Abstract
Gram-negative facultative Aggregatibacter actinomycetemcomitans is an oral pathogen associated with periodontitis. The genetic heterogeneity among A. actinomycetemcomitans strains has been long recognized. This study provides a comprehensive genomic analysis of A. actinomycetemcomitans and the closely related nonpathogenic Aggregatibacter aphrophilus. Whole genome sequencing by Illumina MiSeq platform was performed for 31 A. actinomycetemcomitans and 2 A. aphrophilus strains. Sequence similarity analysis shows a total of 3,220 unique genes across the 2 species, where 1,550 are core genes present in all genomes and 1,670 are variable genes (accessory genes) missing in at least 1 genome. Phylogenetic analysis based on 397 concatenated core genes distinguished A. aphrophilus and A. actinomycetemcomitans. The latter was in turn divided into 5 clades: clade b (serotype b), clade c (serotype c), clade e/f (serotypes e and f), clade a/d (serotypes a and d), and clade e′ (serotype e strains). Accessory genes accounted for 14.1% to 23.2% of the A. actinomycetemcomitans genomes, with a majority belonging to the category of poorly characterized by Cluster of Orthologous Groups classification. These accessory genes were often organized into genomic islands (n = 387) with base composition biases, suggesting their acquisitions via horizontal gene transfer. There was a greater degree of similarity in gene content and genomic islands among strains within clades than between clades. Strains of clade e′ isolated from human were found to be missing the genomic island that carries genes encoding cytolethal distending toxins. Taken together, the results suggest a pattern of sequential divergence, starting from the separation of A. aphrophilus and A. actinomycetemcomitans through gain and loss of genes and ending with the divergence of the latter species into distinct clades and serotypes. With differing constellations of genes, the A. actinomycetemcomitans clades may have evolved distinct adaptation strategies to the human oral cavity.
Keywords: genomic islands, phylogeny, genomic structural variation, genetic variation, aggressive periodontitis, horizontal gene transfer
Introduction
Strain-to-strain heterogeneity in virulence is often observed in periodontal pathogenic species, and it has been evoked to explain the different clinical outcomes from colonization by the same pathogenic species (Neiders et al. 1989; Griffen et al. 1999). Such heterogeneity is a common theme and a focus of numerous studies in microbial pathogenicity (Finlay and Falkow 1989; Kilian et al. 2006). Elucidating the molecular basis of the heterogeneity may provide new insight to the pathogenesis and improve the microbial diagnosis of periodontitis.
Gram-negative facultative Aggregatibacter actinomycetemcomitans has been recognized as important in the etiology of periodontitis (Zambon 1985; Asikainen and Chen 1999; Fine et al. 2007). A number of studies have provided evidence for variable virulence potential among distinct genotypes and serotypes of A. actinomycetemcomitans (Asikainen et al. 1991; Asikainen et al. 1997; Kilian et al. 2006; Haubek et al. 2008; Chen et al. 2010). Since A. actinomycetemcomitans comprises discrete clonal lineages (Poulsen et al. 1994; Kilian et al. 2006), the underlying mechanisms for strain-to-strain variation in virulence (or other phenotypes) may be best understood via comparative genomics or transcriptomics, in which the entire genomes and their expression patterns are examined. Our initial comparative genomic analysis of 14 A. actinomycetemcomitans strains, as well as other studies, showed remarkable variation in large-scale genomic arrangement and gene content among different strains (Kittichotirat et al. 2010; Kittichotirat et al. 2011).
With the revelation of the extent of genomic variation within A. actinomycetemcomitans through comparative genomics, we recognized several unmet needs. First, the numbers of strains included in the previous comparative genomic analysis were unevenly distributed among clades. In particular, strains of serotypes d, e, and f were underrepresented. Second, sequencing and annotation errors are common especially when the technology used involves sequencing by synthesis methods. Data from a different sequencing technology are needed for comparison with the previous sequencing results to correct sequencing and annotation errors. Third, a detailed analysis of gene gain and loss between A. actinomycetemcomitans and the closely related Aggregatibacter aphrophilus, widely considered to have low virulence in periodontitis (Tempro and Slots 1986), may provide new insights into the evolution of A. actinomycetemcomitans as a periodontal pathogen.
In this study, we employed Illumina sequencing technology (San Diego, CA, USA) to resequence 17 previously sequenced strains of A. actinomycetemcomitans and 2 strains of A. aphrophilus. We also sequenced additional 12 strains of A. actinomycetemcomitans. The results were used to assess gene content, to identify accessory genes and genomic islands, to examine the inheritance patterns of the islands, and to evaluate the phylogenetic relationship among A. actinomycetemcomitans strains/lineages and between A. actinomycetemcomitans and A. aphrophilus.
Materials and Methods
Bacteria and Genome Sequencing
Appendix Table 1 lists the 31 A. actinomycetemcomitans strains and 2 A. aphrophilus strains used in this study. With the exception of 4 pairs of A. actinomycetemcomitans strains (SCC1398/SCC4092, I23C/S23A, SCC2303/AAS4a, and SCC393/A160; Sun et al. 2013), the strains were cultured from noncohabitant individuals. In some analyses, 1 strain from each pair was excluded to avoid introducing biases to the results. The new sequencing efforts failed for 2 of the 33 strains, and the previous sequence information for these 2 strains was used in this study.
Details of the sequencing protocols are provided in the Appendix Materials and Methods. The GenBank accession numbers of the 31 assembly results are as follows: SA2149 (AZTT00000000), SC383s (AZTR00000000), SA508 (AZTU 00000000), SA3733 (AZTV00000000), SA3033 (AZTW0000 0000), SA269 (AZTX00000000), SA2200 (AZTY00000000), SA2876 (AZTS00000000), SA3096 (AZTQ00000000), SC936 (AZTP00000000), ANH9776 (AZTZ00000000), SC29R (AZ TO00000000), D17P-3 (ADOA00000000), H5P-1 (AEJK000 00000), SCC1398 (AEJP00000000), SCC4092 (AJMF000000 00), I23C (AEJQ00000000), S23A (AJMH00000000), RHAA-1 (AHGR01000000), D17P-2 (ADOB00000000), SCC2302 (AEJR00000000), AAS4a (AJMG00000000), I63B (AEJL00 000000), SCC393 (AEJN00000000), A160 (AJME00000000), D18P-1 (AEJO00000000), D7S-1 (CP003496), HK1651 (CP0 08984), D11S-1 (CP001733), A. aphrophilus strain ATCC 33389 (AEWB00000000), and NJ8700 (CP009230).
Gene Prediction, Annotation, and Grouping
Details are provided in the Appendix Materials and Methods. Briefly, the NCBI Prokaryotic Genome Annotation Pipeline (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/) was used to process all the assembled sequences. Homologous genes were grouped across multiple genomes as a “gene cluster” based on their sequence homology, as described previously (Kittichotirat et al. 2011).
Phylogenetic Analysis
We selected 397 housekeeping genes (Appendix Table 2) that are present as single copies in all genomes and do not show variation in length of >10% across all isolates. The nucleotide sequences of individual genes were aligned with ClustalW (version 2; European Bioinformatics Institute, Hinxton, UK) with default parameters (Larkin et al. 2007). Gaps were removed from all alignment results, which in turn were concatenated to produce a single alignment (a total length of 335,400 base pairs [bp]). FastTree2 (http://www.microbesonline.org/fasttree/) was then used to create a maximum likelihood tree (Price et al. 2010). A bootstrap analysis was carried out to test the reliability of the tree. Finally, FigTree (version 1.4.0; http://tree.bio.ed.ac.uk/software/figtree/) was used to draw the phylogenetic tree of the strains studied.
Identification of Genomic Islands
Genomic islands of at least 5 kb in length were identified by a stepwise process as described previously (Kittichotirat et al. 2011; Appendix Materials and Methods).
Results
Genome Sequencing
A summary of the sequencing results is provided in Appendix Table 3. Comparison of assembly results based on 454 (454 Life Sciences Corporation, Branford, CT, USA), Illumina, and combined 454 and Illumina data shows that, for most genomes, our combined assembly approach produces better assembly results as indicated by smaller numbers of longer average-sized contigs. The mean of the genome size, the number of protein coding sequences and the coding density were 2,142,199 bp (SD = 114,729), 2,012 (SD = 112), and 85% (SD = 2.7%), respectively. The percentage guanine-cytosine content was 44 ± 0.8, similar to that reported in the previous study (Kittichotirat et al. 2011). The numbers of coding sequences were lower (and the genome sizes smaller) in serotypes b and c (≤1,927) than in other serotypes (≥2,044). Three genome sequences (SA508, SA3733, ANH9776) were found to have poor-quality assemblies with relatively higher numbers of contigs (549 to 787), shorter N50 length (3,625 to 5,568), and smaller-than-expected genome size (1.86 to 1.94 Mbp). The genome sequences of these strains are accessible via GenBank but were excluded in the analyses below, except for the phylogenetic analysis.
Phylogenetic Analysis
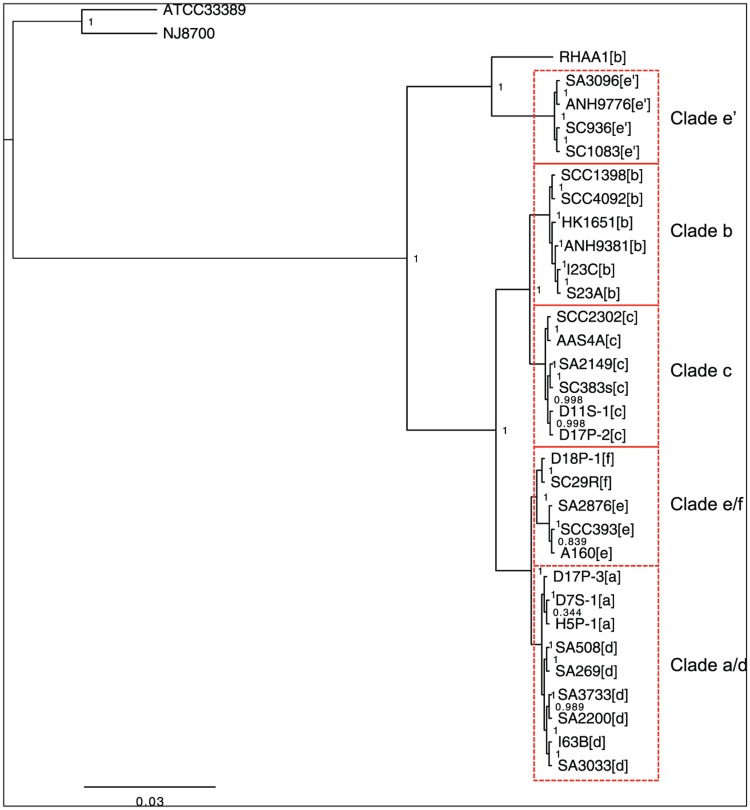
Figure 1 shows the phylogenetic trees built on 397 concatenated core genes (335,400 bp) according to the maximum likelihood method. A. actinomycetemcomitans strains were segregated from the 2 A. aphrophilus strains. A. actinomycetemcomitans RHAA-1, a strain isolated from rhesus monkey, was distinguished from the strains recovered from human. Among A. actinomycetemcomitans strains from human, the pairs of strains recovered from the same individuals (SCC1398/SCC4092, I23C/S23A, SCC2303/AAS4a, SCC393/A160) were closest to one another.
Figure 1.
Phylogenetic tree based on 397 concatenated core genes (335,400 base pairs) of 31 strains of Aggregatibacter actinomycetemcomitans and 2 strains of Aggregatibacter aphrophilus according to the maximum likelihood method. A. actinomycetemcomitans and A. aphrophilus were clearly segregated. RHAA-1 is distinguished from the human strains of A. actinomycetemcomitans. Strains from the same individuals (i.e., SCC1398/SCC4092, I23C/S23A, SCC2302/AAS4a, and SCC393/A160) were closest to one another than to other strains. Five clades, designated as e′, b, c, e/f, and a/d, were distinguished among A. actinomycetemcomitans strains from human. The tree further indicates a greater similarity between clades b and c and between clades a/d and e/f.
Strains of each serotype formed a distinct cluster, with varying degrees of similarity among serotypes. A. actinomycetemcomitans strains from human can be divided into 5 distinct clades, as shown in Figure 1. These include strains of serotype b (designated as clade b), serotype c (clade c), serotypes e and f (clade e/f), and serotypes a and d (clade a/d), with a phylogenetically distinct clade e′ containing serotype e strains. For clarity, serotype e strains of the clade e′ were henceforth referred to as serotype e′ strains.
Variation of Gene Content
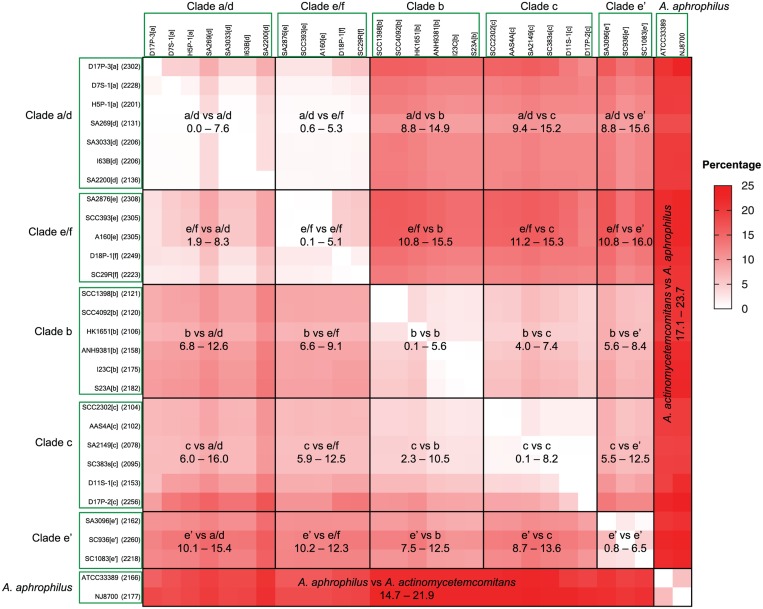
We next compared the gene content of the sequenced A. actinomycetemcomitans and A. aphrophilus. Appendix Figure 1 is a summary of an all-against-all comparison of gene content among strains. The difference in gene content between any 2 strains was analyzed by identifying genes present in one strain (shown by the row label) but absent in the other (shown by the column label) and vice versa. Figure 2 shows the comparison among clades of A. actinomycetemcomitans and between A. actinomycetemcomitans and A. aphrophilus. Five notable findings are highlighted here. First, the differences between the 2 strains recovered from the same individual (i.e., SCC1398/SCC4092, I23C/S23A, SCC2302/AAS4a and SCC393/A160) were 0.1% to 0.4% (Appendix Fig. 1). These strains were expected to exhibit little genetic differences. Therefore, the differences detected here may be in part due to sequencing and/or annotation errors. Second, the differences among all A. actinomycetemcomitans strains were 0% to 16.7%. Even when the nonhuman strain RHAA-1 was excluded, the maximum gene content difference still reached 16% (Appendix Fig. 1). Third, the pattern of gene content differences correlates with the phylogenetically defined clades (Fig. 2). Differences in gene content were much less between strains of the same clade than between strains of different clades. As an example, differences within clade a/d and clade b were 0% to 7.6% and 0.1% to 5.6%, respectively; yet, the differences between clade a/d and clade b were 8.8% to 14.9% and 6.8% to 12.6%. This indicates that gene content differences were fixed with an evolutionary history similar to the gene sequence differences. Fourth, the extent of gene content differences between clades correlates in general with the genetic distances among clades in the phylogenetic tree. This can be easily discerned from the heat map in Figure 2. For example, clade b and clade c were genetically closer to each other than to other clades (Fig. 1). Correspondingly the gene content differences between clade b and clade c were 2.3% to 10.5%, less than between either of these 2 clades and clade e′ (clade b vs. e′: 5.6% to 12.5%; clade c vs. e′: 5.5% to 13.6%), clade a/d (clade b vs. a/d: 6.8% to 14.9%, clade c vs. a/d: 6.0% to 15.2%), or clade e/f (clade b vs. e/f: 6.6% to 15.5%, clade c vs. e/f: 5.9% to 15.3%). Fifth, differences between A. actinomycetemcomitans and A. aphrophilus were 14.7% to 23.7%, indicating a smaller percentage of shared genes between these 2 species.
Figure 2.
Percentage of gene content differences among 5 Aggregatibacter actinomycetemcomitans clades and between A. actinomycetemcomitans and Aggregatibacter aphrophilus. A heat map indicator of percentage difference is provided to the right of the matrices. The number in each box represents the percentage range of gene content between strains of one clade (left) and the other (upper). See Appendix Figure 1 for details of the analysis by strains.
Features of Shared and Nonshared Genes
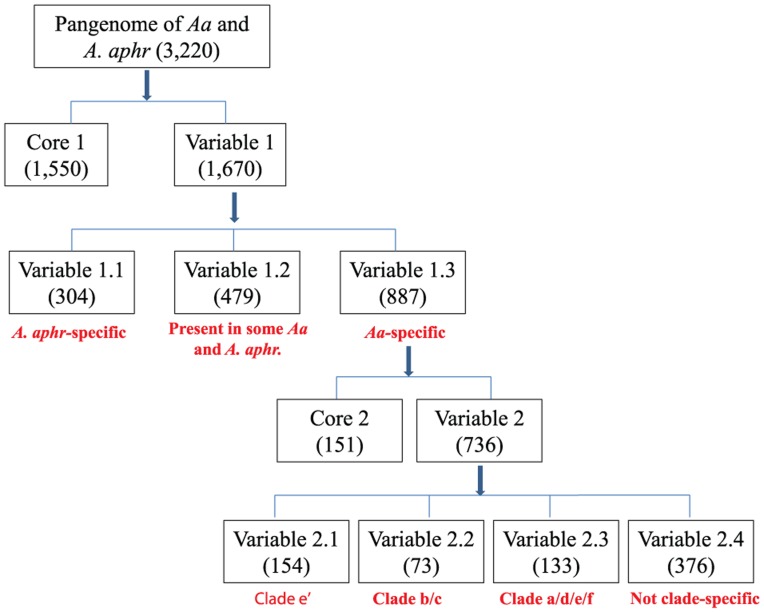
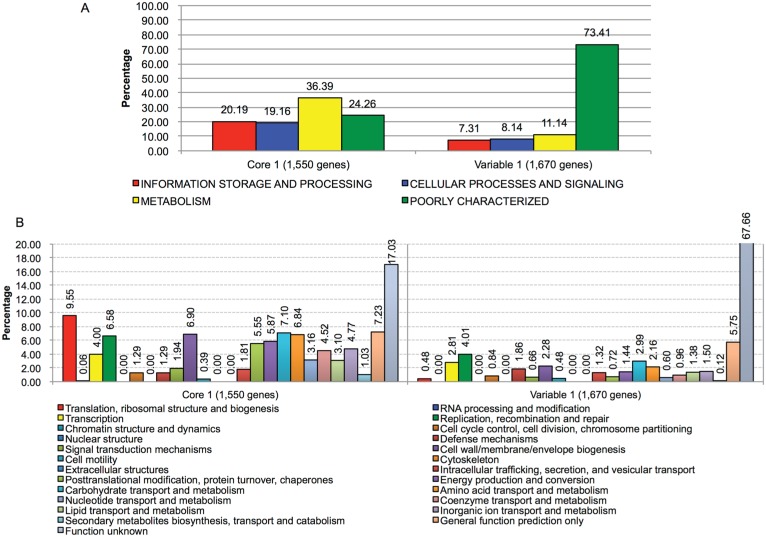
A. actinomycetemcomitans and A. aphrophilus may have evolved from a common ancestral species. Through gain and loss of genes, the organisms diverged to give rise to the modern A. aphrophilus and A. actinomycetemcomitans. We postulated that genes that are preserved throughout evolution may have features distinct from genes that are variably present in the genome. To test this hypothesis, we categorized the 3,220 genes that we identified on the basis of their distribution patterns among A. aphrophilus and A. actinomycetemcomitans as shown in Figure 3. The initial division was into a core 1 group (n = 1,550) found in every strain of both species and a variable 1 group of accessory genes (n = 1,670) found in some but not all strains. Figure 4 illustrates the distribution of the core 1 and variable 1 genes as classified by Clusters of Orthologous Groups. The majority of variable 1 group genes fell into the poorly characterized classification, with relatively lower numbers of genes in the other 3 major Clusters of Orthologous Groups classifications.
Figure 3.
Classification of genes based on their distributions in Aggregatibacter actinomycetemcomitans (Aa) and Aggregatibacter aphrophilus (A. aphr) and within each clade of Aa. These groups are as follows: core 1 (found in all Aa and A. aphr strains), variable 1 (found in some but not all Aa and A. aphr strains), variable 1.1 (found only in A. aphr but not in Aa), variable 1.2 (found in some strains of both species), variable 1.3 (found only in Aa but not in A. aphr), core 2 (found in all Aa), variable 2 (found in some but not all Aa), and variables 2.1 to 2.4 (genes specific to each of clades a/d/e/f, b/c, and e′).
Figure 4.
Clusters of Orthologous Groups gene classification of the core 1 and variable 1 gene groups identified in this study. The core 1 and variable 1 groups are defined in Figure 3. A total of 3,220 genes were identified by whole genome sequencing of Aggregatibacter aphrophilus and Aggregatibacter actinomycetemcomitans. The histograms show the percentage of genes assigned to the major groups (A; upper panel) and/or functional categories (B; lower panel) of Clusters of Orthologous Groups. A higher percentage of genes in variable 1 were assigned to poorly characterized (73.41%) or function unknown (67.66%) than in core 1 (24.26% and 17.08%, respectively).
The variable 1 group was further divided into subgroups (Fig. 3) for Clusters of Orthologous Groups assignments (Appendix Figs. 2–4). In general, the same trend was noted—that is, groups of genes that are variable in distribution among the species or phylogenetic groups are always dominated by poorly characterized genes, many of which may have been acquired by lateral gene transfer (see the description of genomic islands below). This is not a surprise, as many species, including A. aphrophilus and A. actinomycetemcomitans, have been extensively studied with only 1 or at most a few reference genomes. As a result, the functions of variable genes are less likely to be established, as the majority of variable genes are not present in any given isolate. Interestingly, several well-known virulence determinants of A. actinomycetemcomitans, such as cdtABC, ltxCABD, and catalase gene, were found in the group of variable 1, and cdtABC was further assigned to the group of variable 2. This result emphasizes a need for studies that focus on characterizing the functions of variably distributed genes for potential novel virulence determinants.
Genomic Islands
A total of 387 genomic islands were identified in 24 A. actinomycetemcomitans strains (Appendix Table 4). The genomic islands accounted for 10% to 71% (mean = 42%) of the accessory genes in each strain. These 387 genomic islands can be categorized into 165 groups based on at least 80% shared gene content. Hierarchical clustering analysis based on the distribution of genomic islands among strains is in general agreement with the phylogenetic tree based on the core genes (Appendix Fig. 5).
Major Distinctions between Serotype e and e′ Strains
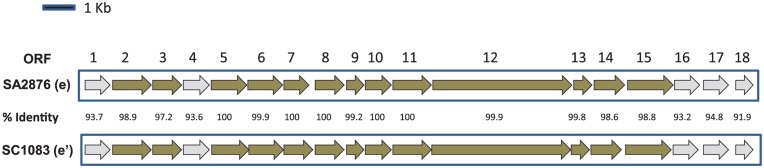
It is interesting to note that strains expressing serotype e–specific antigen were nonetheless divided into 2 phylogenetically distinct clades. Therefore, we further analyzed the previously published serotype e–specific gene cluster and compared notable features between serotype e and e′ strains. Figure 5 shows the nucleotide sequence homology of the serotype-specific gene clusters of 2 representative strains of clade e and clade e′. Within each clade, the nucleotide sequence homology was 99.8% to 100%. In contrast, the homology between clades was <95% nucleotide identity for 5 of the 18 open reading frames (ORFs). Interestingly, the ORFs necessary for the expression of serotype e antigen (ORFs 8 to 15) were highly conserved with a nucleotide sequence identity of 98.8% to 100%. As expected, phylogenetic analysis based on concatenated ORFs of the serotype-specific gene clusters of serotype e and e′ strains in this study also showed a clear distinction between these 2 clades (data not shown).
Figure 5.
Nucleotide sequence homology of serotype-specific gene clusters of serotype e and e′ strains. The 18 open reading frames (ORFs) of the homologous serotype e–specific gene clusters SA2876 (serotype e) and SC1083 (serotype e′) are shown. The percentage identity between the ORFs of the strains is provided in the middle. The ORFs with sequence identify ≤95% are represented by light gray arrows.
None of the serotype e′ strains in this study were found to have the cdtABC operon, which encodes the well-known virulence determinants (cytolethal distending toxins) of A. actinomycetemcomitans. A comparison of this locus between the 2 serotype e′ strains and a cdtABC-positive serotype a strain D7S-1 is provided in Appendix Figure 6. The locus is flanked at one side by a tRNA gene (tRNA-Gly), which is a recognized hot spot for the integration of genomic islands (Blum et al. 1994; Hacker et al. 1997). In strain D7S-1, the cdtABC is carried on a 24-kb genomic island. A 55-bp intergenic region was located in the same locus in strain SC1083. In contrast, in strain SC936, a distinct 34-Kb island was identified in the same locus.
Discussion
A number of clinical studies have supported the idea that not all strains of periodontal pathogenic species are equal in their virulence potentials (Asikainen et al. 1997; Griffen et al. 1999; Haubek et al. 2008). Assessing the molecular basis for such heterogeneity may identify novel strain- or clone-specific pathogenic mechanisms and be used to develop diagnostic and prognostic tests of periodontitis. While A. actinomycetemcomitans is adopted as a model in our study, the results may lead to a general understanding in the evolution of pathogenic mechanisms of periodontopathic species.
Bacteria evolve through point mutations in existing genes as well as through gain and loss of genes (Ochman and Davalos 2006). Some of the changes may provide advantages to the bacteria and become fixed in the genome. Gain of genes via horizontal gene transfer has been shown to be a major driving force in the evolution of bacteria (Lawrence and Ochman 1998; Maurelli et al. 1998; Sokurenko et al. 1999; Lan and Reeves 2000; Ochman et al. 2000). Concomitant with the lack of genetic exchanges among bacterial strains, mutational variants within a bacterial species may gradually diverge into distinct clonal lineages, each with distinct biological properties. Accumulating evidence has suggested that A. actinomycetemcomitans comprises several genetically diverse strains (Poulsen et al. 1994; Kilian et al. 2006). Preliminary evidence (from serotyping or genotyping) pointing to differential association between specific groups of strains and either periodontal health or disease has suggested that there may be variable virulence potentials among strains (Asikainen et al. 1991; Haubek et al. 1995; Asikainen et al. 1997; Haubek et al. 2008).
The aim of this study was to provide a comprehensive genomic analysis of A. actinomycetemcomitans to support future investigations of the adaptation mechanisms, including virulence determinants, of the species in the human oral cavity. Whole genome sequencing has the advantage of providing a thorough catalogue of gene content and, in the case of the closed genomes, information on the genomic arrangement. The results from this study showed a strong consensus among the analyses of nucleotide sequences of the core genes, the whole genome gene content, and the distribution pattern of genomic islands. Collectively, the results suggest a sequential divergence of the 2 species A. aphrophilus and A. actinomycetemcomitans, beginning with the division of A. actinomycetemcomitans into 3 groups represented by clade e′, clade b/c, and clade a/d/e/f, then subsequent division into 5 clades, and a final division into individual serotypes. Since the distribution of genomic islands follows the same general scheme of divergence, these islands appear to have been acquired early in the evolution of A. actinomycetemcomitans, stably fixed in the genomes, and passed on vertically within each lineage. Moreover, the smaller genome sizes of serotype b and c strains may suggest a genome reduction of these 2 clonal lineages.
Poulsen et al. (1994) examined the population structure of A. actinomycetemcomitans, using multilocus enzyme electrophoretic analysis to study the 5 serotypes (a to e) known at the time. The results showed that each of the serotypes a, b, c, d, and e included genetically separated populations of A. actinomycetemcomitans. Two distinct lineages were found within serotype a. Our results are largely in agreement with those of Poulsen et al. (1994). Our phylogenetic analysis based on the concatenated 395 core genes showed that each of serotypes a to e was distinct, which was supported by the analyses of gene content and the distribution of genomic islands. The results also showed greater similarity among serotypes a, d, e, and f and between serotypes b and c. We did not find genetic distinction within serotype a strains. In contrast, we found that the serotype e strains were segregated into 2 groups, 1 of which was designated clade e′. The serotype e′ strains bore the 16S rRNA signature sequence of a group of phylogenetically distinct serotype e strains identified by van der Reijden et al. (2010). Sequence analysis of the serotype e antigen–specific gene clusters of these strains also supported the genetic distinction between serotype e and serotype e′ strains. Interestingly, serotype e′ strains were characterized by the lack of cdtABC, which was found to be carried on a genomic island in various A. actinomycetemcomitans strains. This suggests that serotype e′ strains may be of low virulence potentials.
A typical A. actinomycetemcomitans bacterium contains approximately 2,000 identified genes, but gene content can vary by as much as 16% between strains. Therefore, >300 genes may be found in 1 strain but not in another and vice versa. The remarkable extent of gene content variation may result in distinct phenotypes among A. actinomycetemcomitans strains. Note that in the paper by Socransky et al. (1998), in which different bacterial species were organized into distinct communities, serotypes a and b of A. actinomycetemcomitans appeared to be different in their communities. While serotype a was associated with Green complex bacterial species, serotype b was a stand-alone species. Perhaps different lineages of A. actinomycetemcomitans may have evolved to use different strategies for adaptation to the human oral cavity.
The genes that are variable in their distribution (e.g., group variable 2 in Fig. 3) include known virulence determinants such as cdtABC, but the majority of these genes have no assigned functions. We speculate that some other accessory genes may encode novel virulence determinants. Experiments are underway to explore this possibility.
In conclusion, this study has identified major clonal lineages of A. actinomycetemcomitans that differ in gene content, nucleotide sequence homology, distribution of genomic islands, and presence of virulence determinants. The information provides a logical framework and a resource of detailed sequence data for further investigations into the adaptation and the pathogenesis of A. actinomycetemcomitans in the human oral cavity.
Author Contributions
W. Kittichotirat, R.E. Bumgarner, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript; C. Chen, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This work was supported by the National Institutes of Health (R01 DE012212).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Asikainen S, Chen C. 1999. Oral ecology and person-to-person transmission of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Periodontol 2000. 20:65–81. [DOI] [PubMed] [Google Scholar]
- Asikainen S, Chen C, Saarela M, Saxen L, Slots J. 1997. Clonal specificity of Actinobacillus actinomycetemcomitans in destructive periodontal disease. Clin Infect Dis. 25 Suppl 2:S227–S229. [DOI] [PubMed] [Google Scholar]
- Asikainen S, Lai CH, Alaluusua S, Slots J. 1991. Distribution of Actinobacillus actinomycetemcomitans serotypes in periodontal health and disease. Oral Microbiol Immunol. 6(2):115–118. [DOI] [PubMed] [Google Scholar]
- Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschäpe H, Hacker J. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 62(2):606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wang T, Chen W. 2010. Occurrence of Aggregatibacter actinomycetemcomitans serotypes in subgingival plaque from United States subjects. Molecular Oral Microbiology. 25(3):207–214. [DOI] [PubMed] [Google Scholar]
- Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, McKiernan M, Gunsolley J. 2007. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J Clin Microbiol. 45(12):3859–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Falkow S. 1989. Common themes in microbial pathogenicity. Microbiol Rev. 53(2):210–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Lyons SR, Becker MR, Moeschberger ML, Leys EJ. 1999. Porphyromonas gingivalis strain variability and periodontitis. J Clin Microbiol. 37(12):4028–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Blum-Oehler G, Muhldorfer I, Tschäpe H. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 23(6):1089–1097. [DOI] [PubMed] [Google Scholar]
- Haubek D, Ennibi OK, Poulsen K, Vaeth M, Poulsen S, Kilian M. 2008. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet. 371(9608):237–242. [DOI] [PubMed] [Google Scholar]
- Haubek D, Poulsen K, Asikainen S, Kilian M. 1995. Evidence for absence in Northern Europe of especially virulent clonal types of Actinobacillus actinomycetemcomitans. J Clin Microbiol. 33(2):395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian M, Frandsen EV, Haubek D, Poulsen K. 2006. The etiology of periodontal disease revisited by population genetic analysis. Periodontol 2000. 42:158–179. [DOI] [PubMed] [Google Scholar]
- Kittichotirat W, Bumgarner RE, Asikainen S, Chen C. 2011. Identification of the pangenome and its components in 14 distinct Aggregatibacter actinomycetemcomitans strains by comparative genomic analysis. PLoS One. 6(7):e22420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittichotirat W, Bumgarner R, Chen C. 2010. Markedly different genome arrangements between serotype a strains and serotypes b or c strains of Aggregatibacter actinomycetemcomitans. BMC Genomics. 11:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R, Reeves PR. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8(9):396–401. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. 1998. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci U S A. 95(16):9413–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli AT, Fernández RE, Bloch CA, Rode CK, Fasano A. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci U S A. 95(7):3943–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiders ME, Chen PB, Suido H, Reynolds HS, Zambon JJ, Shlossman M, Genco RJ. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res. 24(3):192–198. [DOI] [PubMed] [Google Scholar]
- Ochman H, Davalos LM. 2006. The nature and dynamics of bacterial genomes. Science. 311(5768):1730–1733. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature. 405(6784):299–304. [DOI] [PubMed] [Google Scholar]
- Poulsen K, Theilade E, Lally ET, Demuth DR, Kilian M. 1994. Population structure of Actinobacillus actinomycetemcomitans: a framework for studies of disease-associated properties. Microbiology. 140(Pt 8):2049–2060. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One. 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr 1998. Microbial complexes in subgingival plaque. J Clin Periodontol. 25(2):134–144. [DOI] [PubMed] [Google Scholar]
- Sokurenko EV, Hasty DL, Dykhuizen DE. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7(5):191–195. [DOI] [PubMed] [Google Scholar]
- Sun R, Kittichotirat W, Wang J, Jan M, Chen W, Asikainen S, Bumgarner R, Chen C. 2013. Genomic stability of Aggregatibacter actinomycetemcomitans during persistent oral infection in human. PLoS One. 8(6):e66472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempro PJ, Slots J. 1986. Selective medium for the isolation of Haemophilus aphrophilus from the human periodontium and other oral sites and the low proportion of the organism in the oral flora. J Clin Microbiol. 23(4):777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Reijden WA, Brunner J, Bosch-Tijhof CJ, van Trappen S, Rijnsburger MC, de Graaff MP, van Winkelhoff AJ, Cleenwerck I, de Vos P. 2010. Phylogenetic variation of Aggregatibacter actinomycetemcomitans serotype e reveals an aberrant distinct evolutionary stable lineage. Infect Genet Evol. 10(7):1124–1131. [DOI] [PubMed] [Google Scholar]
- Zambon JJ. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 12(1):1–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.