Abstract
Background
Somatic embryogenesis is a notable illustration of cell totipotency, by which somatic cells undergo dedifferentiation and then differentiate into somatic embryos. Our previous work demonstrated that pretreatment of immature zygotic embryos with 0.5 M sucrose solution for 72 h efficiently induced somatic embryo initiation in camphor tree. To better understand the molecular basis of somatic embryogenesis induced by osmotic stress, de novo transcriptome sequencing of three tissues of camphor tree (immature zygotic embryos, sucrose-pretreated immature zygotic embryos, and somatic embryos induced from sucrose-pretreated zygotic embryos) were conducted using Illumina Hiseq 2000 platform.
Results
A total of 30.70 G high quality clean reads were obtained from cDNA libraries of the three samples. The overall de novo assembly of cDNA sequence data generated 205592 transcripts, with an average length of 998 bp. 114229 unigenes (55.56 % of all transcripts) with an average length of 680 bp were annotated with gene descriptions, gene ontology terms or metabolic pathways based on Blastx search against Nr, Nt, Swissprot, GO, COG/KOG, and KEGG databases. CEGMA software identified 237 out of 248 ultra-conserved core proteins as ‘complete’ in the transcriptome assembly, showing a completeness of 95.6 %. A total of 897 genes previously annotated to be potentially involved in somatic embryogenesis were identified. Comparative transcriptome analysis showed that a total of 3335 genes were differentially expressed in the three samples. The differentially expressed genes were divided into six groups based on K-means clustering. Expression level analysis of 52 somatic embryogenesis-related genes indicated a high correlation between RNA-seq and qRT-PCR data. Gene enrichment analysis showed significantly differential expression of genes responding to stress and stimulus.
Conclusions
The present work reported a de novo transcriptome assembly and global analysis focused on gene expression changes during initiation and formation of somatic embryos in camphor tree. Differential expression of somatic embryogenesis-related genes indicates that sucrose induced somatic embryogenesis may share or partly share the mechanisms of somatic embryogenesis induced by plant hormones. This study provides comprehensive transcript information and gene expression data for camphor tree. It could also serve as an important platform resource for further functional studies in plant embryogenesis.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2357-8) contains supplementary material, which is available to authorized users.
Keywords: Cinnamomum camphora L, Somatic embryogenesis, RNA-seq, De novo assembly, Comparative transcriptome, Osmotic stress
Background
Camphor tree (Cinnamomum camphora L), a broad-leaved evergreen tree within the Lauraceae family, native to China and Japan, is now cultivated in many countries as an ornamental plant or as a source of camphor [1]. The species is also considered as a medicinal plant in the treatment of muscular strains, inflammation, and rheumatic conditions and has antiseptic properties. Plant regeneration system for camphor tree has been established via direct somatic embryogenesis [2, 3], in which immature zygotic embryos (IZE) were collected and pretreated with 0.5 M sucrose solution before somatic embryo (SE) induction. Sucrose pretreatment, other than various plant hormones, significantly enhanced efficiency of SE initiation from 16.29 to 93.27 % in this recalcitrant species [3].
Somatic embryogenesis is a notable illustration of cell totipotency, by which somatic cells undergo dedifferentiation and then differentiate into somatic embryos [4]. The developmental pathway of somatic embryos shares high similarities at almost all developmental stages to that of their zygotic counterparts, which makes it an attractive model system to study zygotic embryogenesis at molecular, cellular, and tissue levels [5, 6]. Somatic embryogenesis has also been considered as a potential model system for studying developmental mechanism of early embryogenesis [7, 8].
The initiation of SE is a multi-factorial event, in which embryos are derived from vegetative cells by exposing explants to stress conditions or exogenous growth regulators. It is widely recognized that plant hormones, particularly auxin, are the most important factor in stimulating SE initiation, and stress is another factor becoming increasingly recognized in recent years [8]. The stress factors which can stimulate initiation of embryogenic competence include heavy metal ions [9, 10], high temperature [11, 12], explant wounding [13], and high osmotic stress [3, 14–16].
Though initiation of somatic embryogenesis has been observed in many species, the molecular mechanism of triggering vegetative-to-embryogenic transition remains a challenge [17]. It is believed that somatic embryogenesis is a developmental process involving gene expression reprogramming that engages a cascade of genetic triggers turning on or off the expression of specific genes [18, 19]. Analyses of gene expression during somatic embryogenesis can provide information for better understanding of this complicated process.
The patterns of gene regulation during somatic embryogenesis have been investigated in several species, including carrot [20], Arabidopsis [21], alfalfa [22], soybean [23–25], cotton [26, 27], potato [28] and orange [29]. Numerous specifically activated or differentially expressed genes related to somatic embryogenesis have been isolated, such as Somatic Embryogenesis Receptor-like Kinase (SERK), Leafy Cotyledon (LEC), Baby Boom (BBM), and Wuschel (WUS) (reviewed by [30]). Genes controlling early embryogenesis, including Auxin Response Factor19 (ARF19), WUS, LEC1, SERK1 and Heat Shock Protein 17 (HSP17) have been investigated (reviewed by [31]). Molecular basis of stress-induced acquisition of embryogenic competence was described in detail by Karami and Saidi [8]. However, most of these researches have been focused on gene regulation in the process of somatic embryo initiation induced by plant hormones. The role of stress, especially high osmotic stress, in embryogenic culture has not been well characterized at molecular level in plants, including camphor tree.
Camphor tree is a species lacking genome resources and a comprehensive investigation of the global transcription. In an attempt to understand the molecular basis of embryogenic competence acquisition and SE formation in camphor tree, we separately performed de novo transcriptome sequencing of IZE, IZE pretreated with 0.5 M sucrose solution for 72 h (IZE_Suc), and SE obtained from cultured IZE_Suc (SE_5w) using Illumina Hiseq 2000 technology. This work provides new insights into somatic embryogenesis induced by high osmotic stress, and valuable resources for future transcriptomic, genomic and genetic research on camphor tree. The results in the present study also lead to the hypothesis that sucrose induced somatic embryogenesis may share or partly share the mechanisms of somatic embryogenesis induced by plant hormones.
Methods
Plant materials
Immature fruits were collected in late July of 2013 (12–13 weeks after open pollination) from a mature camphor tree in the campus of Huazhong Agricultural University, Wuhan, China. The fruits were surface-sterilized with 0.1 % (w/v) mercuric chloride (HgCl2) solution for 10 min, and then rinsed three times with sterilized distilled water. The fruits were cut open, and IZEs were isolated carefully from the distal end of the fruits (Additional file 1: Figure S1A-C). Meanwhile, IZEs were pretreated with 0.5 M sucrose solution for 72 h. SE_5w were obtained by culturing the sucrose-pretreated IZEs on hormone-free induction medium for five weeks, which contained cotyledonary SE as well as global, heart-shaped and torpedo SE since the asynchronous secondary somatic embryogenesis (Additional file 1: Figure S1D-E) [2]. IZE isolation, sucrose pretreatment, and somatic embryo induction from pretreated IZEs were carried out according to the method previously described [3]. According to our previous results, pretreatment of IZEs with 0.5 M sucrose solution could significantly improve the frequency of SE initiation [3]. In this study, three samples, namely IZE, sucrose-pretreated IZEs (IZE_Suc), and SE_5w, were collected and frozen immediately in liquid nitrogen and then kept at −80 °C for transcriptome analysis. Both samples of IZE and IZE_Suc in the weight of 300 mg were mixed sample pools composed of approximately 2000 embryos about 2 mm in diameter. The sample of SE_5w (300 mg) was also a mixed sample pool containing SEs in different developmental stages, which consisted of about 200 SEs. In addition, stems, fully expanded young leaves, young flowers, and young fruits were collected during March and May. Mature seeds were separated from mature fruits which were collected in November. Roots were obtained from germinated seeds. These samples were also collected and frozen immediately in liquid nitrogen and kept at −80 °C for the experiments of quantitative Real-Time PCR (qRT-PCR). All samples were collected from a single tree in the same year.
RNA isolation
Each frozen sample was ground in a mortar with liquid nitrogen. Total RNA was isolated using Trizol reagent (Invitrogen, Shanghai, China) according to the manufacturer’s instruction. RNA purity was checked using the NanoPhotometer® spectrophotometer (Implen, CA, USA), RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA), and RNA integrity was assessed using the RNA 6000 Nano Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Transcriptome sequencing and de novo assembly
A total amount of 3 μg RNA with RIN value above 8.0 from each of the three samples was respectively used to generate sequencing library using Illumina TruSeq™ RNA Sample Preparation Kit (Illumina, CA, USA) following manufacturer’s recommendations. Six index codes were added to each sample for attributing sequences. The clustering of the coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on an Illumina Hiseq 2000 platform to generate 100 bp paired-end reads.
The clean reads were obtained from raw data by filtering out adapter-only reads, reads containing poly-N, and low quality reads. The values of Q20, Q30, GC-content and sequence duplication level of the clean data were calculated. Clean reads were then assembled with the Trinity program [32]. In order to ensure the quality of assembly, the reads were mapped back to the assembled transcripts using the bowtie aligner by Visualization and Quality Assessment application within Trinity software. The alignment was visualized with Integrated Genomics Viewer (IGV) version 2.3.2 [33]. CEGMA software [34] was used to assess the sequence completeness of the assembly by estimating the presence and completeness of 248 ultra-conserved eukaryotic genes. Profile-hidden Markov model was used to ensure reliability of gene structure.
Gene functional annotation and classification
Unigene sequences were aligned using Blastx with an E-value cut-off of 1.0e-5 (unless stated otherwise) against protein databases, with the priority order of NCBI Nr (non-redundant database), Nt, Swiss-Prot, KEGG (Kyoto Encyclopedia of Genes and Genomes, E-value = 1.0e-10), and KOG/COG (clusters of orthologous groups, E-value = 1.0e-3) if conflicting results were obtained, to retrieve proteins with the highest sequence similarity with the given unigenes along with their functional annotation. Based on the Nr annotation, GO (gene ontology) annotation (E-value = 1.0e-6) was generated using Blast2GO program [35], and GO functional classification was finished using the WEGO software [36].
Analysis of differential gene expression
Gene expression levels were estimated by mapping clean reads to reference set of assembled transcripts using RSEM [37] for each sample. RPKM (reads per kilo bases per million mapped reads) were used as the value of normalized gene expression levels [38]. Pairwise differential expression analysis was done among the three samples using DEGseq [39] R package. P-values were adjusted using the q value method proposed by Storey et al. [40]. A q value < 0.005 and an absolute value of log2 fold_change >1 provided thresholds to determine significant differences in gene expression. Go enrichment analysis (p-value ≤ 0.05) of the differentially expressed genes (DEGs) was performed using GOseq with the Wallenius non-central hyper-geometric distribution model [41] to adjust gene length bias in DEGs. KEGG pathway enrichment analysis of the DEGs was done using KOBAS [42] with the hyper-geometric distribution model. The enrichment p-values were adjusted using the Benjamin and Hochberg method.
Quantitative Real-Time PCR (qRT-PCR) analysis
Analysis of qRT-PCR was performed to validate gene expression results from RNA-seq. Total RNA (3 μg) from each sample was reverse-transcribed into single-stranded cDNA using Prime-ScriptTM RT reagent Kit (TaKaRa, Dalian, China). Mixture of PCR was composed of 10 μl 2× SYBR Premix DimerEraser (TaKaRa, Dalian, China), 1 μl of each primer (Additional file 2: Table S1), and 2 μl of cDNA diluted 1:50. PCR reactions were run on an ABI 7500 Real-Time System (PE Applied Biosystems, CA, USA) under the following conditions: initial incubation at 50 °C for 2 min and 95 °C for 30 s, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Gene expression and standard error were calculated over three biological and two technical replicates.
Results
De novo assembly of camphor tree transcriptome
Genome and transcriptome resources are scarce for gene functional analysis in camphor tree. In this study, de novo transcriptome assembly was performed by merging the valid reads from libraries of three types of camphor embryos, including IZE, IZE_Suc and SE_5w. Sequencing of cDNA libraries from the three samples resulted in 104,771,602, 84,950,386 and 117,117,800 clean reads, representing with 10.48G, 8.50G and 11.72G nucleotides, respectively (Table 1). The statistics of raw reads were also shown in Table 1. For all the sequence data, Q20 percentage was more than 90 %, while Q30 percentage was more than 80 % (Table 1). A total of 306,839,788 clean reads obtained from 319,668,420 raw reads (95.99 %) participated in the assembly (Table 1). The overall de novo assembly of cDNA sequence data generated 205592 transcripts, with an average length of 998 bp. The reads were assembled into 114229 non-redundant unigenes with an average length of 680 bp and an N50 of 1075 bp. All the unigenes were longer than 200 bp in length, 71566 of them (62.65 %) were 200 to 500 bp, and 7375 (6.46 %) were longer than 2 kb (Additional file 3: Table S2, Fig. 1).
Table 1.
Overview of output statistics on camphor tree transcriptome sequencing
| Parameter | IZE | IZE_Suc | SE_5w |
|---|---|---|---|
| The number of total raw reads | 108,397,716 | 89,960,198 | 121,310,506 |
| The number of total clean reads | 104,771,602 | 84,950,386 | 117,117,800 |
| Total base pairs (bp) of clean reads | 10.48G | 8.5G | 11.72G |
| Q20 percentage | 96.19 % | 93.21 % | 96.25 % |
| Q30 percentage | 89.03 % | 82.76 % | 89.50 % |
| N percentage | 0.05 % | 0.05 % | 0.05 % |
| GC percentage | 46.85 % | 45.40 % | 46.12 % |
Q20/30 percentage represents proportion of nucleotides with quality value larger than 20/30, and N percentage represents proportion of unknown nucleotides in clean reads
Fig. 1.
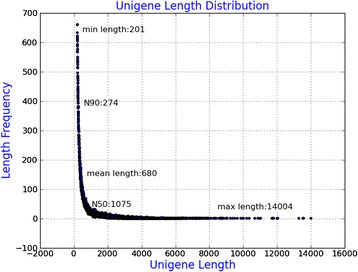
Length distribution of unigenes in the assembled transcriptomes. The x axis shows the lengths of unigenes and the y axis shows the number of unigenes
To assess the quality of the assembled transcripts, the clean reads were mapped back to the assembly, and the results showed that 82.73 % of the clean reads could be mapped back on the assembled transcriptome, among which 47.33 % were mapped uniquely and 35.4 % were mapped more than once. Assessment of assembly completeness by CEGMA software showed 237 out of 248 ultra-conserved core proteins were ‘complete’ in the transcriptome, yielding a completeness of 95.6 %. Five genes were identified as ‘partial’ genes. These results indicated high quality of transcriptome assembly in this study.
Functional annotation of non-redundant unigenes
To annotate the Trinity-assembled unigenes, the 114229 unigenes were subjected to Blastx searches against seven public databases, returning an above cut-off BLAST result (Table 2). A total of 26640 (23.32 %), 10741 (9.4 %), 19352 (16.94 %), 27673 (24.22 %), 32920 (28.81 %) and 11281 (9.87 %) of the unigenes were annotated by Nr, Nt, Swiss-Prot, PFAM, GO and KOG/COG database, respectively (Additional file 4: Table S3). 7742 (6.77 %) of the unigenes were annotated by KEGG database (Additional file 5: Table S4). Among the unigenes, 38020 unigenes (33.28 % of all unigenes) were annotated by at least one database. Of them, 3282 (2.87 %) of the unigenes were simultaneously annotated by all databases.
Table 2.
Annotation of assembled camphor tree unigenes
| Database for annotation | Number of annotated unigenes | Percentage (%) |
|---|---|---|
| Nr | 27752 | 23.33 |
| Nt | 10741 | 9.4 |
| SwissProt | 19352 | 16.94 |
| PFAM | 27673 | 24.22 |
| KOG | 11281 | 9.87 |
| GO | 32920 | 28.81 |
| KEGG | 7742 | 6.77 |
| Annotated in all Databases | 3282 | 2.87 |
| Annotated in at least one Database | 38020 | 33.28 |
| Total queries/unigenes | 114229 | 100 |
To further analyze the BLAST results in Nr database, similarities distribution, E-value distribution, best-hit species distribution, and best-hit species classification were investigated. The results showed that 3330 (12.50 %) of the matched sequences had alignment identities greater than 85 % (Fig. 2a). Only 2661 (9.99 %) of the matched sequences had an E-value higher than 1.0e-10, and 14848 (55.73 %) of them had an E-value lower than 1.0e-50 (Fig. 2b), which indicated high-reliability of the alignment. Among the annotated unigenes, the majority (26330, 98.83 %) matched plants (Fig. 2c). The top three matched plant species were Vitis vinifera (13187, 49.5 %), Populus trichocarpa (2891, 10.85 %), and Ricinus communis (2421, 9.09 %) (Fig. 2d).
Fig. 2.
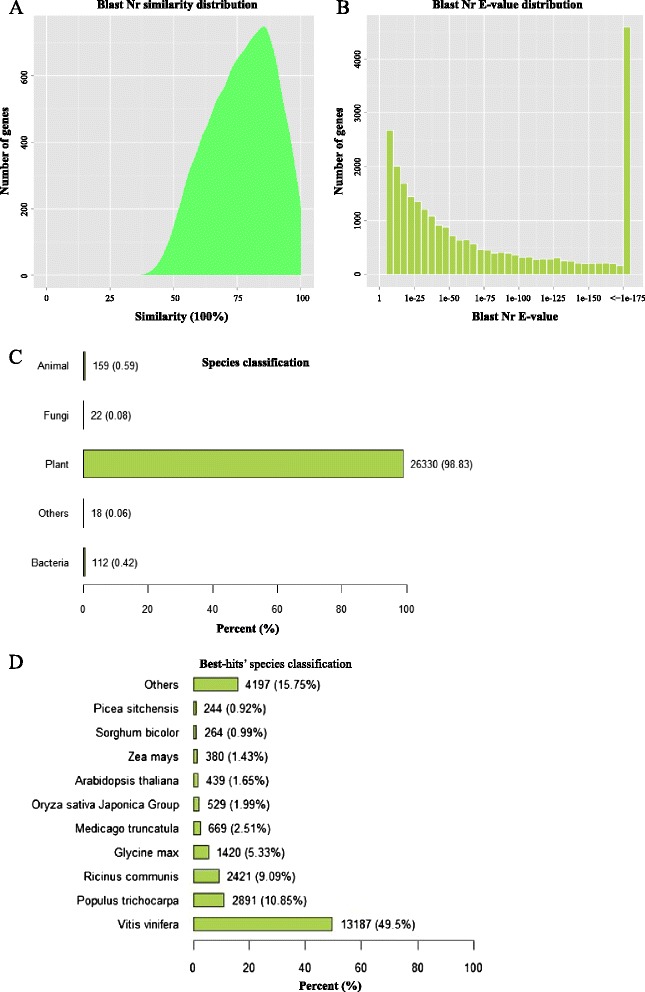
Further analyse of the BLAST results in Nr database. a Similarity distribution; b E-value distribution; c Best hit species distribution; d Best hit species classification
With respect to somatic embryo initial function, 897 unigenes were identified as homologues to the previously annotated genes that are potentially involved in somatic embryogenesis (Additional file 6: Table S5). Among them, Heat-Shock proteins (HSPs, 296 unigenes) were the most dominant group, followed by Chitinase genes (73 unigenes), Ethylene Responsive Factor (ERF, 61 unigenes), Glutathione S-Transferase (GST, 59 unigenes), Auxin Responsive Factor (ARF, 44 unigenes), and C-Repeat Binding Factor/Dehydration-Responsive Element-Binding Protein (CBF/DREB, 41 unigenes) (Additional file 6: Table S5).
Classification of camphor tree unigenes
GO assignment was performed to classify the functions of predicted unigenes. The 32920 unigenes annotated in GO database were categorized into 57 functional groups, belonging to three main GO ontologies: biological process, cellular component, and molecular function (Fig. 3). Among the functional groups, “cellular process”, “metabolic process”, “binding”, “cell”, and “cell part” terms were dominant (Fig. 3).
Fig. 3.
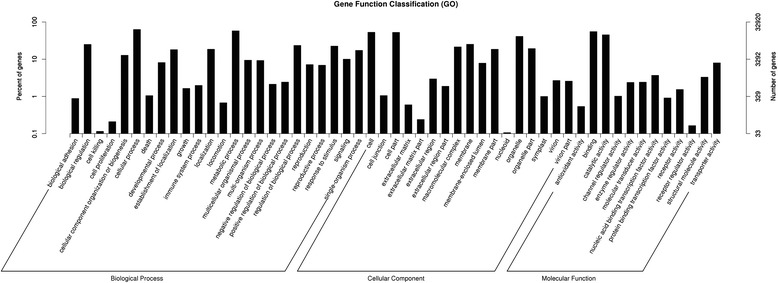
GO classifications of assembled unigenes by Blast2GO in Camphor tree. Unigenes were annotated in three main categories: biological process, cellular component and molecular function. The x-axis indicates the sub-categories and the y-axis indicates the number of unigenes
To further evaluate the function of the assembled unigenes, we searched the annotated unigenes involved in COG. COG annotation yielded 11281 putative proteins in 26 categories (Fig. 4). Among these categories, the cluster for “general functional prediction only” (18.39 %) represented the largest group, followed by “post-translational modification, protein turnover, chaperon” (12.69 %), “signal transduction” (10.08 %) and “transcription” (7.02 %). Clusters for “cell motility” (0.04 %), “unnamed protein” (0.04 %) and “nuclear structure” (0.78 %) were the smallest groups (Fig. 4).
Fig. 4.
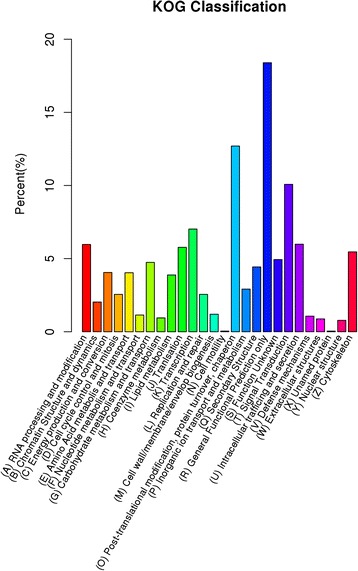
KOG classifications of assembled unigenes in Camphor tree. Out of 114229 de novo assembled unigenes, 11281 were annotated and seperated into 26 categories
KEGG pathways were also searched for biological interpretation of functions of the assembled unigenes. A total of 7742 unigenes were mapped to 31 pathways (Additional file 5: Table S4, Fig. 5). As shown in Fig. 5, the majority of unigenes were classified into pathways for carbohydrate metabolism (794), translation (693), folding, sorting and degradation (641), and energy metabolism (628). In contrast, few unigenes were found in some pathways, for example, signaling molecules and interaction (2), and sensory system (21).
Fig. 5.
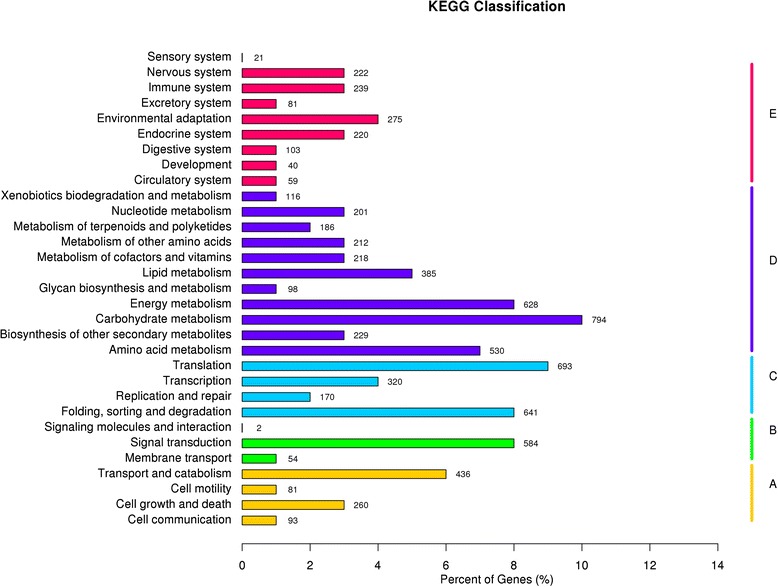
Distribution of the number of genes expressed in various metabolic pathway. a Cellular processes; b Invironmental information processing; c Genetic information processing; d Metabolism; e Organismal systems
Global analysis of gene expression in IZE, IZE_Suc and SE_5w
To characterize the differences of molecular response among IZE, IZE_Suc and SE_5w, expression levels of the assembled unigenes were calculated by RPKM using RSEM software [37]. The three samples showed similar RPKM density distribution (Additional file 7: Figure S2). The results showed that only a small proportion of genes are highly expressed (Table 3). Based on the values of RPKM, approximately 31432 (82.67 % of the 38020, RPKM > 0.3) annotated unigenes showed ubiquitous expression in all the three samples. A total of 44323, 57917 and 49364 genes showed expression (RPKM > 0.3, with 95 % confidence) in IZE, IZE_Suc and SE_5w samples, respectively. 1851 (1.62 %), 1866 (1.63 %) and 1666 (1.46 %) genes (>60 RPKM) were highly expressed in IZE, IZE_Suc and SE_5w, respectively (Table 3). The top 10 most expressed genes in IZE, IZE_Suc and SE_5w had high RPKM values ranging from 2142 to 6688, 1433 to 4452, and 3757 to 23062, respectively. The top 20 most expressed genes from the three libraries are shown in Tables 4, 5 and 6.
Table 3.
Gene expression values for the samples of IZE, IZE_Suc and SE_5w of camphor tree given in RPKM
| RPKM Interval | IZE | IZE_Suc | SE_5w |
|---|---|---|---|
| 0–0.1 | 38793(33.96 %) | 24200(21.19 %) | 35254(30.86 %) |
| 0.1–0.3 | 31113(27.24 %) | 32112(28.11 %) | 29629(25.94 %) |
| 0.3–3.57 | 29099(25.47 %) | 42289(37.02 %) | 34284(30.01 %) |
| 3.57–15 | 8305(7.27 %) | 8180(7.16 %) | 8301(7.27 %) |
| 15–60 | 5068(4.44 %) | 5582(4.89 %) | 5095(4.46 %) |
| >60 | 1851(1.62 %) | 1866(1.63 %) | 1666(1.46 %) |
Table 4.
The top 20 most expressed genes from IZE library
| Gene id | RPKM | Description |
|---|---|---|
| comp93556_c0 | 6688.14 | Serine/threonine protein kinase |
| comp93739_c0 | 5087.09 | Metallothionein-like protein 2a [Nelumbo nucifera] |
| comp96566_c0 | 4146.24 | RhoA GTPase effector DIA/Diaphanous |
| comp110416_c0 | 3628.56 | Xyloglucan endotransglucosylase/hydrolase [Rosa hybrid cultivar] |
| comp96556_c0 | 2666.02 | Polyubiquitin [Cicer arietinum] |
| comp108680_c0 | 2444.65 | AP2/ERF domain-containing transcription factor [Populus trichocarpa] |
| comp100175_c0 | 2423.43 | PREDICTED: uncharacterized protein LOC100255094 [Vitis vinifera] |
| comp101234_c0 | 2370.67 | Late embryogenesis abundant protein [Sesuvium portulacastrum] |
| comp103367_c0 | 2199.46 | Tubulin alpha-7 chain [Medicago truncatula] |
| comp57982_c0 | 2142.17 | Glycine-rich RNA-binding protein GRP1A [Sinapis alba] |
| comp101258_c0 | 2040.98 | Bowman-Birk type proteinase inhibitor [Lupinus albus] |
| comp110065_c1 | 1625.20 | Cytochrome P450, putative [Ricinus communis] |
| comp109418_c0 | 1523.87 | Heat-shock protein, putative [Ricinus communis] |
| comp30496_c0 | 1483.98 | Translationally controlled tumor protein [Hevea brasiliensis] |
| comp97314_c0 | 1478.90 | Hypothetical protein VITISV_035079 [Vitis vinifera] |
| comp101235_c0 | 1448.72 | Annexin [Gossypium hirsutum] |
| comp109861_c0 | 1435.23 | Heat shock cognate 70 kDa protein isoform 1 [Vitis vinifera] |
| comp110306_c3 | 1433.02 | ADP-ribosylation factor [Medicago truncatula] |
| comp110306_c2 | 1345.93 | Umecyanin [Armoracia rusticana] |
| comp109340_c0 | 1341.55 | Cytochrome P450 89A2-like [Vitis vinifera] |
Table 5.
The top 20 most expressed genes from IZE_Suc library
| Gene id | RPKM | Description |
|---|---|---|
| comp87631_c0 | 4452.17 | Alcohol dehydrogenase 1 [Solanum tuberosum] |
| comp98297_c0 | 3790.17 | Thioredoxin H-type [Medicago truncatula] |
| comp57982_c0 | 3444.90 | Glycine-rich RNA-binding protein GRP1A [Sinapis alba] |
| comp93739_c0 | 2837.91 | Metallothionein-like protein 2a [Nelumbo nucifera] |
| comp96556_c0 | 2314.52 | Polyubiquitin [Cicer arietinum] |
| comp96883_c0 | 2051.84 | Probable glutathione S-transferase [Vitis vinifera] |
| comp93556_c0 | 1792.74 | Serine/threonine protein kinase |
| comp103430_c1 | 1727.23 | Probable aquaporin PIP2-5-like [Glycine max] |
| comp110306_c3 | 1515.34 | ADP-ribosylation factor [Medicago truncatula] |
| comp103367_c0 | 1432.87 | Tubulin alpha-7 chain [Medicago truncatula] |
| comp97309_c0 | 1377.05 | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic [Magnolia liliiflora] |
| comp96172_c0 | 1376.65 | Glyceraldehyde-3-phosphate dehydrogenase, partial [Eriobotrya japonica] |
| comp30496_c0 | 1325.62 | Translationally controlled tumor protein [Hevea brasiliensis] |
| comp86932_c0 | 1297.76 | Fructose-bisphosphate aldolase [Persea americana] |
| comp94640_c0 | 1253.98 | Stem-specific protein TSJT1 [Vitis vinifera] |
| comp110306_c2 | 1231.54 | Umecyanin [Armoracia rusticana] |
| comp87522_c0 | 1210.82 | 48 kDa dehydrin-like protein [Cornus sericea] |
| comp110467_c0 | 1204.51 | Conserved hypothetical protein [Ricinus communis] |
| comp86219_c0 | 1202.35 | GTP-binding nuclear protein Ran1 [Solanum lycopersicum] |
| comp108123_c1 | 1194.13 | Pyruvate decarboxylase 1 [Lotus corniculatus] |
Table 6.
The top 20 most expressed genes from SE_5w library
| Gene id | RPKM | Description |
|---|---|---|
| comp108950_c0 | 23061.85 | 7S globulin [Sesamum indicum] |
| comp93739_c0 | 11305.68 | Metallothionein-like protein 2a [Nelumbo nucifera] |
| comp90165_c0 | 10427.33 | -- |
| comp30459_c0 | 8028.17 | Oleosin H-isoform [Ficus pumila var. awkeotsang] |
| comp93572_c2 | 7081.29 | Glutathione S-transferase [Chimonanthus praecox] |
| comp90198_c0 | 6050.84 | Putative defensin 1 [Aquilegia pyrenaica] |
| comp94419_c1 | 5315.23 | -- |
| comp98280_c0 | 4237.01 | -- |
| comp57982_c0 | 3871.00 | Glycine-rich RNA-binding protein GRP1A [Sinapis alba] |
| comp101235_c0 | 3756.90 | Annexin [Gossypium hirsutum] |
| comp93556_c0 | 3306.89 | Serine/threonine protein kinase |
| comp101258_c0 | 2908.99 | Bowman-Birk type proteinase inhibitor [Lupinus albus] |
| comp96556_c0 | 2422.08 | Polyubiquitin [Cicer arietinum] |
| comp79025_c0 | 2222.47 | Pollen allergen Pla o 3 [Platanus orientalis] |
| comp98505_c1 | 2159.77 | 36.4 kDa proline-rich protein [Solanum lycopersicum] |
| comp30496_c0 | 2015.98 | Translationally controlled tumor protein [Hevea brasiliensis] |
| comp30462_c0 | 1943.65 | Translationally-controlled tumor protein homolog [Oryza sativa subsp. Japonica] |
| comp87719_c1 | 1837.68 | Elongation factor 1-alpha [Manihot esculenta] |
| comp90471_c0 | 1715.29 | Chitinase C [Ananas comosus] |
| comp110306_c3 | 1632.62 | ADP-ribosylation factor [Medicago truncatula] |
To characterize the differentially expressed genes (DEGs) during somatic embryogenesis initiation in camphor tree, 3335 DEGs were singled out by comparing the three libraries in pairs (Additional file 8: Figure S3). As shown in Fig. 6a, more genes were down-regulated than up-regulated in the process of somatic embryogenesis. Between each two libraries, IZE_Suc vs IZE, SE_5w vs IZE_Suc and SE_5w vs IZE, 1729, 2027 and 1852 unigenes were differentially expressed, respectively (Additional file 9: Table S6). Of the DEGs, 216 unigenes were differentially expressed in all the three comparisons, 317 unigenes were specifically differential expressed between IZE and IZE_Suc, 598 between SE and IZE_Suc, and 363 between SE and IZE (Fig. 6b). These results indicated that osmotic stress pretreatment of IZE and culture of IZE_Suc on somatic embryo induction medium caused significant differential gene expression.
Fig. 6.
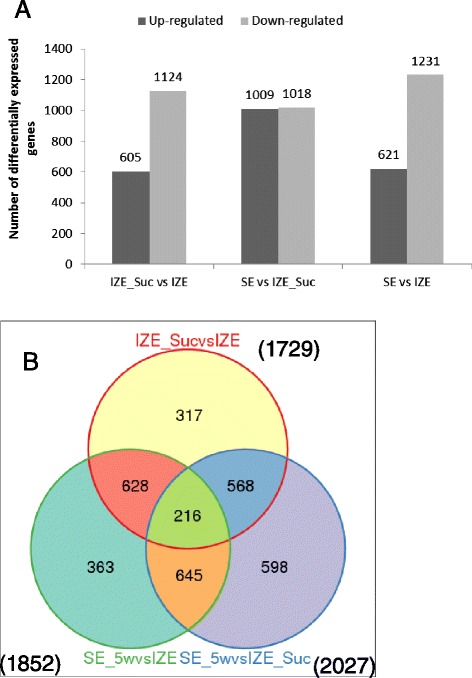
Venn diagram and Histogram of gene numbers differentially expressed during sucrose treatment and somatic embryo formation in camphor tree. a Histogram diagram showing the number of DEGs up- or down-regulated between different libraries; b A Venn diagram for analysis of the number of differentially expressed genes from IZE_Suc vs IZE, SE_5w vs IZE, and SE_5w vs IZE_Suc
To further investigate the expression profiles of the DEGs, they were divided into 6 groups based on the results of K-means clustering (Fig. 7). Group 1 and 2 contained genes positively or negatively modulated during the whole process of somatic embryo initiation and formation. Group 3 contained genes positively modulated during sucrose pretreatment, and then negatively modulated in the process of somatic embryo formation, while group 4 contained genes modulated in the opposite way in both processes. Genes in group 5 were up-regulated after sucrose pretreatment, and then down-regulated during somatic embryo formation, whose expressions were still higher than that of the initial IZE explant. Genes expressed with the opposite pattern of group 5 fell into group 6.
Fig. 7.
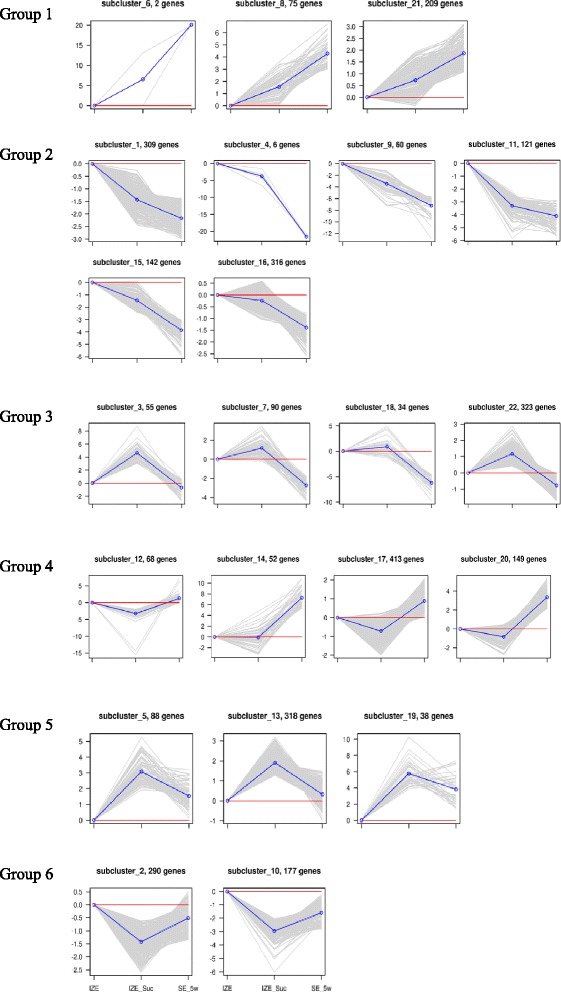
Cluster analysis of differentially expressed genes during somatic embryo induction in camphor tree based on K-means method
Functional annotation of differentially expressed genes
GO enrichment analysis for the DEGs was conducted to characterize the expression changes in the three samples with the whole transcriptome dataset as the background. The DEGs were assigned to 24 GO categories based on biological process (Fig. 8). The result showed that “response to stimulus”, “response to stress”, “response to abiotic stimulus” and “response to chemical stimulus” were among the most highly represented groups in the biological process category in the process of somatic embryogenesis. Other biological processes such as “oxidation-reduction process”, “metabolic process”, and “response to inorganic substance” were also identified. Some GO terms were identified only in specific comparison pair, for example, “carbohydrate metabolic process” and “sucrose metabolic process” were identified only in DEGs of IZE_Suc and IZE, while “response to salt stress”, “response to osmotic stress”, “sulfur compound metabolic process” and “metabolic process” were identified only in DEGs of SE_5w vs IZE_Suc. Among all the assigned DEGs, 1230 genes (SE_5w vs IZE) involved in “metabolic process” fell into the most highly represented group. The top ten GO terms (based on biological process) of up-regulated or down-regulated DEGs in the three comparisons are shown in Additional file 10: Table S7.
Fig. 8.
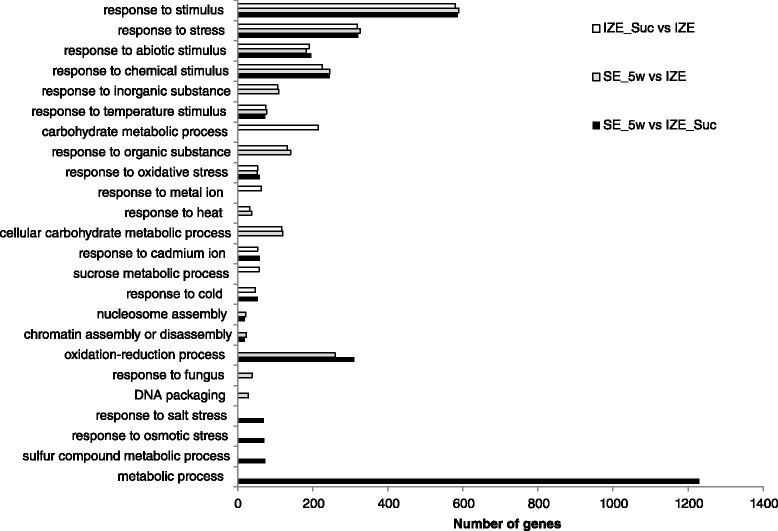
Functional categorization of genes differentially expressed during sucrose pretreatment and somatic embryo iniation based on biological process of Gene Ontology
All the DEGs were mapped to terms in KEGG database to search for metabolic or signal transduction pathways in which genes were significantly enriched, compared with the whole transcriptome background. In total, 3437 DEGs were assigned to 193 KEGG pathways, of which, 1061 DEGs (IZE_Suc vs IZE) were assigned to 172 pathways, 1345 DEGs (SE_5w vs IZE_Suc) to 181 pathways, and 1031 DEGs (SE_5w vs IZE) to 157 pathways, respectively. Pathway enrichment analysis revealed that the annotated changes between IZE_Suc and IZE were mainly involved in glycolysis/gluconeogenesis, starch and sucrose metabolism, plant hormone signal transduction, PPAR signaling pathway, plant-pathogen interaction, while the annotated changes between SE_5w vs IZE were mainly involved in plant hormone signal transduction, flavonoid biosynthesis, antigen processing and presentation, plant-pathogen interaction (Additional file 11: Table S8, Additional file 12: Figure S4).
Confirmation of somatic embryogenesis-related DEGs by qRT-PCR
With regard to the 897 somatic embryogenesis-related genes, expression data were shown in Additional file 13: Table S9 and summarized in Additional file 6: Table S5. To validate gene expression profiles obtained by RNA-seq, 52 DEGs related to somatic embryogenesis were selected for qRT-PCR analysis across eight different tissues of camphor tree: roots, stems, young leaves, young flowers, young fruits, IZE, IZE_Suc and SE_5w. The corresponding primers are listed in Additional file 2: Table S1. Based on the analyzed qRT-PCR data, all the 52 selected unigenes were expressed at varying levels in different tissues (Fig. 9). Seven unigenes, including CDC2_comp95642_c0, ERF_comp108680_c0, SAUR_comp102436_c2, SAUR_comp76441_c0, DREB/CBF_comp85800_c0, SMP_comp103536_c0, AIL_comp98550_c0, were highly expressed in IZE. HSP70_comp97938_c1, H3-1_comp103594_c1, HSP90_comp105220_c2, GH3_comp108441_c1, CEM6_comp87380_c1, H3-1_comp104541_c2, GST_comp104462_c0, HSP70_comp109861_c0, IAA_comp101833_c1, LEA_comp90735_c0, CAM_com96198_c0, CDPK_com99536_0, and GLU_comp95415_c0 showed strong expression in IZE_Suc, but relatively low expression in IZE and SE_5w. Expression of ARF_comp91137_c0, Chitinases_comp85229_c1, Chitinases_comp90471_c0, GST_comp106630_c0, HSP40_comp104158_c0 and GLU_comp107417_c3 were high in SE_5w, while low in IZE and IZE_Suc. These results confirmed that sucrose pretreatment induced differential expression of somatic embryogenesis-related genes, and these DEGs potentially play important roles during somatic embryo induction in C. camphora.
Fig. 9.
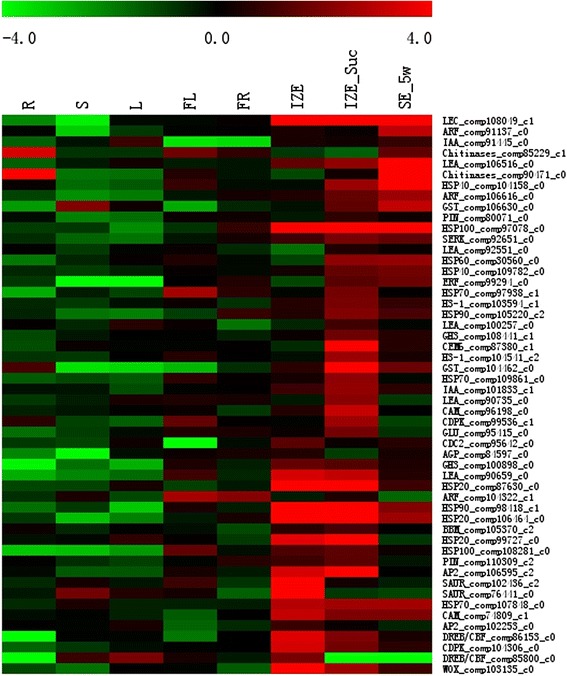
Expression patterns of 52 somatic embryogenesis-related DEGs in different tissues. The bar represents the scale of relative expression levels of DEGs, and the colors indicate relative signal intensities of DEGs. R roots, S stems, L young leaves, FL young flowers, FR young fruits, IZE immature zygotic embryos, IZE_Suc IZE pretreated with 1.0 M sucrose solution, SE_5w somatic embryos obtained from induction medium after culture for 5 weeks
The pearson correlation coefficient was calculated by SPSS to assess the correlation between the platforms of RNA sequencing and qRT-PCR. When the comparisons of IZE_Suc versus IZE, SE_5w versus IZE_Suc, and SE_5w versus IZE were performed, gene expression levels estimated by qRT-PCR were moderately or strongly correlated with RNA-Seq results (R2 = 0.6296, 0.6807 and 0.7439, respectively, correlation is significant at the 0.01 level), indicating the reliability of the RNA-seq analysis (Fig. 10).
Fig. 10.
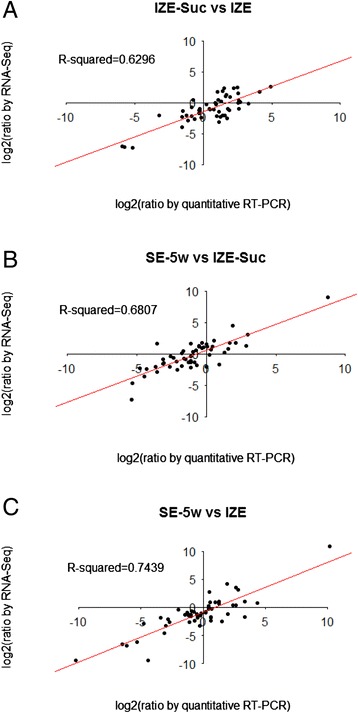
Correlation between RNA-seq and quantitative real-time PCR (qRT-PCR). Comparison of log2 fold change of 52 DEGs related to somatic embryogenesis obtained by RNA-seq and qRT-PCR for IZE_Suc vs IZE (a) SE_5w vs IZE_Suc (b) and SE_5w vs IZE (c) respectively
Discussion
Camphor tree is a non-model organism lacking genome and transcriptome resources. Though somatic embryogenesis-related genes have been extensively characterized in Arabidopsis, none have been published in camphor tree. In addition, the reported transcriptome analyses of somatic embryogenesis were mainly about plant hormone induced systems, few were about stress induced, particularly sucrose stress induced somatic embryogenesis. This study provides a de novo assembled transcriptome and comprehensive gene expression data during somatic embryogenesis induced by sucrose pretreatment. Gene expression changes during SE initiation and formation in camphor tree were focused in this study.
De novo transcriptome analysis of early somatic embryogenesis in camphor tree
Recalcitrance of plant explants to SE induction has long been an impediment to in vitro morphogenesis, which is especially relevant to woody species [43]. In camphor tree, various inducers including different plant hormones and carbon sources, used alone or in combination, failed to initiate efficient SE formation from IZEs, young leaves or flowers. Instead, we found SEs can be efficiently induced from IZEs by sucrose pretreatment without involvement of plant hormones [3]. The previous study indicated sucrose pretreatment other than plant hormones was the key factor for SE initiation in camphor tree. Such kind of direct somatic embryogenic system without exogenous application of plant hormones is invaluable in elucidating early regulatory events in embryo development [44]. As such, the explants of IZE, IZE_Suc, and the obtained SE_5w were selected for this study. Comparative transcriptome analysis of these samples provided an opportunity to examine the molecular aspects underlying early SE development.
Next-generation sequencing technology is especially suitable for gene expression profiling in somatic embryogenesis in such species. As a platform allowing generation of massive amounts of genomic resources rapidly and cost-effectively, this technology has already been used in transcriptome analysis of somatic embryogenesis in cotton [26, 45], hybrid yellow poplar [46], Japanese larch [47], Lycoris aurea [48], Longan [49, 50], and maize [51]. Before this study, only a few number of nucleotide sequences and ESTs of camphor tree were deposited in the NCBI database. Here we report a comprehensive analysis of transcriptome dynamics that may serve as a gene expression profile blueprint in somatic embryogenesis of camphor tree. One of our main goals was to adapt the RNA-Seq technology to this notable development process and to analyze the gene expression profile. De novo transcriptome assembly of the IZE, IZE_Suc and SE_5w by Illumina HiSeq 2000 resulted in a large amount of sequence and gene expression information. Molecular analysis was conducted to gain understanding of key events underlying the process of SE initiation in camphor tree. These sequences provided abundant information for further studies of somatic embryogenesis in camphor tree.
Differentially expressed somatic embryogenesis-related genes during embryogenic initiation
Somatic embryogenesis is a process during which genes were selectively expressed. In the present study, numerous somatic embryogenesis-related genes, including genes responsible for cell cycle and cell wall, hormone-responsive genes, genes in signal transduction pathway in somatic embryogenesis, and transcription factors were differentially expressed during SE induction (Fig. 9). Up-regulated expression of genes from families of GH3, PIN, Indoleacetic Acid-Induced Protein (Aux/IAA), ARF, HSP, Late Embryogenesis Abundant (LEA), CEM6, H3-1, SERK, Calmodulin (CAM), Calcium-Dependent Protein Kinase (CDPK), BBM, Apetala2 (AP2), and ERF was observed after sucrose treatment.
Exposing excised plant tissues to in vitro culture conditions containing high concentrations of auxin is the most used strategy to elicit somatic embryogenesis. Changes of gene expression have been observed in auxin-induced somatic embryogenesis via investigating the role of auxin signaling [52]. Auxin surges occurred in the process of somatic embryogenesis, which resulted in the isolation of several corresponding gene classes, including ARFs, Aux/IAAs, GH3s, PINs, and Small Auxin-up RNAs (SAURs) [30]. In addition, HSPs were also found to be auxin-responsive genes during SE development. Members of the HSP family have been reported to be highly expressed during the initiation of somatic embryogenesis by auxin [53]. Studies have confirmed that some genes of HSPs were expressed during the process of somatic embryo development in carrot [54] and alfalfa [55]. In this study, sucrose pretreatment induced significantly differential expression of transcripts from these gene families. These results indicated that although no auxin was applied for SE induction in camphor tree, the auxin-related genes may still function in SE induction.
LEA genes are abundantly expressed in late zygotic embryogenesis in many plant species [56]. Although LEA genes are well known ABA-inducible, the expression of ABA-inducible genes is not necessarily correlated with the level of ABA [57]. In camphor tree, up-regulated expression of LEA genes were observed in IZE_Suc, indicating osmotic stress is involved in regulating the expression of LEA genes. It is consistent with the result that the synthesis of LEA proteins occurs as soon as embryogenesis is initiated, which requires the application of a stress or exogenous ABA [5].
In addition to differentially expressed genes, various signal transduction pathways for activating or repressing numerous gene sets are also involved in the process of embryogeny acquisition [7]. SERKs, the first one of which was isolated from carrot suspension cultures up to the globular-shaped stage of embryogenesis [58], have been detected and identified in the process of somatic embryogenesis in various species [59–63]. In the present study, a SERK gene was up-regulated in IZE_Suc, indicating that the SERK gene plays a role in mediating SE initiation in camphor tree. CaM and CDPK are two of the three major classes of Ca2+ sensors, which might play an intermediary role during somatic embryogenesis [64, 65]. Genes of CaM and CDPK showed differential expression patterns between IZE and IZE_Suc, suggesting their potential functions in the early stage of somatic embryogenesis in camphor tree.
Several transcription factors, including genes from LEC, BBM, AP2, ERF, DREB, Wuschel-Related Homeobox (WOX) families that play regulatory roles in embryogenic processes [66, 67], were also found up or down regulated in this study, which indicated they might also be critical during somatic embryogenesis.
Differential expression of stress responsive genes during embryogenic initiation
Gene enrichment analysis showed significantly differential expression of genes responding to stress and stimulus (Fig. 8). Treating of explants with 0.5 M sucrose solution exposed the embryos to significantly different conditions from their original environment, which may induce differential expression of stress responsive genes. Plants deploy diverse molecular and cellular mechanisms to survive in stressful environments [68]. Some stress-related genes associated with early stages of SE including GSTs, Germin-like Proteins (GLPs), HSPs, Chitinases and β-1,3-Glucanases [8] were also observed differentially expressed during SE induction in our RNA-seq profiling. Members of GSTs superfamily, playing important roles in the overall natural defense mechanisms in all living organisms, have been shown up-regulated expression during auxin-induced somatic embryogenesis in soybean [25] and cotton [69]. Five GST transcripts present in our DEGs (Additional file 13: Table S9) were also up-regulated in the process of sucrose treatment. β-1,3-Glucanases belong to a large gene family. Members of β-1,3-Glucanases could be induced by pathogen attack or treatment with biotic or abiotic elicitors in plants [70]. Studies on expression of β-1,3-Glucanases during somatic embryogenesis process of Cichorium, spruce, and Araucaria angustifolia suggested that β-1,3-Glucanases may have implications in the somatic embryogenesis process. We found that one β-1,3-Glucanase gene (comp107417_c3) was highly expressed after sucrose treatment, which was also up-regulated during the process of somatic embryo formation (Additional file 13: Table S9).
Comparison of gene expression between somatic and zygotic embryos
SEs undergo a similar developmental program to zygotic embryos [5]. The same set of genes might be operating in both processes to specify embryo development [71]. However, some key differences exist between the two types of embryos, including the lack of surrounding embryo sac and differentiation of endosperm in somatic embryos. It has been observed somatic embryos exhibited more metabolic activity than zygotic embryos at parallel developmental stages in cotton [71]. In this study, the comparison of gene expression in SEs and zygotic embryos showed the number of down-regulated DEGs was much more than that of up-regulated DEGs in SEs. The GO annotations of DEGs revealed that genes response to stimulus or stress were significantly enriched. Secondary SE formation on the culture medium [3] and the results in the present study lead to the speculation that in vitro tissue culture conditions activate or suppress expression of stress responsive genes, which is in consistent with the suggestion that cells of cultured SEs underwent stress stimulation by exogenous compound in vitro, and SEs formation was the outcome of an in vitro adaption process to the culture environment [71].
Conclusions
In this work, de novo assembled transcriptomes of three embyogenic tissues of camphor tree (IZE, IZE_Suc and SE_5w samples) were analyzed and a large amount of sequence information was obtained. Gene expression profiles in the process of SE initiation induced by sucrose treatment and SE formation were investigated. Differential expression of genes potentially functioned in acquisition of embryogenic competence and stress response are of particularly interest. Differential expression of somatic embryogenesis-related genes (IZE_Suc vs IZE) indicates that sucrose induced somatic embryogenesis may share or partly share the mechanisms of somatic embryogenesis induced by plant hormones. This study provides new information about gene expression at early somatic embryogenesis stage, and meanwhile provides comprehensive gene expression data for camphor tree somatic embryogenesis that could serve as an important platform resource for further functional studies in plant embryogenesis.
Availability of supporting data
The Illumina sequence data from this study have been submitted as BioProject ID [PRJNA288748] to the NCBI sequence read archive under the accession number [SRP060394]. All the supporting data are included as additional files.
Acknowledgements
This study is funded by the National Natural Science Foundation of China (31100508), and the Ministry of Education of China (IRT13065).
Abbreviations
- BLAST
Basic local alignment search tool
- CTAB
Cetyltrimethyl ammonium bromide
- DEG
Differentially expressed gene
- GO
Gene ontology
- IZE
Immature zygotic embryo
- KEGG
Kyoto encyclopedia of genes and genomes
- KOG/COG
Clusters of orthologous groups
- NCBI
National center for biotechnology information
- Nr
Non-redundant database
- Nt
Nucleotide database
- qRT-PCR
Quantitative Real-Time PCR
- RIN
RNA integrity number
- RPKM
Reads per kilo bases per million mapped reads
- SE
Somatic embryo
- SPSS
Statistical product and service solutions
Additional files
Samples collected for transcriptome sequencing. (DOCX 470 kb)
Gene specific primers used in qRT-PCR. (XLSX 18 kb)
Length and number distribution of the unigenes. (XLSX 10 kb)
Functional annotation of non-redundant unigenes against Nr, Nt, Swiss-Prot, PFAM, GO and KOG databases. (XLSX 19135 kb)
Functional annotation of non-redundant unigenes against KEGG database. (XLSX 66 kb)
Expression of somatic embryogenesis-related genes in IZE, IZE_Suc and SE_5w in camphor tree based on the results of RNA-Seq. (XLSX 11 kb)
Frequency distribution of IZE, IZE_Suc and SE_5w by RPKM. (DOCX 222 kb)
Volcano plot of differential gene expression in IZE_Suc vs IZE, ZE_5w vs IZE_Suc and SE_5w vs IZE. (DOCX 256 kb)
List of DEGs singled out by comparing the libraries of IZE_Suc vs IZE, SE_5w vs IZE_Suc and SE_5w vs IZE. (XLSX 382 kb)
The top 10 GO terms of up-regulated and down-regulated DEGs in the three comparisons based on biological process. (XLSX 13 kb)
KEGG enrichment analysis of DEGs. (XLSX 19 kb)
Scatterplot of DEG enriched KEGG pathway in IZE_Suc vs IZE, ZE_5w vs IZE_Suc and SE_5w vs IZE. (DOCX 145 kb)
Differentially expressed DEGs associated with somatic embryogenesis. (XLSX 58 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MB and BZ designed the experiment and revised the manuscript. XS contributed to tissue sample collecting, transcriptome sequencing, preliminary data analysis, and manuscript writing. CZ carried out the qRT-PCR analysis, and participated in tissue sample collecting and data analysis. QL participated in tissue sample collecting and data analysis. ZZ contributed to the submission of the Illumina sequence data. All authors read and approved the final version of this manuscript.
Contributor Information
Xueping Shi, Email: xpshi@mail.hzau.edu.cn.
Cuijie Zhang, Email: zhangcuijie125@126.com.
Qinhong Liu, Email: azura007@163.com.
Zhe Zhang, Email: 13618632337@163.com.
Bo Zheng, Email: bo.zheng@mail.hzau.edu.cn.
Manzhu Bao, Email: mzbao@mail.hzau.edu.cn.
References
- 1.Babu KN, Sajina A, Minoo D, John CZ, Mini PM, Tushar KV, et al. Micropropagation of camphor tree (Cinnamomum camphora) Plant Cell Tiss Org. 2003;74(2):179–183. doi: 10.1023/A:1023988110064. [DOI] [Google Scholar]
- 2.Shi XP, Dai XG, Liu GF, Zhang JW, Ning GG, Bao MZ. Cyclic secondary somatic embryogenesis and efficient plant regeneration in camphor tree (Cinnamomum camphora L.) In Vitro Cellular & Developmental Biology-Plant. 2010;46(2):117–125. doi: 10.1007/s11627-009-9272-0. [DOI] [Google Scholar]
- 3.Shi XP, Dai XG, Liu GF, Bao MZ. Enhancement of somatic embryogenesis in camphor tree (Cinnamomum camphora L.): osmotic stress and other factors affecting somatic embryo formation on hormone-free medium. Trees-Structure and Function. 2009;23(5):1033–1042. doi: 10.1007/s00468-009-0345-9. [DOI] [Google Scholar]
- 4.Quiroz-Figueroa FR, Rojas-Herrera R, Galaz-Avalos RM, Loyola-Vargas VM. Embryo production through somatic embryogenesis can be used to study cell differentiation in plants. Plant Cell Tiss Org. 2006;86(3):285–301. doi: 10.1007/s11240-006-9139-6. [DOI] [Google Scholar]
- 5.Dodeman VL, Ducreux G, Kreis M. Zygotic embryogenesis versus somatic embryogenesis. J Exp Bot. 1997;48(313):1493–1509. doi: 10.1093/jexbot/48.313.1493. [DOI] [Google Scholar]
- 6.Willemsen V, Scheres B. Mechanisms of pattern formation in plant embryogenesis. Annu Rev Genet. 2004;38:587–614. doi: 10.1146/annurev.genet.38.072902.092231. [DOI] [PubMed] [Google Scholar]
- 7.Zimmerman JL. Somatic embryogenesis: a model for early development in higher plants. The Plant Cell. 1993;5(10):1411–1423. doi: 10.1105/tpc.5.10.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karami O, Saidi A. The molecular basis for stress-induced acquisition of somatic embryogenesis. Mol Biol Rep. 2010;37(5):2493–2507. doi: 10.1007/s11033-009-9764-3. [DOI] [PubMed] [Google Scholar]
- 9.Kiyosue T, Takano K, Kamada H, Harada H. Induction of somatic embryogenesis in carrot by heavy-metal ions. Canadian Journal of Botany-Revue Canadienne De Botanique. 1990;68(10):2301–2303. doi: 10.1139/b90-293. [DOI] [Google Scholar]
- 10.Ikeda-Iwai M, Umehara M, Satoh S, Kamada H. Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. Plant J. 2003;34(1):107–114. doi: 10.1046/j.1365-313X.2003.01702.x. [DOI] [PubMed] [Google Scholar]
- 11.Kamada H, Kobayashi K, Kiyosue T, Harada H. Stress-induced somatic embryogenesis in carrot and its application to synthetic seed production. In Vitro Cellular & Developmental Biology. 1989;25(12):1163–1166. doi: 10.1007/BF02621268. [DOI] [Google Scholar]
- 12.Kiyosue T, Satoh S, Kamada H, Harada H. Somatic embryogenesis in higher-plants. J Plant Res. 1993;75–82.
- 13.Santarem ER, Pelissier B, Finer JJ. Effect of explant orientation, pH, solidifying agent and wounding on initiation of soybean somatic embryos. In Vitro Cellular & Developmental Biology-Plant. 1997;33(1):13–19. doi: 10.1007/s11627-997-0034-6. [DOI] [Google Scholar]
- 14.Kamada H, Ishikawa K, Saga H, Harada H. Induction of somatic embryogenesis in carrot by osmotic stress. Plant Tissue Cult Lett. 1993;10:38–44. doi: 10.5511/plantbiotechnology1984.10.38. [DOI] [Google Scholar]
- 15.Zhou SJ, Brown DCW. High efficiency plant production of North American ginseng via somatic embryogenesis from cotyledon explants. Plant Cell Rep. 2006;25(3):166–173. doi: 10.1007/s00299-005-0043-z. [DOI] [PubMed] [Google Scholar]
- 16.You XL, Yi JS, Choi YE. Cellular change and callose accumulation in zygotic embryos of Eleutherococcus senticosus caused by plasmolyzing pretreatment result in high frequency of single-cell-derived somatic embryogenesis. Protoplasma. 2006;227(2–4):105–112. doi: 10.1007/s00709-006-0149-3. [DOI] [PubMed] [Google Scholar]
- 17.Karami O, Aghavaisi B, Mahmoudi Pour A. Molecular aspects of somatic-to-embryogenic transition in plants. J Chem Biol. 2009;2(4):177–190. doi: 10.1007/s12154-009-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feher A, Pasternak TP, Dudits D. Transition of somatic plant cells to an embryogenic state. Plant Cell Tiss Org. 2003;74(3):201–228. doi: 10.1023/A:1024033216561. [DOI] [Google Scholar]
- 19.Namasivayam P. Acquisition of embryogenic competence during somatic embryogenesis. Plant Cell Tiss Org. 2007;90(1):1–8. doi: 10.1007/s11240-007-9249-9. [DOI] [Google Scholar]
- 20.Sato S, Toya T, Kawahara R, Whittier RF, Fukuda H, Komamine A. Isolation of a carrot gene expressed specifically during early-stage somatic embryogenesis. Plant Mol Biol. 1995;28(1):39–46. doi: 10.1007/BF00042036. [DOI] [PubMed] [Google Scholar]
- 21.Su YH, Zhao XY, Liu YB, Zhang CL, O’Neill SD, Zhang XS. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009;59(3):448–460. doi: 10.1111/j.1365-313X.2009.03880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantiri FR, Kurdyukov S, Lohar DP, Sharopova N, Saeed NA, Wang XD, et al. The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol. 2008;146(4):1622–1636. doi: 10.1104/pp.107.110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng QL, Perry SE. Alterations in the transcriptome of soybean in response to enhanced somatic embryogenesis promoted by orthologs of AGAMOUS-Like15 and AGAMOUS-Like18. Plant Physiol. 2014;164(3):1365–1377. doi: 10.1104/pp.113.234062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng QL, Zheng YM, Perry SE. AGAMOUS-Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol. 2013;161(4):2113–2127. doi: 10.1104/pp.113.216275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thibaud-Nissen FO, Shealy RT, Khanna A, Vodkin LO. Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol. 2003;132(1):118–136. doi: 10.1104/pp.103.019968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu ZZ, Zhang CJ, Zhang XY, Liu CL, Wu ZX, Yang ZR, et al. Transcriptome profiling reveals auxin and cytokinin regulating somatic embryogenesis in different sister lines of cotton cultivar CCRI24. J Integr Plant Biol. 2013;55(7):631–642. doi: 10.1111/jipb.12073. [DOI] [PubMed] [Google Scholar]
- 27.Yang XY, Zhang XL, Yuan DJ, Jin FY, Zhang YC, Xu J. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 2012;12:110. [DOI] [PMC free article] [PubMed]
- 28.Sharma SK, Millam S, Hedley PE, McNicol J, Bryan GJ. Molecular regulation of somatic embryogenesis in potato: an auxin led perspective. Plant Mol Biol. 2008;68(1–2):185–201. doi: 10.1007/s11103-008-9360-2. [DOI] [PubMed] [Google Scholar]
- 29.Ge XX, Chai LJ, Liu Z, Wu XM, Deng XX, Guo WW. Transcriptional profiling of genes involved in embryogenic, non-embryogenic calluses and somatic embryogenesis of Valencia sweet orange by SSH-based microarray. Planta. 2012;236(4):1107–1124. doi: 10.1007/s00425-012-1661-7. [DOI] [PubMed] [Google Scholar]
- 30.Yang XY, Zhang XL. Regulation of somatic embryogenesis in higher plants. Crit Rev Plant Sci. 2010;29(1):36–57. doi: 10.1080/07352680903436291. [DOI] [Google Scholar]
- 31.Elhiti M, Stasolla C, Wang AM. Molecular regulation of plant somatic embryogenesis. In Vitro Cellular & Developmental Biology-Plant. 2013;49(6):631–642. doi: 10.1007/s11627-013-9547-3. [DOI] [Google Scholar]
- 32.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. [DOI] [PMC free article] [PubMed]
- 33.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genornes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 35.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed]
- 38.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 39.Wang LK, Feng ZX, Wang X, Wang XW, Zhang XG. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 40.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:2. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao XZ, Cai T, Olyarchuk JG, Wei LP. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21(19):3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 43.Rutledge RG, Stewart D, Caron S, Overton C, Boyle B, MacKay J, et al. Potential link between biotic defense activation and recalcitrance to induction of somatic embryogenesis in shoot primordia from adult trees of white spruce (Picea glauca). BMC Plant Biol. 2013;13:116. [DOI] [PMC free article] [PubMed]
- 44.Lakshmanan P, Taji A. Somatic embryogenesis in leguminous plants. Plant Biol. 2000;2(2):136–148. doi: 10.1055/s-2000-9159. [DOI] [Google Scholar]
- 45.Yang X, Zhang X, Yuan D, Jin F, Zhang Y, Xu J. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 2012;12:110. doi: 10.1186/1471-2229-12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T, Chen J, Qiu S, Zhang Y, Wang P, Yang L, et al. Deep sequencing and microarray hybridization identify conserved and species-specific microRNAs during somatic embryogenesis in hybrid yellow poplar. PLoS One. 2012;7(8):e43451. doi: 10.1371/journal.pone.0043451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Zhang SG, Han SY, Li XM, Qi LW. Transcriptome profiling and in silico analysis of somatic embryos in Japanese larch (Larix leptolepis) Plant Cell Rep. 2012;31(9):1637–1657. doi: 10.1007/s00299-012-1277-1. [DOI] [PubMed] [Google Scholar]
- 48.Wang R, Xu S, Jiang Y, Jiang J, Li X, Liang L, et al. De novo sequence assembly and characterization of Lycoris aurea transcriptome using GS FLX Titanium platform of 454 pyrosequencing. PLoS One. 2013;8(4):e60449. doi: 10.1371/journal.pone.0060449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin YL, Lai ZX. Comparative analysis reveals dynamic changes in mirnas and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.) PLoS One. 2013;8(4):e60337. doi: 10.1371/journal.pone.0060337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai ZX, Lin YL. Analysis of the global transcriptome of longan (Dimocarpus longan Lour.) embryogenic callus using Illumina paired-end sequencing. BMC Genomics. 2013;14:561. doi: 10.1186/1471-2164-14-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salvo SAGD, Hirsch CN, Buell CR, Kaeppler SM, Kaeppler HF. Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS One. 2014;9(10):e111407–7. doi: 10.1371/journal.pone.0111407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray WM. Hormonal regulation of plant growth and development. PLoS Biol. 2004;2(9):1270–1273. doi: 10.1371/journal.pbio.0020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitamiya E, Suzuki S, Sano T, Nagata T. Isolation of two genes that were induced upon the initiation of somatic embryogenesis on carrot hypocotyls by high concentrations of 2,4-D. Plant Cell Rep. 2000;19(6):551–557. doi: 10.1007/s002990050772. [DOI] [PubMed] [Google Scholar]
- 54.Pitto L, Schiavo FL, Giuliano G, Terzi M. Analysis of the heat-shock protein pattern during somatic embryogenesis of carrot. Plant Mol Biol. 1983;2(5):231–237. doi: 10.1007/BF01578641. [DOI] [PubMed] [Google Scholar]
- 55.Györgyey J, Gartner A, Németh K, Magyar Z, Hirt H, Heberle-Bors E, et al. Alfalfa heat shock genes are differentially expressed during somatic embryogenesis. Plant Mol Biol. 1991;16(6):999–1007. doi: 10.1007/BF00016072. [DOI] [PubMed] [Google Scholar]
- 56.Dure L, 3rd, Crouch M, Harada J, Ho TH, Mundy J, Quatrano R, et al. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol. 1989;12(5):475–486. doi: 10.1007/BF00036962. [DOI] [PubMed] [Google Scholar]
- 57.Finkelstein RR. Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryogenesis-abundant (lea) gene. Molecular & General Genetics : MGG. 1993;238(3):401–408. doi: 10.1007/BF00291999. [DOI] [PubMed] [Google Scholar]
- 58.Schmidt EDL, Guzzo F, Toonen MAJ, deVries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124(10):2049–2062. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- 59.Talapatra S, Ghoshal N, Sen Raychaudhuri S. Molecular characterization, modeling and expression analysis of a somatic embryogenesis receptor kinase (SERK) gene in Momordica charantia L. during somatic embryogenesis. Plant Cell Tiss Org. 2014;116(3):271–283. doi: 10.1007/s11240-013-0401-4. [DOI] [Google Scholar]
- 60.Ma J, He YH, Wu CH, Liu HP, Hu ZY, Sun GM. Cloning and molecular characterization of a SERK gene transcriptionally induced during somatic embryogenesis in Ananas comosus cv. Shenwan. Plant Mol Biol Rep. 2012;30(1):195–203. doi: 10.1007/s11105-011-0330-5. [DOI] [Google Scholar]
- 61.Perez-Nunez MT, Souza R, Saenz L, Chan JL, Zuniga-Aguilar JJ, Oropeza C. Detection of a SERK-like gene in coconut and analysis of its expression during the formation of embryogenic callus and somatic embryos. Plant Cell Rep. 2009;28(1):11–19. doi: 10.1007/s00299-008-0616-8. [DOI] [PubMed] [Google Scholar]
- 62.Baudino S, Hansen S, Brettschneider R, Hecht VFG, Dresselhaus T, Loerz H, et al. Molecular characterisation of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta (Berlin) 2001;213(1):1–10. doi: 10.1007/s004250000471. [DOI] [PubMed] [Google Scholar]
- 63.Thomas C, Meyer D, Himber C, Steinmetz A. Spatial expression of a sunflower SERK gene during induction of somatic embryogenesis and shoot organogenesis. Plant Physiol Biochem. 2004;42(1):35–42. doi: 10.1016/j.plaphy.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 64.Jansen MA, Booij H, Schel JH, de Vries SC. Calcium increases the yield of somatic embryos in carrot embryogenic suspension cultures. Plant Cell Rep. 1990;9(4):221–223. doi: 10.1007/BF00232184. [DOI] [PubMed] [Google Scholar]
- 65.Kiselev KV, Gorpenchenko TY, Tchernoded GK, Dubrovina AS, Grishchenko OV, Bulgakov VP, et al. Calcium-dependent mechanism of somatic embryogenesis in Panax ginseng cell cultures expressing the rolC oncogene. Mol Biol. 2008;42(2):243–252. doi: 10.1134/S0026893308020106. [DOI] [PubMed] [Google Scholar]
- 66.Jenik PD, Gillmor CS, Lukowitz W. Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Bi. 2007;23:207–236. doi: 10.1146/annurev.cellbio.22.011105.102609. [DOI] [PubMed] [Google Scholar]
- 67.Park S, Harada JJ. Arabidopsis embryogenesis. Methods Mol Biol. 2008;427:3–16. doi: 10.1007/978-1-59745-273-1_1. [DOI] [PubMed] [Google Scholar]
- 68.AbuQamar S, Luo HL, Laluk K, Mickelbart MV, Mengiste T. Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J. 2009;58(2):347–360. doi: 10.1111/j.1365-313X.2008.03783.x. [DOI] [PubMed] [Google Scholar]
- 69.Zeng FC, Zhang XK, Zhu LF, Tu LL, Guo XP, Nie YH. Isolation and characterization of genes associated to cotton somatic embryogenesis by suppression subtractive hybridization and macroarray. Plant Mol Biol. 2006;60(2):167–183. doi: 10.1007/s11103-005-3381-x. [DOI] [PubMed] [Google Scholar]
- 70.Mauch F, Mauch-Mani B, Boller T. Antifungal hydrolases in pea tissue: II. Inhibition of fungal growth by combinations of chitinase and beta-1,3-glucanase. Plant Physiol. 1988;88(3):936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin FY, Hu LS, Yuan DJ, Xu J, Gao WH, He LR, et al. Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: evidence for stress response functions in SE development. Plant Biotechnol J. 2014;12(2):161–173. doi: 10.1111/pbi.12123. [DOI] [PubMed] [Google Scholar]


