Abstract
Objectives
In an attempt α-amylase gene from Pyrococcus woesei was amplified and cloned into a pTYB2 vector to generate the recombinant plasmid pTY- α-amylase.
Methods
Escherichia coli BL21 used as a host and protein expression was applied using IPTG. SDS-PAGE assay demonstrated the 100 kDa protein. Amylolytic activity of proteins produced by transformed E. coli cells was detected by zymography, and the rate of active α-amylase with and without the intein tag in both soluble conditions and as inclusion bodies solubilized by 4M urea were measured.
Results
Amylolytic activity of ∼185,000 U/L of bacterial culture was observed from the soluble form of the protein using this system.
Conclusion
These results indicate that this expression system was appropriate for the production of thermostable α-amylase.
Keywords: amylase, expression, recombinant protein, thermophile
1. Introduction
Pyrococcus woesei is known as an ultra-thermophilic marine archaebacterium that is sulfur-reducing and capable of growing at between 100°C and 103°C. Its cells have a roughly spherical, elongated, and constricted appearance, similar to Thermococcus celer, and frequently occur as diploforms. Cells grown on solid supports have dense tufts of flagellae or pili attached to one pole.
The α-amylase is an enzyme that catalyzes the hydrolysis of α-l,4 glucosidic linkages in polysaccharides of three or more α-1,4-linked d-glucose units to yield glucose and maltose [1]. The α-amylase isolated from the ubiquitous mesophilic soil bacterium Bacillus licheniformis 2, 3, 4 operates optimally at 90°C and pH 6, and requires the addition of calcium (Ca2+) to maintain thermostability [5]. The ideal conditions for starch hydrolysis are pH ∼4.5 and a temperature of 105°C. Given that this enzyme is unstable under these conditions, the pH must be increased to 5.7–6.0 and Ca2+ added [6]. These pH adjustments increase the energetic costs associated with the process. Furthermore, the Ca2+ requirement creates the need for ion-exchange refining to remove added Ca2+ that has a potent inhibitory effect on xylose isomerase, which is used in processing starch hydrolysate to fructose syrup [7]. Given that the majority of industrial processing of starch gelatinization is completed near 100°C, thermostable α-amylases are used. Hyperthermophilic archaeon Pyrococcus woesei producing α-amylase is a good source of such an enzyme, which displays a temperature optimum of 100°C and retains ∼60% of maximal activity in pH ranges of 4.5 to 7.0. Moreover, this enzyme precludes the necessity to add Ca2+ salt to the substrate [7]. In view of the industrial importance of P. woesei α-amylase, we cloned, expressed, and purified P. woesei α-amylase in Escherichia coli.
2. Materials and methods
P. woesei (DSM 3773, Braunschweig, Germany) was cultivated in artificial seawater supplemented with 0.25% soluble starch, 2.5% tryptone, 2% yeast extract, and 0.1% elemental sulfur. The pH of the medium was adjusted to 7.0 with 1M NaOH. Extraction of P. woesei chromosomal DNA and genomic library construction were performed as described previously [8]. E. coli BL21 DE3 plysS cells (Stratagene, La Jolla, CA, USA) transformed with the ligation mixture were plated on Luria-Bertani (LB) agar–ampicillin (100 mg/mL) plates. After 16–20 h of incubation at 37°C, colonies were transferred to a new set of LB-ampicillin plates.
2.1. Construction of expression plasmid
To express the α-amylase protein with its fusion intein protein, the plasmid pTYB2 (New England BioLabs, Beverly, MA, USA) was used. P. woesei chromosomal DNA was used as a source of the α-amylase gene and polymerase chain reaction (PCR) amplification was done with the following primers:
5′-GCTAGCTTGGAGCTTGAAGAGGGAG-3′ and 5′-GAGACCACAATAACTCCATACGGAG-3′ containing recognition sites for restriction endonucleases NheI and XhoI (Fermentas; Vilnius, Lithuania).
The reaction was performed using 10 ng DNA, 2 μL (10 μM) of each primer, 5 μL (10mM) dNTPs, 5 μL 10× PCR buffer [100mM Tris-HCl (pH 8.9), 500mM KCl, 20mM MgCl2, 1% Triton X-100] and High-fidelity PCR Enzyme Mix (Fermentas). After 2 minutes of preliminary heating at 95°C in a thermal cycler, each of the 30 cycles was conducted at 95°C for 0.5 minutes, 56°C for 1 minute, and 72°C for 1 minute, with a final step of 5 minutes at 72°C. The amplified gene was cloned into pTZ57R/T (InsTAclone PCR Cloning Kit; Fermentas), and then subcloned into pTYB2 and transformed into E. coli BL21 (DE3) pLysS cells.
2.2. Purification of α-amylase
Isolation and purification of α-amylase was undertaken as described previously [9]. Briefly, transformed E. coli with pTY-amylase was induced with 1mM isopropyl- β-d-thiogalactopyranoside (IPTG) and incubated for 20 h at 20ºC. The size of the purified expression product according to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis corresponded to the predicted molecular mass of the recombinant protein (100 kDa; Figure 1). The transformed E. coli cells obtained from 0.5 L culture induced with IPTG were sonicated in 50 mL 0.05M phosphate buffer (pH 7.2) containing 1 mL of 50 μmol NaCl and 1 μmol dithiothreitol. The acquired suspension was centrifuged (4°C) at 10,000g for 15 minutes. The E. coli proteins were denatured at 75°C for 30 minutes and then precipitated to obtain recombinant α-amylase. The supernatant containing thermostable α-amylase was then used for further purification. Urea (4M) was used to renature inclusion bodies, and the precipitated enzyme was extracted (2 h, 20°C) from the pellet.
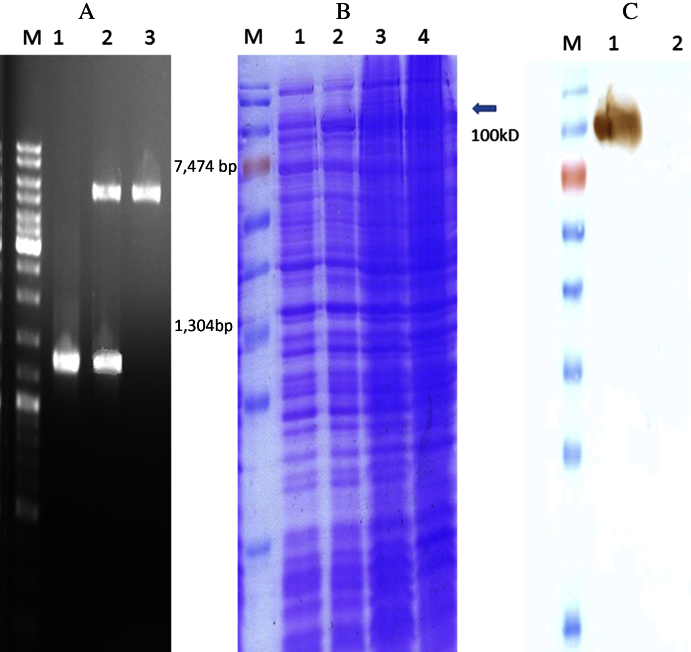
Figure 1.
(A) Digestion of pTY-α-amylase with NheI and XhoI restriction enzymes. Lane 1: pTYB2 digested with NheI and XhoI; lane2: pTY-α-amylase digested with NheI and XhoI; lane 3: PCR product of α-amylase. (B) SDS-PAGE analysis of α-amylase expression. Lane 1: before IPTG induction; lane 2: after IPTG induction; lane 3: 16 hours after IPTG induction; lane 4: 20 hours after IPTG induction. (C) Western blot analysis of α-amylase with anti-intein polyclonal antibody. Lane1: α-amylase; lane 2: lysate of untransformed bacteria. IPTG = isopropyl- β-d-thiogalactopyranoside; M = molecular marker; SDS-PAGE = sodium dodecyl sulfate polyacrylamide gel electrophoresis.
2.3. Enzyme assay
A 1% starch solution in 0.05M acetate buffer (pH 5.5) was used for determining α-amylase activity. Before reaction initiation, the starch (2.5 mL) was preincubated at 80°C, and then the reaction initiated by adding 0.5 mL diluted enzyme solution. After different time periods, cooling on ice was used to stop the reaction. The reducing sugars formed during the reaction were assayed according to the dinitrosalycilic acid method of Bradford [10]. To determine the effect of pH on enzyme activity, 0.05M citrate-phosphate buffers in the pH range of 4.0–8.0 adjusted at 80°C were used. The specific activity of α-amylase (U/mg of protein) was determined as described by Bradford [10].
2.4. Detection of amylolytic activity by electrophoresis
A 12% polyacrylamide gel at room temperature was used to determine amylolytic activity, as described previously [11]. The samples (10 μg of the protein suspension) were dissolved in 0.01M phosphate buffer (pH 7.2) and detected by electrophoresis. The gel was incubated for 15 minutes at 40°C in 0.05M acetate buffer (pH 5.5), then submerged in the same buffer including 1% of soluble starch at 80°C for 15 minutes, and stained with iodine solution (0.01 mol/L I2 in 0.1M KI). Proteins displaying amylolytic activity emerged as bands on a dark brown background.
3. Results
3.1. Construction of pTY-α-amylase
The PCR amplified product of the α-amylase gene, added between NheI and XhoI restriction sites at the 5′ and 3′ termini of the α-amylase gene, respectively, was electrophoresed and the 1304-bp fragment gel purified, T/A cloned in pTZ57R/T, and subcloned into pTY2 to generate pTY-α-amylase expression plasmid. The expression plasmid was first transformed into E. coli TOP 10 cells for vector propagation, and then transformed into E. coli BL21 cells. Presence of the insert in the plasmid preparation from the positive transformants was confirmed by restriction enzyme digestion (Figure 1).
3.2. Expression and purification of α-amylase
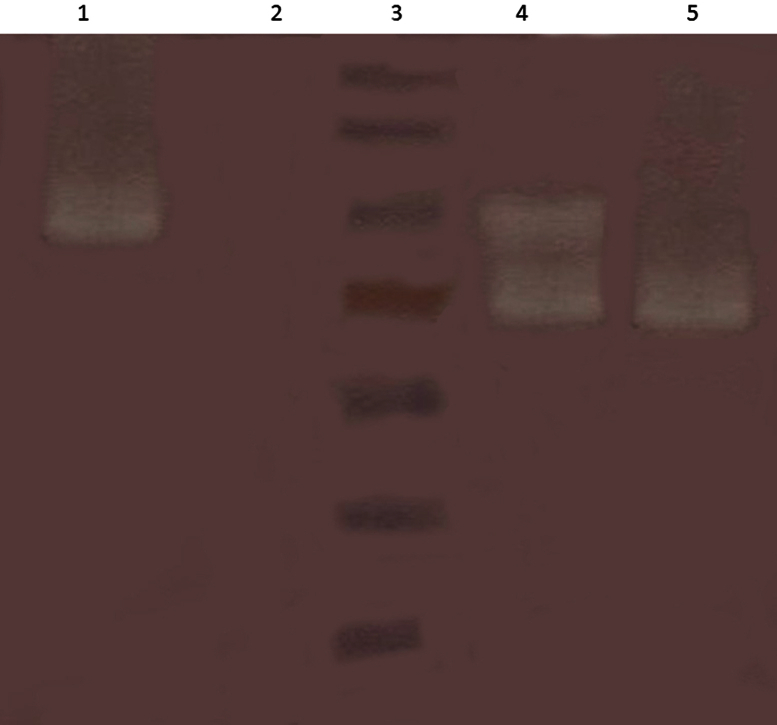
SDS-PAGE analysis of proteins from the culture of E. coli cells carrying pTY-α-amylase and following induction with 1mM IPTG for 20 hours at 20°C showed a clear band at a position corresponding to 100 kDa. Further confirmation was undertaken with western blot analysis using an anti-intein antibody. Amylolytic activity of proteins produced by transformed E. coli cells was detected by zymography using 12% SDS-PAGE gels (Figure 2). Enzymatic activity of the fusion construct of α-amylase and intein was observed in the soluble fraction, as well as in inclusion bodies solubilized by 4M urea. These results showed that heating was unable to cleave the intein fusion from α-amylase, however, the enzyme fused with intein showed activity. Heat treatment of the crude extract effectively removed proteins and can be used as an efficient step for enzyme purification. Most of the α-amylase activity was observed in the cell-free extract, and only 35% of the total enzyme activity was observed in inclusion bodies. The intein fused with the recombinant protein can be partially separated from its fusion partner by heat treatment at ∼85°C in the presence of thiol compounds. Interestingly, the recombinant protein fused with intein also showed enzyme activity (Figure 2).
Figure 2.
Zymography of the amylolytic activity of α-amylase expressed in Escherichia coli BL21 cells transformed with pTYB2α-amyl. Lane 1: lysate of E. coli BL21 cells transformed with pTYB2α-amyl before separation of intein fusion by heat; lane2: lysate of untransformed bacteria; lane 3: supernatant obtained from centrifugation of heated lysate of E. coli BL21 cells transformed with pTYB2α-amyl; lane 4: purified thermostable α-amylase produced by Bacillus licheniformis. M = molecular marker.
3.3. Enzyme activity
Amylolytic activity (∼185,000 ± 8,000 U/L bacterial culture) was observed from the soluble form of the protein, while the solubilized inclusion bodies exhibited similar activity levels. Our findings suggested that the pTYB2 plasmid is an appropriate system to express and produce thermostable α-amylase. No reduction in enzyme activity was observed following mutation of two amino acids in the C- and N-terminal regions of the enzyme.
4. Discussion
α-Amylases are enzymes that hydrolyze starch by cleaving α-l,4-glucosidic bonds. They are among the most important commercial enzymes, having broad applications in starch processing, textiles, brewing and alcohol manufacturing, and other industries. Several α-amylases have been characterized and their genes cloned from microorganisms, plants, and animals. With the exception of two enzymes obtained from eubacteria and archaea [12], α-amylases are considered members of the same α-amylase family, sharing a comparable structure, similar catalytic site, and identical catalytic mechanism [13]. Because starch is soluble beginning at ≥100°C, the only α-amylases used in industry are those that are active at temperatures ≤110°C [14]. The majority of thermostable α-amylases used in industry are purified from B. licheniformis, with optimal temperatures of 90°C and requiring the addition of Ca2+ for thermostability [5]. Hyperthermophilic archaea are increasingly attractive to applied research, given that their enzymes show extreme thermostability 15, 16. Recently, numerous amylolytic enzymes have been obtained from Pyrococcus furiosus 17, 18, 19, 20, P. woesei 9, 18, and Thermococcus profundus [21]. Only the P. furiosus and P. woesei intracellular α-amylase genes were cloned and expressed in E. coli [22] and their sequences shared minimal homology with other α-amylase sequences. Starch hydrolysis at high temperatures requires an α-amylase with improved thermostable characteristics that does not require Ca2+. Expression of the enzyme in E. coli transformed with pTY2-α-amyl led to high levels of soluble enzyme [9]. In this study, we cloned the α-amylase gene of P. woesei in E. coli with two changes in amino acids in comparison of previous work [9].
The activity of the soluble enzyme obtained here (185,000 ± 8,000 U/L of growth medium) was higher that previously reported for native α-amylase from P. woesei and the yield achieved during expression of the recombinant enzyme from P. furious in E. coli (10 g, 1,000 U/L, and 38,000 U/L of induced culture ) 18, 19. The amount of soluble enzyme obtained was similar to that reported by Grzybowska et al (195,000 ± 11,000 U/L of the growth medium) [9]. Our result emphasized that the pTYB2-α-amyl plasmid can be used as an expression system for the production of catalytically active recombinant fusion proteins of thermostable α-amylase and intein in E. coli. In regard to other systems used for P. woesei α-amylase expression, the inclusion bodies described here displayed 85% of total enzyme activity [9]. The soluble form of the active enzyme suggested that this expression system is adequate for the production of thermostable α-amylase, which can be separated from the fusion protein in the presence of thiol compounds by heat treatment at ∼85°C.
In summary, this study emphasized that the pTYB2α-amyl plasmid can be used as an expression system for the production of catalytically active, recombinant, thermostable α-amylase in E. coli, with no impact to enzyme solubility by two amino acid mutations in the C- and N-terminal regions.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License (http://creativecommons.org/licenses/by-nc-nd/4.0) which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Zeng F., Cohen A.C. Comparison of alpha-amylase and protease activities of a zoophytophagous and two phytozoophagous Heteroptera. Comp Biochem Physiol A Mol Integr Physiol. 2000 May;126(1):101–106. doi: 10.1016/s1095-6433(00)00193-8. [DOI] [PubMed] [Google Scholar]
- 2.Matsuzaki H., Yamane K., Yamaguchi K. Hybrid alpha-amylases produced by transformants of Bacillus subtilis. I. Purification and characterization of extracellular alpha-amylases produced by the parental strains and transformants. Biochim Biophys Acta. 1974 Sep;365(1):235–247. doi: 10.1016/0005-2795(74)90268-2. [DOI] [PubMed] [Google Scholar]
- 3.Yuuki T., Nomura T., Tezuka H. Complete nucleotide sequence of a gene coding for heat- and pH-stable alpha-amylase of Bacillus licheniformis: comparison of the amino acid sequences of three bacterial liquefying alpha-amylases deduced from the DNA sequences. J Biochem. 1985 Nov;98(5):1147–1156. doi: 10.1093/oxfordjournals.jbchem.a135381. [DOI] [PubMed] [Google Scholar]
- 4.Saito N. A thermophilic extracellular α-amylase from Bacillus licheniformis. Arch Biochem Biophys. 1973 Apr;155(2):290–298. doi: 10.1016/0003-9861(73)90117-3. [DOI] [PubMed] [Google Scholar]
- 5.Violet M., Meunier J.C. Kinetic study of the irreversible thermal denaturation of Bacillus licheniformis alpha-amylase. Biochem J. 1989 Nov;263(3):665–670. doi: 10.1042/bj2630665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson T.H., Tan X., Frey G. A novel, high performance enzyme for starch liquefaction. Discovery and optimization of a low pH, thermostable alpha-amylase. J Biol Chem. 2002 Jul;277(29):26501–26507. doi: 10.1074/jbc.M203183200. [DOI] [PubMed] [Google Scholar]
- 7.Specka U., Mayer F., Antranikian G. Purification and properties of a thermoactive glucoamylase from Clostridium thermosaccharolyticum. Appl Environ Microbiol. 1991 Aug;57(8):2317–2323. doi: 10.1128/aem.57.8.2317-2323.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghasemi A., Salmanian A.H., Sadeqhifard N. Cloning, expression and purification of Pwo polymerase from Pyrococcus woesei. Iran J Microbiol. 2011 Sep;3(3):118–122. [PMC free article] [PubMed] [Google Scholar]
- 9.Grzybowska B., Szweda P., Synowiecki J. Cloning of the thermostable alpha-amylase gene from Pyrococcus woesei in Escherichia coli: isolation and some properties of the enzyme. Mol Biotechnol. 2004 Feb;26(2):101–110. doi: 10.1385/MB:26:2:101. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Fukusumi S., Kamizono A., Horinouchi S. Cloning and nucleotide sequence of a heat-stable amylase gene from an anaerobic thermophile, Dictyoglomus thermophilum. Eur J Biochem. 1988 May;174(1):15–21. doi: 10.1111/j.1432-1033.1988.tb14056.x. [DOI] [PubMed] [Google Scholar]
- 13.Jespersen H.M., MacGregor E.A., Henrissat B. Starch- and glycogen-debranching and branching enzymes: prediction of structural features of the catalytic (beta/alpha)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J Protein Chem. 1993 Dec;12(6):791–805. doi: 10.1007/BF01024938. [DOI] [PubMed] [Google Scholar]
- 14.Adams M.W. Enzymes and proteins from organisms that grow near and above 100 degrees C. Annu Rev Microbiol. 1993 Oct;47:627–658. doi: 10.1146/annurev.mi.47.100193.003211. [DOI] [PubMed] [Google Scholar]
- 15.Vieille C., Burdette D.S., Zeikus J.G. Thermozymes. Biotechnol Annu Rev. 1996;2:1–83. doi: 10.1016/s1387-2656(08)70006-1. [DOI] [PubMed] [Google Scholar]
- 16.Adams M.W., Perler F.B., Kelly R.M. Extremozymes: expanding the limits of biocatalysis. Biotechnology (N Y) 1995 Jul;13(7):662–668. doi: 10.1038/nbt0795-662. [DOI] [PubMed] [Google Scholar]
- 17.Laderman K.A., Davis B.R., Krutzsch H.C. The purification and characterization of an extremely thermostable alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Biol Chem. 1993 Nov;268(32):24394–24401. [PubMed] [Google Scholar]
- 18.Koch R., Spreinat A., Lemke K. Purification and properties of a hyperthermoactive α-amylase from the archaeobacterium Pyrococcus woesei. Arch Microbiol. 1991 Jun;155(6):572–578. [Google Scholar]
- 19.Dong G., Vieille C., Savchenko A. Cloning, sequencing, and expression of the gene encoding extracellular alpha-amylase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol. 1997 Sep;63(9):3569–3576. doi: 10.1128/aem.63.9.3569-3576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown S.H., Costantino H.R., Kelly R.M. Characterization of amylolytic enzyme activities associated with the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl Environ Microbiol. 1990 Jul;56(7):1985–1991. doi: 10.1128/aem.56.7.1985-1991.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan M.K., Mukund S., Kletzin A. Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science. 1995 Mar;267(5203):1463–1469. doi: 10.1126/science.7878465. [DOI] [PubMed] [Google Scholar]
- 22.Laderman K.A., Asada K., Uemori T. Alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. Cloning and sequencing of the gene and expression in Escherichia coli. J Biol Chem. 1993 Nov;268(32):24402–24407. [PubMed] [Google Scholar]