Abstract
Background
Artificial light at night (ALAN) is a typical feature of urban areas and most organisms living in urban or suburban habitats are exposed to low levels of ALAN. Light is one of the most important environmental cues that organisms use to regulate their activities. Studies have begun to quantify the influence of ALAN on the behavior and ecology of organisms, but research on the effects at the molecular level remains limited. Mosquitoes in the Culex pipiens complex (Diptera, Culicidae) are widespread and abundant in urban areas where they are potential disease vectors. It is thus of particular interest to understand how ALAN may influence biologically and ecologically relevant traits.
Results
We used RNAseq to evaluate the transcriptome response in a Cx. pipiens f. molestus laboratory population that was exposed to near-natural light conditions (light:dark L16:D8 hours, “control”) and ALAN conditions with 3 h of constant low-level light at night (L16 + Llow3:D5 hours, “low-light”). The resulting transcripts were mapped to the reference genome of the closely related Culex quinquefasciatus. Female expression patterns differed significantly between control and treatment conditions at five genes although none showed an absolute fold change greater than two (FC > 2). In contrast, male expression differed at 230 genes (74 with FC > 2). Of these, 216 genes (72 with FC > 2) showed reduced expression in the low-light treatment, most of which were related to gametogenesis, lipid metabolism, and immunity. Of the 14 genes (two with FC > 2) with increased expression, only five had any functional annotation. There was a pronounced sex-bias in gene expression regardless of treatment, with 11,660 genes (51 % of annotated genes; 8694 with FC > 2; 48 % of annotated genes) differentially expressed between males and females, including 14 genes of the circadian clock.
Conclusion
Our data suggest a stronger response to artificial light by males of Cx. pipiens f. molestus than by females, and that a wide range of physiological pathways may be affected by ALAN at the molecular level. The fact that differences in gene expression appear to be sex-specific may have a strong influence at the population level.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2336-0) contains supplementary material, which is available to authorized users.
Keywords: RNAseq, Diptera, Culicidae, Gene expression, Transcriptome, ALAN, LED
Background
Artificial light at night (ALAN) is a typical characteristic of the habitat for organisms living in urban and suburban environments. Along with temperature, light is one of the most important environmental cues for all living things [1], playing a key role in regulating daily (i.e., foraging, photosynthesis) and seasonal (i.e., migration, diapause) activity. As a result, changes to light regimes can play a major role in the ecology and physiology of a wide range of organisms and ecosystems [1, 2]. The addition of ALAN, whereby light of a different spectrum and intensity to natural light is present during naturally dark phases, has been demonstrated to affect metabolism [2, 3], behaviour [4] and circadian clock regulation [5–8] in numerous organisms. Nonetheless, our understanding of the effects of artificial illumination remains limited to a relatively small number of species and ecosystems.
Mosquitoes (Diptera, Culicidae) are an ecologically important group of insects and several species occur in urban and suburban areas in close proximity to humans, where they are exposed to artificial light at night [9–11]. The Culex pipiens complex is a widespread and abundant group of mosquitoes [12] and blood-feeding females act as vectors of vertebrate diseases (e.g. West Nile virus, avian malaria) [13]. Male Cx. pipiens respond directly to light cues and the onset of low light by becoming active and starting to swarm [14]. Although the potential impact of light on circadian rhythms has been studied [8, 15, 16], there remain gaps in understanding whether ALAN evokes changes at the molecular level on processes that may not be directly involved with the circadian clock. Most Cx. pipiens research is focused on females because of their role in disease transmission; however, studies of population-level responses to environmental change must also consider the consequences of artificial light for males. There are pronounced differences in the behaviour and physiology of male and female mosquitoes, e.g. food preference and daily activity [8]. At the molecular level, the only study of which we are aware that reported sex-biased gene expression patterns in adult mosquitoes analysed the malaria vector Anopheles gambiae [17].
A number of physiological pathways could potentially be affected by environmental cues from artificial illumination at night. The genes of the circadian clock are expected to be affected by light because the blue-light receptor cryptochrome-1 [18, 19] receives environmental light cues that are used in clock synchronisation [13]. Rund et al. [8] suggested that a number of additional genes are expressed in diel patterns in response to the surrounding light environment. These genes relate to metabolic detoxification, immunity, and nutrient sensing (e.g. glutathione-S-transferase, serine protease inhibitor and takeout genes, respectively) [8]. This implies that physiological processes not under direct circadian control may nonetheless be influenced by artificial light, with effects discernible at the molecular level.
Here we tested how exposure to low levels of artificial light at night alters gene expression in Cx. pipiens f. molestus by combining controlled laboratory experiments with a near-natural light regime (“control” consisting of Light(L)16 h:Dark(D)8 h) compared to an ALAN light regime (“low-light”, L16h:LLow3h:D5h; Fig. 1) and transcriptome (i.e., the complete set of transcripts in a cell, and their quantity [20]) analysis using RNAseq. The genome of the closely related Cx. quinquefasciatus was used as a reference in our analysis.
Fig. 1.
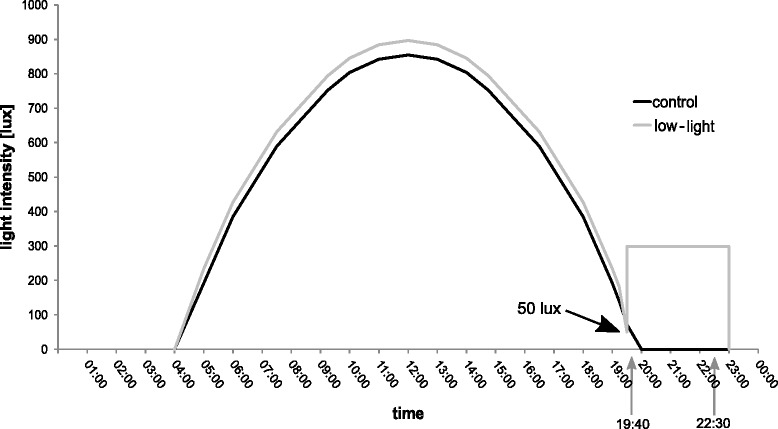
Light regime throughout the experiment. Light intensities, in lux, against clock time throughout the day. The dark line depicts “control” (L16:D8h) and the grey line refers to “low-light” regime. Arrows delimit sampling times that were used as biological replicates after preliminary alanysis (see text). Before “low-light” (L16 + Llow3:D8h) regime reached the constant additional light at night level (300 lux), light intensity smoothly droped to 50 lux. “Control” light regime decreased to darkness uninterrupted
Culex pipiens includes two forms (ecotypes) with partially sympatric distributions, pipiens and molestus. Females of the pipiens form depend on blood meals (anautogeny), and males build mating swarms, whereas females of the molestus form are autogenous and mating can take place in confined spaces. These characteristics make the molestus form highly suited to rearing in the laboratory, allowing for testing of different light regimes under otherwise constant conditions.
Our analysis of the transcriptome revealed a strong sex-bias in gene expression regardless of treatment (approximately 50 % of annotated genes). Females generally exhibited little response to ALAN treatment (up to 5 differentially expressed genes), whereas males exhibited a pronounced response (up to 230 genes), with most changes being reduced expression across a range of gene functions. Our findings suggest that a wide array of physiological pathways may be affected at the molecular level by ALAN.
Results
Sequencing of pooled samples (seven individuals per pool, whole bodies) yielded a total of 195.5 million 100-bp paired-end reads (~400 million total reads) giving an average of ~24.5 million read pairs per sample across eight samples (range 21,589,314 – 28,382,099 read pairs). The reads were mapped to the Culex quinquefasciatus reference genome assembly version CpipJ1.21. Approximately 37–57 % reads aligned to the reference (37–45 % in males, 55–57 % in females) with 17,573 of 22,985 genes receiving at least 1 aligned read. Further annotations, including gene ontology (GO) terms, were obtained from the UniProt-GOA C. quinquefasciatus proteome annotation (see Methods). Because gene expression may show temporal variation [8], we first included treatment and sampling timepoint as factors using negative binomial generalized linear models implemented in the R package DESeq2 version 1.8.2 [21]. Genes were considered differentially expressed when the p-value was <0.05. All p-values reported in the text refer to the p-values adjusted after Benjamini-Hochberg as implemented in DESeq2. Of theses we additionally report the number of genes that showed an absolute 2-fold change in expression (FC > 2).
Only two genes were differentially expressed (FC > 2) according to both treatment and sampling timepoint (in males). Based on this, we ran a second model that treated the two timepoints as replicates and included only treatment as a factor. The results of this second model (i.e. with two replicates per sample) are reported below.
Differential expression testing — artificial light treatment
In response to light treatment 230 (74 > 2FC) genes were differentially expressed in males. Of the genes with >2FC, two were more highly expressed in the “low-light” condition, while 72 genes were more highly expressed in the “control” (Fig. 2a; Additional file 1).
Fig. 2.
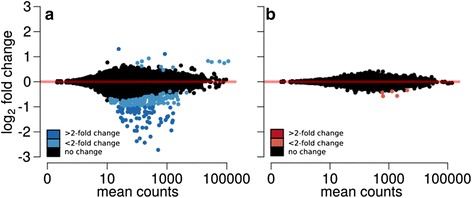
Ratio average plots of differentially expressed genes between treatments. Differentially expressed genes between treatments separately for (a) males and (b) females. The different colours refer to the differences in fold changes (FC). Positive and negative fold changes indicate genes with treatment- and control -biased expression respectively
In contrast, the change in gene expression of females never exceeded the >2FC cut-off although five genes had a p-value < 0.05 (Fig. 2b; Additional file 2).
In males, a striking signature of gametogenesis-related gene expression was identified in the response to artificial light, where expression of genes involved in DNA replication, mitosis, meiosis, spermatogenesis and germ cell proliferation was reduced in artificially lit conditions (Additional file 1). This included a substantial portion of the DNA replication machinery, i.e. all six DNA replication licensing factors (mcm2-7); the components of the origin recognition complex geminin, orc1 and cdt1; DNA polymerase; and an ortholog of fizzy which regulates transition to meiosis. There was clear coordinate expression of several genes with crucial roles in spermatogenesis, such as ance [22] and importin alpha [23]. This includes the major regulator of zygotic genome activation smaug, and the cognate activating kinase pan gu (Additional file 1). Ten additional genes were downregulated that are orthologs of genes that exhibit germ-line stem cell bias in Drosophila [24], as well as a gene encoding a putative uncharacterized protein that possesses a meiosis arrest protein family domain (IPR024768, Additional file 1). Gene ontology (GO) term enrichment analysis also highlighted the signature of gametogenesis with enrichment of GO terms related to gametogenesis (Additional file 3) including “cellular process involved in reproduction in multicellular organism” (GO:0022412), “gamete generation” (GO:0007292), “germ cell development” (GO:0007281), and “regulation of cell cycle” (GO:0051726).
Differential expression of immune genes was also observed, with reduced expression in the artificial light treatment of genes encoding transferrin and orthologs of the Drosophila genes yellow-f and yellow-h which encode dopachrome conversion enzymes that participate in the melanisation response [25]. Three genes encoding proteins with functions in lipid metabolism and transport were also differentially expressed in males in response to artificial light: apolipoprotein and two genes encoding sterol acyltransferases (Additional file 1).
Differential expression testing — sex-biased gene expression
Simultaneous analysis of males and females was performed to test for the main effect of sex across all samples. Independent of treatment, 11,660 genes (ca. 51 % of annotated genes) were differentially expressed between males and females (Fig. 3; Additional file 4).
Fig. 3.
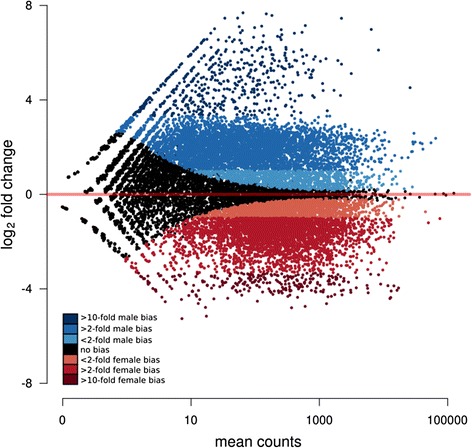
Ratio average plot of differential expression between males and females. The different colours refer to the differences in fold change (FC) sex bias; blue for males, red for females. MB, male-biased; FB, female-biased
Of these, 8694 genes exhibited at least a 2-fold sex bias (48 % of annotated genes). Fourteen genes related to circadian clock function were differentially expressed between males and females (Table 1). The genes photolyase, disc overgrown protein kinase and timeout/timeless-2 were male-biased while the rest were female-biased (Table 1).
Table 1.
circadian clock and related genes differentially expressed in females (negative values) and in males (positive values)
| Gene name | Gene id | Fold change | SE | |
|---|---|---|---|---|
| Canonical clock genes | Clocka | CPIJ002146 | 1.811 | 0.177 |
| Clocka - continued | CPIJ002147 | 1.879 | 0.130 | |
| cryptochrome-1 | CPIJ009455 | 1.310 | 0.126 | |
| photolyaseb | CPIJ017734 | -1.969 | 0.191 | |
| Duplication of cryptochrome-2c | CPIJ015481 | 1.749 | 0.152 | |
| cryptochrome-2 | CPIJ018859 | 1.528 | 0.111 | |
| cycled | CPIJ014938 | 1.512 | 0.195 | |
| period | CPIJ007193 | 1.561 | 0.166 | |
| timeless protein | CPIJ007082 | 1.666 | 0.117 | |
| Genes related to circadian rhythm | timeout/timeless-2e | CPIJ000660 | −2.586 | 0.193 |
| par domain proteinf | CPIJ014920 | 1.729 | 0.135 | |
| discs overgrown protein kinaseg | CPIJ003503 | −1.481 | 0.090 | |
| hypothetical protein | CPIJ016941 | 1.279 | 0.089 | |
| mck1 | CPIJ006114 | 0.384 | 0.121 |
acircadian locomotor output cycles kaput protein (contains artificial break point)
bannotated in reference as cryptochrome 1
cannotated in reference as cryptochrome 1
dcircadian protein clock/arnt/bmal/pas
eparalog of timeless
fhomolog of par domain protein 1 (pdp1) in Drosophila melanogaster
galternative name: doubletime
We compared our overall results to those of Baker et al. [17] who presented a gene expression atlas addressing sex- and tissue-specificity for the mosquito An. gambiae. They reported 1487 female-biased and 1226 male-biased genes, belonging to 1351 and 1030 ortholog groups, respectively. By comparing the expression of genes belonging to ortholog groups that were conserved in An. gambiae, we found that 61 % (female) and 65 % (male) of these groups showed sex bias in Cx. pipiens (based on p-value < 0.05 and >2FC). Analysis of gene ontology terms that were specifically enriched in each sex showed an overlap of a single GO term with An. Gambiae, “ion transport” (GO:0006811) in males (Additional file 5).
Discussion
Despite increasing recognition of the role of ALAN in ecology and behavior [1–3], our understanding of how it affects organisms at the molecular level remains limited. Here we assessed the effects of artificial light on Cx. pipiens, a widespread and abundant mosquito that is prominent in many urban habitats, by examining changes in gene expression in the entire body. This is the first study to provide data on sex-specific gene expression in adult Culex pipiens f. molestus. The majority of studies on mosquitoes focus on females due to their role as disease vector.
We used Cx. quinquefasciatus genome as a reference to map our data. The only other transcriptomic study of Cx. pipiens f. molestus to date found that the relationship between species was sufficiently close to obtain reliable mapping results [26]. Both species belong to the Cx. pipiens complex. They are known to be able to hybridise and the species status of Cx. quinquefasciatus remains debated [13, 27, 28]. This study, however, focussed on searching for ecotype-specific divergence between genes in samples of mixed sexes [20].
The experimental design of our study focused on separate analysis of sexes. There was a very strong sex bias in expression, and males in our laboratory population exhibited a much more pronounced response to the treatment mimicking artificial light at night (“low-light”), with many of the affected genes having functional annotation. This study may serve as foundation for future work by providing whole-body transcriptome data for a widespread mosquito and suggests that ALAN can affect a broad range of physiological pathways at the molecular level. Although we are aware that gene expression varies among tissues [8, 16, 17] we chose this approach because our goal was to obtain an overview of processes potentially affected by artificial light at night. This was done at the expense of tissue-specific responses, but we believe that our results provide an important starting point for more detailed studies concerning gene expression in separate tissues, different developmental stages or different light sources and regimes.
The finding that males and females are affected to a different extent fits with the only other published study of sex-specific gene expression in mosquitoes [17] and suggests possible implications for reproduction biology and, consequently, population-level impacts.
Our experiment was designed to mimic artificial light of the kind generated by street and other outdoor lighting, where the normal transition from natural light (during the day) to darkness at night is altered by an abrupt switching on of light for a period of constant brightness. Veronesi et al. [14] found that certain light intensity thresholds (e.g. 5 lux for Cx. pipiens) function as cues for commencing or ceasing activity. Although the light regime in the treatment never fell below this threshold, our light regime still provides a cue for anticipation of the onset of darkness by its design to mimic the natural rise and fall of light intensity. In contrast, no cue was provided that allows anticipation of the abrupt switch to “artificial low-light” (300 lux in our experiment, an increase of a factor of ~6). We therefore believe the experiments measured a response to artificial light rather than comparing a short day to a long day. However, our choice to deprive individuals of light cues for 48 h prior to the experiment, and thus prohibiting synchronisation to the ambient light environment, constitutes an acute change relative to the darkness experienced before. As a nocturnal species, Cx. pipiens is naturally exposed to varying, albeit low, light intensities produced by moonlight. Studies have suggested that lunar cycles can influence the activity and biting propensity of mosquitoes [29, 30]. There were no cues regarding the lunar cycle in our laboratory setup. Our results are thus likely to be the response to artificial light at night. However, the presence of moonlight in combination with artificial light may produce a different gene expression profile in natural populations, which remains to be addressed in future studies.
Male response to artificial light at night
The response to artificial light at night in males was primarily detected as reduced expression levels of a number of genes in “low-light” treatment conditions, i.e. in males exposed to artificial light at night instead of the (laboratory-simulated) natural progression from daylight to darkness. These down-regulated genes were mainly related to gametogenesis, immunity and lipid metabolism. There is a scarcity of knowledge about the effects of artificial light on mosquitoes. Under natural conditions, decreasing light triggers activity in nocturnal mosquitoes. Individuals begin the search for food and mates, and males begin to swarm. We may speculate that the expression of genes involved in gametogenesis should increase as light levels decrease, but that artificial light inhibited this here. Our findings are preliminary, and we are not aware of any similar studies, so this remains a hypothesis to be tested in the future. Genes involved in lipid metabolism comprised another important group of genes that were less expressed under ALAN. The last food uptake was 12 h prior to sampling which could mean that the carbohydrate reserves had been used up. It may also mean that males in artificially lit environments were less active. Further work should address whether the observed changes in expression of genes related to lipid metabolism are caused by light-mediated reduced activity or a sign of usage due to starvation. Genes involved in immune response also exhibited lower expression. Some immune genes are known to be rhythmically expressed in An. gambiae [8, 16], and this might also be the case for Cx. pipiens. Despite the evidence for the negative influence of artificial light on immune genes and given their cyclic expression patterns, future studies with a 24-h sampling scheme could provide important insights that enable us to fully understand how these two processes interact.
It is well known that some genes exhibit cyclic expression over time [8]. Our initial analysis using ‘treatment’ and ‘timepoint’ resulted in only two differentially expressed genes over time and in response to treatment (in males). Neither of, these genes were detected as differentially expressed when the factor ‘timepoint’ was removed from the model, thus our results using only ‘treatment’ as a factor present a robust estimate for the gene expression level response to the low-light treatment, although this comes at the expense of detecting temporal differences that may occur in the response to light treatment. Future experiments using extended time series sampling under different light regimes may provide more insight into temporal changes in response to ALAN.
Of the genes more highly expressed in the low-light condition in males, the majority encoded conserved hypothetical proteins, i.e. have no clearly defined function. Only the one of the two genes with FC > 2 was annotated, namely as 4-coumarate-CoA ligase 1 (Additional file 1: Table S1). To date, the function of this gene is not known in mosquitoes.
Female response to artificial light at night
It was striking that the different light regimes did not induce detectable changes in gene expression in female Cx. pipiens. One biological explanation is that females in both treatments were inseminated and this may have played a role. Male accessory gland secretion is a powerful modulator of female behaviour and activity [31], which might potentially render females insensitive to light at night. This could have implications for biting propensity, as accessory gland secretion can trigger ovulation and oviposition behaviour. An evaluation of the effect of light at night at different life stages (e.g., virgin, inseminated, or after oviposition) would provide important insights. However, females often mate soon after emergence [32] and thus it is reasonable to assume that the vast majority of adult females in nature are inseminated at a given point in time.
Sex-biased expression
Half of all genes were differentially expressed in males and females, indicating a strong sex-specific pattern of expression regardless of the light treatment. Sex-biased differences in gene expression are known to occur in a number of species and sexual dimorphism (in morphology, behaviour and physiology) is believed to be a main driver of this [33]. Our findings were similar to those of a recent study of the mosquito An. gambiae that reported 72 % of genes to show sex-biased differential expression in whole bodies [17]. In Drosophila, approximately 50 % of genes are sex-biased [34]. By determining ortholog relationships between the sex-biased genes from our study of Cx. pipiens and those in An. gambiae [17] we found a larger overlap of sex-biased genes in males compared to females. This pattern could be explained by greater conservation of male-biased gene expression between Anopheles and Culex. Alternatively, this could reflect differences in ecology of females from these two species. An. gambiae is an obligate blood feeder whereas Cx pipiens f. molestus displays facultative autogeny (a blood meal is not essential). Furthermore, the present study sampled individuals reared on a sugar diet whereas An. gambiae data were derived from blood-engorged females and sugar-fed males [17], potentially contributing to differences in female-biased gene expression.
Among the genes that were differentially expressed in males and females were the circadian clock genes. Although clock-gene expression varies among tissues and our approach was a whole-body sampling, some notable findings suggest areas for more research on the effect of artificial light on clock genes. Male-biased genes related to the clock were generally much more highly expressed compared to females, in which genes were only slightly (although significantly) up-regulated. This suggests that the internal circadian clock system may either be influenced differently in males and females or is inherently different between the sexes, as is the case in Anopheles gambiae [35]. To our knowledge it has not been tested whether this sex-specific circadian rhythm is the case in Culex pipiens. The highly expressed male-biased genes included photolyase. This is a domain of the cryptochrome-1 protein which contains two light-harvesting cofactors and is mainly responsible for DNA repair after UV- and blue-light exposure [36]. timeout/timeless-2 is a paralogue of circadian clock gene timeless and is involved in chromosome stability and light entrainment [37]. disc overgrown protein kinase, in Drosophila also referred to as doubletime. It phosphorylates the period protein and thus contributes to circadian rhythmicity [38]. All of these genes are involved in the perception of light and relate directly to circadian clock function, suggesting a greater potential for the low-light treatment to influence males compared to females, in agreement with our overall findings. Further underlining this potential sex-specific influence is the fact that we found the gene cryptochrome-1 to be more highly expressed as a response to ALAN. The presence of light clearly changed the expression of the gene of paramount importance in synchronisation of the circadian clock to the environment and clock-controlled processes.
Conclusion
In this study we addressed whether artificial light at night similar to that prevalent in urban and suburban settings would affect gene expression in the mosquito Culex pipiens f. molestus. Artificial light at night elicited responses at the molecular level with differing responses in males and females. In males, a significant reduction in expression occurred in genes related to gametogenesis, lipid metabolism and immune response. In contrast there was almost no differential gene expression in females. Taken together these changes potentially lead to dramatic shifts in population dynamics, as reproduction is directly influenced. This might lead to shifts in feeding behaviour of the females, as the search for a blood meal is associated to insemination, and hence vector capacity.
Methods
Mosquitoes used in the experiment originated from a laboratory colony established in 2012 and were reared in a light(L):dark(D) regime of L16h:D8h (hereafter “control”) in a climate chamber with temperature maintained at 26 ± 1 °C and relative humidity at 60–90 %. Adults were kept in mesh cages (60 × 30 × 30 cm) and fed with ~ 10 % saccharose solution offered on cotton pads ad libitum. Light was provided by cool-white LEDs (LED flex SMD, 24VDC, 24 W, 1A, 60 LEDs/m, 500 cm, cool-white single chip, Barthelme GmbH & Co. KG, Nuremberg, Germany). Seven strips consisting of 48 LEDs each were attached to a board (88 × 34 cm) and suspended horizontally over the cages. Light levels in the colony were controlled with custom-made software based on the LabView v 8.5.1 runtime environment (National Instruments Germany GmbH, Munich, Germany). We specified voltage at 15 timepoints over a 24-h period (and at 19 timepoints in the low-light treatment, see below) to which the software fit a hermite spline curve. The result was a smooth change of voltage, and thereby light intensity, over each 24-h period. This allowed us to produce a near-natural light regime with gradually changing light intensities throughout the day (dark: 0 lux, mid-day: 855 and 897 in control and low-light regime, respectively; Fig. 1).
After pupation, individuals were placed into cages and adults were allowed to emerge and mix. We therefore assume that all females were mated. Adults were between 1–8 days old when they were removed from cages with an aspirator and placed in continual darkness for 48 h. This was done to uncouple mosquitoes from the light regime under which they were reared and thus prevent the onset of any processes arising from light-induced anticipation of the time of day. A 10 % saccharose-solution was available ad libitum for the first 36 h and feeding was stopped 12 h prior to the start of the experiment in order to avoid inflated expression of genes involved in digestion relative to other physiological processes. After the 48 h of darkness, adult individuals were exposed to three “days” in one of two different light regimes: “control” (L16h:D8h; the same conditions were used for rearing) and “low-light” (L16 + Llow3h:D5h). The low-light treatment (low-light relative to the maximum light intensity) consisted of a 16-h day as in the rearing cage (described above) followed by a sudden increase to ~300 lux for three additional hours at sunset (Fig. 1). The experiment started on day 1 at 04:00 in both treatments and seven adults of each sex were sampled after 3 days at two timepoints (19:40 and 22:30) for each treatment. Samples were immediately placed in liquid nitrogen, resulting in eight samples. Each of the eight samples consisted of seven pooled individuals (either male or female). “Control” light intensity during sampling was 0-25 lux while “low-light” light intensity was ~300 lux.
Data were analysed first by considering these as independent timepoints and then by combining these and treating timepoints as replicates (see below).
Gene expression levels related to specific metabolic processes can differ among tissues [8, 17]. Because no data on tissue-specificity were available for Cx. pipiens f. molestus, we chose to examine whole bodies in order to investigate whole-organism effects of additional light at night across as broad a range of molecular processes as possible. A disadvantage of our approach was that any tissue-specific differences could not be detected; however, our aim was to gain an overview of gene expression and limit any risk of overestimating the biological significance of results based on tissue-specificity.
RNA was extracted by first adding TRIzol® Reagent (Ambion®, Invitrogen, Carlsbad, USA) to the samples. The tissue was then homogenised on ice with an ULTRA-TURRAX® disperser (IKA®, Staufen, Germany). All remaining steps were carried out according to the manufacturers’ protocol (Ambion®, Invitrogen, Carlsbad, USA) and the RNA-pellet was dissolved in 50 μl RNase-free water (Carl Roth GmbH und Co. KG, Karlsruhe, Germany). The construction of 8 TruSeq cDNA libraries and sequencing on the Illumina HiSeq2000 platform for 207 cycles was performed by LGC Genomics GmbH (Berlin, Germany). The resulting 100-bp paired-end reads are available from the NCBI SRA under BioSample accession PRJNA257052.
The untrimmed read pairs were mapped to the Culex quinquefasciatus reference genome version CpipJ1.21 using the unmasked DNA assembly (3171 supercontigs, totalling 580 Mb, with a Contig N50 of 28 Kb and supercontig N50 size of 486.76 Kb) and corresponding gtf annotation from ensembl metazoa in conjunction with RSEM v1.2.12 [39] and Bowtie 1.0.0 [40]. Bowtie was executed using the default parameters recommended for use with RSEM [39]. Further annotations, including gene ontology terms, were obtained from the UniProt-GOA Cx. quinquefasciatus proteome annotation (ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/). RSEM expects paired reads of uniform length and accounts for quality scores in its statistical model, although it usually does not significantly improve quantification accuracy over a reduced model when using reads with an illumina error profile [39]. Reads were therefore not quality-trimmed.
Differential gene expression in response to the low-light treatment was determined on raw read counts using DESeq2 v1.8.2 in R. We initially specified both treatment and sampling time in the model (design formula: “design = ~treatment + timepoint) and then ran a second analysis using only treatment (“design = ~treatment”). Both models were tested for males and females separately. Sex-specific gene expression has been observed in a range of organisms [17, 33, 41]. We therefore tested for sex-specific differences across all samples using DESeq2 as described above. We specified the design formula as “design = ~ sex + treatment”. This allowed for a contrast of levels for each factor.
DESeq2 uses the negative binomial distribution and a shrinkage estimator to determine variance–mean dependence in mapped read counts and a conditional test for differential expression [26]. Genes with an Benjamini-Hochberg adjusted p-value < 0.05 (implemented in DESeq2) were considered to be differentially expressed. We also report numbers of genes with an absolute fold change greater than 2 (FC > 2) for reference to other studies. Over-representation of Gene Ontology (GO) terms in groups of differentially expressed genes was determined using the GOstats v. 2.14 package [42] for R, by means of a hypergeometric test with a p-value of 0.05 and the GO terms from the CpipJ1.21 proteome annotation as the gene universe. Gene orthology data were obtained from OrthoDB v. 8 [43] and sex-biased Anopheles gambiae gene expression data [17] were downloaded from the Sebida database [44].
Data accessibility
The datasets supporting this article are freely available on the website of the research group: http://monaghanlab.org/data/cxpip_rnaseq/.
Acknowledgments
We thank Janina Kypke for assistance in maintaining the mosquito colony and Jens Rolff and Franz Hölker for their support of the research. Two anonymous referees provided very useful comments and thus helped to greatly improve the manuscript. This is publication number 20 from the Berlin Center for Genomics in Biodiversity Research.
Funding
The study was partly funded by the German Federal Ministry of Education and Research (BMBF) and the Leibniz Association PAKT for Research and Innovation (SAW). PRJ was supported by European Research Council grant 260986.
Abbreviations
- ALAN
artificial light at night
- GO
Gene ontology
- L:D
light:dark cycle
- LED
light-emitting diode
- ncRNA
non-coding RNA
- PC
principal components
- PCA
principal component analysis
- UV
ultra violet
Additional files
Spreadsheet listing all differentially expressed genes in males exposed to the ALAN treatment. Gene = gene ID; base Mean = the mean of the normalized counts for all samples;log2FoldChange = Log 2 fold change in expression low-light relative to control; lfcMLE = unshrunken maximum likelihood estimates of Log2 fold change in expression; lfcSE = Standard Error of log2FoldChange; Stat = Wald statisic; pvalue; padj = p value adjusted after Benjamini-Hochberg; Annotation = functional annotation of the gene; Domain(s) = Interpro domain annotations. Genes that are differentially expressed with an absolute fold change greater than 2 (FC > 2) are shown in bold. (XLS 65 kb)
Spreadsheet listing all differentially expressed genes in females exposed to the ALAN treatment based on adjusted p-values. Gene = gene ID; base Mean = the mean of the normalized counts for all samples; log2FoldChange = Log 2 fold change in expression low-light relative to control; lfcMLE = unshrunken maximum likelihood estimates of Log2 fold change in expression; lfcSE = Standard Error of log2FoldChange; Stat = Wald statisic; pvalue; padj = p value adjusted after Benjamini-Hochberg; Annotation = functional annotation of the gene; Domain(s) = Interpro domain annotations. (XLS 19 kb)
Spreadsheet listing all overrepresented gene ontology terms associated with genes exhibiting decreased expression in males exposed to the ALAN treatment. GOBIP, gene ontology biological process identifier. (XLS 18 kb)
Spreadsheet listing all differentially expressed gene s between male and female mosquitoes. GeneID = gene ID; base Mean = the mean of the normalized counts for all samples;log2FoldChange = Log 2 fold change in expression low-light relative to control; lfcMLE = unshrunken maximum likelihood estimates of Log2 fold change in expression; lfcSE = Standard Error of log2FoldChange; Stat = Wald statisic; pvalue; padj = p value adjusted after Benjamini-Hochberg; Annotation = functional annotation of the gene; Domain(s) = Interpro domain annotations. Genes with an absolute fold change of two are shown in bold. (XLS 2665 kb)
Spreadsheet listing all overrepresented gene ontology terms associated with male-biased genes. GOBIP, gene ontology biological process identifier. (XLS 44 kb)
Footnotes
Ann-Christin Honnen and Paul R. Johnston contributed equally to this work.
Competing interests
The authors do not have competing interests.
Authors’ contributions
AH and MTM conceived the study. AH, PRJ and MTM designed the experiment. AH conducted the experiment and the sampling. AH and PRJ analysed the data and drafted the manuscript. AH, PRJ and MTM contributed to the final manuscript. All authors gave final approval for publication.
References
- 1.Hölker F, Moss T, Griefahn B, Kloas W, Voigt C, Henckel D, et al. The dark side of light: a transdisciplinary research agenda for light pollution policy. Ecol Soc. 2010;15:13. [Google Scholar]
- 2.Longcore T, Rich C. Ecological light pollution. Front Ecol Environ. 2004;2:191–198. doi: 10.1890/1540-9295(2004)002[0191:ELP]2.0.CO;2. [DOI] [Google Scholar]
- 3.Navara KJ, Nelson RJ. The dark side of light at night: physiological, epidemiological, and ecological consequences. J Pineal Res. 2007;43:215–224. doi: 10.1111/j.1600-079X.2007.00473.x. [DOI] [PubMed] [Google Scholar]
- 4.Moore MV, Pierce SM, Walsh HM, Kvalvik SK, Lim JD. Urban light pollution alters the diel vertical migration of Daphnia. Verh Internat Verein Limnol. 2000;27:1–4. [Google Scholar]
- 5.Gentile C, Meireles-Filho ACA, Britto C, Lima JBP, Valle D, Peixoto AA. Cloning and daily expression of the timeless gene in Aedes aegypti (Diptera: Culicidae) Insect Biochem Mol Biol. 2006;36:878–84. doi: 10.1016/j.ibmb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Gentile C, Rivas GBS, Meireles-Filho ACA, Lima JBP, Peixoto AA. Circadian expression of clock genes in two mosquito disease vectors: cry2 is different. J Biol Rhythms. 2009;24:444–451. doi: 10.1177/0748730409349169. [DOI] [PubMed] [Google Scholar]
- 7.Goto SG, Denlinger DL. Short-day and long-day expression patterns of genes involved in the flesh fly clock mechanism: period, timeless, cycle and cryptochrome. J Insect Physiol. 2002;48:803–816. doi: 10.1016/S0022-1910(02)00108-7. [DOI] [PubMed] [Google Scholar]
- 8.Rund SSC, Gentile JE, Duffield GE. Extensive circadian and light regulation of the transcriptome in the malaria mosquito Anopheles gambiae. BMC Genomics. 2013;14:218. doi: 10.1186/1471-2164-14-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vezzani D. Review: artificial container-breeding mosquitoes and cemeteries: a perfect match. Trop Med Int Health. 2007;12:299–313. doi: 10.1111/j.1365-3156.2006.01781.x. [DOI] [PubMed] [Google Scholar]
- 10.Rudolf M, Czajka C, Börstler J, Melaun C, Jöst H, von Thien H, et al. First nationwide surveillance of Culex pipiens complex and Culex torrentium mosquitoes demonstrated the presence of Culex pipiens biotype pipiens /molestus hybrids in Germany. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0071832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Townroe S, Callaghan A. British container breeding mosquitoes: the impact of urbanisation and climate change on community composition and phenology. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker N, Jöst A, Weitzel T. The Culex pipiens complex in Europe. J Am Mosq Control Assoc. 2012;28(4s):53–67. doi: 10.2987/8756-971X-28.4s.53. [DOI] [PubMed] [Google Scholar]
- 13.Meireles-Filho ACA, Kyriacou CP. Circadian rhythms in insect disease vectors. Mem Inst Oswaldo Cruz. 2013;108(Suppl):48–58. doi: 10.1590/0074-0276130438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veronesi R, Gentile G, Carrieri M, Maccagnani B, Stermieri L. Seasonal pattern of activity of Aedes caspius, Aedes detritus, Culex modestus and Culex pipiens in the Po Delta of northern Italy and significance for vector-borne disease risk assessment. J Vector Ecol. 2012;37:49–61. doi: 10.1111/j.1948-7134.2012.00199.x. [DOI] [PubMed] [Google Scholar]
- 15.Das S, Dimopoulos G. Molecular analysis of photic inhibition of blood-feeding in Anopheles gambiae. BMC Physiol. 2008;8:23. doi: 10.1186/1472-6793-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rund SSC, Hou TY, Ward SM, Collins FH, Duffield GE. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proc Natl Acad Sci. 2011;108:E421–E430. doi: 10.1073/pnas.1100584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker DA, Nolan T, Fischer B, Pinder A, Crisanti A, Russell S. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics. 2011;12:296. doi: 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ceriani MF. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 19.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–92. doi: 10.1016/S0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurst D, Rylett CM, Isaac RE, Shirras AD. The drosophila angiotensin-converting enzyme homologue Ance is required for spermiogenesis. Dev Biol. 2003;254:238–247. doi: 10.1016/S0012-1606(02)00082-9. [DOI] [PubMed] [Google Scholar]
- 23.Lindsley DL, Roote J, Kennison JA. Anent the genomics of spermatogenesis in Drosophila melanogaster. PLoS One. 2013;8:e55915. doi: 10.1371/journal.pone.0055915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siddiqui NU, Li X, Luo H, Karaiskakis A, Hou H, Kislinger T, et al. Genome-wide analysis of the maternal-to-zygotic transition in Drosophila primordial germ cells. Genome Biol. 2012;13:R11. doi: 10.1186/gb-2012-13-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kounatidis I, Ligoxygakis P. Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. 2012;2:120075. doi: 10.1098/rsob.120075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price DC, Fonseca DM. Genetic divergence between populations of feral and domestic forms of a mosquito disease vector assessed by transcriptomics. PeerJ. 2015;3:e807. doi: 10.7717/peerj.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattingly PF, Rozeboom LE, Knight KL, Laven H, Drummond FH, Christophers SR, et al. The Culex pipiens complex. Trans R Entomol Soc London. 1951;102:331–342. doi: 10.1111/j.1365-2311.1951.tb00752.x. [DOI] [Google Scholar]
- 28.Mattingly PF. The systematics of the Culex pipiens complex. Bull World Health Organ. 1967;37:257–261. [PMC free article] [PubMed] [Google Scholar]
- 29.Bidlingmayer WL. The effect of moonlight on the flight activity of mosquitoes. Ecology. 1964;45:87–94. doi: 10.2307/1937110. [DOI] [Google Scholar]
- 30.Kampango A, Cuamba N, Charlwood JD. Does moonlight influence the biting behaviour of Anopheles funestus? Med Vet Entom. 2011;25:240–246. doi: 10.1111/j.1365-2915.2010.00917.x. [DOI] [PubMed] [Google Scholar]
- 31.Gillott C. Male accessory gland secretions: modulators of female reproductive physiology and behaviour. Ann Rev Entom. 2003;48:163–184. doi: 10.1146/annurev.ento.48.091801.112657. [DOI] [PubMed] [Google Scholar]
- 32.Spielman A, Leahy MG, Skaff V. Failure of effective insemination of young female Aedes aegypti mosquitoes. J Insect Physiol. 1969;15:1471–1479. doi: 10.1016/0022-1910(69)90168-1. [DOI] [PubMed] [Google Scholar]
- 33.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- 34.Perry JC, Harrison PW, Mank JE. The ontogeny and evolution of sex-biased gene expression in Drosophila melanogaster. Mol Biol Evol. 2014;31:1206–1219. doi: 10.1093/molbev/msu072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rund SSC, Lee SJ, Bush BR, Duffield GE. Strain- and sex-specific differences in daily flight activity and the circadian clock of Anopheles gambiae mosquitoes. J Insect Physiol. 2012;58:1609–1619. doi: 10.1016/j.jinsphys.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, et al. Identification of a new cryptochrome class. structure, function, and evolution. Mol Cell. 2003;11:59–67. doi: 10.1016/S1097-2765(03)00008-X. [DOI] [PubMed] [Google Scholar]
- 37.Benna C, Bonaccorsi S, Wülbeck C, Helfrich-Förster C, Gatti M, Kyriacou CP, et al. Drosophila timeless2 is required for chromosome stability and circadian photoreception. Curr Biol. 2010;20:346–52. doi: 10.1016/j.cub.2009.12.048. [DOI] [PubMed] [Google Scholar]
- 38.Price JL, Blau J, Rothenfluh A, Abodeely M, KLoss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/S0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 39.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsch J, Ellegren H. The evolutionary causes and consequences of sex-biased gene expression. Nat Rev Genet. 2013;14:83–87. doi: 10.1038/nrg3376. [DOI] [PubMed] [Google Scholar]
- 42.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 43.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41:D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:2577–2579. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting this article are freely available on the website of the research group: http://monaghanlab.org/data/cxpip_rnaseq/.


