Polycomb complex member Cbx4 represses nonepidermal lineage and cell cycle inhibitor genes in the epidermal keratinocytes and operates as a direct p63 target, maintaining epithelial identity and proliferative activity in the developing epidermis.
Abstract
During development, multipotent progenitor cells establish lineage-specific programmers of gene activation and silencing underlying their differentiation into specialized cell types. We show that the Polycomb component Cbx4 serves as a critical determinant that maintains the epithelial identity in the developing epidermis by repressing nonepidermal gene expression programs. Cbx4 ablation in mice results in a marked decrease of the epidermal thickness and keratinocyte (KC) proliferation associated with activation of numerous neuronal genes and genes encoding cyclin-dependent kinase inhibitors (p16/p19 and p57). Furthermore, the chromodomain- and SUMO E3 ligase–dependent Cbx4 activities differentially regulate proliferation, differentiation, and expression of nonepidermal genes in KCs. Finally, Cbx4 expression in KCs is directly regulated by p63 transcription factor, whereas Cbx4 overexpression is capable of partially rescuing the effects of p63 ablation on epidermal development. These data demonstrate that Cbx4 plays a crucial role in the p63-regulated program of epidermal differentiation, maintaining the epithelial identity and proliferative activity in KCs via repression of the selected nonepidermal lineage and cell cycle inhibitor genes.
Introduction
During development, tissue differentiation relies on the establishment of specific patterns of gene expression, which is achieved by lineage-specific gene activation and silencing in multipotent stem cells and their progenies (Slack, 2008; Blanpain and Fuchs, 2014). The program of epidermal differentiation in mice begins at about embryonic day 9.5 (E9.5) and results in the formation of an epidermal barrier by E18.5 (Koster and Roop, 2007; Blanpain and Fuchs, 2009). The process of terminal differentiation in epidermal cells is executed by sequential changes of gene expression in the keratin type I/II loci, followed by the onset of expression of the epidermal differentiation complex genes encoding the essential components of the epidermal barrier (Fuchs, 2007). This program is governed by the coordinated involvement of several transcription factors (p63, AP-1, Klf4, Arnt, etc.), signaling pathways (Wnt, Bmp, Hedgehog, EGF, Notch, FGF, etc.), and epigenetic regulators (DNA/histone-modifying enzymes, Polycomb genes, higher order and ATP-dependent chromatin remodelers, and noncoding and microRNAs) that control expression of lineage-specific genes (Khavari et al., 2010; Botchkarev et al., 2012; Frye and Benitah, 2012; Perdigoto et al., 2014).
Among these regulatory molecules, the p63 transcription factor serves as a master regulator of epidermal development and controls expression of a large number of distinct groups of genes (Viganò and Mantovani, 2007; Vanbokhoven et al., 2011; Botchkarev and Flores, 2014; Kouwenhoven et al., 2015). p63 knockout (KO) mice fail to form stratified epithelium and express several epidermis-specific genes (Mills et al., 1999; Yang et al., 1999). In the epidermis, p63 regulates the expression of distinct chromatin-remodeling factors, such as Satb1 and Brg1, which, in turn, control the establishment of specific nuclear positioning and conformation of the epidermal differentiation complex locus required for full activation of keratinocyte (KC)-specific genes during terminal differentiation (Fessing et al., 2011; Mardaryev et al., 2014).
Epigenetic regulators exhibit both activating and repressive effects on chromatin in KCs: the histone demethylase Jmjd3, ATP-dependent chromatin remodeler Brg1, and genome organizer Satb1 promote terminal KC differentiation, whereas the DNA methyltransferase DNMT1, histone deacetylases HDAC1/2, and Polycomb components Bmi1 and Ezh1/2 stimulate proliferation of the progenitor cells via repression of the genes encoding cell cycle inhibitors, as well as inhibiting premature activation of terminal differentiation–associated genes (Sen et al., 2008, 2010; Ezhkova et al., 2009; LeBoeuf et al., 2010; Fessing et al., 2011; Mardaryev et al., 2014).
Polycomb chromatin-remodeling proteins form two complexes (Polycomb repressive complex 1 and 2 or PRC1/2) that compact the chromatin and inhibit transcription by preventing binding of the transcription machinery to gene promoters (Simon and Kingston, 2013; Cheutin and Cavalli, 2014). Recent data reveal that binding of the noncanonical PRC1 complex containing histone demethylase KDM2B, PCGF1, and RING/YY1-binding protein (RYBP) promotes basal ubiquitylation of the H2A at lysine 119 (H2AK119) at unmethylated CpG-rich DNA regions, which is sufficient to recruit the PRC2 complex (Blackledge et al., 2014; Cooper et al., 2014; Kalb et al., 2014). The PRC2 component Ezh1/Ezh2 histone methyltransferase promotes trimethylation of H3K27, followed by targeting of the Cbx proteins as a part of the canonical PRC1 complex to H3K27me3, which result in further increase of the H2AK119 ubiquitylation catalyzed by the PRC1 component Ring1b (Simon and Kingston, 2013; Cheutin and Cavalli, 2014; Perdigoto et al., 2014; Schwartz and Pirrotta, 2014).
In the epidermis, the Polycomb components Bmi1, Ezh1/2, and Jarid2 stimulate proliferation of the progenitor cells via repression of the genes encoding cell cycle inhibitors, including the INK4A–INK4B locus, as well as inhibit premature activation of terminal differentiation–associated genes (Ezhkova et al., 2009; Mejetta et al., 2011). In addition, Ezh1/2 restricts differentiation of the epidermal progenitor cells by repressing the Sox2 gene, which, in turn, promotes Merkel cell–specific differentiation (Bardot et al., 2013).
The Cbx4 gene belongs to the PRC1 family and has a chromodomain interacting with the H3K27me3 histone mark and mediating transcriptional repression together with the PRC2 complex (Li et al., 2007; Luis et al., 2011). Cbx4 also possesses SUMO E3 ligase activity that promotes sumoylation of other proteins, including DNA methyltransferase Dnmt3a, thus providing a link between Polycomb-mediated gene silencing and DNA methylation (Li et al., 2007). In normal human skin, the Cbx4 protein protects epithelial stem cells from senescence through PRC-dependent repression of the Ink4a locus, as well as controls their differentiation through PRC-independent mechanisms (Luis et al., 2011). These data suggest Cbx4 as a critical determinant regulating the activity of stem cells and their progenies in the skin. However, its role in the control of epidermal development and in establishing tissue-specific gene expression patterns still remains unclear.
Here, we show that Cbx4 is a critical determinant regulating the establishment and maintenance of the epidermal differentiation program that represses nonepidermal lineage genes and controls cell proliferation/differentiation in the epidermis. We also show that Cbx4 serves as a direct p63 target and mediates its repressive effects on the expression of nonepidermal lineage genes and selected cell cycle inhibitor genes. These data illustrate how p63 transcription factor operates in concert with epigenetic regulator Cbx4 to establish and maintain the lineage-specific program in differentiating cells and in governing epithelial differentiation/function.
Results
Cbx4 expression is increased in the epidermal KCs during the stratification stage of epidermal development
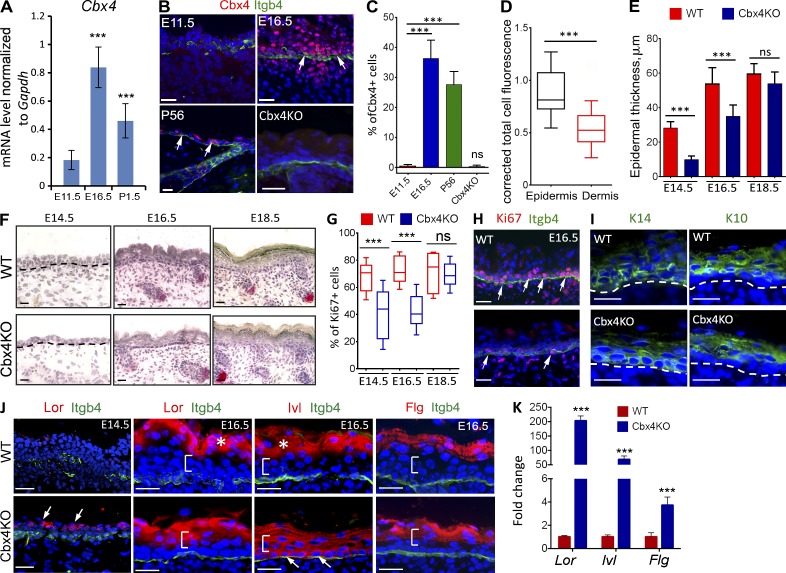
To perform the genome-wide analysis of transcriptomes in epidermal progenitor cells during development, we used laser capture microdissection (LCM) and obtained RNA samples from the basal epidermal layer before and after the onset of epidermal stratification (E11.5 and E16.5, respectively), as well as postnatally at 1.5 d (P1.5) and 8 wk (P56, adult) as described previously (Sharov et al., 2006). Detailed analysis of the transcript levels of different Polycomb group genes revealed their dynamic expression during development of the mouse epidermis (Fig. S1 A and Table S1). Among the PRC1 genes, the Cbx4 transcript and protein levels were strongly increased in the basal KCs during the transition from single-layered epithelium to stratified squamous epidermis (E11.5 vs. E16.5; Fig. 1, A and B; Fig. S1 A; and Table S1). Consistent with previous observations (Ezhkova et al., 2009), the onset of epidermal stratification (E11.5–E16.5) was also characterized by the highest levels of the PRC2 component Ezh2 gene expressions in the basal KCs, followed by its decline at the later stages of epidermal differentiation (E16.5–P1.5; Fig. S1 A and Table S1).
Figure 1.
Cbx4KO mice show a decrease of cell proliferation and premature terminal differentiation in the developing epidermis. (A) Cbx4 transcript expression in epidermal KCs at E11.5, E16.5, and P1.5 (qRT-PCR; mean ± SD; n = 3). (B) Cbx4 immunofluorescence is not detected in the E11.5 epidermis. In comparison to E11.5, Cbx4 levels are increased in basal and suprabasal KCs at E16.5, as well as in postnatal (P56) skin (arrows). Cbx4KO mice show a lack of Cbx4 expression in the epidermis. Integrin-β4 (Itgb4) expression outlines the basement membrane of the epidermis. DAPI counterstain (blue) shows the nuclei. (C) Quantification of Cbx4+ cells in the epidermis at different developmental stages in WT mice and in Cbx4KO skin (mean ± SD). n = 3. (D) Quantification of Cbx4 total cell fluorescence in both epidermal and dermal skin compartments of WT mice at E16.5 (box plot with 5–95% confidential intervals; the whiskers show minimum and maximum values). n = 3. (E and F) Histomorphological analyses of the alkaline phosphatase/hematoxylin-stained skin showed a reduced epidermal thickness in Cbx4KO mice compared with WT mice at E14.5 and E16.5 (mean ± SD). n = 3. Dashed lines separate epidermis and dermis. (G and H) Immunofluorescence detection (H) and quantification (G) of Ki67+ cells (H, arrows) showed reduced cell proliferation in the Cbx4KO epidermis versus WT counterpart at E14.5 and E16.5 (box plot with 5–95% confidential intervals). n = 4. Integrin-β4 expression outlines the basement membrane of the epidermis. (I and J) Immunofluorescence detection of K14 and K10 (I) and late (J) differentiation markers revealed premature onset of the terminal differentiation in the Cbx4KO epidermis (dashed lines indicate the epidermal/dermal border). In Cbx4KO mice, Loricrin protein was detected as early as E14.5, before onset of terminal differentiation in WT mice (J). At E16.5, Loricrin and Filaggrin were detected in early suprabasal cells, whereas Involucrin expression was up-regulated already in the basal progenitor cells (arrows). Brackets and asterisks depict the positions of the spinous and granular epidermal layers, respectively. (K) Increase of the Lor, Ivl, and Flg mRNA levels in the LCM-captured basal epidermal cells of E16.5 Cbx4KO mice versus WT controls (mean ± SD). n = 3. ***, P < 0.001. Bars, 25 µm. ns, not significant.
Interestingly, Cbx4 protein expression was even higher in differentiating suprabasal KCs, whereas significantly lower expression levels were seen in the dermal compartment of the skin (Fig. 1, B and D). In contrast to Cbx4, other PRC1 genes showed a less pronounced increase, or even a decrease, in their expression levels at the early stages of epidermal development (Fig. S1 A). As the epidermis fully matured (P1.5), basal progenitor cells showed a decline in expression of most of the PRC1 genes except Cbx7, Phc1, and Pcgf1 (Fig. S1 A and Table S1). Interestingly, Cbx4 expression levels also decreased in the basal KCs of adult skin compared with E16.5, but were still considerably higher than at E11.5 (Fig. 1 C, Fig. S1 A, and Table S1).
Cbx4 controls KC proliferation and terminal differentiation in the developing epidermis
To uncover the role of Cbx4 in the epidermal progenitor cells during development, we used mice bearing an inactivation allele lacking the first two exons and a 0.9-kb upstream region of the Cbx4 gene disrupting functional Cbx4 protein synthesis (Liu et al., 2013). The Cbx4 homozygous KO mice were not viable and died within 1 h after birth because of some (not yet fully clear) developmental abnormalities, including thymic hypoplasia (Liu et al., 2013). Immunostaining analysis for Cbx4 confirmed the abolished protein expression in the skin of Cbx4KO compared with wild-type (WT) littermates (Fig. 1 B).
To explore the skin phenotype upon Cbx4 ablation, we performed a histological assessment of the stratified skin epithelium at different developmental time points: E14.5, E16.5, and E18.5. At E14.5, Cbx4-null epidermis showed significantly reduced thickness correlated with a decreased number of stratified cellular layers and significant reduction in the number of proliferating Ki67+ cells in the basal layer compared with WT controls (Fig. 1, E–H). Similar to E14.5, Cbx4-null epidermis remained significantly thinner and harbored fewer proliferating basal cells at E16.5, whereas such differences become insignificant at E18.5 (Fig. 1, E–H). At E16.5, the appearance of the senescence-associated marker γ-H2AX was seen in the epidermis of Cbx4KO mice, whereas immunostaining for active caspase 3 showed a lack of any changes in the KC apoptosis between WT and Cbx4KO mice (Fig. S3, E and F). The reduced proliferation of E16.5 basal epidermal KCs was associated with increased expression of the cell cycle inhibitor genes (senescence-linked Ink4a locus p16/p19 and Cdkn1c/p57 gene; Fig. S3, A and B), which are well-known targets of the Polycomb proteins in several tissues, including the skin (Ezhkova et al., 2009; Luis et al., 2011).
As CBX4 was previously shown to prevent terminal differentiation in human epidermal progenitor cells independently of its Polycomb-associated repressive function (Luis et al., 2011), we explored the effect of Cbx4 ablation on the expression of early and late epidermal differentiation genes in the developing mouse epidermis. In the absence of Cbx4, cells of the embryonic ectoderm were able to acquire an epidermal fate and commence a differentiation program as indicated by positive K14 and K10 staining in the basal and suprabasal cells (Figs. 1 I and S4 A). However, Loricrin, a marker of terminally differentiating KCs, was prematurely expressed in the suprabasal cells located just above the basal cells in the Cbx4-deficient epidermis at E14.5, whereas its expression was not detectable in WT epidermis at this stage (Figs. 1 J and S4 A). In E16.5 WT embryos, the most superficial late suprabasal cells underwent terminal differentiation and expressed the key components of the cornified cell envelope such as Loricrin, Involucrin, and Filaggrin (Figs. 1 J and S4 A). In contrast to WT mice, the premature onsets of Loricrin, Involucrin, and Filaggrin expression were seen in the early suprabasal cells in E16.5 Cbx4KO mice (Figs. 1 J and S4 A). Immunofluorescence data on the expression of the markers of terminally differentiated KCs were in concordance with quantitative real time–PCR (qRT-PCR) data showing the markedly elevated levels of the Lor, Inv, and Flg transcripts in the E16.5 epidermis of Cbx4KO mice versus WT controls (Fig. 1 K). Thus, Cbx4 suppresses the expression of terminal differentiation–associated genes in the basal progenitor cells during murine epidermal development.
Cbx4 represses nonepidermal lineage genes in KCs during epidermal development
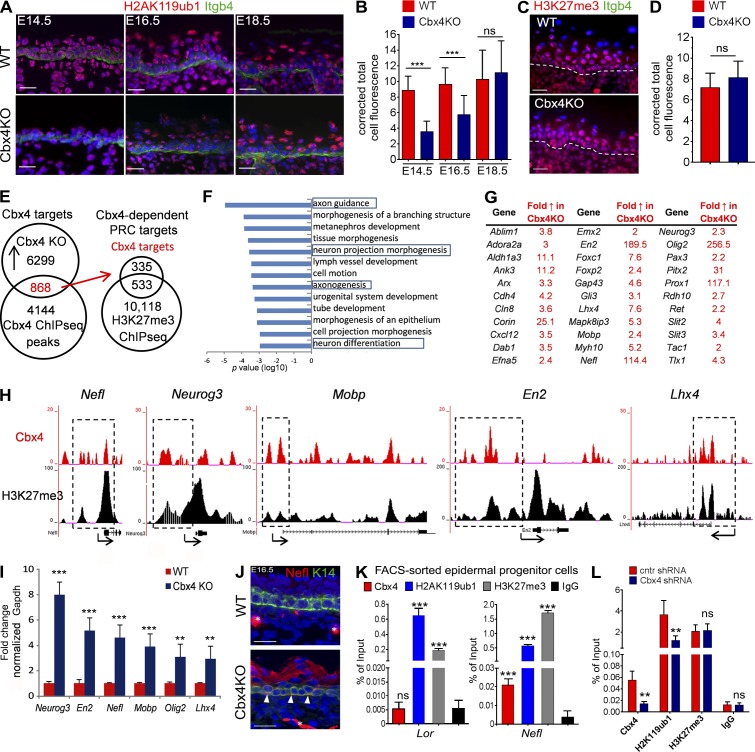
Next, we analyzed the distribution of H2AK119Ub and H3K27me3, two key histone modifications associated with Polycomb-mediated gene repression, in the epidermis of Cbx4KO mice. According to the model of Polycomb-mediated gene silencing, a basal level of H2AK119 ubiquitylation at unmethylated CpG-rich DNA regions is maintained by noncanonical PRC1 complexes lacking the Cbx proteins, whereas one of the Cbx family members serving as a part of the canonical PRC1 complex is only involved in the subsequent step in the Polycomb-mediated gene silencing and is required for binding to H3K27me3, followed by the Ring1b-mediated further increase of the H2AK119 ubiquitylation over its basal level (Simon and Kingston, 2013; Cheutin and Cavalli, 2014; Perdigoto et al., 2014; Schwartz and Pirrotta, 2014).
In contrast to WT mice, epidermal cells in E14.5 and E16.5 Cbx4KO mice showed a significantly decreased level of H2AK119ub1 expression, which we considered as a basal level (Fig. 2, A and B). However, the expression levels of H3K27me3 were not changed in the epidermis of Cbx4KO mice compared with WT controls (Fig. 2, C and D). Also, expression of the histone methylase Kdm2b, a constituent of the noncanonical PRC1 complex (Schwartz and Pirrotta, 2014), was increased in the epidermis of Cbx4KO mice, whereas expression of Ring1b forming the canonical PRC1 complex together with Cbx4 and catalyzing the H2AK119 ubiquitylation was not changed (Fig. S2, A and B). Importantly, Cbx4 ablation changed neither H2AK119ub1 nor H3K27me3 expression in the dermal cells (Fig. S2 C), thus suggesting epidermis-specific effects of Cbx4 on gene repression in the skin. These data were consistent with the data showing the significantly increased Cbx4 expression levels in the epidermis versus dermis in WT mice (Fig. 1 D) and suggested that, similar to other cell lineages, Cbx4 most likely mediates the recruitment of the canonical PRC1 complex to H3K27me3, followed by further ubiquitylation of H2AK119 over its basal levels mediated by PRC1 in epidermal cells.
Figure 2.
Activation of nonepidermal lineage genes in Cbx4-deficient epidermis. (A and B) Distribution and quantification of H2AK119ub1 immunofluorescence in WT and Cbx4KO skin. H2AK119ub1 is markedly reduced in the Cbx4KO epidermis at E14.5 and E16.5, whereas its expression is restored by E18.5. (C and D) Lack of changes in the cutaneous H3K27me3 expression between WT and Cbx4KO mice. Dashed lines separate epidermis and dermis. (E) Overlap of the Cbx4 and H3K27me3 ChIP-seq data with the expression arrays obtained from the epidermis of Cbx4KO versus WT mice depicts the direct Polycomb-dependent Cbx4 target genes in KCs that are up-regulated upon Cbx4 ablation. (F) Ontology of the Cbx4 target genes using DAVID bioinformatic resources. The bars represent the log10 p-values of each category. (G) Selected Cbx4 target genes involved in neuronal development and/or functioning. (H) ChIP-seq tracks depicting Cbx4 and H3K27me3 binding to the selected target genes (a single representative experiment out of two repeats is shown). Dashed outlines depict the corresponding gene regulatory regions. (I) qRT-PCR validation of the selected Cbx4 target genes up-regulated in the basal progenitor cells from Cbx4KO mice versus WT controls (mean ± SD). n = 3. (J) Ectopic Nefl protein expression in the Cbx4KO epidermis (arrowheads) compared with WT counterparts. Asterisks denote Nefl+ dermal nerve fibers in the WT and Cbx4KO mice. (K) ChIP-qPCR shows Cbx4 binding to Nefl but not to Lor promoter regions. Both Lor and Nefl are enriched in H2AK119ub1 and H3K27me3 histone marks (mean ± SD). n = 3. (L) Cbx4 knockdown in primary mouse KCs with shRNA decreases Cbx4 binding and reduces the H2AK119ub1 enrichment on the Nefl promoter region (mean ± SD). n = 3. **, P < 0.01; ***, P < 0.001. Bars, 25 µm. ns, not significant.
Our data also demonstrate that the expression levels of H2AK119ub1 in the epidermis of E18.5 Cbx4KO and WT mice became quite similar (Fig. 2, A and B), which suggests that other Cbx proteins (Fig. S1 C) might substitute for Cbx4 in the canonical PRC1 complex in Cbx4KO mice. These data are also quite consistent with the data demonstrating that maximal differences in the epidermal thickness and proliferation between WT and Cbx4KO mice are seen at E16.5, whereas such differences become insignificant at E18.5 (Fig. 1, E–H).
To determine the molecular targets of Cbx4 in the epidermal KCs during development, we correlated a microarray transcript profiling from E16.5 LCM-captured Cbx4-deficient basal cells with Cbx4 chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) analyses performed on primary epidermal KCs isolated from E16.5 WT mice. About 13.8% of genes up-regulated in the basal epidermal cells from Cbx4KO mice showed Cbx4 ChIP-seq binding, thus likely representing direct Cbx4 targets in KCs (Fig. 2, E–G). Because the Cbx4 chromodomain binds to H3K27me3, which results in a further increase of the H2AK119 ubiquitylation over its basal levels catalyzed by the PRC1 component Ring1b (Li et al., 2007; Luis et al., 2011), we merged data for direct Cbx4 targets and ChIP-seq data for H3K27me3 and found that 60.9% of Cbx4 direct target genes in primary epidermal KCs show peaks for both Cbx4 and H3K27me3 within 100 kb of their transcription start sites (Fig. 2 E and Table S5). Furthermore, 12.1% of these genes showed the overlapping ChIP-seq peaks for Cbx4 and H3K27me3, thus pointing to these genes as direct targets for Cbx4-mediated repression (Table S5).
The DAVID bioinformatics tool for gene ontology functional annotation revealed an enrichment of the genes involved in the control of neuronal development among the Cbx4 targets (Fig. 2, F and G; and Table S5). In primary epidermal KCs, several neuronal and other lineage-specific genes (Nefl, Neurog3, Mobp, En2, and Lhx4) contained both Cbx4 and H3K27me3 ChIP-seq peaks in their regulatory regions (Fig. 2 H). Validation by qRT-PCR showed increased transcript levels of the selected Cbx4 target genes in the Cbx4-null epidermal cells (Fig. 2 I), including the Nefl and Mobp genes encoding a neurofilament light polypeptide and myelin-associated oligodendrocyte basic protein, structural components of the neurons, and Schwann cells, respectively (Montague et al., 1997; Jordanova et al., 2003). In addition, expression of the Neurog3, Olig2, En2, and Lhx4 genes, encoding the corresponding transcription factors controlling the development of nervous system and some other non–KC cell lineages (Hobert and Westphal, 2000; Wallén and Perlmann, 2003; Pelling et al., 2011; Mitew et al., 2014), was markedly increased in the epidermis of Cbx4KO mice compared with WT controls (Fig. 2 I).
Interestingly, Nefl transcript levels showed an inverse correlation with Cbx4 expression in the WT epidermis during E11.5–E16.5 (Figs. 1 A, 2 I, and S1 C). Further analysis revealed that, in addition to the dermal nerve fibers, an ectopic expression of the Nefl protein was seen in both basal and suprabasal epidermal layers of E16.5 Cbx4KO mice, whereas an exclusively neuronal Nefl expression was seen in WT mice (Fig. 2 J). Furthermore, ChIP–quantitative PCR (ChIP-qPCR) analyses showed enrichments for Cbx4, H2AK119ub1, and H3K27me3 at the regulatory region of the Nefl gene, whereas a lack of Cbx4 enrichment was seen at the regulatory region of the Lor gene that did not belong to the category of the Cbx4 direct targets and served as a control in this experiment (Fig. 2 K). In addition, shRNA-mediated Cbx4 gene silencing resulted in a significant decrease in the Cbx4 and H2AK119ub1 levels at the regulatory region of the Nefl gene in primary epidermal KCs, whereas the H3K27me3 levels were not changed (Figs. 2 L and S2 D). These data were consistent with the data demonstrating the decrease in the H2AK119ub1 levels in the epidermis of Cbx4KO versus WT mice (Fig. 2, A and B) and suggest that Cbx4 as a part of the canonical PRC1 complex facilitates repression of its target Nefl gene. However, Cbx4 ablation did not affect Merkel cell numbers in the developing epidermis or expression of the transcription factors (Atoh1 and Sox2) involved in their differentiation (Fig. 4, B and C; and Table S5; Van Keymeulen et al., 2009; Bardot et al., 2013; unpublished data).
Figure 4.
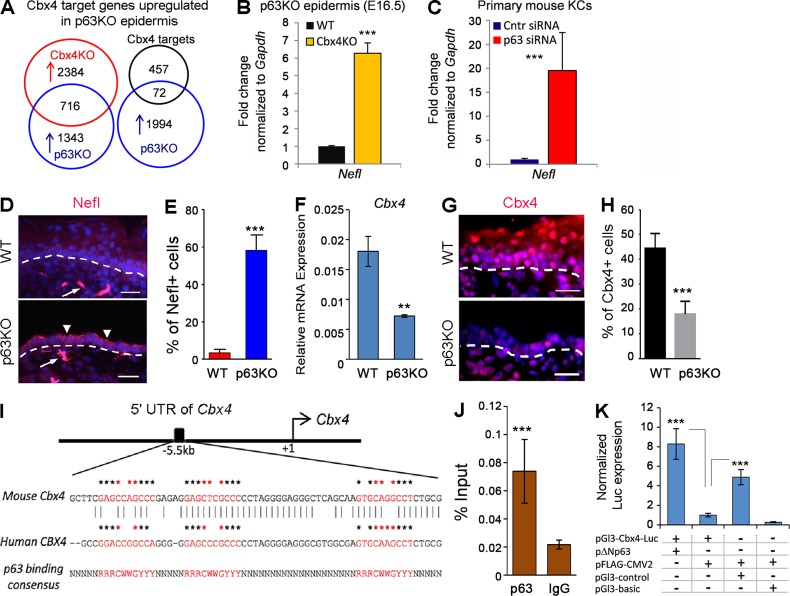
Cbx4 serves as a direct p63 target in epidermal KCs. (A) Overlap of the Cbx4 target genes commonly up-regulated in laser-captured epidermis of E16.5 Cbx4KO and p63KO mice. (B and C) qRT-PCR validation of Nefl gene up-regulation in the epidermal progenitor cells in p63KO mice (B) and primary epidermal KCs expressing p63 shRNA (C; mean ± SD). n = 3. (D) Increased Nefl protein expression in the epidermis of p63 mutant mice (arrowheads). Nefl-expressing dermal nerve fibers are shown by arrows. Dashed lines separate epidermis and dermis. (E) The number of Nefl+ epidermal cells is significantly increased in p63KO mice (mean ± SD). n = 3. (F and G) Cbx4 transcript (F) and protein (G) are markedly down-regulated in the epidermis of p63KO mice (mean ± SD). n = 3. Dashed lines separate epidermis and dermis. (H) A significant reduction in the number of Cbx4+ cells in p63KO skin compared with WT skin (mean ± SD). n = 3. (I) Conserved putative p63-binding sites are identified by the PatSearch tool in the mouse and human Cbx4 5′ UTR. Asterisks indicate consensus sequences for p63-binding sites, and core sequence elements are indicated by red asterisks. (J) ChIP-qPCR showing p63 binding to the identified region in the mouse Cbx4 5′ UTR. (K) A 2.4-kb region in the Cbx4 5′ UTR increases minimal SV40-promoter activity (pGl3-Cbx4-luc) upon coexpression with a ΔNp63-expressing construct compared with an empty plasmid (pFLAG-CMV2). Bars, 25 µm. **, P < 0.01; ***, P < 0.001.
Chromodomain- and SUMO E3 ligase–dependent Cbx4 activities differentially regulate proliferation, differentiation, and expression of nonepidermal genes in KCs
As Cbx4 uniquely possesses, among PRC1-associated Cbx subunits, both Polycomb and non-Polycomb SUMO E3 ligase activities (Li et al., 2007; Luis et al., 2011), we next questioned whether its effects on epidermal proliferation, differentiation, and repression of nonepidermal genes seen during murine skin development are associated with one of these Cbx4 activities. We infected primary epidermal KCs isolated from newborn WT mice with retroviral constructs expressing either WT Cbx4 (Cbx4WT), chromodomain-mutated Cbx4 (Cbx4CDM), or E3 ligase–deficient Cbx4 (Cbx4ΔSIM; Luis et al., 2011).
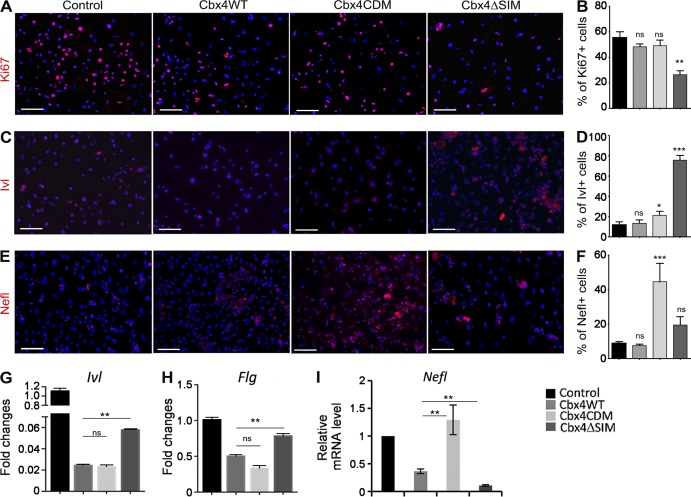
Interestingly, transfection of cells with Cbx4WT or Cbx4CDM constructs did not show any effect on the number of proliferating Ki67+ cells, whereas a prominent decrease in proliferation was seen in the cells expressing the E3 ligase–deficient Cbx4ΔSIM construct (Fig. 3, A and B). These data were consistent with the data showing the decrease of epidermal proliferation in Cbx4KO mice (Fig. 1, G and H) and suggest that the effects of Cbx4 on proliferation in cultured KCs are mediated predominantly by its E3 ligase domain.
Figure 3.
SUMO E3 ligase– and chromodomain-dependent activities of Cbx4 in the epidermal progenitor cells. (A–F) Immunofluorescent detection and quantification of Ki67 (A and B), Ivl (C and D), and Nefl (E and F) expression in the primary mouse KCs infected with Cbx4WT-, Cbx4CDM-, and Cbx4ΔSIM-expressing retroviruses. The number of Ki67+-proliferating cells is significantly reduced, whereas Ivl expression is markedly increased in Cbx4ΔSIM-expressing KCs. In contrast, Nefl expression is up-regulated in Cbx4CDM-expressing cells (mean ± SD). n = 3. Bars, 50 µm. (G and H) qRT-PCR shows increased Ivl (G) and Flg (H) transcription in Cbx4ΔSIM-expressing cells (mean ± SEM). n = 3. (I) Increased levels of the Nefl transcripts in primary mouse KCs infected with Cbx4CDM-expressing retrovirus (mean ± SEM). n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant.
Given that Cbx4 suppresses senescence in human epidermal KCs (Luis et al., 2011), we examined the mechanisms underlying the Cbx4-dependent regulation of the senescence-linked cell cycle inhibitor genes and performed ChIP-qPCR analyses that revealed Cbx4 binding to the Cdkn2a (p16/p19) regulatory regions in primary epidermal KCs (Fig. S3 C). Transfection of primary epidermal KCs with Cbx4WT or Cbx4CDM constructs showed that the Cbx4 chromodomain is involved in regulating the expression of Cdkn2a and Cdkn1c (p57; Fig. S3 D). Together with our data demonstrating an increase in the expressions of p16/p19, p57, and the senescence marker γ-H2AX in the epidermis of Cbx4KO mice versus WT controls (Fig. S3, A, B, and E), these data demonstrated an involvement of the Cbx4 chromodomain in the control of expression of the senescence-associated p16/p19 and p57 genes. These data were in full concordance with the data of Luis et al. (2011) that show similar effects of the Cbx4 chromodomain on the expression of senescence-associated genes in human epidermal KCs.
To assess whether targeting of Cbx4 in the primary KCs affects cell differentiation, we analyzed the expression of two established terminal differentiation–associated markers (Involucrin and Filaggrin) after infection of cells with the WT or mutated Cbx4-expressing retroviruses. Cbx4WT-expressing cells showed a decrease in protein and gene expression of Involucrin in comparison with control cells (Fig. 3, C and D). However, Cbx4ΔSIM-treated KCs showed a prominent increase of Involucrin protein and gene expression compared with Cbx4WT- or Cbx4CDM-treated cells (Fig. 3, C, D, and G). Furthermore, transcripts of Flg were also increased upon Cbx4ΔSIM overexpression compared with the Cbx4WT and Cbx4CDM constructs (Fig. 3 H). Thus, similar to human epidermal progenitor cells (Luis et al., 2011), expression of terminal differentiation markers in mouse KCs occurs predominantly under the control of non-Polycomb SUMO E3 ligase activity of Cbx4.
To further elucidate which of the Cbx4 activities were likely responsible for repression of the nonepidermal lineage genes, we analyzed Nefl expression in the response to overexpression of the Cbx4 constructs harboring either chromodomain- or SUMO E3 ligase–mutated domains. A marked increase of both Nefl transcript and protein expression was detected in the Cbx4CDM-expressing cells compared with Cbx4WT and Cbx4ΔSIM (Fig. 3, E, F, and I). Thus, Cbx4 represses neuronal gene Nefl in epidermal KCs predominantly via the Polycomb-mediated inhibitory pathway.
Cbx4 is a direct p63 target mediating its inhibitory activity on nonepidermal genes in KCs
p63 is a major transcriptional regulator of epidermal development with multiple functions, including a direct positive transcriptional regulation of epidermal genes such as keratins and adhesion molecules (Koster and Roop, 2007; Vanbokhoven et al., 2011; Botchkarev and Flores, 2014; Kouwenhoven et al., 2015). A recent study has also highlighted an important role for p63 in suppression of nonepidermal lineage genes (De Rosa et al., 2009). To test whether p63-dependent suppression of the nonepidermal genes is mediated by Cbx4, we compared the transcriptomes of epidermal cells isolated by LCM from E18.5 Cbx4KO and p63KO embryos. A significant portion (34.7%) of the genes (716 out of 2,059) up-regulated in the p63-deficient KCs were also up-regulated in Cbx4KO basal epidermal cells, suggesting a functional link between p63 and Cbx4 (Fig. 4 A and Table S2). Furthermore, ChIP-seq analyses showed that 72 of these genes, including the Nefl gene, served as direct Cbx4 targets in KCs (Fig. 4 A and Table S2). The expression changes of the Nefl gene were further validated by qRT-PCR in p63-deficient KCs isolated from p63KO embryos and p63 siRNA–treated primary mouse KCs (Fig. 4, B and C). Similar to Cbx4KO epidermal cells, p63-deficient skin epithelial cells showed strongly up-regulated Nefl protein expression compared with WT cells (Fig. 4, D and E), further substantiating functional cooperation between p63 and Cbx4 in the repression of nonepidermal genes in the developing epidermal KCs.
To further investigate the relationships between p63 and Cbx4, we analyzed Cbx4 expression in p63KO embryonic skin. Both the Cbx4 transcript and protein levels were markedly reduced in the p63-deficient KCs (Fig. 4, F–H). Interestingly, in p63KO dermal cells, Cbx4 protein expression was detectable at similar levels to control mice (Fig. 4 G), suggesting a KC-specific Cbx4 regulation by p63 transcription factor. By using a PatSearch tool (Grillo et al., 2003), we identified a highly conserved region ∼5.5 kb upstream of the Cbx4 transcription start site containing several putative p63-binding sites (Fig. 4 I). Furthermore, ChIP revealed binding of the p63 transcription factor to the identified region in primary mouse KCs in vivo (Fig. 4 J). We cloned a 2.4-kb Cbx4 5′ UTR fragment, containing the putative p63-binding sites as an enhancer, into the pGl3 promoter plasmid (pGl3-Cbx4-luc) and coexpressed it with either ΔNp63-expressing or empty vector in the immortal KC HaCaT cell line. A marked increase in luciferase activity was seen in cells coexpressing pGl3-Cbx4-luc with pΔNp63 vectors compared with a control plasmid (pFLAG-CMV2; Fig. 4 K), thus showing a direct transcriptional regulation of Cbx4 by p63 in KCs.
Ectopic Cbx4 expression partially rescues the epidermal phenotype in embryonic skin explants of p63-deficient mice
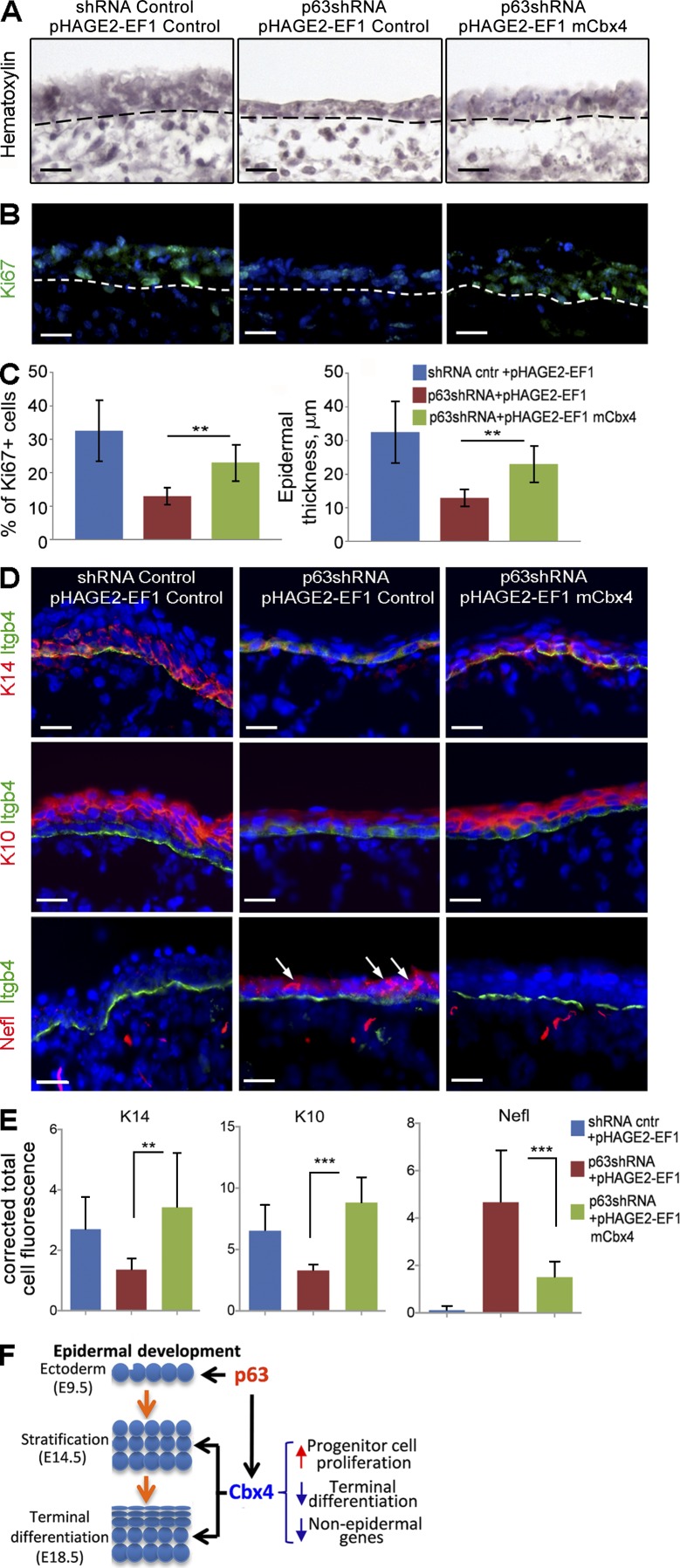
To assess whether the restoration of Cbx4 expression is capable of partially rescuing the skin phenotype of p63KO mice, E13.5 skin explants isolated from p63 heterozygous (p63+/−) mice were cultured ex vivo and infected with p63 shRNA–expressing lentiviruses in combination with either Cbx4-expressing or control lentiviruses (Fig. 5, A–E). Compared with the p63+/− skin explants treated with control viruses, treatment with p63 shRNA in combination with control lentiviruses resulted in a marked decrease of the epidermal thickness, cell proliferation, and expressions of epidermal keratins K14 and K10, thus reproducing in part the skin phenotype of p63KO mice (Fig. 5, A–E). However, cotreatment of p63+/− skin samples with p63 shRNA– and Cbx4-expressing lentiviruses resulted in a significant increase of epidermal thickness, cell proliferation, and K14/K10 expression compared with the samples treated with p63 shRNA and control lentiviruses (Fig. 5, A–E). The restoration of epidermal proliferation after ectopic Cbx4 expression was consistent with data showing reduced cell proliferation in the epidermis of Cbx4KO mice compared with WT controls (Fig. 1, G and H). Furthermore, the Nefl protein was strongly decreased in the epithelium of p63-deficient embryonic skin explants transduced with Cbx4 (Fig. 5, D and E), suggesting that Cbx4 is indeed capable of inhibiting its expression in epidermal KCs ex vivo. Therefore, Cbx4 plays a role in mediating the p63-dependent program of gene repression during epidermal development and is capable of partially restoring the epidermal phenotype of p63-deficient mice.
Figure 5.
Cbx4 partially rescues an epidermal phenotype in p63KO mice. (A and B) Hematoxylin/AP staining (A) and immunodetection of Ki67+-proliferating cells (B) in E13.5 ex vivo embryonic skin explants of heterozygous p63+/−KO mice treated with p63 shRNA– and Cbx4-expressing lentiviruses. Dashed lines separate epidermis and dermis. (C) Cbx4 overexpression in p63-deficient epidermal cells significantly increases epidermal thickness and cell proliferation (mean ± SD). n = 3. (D and E) Quantitative immunofluorescence stainings with anti-K14, anti-K10, and anti-Nefl antibodies show down-regulation of K14/K10 and up-regulation of Nefl (arrows) in the epithelium of p63-deficient skin explants. Cotreatment of the explants with p63 shRNA– and Cbx4-expressing lentiviruses results in up-regulation of K14/K10 expression and down-regulation of Nefl expression in p63-deficient epidermis. Integrin-β4 (Itgb4) expression outlines the basement membrane of the epidermis. DAPI counterstain (blue) shows the nuclei (mean ± SD). n = 3. **, P < 0.01; ***, P < 0.001. (F) Scheme summarizing the p63-/Cbx4-mediated effects on gene repression in the developing epidermis. Bars, 25 µm.
Discussion
Polycomb-dependent transcriptional repression is a powerful mechanism that shapes gene expression programmers in essentially all cell types of living organisms during development and postnatal homeostasis, whereas alterations in the patterns of Polycomb-dependent gene repression contribute to many pathological conditions including carcinogenesis (Simon and Kingston, 2013). Polycomb PRC1 and PRC2 are multiprotein complexes, and despite the fact that the role of some Polycomb genes, such as Ezh1/2, in execution of terminal differentiation programmers in the epidermis and hair follicles was shown previously (Ezhkova et al., 2009, 2011), the effects of the other PRC1/2 components on the establishment of lineage-specific patterns of gene repression during skin development and epidermal differentiation remain unclear. Here, we show that PRC1 component Cbx4 plays a unique role in the establishment and maintenance of the KC lineage identity and represses nonepidermal lineage (neuronal) genes in the epidermal progenitor cells, as well as controls proliferation and inhibits premature differentiation in basal epidermal KCs.
We demonstrate that among different PRC1 genes, Cbx4 shows the most prominent changes in its expression in the embryonic epidermis during its stratification, where Cbx4 is seen in basal and suprabasal KCs. Genetic Cbx4 ablation results in a marked decrease of epidermal thickness and proliferation, as well as in the premature appearance of the terminal differentiation markers in the immediate suprabasal epidermal cells. Interestingly, Ezh2KO mice also show a decrease of epidermal proliferation and premature terminal differentiation (Ezhkova et al., 2009). Similar to Ezh2KO mice (Ezhkova et al., 2009), Cbx4-null mice show an increase of the p16/p19 and p57 cell cycle inhibitor genes in the epidermis, which, at least in part, explains the stimulatory effects of Cbx4 on epidermal proliferation. However, in contrast to Ezh2KO mice, Cbx4 genetic ablation results in a marked decrease of the epidermal thickness, suggesting that Cbx4 plays a unique yet partially overlapping role with Ezh2 in the control of epidermal development.
The decreased cell proliferation observed in the epidermis upon Cbx4 ablation is consistent with our previous results that show reduced cell proliferation and thymus hypoplasia in Cbx4KO mice (Liu et al., 2013). In contrast to other Cbx proteins, Cbx4 uniquely possesses both Polycomb and non-Polycomb SUMO E3 ligase activities (Li et al., 2007; Luis et al., 2011). We show here that in cultured primary epidermal KCs, only the non-Polycomb E3 ligase–deficient Cbx4 (Cbx4ΔSIM) is capable of inhibiting proliferation, suggesting that the SUMO E3 ligase activity of Cbx4 contributes to the alterations of epidermal proliferation in Cbx4KO mice.
Interestingly, KCs treated with Cbx4ΔSIM also show a marked increase in the expression of terminal differentiation–associated markers, such as Involucrin or Filaggrin, thus suggesting that non-Polycomb SUMO E3 ligase Cbx4 activity might also inhibit the premature activation of terminal differentiation genes in the epidermis. These data are consistent with data demonstrating an involvement of SUMO E3 ligase Cbx4 activity in inhibiting differentiation in human epidermal progenitor cells (Luis et al., 2011). However, additional analyses are required to fully understand the mechanisms underlying the effects of the SUMO E3 ligase domain of Cbx4 in regulating gene expression in KCs.
Our data also reveal that the Cbx4 chromodomain is involved in the repression of the senescence-associated Cdkn2a gene, whereas Cbx4 ablation results in the accumulation of H2AX as one of the senescence markers (Turinetto and Giachino, 2015) in epidermal KCs. These data are in full concordance with the data of Luis et al. (2011) that show similar effects of the Cbx4 chromodomain on the expression of senescence-associated genes in human epidermal KCs.
Histone methyltransferase Ezh2 catalyzing H3K27 trimethylation plays an important role in the prevention of premature activation of terminal differentiation genes in basal epidermal KCs (Ezhkova et al., 2009). The Cbx4 chromodomain mediates the targeting of the canonical PRC1 complex to H3K27me3, leading to the increase of H2AK119 ubiquitylation over its basal levels catalyzed by the PRC1 component Ring1b (Simon and Kingston, 2013; Cheutin and Cavalli, 2014; Perdigoto et al., 2014; Schwartz and Pirrotta, 2014). Although the levels of H3K27me3 were not altered in the epidermis of Cbx4KO mice, Cbx4 ablation resulted in a marked decrease in H2AK119Ub in the epidermis (presumably to its basal levels) compared with WT controls. The fact that Cbx4-null mice showed reduced levels of H2AK119Ub only in the epidermis, whereas H2AK119Ub levels in the dermal cell were not altered upon Cbx4 ablation compared with WT controls, points to the specificity of this interaction.
Interestingly, Cbx4 ablation is also accompanied by an increase of epidermal expression of Kdm2b histone demethylase that forms a noncanonical PRC1 complex together with PCGF1 and RYBP and promotes basal ubiquitylation of H2AK119 at unmethylated CpG-rich DNA regions (Blackledge et al., 2014; Cooper et al., 2014; Kalb et al., 2014). These data suggest that interaction of the noncanonical PRC1 complex with gene targets is likely not altered in the epidermis of Cbx4KO mice and that the Cbx4 chromodomain is specifically involved in the execution of the canonical PRC1-mediated gene repression in the epidermis via interaction with H3K27me3 followed by H2AK119 ubiquitylation.
However, a correlation between the Cbx4 and Ezh1/2 ChIP-seq targets in epidermal KCs might help in addressing the roles for PRC1 and PRC2 complexes in repressing the activities of the distinct gene groups in KCs. Our data reveal a contribution of the Cbx4 chromodomain to the repression of non-KC lineage (neuronal) genes in the epidermis, which is quite consistent with the data obtained from the Ezh2KO mice (Ezhkova et al., 2009). By merging the microarray data obtained from the epidermis of Cbx4KO mice with Cbx4 ChIP-seq and H3K27me3 ChIP-seq data, we demonstrate an enrichment of the genes associated with nervous system development among the direct Cbx4 targets in KCs. We show that several key genes encoding structural components of the neurons and Schwann cells (Nefl and Mobp) and transcription factors involved in the control of nervous system development (Neurog3, Olig2, En2, and Lhx4) are up-regulated in the epidermis of Cbx4KO mice compared with WT controls.
Epidermal and neuronal lineages share the common ectodermal origin (Patthey and Gunhaga, 2011; Benitah and Frye, 2012), and mechanisms that control gene repression during specification of these two lineages in the embryo are unclear. Ezh2 is strongly expressed in neural stem cells and is down-regulated during their differentiation into neurons and astrocytes, whereas its expression is increased during neural stem cell differentiation into the oligodendrocyte lineage (Sher et al., 2008). Analyses of the Ezh2 targets in neural stem cells (Sher et al., 2012) reveal that several genes encoding transcription factors involved in the control of nervous system development (En2, Lhx4, Olig2, Pax3, and Pitx2) also represent the direct Cbx4 targets in epidermal KCs. These data support an idea that Cbx4, through its chromodomain, and Ezh2 operate in concert to repress neuronal genes and maintain the epithelial fate in the epidermal progenitor cells, whereas Ezh2 also represses their differentiation into the Merkel cell lineage independently of Cbx4 (Bardot et al., 2013).
Specification and differentiation of the epidermal progenitor cells during development is supported by the p63 transcription factor that serves as a master regulator of epidermal development (Koster and Roop, 2007; Vanbokhoven et al., 2011; Botchkarev and Flores, 2014; Kouwenhoven et al., 2015). Increased evidence demonstrates that p63 regulates several components of the epigenetic machinery (Brg1, Lsh, and Satb1) to regulate chromatin remodeling and establish the epidermal differentiation program (Fessing et al., 2011; Keyes et al., 2011; Mardaryev et al., 2014).
We show here that p63 also operates as a direct upstream regulator of Cbx4, which mediates its effects on cell proliferation and is involved in the repression of non-KC lineage genes. We showed previously that p63 directly interacts with Cbx4 in thymocytes, suggesting a possibility for both proteins to form a transcription complex to regulate expression of target genes (Liu et al., 2013). Consistently, comparison of the microarray data from the skin epithelium of p63KO mice and epidermis of Cbx4KO mice reveals that ∼35% of the genes up-regulated in the skin epithelium of p63KO mice are also up-regulated in the epidermis of Cbx4 mutants, whereas ∼10% of these genes represent direct Cbx4 targets in KCs. Furthermore, we show that Cbx4 expression is markedly decreased in the skin epithelium of p63KO mice, p63 shows binding to the Cbx4 regulatory region and stimulates Cbx4 promoter activity, and Cbx4 is capable of partially rescuing alterations of KC proliferation upon p63 ablation in skin organ cultures.
Collectively, these data provide compelling evidence that Cbx4 operates as an important part of the p63-regulated program of establishing and maintaining epithelial cell fate in epidermal progenitors and that Cbx4-mediated repression of non-KC lineage genes serves as a crucial component of this program (Fig. 5 F). However, additional analyses are required to fully understand the complexity of Cbx4-dependent gene repression in epidermal KCs. In particular, the role of Cbx4 in the repression of other lineage-specific genes, including mesodermal genes in KCs, and the potential contribution of these genes in the control of epithelial–mesenchymal transition need to be carefully explored. Also, the roles of other members of the Cbx family, including Cbx6 and Cbx7, which might substitute for Cbx4 in the canonical PRC1 complex and partially compensate for the effects of Cbx4 ablation in epidermal KCs, need to be clarified.
Because skin contains a large variety of the progenitor cells for distinct cell lineages (epithelial, neuronal, and mesenchymal; Hunt et al., 2009), future research in this direction will shed light on the role of distinct Cbx family members in the control of progenitor cell differentiation and/or reprogramming isolated from the skin. This will also help to design novel approaches for modulating gene expression programmers in healthy and diseased skin via targeting the activity of Cbx genes.
Materials and methods
Animals and tissue collection
All animal studies were performed under protocols approved by a Home Office Project License. C57BL/6 mice were purchased from Charles River. Skin samples were collected from mice at distinct days of embryonic and postnatal development (E11.5, E14.5, E16.5, E18.5, P1.5, and P56). p63KO embryos were obtained by breeding p63+/− animals purchased from The Jackson Laboratory. Generation of Cbx4KO mice was described previously (Liu et al., 2013). In brief, the N-terminal region of the Cbx4 gene including the first two exons and a 0.9-kb upstream region was targeted for Cbx4 gene disruption. The null allele was gained upon Cre-loxP excision by crossing mice carrying the floxed allele with EIIa-Cre transgenic mice to achieve global gene KO, and the mice were bred on the C57BL/6J-129Sv genetic background. Genotyping of mice was performed using PCR, as described previously (Liu et al., 2013). For each developmental stage, six to seven samples were collected. Tissue samples were snap frozen in liquid nitrogen, embedded into optimal cutting temperature medium (VWR), and processed for immunofluorescent or LCM and microarray analyses as described in the following paragraph.
LCM, microarray, and qRT-PCR analysis
LCM of mouse basal KCs at selected stages of development was performed followed by RNA isolation, amplification, and microarray analysis as previously described (Sharov et al., 2006, 2009). In brief, 8-µm-thick frozen skin cryosections were dehydrated to preserve RNA integrity, and LCM was performed from the epidermis of WT, Cbx4KO, and p63KO mice at E11.5, E16.5, P0.5, and P56 using an LCM system (Arcturus; Life Technologies). Total RNAs were isolated using an RNA isolation kit (PicoPure; Life Technologies) followed by two rounds of linear RNA amplification using an RNA amplification kit (RiboAmp; Life Technologies). Equal amounts of RNA from each sample were labeled by Cy3 using a fluorescent labeling kit (Agilent Technologies). Quality and size distribution of the targets was determined by an RNA 6000 Nano Lab-on-a-chip assay (Agilent Technologies), and quantification was determined using a microscale spectrophotometer (NanoDrop). After one round of linear amplification in all analyses, Universal Mouse Reference RNA (Agilent Technologies) was used as a control. All microarray analyses were performed by MOgene, LC, using the 41K Whole Mouse Genome 60-mer Oligo Microarray kit (Agilent Technologies). All microarray data were normalized to the corresponding data obtained from the reference RNA. Two independent datasets were obtained from WT and transgenic mice, and p-values were calculated by Feature Extraction software (version 7.5; Agilent Technologies) using distribution of the background intensity values to signal intensity and using a Student’s t test.
For qRT-PCR analysis of RNA isolated and amplified after LCM, PCR primers were designed on Beacon Designer software (PREMIER Biosoft International; Table S3). RT-PCR was performed using iQ SYBR green Supermix and the MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad Laboratories). Differences between samples and controls were calculated using the Gene Expression Macro program (Bio-Rad Laboratories) based on the ΔΔCt equitation method with glyceraldehyde 3-phosphate dehydrogenase and peptidylprolyl isomerase A genes as internal housekeeping controls.
Plasmids
Cbx4WT and Cbx4-mutated retroviral plasmids (Cbx4CDM and Cbx4ΔSIM) were previously validated (Luis et al., 2011). In brief, Cbx4 cDNA was cloned into pMSCV and pMSCV-Puro-IRES-GFP. The chromodomain was mutated using the QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) to create the F11A and W35L double mutants. In Cbx4ΔSIM plasmid (a gift from D. Wotton, University of Virginia, Charlottesville, VA), Cbx4 cDNA was mutated to harbor internal deletion of nucleotides at the 784–795 and 1,381–1,404 positions, corresponding to both SIM1 (IVIV) and SIM2 (EVILLDSD) amino acid sequences, respectively (Merrill et al., 2010). Cbx4 knockdown was done using a validated shRNA construct (5′-CCGGCGACACCAGTAACCTTGGTATCTCGAGATACCAAGGTTACTGGTGTCGTTTTTG-3′) cloned into pLKO.1-GFP lentiviral plasmid (Morey et al., 2012). pGIPZ–p63 shRNA lentiviral plasmid (V3LMM_508694; GE Healthcare) contains a 5′-TGATCTTCAGCAACATCTC-3′ sequence and was used to target p63. pHAGE2-EF1-mCbx4 lentiviral plasmid was produced by cloning PCR-amplified mouse Cbx4 cDNA into pHAGE2-EF1 vector (a gift from G. Mostoslavsky, Boston University, Boston, MA).
Production of the Cbx4 and p63 expression and knockdown viruses
Cbx4WT-, Cbx4CDM-, and Cbx4ΔSIM-expressing retroviruses were produced by transfection of Phoenix-E/HEK293T packaging cells. Cbx4 shRNA– and p63 shRNA–expressing lentiviruses were produced by cotransfection of HEK293T cells with pLKO.1-EGFP-Cbx4 shRNA or pGIPZ–p63 shRNA and packaging (psPAX2) and envelope (pMD2.G) plasmids. For Cbx4-expressing lentiviruses, HEK293T cells were cotransfected with pHAGE2-EF1-mCbx4 and helper plasmids (pTAT, pREV, pHagp2(GAG/Pol), and pVSV-G) as published elsewhere (Mostoslavsky et al., 2005). For Cbx4-mutated retroviruses, Phoenix-E cells were transfected with pMSCV-PIG-CBX4-WT, pMSCV-PIG-CBX4-CDM, and pMSCV-PIG-CBX4-ΔSIM plasmids as published previously (Luis et al., 2011). Cell culture medium containing viruses was collected 24 h, 48 h, and 78 h after transfection, followed by precipitation of the viral particles using PEG-it solution (System Biosciences) as per the manufacturer’s protocol.
Primary mouse KC culture
Epidermis from newborn C57BL/6 was separated from dermis by overnight digestion with 0.25% trypsin at 4°C. Epidermal sheath was added to prechilled, supplemented, low-calcium KC culture medium (MEM, 4% FBS, 0.05-mM CaCl2, 0.4 µg/ml hydrocortisone, 5 µg/ml insulin, 10 mg/ml EGF, 10−10-M cholera toxin, 2 × 10−9 T3, 2-mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin). To make a single-cell suspension, the epidermal tissue was chopped with scissors and triturated, followed by filtering through a 70-µM silicon strainer and plating onto collagen-coated plates and coverslips. Primary cells were grown in the KC culture medium at 33°C and 5% CO2 until 60–70% confluent and infected with the retroviral/lentiviral particles. Cells were collected for subsequent qRT-PCR or immunofluorescent analyses 48 h after infection.
FACS
A single-cell suspension of newborn epidermal KCs was prepared as described in the previous paragraph. To ensure analysis and sorting of viable cells with intact chromatin, KCs were stained with UV Live/Dead Fixable Dye (Life Technologies) for 30 min on ice before fixation with 1% PFA for 10 min at RT. Fixed cells were labeled with CD49f-phycoerythrin and Sca-1–FITC antibodies (Table S4) for 1 h on ice. CD49f+/Sca-1+ basal KCs were gated after exclusion of dead cells and sorted on a cell sorter (MoFlo XDP; Beckman Coulter) as described elsewhere (Jensen et al., 2010). Sorted cells were pelleted at 2,000 g and stored at −80°C.
Luciferase reporter assay
To make the pGl3-Cbx4-luc plasmid, a 2.4-kb Cbx4 enhancer region (−5,954/−3,523 bp upstream of the Cbx4 transcription start site) was PCR amplified from genomic DNA and cloned into the Luciferase reporter plasmid pGl3 promoter (Promega) at BamHI–SalI sites using the In-Fusion HD Cloning System (Takara Bio Inc.). For the reporter assay, HaCaT cells were seeded into white-bottomed/white-walled 96-well plates at 10,000 cells/well, and transfections were performed after overnight incubation at 37°C. Cells were transfected with 200 ng DNA total plus 10 ng pRLTK Renilla Control vector (Promega): 100 ng pGl3-Cbx4-luc and 100 ng pΔNp63 or empty pFLAG-CMV2 vectors. After 48 h of incubation at 37°C, cells were washed with PBS, and the assay was performed using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol. Firefly luciferase activity was normalized against the Renilla luciferase activity, and the data represent three independent triplicates.
Histology and immunohistochemistry
For histology and histomorphometry, 8-µm cryosections were fixed in 4% PFA for 10 min at RT followed by hematoxylin staining and AP activity visualization using histoenzymatic reaction (Botchkareva et al., 1999, 2000; Ahmed et al., 2011, 2014). Epidermal thickness was measured in 10 microscopic fields using ImageJ software (National Institutes of Health). For immunohistochemistry, 8-µm cryosections were fixed in 4% PFA for 10 min at RT and incubated with primary antibodies (Table S4) overnight at 4°C, followed by application of corresponding Alexa Fluor 488 and Cy-555–coupled secondary antibodies (1:200; Life Technologies) for 60 min at 37°C. Cell nuclei were visualized with 4′6′-diamidino-2-phenylindol. Images were acquired on a microscope (Eclipse 50i; Nikon) with 20×/0.50 NA and 40×/0.75 NA Plan Fluor objectives (Nikon) using a digital camera (ExiAqua; QImaging) and Image-Pro Express 6.3 image analysis software (QImaging).
Comparative analysis of Ki67+, Ivl+, Nefl+, and Cbx4+ cells was done by evaluating the ratio of the number of positive cells to DAPI+ cells, as descried previously (Sharov et al., 2003, 2005; Mardaryev et al., 2011). For quantification of Ki67+ and Cbx4+ cells in tissue cryosections, 10 microscopic fields (20×) from three WT and Cbx4KO skin sections were included in the analysis. For quantification of Ki67+, Ivl+, and Nefl+ cells cultured in vitro, five microscopic fields (20×) taken from the center and each of the four sides of the well were used in the analysis. Data were pooled, the means ± SD were calculated, and statistical analysis was performed using a two-tailed t test (α = 0.05).
Immunofluorescence intensity was determined using ImageJ software as described previously (McCloy et al., 2014). In brief, red or green fluorescent signals were collected from experimental tissues in RGB format using the same exposure conditions. To measure the fluorescence intensity at each pixel, the RGB images were converted to 8-bit grayscale format. Regions of interest of distinct size were selected within WT or Cbx4KO epidermis and dermis, and the corrected values of total cell fluorescence were calculated for each selected area using the following formula: corrected total cell fluorescence = integrated density − (area of selected cell × mean fluorescence of background readings). For pairwise comparisons, a two-tailed t test (α = 0.05) was used. Where multiple samples were compared, a one-way analysis of variance (ANOVA) was used, followed by the Newman–Keuls test (α = 0.05).
ChIP-seq and ChIP-qPCR assays
ChIP was performed using epidermal KCs isolated from newborn mouse skin and anti-H3K27me3, anti-Cbx4, and anti-H2AK119ub1 antibodies (Table S5). In brief, a single-cell suspension of mouse epidermal KCs was prepared after overnight digestion in 5 mg/ml dispase (Roche) at 4°C, followed by a 5-min treatment with 0.2% trypsin (Life Technologies). Basal epidermal KCs were sorted by FACS and further processed using a ChIP-IT High Sensitivity kit (Active Motive). 5 × 106 PFA-fixed cells were used as a starting material. Indexed ChIP-seq libraries were generated using NEBNext reagents (New England Biolabs, Inc.), and ChIP libraries were sequenced on the HiSeq 2500 platform (Illumina), producing 30–70 million reads per library. Sequencing reads were aligned to the mm9 mouse genome assembly (University of California, Santa Cruz, Genome Browser). Specific areas of binding were identified with Sicer using default settings. For ChIP-qPCR, precipitated DNA was analyzed using primers designed for the promoter regions of Cbx4, Lor, Nefl, and Cdkn2a genes (Table S3). ChIP-qPCR data were pooled, means ± SD were calculated, and statistical analysis was performed using a one-way ANOVA test as described previously (Mardaryev et al., 2011).
Embryonic tissue culture
Whole dorsal skin culture from E13.5 p63+/− embryos were prepared and maintained as previously described (Botchkarev et al., 1999). In brief, skin samples were dissected from embryos and cultured in the 6-well plates containing Williams’ medium E. Within 2 h after the culture preparation, tissue samples were infected with p63 shRNA lentivirus in combination with either Cbx4-expressing or control lentiviruses and the addition of 10 µg/ml polybrene (Sigma-Aldrich) for 48 h. The tissue was frozen and embedded into optimal cutting temperature medium for subsequent immunofluorescent and qRT-PCR analyses.
Online supplemental material
Fig. S1 shows expression of the PRC1/PRC2 and Nefl genes in mouse epidermis during development. Fig. S2 shows characterization of the changes in gene expression in the epidermis of Cbx4KO mice and in primary KCs after Cbx4 knockdown. Fig. S3 shows analyses of the senescence and apoptotic markers in WT and Cbx4-deficient skin. Fig. S4 shows the details of the epidermal and Merkel cell differentiation in Cbx4KO and WT mice. Table S1 shows expression of the PRC1 and PRC2 genes in epidermal KCs at different stages of epidermal development. Table S2 shows Cbx4 target genes up-regulated in the skin epithelium of p63KO mice. Tables S3 and S4 present a list of primers used for qRT-PCR analyses and a list of antibodies used in this study, respectively. Table S5 shows Cbx4 target genes in epidermal KCs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506065/DC1.
Supplementary Material
Acknowledgments
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BB/K010050/1), the Medical Research Council (MRC; MR/M010015/1), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR049778 and AR064580), and EpiGeneSys to V.A. Botchkarev; from MRC (G1000846) to C.A. Jahoda; from the European Research Council, the Association for International Cancer Research, the Foundation La Marató, the Spanish Ministry of Economy and Development, the Foundation Vencer el Cancer, the Government of Cataluña (SGR and Mario Salviá), the Foundation Botín, and the Institute for Research in Biomedicine Barcelona to S.A. Benitah; and from the National Science Foundation of China (81461138034) to G.-L. Xu.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- ChIP
- chromatin immunoprecipitation
- ChIP-qPCR
- ChIP–quantitative PCR
- KC
- keratinocyte
- KO
- knockout
- LCM
- laser capture microdissection
- qRT-PCR
- quantitative real time–PCR
- WT
- wild type
References
- Ahmed M.I., Mardaryev A.N., Lewis C.J., Sharov A.A., and Botchkareva N.V.. 2011. MicroRNA-21 is an important downstream component of BMP signalling in epidermal keratinocytes. J. Cell Sci. 124:3399–3404. 10.1242/jcs.086710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M.I., Alam M., Emelianov V.U., Poterlowicz K., Patel A., Sharov A.A., Mardaryev A.N., and Botchkareva N.V.. 2014. MicroRNA-214 controls skin and hair follicle development by modulating the activity of the Wnt pathway. J. Cell Biol. 207:549–567. 10.1083/jcb.201404001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardot E.S., Valdes V.J., Zhang J., Perdigoto C.N., Nicolis S., Hearn S.A., Silva J.M., and Ezhkova E.. 2013. Polycomb subunits Ezh1 and Ezh2 regulate the Merkel cell differentiation program in skin stem cells. EMBO J. 32:1990–2000. 10.1038/emboj.2013.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitah S.A., and Frye M.. 2012. Stem cells in ectodermal development. J. Mol. Med. 90:783–790. 10.1007/s00109-012-0908-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge N.P., Farcas A.M., Kondo T., King H.W., McGouran J.F., Hanssen L.L., Ito S., Cooper S., Kondo K., Koseki Y., et al. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 157:1445–1459. 10.1016/j.cell.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., and Fuchs E.. 2009. Epidermal homeostasis: A balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10:207–217. 10.1038/nrm2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., and Fuchs E.. 2014. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science. 344:1242281 10.1126/science.1242281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev V.A., and Flores E.R.. 2014. p53/p63/p73 in the epidermis in health and disease. Cold Spring Harb. Perspect. Med. 4:301–312. 10.1101/cshperspect.a015248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev V.A., Peters E.M., Botchkareva N.V., Maurer M., and Paus R.. 1999. Hair cycle-dependent changes in adrenergic skin innervation, and hair growth modulation by adrenergic drugs. J. Invest. Dermatol. 113:878–887. 10.1046/j.1523-1747.1999.00791.x [DOI] [PubMed] [Google Scholar]
- Botchkarev V.A., Gdula M.R., Mardaryev A.N., Sharov A.A., and Fessing M.Y.. 2012. Epigenetic regulation of gene expression in keratinocytes. J. Invest. Dermatol. 132:2505–2521. 10.1038/jid.2012.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkareva N.V., Botchkarev V.A., Chen L.H., Lindner G., and Paus R.. 1999. A role for p75 neurotrophin receptor in the control of hair follicle morphogenesis. Dev. Biol. 216:135–153. 10.1006/dbio.1999.9464 [DOI] [PubMed] [Google Scholar]
- Botchkareva N.V., Botchkarev V.A., Welker P., Airaksinen M., Roth W., Suvanto P., Müller-Röver S., Hadshiew I.M., Peters C., and Paus R.. 2000. New roles for glial cell line-derived neurotrophic factor and neurturin: Involvement in hair cycle control. Am. J. Pathol. 156:1041–1053. 10.1016/S0002-9440(10)64972-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T., and Cavalli G.. 2014. Polycomb silencing: From linear chromatin domains to 3D chromosome folding. Curr. Opin. Genet. Dev. 25:30–37. 10.1016/j.gde.2013.11.016 [DOI] [PubMed] [Google Scholar]
- Cooper S., Dienstbier M., Hassan R., Schermelleh L., Sharif J., Blackledge N.P., De Marco V., Elderkin S., Koseki H., Klose R., et al. 2014. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Reports. 7:1456–1470. 10.1016/j.celrep.2014.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa L., Antonini D., Ferone G., Russo M.T., Yu P.B., Han R., and Missero C.. 2009. p63 Suppresses non-epidermal lineage markers in a bone morphogenetic protein-dependent manner via repression of Smad7. J. Biol. Chem. 284:30574–30582. 10.1074/jbc.M109.049619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Pasolli H.A., Parker J.S., Stokes N., Su I.H., Hannon G., Tarakhovsky A., and Fuchs E.. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 136:1122–1135. 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Lien W.H., Stokes N., Pasolli H.A., Silva J.M., and Fuchs E.. 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 25:485–498. 10.1101/gad.2019811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessing M.Y., Mardaryev A.N., Gdula M.R., Sharov A.A., Sharova T.Y., Rapisarda V., Gordon K.B., Smorodchenko A.D., Poterlowicz K., Ferone G., et al. 2011. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J. Cell Biol. 194:825–839. 10.1083/jcb.201101148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M., and Benitah S.A.. 2012. Chromatin regulators in mammalian epidermis. Semin. Cell Dev. Biol. 23:897–905. 10.1016/j.semcdb.2012.08.009 [DOI] [PubMed] [Google Scholar]
- Fuchs E. 2007. Scratching the surface of skin development. Nature. 445:834–842. 10.1038/nature05659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo G., Licciulli F., Liuni S., Sbisà E., and Pesole G.. 2003. PatSearch: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 31:3608–3612. 10.1093/nar/gkg548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., and Westphal H.. 2000. Functions of LIM-homeobox genes. Trends Genet. 16:75–83. 10.1016/S0168-9525(99)01883-1 [DOI] [PubMed] [Google Scholar]
- Hunt D.P., Jahoda C., and Chandran S.. 2009. Multipotent skin-derived precursors: From biology to clinical translation. Curr. Opin. Biotechnol. 20:522–530. 10.1016/j.copbio.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Jensen K.B., Driskell R.R., and Watt F.M.. 2010. Assaying proliferation and differentiation capacity of stem cells using disaggregated adult mouse epidermis. Nat. Protoc. 5:898–911. [DOI] [PubMed] [Google Scholar]
- Jordanova A., De Jonghe P., Boerkoel C.F., Takashima H., De Vriendt E., Ceuterick C., Martin J.J., Butler I.J., Mancias P., Papasozomenos S.Ch., et al. 2003. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain. 126:590–597. 10.1093/brain/awg059 [DOI] [PubMed] [Google Scholar]
- Kalb R., Latwiel S., Baymaz H.I., Jansen P.W., Müller C.W., Vermeulen M., and Müller J.. 2014. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 21:569–571. 10.1038/nsmb.2833 [DOI] [PubMed] [Google Scholar]
- Keyes W.M., Pecoraro M., Aranda V., Vernersson-Lindahl E., Li W., Vogel H., Guo X., Garcia E.L., Michurina T.V., Enikolopov G., et al. 2011. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 8:164–176. 10.1016/j.stem.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari D.A., Sen G.L., and Rinn J.L.. 2010. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle. 9:3880–3883. 10.4161/cc.9.19.13385 [DOI] [PubMed] [Google Scholar]
- Koster M.I., and Roop D.R.. 2007. Mechanisms regulating epithelial stratification. Annu. Rev. Cell Dev. Biol. 23:93–113. 10.1146/annurev.cellbio.23.090506.123357 [DOI] [PubMed] [Google Scholar]
- Kouwenhoven E.N., van Bokhoven H., and Zhou H.. 2015. Gene regulatory mechanisms orchestrated by p63 in epithelial development and related disorders. Biochim. Biophys. Acta. 1849:590–600. [DOI] [PubMed] [Google Scholar]
- LeBoeuf M., Terrell A., Trivedi S., Sinha S., Epstein J.A., Olson E.N., Morrisey E.E., and Millar S.E.. 2010. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev. Cell. 19:807–818. 10.1016/j.devcel.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Zhou J., Liu P., Hu J., Jin H., Shimono Y., Takahashi M., and Xu G.. 2007. Polycomb protein Cbx4 promotes SUMO modification of de novo DNA methyltransferase Dnmt3a. Biochem. J. 405:369–378. 10.1042/BJ20061873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Liu Y.F., Du Y.R., Mardaryev A.N., Yang W., Chen H., Xu Z.M., Xu C.Q., Zhang X.R., Botchkarev V.A., et al. 2013. Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development. 140:780–788. 10.1242/dev.085035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis N.M., Morey L., Mejetta S., Pascual G., Janich P., Kuebler B., Cozutto L., Roma G., Nascimento E., Frye M., et al. 2011. Regulation of human epidermal stem cell proliferation and senescence requires polycomb-dependent and -independent functions of Cbx4. Cell Stem Cell. 9:233–246. 10.1016/j.stem.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Mardaryev A.N., Meier N., Poterlowicz K., Sharov A.A., Sharova T.Y., Ahmed M.I., Rapisarda V., Lewis C., Fessing M.Y., Ruenger T.M., et al. 2011. Lhx2 differentially regulates Sox9, Tcf4 and Lgr5 in hair follicle stem cells to promote epidermal regeneration after injury. Development. 138:4843–4852. 10.1242/dev.070284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev A.N., Gdula M.R., Yarker J.L., Emelianov V.U., Poterlowicz K., Sharov A.A., Sharova T.Y., Scarpa J.A., Joffe B., Solovei I., et al. 2014. p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development. 141:101–111. 10.1242/dev.103200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy R.A., Rogers S., Caldon C.E., Lorca T., Castro A., and Burgess A.. 2014. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 13:1400–1412. 10.4161/cc.28401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejetta S., Morey L., Pascual G., Kuebler B., Mysliwiec M.R., Lee Y., Shiekhattar R., Di Croce L., and Benitah S.A.. 2011. Jarid2 regulates mouse epidermal stem cell activation and differentiation. EMBO J. 30:3635–3646. 10.1038/emboj.2011.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill J.C., Melhuish T.A., Kagey M.H., Yang S.H., Sharrocks A.D., and Wotton D.. 2010. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS One. 5:e8794 10.1371/journal.pone.0008794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., and Bradley A.. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 398:708–713. 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- Mitew S., Hay C.M., Peckham H., Xiao J., Koenning M., and Emery B.. 2014. Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience. 276:29–47. 10.1016/j.neuroscience.2013.11.029 [DOI] [PubMed] [Google Scholar]
- Montague P., Dickinson P.J., McCallion A.S., Stewart G.J., Savioz A., Davies R.W., Kennedy P.G., and Griffiths I.R.. 1997. Developmental expression of the murine Mobp gene. J. Neurosci. Res. 49:133–143. [DOI] [PubMed] [Google Scholar]
- Morey L., Pascual G., Cozzuto L., Roma G., Wutz A., Benitah S.A., and Di Croce L.. 2012. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 10:47–62. 10.1016/j.stem.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Mostoslavsky G., Kotton D.N., Fabian A.J., Gray J.T., Lee J.S., and Mulligan R.C.. 2005. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol. Ther. 11:932–940. 10.1016/j.ymthe.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Patthey C., and Gunhaga L.. 2011. Specification and regionalisation of the neural plate border. Eur. J. Neurosci. 34:1516–1528. 10.1111/j.1460-9568.2011.07871.x [DOI] [PubMed] [Google Scholar]
- Pelling M., Anthwal N., McNay D., Gradwohl G., Leiter A.B., Guillemot F., and Ang S.L.. 2011. Differential requirements for neurogenin 3 in the development of POMC and NPY neurons in the hypothalamus. Dev. Biol. 349:406–416. 10.1016/j.ydbio.2010.11.007 [DOI] [PubMed] [Google Scholar]
- Perdigoto C.N., Valdes V.J., Bardot E.S., and Ezhkova E.. 2014. Epigenetic regulation of epidermal differentiation. Cold Spring Harb. Perspect. Med. 4:281–300. 10.1101/cshperspect.a015263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y.B., and Pirrotta V.. 2014. Ruled by ubiquitylation: A new order for polycomb recruitment. Cell Reports. 8:321–325. 10.1016/j.celrep.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Sen G.L., Webster D.E., Barragan D.I., Chang H.Y., and Khavari P.A.. 2008. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 22:1865–1870. 10.1101/gad.1673508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.L., Reuter J.A., Webster D.E., Zhu L., and Khavari P.A.. 2010. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 463:563–567. 10.1038/nature08683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A.A., Li G.Z., Palkina T.N., Sharova T.Y., Gilchrest B.A., and Botchkarev V.A.. 2003. Fas and c-kit are involved in the control of hair follicle melanocyte apoptosis and migration in chemotherapy-induced hair loss. J. Invest. Dermatol. 120:27–35. 10.1046/j.1523-1747.2003.12022.x [DOI] [PubMed] [Google Scholar]
- Sharov A.A., Fessing M., Atoyan R., Sharova T.Y., Haskell-Luevano C., Weiner L., Funa K., Brissette J.L., Gilchrest B.A., and Botchkarev V.A.. 2005. Bone morphogenetic protein (BMP) signaling controls hair pigmentation by means of cross-talk with the melanocortin receptor-1 pathway. Proc. Natl. Acad. Sci. USA. 102:93–98. 10.1073/pnas.0408455102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A.A., Sharova T.Y., Mardaryev A.N., Tommasi di Vignano A., Atoyan R., Weiner L., Yang S., Brissette J.L., Dotto G.P., and Botchkarev V.A.. 2006. Bone morphogenetic protein signaling regulates the size of hair follicles and modulates the expression of cell cycle-associated genes. Proc. Natl. Acad. Sci. USA. 103:18166–18171. 10.1073/pnas.0608899103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A.A., Mardaryev A.N., Sharova T.Y., Grachtchouk M., Atoyan R., Byers H.R., Seykora J.T., Overbeek P., Dlugosz A., and Botchkarev V.A.. 2009. Bone morphogenetic protein antagonist noggin promotes skin tumorigenesis via stimulation of the Wnt and Shh signaling pathways. Am. J. Pathol. 175:1303–1314. 10.2353/ajpath.2009.090163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F., Rössler R., Brouwer N., Balasubramaniyan V., Boddeke E., and Copray S.. 2008. Differentiation of neural stem cells into oligodendrocytes: Involvement of the polycomb group protein Ezh2. Stem Cells. 26:2875–2883. 10.1634/stemcells.2008-0121 [DOI] [PubMed] [Google Scholar]
- Sher F., Boddeke E., Olah M., and Copray S.. 2012. Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PLoS One. 7:e40399 10.1371/journal.pone.0040399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., and Kingston R.E.. 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 49:808–824. 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack J.M. 2008. Origin of stem cells in organogenesis. Science. 322:1498–1501. 10.1126/science.1162782 [DOI] [PubMed] [Google Scholar]
- Turinetto V., and Giachino C.. 2015. Multiple facets of histone variant H2AX: A DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 43:2489–2498. 10.1093/nar/gkv061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbokhoven H., Melino G., Candi E., and Declercq W.. 2011. p63, a story of mice and men. J. Invest. Dermatol. 131:1196–1207. 10.1038/jid.2011.84 [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Mascre G., Youseff K.K., Harel I., Michaux C., De Geest N., Szpalski C., Achouri Y., Bloch W., Hassan B.A., and Blanpain C.. 2009. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J. Cell Biol. 187:91–100. 10.1083/jcb.200907080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viganò M.A., and Mantovani R.. 2007. Hitting the numbers: The emerging network of p63 targets. Cell Cycle. 6:233–239. 10.4161/cc.6.3.3802 [DOI] [PubMed] [Google Scholar]
- Wallén A., and Perlmann T.. 2003. Transcriptional control of dopamine neuron development. Ann. N. Y. Acad. Sci. 991:48–60. 10.1111/j.1749-6632.2003.tb07462.x [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., and McKeon F.. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 398:714–718. 10.1038/19539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.