Abstract
Epigenetic regulators are essential for cell lineage choices during development. In this issue, Mardaryev et al. (2016. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201506065) show that Polycomb subunit Cbx4 acts downstream of transcriptional regulator p63 to maintain epidermal progenitor identity and proliferation in the developing epidermis via Polycomb-dependent and -independent SUMO E3 ligase activities.
The epidermis provides an essential barrier between the body and the environment. Epidermal progenitors, which are located in the basal layer in direct contact with the basement membrane, proliferate and move upward to give rise to the nonproliferative, differentiated cells of the suprabasal layers (Blanpain and Fuchs, 2009). The process of epidermal differentiation occurs in a stepwise manner, characterized by the activation or repression of unique sets of genes as basal cells differentiate through the epidermal layers.
Polycomb group proteins are central epigenetic transcriptional repressors that have been shown to control stem cell (SC) identity and differentiation in many developmental systems (Surface et al., 2010; Aloia et al., 2013). Polycomb activity is generally divided between two multisubunit complexes known as the Polycomb repressive complex 1 (PRC1) and 2 (PRC2), which facilitate transcriptional repression via histone modifications and chromatin compaction. PRC1 catalyzes histone H2A monoubiquitylation (H2AK119ub1), whereas PRC2 catalyzes histone H3 lysine 27 trimethylation (H3K27me3). A canonical PRC1 complex contains one of five different Cbx protein subunits (Cbx2/4/6/7/8), which recognizes H3K27me3 via its chromodomain and enables PRC1 recruitment to its target genes (Simon and Kingston, 2013).
Loss-of-function studies have highlighted the pivotal role of the PRC2 complex in skin development. Loss of Polycomb component Ezh2 in the epidermis results in the derepression of epidermal differentiation genes, leading to accelerated barrier formation (Ezhkova et al., 2009). Knocking out both essential Ezh1/2 subunits in embryonic epidermal progenitors results in the complete ablation of PRC2 activity and subsequent loss of the H3K27me3 histone mark, leading to impaired hair follicle development and a dramatic expansion of the Merkel cell lineage (Ezhkova et al., 2011; Bardot et al., 2013). Loss of expression of Jarid2, a PRC2 accessory protein, was shown to lead to thickening of the epidermis, a mild reduction in cell proliferation, and a delay in activation of the hair growth (Mejetta et al., 2011). Although the PRC1 complex has not been studied in the skin in vivo, PRC1 subunit Cbx4 has been shown to maintain quiescence and inhibit differentiation of human epidermal keratinocytes in vitro through Polycomb-dependent and -independent functions (Luis et al., 2011).
To uncover regulators of epidermal development and homeostasis, Mardaryev et al. performed transcriptional profiling of epidermal basal cells obtained via laser capture microdissection at different time points from embryogenesis through adulthood in mice. Interestingly, the analysis revealed dynamic expression of Polycomb group genes. Indeed, Mardaryev et al. (2016) noticed that Cbx4 transcript and protein levels were strongly increased both in basal keratinocytes during the onset of epidermal stratification and in differentiating keratinocytes. This pattern of Cbx4 expression prompted them to investigate the role of Cbx4 in the control of epidermal development by analyzing Cbx4-null mice.
During the early stages of epidermal development, Cbx4-null epidermis showed significantly reduced thickness, which was attributed to reduced proliferation of the basal cells, as assessed by Ki67 staining in wild-type and knockout animals. These changes in Cbx4-deficient cells were associated with the increased expression of the cell cycle inhibitors Cdkn2a/Arf and Cdkn1c, which are known to be direct targets of Polycomb-mediated repression (Agherbi et al., 2009; Yang et al., 2009; Ezhkova et al., 2011). These marked changes in basal cell proliferation and epidermal thickness diminished as epidermal development progressed and were not observed in newborn Cbx4-knockout skins. Interestingly, these transient effects on cell proliferation and epidermal thickness correlated with a temporal reduction in the levels of the PRC1-dependent H2AK119ub1 histone mark in Cbx4-null epidermis, but not in dermal cells. H2AK119ub1 levels were restored by embryonic day 18.5, concomitant with a significant up-regulation of Cbx6 in Cbx4-null epidermis. Thus, it seems that other Cbx proteins might be able to compensate for the absence of Cbx4, and careful characterization of the other Cbx proteins in the skin epidermis will be necessary to determine the mechanisms behind this compensatory effect.
Apart from defects in cell proliferation, loss of Cbx4 in the epidermis also caused a derepression of both epidermal differentiation genes and non-epidermal neuronal lineage genes. Chromatin immunoprecipitation (ChIP)–sequencing analysis of Cbx4 gene targets as well as ChIP-sequencing data for H3K27me3 and microarray transcript profiling from Cbx4-deficient basal cells revealed that whereas neuronal development genes, such as Nefl and Mobp, were direct targets of Cbx4, epidermal differentiation genes were not. Mardaryev et al. (2016) focused on Nefl to continue their characterization. They showed via shRNA-mediated Cbx4 silencing in primary epidermal keratinocytes and via ChIP–quantitative PCR analysis that Cbx4 directly represses the expression of this neuronal gene target. Altogether, the authors conclude that Cbx4 represses nonepidermal (neuronal) lineage genes in keratinocytes during epidermal development.
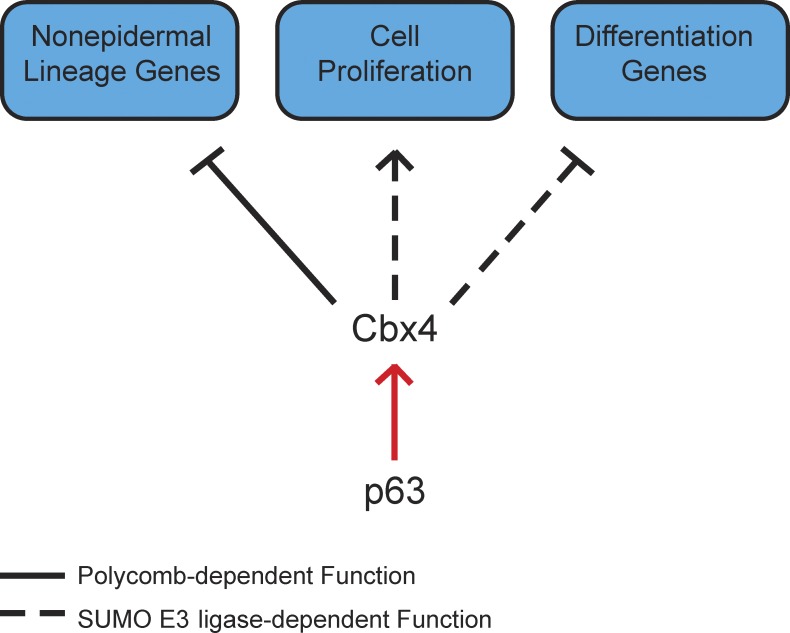
Cbx proteins differ greatly in their structures and sequences, and Cbx4 uniquely possesses both Polycomb and non-Polycomb SUMO E3 ligase activities (Roscic et al., 2006). To elucidate the molecular mechanisms by which Cbx4 promotes proliferation and repression of nonepidermal genes, Mardaryev et al. (2016) used retroviral constructs to overexpress mutated forms of Cbx4 lacking either the chromodomain (Polycomb-dependent function) or the SUMO E3 ligase domain (Polycomb-independent functions) in primary epidermal keratinocytes. These experiments demonstrated that Cbx4’s repression of nonepidermal genes relies on its chromodomain and can be attributed to its function within the canonical PRC1 complex. In contrast, Cbx4’s control of keratinocyte proliferation is Polycomb independent and requires its SUMO E3 ligase activity (Fig. 1). These results are consistent with previous findings indicating that the SUMO E3 ligase activity of Cbx4 controls the proliferation of human primary keratinocytes (Luis et al., 2011). It will be of great interest to identify the pro-proliferative proteins that are SUMOylated by Cbx4 in basal cells.
Figure 1.
Cbx4-mediated control in the skin epidermis. Mardaryev et al. (2016) show that PRC1 member Cbx4 acts as a direct downstream target of p63 to maintain cell identity and proliferation in the epidermis by Polycomb-dependent and SUMO E3 ligase–dependent (Polycomb-independent) functions.
The ability to execute precise cell specification and developmental programs largely relies on tissue-specific transcription factors. The transcription factor p63 is a master regulator of the epidermal lineage that controls multiple processes, including ectoderm specification, basal cell proliferation, and epidermal differentiation (Botchkarev and Flores, 2014). In addition, p63 can function as a transcriptional repressor of nonepidermal lineage genes (De Rosa et al., 2009). Strikingly, Mardaryev et al. (2016) found that many genes up-regulated in p63-null epidermis were also up-regulated in Cbx4-null epidermis, suggesting a functional link between p63 and Cbx4. Further molecular studies identified Cbx4 as a direct downstream target of p63 that mediates its effects on epidermal cell proliferation and the repression of nonepidermal lineage genes during epidermal differentiation. Indeed, the researchers observed that Cbx4 levels were markedly reduced in p63-null embryonic skin, whereas they were not affected in the dermis. The in silico prediction of p63 binding sites upstream of the Cbx4 transcription start site were confirmed by ChIP analysis, which revealed that p63 binds to this genomic region in primary mouse keratinocytes. In vitro reporter assays additionally validated the direct transcriptional regulation of Cbx4 by p63, and the researchers lastly showed that ectopic Cbx4 expression partially rescued the epidermal phenotype in embryonic skin explants from p63+/− mice that were treated with p63 shRNA-expressing lentiviruses.
Collectively, the data show that Cbx4 plays a unique role in the epidermal lineage by repressing nonepidermal lineage (neuronal) genes in epidermal progenitor cells via its canonical PRC1 function, as well as by controlling proliferation of basal epidermal keratinocytes and inhibiting their premature differentiation via its Polycomb-independent SUMO E3 ligase activity (Fig. 1). The demonstration that Cbx4 represses neuronal lineage genes but does not directly target epidermal differentiation genes is intriguing, as genes from both neuronal and epidermal lineages (which share an ectodermal origin) were shown to be direct targets of the PRC2 complex and are up-regulated upon loss of Ezh1/2 (Ezhkova et al., 2009, 2011). Whereas PRC2-mediated repression is direct, as epidermal differentiation genes carry the PRC2-dependent H3K27me3 mark, Cbx4 repression is indirect and requires Cbx4’s SUMO E3 ligase activity. Moreover, ablation of the PRC2 subunit Ezh2 in epidermal progenitors affects the timing of differentiation and accelerates skin barrier formation (Ezhkova et al., 2009), whereas Mardaryev et al. (2016) show that the loss of Cbx4 leads to the aberrant expression of differentiation genes in the basal and spinous layers. Additionally, the loss of Ezh1/2 in epidermal progenitors also affected the Merkel and hair follicle lineages, whereas Cbx4-null effect was restricted to the epidermis. Overall, these results suggest that Cbx4 plays unique and only partially overlapping roles with Ezh1/2 in the control of epidermal development.
Another intriguing observation in this work is that the reduction in H2AK119ub1 levels was restored to normal levels in Cbx4-null epidermis at late developmental time points. The authors concomitantly document the varying levels of several Cbx subunits, suggesting that other subunits might compensate for the lack of Cbx4 at specific times. Careful and detailed molecular characterization of the other Cbx proteins in the skin epidermis will be necessary to determine the mechanisms behind this compensatory effect. Recent studies by Morey et al. (2012, 2015) have shown that the subunit composition and the effects of canonical PRC1 complexes are cell type specific. For example, during the differentiation of embryonic stem cells into early cardiac mesoderm precursors, the canonical PRC1 complex exchanges Cbx7 and Phc1 for Cbx2 and Phc2, respectively (Morey et al., 2015). Similarly, Cbx7 plays a role in the maintenance of ES cell pluripotency, whereas Cbx2 and Cbx4 play specific roles in embryonic stem cell differentiation (Morey et al., 2012). Based on these examples, the distinct roles of each PRC1 subunit in specific cellular contexts, the extent to which Polycomb subunits can compensate for one another, and the determination of whether the observed phenotypes are PRC1 dependent or independent will require future investigation.
Lastly, Mardaryev et al. (2016) convincingly demonstrate that Cbx4 is a novel critical mediator of the p63 regulatory network. Cbx4 was previously found to directly interact with p63 in thymocytes (Liu et al., 2013), suggesting that these factors may act together to control tissue development. Additional analyses will be required to investigate the involvement of the p63/Cbx4 network in the repression of lineage-specific genes other than those of the neuronal lineage. Moreover, as epidermal proliferation was restored to normal during later stages of development in the Cbx4 knockout, this work opens the way for future studies to determine whether other p63 targets or members of the Cbx family partially compensate for the loss of Cbx4 function.
Acknowledgments
The work in E. Ezhkova’s laboratory is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases/National Institutes of Health (R01 AR063724 and R00 AR057817) and NYSTEM (N11G-152) grants.
The authors declare no competing financial interests.
References
- Agherbi H., Gaussmann-Wenger A., Verthuy C., Chasson L., Serrano M., and Djabali M.. 2009. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS One. 4:e5622 10.1371/journal.pone.0005622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L., Di Stefano B., and Di Croce L.. 2013. Polycomb complexes in stem cells and embryonic development. Development. 140:2525–2534. 10.1242/dev.091553 [DOI] [PubMed] [Google Scholar]
- Bardot E.S., Valdes V.J., Zhang J., Perdigoto C.N., Nicolis S., Hearn S.A., Silva J.M., and Ezhkova E.. 2013. Polycomb subunits Ezh1 and Ezh2 regulate the Merkel cell differentiation program in skin stem cells. EMBO J. 32:1990–2000. 10.1038/emboj.2013.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., and Fuchs E.. 2009. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10:207–217. 10.1038/nrm2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev V.A., and Flores E.R.. 2014. p53/p63/p73 in the epidermis in health and disease. Cold Spring Harb. Perspect. Med. 4:4 10.1101/cshperspect.a015248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa L., Antonini D., Ferone G., Russo M.T., Yu P.B., Han R., and Missero C.. 2009. p63 Suppresses non-epidermal lineage markers in a bone morphogenetic protein-dependent manner via repression of Smad7. J. Biol. Chem. 284:30574–30582. 10.1074/jbc.M109.049619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Pasolli H.A., Parker J.S., Stokes N., Su I.H., Hannon G., Tarakhovsky A., and Fuchs E.. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 136:1122–1135. 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Lien W.H., Stokes N., Pasolli H.A., Silva J.M., and Fuchs E.. 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 25:485–498. 10.1101/gad.2019811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Liu Y.F., Du Y.R., Mardaryev A.N., Yang W., Chen H., Xu Z.M., Xu C.Q., Zhang X.R., Botchkarev V.A., et al. 2013. Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development. 140:780–788. 10.1242/dev.085035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis N.M., Morey L., Mejetta S., Pascual G., Janich P., Kuebler B., Cozutto L., Roma G., Nascimento E., Frye M., et al. 2011. Regulation of human epidermal stem cell proliferation and senescence requires polycomb-dependent and -independent functions of Cbx4. Cell Stem Cell. 9:233–246. 10.1016/j.stem.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Mardaryev A.N., Liu B., Rapisarda V., Poterlowicz K., Malashchuk I., Rudolf J., Sharov A.A., Jahoda C.A., Fessing M.Y., Aznar-Benitah S., et al. 2016. Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J. Cell Biol. 10.1083/jcb.201506065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejetta S., Morey L., Pascual G., Kuebler B., Mysliwiec M.R., Lee Y., Shiekhattar R., Di Croce L., and Benitah S.A.. 2011. Jarid2 regulates mouse epidermal stem cell activation and differentiation. EMBO J. 30:3635–3646. 10.1038/emboj.2011.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L., Pascual G., Cozzuto L., Roma G., Wutz A., Benitah S.A., and Di Croce L.. 2012. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 10:47–62. 10.1016/j.stem.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Morey L., Santanach A., Blanco E., Aloia L., Nora E.P., Bruneau B.G., and Di Croce L.. 2015. Polycomb regulates mesoderm cell fate-specification in embryonic stem cells through activation and repression mechanisms. Cell Stem Cell. 17:300–315. 10.1016/j.stem.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Roscic A., Möller A., Calzado M.A., Renner F., Wimmer V.C., Gresko E., Lüdi K.S., and Schmitz M.L.. 2006. Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol. Cell. 24:77–89. 10.1016/j.molcel.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Simon J.A., and Kingston R.E.. 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 49:808–824. 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface L.E., Thornton S.R., and Boyer L.A.. 2010. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 7:288–298. 10.1016/j.stem.2010.08.004 [DOI] [PubMed] [Google Scholar]
- Yang X., Karuturi R.K., Sun F., Aau M., Yu K., Shao R., Miller L.D., Tan P.B., and Yu Q.. 2009. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS One. 4:e5011 10.1371/journal.pone.0005011 [DOI] [PMC free article] [PubMed] [Google Scholar]