Abstract
Lipid droplets (LDs) are sometimes found in the nucleus of some cells. In this issue, Ohsaki et al. (2016. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201507122) show that the nuclear membrane, promyelocytic leukemia bodies, and the protein PML-II play a role in nuclear LD formation, suggesting functional relationships between these structures.
Lipid droplets (LDs) are organelles that store lipids as reservoirs of metabolic energy and membrane lipid precursors. The cell biology of LDs as cellular organelles is only beginning to be unraveled (Thiam et al., 2013; Pol et al., 2014; Gao and Goodman, 2015). LDs can be found in most eukaryotic cells. Most prominent are LDs in adipocytes, which make up most of the cellular volume, but other metabolically active cell types, such as liver or muscle cells, also have abundant LDs. LDs are unusual organelles in that they are bound by a monolayer of surface phospholipids, into which specific proteins are embedded, such as perilipins and metabolic enzymes (Thiam et al., 2013).
LDs are generally considered to be cytoplasmic organelles. They are formed from the ER (Pol et al., 2014), where the synthesis of neutral lipids occurs, such as triacylglycerols (TGs) by DGAT1 or DGAT2 enzymes or sterol esters by ACAT1 or ACAT2 enzymes (Buhman et al., 2001; Wilfling et al., 2014a). After initial LDs (iLDs) are formed, a subset of them recruit enzymes via ER–LD membrane bridges and acquire the capacity to locally synthesize TGs, converting them to expanding LDs (eLDs; Wilfling et al., 2013). eLD formation requires the Arf1/COP-I proteins to recruit TG synthesis enzymes (Wilfling et al., 2014b), but other aspects of this process remain unclear.
Many LD researchers have also observed that LDs appear to localize to the cell nucleus (Hillman and Hillman, 1975; Layerenza et al., 2013; Uzbekov and Roingeard, 2013). However, the presence of nuclear LDs has seemed somewhat random among cell types, and it has not been clear whether such LDs are located within the nucleoplasm or on the cytoplasmic side of invaginations into the nuclear envelope.
In this issue, Ohsaki et al. elegantly use confocal and electron microscopy to investigate nuclear LDs and make a series of surprising discoveries. Using serial section electron microscopy, they convincingly show that nuclear LDs are indeed localized within the nucleoplasm of a variety of human and mammalian hepatocyte cell lines. Consistent with previous observations, nuclear LDs were not found in all cell types and were scarcely found in HeLa cells, fibroblasts, and differentiated adipocytes. In hepatocytes, nuclear LDs appeared to have a distinct but overlapping protein composition compared with cytoplasmic LDs, differing for instance in the types of perilipin proteins bound to LD surfaces.
Light and electron microscopy analyses showed that the nuclear LDs were closely associated with protrusions of the inner nuclear envelope membrane and with nuclear structures known as promyelocytic leukemia (PML) bodies. PML bodies (also known as nuclear dots or nuclear bodies) are one of several nuclear domains that are marked by specific proteins, including nucleoli, Cajal bodies, nuclear speckles, and nuclear paraspeckles. The function of PML bodies is somewhat of an enigma, but they may be involved in modulating specific stress responses in the nucleus (Lallemand-Breitenbach and de Thé, 2010). The expression of PML-II, one isoform of the prominent PML protein in PML bodies, correlated with the presence of nuclear LDs in knockdown/overexpression experiments. Interestingly, overexpression of a mutant PML-II protein that does not target the nuclear envelope fails to induce the increase in nuclear LDs seen after overexpression of the WT protein, suggesting that the ability of PML-II to induce nuclear LD formation is associated with its binding to the nuclear envelope. Furthermore, overexpression of PML-II in cell types in which the protein does not distribute along the nuclear envelope also failed to increase the amounts of nuclear LDs. The discovery that PML-II is intimately linked to the formation of nuclear LDs now provides a molecular handle to study nuclear LD biology and its cell type specificity.
Lastly, the researchers explored the contribution of other nuclear proteins to nuclear LD formation. Although knockdown of lamins or of inner nuclear membrane proteins did not impact nuclear LDs, knockdown of SUN proteins increased the proportion of nuclear LDs and intranuclear membranes. PML-II depletion prevented the increase of nuclear LDs after SUN protein knockdown, suggesting that SUN proteins act upstream of PML-II. The mechanistic basis for this is still unclear but might involve SUN protein–mediated control of membrane interactions with chromatin (Turgay et al., 2014).
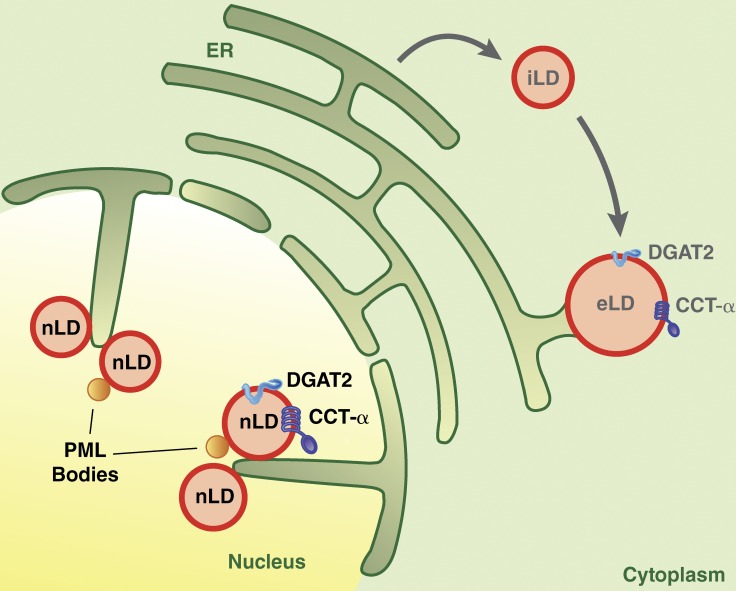
Results from Ohsaki et al. (2016) suggest that nuclear LDs appear to be most closely related to eLDs found in the cytoplasm (Fig. 1). Similar to cytoplasmic eLDs, nuclear LDs were found to colocalize with the TG synthesis enzyme DGAT2 and its substrates, which should enable them to expand by locally synthesizing TG. They also colocalized with CCT-α, the rate-limiting enzyme of phosphatidylcholine (PC) synthesis, which shuttles between the nucleus and the cytoplasm and can bind to and become activated at eLDs when there is insufficient PC to cover their surfaces (Krahmer et al., 2011). The presence of these specific proteins and the possible connection of nuclear LDs to the inner nuclear membrane suggest nuclear LDs are analogous to eLDs in the cytoplasm.
Figure 1.
Model of cytoplasmic and nuclear LDs. iLDs are formed from the ER. A subset of iLDs can be converted to eLDs via establishment of ER–LD membrane bridges and relocalization of TG synthesis enzymes, such as DGAT2, to their surfaces. CCT-α binds to eLDs with a relative deficiency of PC on their surfaces, where it is activated and catalyzes PC synthesis. Ohsaki et al. (2016) show that nuclear LDs (nLD) form in association with invaginations of the inner nuclear membrane and also are marked by DGAT2 and CCT-α. Nuclear LDs are found in close proximity to PML bodies and may depend on PML proteins for formation.
The important discoveries by Ohsaki et al. (2016) elicit many new questions concerning both LD and PML biology. Primarily, it will be important to decipher the cellular function of nuclear LDs. Do nuclear LDs provide lipid stores for nuclear membrane expansion or lipid signaling? Are they storage sites for histones, as they are in the fly embryo (Li et al., 2012)? Do they provide a nuclear platform for misfolded or unfolded hydrophobic proteins? Are they detoxification sites for hydrophobic substances in the liver? And what is the functional meaning of the association between nuclear LDs and PMLs? Is PML-II involved in their biogenesis? Answers to these questions will provide exciting insights into the pathways of cellular lipid storage.
Acknowledgments
We thank Gary Howard for editorial assistance and Kim Cordes for graphics assistance.
The authors declare no competing financial interests.
References
- Buhman K.K., Chen H.C., and Farese R.V. Jr. 2001. The enzymes of neutral lipid synthesis. J. Biol. Chem. 276:40369–40372. 10.1074/jbc.R100050200 [DOI] [PubMed] [Google Scholar]
- Gao Q., and Goodman J.M.. 2015. The lipid droplet—a well-connected organelle. Front. Cell Dev. Biol. 3:49 10.3389/fcell.2015.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman N., and Hillman R.. 1975. Ultrastructural studies of tw32/tw32 mouse embryos. J. Embryol. Exp. Morphol. 33:685–695. [PubMed] [Google Scholar]
- Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H.W., Schmidt-Supprian M., Vance D.E., Mann M., et al. 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14:504–515. 10.1016/j.cmet.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., and de Thé H.. 2010. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2:a000661 10.1101/cshperspect.a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layerenza J.P., González P., García de Bravo M.M., Polo M.P., Sisti M.S., and Ves-Losada A.. 2013. Nuclear lipid droplets: A novel nuclear domain. Biochim. Biophys. Acta. 1831:327–340. 10.1016/j.bbalip.2012.10.005 [DOI] [PubMed] [Google Scholar]
- Li Z., Thiel K., Thul P.J., Beller M., Kühnlein R.P., and Welte M.A.. 2012. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr. Biol. 22:2104–2113. 10.1016/j.cub.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y., Kawai T., Yoshikawa Y., Cheng J., Jokitalo E., and Fujimoto T.. 2016. PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 10.1083/jcb.201507122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A., Gross S.P., and Parton R.G.. 2014. Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 204:635–646. 10.1083/jcb.201311051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam A.R., Farese R.V. Jr., and Walther T.C.. 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14:775–786. 10.1038/nrm3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y., Champion L., Balazs C., Held M., Toso A., Gerlich D.W., Meraldi P., and Kutay U.. 2014. SUN proteins facilitate the removal of membranes from chromatin during nuclear envelope breakdown. J. Cell Biol. 204:1099–1109. 10.1083/jcb.201310116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov R., and Roingeard P.. 2013. Nuclear lipid droplets identified by electron microscopy of serial sections. BMC Res. Notes. 6:386 10.1186/1756-0500-6-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A., Cheng J.X., Graham M., Christiano R., Fröhlich F., et al. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24:384–399. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Haas J.T., Walther T.C., and Farese R.V. Jr. 2014a Lipid droplet biogenesis. Curr. Opin. Cell Biol. 29:39–45. 10.1016/j.ceb.2014.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Thiam A.R., Olarte M.J., Wang J., Beck R., Gould T.J., Allgeyer E.S., Pincet F., Bewersdorf J., Farese R.V. Jr., and Walther T.C.. 2014b Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife. 3:e01607 10.7554/eLife.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]