Abstract
Background
Animal and human studies indicate that GABBR1, encoding the GABAB1 receptor subunit, and SLC6A1, encoding the neuronal GABA transporter GAT1, play a role in addiction by modulating synaptic GABA. Therefore variants in these genes might predict risk/resilience for alcoholism.
Methods
This study included three populations that differed by ethnicity and alcoholism phenotype: African American (AA) men: 401 treatment-seeking inpatients with single/comorbid diagnoses of alcohol and drug dependence, 193 controls; Finnish Caucasian men: 159 incarcerated alcoholics, half with comorbid ASPD, 181 controls; a community sample of Plains Indian (PI) men and women: 239 alcoholics, 178 controls. Seven GABBR1 tag SNPs were genotyped in the AA and Finnish samples; rs29220 was genotyped in the PI for replication. Also, a uniquely African, functional SLC6A1 insertion promoter polymorphism (IND) was genotyped in the AAs.
Results
We found a significant and congruent association between GABBR1 rs29220 and alcoholism in all three populations. The major genotype (heterozygotes in AAs, Finns) and the major allele in PIs were significantly more common in alcoholics. Moreover, SLC6A1 IND was more abundant in controls, i.e. the major genotype predicted alcoholism. An analysis of combined GABBR1 rs29220 and SLC6A1 IND genotypes showed that rs29220 heterozygotes, irrespective of their IND status, had an increased risk for alcoholism whereas carriers of the IND allele and either rs29220 homozygote were more resilient.
Conclusions
Our results show that with both GABBR1 and SLC6A1, the minor genotypes/alleles were protective against risk for alcoholism. Finally, GABBR1 rs29220 might predict treatment response/adverse effects for baclofen, a GABAB receptor agonist.
Keywords: GABBR1, GABAB receptor subunit, SLC6A1, GAT1, alcohol use disorders
INTRODUCTION
Addiction is caused in part by an imbalance in neuronal excitation and inhibition, which is largely due to alterations in transmission of excitatory glutamate and inhibitory gamma-aminobutyric acid (GABA). Metabotropic GABAB receptors (GABABRs) are abundantly expressed at inhibitory and excitatory synapses throughout the brain and play an important role in modulating synaptic transmission by their presynaptic inhibitory effects on calcium channels and postsynaptic activating effects on potassium channels (Bettler et al., 2004; Chalifoux and Carter 2010, 2011). Presynaptic GABABRs repress Ca2+ influx and thereby GABA release. GABABRs are G protein-coupled receptors that are heterodimers of GABAB1 and GABAB2 subunits. The GABAB1 subunit, encoded by the GABBR1 gene, is necessary for activation by agonists including GABA (Chalifoux and Carter 2011). The principal neuronal GABA transporter 1 (GAT1), encoded by the SLC6A1 gene, mediates uptake of synaptic GABA and in a complex fashion regulates GABABR mediated signaling (Gonzalez-Burgos, 2010).
In an earlier RNA-Seq study of GABAergic gene expression in human postmortem hippocampus we found that GABBR1 was down-regulated in alcoholics and cocaine addicts relative to controls, potentially resulting in increased synaptic GABA (Enoch et al., 2012). The involvement of GABBR1 in alcohol-related traits is also supported by other genome- and transcriptome studies (Farris et al., 2014; Flatscher-Bader et al., 2005; Zhao et al., 2012). Evidence from studies in rodents shows that GABABR up-regulation by agonists such as baclofen results in a reduction of drug-related behaviors including alcohol consumption, relapse-like drinking and re-instatement of cocaine seeking behavior (Liang et al., 2006; Maccioni and Colombo, 2009; Vlachou et al., 2010). Moreover, baclofen has been shown to decrease alcohol consumption, craving and severity of alcohol withdrawal symptoms in humans (Addolorato et al., 2009; Colombo et al., 2004; Morley et al., 2014; Tyacke et al., 2010). By inference, it could be speculated that down-regulation of GABABRs resulting in increased synaptic GABA may increase the rewarding effects of drugs of abuse and thereby increase vulnerability for addiction. Our RNA-Seq study also showed that in both alcoholics and cocaine addicts compared with controls there was down-regulation of SLC6A1. Decreased levels of GAT1 could result in increased synaptic GABA levels, as has been shown to occur in GAT1 deficient mice (Jensen et al., 2003). Therefore since GABBR1 and perhaps SLC6A1 may play a role in the addictive process, we hypothesized that variants in these genes might predict risk for, or resilience against, addiction.
In the current study, the primary analyses of GABBR1 SNPs and haplotypes were conducted in a group of treatment-seeking African American (AA) men with single and comorbid diagnoses of alcohol dependence (AD), cocaine dependence (CD) and heroin dependence (HD). The replication dataset was a group of incarcerated Finnish Caucasian men with diagnoses of alcohol use disorders (AUD), 58% of whom also had diagnoses of antisocial personality disorder (ASPD). We selected a second replication dataset to test our finding with the GABBR1 SNP rs29220. This was a community sample of Plains Indian (PI) men and women with diagnoses of AUD. Finally, we genotyped a functional SLC6A1 insertion promoter polymorphism, unique to African ancestry, in the AA sample.
PARTICIPANTS AND METHODS
African Americans
A detailed description of this sample has previously been provided (Roy et al., 2012). All participants gave written informed consent to the study that was approved by the Institutional Review Boards (IRBs) of the Department of Veteran Affairs New Jersey Healthcare System (VANJHCS) and the University of Medicine and Dentistry, New Jersey Medical School (UMDNJ).
Patients were recruited from the Substance Abuse Treatment Program at the VANJHCS. All were abstinent. There were 449 patients (401 men) who had been genotyped. Patients were interviewed using the substance abuse section of the Structured Clinical Interview for DSM-IV (Spitzer et al., 1995) to establish lifetime diagnoses. Of the male patients, 44% had single diagnoses of: AD (18%), CD (15%) or HD (11%). Other patients had dual dependencies: AD + CD (24%), CD + HD (11%) and AD + HD (8%), and a further 13% were dependent on all three substances. Their mean (SE) age was 45.2 (0.4) years.
Controls were recruited from the same geographical area as the patients; approximately half were diabetic outpatients at an ophthalmology clinic at UMDNJ and half were recruited from churches and a blood bank in Newark, NJ. Controls were interviewed about substance abuse/dependence and were free of any lifetime alcohol or drug use disorder. There were 470 controls (193 men) with genotypes available. Their mean (SE) age was 34.3 (0.4) years.
Primary analyses
Primary analyses were conducted in men only in order to avoid gender imbalance in cases and controls. The total dataset and the subset of female controls were used in secondary analyses for the purpose of replication.
Finnish Caucasians
The sample from Helsinki, Finland, has been described in detail elsewhere (Enoch et al., 2009; Lappalainen et al., 1998). All participants gave written informed consent to the study that was approved by the IRBs of the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute of Mental Health, the University of Helsinki, and the University of Helsinki Central Hospital, Helsinki, Finland.
The genotyped sample included 159 male incarcerated alcoholic criminal offenders (93 with comorbid ASPD) and 181 male population controls. The Structured Clinical Interview for DSM-III-R was administered to both alcoholics and controls and blind-rated DSM-III-R psychiatric diagnoses were obtained. Their mean (SE) ages were: alcoholics, 33.5 (0.8) yrs, controls: 35.2 (0.8) yrs.
Plains American Indians
Volunteers were recruited from a PI tribe living in rural Oklahoma. Full details can be found in Enoch et al., 2009. All participants gave written informed consent to the study that was approved by the NIAAA IRB and the PI Tribal Council. Blind-rated DSM-III-R lifetime psychiatric diagnoses were derived for all participants from the Schedule for Affective Disorders and Schizophrenia-Lifetime Version. A total of 239 individuals with AUD were genotyped for GABBR1 rs29220 (mean (SE) age = 41.7 (0.9), 46% women) together with 178 non-alcoholics (mean (SE) age = 42.4 (1.1), 70% women).
Genotyping
Genomic DNA was isolated from blood for the AA sample and from EBV transformed lymphocytes for the Finnish and PI samples.
GABBR1
The AA and Finnish samples were genotyped for seven tag SNPs across GABBR1 based on the HapMap African sample (Table 1). PIs were genotyped for rs29220 for replication purposes. Genotyping was undertaken using Taqman assays (Life Technologies). PCR was performed using the following parameters: 95°C for 10 min, followed by 40 cycles of 92°C for 15 sec, 60°C for 60 sec, with a final hold at 4°C. Genotypes were read using the allelic discrimination protocol on the 7900HT Sequence Detection system (Applied Biosystems).
TABLE 1.
Description of GABBR1 SNPS
| GABBR1 | Current study MAFs | HapMap samples MAFs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNPS | Chr Pos* | Gene Location |
Base | AA | Finns | PI | YRI | CEU | HCB | JPT |
| rs3025643 | 29602178 | intergenic | C > T | 0.099 | 0.288 | 0.083 | 0.350 | 0.111 | 0.133 | |
| rs2267633 | 29603064 | UTR-3 | A > G | 0.109 | 0.047 | 0.108 | 0.100 | 0.244 | 0.144 | |
| rs29267 | 29607226 | intron | C > T | 0.109 | 0.047 | 0.102 | 0.097 | 0.256 | 0.174 | |
| rs29230 | 29608616 | cds Phe/Phe | T > C | 0.168 | 0.080 | 0.119 | 0.133 | 0.279 | 0.174 | |
| rs29259 | 29610161 | intron | T > C | 0.166 | 0.086 | 0.125 | 0.167 | 0.278 | 0.144 | |
| rs29225 | 29613264 | cds Ser/Ser | T > C | 0.099 | 0.057 | 0.075 | 0.075 | 0.163 | 0.017 | |
| rs29220 | 29621889 | intron | C > G | 0.359 | 0.427 | 0.388 | 0.307 | 0.181 | 0.409 | 0.369 |
Genome build GRCh38
Chr Pos = location on chromosome; MAF = minor allele frequency; cds = coding SNP Current study samples: AA = African Americans; Finns = Finnish Caucasians; PI = Plains American Indians
HapMap samples: YRI = Africans; CEU = Caucasians; HCB = Chinese; JPT = Japanese
For the AA dataset, the genotype completion rate was 0.995. The proportion of replicate genotyping was 0.16 and the genotyping error rate ranged from 0.01–0.02 across SNPs. Other than rs29220, all SNPs were in Hardy Weinberg equilibrium (HWE) (p >0.43). Rs29220 was in HWE in male participants (p = 0.09) but not the total group of men and women (p = 0.01).
For the Finnish dataset, the genotype completion rate was 0.997. The proportion of replicate genotyping was 0.10 and the genotyping error rate ranged from 0–0.02. All SNPs was in HWE (p >0.08).
For the PI dataset, the genotype completion rate for rs29220 was 0.931. The proportion of replicate genotyping was 0.08 and the genotyping error rate was zero. Rs29220 was in HWE (p = 0.26).
SLC6A1
A 21-bp insertion polymorphism in the SLC6A1 promoter region creates a second tandem copy of the sequence that results in an enhancer element, thereby increasing gene expression (Hirunsatit et al., 2007, 2009). The genotyping methodology is provided in supplementary materials.
Assessment of population stratification using ancestry informative markers
The AA, Finnish and PI samples had been previously genotyped for 186 ancestry informative markers (AIMs) on an ‘addictions array’ using the Illumina GoldenGate platform (Hodgkinson et al., 2008). As previously described, the AIMS genotypes from our samples were run against the 51 worldwide populations represented in the HGDP-CEPH Human Genome Diversity Panel and individual ethnic factor scores were computed in order to identify population substructure (Roy et al., 2012).
Statistical Analyses
Logistic regression analyses were undertaken using JMP 11 software. Backward stepwise regression was performed with variables (age, ethnic factor scores, sex) being eliminated from the model in an iterative process. Genotypes were entered individually into the regression analysis rather than as coded 0, 1, 2 since the former is a more conservative approach. Logistic regression models with nominal variables yielded likelihood ratio (L-R) χ2 results.
Haplotype frequencies were estimated using a Bayesian approach implemented with PHASE (Stephens et al., 2003). Haploview version 2.04 Software (Whitehead Institute for Biomedical Research, USA) was used to produce linkage disequilibrium (LD) matrices (Figure S1). Since rare and uncommon haplotypes are subject to estimation errors because of increased sampling variance, all analyses were conducted with haplotypes ≥ 1% frequency.
RESULTS
GABBR1: African American male sample
Haplotype analyses
One haplotype block spanned GABBR1 (Figure S1). This block included 6 haplotypes (> 0.010 frequency) (Figure S2): H1 (0.376), H2 (0.355), H3 (0.098), H4 (0.066), H5 (0.057) and H6 (0.042). A logistic regression analysis with ‘any addiction’ vs. controls as the dependent variable and with the 6 haplotypes and the African ethnic factor score included as independent variables did not show a global haplotype effect (χ2 = 7.4, 5df, p = 0.19). Likewise, there was no global haplotype effect for total AD (p = 0.45), total CD (p = 0.14) or total HD (p = 0.51). It is noteworthy that within the logistic regression analysis the African ancestry score had a significant effect on ‘any addiction’ (p < 0.0001) thus validating the requirement for the use of AIMS.
SNP analyses
As can be seen from Table 1, with the exception of rs29220 (minor allele frequency (MAF) = 0.359) the SNP MAF was sufficiently low to necessitate the grouping of heterozygotes and minor homozygotes. Entering all 7 SNPs into the logistic regression model with ‘any addiction’ as the dependent variable rendered the model unstable. Therefore, SNPs were initially entered individually into the model. There were significant associations for rs29220 (p = 0.009, 2df), the adjacent rs29225 (p = 0.035, 1df) and a trend association for the adjacent rs29259 (p = 0.052, 1 df). However when these 3 SNPs were entered simultaneously into the model, only rs29220 showed a significant association with ‘any addiction’ (p = 0.0006, 2df). Subsequent analyses revealed the same association between rs29220 and total AD vs. no addiction (p = 0.0048, N = 447), total CD vs. no addiction (p = 0.0045, N = 445) and total HD vs. no addiction (p = 0.0329, N = 367).
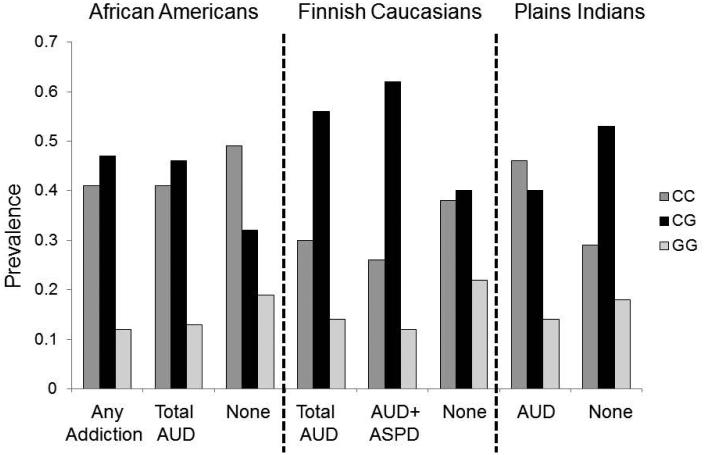
The allele frequencies were identical in men with any addiction and male controls (MAF = 0.35). The genotype distribution of men with any addiction was in HWE (p = 0.48) (Table 2). In contrast, the genotype distribution in male controls was significantly out of HWE (p = 0.0001). As shown in Table 2 and Figure 1, this was due to a significant deficit of heterozygotes and an excess of both homozygotes in controls. This decrease in heterozygotes was also seen in the sample of 277 female controls (HWE p = 0.03). We provide detailed evidence in the Discussion to support our view that deviation from HWE in the controls was not due to methodological error or for technical reasons.
TABLE 2.
GABBR1 rs29220 genotype frequencies in the three study samples
| Sample | GABBR1 rs29220 genotypes | N | MAF | HWE | ||
|---|---|---|---|---|---|---|
| CC | GC | GG | p value | |||
| AA men, any addiction | 166 (0.41) | 189 (0.47) | 46 (0.12) | 401 | 0.35 | 0.48 |
| AA men, total AUD | 105 (0.41) | 117 (0.46) | 32 (0.13) | 254 | 0.36 | 0.95 |
| AA men, controls | 95 (0.49) | 62 (0.32) | 36 (0.19) | 193 | 0.35 | 0.0001 |
| AA women, controls | 110 (0.40) | 115 (0.42) | 52 (0.18) | 277 | 0.39 | 0.03 |
| Finns, all AUD | 47 (0.30) | 89 (0.56) | 23 (0.14) | 159 | 0.43 | 0.09 |
| Finns, AUD + ASPD | 24 (0.26) | 58 (0.62) | 11 (0.12) | 93 | 0.44 | 0.01 |
| Finns, controls | 68 (0.38) | 73 (0.40) | 40 (0.22) | 181 | 0.42 | 0.03 |
| PI all AUD | 112 (0.47) | 95 (0.40) | 32 (0.13) | 239 | 0.33 | 0.11 |
| PI controls | 52 (0.29) | 94 (0.53) | 32 (0.18) | 178 | 0.44 | 0.35 |
N (frequency) for each genotype; MAF = minor allele frequency; HWE = Hardy-Weinberg Equilibrium AA = African Americans; Finns = Finnish Caucasians; PI = Plains American Indians AUD = alcohol use disorders (for AA men this includes single and comorbid diagnoses); ASPD = antisocial personality disorder
FIGURE 1. Association of GABBR1 rs29220 genotypes with addiction / alcoholism in three populations.
Sample sizes of rs29220 genotypes in three datasets:
African American men:
Any addiction: CC, n = 166, CG, n = 189, GG, n = 46
Total group with alcohol use disorders (AUD): CC, n = 105, CG, n = 117, GG, n = 32
None (no addiction): CC, n = 95, CG, n = 62, GG, n = 36
Finnish Caucasian men:
Total group with AUD: CC, n = 47, CG, n = 89, GG, n = 23
Subgroup of AUD with comorbid ASPD (AUD + ASPD): CC, n = 24, CG, n = 58, GG, n = 11
None (no AUD or ASPD): CC, n = 68, CG, n = 73, GG, n = 40
Plains Indian men and women:
AUD: CC, n = 112, CG, n = 95, GG, n = 32
None (no AUD): CC: n = 52, CG: n = 94, GG: n = 32
GABBR1: Finnish Caucasian male sample
Haplotype analyses
One haplotype block spanned GABBR1 (Figure S1) in Finns. This block included the same 6 haplotypes as in the AA sample but the frequencies differed: H1 (0.203), H2 (0.424), H3 (0.286), H4 (0.026), H5 (0.037) and H6 (0.020) (Figure S2). Similar to the AA sample, there was no haplotype association with AUD (global p = 0.77). There was no global haplotype association with the subset of alcoholics with comorbid ASPD (global p = 0.19).
SNP analyses -- replication of association with rs29220
Similar to the AA sample, rs29220 was associated with AUD (χ2 = 8.7, 2df, p = 0.013, N = 340) and the effect was greater in the subset of alcoholics with ASPD: χ2 = 11.9, 2df, p = 0.003, N = 274.
As can be seen in Table 2 and Figure 1, there was an excess of heterozygotes in alcoholics and a relative deficit of both homozygous genotypes. This was more marked in the alcoholics with ASPD (N = 93) who had an excess of heterozygotes (HWE p = 0.013) whereas controls (N = 181) had a deficit of heterozygotes (HWE p = 0.026). Allele frequencies in alcoholics with ASPD were similar to those in controls (0.44 vs. 0.42).
Consistent with the AA sample, there were also no significant associations between the other six GABBR1 SNPs and AUD (p > 0.51). However, only rs3025643 (MAF = 0.29) was sufficiently powered; the MAF of the other SNPs was less than 0.09.
GABBR1: Plains Indian sample
Only SNP rs29220 was genotyped in the PI sample of men and women to determine whether the associations in the AA and Finnish samples could be replicated. There was a significant genotype association with AUD (χ2 = 12.9, 2df, p = 0.0015, N = 417) (Table 2). The major C allele was more common in alcoholics than in controls: 0.67 vs. 0.56, χ2 = 10.2, 1df, p = 0.001.
SLC6A1 promoter insertion polymorphism, unique to African ancestry
African American male sample
The one homozygote was grouped with the heterozygotes in analyses. We had hypothesized that the SLC6A1 promoter insertion (IND) polymorphism would be associated with AD and/or CD. Since a preliminary analysis revealed that there was no association with total AD (p = 0.32) or total CD (p = 0.84) we conducted analyses in the patients with single diagnoses of AD (N = 61) or CD (N = 52) compared with controls with no addiction (N = 180). The IND allele was more common in controls than in AD (0.28 vs. 0.13) (L-R χ2 = 4.8, 1df, p = 0.028; effect of African ancestry: p = 0.050) however this result was only nominally significant (Bonferroni p value threshold = p < 0.013). There was no association with CD (p = 0.26).
African American total sample
The same results were obtained in the total sample of AA men and women that included a modest increase in patients with single diagnoses of AD (N = 87) and CD (N = 67) and a far greater number of controls (N = 470). Within the total model, sex (p < 0.0001) and African ancestry (p = 0.033) had independent effects on AD, as did SLC6A1 genotype: the IND allele was more common in controls than in AD (0.28 vs. 0.13) (L-R χ2 = 5.8, 1df, p = 0.016), with no association with CD (p = 0.18).
Exploratory analysis: combined effect of GABBR1 rs29220 and SLC6A1 INS promoter polymorphism
We performed an exploratory analysis of the combined effects of the SLC6A1 insertion polymorphism (coded as IND for insertion (minor allele), N for major allele) and GABBR1 rs29220 genotypes coded as CC, CG, GG, where allele G is the minor allele. In order to increase power we performed the analysis in the whole AA dataset that includes men and women and looked at the effects of combined genotypes on total AD (Table 3). In the whole sample, the SLC6A1 indel allele had a trend towards increased frequency in controls vs. all alcoholics (0.28 vs. 0.23, 1.22 fold change, L-R χ2 = 2.61, 1df, p = 0.106, whole model variance = 0.18). The association of rs29220 with total AD was significant (L-R χ2 = 8.9, 2df, p = 0.012, whole model variance = 0.19); this was due to a deficit of heterozygotes in controls.
TABLE 3.
Combined effects of the SLC6A1 functional indel promoter polymorphism and GABBR1 rs29220 on alcohol dependence in the African American sample of men and women
|
SLC6A1 IND + GABBR1 rs29220 |
M+F all AD N (freq) |
M+F controls N (freq) |
Fold increase |
|---|---|---|---|
|
| |||
| IND/CC | 18 (0.071) | 60 (0.139) | 1.96 |
| IND/GG | 7 (0.028) | 25 (0.058) | 2.07 |
| N/GG | 23 (0.091) | 58 (0.134) | 1.47 |
|
| |||
| total | 48 (0.190) | 143 (0.331) | 1.74 |
|
| |||
| IND/CG | 29 (0.115) | 38 (0.088) | 1.31 |
| N/CC | 89 (0.353) | 130 (0.301) | 1.17 |
| N/CG | 86 (0.341) | 121 (0.280) | 1.22 |
|
| |||
| total | 204 (0.809) | 289 (0.669) | 1.22 |
The SLC6A1 promoter indel polymorphism is coded as IND (insertion polymorphism, minor allele), N (major allele) and GABBR1 rs29220 genotypes are coded as CC, CG, GG, where allele G is the minor allele.
All AD = total group of patients with alcohol dependence; controls have no addiction
The genotype groups IND/CC, IND/GG, N/GG were more common in controls (0.33, n = 143) than in patients with AD (0.19, n = 48) whereas the genotype groups IND/CG, N/CC, N/CG, were more common in patients (0.81, n = 204) than in controls (0.67, n = 289) (L-R χ2 = 16.0, 5df, p = 0.007, with independent effects of sex (p <0.001) and African ancestry (p = 0.042); whole model variance = 0.20). As mentioned earlier, in the whole dataset the IND allele was 1.2 times more common in controls than alcoholics. However individuals with the IND allele and either rs29220 homozygous genotype were twice as likely to be non-alcoholics (Table 3). In contrast, rs29220 heterozygotes, irrespective of their IND status, were 1.24 times more likely to be alcoholics (Table 3).
DISCUSSION
Since there is evidence from animal studies, treatment studies in humans and human brain transcriptome (Enoch et al., 2012) that GABBR1 and SLC6A1 play a role in the addictive process, we tested the hypothesis that variants in these genes might predict risk or resilience for addiction. We found an association between the GABBR1 SNP rs29220 and alcoholism in three independent populations that differ not only in terms of ethnicity but also alcoholism phenotype. The AA male alcoholics were treatment-seeking inpatients and many had comorbid CD and HD. The Finnish male alcoholics were incarcerated; the subgroup with comorbid ASPD are likely to be more severe alcoholics. The PI men and women were a community sample. A major strength of our study is that we found similar results in these three ethnically and phenotypically diverse samples. There was a common finding in that it was the major rs29220 genotype (heterozygotes in AAs and Finns) and the major allele in PIs that were associated with risk for alcoholism. Moreover, the SLC6A1 IND allele, unique to African ancestry, predicted resilience being more common in controls, although a caveat to this finding is that this result was nominally significant and did not quite reach the threshold for Bonferroni correction. In other words, with both GABBR1 and SLC6A1, the risk genotype for this common heterogeneous disorder is more abundant than the protective genotype. Our exploratory analysis of the combined effects of GABBR1 rs29220 and SLC6A1 IND polymorphism showed that rs29220 heterozygotes, irrespective of their IND allele status, had an increased risk for alcoholism whereas individuals with the IND allele and either of the rs29220 homozygous genotypes were more resilient.
The SNP rs29220 is located in intron 9 of GABBR1 and, according to the UCSC genome browser (http://genome.ucsc.edu/) is within all transcripts however there is no evidence that it is functional. In contrast, the SLC6A1 21 bp insertion polymorphism creates an enhancer element that potentiates promoter activity (Hirunsatit et al., 2009) that is likely to result in increased GAT1, and by inference decreased synaptic GABA. This could be protective because increased synaptic GABA may increase the rewarding effects of drugs of abuse.
An unusual finding of our study was that, unlike the other 6 GABBR1 SNPs, rs29220 genotypes distinctly deviated from HWE in AA controls, both men and women, with a deficit of heterozygotes and an excess of both homozygous genotypes (men: p = 0.0001; women p = 0.03). In contrast, the patients’ genotypes were in HWE. The Finnish controls’ rs29220 genotypes were also out of HWE, likewise with a deficit of heterozygotes and an excess of both homozygotes (p = 0.03). Needless to say, genotyping error must first be ruled out. Visual inspection of the Taqman genotype discrimination plots was performed blind to diagnosis (cases and controls were inter-mingled on the DNA plates) and showed no evidence of genotyping error or differing intensities of genotype groups (Figures S3, S4). Poor genotyping completion rates could bias the proportion of genotypes because heterozygotes are more likely to drop out, but the completion rates were high (≥ 0.995 in AAs, Finns). Moreover, the excess of heterozygotes in the Finnish alcoholics was not due to Taqman genotyping because there was excellent clustering (Figure S4). There might be an unknown SNP within the region of the Taqman primer or probe that is common in controls and rare in addicted individuals and might differentially affect amplification. However, inspection of the UCSC Genome browser shows that there is no SNP with a frequency > 1% within the 501 bp region that encompasses rs29220. Neither are there any microsatellites or repeat regions. There is an outside possibility of an insertion/deletion polymorphism in AA and Finnish, but not PI, controls that is closely adjacent to, and in LD with the rs29220 G allele. However, this would result in differing intensity of homozygous genotypes on the Taqman plots which was not seen (Figures S3, S4).
The excess of rs29220 homozygosity seen in AA controls could theoretically be due to population substructure. Thus, if two or more subpopulations have different allele frequencies, the overall heterozygosity is reduced -- the so-called ‘Wahlund’ effect. The MAF range in the seven 1000 Genome African populations is 0.31–0.37 and that in the five Caucasian populations is 0.20-0.44 (Table S1). Therefore European admixture in the AAs is unlikely to result in significant subpopulation structure. To test for this, we divided the AA male controls across the median African ancestry of 0.83; the two groups had an excess of both homozygotes and deviated from HWE: < 0.83 African ancestry (N=87), p = 0.01, MAF = 0.33; ≥ 0.83 ancestry (N=92), p = 0.0003, MAF = 0.38; i.e. deviation from HWE in controls is not due to population substructure. Finally, although the 1000 Genome datasets include population samples and not psychiatrically-screened controls, we analyzed the seven African populations; rs29220 genotypes were in HWE in six populations however the Yorubans (N=108), from whom our AA sample may be derived, show a similar but trend effect of increased homozygosity (observed/expected: CC: 52/49; CG: 41/44; GG: 15/11, HWE p = 0.15. This deviation from HWE might be significant in a larger sample.
Another consideration is that the AA controls might be uniquely different; for example, half were diabetic. GABA and GABABRs are located throughout the gastrointestinal tract where they have many roles including modulation of insulin release from pancreatic islet cells (Hyland and Cryan, 2010; Purwana et al., 2014). However it is not obvious how rs29220 homozygosity might increase risk for diabetes, and moreover the Finnish control genotypes similarly deviated from HWE.
With no obvious technical or methodological explanation for similar deviation from HWE in AA and Finnish controls, might there be a biological explanation? GABBR1 maps to chromosome 6p21.3 and is located within an area of extensive LD. Therefore rs29220 might be linked with a functional SNP. The rs29220 MAFs are high across all 26 worldwide 1000 Genome populations implying that this is a balanced polymorphism and both homozygotes might be advantageous, as is the case for the common, functional polymorphism, COMT Val158Met (Zhu et al., 2004). However within the context of the currently available data it is not possible to comment further, or to speculate as to mechanisms.
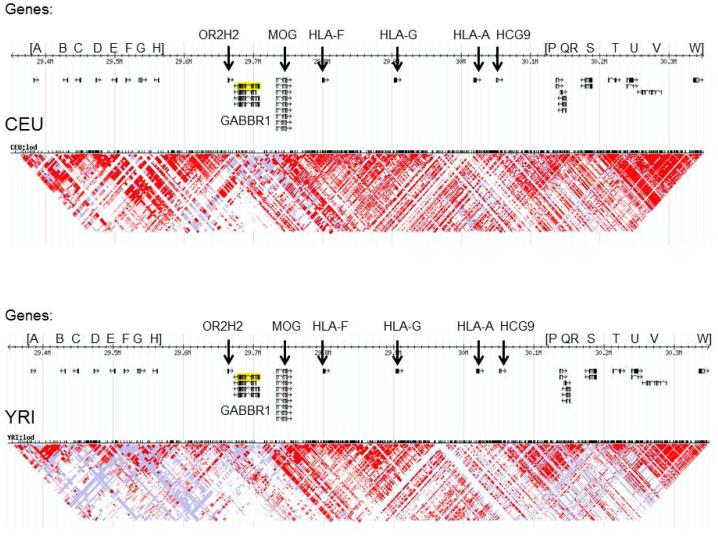
GABBR1 is located between an olfactory receptor gene cluster downstream and the HLA class I region upstream. Figure 2 indicates that in both the Caucasian (CEU) and African (YRI) HapMap populations, GABBR1 is not in LD with the olfactory gene cluster however it is in LD with the closely adjacent downstream OR2H2 olfactory receptor gene. Unlike in the YRI sample, LD in the CEU sample extends upstream from GABBR1 to include MOG and at least the HLA-F gene and, as shown later, with 2 SNPs in HLA-G. The myelin oligodendrocyte glycoprotein gene, MOG, may have relevance for alcoholism (Manzardo et al, 2015). This gene includes a potentially functional missense variant rs2857766 Val17Leu that has a high frequency (0.30) in the CEU sample. The HLA-G gene is a prime example of balancing selection (Gineau et al., 2015; Sabbagh et al., 2014). In the CEU sample, the GABBR1 SNP rs29220 is not in LD with SNPs across HLA-G with the exception of two intronic SNPs, rs6932888 and rs6932596 (Figure S5) with a frequency of 0.40. However, it should be noted that in the YRI sample, there is no LD between rs29220 and these two HLA-G SNPs and moreover, in the YRI sample GABBR1 is not in LD with any upstream genes. Thus the African data narrows the region of interest and implies that any functional SNP for which rs29220 might be a tag should be located in GABBR1 or in OR2H2. The UCSC genome browser identifies no known functional SNPs within GABBR1. However, OR2H2 includes rs1233387, a missense variant (Ala48Val) that has similar frequencies as rs29220 in HapMap samples but an analysis of 1000 Genomes data (not shown) reveals that the minor alleles of rs29220 and rs1233387 are never on the same haplotype background in the CEU and YRI populations and are therefore independent of each other. Therefore at this time information is lacking on the linkage of rs29220 with a functional SNP.
FIGURE 2. Linkage disequilibrium and genes (A to W) in regions upstream and downstream of GABBR1 (chromosome 6p21.3) in the HapMap CEU (Caucasian) and YRI (African) populations.
Genes downstream (to left) of GABBR1:
[A = OR5U1; B = OR5V1; C = OR12D3; D = OR12D2; E = OR11A1; F = ORI0C1; G = OR2H1 (A – G are an olfactory receptor gene cluster).; H = MASIL];
OR2H2
Genes upstream (to right) of GABBR1:
MOG; HLA-F; HLA-G; HLA-A; HCG9
[P = ZNRD1; Q = PPP1R11; R = RNF39; S = TRIM31; T = TRIM40; U = TRIM15; V = TRIM26; W = FLJ45422]
Distance from GABBR1 to: OR2H2 = 13.3 kb; MOG = 23.8 kb; HLA-F = 90 kb.
Distance across whole region shown in the figure is approximately 867 kb.
In conclusion, we have shown that the major genotype/allele of GABBR1 rs29220 and the SLC6A1 indel polymorphism predict alcoholism, whereas carriers of the minor genotypes/allele are more resilient. GABBR1 rs29220 is likely to be a tag for a common functional SNP and could therefore be a predictor for response to baclofen or its adverse effects in the treatment of alcoholism.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute on Alcohol Abuse and Alcoholism, NIH and in part by grant RO1 DA 10336-02 to AR from the National Institute of Drug Abuse, NIH.
Footnotes
DISCLOSURE /CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Addolorato G, Leggio L, Cardone S, Ferrulli A, Gasbarrini G. Role of the GABA(B) receptor system in alcoholism and stress: focus on clinical studies and treatment perspectives. Alcohol. 2009;43:559–563. doi: 10.1016/j.alcohol.2009.09.031. [DOI] [PubMed] [Google Scholar]
- Bettler B, Kaupmann K, Mosbacher J, Gassmann M. Molecular structure and physiological functions of GABA(B) receptors. Physiol Rev. 2004;84:835–867. doi: 10.1152/physrev.00036.2003. [DOI] [PubMed] [Google Scholar]
- Chalifoux JR, Carter AG. GABAB receptors modulate NMDA receptor calcium signals in dendritic spines. Neuron. 2010;66:101–113. doi: 10.1016/j.neuron.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalifoux JR, Carter AG. GABAB receptor modulation of synaptic function. Curr Opin Neurobiol. 2011;21:339–344. doi: 10.1016/j.conb.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo G, Addolorato G, Agabio R, Carai MA, Pibiri F, Serra S, Vacca G, Gessa GL. Role of GABA(B) receptor in alcohol dependence: reducing effect of baclofen on alcohol intake and alcohol motivational properties in rats and amelioration of alcohol withdrawal syndrome and alcohol craving in human alcoholics. Neurotox Res. 2004;6:403–414. doi: 10.1007/BF03033315. [DOI] [PubMed] [Google Scholar]
- Enoch M-A, Hodgkinson CA, Yuan Q, Albaugh B, Virkkunen M, Goldman D. GABRG1 and GABRA2 haplotypes as independent predictors for alcoholism in two populations. Neuropsychopharmacology. 2009;34:1245–1254. doi: 10.1038/npp.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M-A, Zhou Z, Kimura M, Mash DC, Yuan Q, Goldman D. GABAergic Gene Expression in Postmortem Hippocampus from Alcoholics and Cocaine Addicts; Corresponding Findings in Alcohol Naïve P and NP Rats. PLOS ONE. 2012;7(1):e29369. doi: 10.1371/journal.pone.0029369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. Mol Psychiatry. 2014 Dec 2; doi: 10.1038/mp.2014.159. doi: 10.1038/mp.2014.159. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Gineau L, Luisi P, Castelli EC, Milet J, Courtin D, Cagnin N, Patillon B, Laayouni H, Moreau P, Donadi EA, Garcia A, Sabbagh A. Balancing immunity and tolerance: genetic footprint of natural selection in the transcriptional regulatory region of HLA-G. Genes Immun. 2015;16:57–70. doi: 10.1038/gene.2014.63. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Burgos G. GABA transporter GAT1: a crucial determinant of GABAB receptor activation in cortical circuits? Adv Pharmacol. 2010;58:175–204. doi: 10.1016/S1054-3589(10)58008-6. [DOI] [PubMed] [Google Scholar]
- Hirunsatit R, Ilomäki R, Malison R, Räsänen P, Ilomäki E, Kranzler HR, Kosten T, Sughondhabirom A, Thavichachart N, Tangwongchai S, Listman J, Mutirangura A, Gelernter J, Lappalainen J. Sequence variation and linkage disequilibrium in the GABA transporter-1 gene (SLC6A1) in five populations: implications for pharmacogenetic research. BMC Genet. 2007;8:71. doi: 10.1186/1471-2156-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirunsatit R, George ED, Lipska BK, Elwafi HM, Sander L, Yrigollen CM, Gelernter J, Grigorenko EL, Lappalainen J, Mane S, Nairn AC, Kleinman JE, Simen AA. Twenty-one-base-pair insertion polymorphism creates an enhancer element and potentiates SLC6A1 GABA transporter promoter activity. Pharmacogenet Genomics. 2009;19:53–65. doi: 10.1097/FPC.0b013e328318b21a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, Binder EB, Cubells J, Ehlers CL, Gelernter J, Mann J, Riley B, Roy A, Tabakoff B, Todd RD, Zhou Z, Goldman D. Addictions Biology: Haplotype-Based Analysis for 130 Candidate Genes on a Single Array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland NP, Cryan JF. A Gut Feeling about GABA: Focus on GABA(B) Receptors. Front Pharmacol. 2010;1:124. doi: 10.3389/fphar.2010.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K, Chiu CS, Sokolova I, Lester HA, Mody I. GABA transporter-1 (GAT1)-deficient mice: differential tonic activation of GABAA versus GABAB receptors in the hippocampus. J Neurophysiol. 2003;90:2690–2701. doi: 10.1152/jn.00240.2003. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Long JC, Eggert M, Ozaki N, Robin RW, Brown GL, Naukkarinen H, Virkkunen M, Linnoila M, Goldman D. Linkage of antisocial alcoholism to the serotonin 5-HT1B receptor gene in 2 populations. Arch Gen Psychiatry. 1998;55:989–994. doi: 10.1001/archpsyc.55.11.989. [DOI] [PubMed] [Google Scholar]
- Liang JH, Chen F, Krstew E, Cowen MS, Carroll FY, Crawford D, Beart PM, Lawrence AJ. The GABA(B) receptor allosteric modulator CGP7930, like baclofen, reduces operant self-administration of ethanol in alcohol-preferring rats. Neuropharmacology. 2006;50:632–639. doi: 10.1016/j.neuropharm.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Maccioni P, Colombo G. Role of the GABA(B) receptor in alcohol-seeking and drinking behavior. Alcohol. 2009;43:555–558. doi: 10.1016/j.alcohol.2009.09.030. [DOI] [PubMed] [Google Scholar]
- Manzardo AM, McGuire A, Butler MG. Clinically relevant genetic biomarkers from the brain in alcoholism with representation on high resolution chromosome ideograms. Gene. 2015;560:184–194. doi: 10.1016/j.gene.2015.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley KC, Baillie A, Leung S, Addolorato G, Leggio L, Haber PS. Baclofen for the Treatment of Alcohol Dependence and Possible Role of Comorbid Anxiety. 2014 doi: 10.1093/alcalc/agu062. See comment in PubMed Commons belowAlcohol Alcohol pii: agu062. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwana I, Zheng J, Li X, Deurloo M, Son DO, Zhang Z, Liang C, Shen E, Tadkase A, Feng ZP, Li Y, Hasilo C, Paraskevas S, Bortell R, Greiner DL, Atkinson M, Prud'homme GJ, Wang Q. GABA promotes human β-cell proliferation and modulates glucose homeostasis. Diabetes. 2014;63:4197–4205. doi: 10.2337/db14-0153. [DOI] [PubMed] [Google Scholar]
- Roy A, Hodgkinson CA, Deluca V, Goldman D, Enoch MA. Two HPA axis genes, CRHBP and FKBP5, interact with childhood trauma to increase the risk for suicidal behavior. J Psychiatric Res. 2012;46:72–79. doi: 10.1016/j.jpsychires.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh A, Luisi P, Castelli EC, Gineau L, Courtin D, Milet J, Massaro JD, Laayouni H, Moreau P, Donadi EA, Garcia A. Worldwide genetic variation at the 3' untranslated region of the HLA-G gene: balancing selection influencing genetic diversity. Genes Immun. 2014;15:95–106. doi: 10.1038/gene.2013.67. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Williams JBW, Gibbon M. Structured Clinical Interview for DSM-IV (SCID) New York State Psychiatric Institute, Biometrics Research; New York: 1995. [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyacke RJ, Lingford-Hughes A, Reed LJ, Nutt DJ. GABAB receptors in addiction and its treatment. Adv Pharmacol. 2010;58:373–396. doi: 10.1016/S1054-3589(10)58014-1. [DOI] [PubMed] [Google Scholar]
- Vlachou S, Markou A. GABAB receptors in reward processes. Adv Pharmacol. 2010;58:315–371. doi: 10.1016/S1054-3589(10)58013-X. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Guo AY, van den Oord EJ, Aliev F, Jia P, Edenberg HJ, Riley BP, Dick DM, Bettinger JC, Davies AG, Grotewiel MS, Schuckit MA, Agrawal A, Kramer J, Nurnberger JI, Jr, Kendler KS, Webb BT, Miles MF. Multi-species data integration and gene ranking enrich significant results in an alcoholism genome-wide association study. BMC Genomics. 2012;13(Suppl 8):S16. doi: 10.1186/1471-2164-13-S8-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Lipsky RH, Xu K, Ali S, Hyde T, Kleinman J, Akhtar LA, Mash DC, Goldman D. Differential expression of human COMT alleles in brain and lymphoblasts detected by RT-coupled 5' nuclease assay. Psychopharmacology (Berl) 2004;177:178–184. doi: 10.1007/s00213-004-1938-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.