision approach to automatically segment and track neuronal morphodynamics in time-lapse datasets. Feature extraction then quantifies dynamic neurite outgrowth phenotypes. We identify a set of stereotypic neurite outgrowth morphodynamic behaviors in a cultured neuronal cell system. Systematic RNA interference perturbation of a Rho GTPase interactome consisting of 219 proteins reveals a limited set of morphodynamic phenotypes. As proof of concept, we show that loss of function of two distinct RhoA-specific GTPase-activating proteins (GAPs) leads to opposite neurite outgrowth phenotypes. Imaging of RhoA activation dynamics indicates that both GAPs regulate different spatiotemporal Rho GTPase pools, with distinct functions. Our results provide a starting point to dissect spatiotemporal Rho GTPase signaling networks that regulate neurite outgrowth.
Introduction
Neurite outgrowth is a prerequisite step to produce the axons and dendrites that wire the adult brain. Neuronal cell migration is crucial for brain morphogenesis. In cell culture, neurite outgrowth is highly dynamic and involves a series of dynamic morphogenetic processes (MPs) such as neurite initiation, elongation, branching, growth cone motility, and collapse (da Silva and Dotti, 2002). Cultured neuronal cells are also highly motile, which might reflect the process of neuronal migration (Nadarajah and Parnavelas, 2002). Specific spatiotemporal signaling networks control the cytoskeletal, trafficking, and adhesion dynamics required for each MP to occur. Because neurite outgrowth and cell migration use overlapping cellular machineries, it is conceivable that some molecular components might regulate both processes. These MPs fluctuate on length and time scales of tens of microns and minutes to hours, which have been missed in steady-state neurite outgrowth measurements, most often at a late differentiation stage. Identification of MP-specific signaling networks thus requires the quantification their morphodynamics.
Rho GTPases are key regulators of the cytoskeletal dynamics that regulate neuronal cell morphogenesis (da Silva and Dotti, 2002). Their activity is tightly controlled in time and space by guanine nucleotide-exchange factors (GEFs; Rossman et al., 2005) and GTPase-activating proteins (GAPs; Moon and Zheng, 2003) that activate and deactivate GTPases, respectively. This regulation, as well as coupling of Rho GTPases to specific downstream effectors, dictates their cytoskeletal output at any given subcellular localization (Pertz, 2010). Current models state that Rac1 and Cdc42 regulate neurite outgrowth, whereas RhoA controls neurite collapse (da Silva and Dotti, 2002). However, multiple GEFs, GAPs, and effectors are ubiquitously expressed by cells and outnumber their cognate Rho GTPases (Moon and Zheng, 2003; Rossman et al., 2005). This raises the question of the significance of this signaling complexity.
We present NeuriteTracker, a computer vision (CV) platform to track neuronal morphodynamics from high-content time-lapse imaging datasets. Automatic extraction of a large set of morphological and morphodynamic features, coupled with adequate statistical analysis, can then quantify the dynamics of neuronal morphogenesis. Our pipeline identifies distinct, stereotyped morphodynamic phases during neuronal cell morphogenesis and quantifies a set of morphodynamic phenotypes in a siRNA screen targeting a candidate Rho GTPase interactome. This provides insight into the spatiotemporal Rho GTPase signaling networks regulating distinct MPs. As proof of concept for our screen, we show that two RhoA-specific GAPs regulate two distinct spatiotemporal RhoA signaling networks controlling different cytoskeletal outputs. Our data provide an initial resource to study the complex spatiotemporal Rho GTPase signaling networks that regulate neuronal cell morphogenesis.
Results
High-content live-cell imaging pipeline
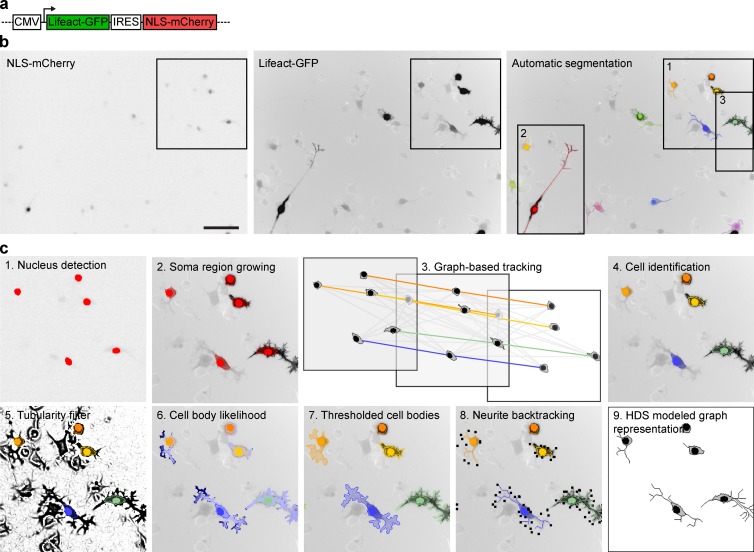
To study neuronal dynamics, we used neuronal-like mouse N1E-115 neuroblastoma cells. To visualize cell morphology, we used a bicistronic vector that expresses Lifeact-GFP, a fusion of GFP with the F-actin binding peptide Lifeact (Riedl et al., 2008), and a nuclear localization NLS-mCherry fusion, which labels the nucleus for cell detection (Fig. 1 a). This construct can be expressed at a high level without affecting neurite outgrowth (Fig. S1, a and b) and provides homogeneous high contrast on neurites and somata for imaging with air objectives (Fig. 1 b and Video 1). An additional benefit of our reporter is that it labels F-actin–containing branches not apparent using the classic microtubule staining used in neurite outgrowth assays. To perturb different signaling molecules, we cotransfected our reporter plasmid with siRNAs in nondifferentiated cells. These cells were subsequently differentiated by serum starvation, replated on laminin-coated coverslips, and allowed to extend neurites for 19.6 h (Fig. S1 c). As proof of concept, we evaluated knockdown (KD) of previously characterized proteins. MKK7 KD leads to highly unstable, short neurites owing to loss of microtubule bundling in the neurite shaft (Feltrin et al., 2012). RhoA KD leads to increased neurite outgrowth (Hirose et al., 1998) through loss of growth cone collapse controlled by RhoA (Fritz et al., 2013). SrGAP2 KD leads to stabilization of the growth cone and soma lamellipodial veils, increasing neurite outgrowth and branching, as well as cell spreading and migration (Pertz et al., 2008). Our protocol enabled efficient KD of these proteins, which recapitulated the expected phenotypes (Fig. S1, d–f). We also identified a nontargeting siRNA that did not affect neurite outgrowth (Fig. S1 g). To perform high-content live-cell imaging, we optimized our microscope setup for fast, two-color imaging of multiple wells. Time-lapse analysis started 3 h postplating, a time point at which initial neurites already protruded, rendering cells resistant to phototoxicity. An automated image analysis routine was then used to identify 10 fields of view per well containing an adequate number of cells exhibiting a specific range in fluorescence intensities and cell areas (Fig. S1, h–j). We then time-lapsed neurite outgrowth dynamics in 10 fields of view per well across a 24-well plate with 12-min time resolution for a total of 19.6 h.
Figure 1.
Tracking and segmentation of neuronal morphodynamics. (a) Schematics of Lifeact-GFP/NLS-mCherry expression vector. (b) Raw NLS-mCherry channel (left), raw Lifeact-GFP channel (middle), and Lifeact-GFP channel (right) with superimposed nucleus, soma, and neurite segmentations color-coded according to cell identity. Bar, 100 µm. Fluorescent images are shown in inverted black-and-white contrast. (c) Different segmentation steps highlighted in magnification from inset 1 from panel b.
NeuriteTracker pipeline to quantify neuronal dynamics
To analyze the time-lapse datasets, we engineered NeuriteTracker, an automated CV pipeline to segment and track soma and neurite morphodynamics described in Materials and methods (Fig. 1, b and c; and Video 2). First, mCherry-NLS–labeled nuclei and their associated somata were identified using maximally stable extremal regions and a fast marching region growing step (Fig. 1 c, steps 1 and 2). Second, a graph-association tracking method was applied over the time-lapse data to provide a unique label for each tracked cell over time (Fig. 1 c, steps 3 and 4). Neurites were extracted for each cell by applying a calibrated Hessian-based filter, computing a geodesic distance combining intensity and geometric distance, and applying a threshold (Fig. 1 c, steps 5–7). Candidate terminals were identified, and neurite filaments were detected by backtracing and reconstruction with a minimal spanning tree (Fig. 1 c, step 8). Again applying the graph association to the neurites yielded automatic segmentation and tracking of individual cell nuclei, soma, and neurites (Fig. 2 a and Video 2). Our pipeline provided robust results across a wide variety of fluorescence intensities that result from transient transfection and was evaluated against human annotated ground-truth datasets (Fig. S2 and Videos 3 and 4).
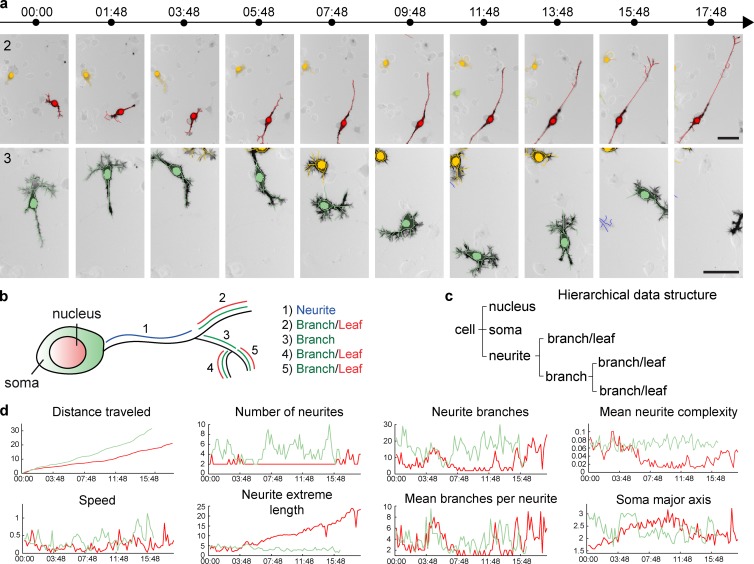
Figure 2.
Feature extraction from time-lapse datasets. (a) Example of segmentation of two cells from a time-lapse dataset (from insets 2 and 3 in Fig. 1 b). Nucleus, soma, and neurite segmentations are color-coded for cell identity. Time, h:min. Bar, 50 µm. (b) Schematics describing neuronal cell morphology, and how different neuronal parameters are extracted for definition of neuronal arborescence. (c) Example of the HDS that models the schematic cell in b. (d) Temporal evolution of a set of features for the two cells shown in panel a.
To describe the segmented cells, we defined a series of parameters that describe nucleus, soma, and neurite morphology, the neurite being modeled as a tree arborescence (Figs. 2 b and S3; also see Materials and methods, Definition of nuclei and soma parameters and Definition of neurite parameters). These data were then organized in a hierarchical data structure (HDS) that models the neurite, as well as the nucleus and soma geometries, at a given time point (Fig. 2 c). A graph representation of this HDS can then be rendered (Fig. 1 c, step 9). Finally, we used these parameters and the HDS to extract an as-comprehensive-as-possible set of features that describe nuclei, soma, and neurite morphology and morphodynamics. For each cell, 28 “static” features that provide measurements relevant to cell morphology at one specific time point were extracted from the HDS and stored in a feature vector as described in Materials and methods (Definition of static features). The dynamic evolution of some of these features is shown in Fig. 2 d. Furthermore, we computed five layers of dynamic features, which provide summaries of the temporal evolution of static features on a per-cell basis (described in Materials and methods, Definition of dynamic features). These include the mean value of static features through time (1), the time a feature expands/contracts (2/3), the number of inflection points during feature evolution (4), and the frequency of the expansion of a feature (5). This provides a comprehensive description of neurite and soma morphodynamics. Importantly, these different layers of dynamic features are to some extent redundant, and a selection of a specific set of features can be required to avoid redundancy during their analysis.
We also used our CV pipeline to track and segment neuronal morphodynamics of Lifeact-GFP/NLS-mCherry–transfected Pheochromocytoma PC-12 cells plated on collagen and stimulated with nerve growth factor. The raw images, their superimposed HDS graph representations, and the temporal evolution of some extracted features are shown in Fig. S4 (a–c) and indicate that our CV pipeline is sufficiently versatile to be applied to different neuronal-like cell types.
We provide access to the MATLAB code of NeuriteTracker through the Github platform at https://github.com/sgbasel/neuritetracker. Analysis of different neuronal morphologies requires the tuning of a small set of parameters, which can be conveniently performed using a graphical user interface. Ultimately, NeuriteTracker provides both the extracted features in the HDS format in a CSV file, as well as a graph representation of the HDS. A test time-lapse dataset and manual are also provided.
Functional analysis of a Rho GTPase signaling network
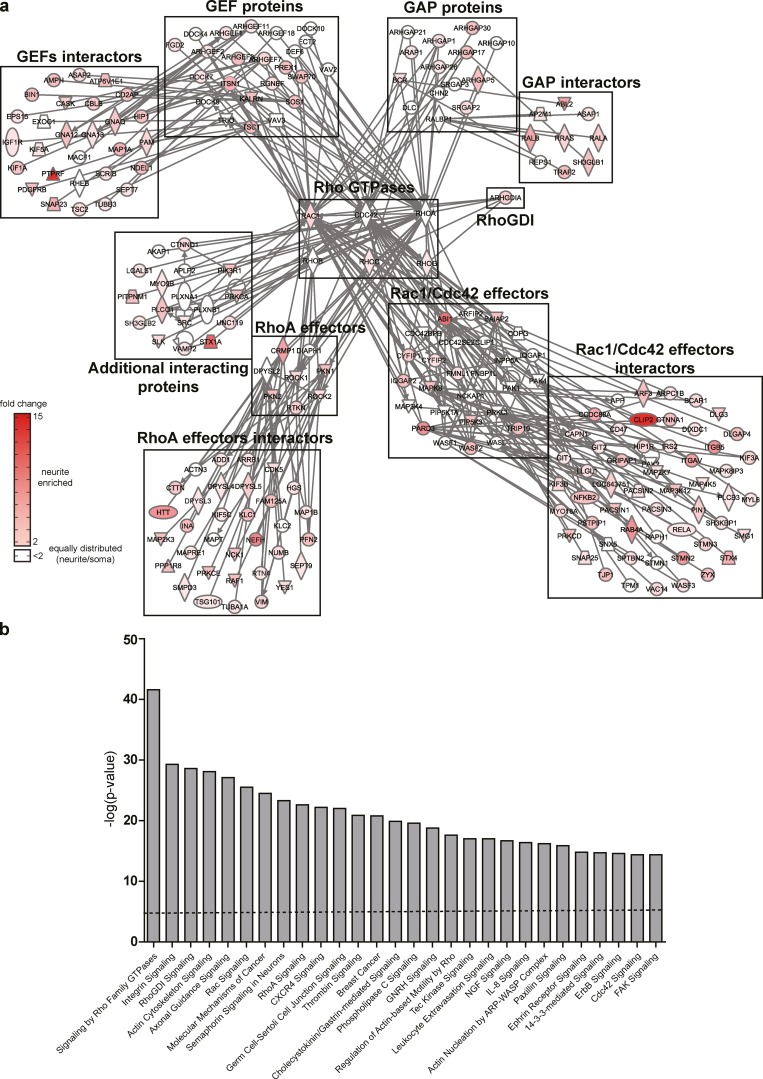
As a proof of concept to validate NeuriteTracker, we performed an siRNA screen targeting a potential Rho GTPase interactome. We used a bioinformatics approach to mine a proteomics dataset of purified neurite and soma fractions of N1E-115 cells (Pertz et al., 2008) to identify GEFs, GAPs, effectors, and their interacting proteins that potentially interact with a panel of Rho GTPases (Fig. 3 a; described in Materials and methods). This revealed a complex interactome consisting of 219, mostly neurite-enriched, proteins. Gene ontology analysis revealed a large coverage of cellular processes relevant to adhesion and cytoskeletal dynamics and axonal guidance (Fig. 3 b). We used three distinct siRNAs per gene, leading to a total of 657 perturbations (siRNA sequences are described in Table S1). These were distributed among 32 experiments, with each 24-well plate containing one negative, nontargeting and one positive, MKK7-targeting siRNA control. For each of these 721 wells (657 perturbations + 64 controls), we acquired 10 fields of view, therefore accounting for a total of 7,210 time-lapse movies. We then developed a statistical analysis pipeline to analyze this dataset.
Figure 3.
Identification of a candidate Rho GTPase signaling network. (a) Rho GTPase protein interaction network as determined by the strategy described in Materials and methods. Different families of proteins known to interact with six expressed Rho GTPases are shown. Proteins are named by their official gene symbol according to Entrez Gene. Relative neurite protein enrichment is color-coded according to the color scale bar at the bottom of the figure. Plain lines represent direct protein–protein interaction. Icons are color-coded according to the color scale bar. (b) Gene Ontology analysis of the most significant canonical pathways present in the protein network shown in panel a. Dotted line represents significance threshold as measured by Fisher’s exact test (P < 0.05).
Morphodynamic signature extraction
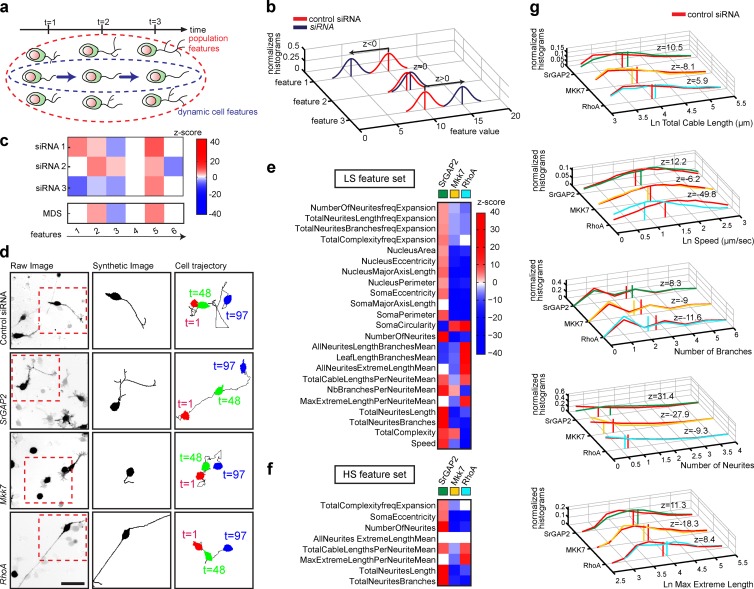
To quantify the effect of perturbations on neuronal dynamics, we considered 28 static features measured on the whole-cell population at all the different time points of a time-lapse dataset. This provides the distribution of feature values throughout the whole lifetime of the cell population (Fig. 4 a, population-level features). We also considered the “frequency expansion” feature set that consists of 18 features that summarize the dynamics of single cells throughout the movie (Fig. 4 a, dynamic cell-level features).
Figure 4.
Morphodynamic signature extraction. (a) Schematics of two distinct feature analysis levels. Static population-level and dynamic cell-level features are considered. (b) Feature extraction procedure. z-score vector defines statistical significance, penetrance, and polarity of measured feature distributions of control compared with siRNA-treated cells. (c) Deconvolution of RNAi OFF-target effects. For each given feature: (1) if the three siRNA perturbations lead to a z-score with identical polarity, the mean z-score is computed; (2) if two of three siRNA perturbations lead to a z-score with identical polarity, the mean z-score for the two features with the same polarity is computed; (3) if the three siRNA perturbations yield three different z-score polarities, then the z feature score is set to 0. (d) Raw image (left, inverted black-and-white contrast), HDS graph representation (middle), and cell migration tracks (right) for SrGAP2, MKK7, and RhoA siRNA perturbations are shown. Bar, 50 µm. (e and f) Gene-associated MDSs represented by a color map. (e) LS feature set; (f) HS feature set. z-score scale is also shown. (g) Normalized histograms of representative feature distribution profiles. The data were normalized dividing each histogram value by the histogram area. Vertical bars represent the mean. The z-score associated with each feature is also shown.
For each feature, we used a z-test to compare feature value distributions of control and siRNA-transfected cells (Fig. 4 b). The z-score polarity indicates if a specific feature value augments or decreases when control and KD cells are compared. All the details relevant to our data analysis pipeline are explained in Materials and methods. To mitigate any potential off-target effects, we computed mean z-scores from the three siRNAs, according to the rule that if at least two of three siRNAs produce the same phenotype for a given feature (Echeverri et al., 2006), then this phenotype is on target (Fig. 4 c). This approach provides a mean z-score vector that indicates penetrance and polarity for each feature in response to a siRNA perturbation (Lefort et al., 2015). We refer to this z-score vector as the morphodynamic signature (MDS) of the target gene.
We then reduced the feature space by two successive steps. First, we evaluated the feature robustness at different stringency levels by comparing the MDS of the MKK7-KD positive control phenotype across the 32 independent performed experiments. Specifically, we selected features that exhibited a statistically relevant z-score with identical polarity in at least 65% (low stringency [LS]) or 90% (high stringency [HS]) of the 32 MKK7 KD control experiments. Second, we selected features that have high information content; we retained features that exhibit a statistically significant z-score in at least 40% of the 219 genes across the siRNA screen. This especially affected “dynamic” features, because those require integration of a large number of measurements over time, leading to a lower measurement sample size than static features. This reduced the feature space to 32%/78% (LS feature set) and 75%/95% (HS feature set) static/dynamic features, providing robust feature sets that are highly informative about neuronal morphology and dynamics.
We evaluated the validity of our pipeline by extracting MDSs associated with our previously characterized SrGAP2, MKK7, and RhoA KD phenotypes (Figs. 4 d and S1 e). This revealed that our approach successfully captures morphological and morphodynamic phenotypes (Fig. 4, e and f). SrGAP2 KD led to increased neurite number, length, and branching, as well as increased soma migration, which further correlate with increased cell and nucleus eccentricity. As expected, MKK7 KD led to reduced frequency of neurite extension events, short neurites, and low soma motility. RhoA KD led to reduced collapse frequency, producing longer neurites, with reduced branching. Some of these features can be easily assessed by visual inspection of static images (neurite length/branching) or time-lapse datasets (soma speed). Others, such as soma circularity, are more subtle and can be detected only by our CV approach. Plotting feature distribution in the different perturbed states indicates the cellular heterogeneity associated with our cell system (Fig. 4 g).
siRNA screen analysis
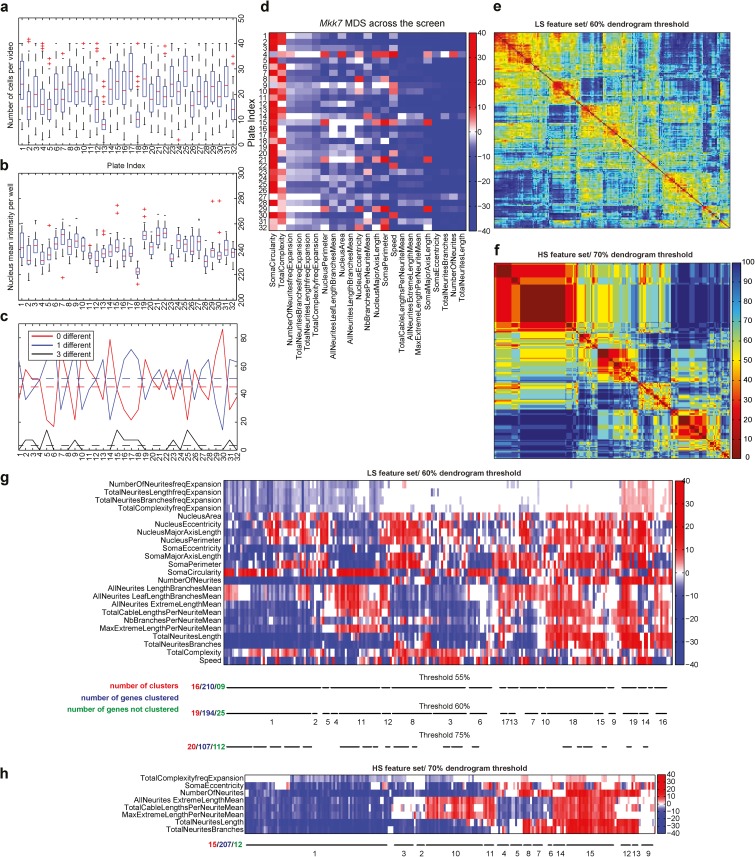
A set of quality-control metrics showed that a similar number of cells per field of view and NLS-mCherry fluorescence intensities occurred in the different experimental plates (Fig. 5, a and b). Quantification of off-target effects across the features, throughout the screen, revealed that 43% of the measurements led to identical z-score polarities for all three siRNAs; 52% of the measurements had two of three siRNAs yielding identical z-score polarities, whereas in only 5% of the cases, one siRNA led to a statistically nonsignificant z-score and the two other yielded two distinct polarities. Further, this low level of off-target effect was not biased to a specific set of experimental plates (Fig. 5 c). Finally, the positive MKK7-KD control associated with each plate yielded a stable MDS across the different experimental plates (Fig. 5 d), indicating the stability of our pipeline. Each siRNA perturbation led to a quantifiable MDS with a mean z-score of 1.2 ± 1.9 for the least penetrant and 40.9 ± 21.6 for the most penetrant phenotype. Even in the least penetrant phenotypes, a subset of features displayed statistically relevant z-scores.
Figure 5.
siRNA screen quality controls and MDS hierarchical clustering. (a) Boxplots quantification of number of cells per field of view across 32 independent experiments. Median (red line), interquartile range (box), and domain (dashed line) are represented. The outliers are plotted individually. (b) Boxplot quantification of NLS-mCherry mean fluorescence intensity across 32 independent experiments. Median (red line), interquartile range (box), and domain (dashed line) are represented. The outliers are plotted individually. (c) Per-plate off-target effect analysis across 32 independent experiments. The analysis takes in consideration the off-target effect of the LS feature set. Black lines indicate the three siRNAs generate different phenotypes. Blue line indicates that two siRNAs generate the same phenotype, but the phenotype of the third siRNA is different. Red lines indicate that the three siRNAs generate the same phenotype. For each plate, the proportion of each case is represented by continuous lines. The dashed line represents the mean value of the case across the whole screen. (d) MDS of MKK7-KD cells across 32 independent experiments. z-score vector maps are shown for each experimental plate. Right bar indicates z-score color code. (e and f) L0 distances comparing all the MDS to each other for low- (e) and high-stringency (f) feature sets. L0 distances were normalized to range between 0 and 100 (percentage). L0 distances are color-coded so that warm colors indicate low distances and cold colors indicate high distances. (g) MDSs hierarchical clustering using the LS feature set. MDS z-scores are color-coded according to the color scale. Identified clusters for different dendrogram thresholds are shown by black lines. The number of clusters, genes clustered, and nonclustered genes are also indicated. (h) MDSs hierarchical clustering using the HS feature set with a dendrogram threshold of 70%.
To identify sets of gene perturbations that lead to common MDSs, we binarized z-score vectors to give equal weight to all features and performed bottom-up, hierarchical clustering. Similar clustering results were observed whether z-score matrices were binarized or not (e.g., the z-score value was taken into account). Iterative evaluation of different dendrogram thresholds was performed by visual inspection of the quality of the clusters (Fig. 5, e–h), using HDS representations to evaluate the similarity of penetrant phenotypes for different gene perturbations within a cluster. The rationale to identify cells with penetrant phenotypes is mentioned in the next paragraph. For the LS feature set, a good compromise was obtained with a 60% dendrogram threshold, meaning that 60% of the features of the MDSs within a cluster have the same polarity. This led to the identification of 19 phenoclusters that comprise 194 of 219 genes (Fig. 5, e and g). Using the HS feature set led to the identification of 15 clusters that comprise 207 of the 219 genes with a 70% dendrogram threshold (Fig. 5, f and h). Any of the nonclustered genes also led to phenotypes, but these have to be inspected on a case-by-case basis.
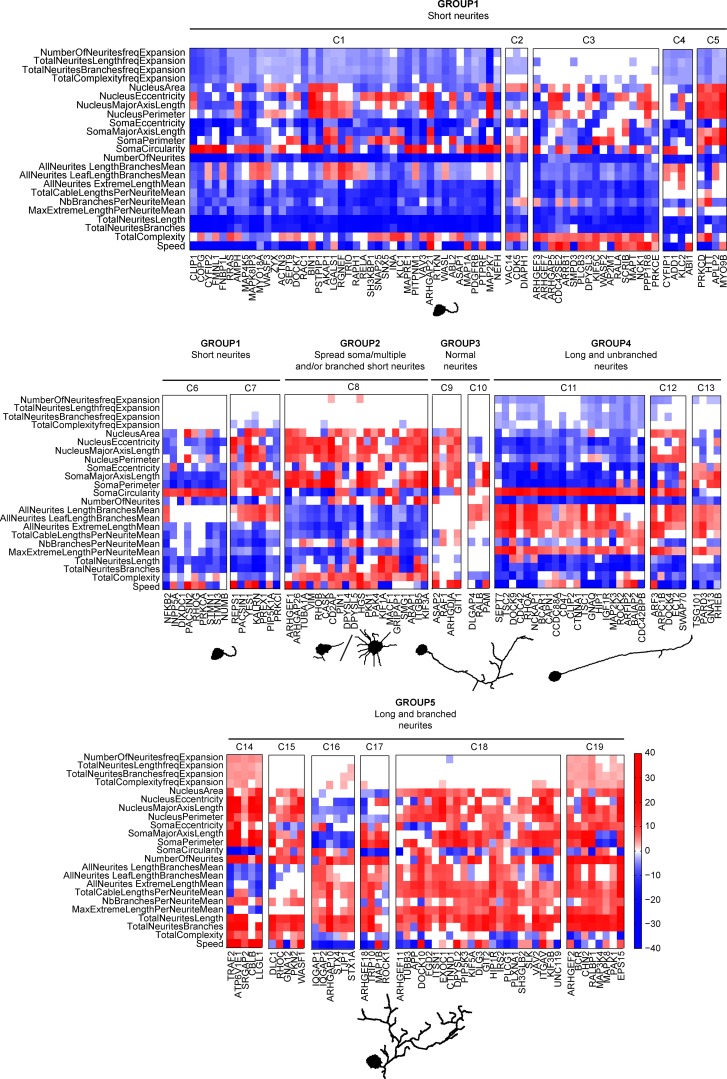
For intuitive display, we manually sorted phenoclusters according to the overall polarity of their MDSs (Figs. 6 and S4 d). We then described the phenoclusters at different complexity levels and focused first on static, then on dynamic phenotypes. Phenotypes that can be explained by static features led us to group the different clusters in five large phenotypic categories: short neurites (group 1: C1–C7), spread soma multiple and/or branched short neurites (group 2: C8), normal neurites with soma phenotypes (group 3: C9–C10), long/unbranched neurites (group 4: C11–C13), and long/branched neurites (group 5: C14–C19). To provide a visual representation of these phenotypic categories at the static level, we used a mathematical model that selects specific cells with a highly penetrant phenotype according to three visually intuitive features (number of neurites, number of branches, and maximum extreme length per neurite mean). These are then represented using HDS-modeled graph representations, providing a representative example of the phenotype (Fig. 6, Fig. S4 d, and Video 5). Within these static-level phenotype groups, the different subclusters are characterized by additional static features that are mostly relevant to nucleus and soma morphology and that are less visually evident. Dynamic features such as cell migration (speed) and features relevant to neurite outgrowth dynamics (total neurite length frequency expansion, etc.) then discriminate further clusters. Representative dynamic phenotypes were selected using our mathematical model using the same three features set but additionally including the total neurite length frequency expansion feature (Videos 6 and 7). Phenoclusters computed using LS and HS feature sets were similar, except for some sensitive phenotypes observed within cluster 8 (LS feature set; Figs. 6 and S4 d). The lower amount of clusters when the HS feature set is used can mostly be explained by the loss of the cell migration feature. Schematics and statistics of the whole workflow of our pipeline are described in Fig. S5. A web application based on the openBIS data management platform (Bauch et al., 2011) illustrates the static and dynamic phenotypes associated to each siRNA perturbation using the LS feature set (https://mat-openbis.dbm.unibas.ch:8443/openbis/webapp/sinergia/). This platform provides access to the z-score vector associated with each siRNA perturbation, as well as to the mean MDS. 10 representative images, as well as 10 representative time-lapse datasets of penetrant phenotypes (selected using the approach described at the beginning of this paragraph), and their matched experimental controls are also provided.
Figure 6.
Hierarchical clustering identifies MDS phenoclusters. Hierarchical clustering of MDSs using the LS feature set. Phenotypic clusters were manually sorted by the overall polarity of their MDSs. Representative images of major phenotype classes comprising multiple clusters were identified by a mathematical model described in Materials and methods section Identification of phenocluster representative cells. These representative images do not take into account any dynamic feature.
Characterization of stage-specific neuronal dynamics
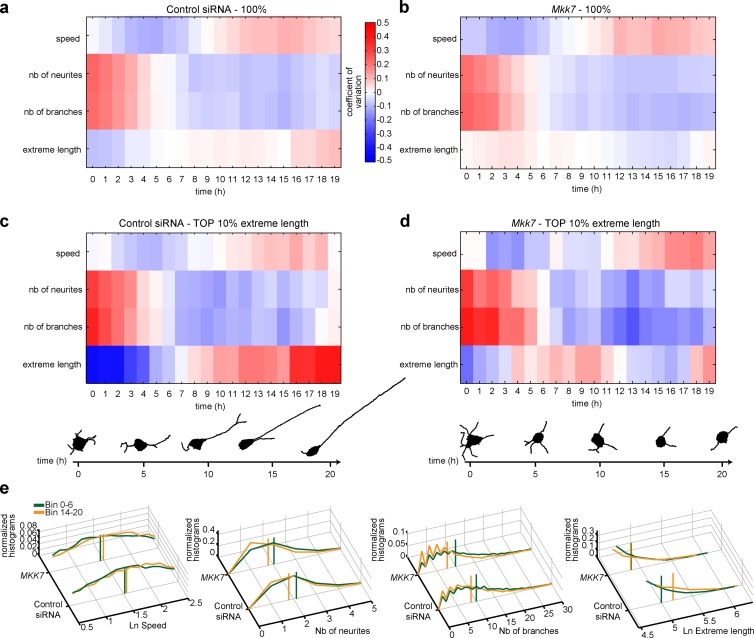
Neuronal differentiation involves multiple temporal episodes with distinct morphodynamic behaviors that have until now been described only qualitatively (da Silva and Dotti, 2002). To get quantitative insight into neuronal morphodynamics associated with different temporal episodes, we took advantage of the large control and MKK7-KD dataset from our siRNA screen dataset. We studied MDSs associated with sequential 1-h episodes from our 20-h time-lapse datasets in control cells. For scale-free representation, we plotted the temporal evolution of the coefficient of variation of a panel of features. Because only a fraction of the cell population extends robust neurites, we considered the whole population but also gated on the 10% of cells with the most robust neurite outgrowth (“all neurites extreme length mean feature”). At the whole-cell population level (Fig. 7 a), robust initiation of multiple short and branched neurites is observed at the onset of differentiation and then decreases during a first phase that lasts 7 h. This period is also associated with a low soma motility state. After 7 h, a small cell population extends a lower number of long unbranched neurites (Fig. 7 c), and soma motility steadily increases during the remaining period studied. We used a mathematical model to identify cells that represent the mean static phenotypes relevant to select temporal bins of the 10% cells with longest neurites (Fig. 7 c). These results quantitatively define two distinct phases with different morphodynamic behaviors during the neurite outgrowth process.
Figure 7.
Temporal analysis of neuronal dynamics in control and MKK7-KD cells. (a–d) Control and MKK7-KD time-lapse sequences from multiple experimental plates were divided in 19 1-h bins. Coefficient of variation of selected features for each temporal bin for control (a and c) and MKK7 KD (b and d) cells. Measurements for the whole-cell population (a and b) and the 10% of cells with the highest neurite extreme length (c and d) are shown. Representative HDS graph representations of cells from the tenth percentile at the 1-, 5-, 10-, 15-, and 20-h time points have been identified using a mathematical model (Materials and methods; c and d). (e) Feature distributions at the two different time point intervals (1–6 h and 14–20 h). Vertical bars display distribution means.
We then applied the same approach to study the short, unstable neurites MKK7-KD phenotype (Fig. 7, b and d). Although short neurites were observed in phase 2, temporal analysis surprisingly revealed a burst of neurite outgrowth in phase 1 compared with control cells, which previously had eluded our visual inspection of time-lapse movies. Other parameters such as neurite and branch numbers, as well as cell migration speed, were not affected. These results suggest that MKK7 regulates two distinct functions, one occurring at an early and the other at a late phase during neurite outgrowth. Feature distribution plots again represent the heterogeneous, but statistically significant, behavior of cell populations (Fig. 7 e).
Spatiotemporal regulation of RhoA activation by two RhoA-specific different GAPs
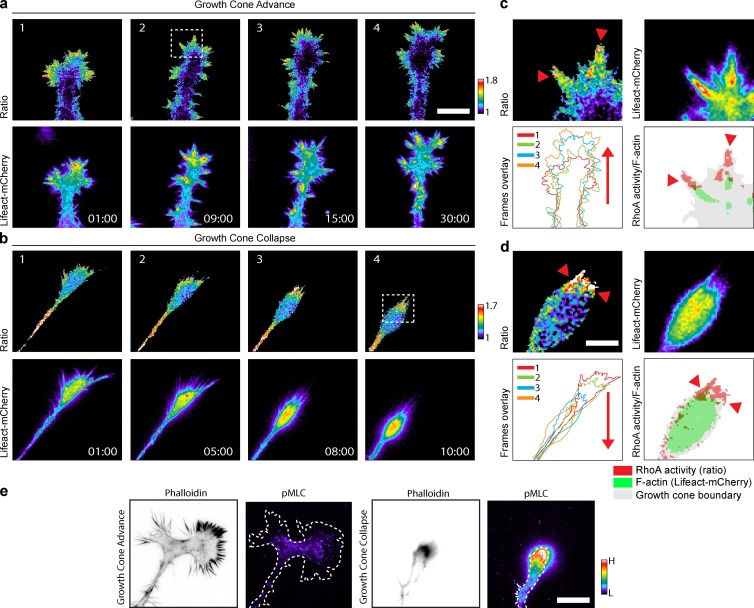
As a proof of concept for the validity of our siRNA screen, we sought to explore some relevant neurite outgrowth phenotypes with respect to spatiotemporal Rho GTPase activation dynamics. We focused on an apparently paradoxical result of our screen that is centered around RhoA and two RhoA-specific GAPs: ARHGAP5 (p190RhoGAP; Ridley et al., 1993; Arthur and Burridge, 2001) and DLC1 (deleted in liver cancer; Kim et al., 2009; Braun and Olayioye, 2015). Consistently with the classic model (Hirose et al., 1998), we observed that RhoA KD led to longer neurites, which results in decreased neurite collapse events (Fig. 6, cluster 11). Furthermore, as expected, ARHGAP5 KD, which is expected to augment RhoA activity, led to short neurites owing to increased neurite collapse (Fig. 6, cluster 3). In marked contrast, DLC1 KD, which is also expected to augment RhoA activity, surprisingly led to an increase in neurite outgrowth (Fig. 6, cluster 15). As we had previously shown (Fritz et al., 2013), two distinct spatiotemporal RhoA activation pools, most likely involving different signaling complexes, are associated with growth cone advance and collapse in N1E-115 cells. We therefore hypothesized that each GAP regulates one of the specific RhoA signaling complexes. During growth cone advance, RhoA activity localizes to the tip of filopodial F-actin bundles (Fig. 8, a and c; and Video 8). In contrast, RhoA is activated in retraction fibers at the tip of spontaneously collapsing growth cones (Fig. 8, b and d; and Video 8). Importantly, RhoA activity correlates only with its downstream cytoskeletal output, myosin light chain phosphorylation (pMLC) during growth cone collapse (Fig. 8 e), suggesting that this specific process is regulated by Rho kinase. In contrast, low pMLC signal was observed during growth cone advance, suggesting that RhoA regulates a distinct effector pathway involved in filopodium formation.
Figure 8.
Dynamics of RhoA activation in advancing and collapsing growth cones. (a and b) Time series showing RhoA activation in advancing and collapsing growth cones from differentiated N1E-115 cells cotransfected with the RhoA2G FRET probe and pLenti-Lifeact-mCherry. (top) Ratio (RhoA activation) images. (bottom) Lifeact-mCherry (F-actin) images. Time scale, min:s. Bar, 10 µm. (c and d, top left and right) High-magnification images of advancing and collapsing growth cones outlined in the top rows of panels a and b. Red arrowheads point to the RhoA activity zone. (bottom left) Overlay of the time series frames showed in the top rows of panels a and b. Red arrows indicate the advance or collapse direction. (bottom right) Overlay images of the top-left and right panels of c and d highlight the RhoA activity zone (red) over the filopodia F-actin bundles (green). Red arrowheads point to the RhoA activity zone. Bar, 2 µm. (e) High-resolution images of advancing and collapsing growth cones from differentiated N1E-115 cells stained for phalloidin and pMLC are shown. Bar, 10 µm.
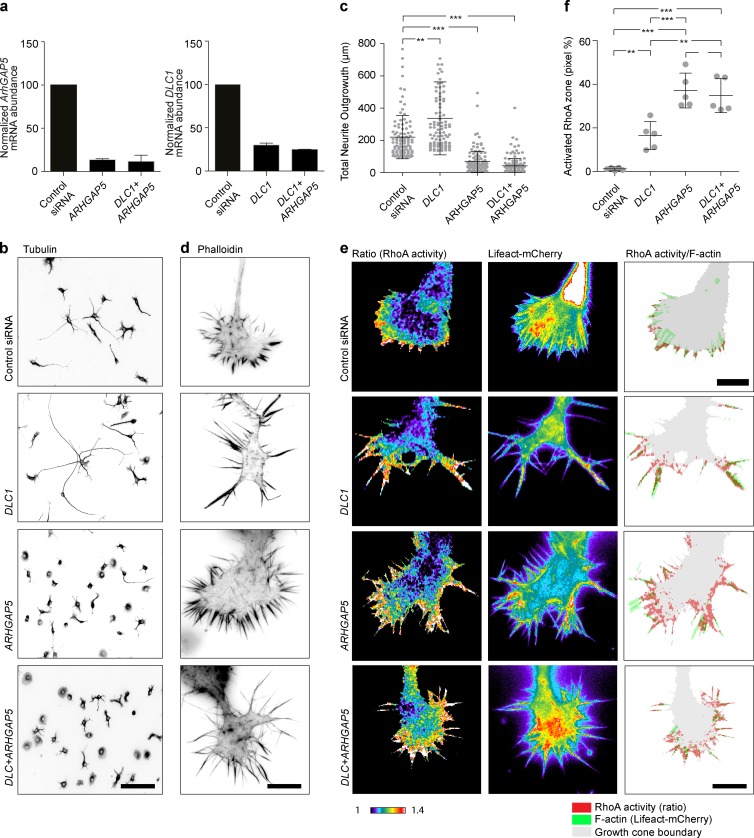
To explore the possibility that DLC1 and ARHGAP5 regulate two distinct spatiotemporal pools of RhoA activity, we performed a series of experiments in which we knocked down DLC1, ARHGAP5, or both. For these experiments, we selected the single siRNAs of which the feature vector most resembled the mean MDS. Quantitative PCR indicated potent reduction of DLC1 and ARHGAP5 messages in both single and double KD (Fig. 9 a). Quantification of steady-state neurite outgrowth levels recapitulated the results observed using our CV approach: DLC1 KD led to increased whereas ARHGAP5 KD led to reduced neurite outgrowth (Fig. 9, b and c). Combined DLC1/ARHGAP5 KD led to reduced neurite outgrowth, indicating that, at this specific length scale, ARHGAP5 loss of function is dominant over DLC1 loss of function. Because RhoA spatiotemporal signaling patterns are evident only when the growth cone is examined (see Fig. 8 and Fritz et al., 2013), we zoomed in on the cytoskeletal and spatiotemporal RhoA activation phenotypes at this specific length scale (Fig. 9, d–f). DLC1 KD cells exhibited longer filopodia that extended out of the growth cone veil (Fig. 9 d). Rather than being focused at the tip of filopodial F-actin bundles, RhoA was activated throughout the filopodium (Fig. 9, e and f). This suggests that DLC1 functions by locally deactivating RhoA to shape its focused activity pattern at the tip of F-actin bundles. In DLC1 KD cells, the aberrant RhoA activation pattern then leads to longer and more stable filopodia, which favor steady growth cone motility (Video 9). Ultimately, this can explain the increased neurite outgrowth phenotype observed at the length scale of a whole cell. ARHGAP5 KD cells did not display a specific filopodium phenotype as with DLC1 KD (Fig. 9 d). Instead, rather than being confined to filopodia, aberrant RhoA activity was potently upregulated not only throughout filopodia structures but also within the growth cone (Fig. 9, e and f). Furthermore, ARHGAP5 KD growth cone filopodia exhibited “frozen” morphodynamics, suggestive of excessive actomyosin contractility (Video 9). This aberrant cytoskeletal state then most likely is highly permissive for the increased growth cone collapse frequency observed at larger time scales. Double KD of ARHGAP5 and DLC1 revealed a hybrid phenotype (Fig. 9, d–f). Long filopodia, with activated RhoA throughout the filopodium, were observed as for the DLC1 KD phenotype. Additional RhoA activity throughout the growth cone body was observed as for the ARHGAP5 phenotype. The latter pool of aberrant RhoA activity might then lead to excessive contractility that favors growth cone collapse, ultimately leading to short neurites in double-KD cells (Fig. 9, b and c). These results strongly suggest that DLC1 and ARHGAP5 respectively regulate filopodia formation and growth cone advance, versus actomyosin contractility and growth cone collapse.
Figure 9.
Characterization of DLC1, ARHGAP5, and double-KD phenotypes. (a) Quantitative RT-PCR of DLC1, ARHGAP5, and double DLC1 and ARHGAP5 KD efficiencies in N1E-115 cells. Mean ± standard deviations from four replicates are shown. (b and c) Quantification of global neurite outgrowth phenotypes of the three perturbations. (b) Representative images of α-tubulin–stained N1E-115 KD cells. Inverted black-and-white contrast. (c) Quantification of total neurite outgrowth. Images were segmented and quantified using the Metamorph neurite outgrowth plugin. The single siRNAs that produced the z-score vector with lowest distance to the mean MDS were used for KD. Population mean ± standard deviations are shown for the whole-cell population (n = 150 cells). One-way ANOVA with Bonferroni’s multiple comparison test was used. ***, P < 0.0001. (d) High-resolution growth cone images of phalloidin-stained, control, and KD growth cones. All experiments were performed and stained simultaneously and acquired with identical acquisition settings. (e) Spatiotemporal RhoA activation patterns in response to the three perturbations. Ratio (RhoA activation), Lifeact-mCherry (F-actin), and overlay representative images of both signals are shown. Cells were cotransfected with the RhoA2G FRET probe, a Lifeact-mCherry construct, and the indicated siRNA. Ratio and F-actin images are color-coded for signal intensity. Ratio images have been scaled identically across experiments. Overlay images highlight the RhoA activity zone (red) over the filopodia F-actin bundles (green). (f) Quantification of RhoA activation in the three perturbed states. The RhoA activity zone was calculated as the pixel percentage of the growth cone RhoA activity area over the growth cone p-domain area. Population mean ± standard deviation are shown for n = 5 cells/sample. One-way ANOVA with Bonferroni's multiple comparison test was used. ***, P < 0.0001. Bars: (b) 100 µm; (d) 10 µm; (e) 10 µm.
Discussion
We present NeuriteTracker, a robust CV pipeline to track morphodynamics of neurons transfected with a genetically encoded, nonperturbing, fluorescent morphology reporter that can be time-lapsed for hours using air objectives in different neuronal-like cell types (Fig. S4, a–c). Automatic identification of multiparametric MDSs provides the throughput to analyze large time-lapse datasets and detects complex dynamic phenotypes that are difficult to discern by human visual inspection. The benefit of using dynamic features is demonstrated by the finding that they stratify phenotypes in additional clusters when only static features are considered during the analysis of our siRNA screen. Transiently occurring phenotypes can also be identified when specific temporal episodes are taken into account (Fig. 7, c and d), at the expense, however, that larger datasets are required to obtain adequate statistical power. Our approach identified two discrete neuronal differentiation episodes with specific morphodynamic states in an unbiased fashion (Fig. 7, a and b). Furthermore, we analyzed a Rho GTPase interactome that provides substantial coverage of the cytoskeleton regulating machinery controlling neurite outgrowth, with all candidates yielding a quantifiable phenotype. Our unbiased approach that uses unsupervised clustering of the MDSs uncovers a relatively small phenotypic space that is already visible using a small feature set (LS feature set, 19 clusters; HS feature set, 15 clusters). The benefit of using a larger feature set is then simply that it provides more information about the phenotypes. Our results are consistent with the recent finding of a similarly small phenotypic space in a perturbation screen targeting cell morphology in Drosophila melanogaster hemocytes (Yin et al., 2013). Importantly, cell migration was also impacted by many gene perturbations, revealing that the cytoskeletal regulation machinery regulating neurite outgrowth and cell migration is to a large extent shared.
Current models of Rho GTPase function in neurite outgrowth posit that Rac1/Cdc42 regulate neurite extension, whereas RhoA controls neurite retraction (Hirose et al., 1998; da Silva and Dotti, 2002; Govek et al., 2005). However, such simple models are not consistent with the apparent complexity of Rho GTPase signaling in which GEFs (Rossman et al., 2005), GAPs (Moon and Zheng, 2003), and effectors (Pertz, 2010) outnumber their cognate Rho GTPases. The rationale behind our screen was to explore the significance of the large potential Rho GTPase interactome we had previously unveiled using a proteomics approach (Pertz et al., 2008; Fig. 3). Indeed, our screen revealed that the different proteins of this potential Rho GTPase interactome are all important for the neurite outgrowth process. Thus, our results suggest a complex scenario in which multiple spatiotemporal Rho GTPase signaling networks, most likely consisting of GEFs, GAPs, GTPases, effectors, and their associated proteins, fine-tune different MPs.
Our proof-of-concept study in which we perturbed two RhoA-specific GAPs, which surprisingly led to two distinct global neurite outgrowth phenotypes, illustrates how this spatiotemporal signaling complexity will have to be tackled in the future. Our data suggest that the two RhoA-specific GAPs regulate two distinct spatiotemporal RhoA signaling complexes, one involved in regulation of filopodium formation during growth cone advance and the other involved in the generation of contractility necessary for growth cone collapse. Our data suggest that ARHGAP5 is involved in the regulation of growth cone collapse because its perturbation leads to aberrant RhoA activity not only within unprotrusive filopodia but also within the body of the growth cone, which further correlates with frozen growth cone morphodynamics and ultimately with short neurites. This conceptually fits well with the function of RhoA in growth cone collapse, as exemplified by decreased instance of neurite retraction leading to longer neurites in the RhoA KD phenotype. Given the different global neurite outgrowth phenotype induced by DLC1 KD, we propose that DLC1 regulates a RhoA-dependent function distinct from that of ARHGAP5. Our data strongly suggest that DLC1 is involved in the fine spatial regulation of the focused RhoA activation pool at the tip of F-actin bundles, which most likely involves reaction–diffusion systems as previously documented for RhoA (Bement and von Dassow, 2014). This specific spatiotemporal RhoA activity pool might serve to locally position the formin mDia1 to polymerize F-actin to enable filopodia extension (Goh et al., 2012). DLC1 loss of function might then compromise RhoA deactivation, leading to widening of the RhoA activation domain. This in turn might lead to aberrant formin regulation and longer and more stable filopodia that favor growth cone advance, ultimately leading to increased neurite outgrowth. Our ARHAGP5/DLC1 double-KD data clearly show a hybrid phenotype with characteristic features from both single KDs, strongly suggesting that that both GAPs specifically regulate the two distinct cytoskeletal processes of growth cone advance and collapse.
These observations have important consequences for the interpretation of our global neurite outgrowth phenotypes. Many of the neurite outgrowth phenotypes observed in our siRNA screen might emerge from aberrant spatiotemporal Rho GTPase signaling patterns occurring at the micrometer-length scale at which the growth cone functions, which might then propagate at the whole-cell scale. The information-rich, multiparametric phenotypic signatures extracted by our CV pipeline therefore do not allow to unambiguously decipher the spatiotemporal complexity of Rho GTPase signaling. Deconvolving this molecular complexity of Rho GTPase signaling will therefore require evaluation of molecular perturbations at multiple time and length scales.
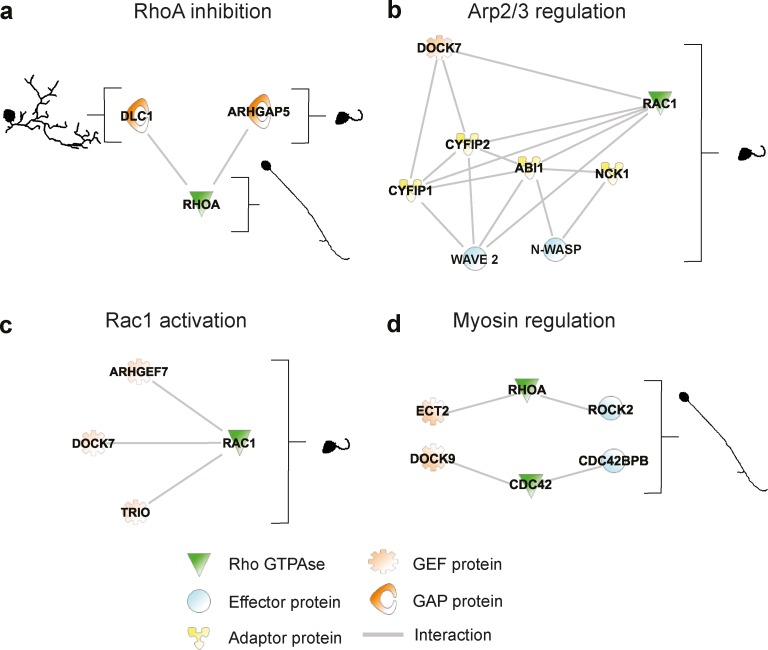
Finally, we nonexhaustively discuss some potential Rho GTPase signaling networks suggested by our screen (Fig. 10). The apparently conflictual phenotypes caused by DLC1 and ARHGAP5 KD, which can be understood from the experiments performed in Fig. 9, are illustrated in Fig. 10 a. Our approach detects a continuum of similar phenotypes for KD of members of a well-known Rac1 effector pathway, consisting of the effectors Wave2 (WASF2) and N-WASP (WASL), as well as additional interactors known to regulate the Arp2/3 complex (Machesky et al., 1999; Banzai et al., 2000; Bradley and Koleske, 2009; Tahirovic et al., 2010), of which loss of function leads to short neurites (Fig. 10 b). Surprisingly, the Rac1 “short neurites” KD phenotype (group 1: clusters 1 and 3) is also phenocopied by multiple Rac1-specific GEFs: KD of GEFs such as ARHGEF7 (ten Klooster et al., 2006; β-PIX), DOCK7 (Pinheiro and Gertler, 2006), or Trio (Fig. 10 c; Bellanger et al., 2000). This suggests the existence of multiple complex spatiotemporal Rac1 pathways that regulate neurite protrusion. Consistently with the classic model in which RhoA regulates myosin contractility to enable collapse (Hirose et al., 1998), ROCK2 KD, a RhoA-effector protein regulating myosin light chain kinase (MLCK; Amano et al., 1996; Matsui et al., 1996; Borisoff et al., 2003; Govek et al., 2005), phenocopies RhoA KD. This is also true for Ect2 KD, a GEF that can activate RhoA (Fig. 10 d; Schmidt and Hall, 2002; Tsuji et al., 2011), suggesting that Ect2, RhoA, and ROCK2 might function together to promote neurite collapse. However, this is not phenocopied by KD of other RhoA-specific GEFs (ARHGEF3: C3; TRIO: C1; ARHGEF11 and ARHGEF18: C18; and ARHGEF2: C19), suggesting additional mechanisms of spatiotemporal RhoA activation. Another novel finding was that KD of Cdc42, DOCK9 (a Cdc42-specific GEF; Lin et al., 2006) and Cdc42BPB (a Cdc42 effector also called myotonin-related dystrophyn kinase [MRCK]; Leung et al., 1998; Chen et al., 1999) phenocopy RhoA KD (Fig. 10 d). Because MRCK also regulates MLCK and contractility, a DOCK9, Cdc42, and MRCK signaling network might therefore also regulate growth cone collapse. It has been shown that the simultaneous phosphorylation of MLC, induced by RhoA and Cdc42 via ROCK and MRCK, is crucial to promote actomyosin contractility and guarantee proper migration (Wilkinson et al., 2005). However, this cooperative mechanism between these two GTPases has never been shown in neuronal cells. These results suggest that RhoA and Cdc42 may cooperate to regulate actomyosin contractility to control growth cone collapse and neurite retraction. Our CV approach provides a first estimate of the complexity of a spatiotemporal Rho GTPase signaling network during neuronal morphodynamics. This provides an initial resource that will allow researchers to gate on relevant morphodynamics processes to dissect these complex spatiotemporal networks at multiple time and length scales.
Figure 10.
Selected potential Rho GTPase functional networks identified in the RNAi screen. Potential Rho GTPase signaling networks were identified using Ingenuity Pathway Analysis based on high-confidence protein–protein interactions or literature-based knowledge. Representative static phenotypes corresponding to the RNAi perturbations are shown. Each solid line represents a direct protein–protein interaction. A legend identifies the functions of the proteins in the network. (a) Multiple functions for distinct RhoA-specific GAPs. (b) Rac1-dependent effector pathway controlling the Arp2/3 complex. KD of these proteins all lead to a “short neurites” phenotype (group 1: clusters 1, 3, and 4). (c) Potential regulation of Rac1 by multiple Rac1-specific GEFs. (d) Potential Rho GTPase signaling networks regulating myosin light chain. All phenotypes correspond to a “long and unbranched neurites” phenotype (group 4 clusters 11 or 12).
Materials and methods
Overview of wetlab experiments
DNA constructions
For simultaneous expression of Lifeact-GFP and NLS-mCherry, we created a bicistronic expression vector consisting of Lifeact-GFP, an IRES sequence (derived from a pIRES vector; Clontech), and NLS-mCherry (Fig. 1 a). This construct was built using InFusion technology (Clontech) and was cloned in a pcDNA3.1(+) eukaryotic expression vector (Invitrogen).
Accession numbers
The accession number for the pcDNA Lifeact-GFP/NLS-mCherry construct is Addgene 69058.
Cell culture and transfection
N1E-115 neuroblastoma cells (American Tissue Culture Collection) were cultured in DMEM supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin/streptomycin. For differentiation, N1E-115 cells were starved for 24 h in serum-free Neurobasal medium (Invitrogen) supplemented with 1% l-glutamine and 1% penicillin/streptomycin. For the double siRNA-mediated KD and plasmid transfection, cells were transfected as previously described (Chong et al., 2006), using 400 ng of the plasmid pcDNA-Lifeact-GFP-IRES-NLS-mCherry and 20 pmol Stealth Select siRNAs (Invitrogen). 1 µl of Transfectin (Bio-Rad) was used as transfection reagent. 48 h posttransfection, cells were starved in Neurobasal medium. 72 h posttransfection, cells were detached with Puck’s saline and replated on a glass-bottom 24-multiwell plate (MatTek) coated with 10 µg/ml laminin (Millipore). Approximately 3 h after seeding, cells were immediately used for time-lapse imaging. For transfection of both siRNAs and plasmids, cells were transfected as previously described (Feltrin et al., 2012). PC12 Neuroscreen-1 subclone cells (Chen et al., 2012) were cultured in DMEM with 1% l-glutamine, supplemented with 10% horse serum, 5% FBS, and 1% penicillin/streptomycin. Cells were culture on 50 µg/ml collagen solution from bovine skin (Sigma-Aldrich). For differentiation, PC12 Neuroscreen-1 cells were starved for 24 h in DMEM with 1% l-glutamine supplemented with 1% glucose, 1% penicillin/streptomycin, and 1% horse serum; 50 ng/ml NGF was added to the medium to induce differentiation. For PC12 transfection, cells were transfected as previously described (Chong et al., 2006), using 400 ng of the plasmid pcDNA-Lifeact-GFP-IRES-NLS-mCherry. 0.8 µl of Transfectin (Bio-Rad) was used as transfection reagent. 24 h posttransfection, cells were starved in DMEM medium. 72 h posttransfection, cells were gently detached using a cell scraper and replated on a glass-bottom 24-multiwell plate (MatTek) coated with 50 µg/ml collagen solution from bovine skin (Sigma-Aldrich).
Immunofluorescence
N1E-115 cells were washed in PBS, fixed in PBS containing 4% PFA (Sigma-Aldrich) for 10 min, and permeabilized in PBS containing 1% Triton X-100 for 2 min. Coverslips were washed in PBS for 20 min and blocked in 2% BSA and 0.1% Triton X-100 in PBS for 15 min. Cells were stained with primary antibodies for 1 h (Alpha Tubulin Clone DM1A; Sigma-Aldrich), phospho–myosin light chain 2 (Cell Signaling Technology), and then with secondary antibodies for 30 min (Alexa Fluor 555–labeled phalloidin, Alexa Fluor 546 secondary antibody, and DAPI for 30 min; Invitrogen).
Microscopy and image acquisition
All experiments were imaged using an inverted Eclipse Timicroscope (Nikon) controlled by Metamorph software (Universal Imaging). Laser-based autofocus, a CFI Plan Apo Lambda 10× (NA 0.45; Nikon) objective, and a CoolSnap HQ2 camera (Roper Scientific) were used throughout. For live-cell imaging experiments, rapid switching between the GFP and mCherry channels was performed using two independent LED lamps with excitation at 470 and 585 nm (Cool LED) with a multibandpass EGFP/mCherry filter cube (Chroma Technology Corporation). All experiments were performed in serum-free Neurobasal medium (Invitrogen) supplemented with 1% l-glutamine and 1% penicillin/streptomycin. For measurements of neurite outgrowth at steady state, differentiated, siRNA-transfected cells were replated on 18-mm glass coverslips coated with 10 µg/ml laminin. 24 h after plating, the cells were fixed and stained with DAPI and an antitubulin antibody. DAPI and tubulin images were acquired in a 5 × 5 matrix of fields of view. Image analysis was performed using the Metamorph neurite outgrowth plugin (Universal Imaging). Selected fields of view were then acquired using a CFI Plan Apo Lambda 10× (NA 0.45) objective (Nikon).
For time-lapse imaging of neurite outgrowth dynamics in cells expressing Lifeact-GFP and NLS-mCherry, 25,000 transfected N1E-115 cells were replated on a glass-bottom, 24-well plate (MatTek) coated with 10 µg/ml laminin. 4 h after plating, 10 selected fields of view per well were imaged in Neurobasal medium (Invitrogen) in a heated closed chamber using a CFI Plan Apo Lambda 10× (NA 0.45) objective (Nikon). Selection of fields of view was performed using Metamorph software. For that purpose, the GFP channel was acquired in a 7 × 7 matrix of fields of view for each of the 24 wells. A macro was then used postacquisition to select 10 fields of view containing an adequate number of cells exhibiting a specific range in fluorescence intensities and cell areas. This ensured that each field of view displayed a similar number of cells and fluorescence intensities and excluded fields of view with cellular debris. The selected fields of view were automatically translated in a stage position list, which was used for the time-lapse experiment. This resulted in acquisition of 240 fields of view in GFP and mCherry channels throughout the 24-well plate every 12 min for a total of 19.6 h.
For imaging of F-actin and/or RhoA activation dynamics, N1E-115 cells were imaged using a Plan Apo VC 60× (NA 1.4) objective (Nikon) in Neurobasal medium (Invitrogen). Förster resonance energy transfer (FRET) imaging was performed as described before (Fritz et al., 2013). Ratio imaging analysis was performed with Biosensor Processing Software 2.1 (http://lccb.hms.harvard.edu/software.html). Images were thresholded on each channel; shade, background, and photobleach corrected; masked; and registered before ratios were calculated. Ratio images are color-coded so that warm and cold colors represent high and low biosensor activity, respectively.
For time-lapse imaging of neurite outgrowth dynamics in PC-12 Neuroscreen-1 cells, 80,000 transfected Neuroscreen-1 cells were replated on a glass-bottom, 24-well plate (MatTek) coated with 50 µg/ml collagen solution from bovine skin (Sigma-Aldrich). 30 min after seeding, 50 ng/ml NGF was added to the medium to induce differentiation. After 6 h, selected fields of view per well were imaged in DMEM medium (Invitrogen) plus NGF in a heated closed chamber using a CFI Plan Apo Lambda 10× (NA 0.45) objective (Nikon).
Identification of a Rho GTPase signaling network
To identify a Rho GTPase signaling network, we took advantage of our recent proteomic analysis of purified neurite and soma fractions of N1E-115 cells (Pertz et al., 2008). This dataset provides spatial information about the subcellular localization (neurite/soma) for 4,855 proteins through the measurement of relative protein enrichment in purified neurite and soma fractions. Using Ingenuity Pathways Analysis (IPA) software, we first identified Rho GTPases expressed in our cell system (Rac1, Cdc42, RhoA, RhoB, RhoC, and RhoG). We then used these as “bait” to identify proteins that are documented to interact directly with these Rho GTPases by interrogating the Ingenuity Pathways Knowledge Base, which is a systemwide database of biological pathways created from multiple relationships of proteins, genes, and diseases. Only considering proteins identified in our proteomics screen to be neurite enriched or equally distributed in neurite and soma fractions, we then manually sorted these proteins in GEFs, GAPs, RhoA effectors, Rac1 and Cdc42 effectors, and additional interacting proteins. In a second round, we then used these different protein groups as baits to identify additional interactors that might work upstream of GEFs and GAPs or downstream of effectors. This yielded a Rho GTPase signaling network of 219 proteins (Fig. 3 a), in which most proteins are significantly enriched in the neurite. We used Ingenuity analysis pathway software to perform a Gene Ontology analysis of the protein ensemble in this signaling network (Fig. 3 b). As expected, we observed enrichment of Rho GTPase, adhesion, cytoskeleton, and axonal Gene Ontology terms. Our approach reveals the existence of a neurite-enriched, Rho GTPase signaling network of unanticipated complexity.
Overview of CV analysis
We developed a CV pipeline to segment and track somata and neurites in time-lapse datasets. It extracts and stores representative models for each cell over time. The pipeline is summarized in Fig. 1 c. From these models, we extract features that are used to characterize the morphodynamic phenotype of the cell. The resulting data are stored in an HDS representing the cell (Fig. 2 b). For a cell detected at a specific time point, the top of the hierarchy represents the detected cell, with subcellular components nucleus, soma, and one or more neurite nodes (Fig. S3). The neurite is the trunk attached to the soma (Fig. 2 b, blue segment), and this can be followed by a tree structure of branches, which are any intersection points between neurite elements (Fig. 2 b, green segments), and leaf nodes, which are branches situated at the extremity of the neurite arborescence (Fig. 2 b, red segments). The CV algorithm first segments the nucleus and soma of each cell. Detected cells in each frame of the movie are linked using a graph-based tracking algorithm to identify individual cells over time. Next, neurites arbors are extracted for each detection by segmenting the full cell body and reconstructing tree structures corresponding to the neurites. Finally, detected neurites are tracked over time using a similar method as for the somata, and the tracked cell is stored in the HDS data structure. Validation of soma and neurite segmentation and tracking against human annotated ground truth is also shown.
Nucleus segmentation
We extracted a set of nucleus detections {d1,...,dk} from the NLS-mCherry channel of every image (Fig. 1 c) using the MSER (Maximally Stable Extremal Regions) feature detector (Matas et al., 2004) from the VLFeat library followed by a morphological filling operation. The MSER detector is a robust method to find regions that are stable over a wide range of thresholds while remaining insensitive to contrast change. The minimum and maximum size parameters were fixed at 70 and 170; all other parameters used default settings.
Soma region growing
Using the segmented nuclei as seed regions, somata segmentation is accomplished using a region growing and competition algorithm on the Lifeact-GFP channel (Fig. 1 c). For every pixel in the image, we compute the geodesic distance to each nucleus, combining the local intensity differences and the Euclidean distance. For each detected nucleus dk, we defined a potential:
where and are the mean and standard deviation of the intensities of the pixels describing dk, respectively, and β is a multiplicative factor of representing a tolerance of variation between the local soma intensity and the local background. The geodesic distance Uk associated with nucleus dk is defined such that it increases with geometric or intensity distance from the nucleus, as the solution of the Eikonal equation such that . A distance map to all detected nuclei is given by , which we approximate by computing the solution of such that ; for all k where
and the index of the closest region k is decided during the propagation of a variant of the Fast Marching algorithm. The main advantage of this approach over the original (Benmansour and Cohen, 2009) is that the potential values are decided during the propagation. The somata segmentations sk are obtained by thresholding , where {Rk} is the Voronoi tessellation associated with U the set of nuclei {dk}, the Euclidean distance to the set of nuclei is denoted D, and the geodesic and Euclidean thresholds are denoted Tg and Te, respectively. For all our experiments, we set parameter values A = 1e7, β = 1.5, Tg = 2e − 6, and Te = 7.
Detection filtering
Cell detections are filtered before tracking is performed to retain the most reliable detections. Detections too close to the image boundary are ignored (distance from nucleus centroid <10 pixels). The minimum tolerated nuclei circularity is 0.2, and the maximum is 0.85.
Cell tracking and identification
For each time-lapse sequence, an algorithm searches through the full set of cell detections and iteratively associates the most similar cells to track them over time (Fig. 1 c). We construct a graph G = (C,E), where each node corresponds to a detection. For each cell in time step t, edges are formed between and all past and future detections within a time window W. A weight we measuring the similarity between each pair of cell detections is assigned to each edge. The weight takes into account spatial distance, temporal distance, and differences in shape between the two cells. The weight is given by , where connects and , and is a shape feature vector containing ’s area, perimeter, mean intensity, and major and minor axis lengths of a fitted ellipse. f evaluates differences between a feature a extracted from and as
According to this formulation, the tracking solution corresponds to the set of edges that minimizes the cost .To perform the minimization, we use a graph association tracking algorithm that iteratively selects an edge with minimum cost and adds it to the tracking set until a cost threshold is reached (see below). The output of this algorithm is a set of lists of cell detections corresponding to the instantaneous location of each neuron over time. Using these lists, we assign identities to the detections belonging to each neuron.
Neurite segmentation and association
Neurites are detected for each neuron using a three-step process. First, the image is partitioned into regions, one assigned to each tracked cell. The probability of a pixel u being assigned a label Jt(u) corresponding to neuron i is deduced using Bayes’ rule:
where It is the image at time is the set of cells present in it, and P(Jt(u)) is a uniform distribution. The numerator is modeled as the probability of the path L that connects maximally the pixel u to the cell
where lr are indicator variables for the locations forming the path L. Optimal maxima can be found by minimizing the negative likelihood using geodesic shortest path, which produces a partitioning of the image into connected components assigned to each cell. Next, a tubularity measurement is computed for each pixel (Sethian et al., 1999) to identify likely neurites. Cells that were detected but not tracked have a suppressed tubularity value to avoid false positives on the cell boundary. A sigmoid function is fitted to the tubularity score using maximum likelihood to obtain a potential Pt. A front propagation Fast Marching algorithm is simultaneously launched from all cells (Benmansour and Cohen, 2009), which solves the Eikonal equation and . This yields a geodesic distance map Ut which serves as the cell body likelihood for each neuron (Fig. 1 c), the associated tessellation Vt, and the map of the Euclidian lengths L of the geodesics. The geodesic distance map is thresholded with a soft threshold Ts, yielding cell bodies for each neuron (Fig. 1 c). Candidate terminal points for the neurites are identified as the local maxima of Lt in each thresholded region. Candidate neurite tracings are found by back-propagating the geodesic distance Ut from the local maxima. Only those points for which Ut > Th is above a hard threshold Th are retained. The idea is that even if the candidate endpoints detected in the previous step are not necessarily all correct, they will quickly converge to interesting elongated structures during the back propagation. Finally, a graph representing a each full neurite tree is constructed as a Minimum Spanning Tree of all back-propagated points, starting from a root touching the soma (Fig. 1 c). Neurites less than 30 pixels in length are removed.
Neurite tracking
Neurites are tracked using the same graph-association algorithm used to track cells. Instead of nucleus centroids, the positions of the centroids of the neurite trees are tracked. Edges connect neurites that emanate from the same cell. The weight we of an edge connecting two neurites and is assigned according to spatial distance and a shape measure:
where wTCL, wCentroid, and wSomaContract are the weights, TotalCableLength(N) is the total cable length of a neurite N, Centroid(N) is its centroid, SomaContact(N) is its contact point with the soma, and
Unlike the weights used for cell tracking, the neurite weights ignore temporal distance because each neurite is already associated to a tracked cell body.
Graph association tracking
We construct a graph G = (C,E) where each node corresponds to a detection. For each cell in time step t, edges are formed between and all past and future detections within a time window W. A weight we is assigned to each edge according to spatial and temporal distances, and a shape measure where connects and , and is a shape feature vector containing ’s area, perimeter, mean intensity, and major and minor axis lengths of a fitted ellipse. f evaluates differences between a feature a extracted from and as
To minimize this cost function, we adopt a greedy selection algorithm that iteratively selects an edge with minimum cost and adds it to the set E′, removing future and past connections from the detections connects. The algorithm iterates until the minimum cost is greater than a threshold T. The track for neuron is extracted from E′ by traversing the graph (C, E′) and appending linked nucleus detections to Xi.
Algorithm 1. Greedy tracking association
Start with an empty set E′.
(1) Repeat. Find edge with minimum cost . Add to E′, linking detections and . Remove from E.
(2) If t1 < t2, then: remove edges between and future detections (where t > t1) from E. Remove edges between and past detections (where t < t2) from E.
(3) Else, remove edges between and past detections (where t < t1) from E. Remove edges between and future detections (where t > t2) from E.
(4) End if, until .
Soma segmentation validation
The ability of our method to segment nuclei and somata from static images was evaluated using a set of three hand-annotated movies selected randomly from the screen. The data were annotated according to the following protocol. First, annotators should label only cells that appear in the video for a sufficient amount of time (five frames). Second, they should label only cells with good fluorescent reporter expression. Finally, annotators should carefully trace the nucleus and soma of cells that fit this criterion. We then used our software to automatically segment the same cells that were annotated by experts. For the quality control analysis, the annotation and automatic segmentation areas in each frame are divided into regions and assigned a label from the following list: (a) OK: the nucleus/soma is correctly segmented; (b) FP (false positive): area of nucleus/soma segmentation that does not exist in the annotation; (c) FN (false negative): area of the nucleus/soma annotation that is not recovered by the segmentation.
The results of the validation can be found in Fig. S2 and Video 3. In total, 790 annotations outlining the nucleus and somata were collected from the movies and evaluated.
The automatic nucleus segmentation correctly recovered 94.5% of the area annotated as nuclei by the experts. 35.2% of the area of the segmentation fell outside of the annotation and was considered to be false positive. 5.5% of the area of the annotation was not recovered by the segmentation and was considered false negative. In general, the nucleus segmentation was very accurate but tuned to be slightly larger than the annotation, which explains the difference between the false-positive and false-negative rates.
The automatic soma segmentation correctly recovered 81.1% of the area annotated as soma by the experts. 19.1% of the segmentation area fell outside of the annotation, and was considered to be false positive. 18.9% of the area of the annotation was not recovered by the segmentation and was considered false negative.
Cell-tracking validation
To evaluate the ability of our method to track cells over time, we annotated 20 videos chosen randomly from the screen data. The data were annotated according to the following protocol. Any cell that our software attempted to track for more than five frames was labeled and added to the ground truth. Labeling a cell consisted of placing a bounding box around the soma for every frame in which the cell is present in the video. We compared these annotations to the soma segmentations recovered by our method using a standard tracking evaluation procedure (Smith et al., 2008). In each frame, every cell is assigned one or more states from the following list: (a) OK: the cell is correctly tracked; (b) FP (false positive): a cell was detected that does not exist in the annotation; FN (false negative): a cell appearing in the annotation was not detected; (c) MT (multiple tracker): the area of cell in the annotation was shared among multiple tracked cells; MO (multiple objects): the area of a tracked cell covered more than one annotated cells; (d) FI (false identification): an error was made in maintaining the identity of a cell over time (e.g., a detected cell label switched from one annotated cell to another).
A correct detection occurs if a labeled detection sufficiently overlaps the correct corresponding annotation. To establish correspondences between detections and annotations, we use the coverage test which checks the F1-measure of the overlap between the area of the detection and the annotation:
where is the intersection of the detection d with the annotation a, and is the cardinality operation. If the overlap is greater than a threshold F1 > 0.33, an association is made between the detection and the annotation.
The results of the validation can be found in Fig. S2 and Video 4. In total, 58,064 total cell detections were evaluated. The automatic tracking was correct in more than 90% of the cases. The most common errors were false detections (6.8%), followed by false identifications (2.6%). The remaining errors all occurred with <1% frequency.
Neurite validation
To evaluate the ability of our method to recover neurites, we compared automatic neurite detections against 128 hand-annotated cells selected randomly from the screen. Each annotation was performed using the TrackEM2 plugin for ImageJ (Schneider et al., 2012). The protocol for the evaluation is a slightly modified version of the TREES protocol (Cuntz et al., 2010), where a hard decision is made as to whether a pixel belonging to a neurite is correctly detected instead of a soft matching score. Each detected neurite pixel must fall within a specified distance (σ = 4.5 pixels) of an annotated neurite pixel to be counted as correct. If it is outside of this range, it is considered a false detection. If no neurite detection falls within the specified distance of an annotation, the annotation is considered a missed detection. The results of our evaluation show that the neurite missed detection rate is just below 30% (Fig. S2 m). The false detection rate is ∼27%. The disagreement between the automatic method and the annotator might seem high, but it is in line with the disagreement between annotators (Fig. S2 n).
Overview of the statistical analysis
Features are extracted from the HDSs representing tracked cells and stored in a matrix format. Next, a feature profile is created for each siRNA, and the profiles for siRNAs corresponding to the same gene are combined to mitigate off-target effects and build a phenotypic signature for that gene. A robust subset of the complete set of collected features is selected and used for phenotypic clustering. A mathematical model allows automatic selection of representative cells as prototypical examples of each phenocluster. Finally, we perform a temporal analysis comparing the evolution of various features characterizing cell behavior over time.
Definition of features describing soma and neurite morphodynamics
To generate a list of features that describe cell morphology and morphodynamics, we first defined a series of parameters that describe cell components, such as the nucleus, soma and neurite, at a given time point (see the following paragraphs). In a second step, these parameters are then used to compute static and dynamic features.
Definition of nuclei and soma parameters
Area: area of a nucleus or a soma. Perimeter: perimeter of a nucleus or a soma. Major axis length and minor axis length: an ellipse is fitted through the nucleus and the soma. Major axis A and a minor axis B of the ellipse are measured. Orientation: orientation of the major axis length A of the fitted ellipse (radians). Eccentricity: eccentricity of the fitted ellipse: . Circularity: distance measured between a pure circle and the real shape of the nucleus or the soma.
Definition of neurite parameters
We modeled the neurite arborescence using different elements. A neurite is considered as the trunk of a tree. We define a tree root as the attachment point between the neurite and the soma, a branch point as any intersection point between neurite elements, and the extreme point as the extremities of the arborescence. The root is the neurite segment between the tree root and the first branch point, a branch is any elementary part of the tree, and a leaf is a branch situated at the extremity of the neurite arborescence. Using this notation, we extracted a series of parameters that describe the different components of a neurite, and that are shown below. A schematic representation of these parameters is shown in Fig. S3.
Extreme length: distances between the tree root and the extreme points. Total cable length: sum of the lengths of the root and the branches. Complexity: number of branches in the tree divided by the total cable length of the tree. Number of neurites/branches: number of roots or branches belonging to a cell. Branches length: total length of the branches of a cell. Leaf branches length: total length of the leaves of a cell.
Definition of static features
Using the nuclei, soma, and neurite parameters, we created a set of static features, which are described below. Nucleus area: nucleus area of the cell per frame, in pixels. Nucleus eccentricity: nucleus eccentricity of the cell per frame. Nucleus major axis length: nucleus major axis length of the cell per frame. Nucleus minor axis length: nucleus minor axis length of the cell per frame. Nucleus orientation: nucleus orientation of the cell per frame. Nucleus perimeter: nucleus perimeter of the cell per frame. Nucleus circularity: nucleus circularity of the cell per frame. Soma area: soma area of the cell per frame. Soma eccentricity: soma eccentricity of the cell per frame. Soma major axis length: soma major axis length of the cell per frame. Soma minor axis length: soma minor axis length of the cell per frame. Soma orientation: soma orientation of the cell per frame. Soma perimeter: soma perimeter of the cell per frame. Soma circularity: soma circularity of the cell per frame. Number of neurites: number of roots of the cell per frame. All neurites length branches mean: mean branch length of the cell per frame. All neurites leaf length branches mean: mean leaf length of the cell per frame. All neurites extreme length mean: mean extreme length of the cell per frame. Total cable lengths per neurite mean: mean neurite total cable length of the cell per frame. Complexity per neurite mean: mean neurite complexity of the cell per frame. Number of branches per neurite mean: mean number of branches per neurite of the cell per frame. Max extreme length per neurite mean: mean maximum extreme length per neurite of the cell per frame. Total neurites length: sum of the total cable length of each neurite of the cell per frame. Total neurites branches: sum of the number of branches of each neurite of the cell per frame. Total complexity: “total neurites branches” divided by “total neurites length” per frame. Distance traveled: distance traveled by the soma of the cell between two consecutive frames. Speed: instantaneous speed of the cell between two consecutive frames. Acceleration: instantaneous acceleration of the cell between two consecutive frames.
Definition of dynamic features
For the dynamic cell features, starting from the initial set of static features, we created five layers of dynamic cell features such as mean value of the feature through time, time a feature expands/contracts, number of inflection points in the feature evolution, and frequency of expansion (freq expansion) of a feature. Please note that to avoid redundancy, only a subset of dynamic features (freq expansion) was taken in account during the analysis of our siRNA screen.
Next, we provide the description of the four layers of dynamic cell features. Note that in the example we only listed the freq expansion features; however, the same set of features is also valid for the other four layers.
Let F(t) be the feature value at time t and let ΔF(t) = F(t) − F(t − 1) be the feature difference between the two consecutive frames t − 1 and t.
Mean: The mean value of the feature through the entire movie. This gives rise to the following features. Time expanding: the number of times where ΔF(t) is positive, which is the number of times where the feature changes positively. Time contracting: number of times where ΔF(t) is negative which is the number of times where the feature changes negatively. Number inflection points: number of times where ΔF(t) switches from a positive value to a negative value and from a negative value to a positive value. Freq expansion: number of times where ΔF(t) is positive divided by the number of frames where the cell appears. This is the number of times per second where the feature changes positively.
This gives rise to the following features. Nucleus area freq expansion: number of times per second where the nucleus area of the cell changes positively. Nucleus eccentricity freq expansion: number of times per second where the nucleus eccentricity of the cell changes positively. Nucleus major axis length freq expansion: number of times per second where the nucleus major axis length neurites of the cell changes positively. Nucleus minor axis length freq expansion: number of times per second where the nucleus minor axis length neurites of the cell changes positively. Nucleus orientation freq expansion: number of times per second where the nucleus orientation neurites of the cell changes positively. Nucleus perimeter freq expansion: number of times per second where the nucleus perimeter of the cell changes positively. Nucleus Circularity Freq Expansion: Number of times per second where the nucleus circularity of the cell changes positively. Soma area freq expansion: number of times per second where the soma area of the cell changes positively. Soma eccentricity freq expansion: number of times per second where the soma eccentricity of the cell changes positively. Soma major axis length freq expansion: number of times per second where the soma major axis length of the cell changes positively. Soma minor axis length freq expansion: number of times per second where the soma minor axis length of the cell changes positively. Soma orientation freq expansion: number of times per second where the soma orientation of the cell changes positively. Soma perimeter freq expansion: number of times per second where the soma perimeter of the cell changes positively. Soma circularity freq expansion: number of times per second where the soma circularity of the cell changes positively. Number of neurites freq expansion: number of times per second where the number of neurites of the cell changes positively. Total neurites length freq expansion: number of times per second where the total neurites length of the cell changes positively. Total neurites branches freq expansion: number of times per second where the total neurites branches of the cell changes positively. Total complexity freq expansion: number of times per second where the total complexity of the cell changes positively.
Data formatting
Data extracted by the segmentation and tracking steps are stored in a nonvectorial form, which must be converted into a matrix for statistical analysis. The analysis is performed on two groups of features: “population” features, which provide instantaneous characterizations of a cell or groups of cells; and “dynamic cell” features, which characterize a particular cell over time (Fig. 4 a). The population features account for several static shape measurements. The population tracking data structure takes the form movie(m).cell(c).time(t).feature(f), where is the movie index, is the cell index, is the time index, and is the feature index. For analysis, it is aggregated and converted to a matrix with rows and F columns; i.e., one row per movie/cell/frame and one column per feature. The dynamic cell features capture both shape and c):feature(f) is aggregated and organized into a matrix with rows and F columns; i.e., one row per movie/cell and one column per feature.
Single siRNA profiles
To characterize the phenotype associated with a perturbation, we create a profile for each siRNA using measurements extracted from the movies. Each siRNA profile measures whether the siRNA perturbation increased or decreased various measurements with respect to the control (Fig. 4 b). A siRNA profile is a vector of same dimension as the feature vector described above, where and denotes the number of features. It is constructed by performing a -test with 5% significance, comparing for each feature the perturbation population against the control population. If the statistical test indicates that a significant difference exists, the siRNA profile entry for that feature takes the value of the z-score. If the difference is insignificant, the profile takes a value of zero. More specifically, for a feature f the z-score is given by and is the result of the statistical test. If the mean of the two distributions are significantly different the result is , otherwise . The siRNA profile is given by , where rf denotes a signed distance measure between the distribution of the control and the perturbed feature. If rf > 0, the polarity is positive and the mean feature value is higher for the siRNA than for the control. If rf < 0, the polarity is negative and the mean feature value is lower for the siRNA than for the control. If rf = 0, the polarity is null and the mean feature value is the same for the siRNA and the control.
Morphodynamic signatures
To mitigate siRNA off-target effects, we create a gene-specific MDS by “averaging” the profiles of the three siRNAs targeting that gene according to the rules described below (Fig. 4 c). As previously proposed (Echeverri et al., 2006), if at least two of three siRNAs produce the same phenotype, this phenotype can be considered on target. We applied this rule to each independent feature, if the feature had the same polarity for two of three siRNAs (positive, negative, or null), it was considered on target. If all three siRNA polarities agree, the MDS score is the mean of the -scores. If two of three siRNA polarities agree, the MDS is the mean z value of the two siRNAs profiles with the same polarity. In all other cases, we set the z MDS score to zero. Formally, let be a siRNA profile, where F denotes the number of features, and indexes the three siRNAs . Let be a binary value and be the number of siRNA perturbations resulting in a positive polarity or negative polarity . The MDS is given by the mean siRNA profile of those that have the same polarity:
where
For instance, for a given feature, if two siRNAs yield significantly positive z-scores and if the third siRNA molecule yields a negative z-score, then the feature f value of the gene profile is the mean z-score of the two first RNA molecules. In Table 1, we provide an example of how the MDS is computed from three siRNA feature profiles.
Table 1. An example of how the MDS is computed from three siRNA feature profiles.
| Profiles | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| siRNA1 | 0 | 0 | 0 | 0 | 0 | −15 | 10 | 0 | 2 | −12 |
| siRNA2 | 0 | 0 | 0 | 5 | 8 | 8 | 8 | −6 | −11 | −10 |
| siRNA3 | 0 | 10 | −5 | −8 | 10 | 4 | 6 | −4 | −15 | −8 |
| MDS | 0 | 0 | 0 | 0 | 9 | 6 | 8 | −5 | −13 | −10 |
To determine whether a feature is robust, we checked whether the z-score of a feature for the positive MKK7 KN control is consistent across at least 65% (LS features) or 90% (HS features) for the 32 experimental plates (compared with the negative, nontargeting control). Formally, let F be the full set of collected features. We reduce this to a subset such that if, and only if:
where P is the number of plates, is the siRNA profile of the MKK7 control in the plate p, and is a threshold value. By setting S = 0.65 (LS) and 0.9 (HS), we reduce the size of the feature space to 23 (LS) and 8 (HS) features.
Phenocluster identification
We used bottom-up hierarchical clustering (Bakal et al., 2007; Yin et al., 2013) to identify sets of genes that share common MDSs. To accomplish this, we first defined a distance metric D(i,j) allowing us to compare pairs of MDSs gi and gj. We used a straightforward method based on the MDS polarity. The proposed distance depends on the signed distance between the gene profiles: D(i,j) = D(sign(gi),sign(gj)), where if. In other words, two genes have a similar phenotype if their features have the same polarity. This can be computed by using the L0 distance:
which gives a percentage of features that are matching between two genes. Evaluation of the distance between two clusters was performed by computing the L0 distance between the two mean points of the clusters. We set the minimum number of genes per cluster to three, with the consequence that if a cluster is composed of fewer than three genes, we consider these genes as unclustered. The number of clusters also depends on the dendrogram threshold, which gives the minimum percentage of matching features in each cluster. This percentage is directly connected to the L0 distance. If this threshold is too high, there are few clusters; if the threshold is too low, the clusters are too noisy. We came to a compromise by iteratively testing different threshold values, visually evaluating the quality of both the clusters and the identified phenotypes by inspection of HDS graph representations of cells identified using a mathematical model to select penetrant phenotypes. The mathematical model is described below. We then chose the 60% (LS feature set) and 70% (HS feature set) threshold, implying that 60% or 70% of the features match in a given cluster.
Identification of phenocluster representative cells
For each phenocluster, we identified representative cells with a highly penetrant phenotype. We then provide static or time-lapse datasets using graph representations of the HDS extracted for these cells (Video 5). For that purpose, we first identified the single siRNA profile that has the highest similarity with the mean MDS. We then selected the single siRNA profile with the highest number of on-target features. We then selected cells with a highly penetrant phenotype. More specifically, for a given gene i, we looked for a cell , where , such that this cell has features strongly consistent with the centroid . The optimization formulation is:
where is the standard deviation of the feature f (computed from all the data), is the centroid value of the feature f for the control, sign if the gene profile is positive, and sign otherwise. For this analysis, we focused on a subset of features that are easily visually interpretable. Specifically, we used the following features: number of neurites, total cable length per neurite mean, and number of branches.
Temporal analysis of neurite outgrowth dynamics
We used our large amount of time-lapse datasets of control and MKK7-KD cells (320 movies each acquired in 32 independent experiments) to analyze the morphodynamic behavior associated with specific temporal episodes during the neurite outgrowth process. For this, we divided each time-lapse series (of ∼20 h) into 20 temporal bins and computed the mean of the feature value in each bin. To compare the temporal evolution of a feature, we normalized the feature value by its variance. Let where , be the values of a feature for all the cells of a control. We denote by μ and σ2 the mean and variance of A, respectively. Let , where is a subset such that is restricted in the temporal bin b. Similarly, we denote by μb and the mean and variance of Ab, respectively. We normalized the data as follows:
Additionally, we illustrated this temporal analysis by selecting representative cells in Fig. 5 (A and B). Each cell is automatically selected from a model. Let be a model for a targeted cell, where is chosen regarding the variation of μb in the analysis of Fig. 5 (A and B). The method consists of selecting the cell , where , such that . To filter the cells without neurite, we gated on the 10% cells with longest neurites.
Online supplemental material
Fig. S1 shows high-content live-cell imaging platform and RNAi pipeline. Fig. S2 shows an evaluation of the automatic soma and neurite segmentation and soma tracking. Fig. S3 shows the definition of parameters for feature extraction. Fig. S4 shows PC-12 cell neuronal differentiation tracking and segmentation and hierarchical clustering of MDSs using the HS feature sets. Fig. S5 shows a summary of workflow and statistics of the RNAi screen. Video 1 is raw movies of Lifeact-GFP/NLS-mCherry channels in N1E-115 cells. Video 2 shows dynamic visualization of different steps in the computer vision pipeline. Video 3 shows nuclei and soma segmentation validation. Video 4 shows soma detection and tracking validation. Video 5 shows HDS-modeled graph representations of five global phenotypical classes. Video 6 shows HDS-modeled graph representations of MDSs of dynamic short neurite phenotypes in the LS map. Video 7 shows HDS-modeled graph representations of MDSs of dynamic long and branched neurite phenotypes in the LS map. Video 8 shows RhoA activation dynamics in advancing and collapsing growth cone in N1E-115 cells. Video 9 is high-resolution time-lapse imaging of DLC1 and ARHGAP5 KD growth cones in N1E-115 cells. Table S1 lists a description of all siRNAs in the RNAi library. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506018/DC1.
Supplementary Material
Acknowledgments
We are grateful to Chris Bakal for constructive comments on the manuscript.
This work was supported by a Swiss National Science Foundation Sinergia grant to O. Pertz, P. Fua, and F. Fleuret and by Swiss National Science Foundation and International Foundation for Research in Paraplegia grants to O. Pertz.
The authors declare no competing financial interests.
Author contributions: O. Pertz and K. Smith initially conceived the study; L. Fusco performed all experiments; K. Smith, F. Benmansour, G. Gonzalez, and P. Fua led the image segmentation effort; R. Lefort and F. Fleuret led the statistical analysis effort; C. Barillari and B. Rinn designed the OpenBis-based image database; O. Pertz and L. Fusco designed and interpreted the FRET validation experiments. O. Pertz, K. Smith, L. Fusco, and R. Lefort wrote the paper.
Footnotes
Abbreviations used in this paper:
- CV
- computer vision
- FRET
- Förster resonance energy transfer
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide-exchange factor
- HDS
- hierarchical data structure
- HS
- high stringency
- KD
- knockdown
- LS
- low stringency
- MDS
- morphodynamic signature
- MLCK
- myosin light chain kinase
- MP
- morphogenetic process
- MRCK
- myotonin-related dystrophyn kinase myosin light chain kinase
- pMLC
- myosin light chain phosphorylation
References
- Amano M., Ito M., Kimura K., Fukata Y., Chihara K., Nakano T., Matsuura Y., and Kaibuchi K.. 1996. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271:20246–20249. 10.1074/jbc.271.34.20246 [DOI] [PubMed] [Google Scholar]
- Arthur W.T., and Burridge K.. 2001. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol. Biol. Cell. 12:2711–2720. 10.1091/mbc.12.9.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C., Aach J., Church G., and Perrimon N.. 2007. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 316:1753–1756. 10.1126/science.1140324 [DOI] [PubMed] [Google Scholar]
- Banzai Y., Miki H., Yamaguchi H., and Takenawa T.. 2000. Essential role of neural Wiskott-Aldrich syndrome protein in neurite extension in PC12 cells and rat hippocampal primary culture cells. J. Biol. Chem. 275:11987–11992. 10.1074/jbc.275.16.11987 [DOI] [PubMed] [Google Scholar]
- Bauch A., Adamczyk I., Buczek P., Elmer F.J., Enimanev K., Glyzewski P., Kohler M., Pylak T., Quandt A., Ramakrishnan C., et al. 2011. openBIS: a flexible framework for managing and analyzing complex data in biology research. BMC Bioinformatics. 12:468 10.1186/1471-2105-12-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger J.M., Astier C., Sardet C., Ohta Y., Stossel T.P., and Debant A.. 2000. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat. Cell Biol. 2:888–892. 10.1038/35046533 [DOI] [PubMed] [Google Scholar]
- Bement W.M., and von Dassow G.. 2014. Single cell pattern formation and transient cytoskeletal arrays. Curr. Opin. Cell Biol. 26:51–59. 10.1016/j.ceb.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmansour F., and Cohen L.. 2009. Fast object segmentation by growing minimal paths from a single point on 2D or 3D images. J. Math. Imaging Vis. 33:209–221. 10.1007/s10851-008-0131-0 [DOI] [Google Scholar]
- Borisoff J.F., Chan C.C., Hiebert G.W., Oschipok L., Robertson G.S., Zamboni R., Steeves J.D., and Tetzlaff W.. 2003. Suppression of Rho-kinase activity promotes axonal growth on inhibitory CNS substrates. Mol. Cell. Neurosci. 22:405–416. 10.1016/S1044-7431(02)00032-5 [DOI] [PubMed] [Google Scholar]
- Bradley W.D., and Koleske A.J.. 2009. Regulation of cell migration and morphogenesis by Abl-family kinases: emerging mechanisms and physiological contexts. J. Cell Sci. 122:3441–3454. 10.1242/jcs.039859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A.C., and Olayioye M.A.. 2015. Rho regulation: DLC proteins in space and time. Cell. Signal. 27:1643–1651. 10.1016/j.cellsig.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Chen J.Y., Lin J.R., Cimprich K.A., and Meyer T.. 2012. A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision. Mol. Cell. 45:196–209. 10.1016/j.molcel.2011.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.Q., Tan I., Leung T., and Lim L.. 1999. The myotonic dystrophy kinase-related Cdc42-binding kinase is involved in the regulation of neurite outgrowth in PC12 cells. J. Biol. Chem. 274:19901–19905. 10.1074/jbc.274.28.19901 [DOI] [PubMed] [Google Scholar]
- Chong K.W., Lee A.Y., Koay E.S., Seet S.J., and Cheung N.S.. 2006. pH dependent high transfection efficiency of mouse neuroblastomas using TransFectin. J. Neurosci. Methods. 158:56–63. 10.1016/j.jneumeth.2006.05.017 [DOI] [PubMed] [Google Scholar]
- Cuntz H., Forstner F., Borst A., and Häusser M.. 2010. One rule to grow them all: a general theory of neuronal branching and its practical application. PLOS Comput. Biol. 6:e1000877 10.1371/journal.pcbi.1000877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva J.S., and Dotti C.G.. 2002. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat. Rev. Neurosci. 3:694–704. 10.1038/nrn918 [DOI] [PubMed] [Google Scholar]
- Echeverri C.J., Beachy P.A., Baum B., Boutros M., Buchholz F., Chanda S.K., Downward J., Ellenberg J., Fraser A.G., Hacohen N., et al. 2006. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 3:777–779. 10.1038/nmeth1006-777 [DOI] [PubMed] [Google Scholar]
- Feltrin D., Fusco L., Witte H., Moretti F., Martin K., Letzelter M., Fluri E., Scheiffele P., and Pertz O.. 2012. Growth cone MKK7 mRNA targeting regulates MAP1b-dependent microtubule bundling to control neurite elongation. PLoS Biol. 10:e1001439 10.1371/journal.pbio.1001439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz R.D., Letzelter M., Reimann A., Martin K., Fusco L., Ritsma L., Ponsioen B., Fluri E., Schulte-Merker S., van Rheenen J., and Pertz O.. 2013. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci. Signal. 6:rs12 10.1126/scisignal.2004135 [DOI] [PubMed] [Google Scholar]
- Goh W.I., Lim K.B., Sudhaharan T., Sem K.P., Bu W., Chou A.M., and Ahmed S.. 2012. mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J. Biol. Chem. 287:4702–4714. 10.1074/jbc.M111.305102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek E.E., Newey S.E., and Van Aelst L.. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19:1–49. 10.1101/gad.1256405 [DOI] [PubMed] [Google Scholar]
- Hirose M., Ishizaki T., Watanabe N., Uehata M., Kranenburg O., Moolenaar W.H., Matsumura F., Maekawa M., Bito H., and Narumiya S.. 1998. Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J. Cell Biol. 141:1625–1636. 10.1083/jcb.141.7.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.Y., Vigil D., Der C.J., and Juliano R.L.. 2009. Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in regulation of the cytoskeleton and cell motility. Cancer Metastasis Rev. 28:77–83. 10.1007/s10555-008-9167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort R., Fusco L., Pertz O., and Fleuret F.. 2015. Machine learning-based tools to model and to remove the off-target effect. Pattern Anal. Appl. 10.1007/s10044-015-0469-z [DOI] [Google Scholar]
- Leung T., Chen X.Q., Tan I., Manser E., and Lim L.. 1998. Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol. Cell. Biol. 18:130–140. 10.1128/MCB.18.1.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Yang W., Baird D., Feng Q., and Cerione R.A.. 2006. Identification of a DOCK180-related guanine nucleotide exchange factor that is capable of mediating a positive feedback activation of Cdc42. J. Biol. Chem. 281:35253–35262. 10.1074/jbc.M606248200 [DOI] [PubMed] [Google Scholar]
- Machesky L.M., Mullins R.D., Higgs H.N., Kaiser D.A., Blanchoin L., May R.C., Hall M.E., and Pollard T.D.. 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA. 96:3739–3744. 10.1073/pnas.96.7.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matas J., Kaiser D.A., Blanchoin L., May R.C., Hall M.E., and Pollard T.D.. 2004. Robust Wide baseline Stereo from Maximally Stable Extremal Regions. British Machine Vision Computing. 22:761–767. 10.1016/j.imavis.2004.02.006 [DOI] [Google Scholar]
- Matsui T., Amano M., Yamamoto T., Chihara K., Nakafuku M., Ito M., Nakano T., Okawa K., Iwamatsu A., and Kaibuchi K.. 1996. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J. 15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- Moon S.Y., and Zheng Y.. 2003. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 13:13–22. 10.1016/S0962-8924(02)00004-1 [DOI] [PubMed] [Google Scholar]
- Nadarajah B., and Parnavelas J.G.. 2002. Modes of neuronal migration in the developing cerebral cortex. Nat. Rev. Neurosci. 3:423–432. 10.1038/nrn845 [DOI] [PubMed] [Google Scholar]
- Pertz O. 2010. Spatio-temporal Rho GTPase signaling—Where are we now? J. Cell Sci. 123:1841–1850. 10.1242/jcs.064345 [DOI] [PubMed] [Google Scholar]
- Pertz O.C., Wang Y., Yang F., Wang W., Gay L.J., Gristenko M.A., Clauss T.R., Anderson D.J., Liu T., Auberry K.J., et al. 2008. Spatial mapping of the neurite and soma proteomes reveals a functional Cdc42/Rac regulatory network. Proc. Natl. Acad. Sci. USA. 105:1931–1936. 10.1073/pnas.0706545105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro E.M., and Gertler F.B.. 2006. Nervous Rac: DOCK7 regulation of axon formation. Neuron. 51:674–676. 10.1016/j.neuron.2006.08.020 [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Self A.J., Kasmi F., Paterson H.F., Hall A., Marshall C.J., and Ellis C.. 1993. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 12:5151–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman K.L., Der C.J., and Sondek J.. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180. 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- Schmidt A., and Hall A.. 2002. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16:1587–1609. 10.1101/gad.1003302 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethian J., Linsley P.S., Lum L., Ma Y., Mathey-Prevot B., Root D.E., and Sabatini D.M.. 1999. Level Set Methods and Fast Marching Methods Evolving Interfaces in Computational Geometry, Fluid Mechanics, Computer Vision, and Materials Science. Cambridge University Press, Cambridge, UK. 400 pp. [Google Scholar]
- Smith K., Ba S., Gatica-Perez D., and Odobez J.M.. 2008. Tracking the visual focus of attention for a varying number of wandering people. IEEE Trans. Pattern Anal. Mach. Intell. 30:1212–1229. [DOI] [PubMed] [Google Scholar]
- Tahirovic S., Hellal F., Neukirchen D., Hindges R., Garvalov B.K., Flynn K.C., Stradal T.E., Chrostek-Grashoff A., Brakebusch C., and Bradke F.. 2010. Rac1 regulates neuronal polarization through the WAVE complex. J. Neurosci. 30:6930–6943. 10.1523/JNEUROSCI.5395-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Klooster J.P., Jaffer Z.M., Chernoff J., and Hordijk P.L.. 2006. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J. Cell Biol. 172:759–769. 10.1083/jcb.200509096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T., Higashida C., Yoshida Y., Islam M.S., Dohmoto M., Koizumi K., and Higashida H.. 2011. Ect2, an ortholog of Drosophila’s pebble, negatively regulates neurite outgrowth in neuroblastoma × glioma hybrid NG108-15 cells. Cell. Mol. Neurobiol. 31:663–668. 10.1007/s10571-011-9668-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S., Paterson H.F., and Marshall C.J.. 2005. Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat. Cell Biol. 7:255–261. 10.1038/ncb1230 [DOI] [PubMed] [Google Scholar]
- Yin Z., Sadok A., Sailem H., McCarthy A., Xia X., Li F., Garcia M.A., Evans L., Barr A.R., Perrimon N., et al. 2013. A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nat. Cell Biol. 15:860–871. 10.1038/ncb2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.