Abstract
5′-(2-Phosphoryl-1,4-dioxobutane) (DOB) is an oxidized abasic site that is produced by several antitumor agents and γ-radiolysis. DOB reacts reversibly with a dA opposite the 3′-adjacent nucleotide to form DNA interstrand cross-links (ICLs), genotoxic DNA lesions that can block DNA replication and transcription. Translesion synthesis (TLS) is an important step in several ICL repair pathways to bypass unhooked intermediates generated by endonucleolytic incision. The instability of DOB-ICLs has made it difficult to learn about their TLS-mediated repair capability and mutagenic potential. We recently developed a method for chemically synthesizing oligonucleotides containing a modified DOB-ICL analogue. Herein, we examined the capabilities of several highly relevant eukaryotic TLS DNA polymerases (pols), including human pol η, pol κ, pol ι, pol ν, REV1, and yeast pol ζ, to bypass this DOB-ICL analogue. The prelesion, translesion, and postlesion replication efficiency and fidelity were examined. Pol η showed moderate bypass activity when encountering the DOB-ICL, giving major products one or two nucleotides beyond the cross-linked template nucleotide. In contrast, DNA synthesis by the other pols was stalled at the position before the cross-linked nucleotide. Steady-state kinetic data and liquid chromatography–mass spectrometry sequencing of primer extension products by pol η unambiguously revealed that pol η-mediated bypass is highly error-prone. Together, our study provides the first set of in vitro evidence that the DOB-ICL is a replication-blocking and highly miscoding lesion. Compared to several other TLS pols examined, pol η is likely to contribute to the TLS-mediated repair of the DOB-ICL in vivo.
Graphical Abstract
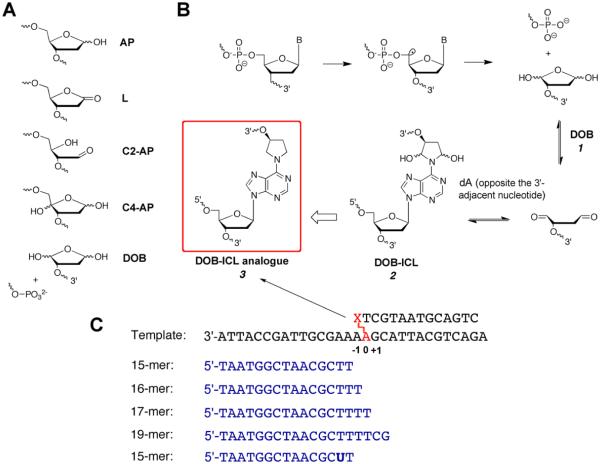
Abasic sites (AP) are the most prevalent endogenous DNA lesions with a steady-state level of approximately 30000 abasic lesions per cell each day.1,2 AP sites are derived from spontaneous depurination and depyrimidination, as well as DNA glycosylase reactions during base excision repair (BER).2 Other abasic sites arise following abstraction of a hydrogen atom from the 2′-deoxyribose ring upon γ-radiolysis and exposure to a number of cytotoxic antitumor agents, such as bleomycin and the enediynes.3,4 These oxidized abasic lesions include 5′-(2-phosphoryl-1,4-dioxobutane) (DOB), C4′-oxidized abasic site (C4-AP), C2′-oxidized abasic sites (C2-AP), and 2-deoxyribonolactone (L) (Figure 1A), resulting from abstraction of a hydrogen atom from positions C5′, C4′, C2′, and C1′, respectively. Although all abasic sites lack a Watson–Crick base, AP and oxidized abasic sites each contribute difierently to mutagenicity in Escherichia coli, most likely because of their specific effects on nucleic acid structure and interactions with polymerases.5–10
Figure 1.
(A) Structures of abasic lesions. (B) Formation of oxidized abasic lesion 5′-(2-phosphoryl-1,4-dioxobutane) (DOB) and its subsequent cross-linking with dA opposite a 3′-adjacent thymidine.13 DOB (1) reversibly reacts with a dA opposite the 3′-adjacent nucleotide to form DNA interstrand cross-links (DOB-ICL, 2). The chemically stable analogue is shown as structure 3. (C) Sequences of the template and primers used in this study.
DOB is formed under O2-defficient conditions via C5′ oxidation that simultaneously yields a single-strand break resulting in a tandem DNA lesion (Figure 1B). The 1,4-dicarbonyl functional group renders DOB very reactive and potentially cytotoxic. DOB reacts readily with lysine residues of DNA polymerase (pol) β or pol λ and irreversibly inhibits these enzymes, which could be deleterious to the base excision repair (BER) pathway (a major pathway for repairing AP).11,12 In addition, DOB reversibly reacts with a dA opposite the 3′-adjacent nucleotide on a complementary strand to form a DNA interstrand cross-link (ICL) in vitro (Figure 1B).13 ICLs are considered to be extremely cytotoxic lesions that block a variety of DNA metabolisms.14,15 Direct characterization of the toxic effect of DOB-derived ICLs is dificult because of the reversibility of the reaction and the short half-life (t1/2 = 11.2 h) of the DOB-ICL lesion.13 To better understand the biological effect of DOB-induced ICLs, we recently synthesized site-specifically modified oligonucleotides containing a DOB-ICL analogue (hereafter termed DOB-ICL).16 We found that misrepair of DOB-ICL by the model nucleotide excision repair enzyme UvrABC results in the formation of a double-strand break (the most deleterious form of DNA damage).17
In mammalian cells, ICL repair involves sequential endonucleolytic excision of the lesion from one strand and then the other.15 In both replication-dependent and -independent ICL repair pathways, translesion synthesis (TLS) is a critical step for bypassing the unhooked intermediates digested from the nontemplate strand or for conducting postreplication gap-filling DNA synthesis.14,15 TLS has been extensively studied for lesions with base modifications; however, the translesion synthesis activities of TLS pols toward ICLs have been studied in only a limited number of instances.18,19 Previous investigations focused on ICLs formed from antitumor reagents, such as psoralen,20 mitomycin C,14 or nitrogen mustard21 and endogenously produced lipid peroxidation products.22–24 These studies have led to the understanding that translesion synthesis activities of various TLS pols are highly dependent on the structure of the ICL lesion and the size of the unhooked intermediates.14,15
Abasic lesion-induced ICLs may be a critically important class of endogenous ICLs considering the abundance of AP.15 Gates and colleagues were the first to unambiguously characterize an AP-induced ICL formed between an abasic lesion and the exocyclic amino group of dG or dA on a complementary strand.25–29 ICLs from DOB and C4-AP were uncovered shortly thereafter, which involve C4-AP or DOB condensed with the N6-amino group of dA on a complementary strand.13,30,31 Using LC–MS, Ravanat and colleagues were the first to identify oxidized abasic lesion-derived ICLs in cellular DNA, whereby an ICL formed between C4-AP and the N4-amino group of dC was detected in cells treated with radiation or bleomycin as well as untreated cells.32 The fact the C4-AP-dervied ICL is present in control cells suggests that endogenous reactive oxygen species are potential sources of oxidized abasic lesion-induced ICLs formed in vivo.32 It remains challenging to detect and measure ICLs in biological samples because many are unstable and exist in low abundance in viable cells.15
To date, little is known about the translesion DNA synthesis activity toward abasic lesion-induced ICLs, and data on the mutagenic properties of these ICLs are lacking. Herein, we used the previously developed strategy to synthesize an oligonucleotide containing a site-specifically modified DOB-ICL analogue.16 This DOB-ICL models an intermediate produced by endonucleolytic incision. Using this substrate, we thoroughly characterized the bypass activities of several important TLS DNA polymerases (pols), including human pol η, pol ι, pol κ, pol ν, REV1, and yeast pol ζ. We found that DOB-ICL is a strong block to the translesion synthesis activities of these enzymes. Among all pols examined, pol η exhibited the highest bypass and postlesion extension activities. Our results suggest that DOB-ICL is highly miscoding and primarily induces the incorporation of dATP opposite the template cross-linked dA. This study provides important insight into the biological consequences of DOB-ICLs.
EXPERIMENTAL PROCEDURES
Materials
All commercial chemicals were from Sigma-Aldrich (St. Louis, MO) or Alfa Aeser (Ward Hill, MA) and were of the highest quality available. Unlabeled dNTPs, T4 polynucleotide kinase, and uracil DNA glycosylase (UDG) were from New England Biolabs (Ipswich, MA). [γ-32P]ATP (specific activity of 4500 Ci mmol−1) was from MP Biomedicals (Santa Ana, CA). Unmodified oligonucleotides were synthesized and gel-purified by Integrated DNA Technologies (Coralville, IA). The oligonucleotide containing a site-specifically modified DOB-ICL analogue was synthesized and purified as described previously.16 The catalytic fragments of human Y-family polymerases pol ι (1–420),33 pol η (1–432),34 pol κ (19–526),35 and REV1 (330–833)36 were purified following previously published protocols; expression vectors were kindly provided by F. P. Guengerich (Vanderbilt University School of Medicine, Nashville, TN). The human DNA polymerase ν expression vector was a generous gift from R. D. Wood (University of Texas M. D. Anderson Cancer Center, Houston, TX), and the full-length recombinant pol ν was purified as described previously.37 Yeast DNA polymerase ζ (pol ζ, two-subunit REV3/REV7 complex) was from Enzymax (Lexington, KY).
Primer Extension Assays
A 15-, 16-, 17-, or 19-mer primer was 5′-[γ-32P]ATP end-labeled and annealed to a 30-mer unmodified or ICL-bearing oligomer (sequences shown in Figure 1). Assays were performed according to a published procedure.38 Specifically, full-length primer extension experiments were conducted at 37 °C with 80 nM duplex DNA (with or without ICL), 80 nM DNA polymerase, four dNTPs at 100 μM each, 4% (v/v) glycerol, 5 mM DTT, 50 mM NaCl, 5 mM MgCl2, and 100 μg mL−1 bovine serum albumin (BSA) in 50 mM Tris-HCl (pH 7.4 at 25 °C). Assays with pol ζ contained 10% glycerol and 90 mM KCl. Single-nucleotide incorporation assays were conducted under steady-state kinetic conditions using 80 nM duplex DNA, 0.5–20 nM DNA polymerase, and varying concentrations of a single dNTP. Reactions were quenched with 50 mM ethylenediaminetetraacetic acid (EDTA, pH 8.0) in 95% formamide (v/v) followed by denaturation at 90 °C. Products were resolved using 18% acrylamide (w/v) gel electrophoresis containing 7 M urea. Results were visualized using a phosphorImaging system (GE healthcare, Typhoon FLA7000) and analyzed with ImageQuant. Data were fit to the Michaelis–Menten equation to obtain kcat and Km using Prism (GraphPad, San Diego, CA).
NanoLC–MS Analysis of Full-Length Extension Products by Pol η
A 15-mer primer 5′-TAATGGCTAACGC-(dU)T-3′ was annealed to a regular or an ICL-containing template oligomer at a 1:1.2 molar ratio. Reaction conditions were similar to those used in full-length primer extension assays except for the following final concentrations: pol η, 1 μM; primer–template complex, 2 μM; glycerol, 2% (v/v); four dNTPs (100 μM each) in a total volume of 50 μL. Polymerization reactions were allowed to proceed for 3 h at 37 °C and terminated with 10 mM EDTA (final concentration). The resulting products were incubated with 10 units of UDG overnight followed by incubation at 90 °C for 1 h in the presence of 0.25 M piperidine.39 The fragmented oligomers were cleaned using a C18 SampliQ solid-phase extraction cartridge (Agilent Technologies). The fractions containing oligomer were dried under vacuum and suspended in 40 μL of water.
NanoLC–MS/MS analysis was performed on a nanoAcquity ultraperformance liquid chromatography system (Waters Corp.) connected to a Finnigan LTQ XL mass spectrometer (Thermo Scientific Corp.). Data were collected under negative ionization mode. The column was a PicoChip column (75 μm inside diameter, 105 mm bed length, and a 15 μm tip, New Objective, Woburn, MA) packed with Reprosil-PUR C18 (3 μm, 120 Å) chromatography medium. Chromatography mobile phase A was 400 mM 1,1,1,3,3,3-hexafluoro-2-propanol in water (pH adjusted to 7.0 with triethylamine), and mobile phase B was methanol. The following gradient program was used at a flow rate of 250 nL min−1: from 0 to 5 min, maintained at 98% A/2% B (v/v); from 5 to 35 min, linear gradient to 30% B (v/v); from 35 to 45 min, linear gradient to 50% B; from 45 to 55 min, held at 50% B; from 55 to 65 min, linear gradient to 2% B; from 65 to 95 min, held at 2% B to re-equilibrate the column. A 3 μL aliquot was injected onto the column. Nanoelectrospray conditions were as follows: ionization voltage, 3 kV; capillary temperature, 300 °C; capillary voltage, −45 V; tube lens voltage, −110 V. MS/MS conditions were as follows: normalized collision energy, 35%; activation Q, 0.250; activation time, 30 ms. Product ion spectra were acquired over the range of m/z 300–2000. The most abundant species was used for CID analysis. Theoretical CID fragmentation of oligonucleotides was calculated using the Mongo Oligo mass calculator version 2.06 (hosted by the University at Albany, State University of New York, Albany, NY).40 The relative yields of extension products were estimated on the basis of the peak areas in extracted ion chromatograms. The sum of the peak area was used for multiply charged species. The peak area ratio of total products to residual primer with a regular duplex was set to 100% (based on the complete primer extension shown in Figure 2A, pol η, Reg). The peak area ratio of each product/residual primer with an ICL-containing duplex was normalized to the peak area ratios found with an undamaged duplex (Table 3).
Figure 2.
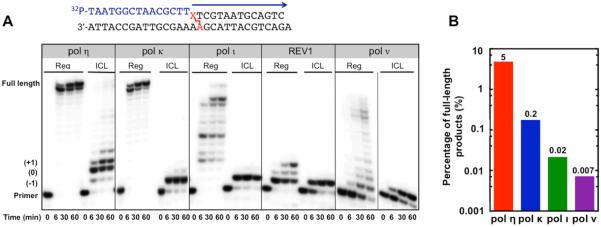
Translesion DNA synthesis across DOB-ICL catalyzed by pol η, pol κ, pol ι, REV1, or pol ν. (A) Assays were performed at 37 °C with 80 nM 15/30-mer duplex (with or without ICL), dNTP mixtures at 100 μM each, 80 nM polymerase, 4% (v/v) glycerol, 5 mM DTT, 50 mM NaCl, 5 mM MgCl2, and 100 μg mL−1 bovine serum albumin (BSA) in 50 mM Tris-HCl (pH 7.4). Reaction mixtures with pol ν contained 10% glycerol and 90 mM KCl. (B) The percentages of full-length products were 5, 0.2, 0.02, and 0.007% for pol η, pol κ, pol ι, and pol ν, respectively. The reaction time was 60 min; the percentage was calculated using the intensity of the full-length product divided by the total intensities of all products and the remaining substrate.
Table 3.
Summary of Pol η-Catalyzed Bypass Products from LC-MS/MS Analysisa
|
|
Normalized % of products |
Nucleotide opposite the 0 position |
|
|---|---|---|---|
| Reg | TTTCGTAATGCAGTCT | 57 | T (correct) |
| TTTCGTAATGCAGTCTA | 43 | ||
| DOB-ICL | TTTCGTAATGCAGTCT | 4 | T (correct) |
| TTTCGTAATGCAGTCTA | 1 | ||
| TTACGTCTTAA | 2 | A (mutagenic) | |
| TTACGTATCAA | 1 | ||
The results were obtained using a regular or ICL-containing duplex. Percents of products were estimated on the basis of the peak area in extracted ion chromatograms. Total product/residual primer ratios obtained with a regular duplex were considered as 100%. Products from ICL bypass were normalized to the product ratio of an undamaged duplex. Nucleotides inserted opposite the cross-linked dA (0 position) are underlined in the extended products.
RESULTS
Biological Relevance of the DOB-ICL-Bearing Substrate
The structurally similar C4-AP lesion forms ICLs in vitro and in cells.30–32 Although DOB-ICL has not yet been detected in cellular DNA, examination of its TLS-mediated repair capability and mutagenic potential is useful given the dearth of information about this and other ICLs involving abasic lesions. Consequently, we synthesized a site-specifically modified oligomer containing a chemically stable DOB-ICL analogue for detailed biochemical characterizations (structure shown in Figure 1B).16 The ICL-harboring template consisted of a 30-mer oligomer template and a 15-mer overhang covalently linked with the N6 atom of a dA on the template strand (Figure 1C). The linkage between the DOB residue and the N6-amino group of dA has been shown to form in vitro.13,30 The 15-mer oligomer overhang mimicked a product containing a single-strand break at the 5′-adjacent nucleotide of DOB (mechanisms of formation shown in Figure 1B) and an 3′-end formed after endonucleolytic processing. The length of the 15-mer oligomer is within the estimated size of an unhooked fragment (4–17 nucleotides).22,41 Therefore, our DOB-ICL-bearing substrate serves as a reasonable model for examination of the bypass capability of TLS pols.
Pol η, but Not Pol ι, Pol κ, Pol ν, or REV1, Bypasses DOB-ICL
In vertebrates, ICLs are repaired in and outside the S phase of the cell cycle.15 The former pathway is triggered by the stalling of a replication fork and involves endonucleases, TLS pols, Fanconi anemia (FA) proteins, and homologous recombination (HR) factors.42 Bypass of unhooked intermediates via TLS is an important step for creating a DNA duplex that is amenable to HR or for gap-filling synthesis in a replication-independent repair pathway.15 Using a DOB-ICL-containing substrate, we tested the bypass capabilities of several important TLS pols, including all four human Y-family DNA polymerases, pol η, pol ι, pol κ, and REV1, as well as an A-family DNA polymerase, pol ν. These TLS pols were reported to have bypass activities in vitro or in vivo toward other types of ICLs with difierent chemical linkages.20,21,23,24 Primer extension assays were performed in the presence of four dNTPs with a DOB-ICL-containing duplex in comparison with a regular duplex (one containing only native nucleotides). DOB-ICL strongly inhibited DNA synthesis of all five TLS pols (Figure 2A). Pol η exhibited translesion and postlesion synthesis activities higher than those of the other four TLS pols, although the full-length extension products were only 5% for pol η relative to 0.2% for pol κ, 0.02% for pol ι, and 0.007% for pol ν after a 60 min reaction (Figure 2B). Pol η was able to moderately bypass the DOB-ICL lesion, producing 17-mer (+2 nt, 33%), 18-mer (+3 nt, 32%), and 19-mer (+4 nt, 12%) oligomers as major products after a 60 min reaction. Pol κ and pols ι weakly bypassed the cross-link and produced nearly no products extended to the 0 position or beyond (−1, 0, and +1 positions are illustrated in Figure 1C). DNA replication by pol ν or REV1 stalled at the −1 position, indicating that DOB-ICL impeded bypass by these polymerases even more strongly. Overall, these results demonstrated that pol η had modest bypass TLS activity across the DOB-ICL lesion, but the other pols examined are unable to bypass the cross-link under these conditions.
Translesion Synthesis across DOB-ICL Was Inefficient and Error-Prone
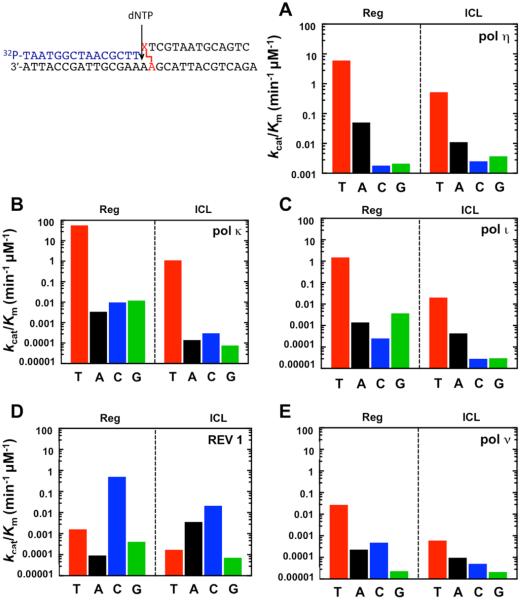
Translesion synthesis can be error-prone and presents a major source of DNA damage-induced mutations.43 On the basis of the mutagenicity of AP and the size of an ICL relative to other alkylated DNA lesions, we hypothesized that the DOB-ICL lesion is highly miscoding. To test this hypothesis, we determined the Michaelis constant (Km), turnover number (kcat), and misincorportation frequency (f) of nucleotide incorporation steps prior to, opposite, and after the cross-linked nucleotide. These biochemical parameters allowed us to quantitatively evaluate the bypass efficiency and mutagenic potential of the DOB-ICL lesion. According to the primer extensions observed (Figure 2A), all five pols facilely incorporated a nucleotide at the −1 position. The catalytic efficiency (kcat/Km) of correct nucleotide (dTTP) incorporation across the −1 position (Figure 3 and Table S1 of the Supporting Information) revealed that relative to a regular dA:dTTP pair, the DOB-ICL-harboring duplex reduced DNA replication efficiency of pols η, ι, κ, and ν and REV1 by 10-, 75-, 50-, 44-, and 9-fold, respectively. For REV1, a 24-fold decrease in its deoxycytidyl transferase efficiency was also observed. The decrease in enzymatic efficiency is likely due to the steric hindrance imposed by the DOB residue as polymerases approach the cross-linked site. The misincorporation frequency ( f), an indication of replication fidelity, of all the pols at the −1 position was comparable to or slightly lower than that of a regular duplex, implying that DNA replication at the −1 position was largely error-free. Together, these data showed that at the −1 position, the presence of DOB-ICL compromised DNA replication efficiency but did not significantly alter the fidelity of DNA replication by TLS pols.
Figure 3.
Catalytic efficiencies (kcat/Km,dNTP) of single-nucleotide incorporation at the −1 position catalyzed by (A) pol η, (B) pol κ, (C) pol ι, (D) REV1, and (E) pol ν. The DNA substrate (with 16-mer primer shown in Figure 1B) was either a regular duplex (Reg) or an ICL-containing duplex (ICL). kcat and Km,dNTP were obtained under steady-state kinetic conditions. Assays were performed at 37 °C with 60 nM 15/30-mer duplex, 0.5–20 nM polymerase, varying dNTP concentrations, 4% (v/v) glycerol, 5 mM DTT, 50 mM NaCl, 5 mM MgCl2, and 100 μg mL−1 bovine serum albumin (BSA) in 50 mM Tris-HCl (pH 7.4). Assays with pol ν contained 10% glycerol. The correct nucleotide was dTTP (red). Complete data are listed in Table S1 of the Supporting Information.
On the basis of the weak primer extension activity at the 0 position seen for pol κ, pol ι, pol ν, and REV1 (Figure 2), we hypothesized that a more significant decrease in replication efficiency occurs at the 0 (translesion) position during bypass. To test this, we analyzed replication efficiency and misincorporation frequency at the 0 position by pols η, κ, and ι (three TLS pols showed bypass activities across DOB-ICL in Figure 2A). A 16-mer oligomer with a 3′-dT was hybridized with a regular or an ICL-containing template based on our findings that dTTP was the nucleotide preferred by these pols at the −1 position (Figure 2 and Table S1 of the Supporting Information). Drastic decreases in the catalytic efficiency for incorporation of the correct dTTP were observed (Table 1), including 4 × 104-fold for pol η, 4 × 106-fold for pol κ, and 5 × 105-fold for pol ι, consistent with the stalling of DNA replication at the −1 position (Figure 2A). Notably, the bypass by pol η and pol κ was highly error-prone. The catalytic efficiencies for incorporation of dATP and dGTP by pol η were 16- and 5-fold higher, respectively, than that of the correct nucleotide, dTTP; dCTP was incorporated at approximately the same rate as dTTP. For pol κ, the three incorrect nucleotides were misinserted approximately as efficiently as the correct dTTP. On the other hand, pol ι seemed to prefer the correct base even in the presence of DOB-ICL, although replication by pol ι was extremely slow (the catalytic efficiency of dTTP was 27-fold lower than that with pol η). The misincorporation frequencies for incorrect nucleotides of pol ι were in the range of 0.01 to 0.001, which were 2 to 3 orders of magnitude higher relative to a regular template. Nevertheless, pol ι was more faithful than pol η and pol κ during bypass, indicating that pol ι may play a role in the error-free bypass of DOB-ICL. Overall, translesion synthesis across the DOB-ICL lesion was very inefficient, and highly error-prone for pol η and pol κ.
Table 1.
Lesion Bypass Activities of Human TLS Pols at the 0 Position (illustrated in Figure 1B) with a Regular (Reg) or DOB-ICL-Containing (ICL) Duplexa
| TLS pol |
template | nucleotide | kcat (min−1) | Km,dNTP (μM) |
kcat/Km,dNTP
(min−1 μM−1) |
fb | decrease in efficiency relative to a regular duplexc |
|---|---|---|---|---|---|---|---|
| pol η | Reg | dTTP | 3.2 ± 0.3 | 0.80 ± 0.33 | 4.1 ± 1.7 | ||
| dATP | 3.1 ± 0.1 | 330 ± 20 | (9.4 ± 0.7) × 10−3 | 2.3 × 10−3 | |||
| dCTP | 1.7 ± 0.0 | 110 ± 9 | (1.6 ± 0.1) × 10−2 | 4.0 × 10−3 | |||
| dGTP | 0.64 ± 0.03 | 69 ± 13 | (9.3 ± 1.7) × 10−3 | 2.3 × 10−3 | |||
| ICL | dTTP | 0.12 ± 0.02 | 1100 ± 300 | (1.1 ± 0.3) × 10−4 | 4 × 104-fold | ||
| dATP | 0.37 ± 0.01 | 200 ± 20 | (1.8 ± 0.2) × 10−3 | 16 | |||
| dCTP | 0.023 ± 0.002 | 230 ± 70 | (1.0 ± 0.3) × 10−4 | 9.0 × 10−1 | |||
| dGTP | 0.083 ± 0.006 | 150 ± 40 | (5.5 ± 1.5) × 10−4 | 4.9 | |||
| pol η | Reg | dTTP | 85 ± 9 | 6.1 ± 1.5 | 14 ± 4 | ||
| dATP | 0.047 ± 0.007 | 580 ± 230 | (8.1 ± 3.4) × 10−5 | 5.8 × 10−6 | |||
| dCTP | 3.0 ± 0.2 | 790 ± 150 | (3.8 ± 0.8) × 10−3 | 2.7 × 10−4 | |||
| dGTP | 0.15 ± 0.02 | 780 ± 270 | (1.9 ± 0.7) × 10−4 | 1.4 × 10−5 | |||
| ICL | dTTP | 0.0021 ± 0.0002 | 570 ± 140 | (3.6 ± 1.0) × 10−6 | 4 × 106-fold | ||
| dATP | 0.0052 ± 0.0004 | 620 ± 100 | (8.4 ± 1.5) × 10−6 | 2.3 | |||
| dCTP | 0.0012 ± 0.0001 | 240 ± 30 | (5.3 ± 0.6) × 10−6 | 1.4 | |||
| dGTP | 0.0019 ± 0.0002 | 510 ± 150 | (3.8 ± 1.2) × 10−6 | 1.0 | |||
| pol t | Reg | dTTP | 23 ± 2 | 11 ± 1 | 2.1 ± 0.3 | ||
| dATP | 0.021 ± 0.002 | 340 ± 100 | (6.1 ± 1.9) × 10−5 | 2.9 × 10−5 | |||
| dCTP | 0.30 ± 0.03 | 120 ± 40 | (2.5 ± 0.9) × 10−3 | 1.2 × 10−3 | |||
| dGTP | 0.014 ± 0.002 | 2300 ± 600 | (6.2 ± 2.0) × 10−6 | 2.9 × 10−6 | |||
| ICL | dTTP | 0.0025 ± 0.0005 | 610 ± 300 | (4.1 ± 2.1) × 10−6 | 5 × 105-fold | ||
| dATP | 0.00073 ± 0.00014 | 2600 ± 800 | (2.8 ± 1.0) × 10−7 | 6.8 × 10−2 | |||
| dCTP | ~0.000016d | >3000 | <5.3 × 10−8 | <1.3 × 10−2 | |||
| dGTP | ~0.000037d | >3000 | <1.2 × 10−8 | <2.9 × 10−3 |
The correct nucleotide is shown in italics. Steady-state kinetic assays of single-nucleotide incorporation were performed at 37 °C with 80–100 nM 16/30-mer duplex, varying dNTP concentrations, 0.5–50 nM polymerase, 4% (v/v) glycerol, 5 mM DTT, 50 mM NaCl, 5 mM MgCl2, and 100 μg mL−1 bovine serum albumin (BSA) in 50 mM Tris-HCl (pH 7.4).
Misinsersion frequency f = (kcat/Km,dNTP)incorrect/(kcat/Km,dNTP)correct.
Decrease in catalytic efficiency relative to an undamaged duplex is calculated from (kcat/Km,dNTP)undamaged,dTTP/(kcat/Km,dNTP)ICL,dTTP.
Because of the extremely low conversion, the kcat of incorporation of dCTP and dGTP by pol t with an ICL-bearing duplex was estimated by using 2 mM nucleotide, and catalytic efficiency was estimated using a Km of ~3000 μM.
Pol η, but Not Yeast Pol ζ, Was Able To Perform Postlesion Synthesis
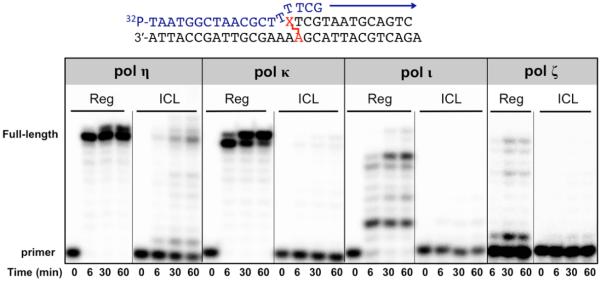
In eukaryotes, translesion DNA synthesis is achieved by one or two TLS DNA polymerases in concert before a replicative polymerase reclaims responsibility for copying the template downstream of the lesion.44 We sought to address whether pol η can extend the mispaired primer–template substrate and whether a second polymerase may contribute to the extension steps beyond the cross-linked nucleotide. As described above, pol η incorporated the incorrect dATP 16-fold more efficiently than the correct dTTP at the 0 position (Table 1); we designed a primer with an incorrect dA mispaired with the cross-linked dA of the template as well as a primer with a correct dT paired with the template dA and examined the extension pattern using human pol η or Saccharomyces cerevisiae pol ζ. The yeast pol ζ two-subunit complex (REV3/REV7) was used because of the unavailability of recombinant mammalian pol ζ. In the case of the DOB-ICL-bearing duplex, pol η readily extended the primer with a paired or mispaired primer–template substrate (Figure 4), suggesting that pol η tolerates the bulky cross-link during catalysis. In contrast, yeast pol ζ, a well-known “extender” for distorted substrates or mispaired primer termini,45 exhibited nearly no polymerase activity.
Figure 4.
Full-length extension by human pol η or yeast pol ζ (REV3/REV7) from paired (17T) or mispaired (17A) primer–template substrate. A duplex (with 17-mer primer shown in Figure 1B) was either a regular (Reg) or ICL-harboring (ICL) duplex. Experiments were conducted at 37 °C with 80 nM duplex DNA, 80 nM DNA polymerase, four dNTPs at 100 μM each, 4% (v/v) glycerol, 5 mM DTT, 50 mM NaCl, 5 mM MgCl2, and 100 μg mL−1 bovine serum albumin (BSA) in 50 mM Tris-HCl (pH 7.4 at 25 °C). Assays with pol ζ contained 10% glycerol and 90 mM KCl.
We further characterized the specificity of incorporation of a nucleotide by pol η at the +1 position to better understand the change in catalytic efficiency and replication fidelity during postlesion DNA synthesis. With a paired primer–template substrate (Reg-17T or ICL-17T in Table 2), pol η preferred to insert a correct C opposite the +1 position of the template, and the catalytic efficiency with an ICL-bearing substrate was 4000-fold lower than that of a regular duplex. Pol η retained its replication fidelity. The misincorporation frequency when presented with the DOB-ICL substrate is similar to that observed when acting on a nonadducted duplex. When encountering a mispaired primer terminus (Reg-17A or ICL-17A in Table 2), pol η maintained the preference of inserting a correct C at the +1 position. In the case of an ICL-containing duplex, the catalytic efficiency of dCTP [(6.0 ± 0.8) × 10−3 min−1 μM−1] with a mispaired substrate was similar to that of a paired substrate [(3.8 ± 0.6) × 10−3 min−1 μM−1], consistent with our observations that pol η readily extended the mispaired primer–template duplex (Figure 4) and with previous findings that pol η catalyzed extension from mismatched nucleotides opposite a cis-syn thymine-thymine dimer (TT dimer).46
Table 2.
| TLS pol |
template | nucleotide | kcat (min−1) | Km,dNTP (μM) |
kcat/Km,dNTP
(min−1 μM−1) |
fb | decrease in efficiency relative to a regular duplexc |
|---|---|---|---|---|---|---|---|
| pol η | Reg-17T | dCTP | 36 ± 3 | 2.3 ± 0.5 | 15 ± 4 | ||
| dATP | 6.4 ± 0.2 | 180 ± 20 | (3.5 ± 0.3) × 10−2 | 2.3 × 10−3 | |||
| dTTP | 8.6 ± 0.2 | 190 ± 10 | (4.5 ± 0.3) × 10−2 | 2.9 × 10−3 | |||
| dGTP | 1.1 ± 0.0 | 12 ± 3 | (9.9 ± 2.4) × 10−2 | 6.6 × 10−3 | |||
| ICL-17T | dCTP | 0.85 ± 0.04 | 220 ± 30 | (3.8 ± 0.6) × 10−3 | 4 × 103-fold | ||
| dATP | 0.042 ± 0.008 | 2600 ± 800 | (1.6 ± 0.6) × 10−5 | 4.3 × 10−3 | |||
| dTTP | 0.0063 ± 0.0007 | 1400 ± 200 | (4.3 ± 0.9) × 10−6 | 1.1 × 10−3 | |||
| dGTP | 0.0017 ± 0.0002 | 860 ± 20 | (1.9 ± 0.5) × 10−6 | 5.1 × 10−4 | |||
| Reg-17A | dCTP | 3.5 ± 0.2 | 4.0 ± 0.6 | 0.89 ± 0.14 | |||
| dATP | 1.3 ± 0.0 | 88 ± 14 | (1.5 ± 0.2) × 10−2 | 1.7 × 10−2 | |||
| dTTP | 1.6 ± 0.0 | 110 ± 10 | (1.5 ± 0.2) × 10−2 | 1.7 × 10−2 | |||
| dGTP | 0.93 ± 0.03 | 54 ± 12 | (1.7 ± 0.4) × 10−2 | 1.9 × 10−2 | |||
| ICL-17A | dCTP | 0.55 ± 0.01 | 160 ± 20 | (6.0 ± 0.8) × 10−3 | 1.5 × 102-fold | ||
| dATP | 0.046 ± 0.001 | 390 ± 40 | (2.9 ± 0.3) × 10−4 | 4.8 × 10−2 | |||
| dTTP | 0.021 ± 0.001 | 91 ± 13 | (5.3 ± 0.5) × 10−5 | 8.8 × 10−3 | |||
| dGTP | 0.035 ± 0.001 | 180 ± 20 | (1.9 ± 0.2) × 10−4 | 3.2 × 10−3 |
The primer terminal nucleotide is either a correct dT or incorrect dA. The correct nucleotide to be paired with the subsequent template base is shown in italics. The assays were performed at 37 °C with 80 nM 17/30-mer duplex, varying dNTP concentrations, 0.5-40 nM pol η, 4% (v/v) glycerol, 5 mM DTT, 50 mM NaCl, 5 mM MgCl2, and 100 μg mL−1 bovine serum albumin (BSA) in 50 mM Tris-HCl buffer (pH 7.4).
Misinsersion frequency f = (kcat/Km,dNTP)incorrect/(kcat/Km,dNTP)correct.
Decrease in catalytic efficiency relative to an undamaged duplex is calculated from (kcat/Km,dNTP)undamaged,dCTP/(kcat/Km,dNTP)ICL,dCTP.
Pol η efficiently incorporated four nucleotides on a 15-mer primer (Figure 2A), correlating to both translesion and extension steps during bypass. To further investigate the activity of a second polymerase for subsequent extension, we used a 19-mer primer to mimic an intermediate from pol η-catalyzed insertion and tested the primer extension capability of pol η, pol ι, pol κ, and yeast pol ζ (Figure 5). Pols ι, κ, and ζ exhibited no bypass activity under this condition, whereas pol η converted 30% of the primer to full-length (or longer) products in a 60 min reaction. Together, our data demonstrated that yeast pol ζ was unable to extend either a 17-mer primer–template duplex or a 19-mer primer–template duplex when DOB-ICL is present. Pol η, on the other hand, showed activities for both translesion and postlesion syntheses. These results suggest that pol η was able to conduct the bypass reaction and produce mutagenic products in vitro.
Figure 5.
Postlesion DNA synthesis by human pol η, pol κ, or pol ι or yeast pol ζ (REV3/REV7) with a duplex containing a 19-mer primer shown in Figure 1B. The DNA subtrate was either a regular (Reg) or ICL-harboring (ICL) duplex. Experiments were conducted at 37 °C with 80 nM duplex DNA, 80 nM DNA polymerase, four dNTPs at 100 μM each, 4% (v/v) glycerol, 5 mM DTT, 50 mM NaCl, 5 mM MgCl2, and 100 μg mL−1 bovine serum albumin (BSA) in 50 mM Tris-HCl (pH 7.4 at 25 °C). Assays with pol ζ contained 10% glycerol and 90 mM KCl.
LC–MS Sequencing of Extension Products by Pol η
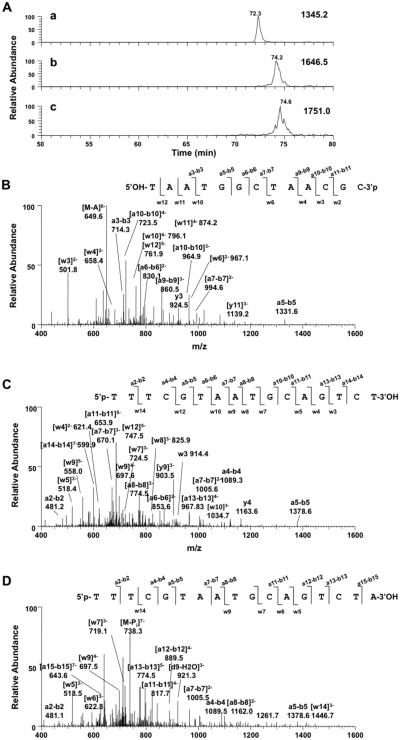
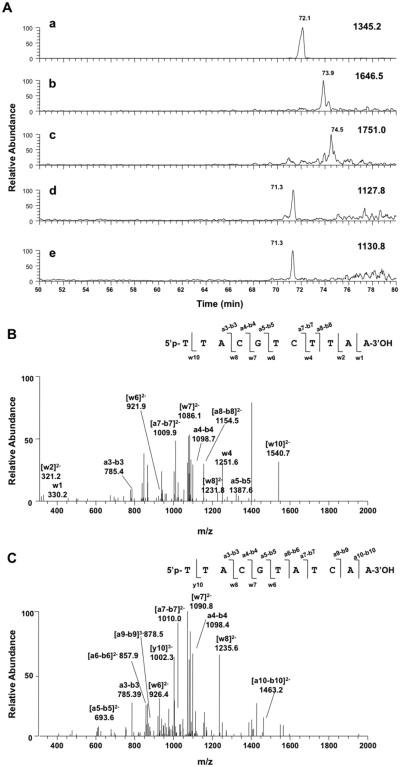
Results from primer extension assays (Figures 2, 4, and 5) provided insight into the extension capabilities of various pols but did not reveal the identity or the yield of mutagenic products. Miscoding frequencies obtained from steady-state kinetic assays are useful in correlating lesion-induced mutagenesis with a certain mutation pattern47,48 but cannot reveal frameshift mutations. LC–MS sequencing of polymerization products is a powerful method for detecting mutagenic products, especially deletions or insertions.49,50 LC–MS analysis can provide valuable information that is complementary to the gel electrophoretic and steady-state kinetic experiments. To gain further insight into the fidelity of DOB-ICL bypass, we performed pol η-catalyzed primer extension reactions in the presence of four dNTPs at physiological concentrations (100 μM for each dNTP)51 using a regular or an adducted duplex. Products were then sequenced using nanoLC–MS/MS to identify point mutations and/or deletions. With a regular duplex, pol η extended the primer with high fidelity and produced approximately 60% of full-length products and the other 40% with an extra A added at the 3′-end, which is commonly seen during in vitro polymerizations (Table 3).50 These results are consistent with earlier observations from electrophoretic analysis (Figure 2A) that pol η readily extends the primer to a full-length product in a regular duplex. The sequences of polymerase-extended products were confirmed by correlating the MS/MS fragmentation patterns and the retention times of extracted ion chromatograms with those obtained for authentic standards (Figure 6 and Figure S2 of the Supporting Information).
Figure 6.
LC–MS analysis of pol η-catalyzed primer extension with an undamaged duplex. (A) Extracted ion chromatograms of (a) the residual primer (m/z 1345.2), (b) a full-length product (m/z 1646.5), and (c) a product with an extra A at the 3′-end (m/z 1751.0). Collision-induced dissociation spectra from LC–MS/MS analysis are shown in panels B (product ions of [M − 6H+]6− 672.0, same species as [M − 3H+]3− 1345.2), C (product ions of [M − 6H+]6− 616.7, same species as [M − 3H+]3− 1646.5), and D (product ions [M − 6H+]6− 656.0, same species as [M − 3H+]3− 1751.0) with product ions that matched theoretical fragmentation patterns labeled. Reaction mixtures contained 1 μM pol η, 2 μM primer–template complex, 2% (v/v) glycerol, four dNTPs (100 μM each), 5 mM DTT, 50 mM NaCl, 5 mM MgCl2, and 50 μg mL−1 BSA in a total volume of 50 μL.
In the case of translesion DNA synthesis, pol η produced error-free products that are full-length or with an extra A at the 3′-end from a DOB-ICL duplex (both contained the correct dT opposite the cross-linked template dA). However, the yields of these products were only ~5% of those of the comparable products from the undamaged substrate, in concert with the low yield of full-length products shown in Figure 2B. The retention times of these products correlated well with those resulting from an undamaged duplex, as well as authentic standards (Figures 6 and 7 and Figure S2 of the Supporting Information). The fragmentation patterns of full-length products obtained with an ICL-bearing duplex are similar to those in panels C and D of Figure 6 and are shown in Figure S1 of the Supporting Information. In addition, two highly mutagenic products were observed during pol η-catalyzed ICL bypass (Figure 7B,C). These products contained an A opposite the cross-linked dA on the template (0 position), consistent with the highest miscoding frequency observed for dATP at the 0 position in steady-state kinetic analysis (Table 1). Moreover, these partially extended products (11-mer instead of 16-mer oligomer for full-length products) contained another four misincorporated nucleotides at the 3′-end, i.e., -TTAA-3′ for one and -TCAA-3′ for the other. The chromatographic retention times and MS/MS fragmentation patterns of the partially extended products correlate well with those of authentic standards (Figure 7 and Figure S3 of the Supporting Information). Although the normalized percentage was only 3%, these highly miscoded products account for nearly half of the all extended primers observed during DOB-ICL bypass under our experimental conditions (Table 3).
Figure 7.
LC–MS analysis of pol η-catalyzed primer extension with an ICL-contaning duplex. (A) Extracted ion chromatograms of (a) residual primer (m/z 1345.2, [M − 3H+]3–), (b) a full-length product (m/z 1646.5, [M − 3H+]3−), (c) a product with an extra A at the 3′-end (m/z 1751.0, [M − 3H+]3−), and (d and e) two truncated products at m/z 1127.8 ([M − 3H+]3−) and m/z 1130.8 ([M − 3H+]3−), respectively. Collision-induced dissociation spectra from LC–MS/MS analysis are shown in panels B (product ions from m/z 1127.8) and C (product ions from m/z 1130.8) with observed ions that matched the theoretical fragmentation patterns labeled. Reaction conditions were the same as those described in the legend of Figure 6.
Overall, results from the LC–MS analysis were in agreement with data from gel electrophoretic and steady-state kinetic analyses. Several key observations are summarized as follows. (1) Bypass of DOB-ICL by human TLS pols was inefficient for pols η, ι, κ, ν, and REV1, and the highest yield of full-length products was observed with pol η. (2) Opposite the cross-linked dA, DOB-ICL is highly miscoding and induced dA:dA mispairing. (3) Pol η, but not yeast pol ζ, was able to extend the primer with a paired or mispaired terminus. (4) With the current DOB-ICL model tested, mutagenic nucleotide insertions three nucleotides downstream of the cross-linked site were also observed.
DISCUSSION
ICLs derived from abasic sites could be a tremendously important class of ICLs formed endogenously, because of the abundance of abasic lesions.15 Previous studies have focused on ICLs formed by the DNA cross-linking reagents, such as psoralen,20 mitomycin C,14 or nitrogen mustard,21 and bis-electrophile acrolein.22–24 However, little is known about the potential deleterious effect imposed by oxidized abasic site-induced ICLs. A recent study from the Gates lab demonstrated that a chemically reversible dA abasic lesion cross-linked via a Schiff base linkage blocks the DNA replication of a model replicative DNA polymerase, bacteriophage φ29.52 This study showed that a chemically reversible dA–AP cross-link behaved like an irreversible, covalent DNA–DNA cross-link during DNA replication. The half-life of DOB-ICL (11.2 h) under physiological conditions13 could be long enough for DOB-ICL to act as a replication-blocking and highly miscoding lesion.
To test this hypothesis, we constructed an oligonucleotide with a site-specifically modified, chemically stable DOB-ICL analogue and characterized the bypass activities of several important human TLS pols (η, ι, κ, ν, and REV1) and yeast pol ζ. Our data suggest that among the TLS pols examined, pol η is most likely to contribute to the TLS-mediated repair of DOB-ICL in vivo. Given the current two-polymerase bypass model44 and our observation that pol η primarily catalyzes the formation of products extended to +1 and +2 positions (Figure 2), we propose that pol η is an important enzyme in the insertion steps of TLS-mediated repair of DOB-ICL. The importance of pol η in TLS of DOB-ICL is consistent with previous in vitro and in vivo studies with difierent ICL substrates. In vitro pol η bypasses a number of structurally distinct ICLs, such as acrolein-derived N2-G-N2-G ICLs24 and ICLs caused by cisplatin and nitrogen mustards.21 Using plasmid-borne psoralen and mitomycin C-derived ICLs in human cell cultures, pol η has been shown to be involved but not essential in replication-independent ICL repair.53,54 It is well documented that TLS pols act redundantly during lesion bypass;55,56 thus, we cannot exclude the possibility of pol ι and pol κ serving as backups for pol η in DOB-ICL repair in vivo.
Other TLS pols tested exhibited either minimal (pol ι and pol κ) or no (REV1 and pol ν) bypass activities. Pol κ showed low activities in replicating across and past the DOB-ICL substrate (Figures 2 and 5). Relative to that of a regular duplex, a drastic decrease (4 × 106-fold) in the catalytic efficiency of dTTP (opposite the cross-linked dA) was observed for pol κ with an ICL-containing duplex (Table 1), suggesting that DOB-ICL was a poor substrate for pol κ-catalyzed replication. On the other hand, pol κ is known for its ability to replicate through bulky N2-G adducts, such as those derived from genotoxic compounds benzo[a]pyrene and benz[a]anthracene.57,58 In keeping with its role in replicating past N2-G adducts (minor groove adducts), pol κ also efficiently replicates past the acrolein-derived N2-G-N2-G ICLs in vitro.22 Recently, using a knock-in mouse model that express catalytically inactive pol κ, Takeiri et al. reported in vivo evidence that pol κ is responsible for error-free bypass across ICLs induced by mitomycin C (a type of N2-G-N2-G ICL).59 In contrast, only moderate bypass activities of pol κ were observed in vitro toward N7-G-N7-G (major groove lesions) ICLs derived from cisplatin and nitrogen mustard.21 Therefore, given that DOB-ICL is a major groove lesion, it is not surprising that pol κ showed low bypass activity.
Results with pol ι were similar to those obtained with pol κ: minimal bypass and extension activities were observed with pol ι (Figures 2 and 5). On the basis of the structural evidence that pol ι rotates the template purines into the syn conformation to facilitate DNA synthesis,60 it is unlikely that a cross-link can undergo such a conformational change. Indeed, in vitro results showed that pol ι incorporates a correct dCTP opposite the acrolein-derived N2-G-N2-G ICL but could not replicate past the lesion.24 We found that pol ι was the most accurate enzyme for incorporation of a nucleotide opposite the cross-linked dA (Table 1). Together with its previously suggested roles, pol ι could be involved in error-free nucleotide insertion opposite the DOB-ICL lesion but is unlikely to be involved in extension steps.
Although the exact physiological function of pol ν is still under investigation, it is another TLS pol that might be involved in DNA interstrand cross-link and DNA–protein cross-link repair.61 In vitro experiments suggested a role for pol ν in bypassing major groove ICL adducts.23 Studies with human cells have demonstrated that siRNA-mediated knock-down of pol ν sensitized cells to a DNA cross-linking agent mitomycin C.62,63 We found that pol ν could not replicate past the DOB-ICL lesion (Figure 2). This is difierent from previous reports in which pol ν bypassed major groove adducts, N6-A-N6-A ICLs and N6-dA-protein cross-links,23 which once again emphasizes the importance of ICL structures in affecting bypass outcomes.
REV1 maintained a 2′-deoxycytidyl transferase activity at the −1 position of the DOB-ICL-containing template (Figure 2), with a 3-fold decrease in catalytic efficiency of dCTP relative to that of a regular duplex (Table S1). However, insertion of dCTP opposite dA at the −1 position is mutagenic, suggesting that it is unlikely that the catalytic activity of REV1 contributes to the error-free bypass of DOB-ICL in vivo. This property of DOB-ICL is distinct from the behavior of REV1 when it encounters other DNA ICLs. For instance, the dCTP transferase activity of REV1 has been suggested to contribute to error-free bypass of ICLs cross-linked with a template dG.15 Perhaps the most well-known function of REV1 is its role as a scafflold protein during TLS. For example, the coordination of REV1 and pol ζ is particularly necessary in replication-dependent and -independent ICL repair pathways.64 This is supported by the observations that REV1-, REV3-, or REV7-defficient mammalian cells are uniquely sensitive to cisplatin, nitrogen mustard, and mitomycin C treatment.65–68 Studies with Xenopus egg extracts also demonstrated the importance of pol ζ and REV1 in replication-dependent ICL repair.69
Pol ζ is considered as an efficient “extender” in bypassing a number of DNA lesions.45,70 In addition to the importance of pol ζ in ICL repair demonstrated by in vivo experiments,45,70 yeast pol ζ extends a primer from a mismatched terminal nucleotide positioned opposite a cross-linked dG in an N2-G-N2-G ICL in vitro.22 Yeast and human pol ζ are involved in TLS-mediated repair of ICLs formed by psoralen or mitomycin C.70 To explore the role of pol ζ as an extension polymerase, we used S. cerevisiae pol ζ two-subunit complex (REV3/REV7) to test the capability of pol ζ to extend from a correctly or incorrectly paired primer terminal nucleotide opposite cross-linked dA in a DOB-ICL-containing duplex. With the current DOB-ICL model, yeast pol ζ did not show any primer extension activity, whereas pol η extended the primer to full-length products regardless of primer–template base pairing (Figure 4). The fact that yeast pol ζ is able to extend the primer–template terminus in N2-G-N2-G ICL but not DOB-ICL underscores the importance of the structure of the DNA damage and the length of the unhooked intermediate in determining the repair capabilities of enzymes.71 In addition, other accessory proteins might be required to facilitate the bypass in vivo.45 Consequently, additional studies are needed to establish the potential role of human pol ζ in DOB-ICL repair.
Our steady-state kinetic analysis and LC–MS sequencing studies demonstrate that DOB-ICL is highly miscoding (Tables 1 and 3). Pol η-mediated primer extension yielded 50% of the products containing a dA opposite the cross-linked template dA, which is indicative of the potential to induce A to T transversion mutations. Our electrophoretic analysis demonstrated that pol η produced partially extended primers. The LC–MS results confirmed this observation and revealed that the partially extended primers (four nucleotides shorter than the full-length products) contained randomly incorporated nucleotides at the 3′-end. Although no frameshift mutations were observed, DOB-ICL-induced mutagenic products account for a significant portion (50%) of those formed during pol η-catalyzed translesion DNA synthesis. Although the structural basis for miscoding is unknown, we speculate that disruption in base pairing due to cross-linking of the N6-amino group of dA with the abasic site on the opposing strand is involved.
In summary, we have used the DOB-ICL lesion as a prototypical lesion to characterize the mutagenic properties of oxidized abasic site-induced ICLs. DOB-ICL impeded translesion DNA synthesis and induced mutagenic bypass products. Together with the important role of pol η in ICL repair suggested by a recent proteomic analysis,72 we propose that pol η is likely to contribute to the replication-dependent repair of DOB-ICL in vivo in concert with other accessory and backup proteins. Highly sensitive and specific techniques are warranted to demonstrate the formation and relative abundance of a spectrum of abasic lesion-induced ICLs in biological samples.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Profs. F. Peter Guengerich and Richard D. Wood for providing expression plasmids of human TLS pols. We thank Prof. Kei-ichi Takata for helpful discussions about the expression and purification of pol ν.
Funding
This work was supported in part by Central Michigan University start-up funds (to L.Z.), a Central Michigan University Faculty Research and Creative Endeavors Award (to L.Z.), the U.S. Army Research Office (Grant W911NF-15-1-0140 to L.Z.), ResearchRewards from TriLink Biotechnologies (to L.Z.), and National Institute of General Medical Sciences Grant GM-063028 (to M.M.G.).
ABBREVIATIONS
- AP
abasic sites
- BER
base excision repair
- BSA
bovine serum albumin
- C2-AP
C2′-oxidized abasic sites
- C4-AP
C4′-oxidized abasic sites
- DOB
5′-(2-phosphoryl-1,4-dioxobutane)
- DTT
dithiothreitol
- HR
homologous recombination
- ICL
DNA interstrand cross-link
- L
2-deoxyribonolactone
- LC–MS
liquid chromatography–mass spectrometry
- NER
nucleotide excision repair
- pol
DNA polymerase
- TLS
translesion synthesis
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b01027.
Steady-state kinetic parameters for single-base incorporation at the −1 position (Table S1) and nanoLC-MS/MS analysis of pol η-catalyzed bypass products and authentic standards (Figures S1–S3). (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Swenberg JA, Lu K, Moeller BC, Gao LN, Upton PB, Nakamura J, Starr TB. Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. 2011;120:S130–S145. doi: 10.1093/toxsci/kfq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Nakamura J, Mutlu E, Sharma V, Collins L, Bodnar W, Yu R, Lai Y, Moeller B, Lu K, Swenberg J. The endogenous exposome. DNA Repair. 2014;19:3–13. doi: 10.1016/j.dnarep.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Pitié M, Pratviel G. v. Activation of DNA carbon–hydrogen bonds by metal complexes. Chem. Rev. 2010;110:1018–1059. doi: 10.1021/cr900247m. [DOI] [PubMed] [Google Scholar]
- (4).Greenberg MM. Abasic and oxidized abasic site reactivity in DNA: enzyme inhibition, cross-linking, and nucleosome catalyzed reactions. Acc. Chem. Res. 2014;47:646–655. doi: 10.1021/ar400229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- (6).Kroeger KM, Goodman MF, Greenberg MM. A comprehensive comparison of DNA replication past 2-deoxyribose and its tetrahydrofuran analog in Escherichia coli. Nucleic Acids Res. 2004;32:5480–5485. doi: 10.1093/nar/gkh873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kroeger KM, Jiang YL, Kow YW, Goodman MF, Greenberg MM. Mutagenic effects of 2-deoxyribonolactone in Escherichia coli. An abasic lesion that disobeys the A-rule. Biochemistry. 2004;43:6723–6733. doi: 10.1021/bi049813g. [DOI] [PubMed] [Google Scholar]
- (8).Kroeger KM, Kim J, Goodman MF, Greenberg MM. Effects of the C4′-oxidized abasic site on replication in Escherichia coli. An unusually large deletion is induced by a small lesion. Biochemistry. 2004;43:13621–13627. doi: 10.1021/bi048337r. [DOI] [PubMed] [Google Scholar]
- (9).Kroeger KM, Kim J, Goodman MF, Greenberg MM. Replication of an oxidized abasic site in Escherichia coli by a dNTP-stabilized misalignment mechanism that reads upstream and downstream nucleotides. Biochemistry. 2006;45:5048–5056. doi: 10.1021/bi052276v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Huang H, Greenberg MM. Hydrogen bonding contributes to the selectivity of nucleotide incorporation opposite an oxidized abasic lesion. J. Am. Chem. Soc. 2008;130:6080–6081. doi: 10.1021/ja801715c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Guan L, Bebenek K, Kunkel TA, Greenberg MM. Inhibition of short patch and long patch base excision repair by an oxidized abasic site. Biochemistry. 2010;49:9904–9910. doi: 10.1021/bi101533a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Guan L, Greenberg MM. Irreversible inhibition of DNA polymerase β by an oxidized abasic lesion. J. Am. Chem. Soc. 2010;132:5004–5005. doi: 10.1021/ja101372c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Guan L, Greenberg MM. DNA interstrand cross-link formation by the 1,4-dioxobutane abasic lesion. J. Am. Chem. Soc. 2009;131:15225–15231. doi: 10.1021/ja9061695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ho TV, Schärer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ. Mol. Mutagen. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- (15).Clauson C, Schärer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor Perspect. Biol. 2013;5:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ghosh S, Greenberg MM. Synthesis of cross-linked DNA containing oxidized abasic site analogues. J. Org. Chem. 2014;79:5948–5957. doi: 10.1021/jo500944g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ghosh S, Greenberg MM. Nucleotide excision repair of chemically stabilized analogues of DNA interstrand cross-links produced from oxidized abasic sites. Biochemistry. 2014;53:5958–5965. doi: 10.1021/bi500914d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Smith LA, Makarova AV, Samson L, Thiesen KE, Dhar A, Bessho T. Bypass of a psoralen DNA interstrand cross-link by DNA polymerases β, ι, and κ in vitro. Biochemistry. 2012;51:8931–8938. doi: 10.1021/bi3008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Schärer OD. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 2011;39:7455–7464. doi: 10.1093/nar/gkr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase κ in the processing of N2-N2-guanine interstrand cross-links. J. Biol. Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yamanaka K, Minko IG, Takata K.-i., Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel enzymatic function of DNA polymerase ν in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links. Chem. Res. Toxicol. 2010;23:689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Klug AR, Harbut MB, Lloyd RS, Minko IG. Replication Bypass of N2-Deoxyguanosine Interstrand Cross-Links by Human DNA Polymerases η and ι. Chem. Res. Toxicol. 2012;25:755–762. doi: 10.1021/tx300011w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dutta S, Chowdhury G, Gates KS. Interstrand cross-links generated by abasic sites in duplex DNA. J. Am. Chem. Soc. 2007;129:1852–1853. doi: 10.1021/ja067294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Johnson KM, Price NE, Wang J, Fekry MI, Dutta S, Seiner DR, Wang Y, Gates KS. On the formation and properties of interstrand DNA–DNA cross-links forged by reaction of an abasic site with the opposing guanine residue of 5′-CAp sequences in duplex DNA. J. Am. Chem. Soc. 2013;135:1015–1025. doi: 10.1021/ja308119q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Price NE, Johnson KM, Wang J, Fekry MI, Wang Y, Gates KS. Interstrand DNA–DNA cross-link formation between adenine residues and abasic sites in duplex DNA. J. Am. Chem. Soc. 2014;136:3483–3490. doi: 10.1021/ja410969x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Catalano MJ, Liu S, Andersen N, Yang Z, Johnson KM, Price NE, Wang Y, Gates KS. Chemical structure and properties of interstrand cross-links formed by reaction of guanine residues with abasic sites in duplex DNA. J. Am. Chem. Soc. 2015;137:3933–3945. doi: 10.1021/jacs.5b00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Price NE, Catalano MJ, Liu S, Wang Y, Gates KS. Chemical and structural characterization of interstrand cross-links formed between abasic sites and adenine residues in duplex DNA. Nucleic Acids Res. 2015;43:3434–3441. doi: 10.1093/nar/gkv174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Sczepanski JT, Jacobs AC, Greenberg MM. Self-promoted DNA interstrand cross-link formation by an abasic site. J. Am. Chem. Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- (31).Sczepanski JT, Jacobs AC, Majumdar A, Greenberg MM. Scope and mechanism of interstrand cross-link formation by the C4′-oxidized abasic aite. J. Am. Chem. Soc. 2009;131:11132–11139. doi: 10.1021/ja903404v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Regulus P, Duroux B, Bayle P-A, Favier A, Cadet J, Ravanat J-L. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Pence MG, Choi JY, Egli M, Guengerich FP. Structural basis for proficient incorporation of dTTP opposite O6-methylguanine by human DNA polymerase iota. J. Biol. Chem. 2010;285:40666–40672. doi: 10.1074/jbc.M110.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Choi J-Y, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J. Mol. Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- (35).Zhao L, Pence MG, Eoff RL, Yuan S, Fercu CA, Guengerich FP. Elucidation of kinetic mechanisms of human translesion DNA polymerase κ using tryptophan mutants. FEBS J. 2014;281:4394–4410. doi: 10.1111/febs.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the Human Rev1-DNA-dNTP Ternary Complex. J. Mol. Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Takata K.-i., Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- (38).Xu W, Ouellette AM, Wawrzak Z, Shriver SJ, Anderson SM, Zhao L. Kinetic and structural mechanisms of (5′S)-8,5′-cyclo-2′-deoxyguanosine-induced DNA replication stalling. Biochemistry. 2015;54:639–651. doi: 10.1021/bi5014936. [DOI] [PubMed] [Google Scholar]
- (39).Zhao L, Pence MG, Christov PP, Wawrzak Z, Choi J-Y, Rizzo CJ, Egli M, Guengerich FP. Basis of miscoding of the DNA adduct N2, 3-ethenoguanine by human Y-family DNA polymerases. J. Biol. Chem. 2012;287:35516–35526. doi: 10.1074/jbc.M112.403253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Rozenski J, McCloskey JA. SOS: A simple interactive program for ab initio oligonucleotide sequencing by mass spectrometry. J. Am. Soc. Mass Spectrom. 2002;13:200–203. doi: 10.1016/S1044-0305(01)00354-3. [DOI] [PubMed] [Google Scholar]
- (41).Fisher LA, Bessho M, Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF-ERCC1 complex in vitro. J. Biol. Chem. 2008;283:1275–1281. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- (42).Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair. 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- (44).Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one-or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- (45).Makarova AV, Burgers PM. Eukaryotic DNA polymerase ζ. DNA Repair. 2015;29:47–55. doi: 10.1016/j.dnarep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase η. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zhao L, Christov PP, Kozekov ID, Pence MG, Pallan PS, Rizzo CJ, Egli M, Guengerich FP. Replication of N2,3-ethenoguanine by DNA polymerases. Angew. Chem., Int. Ed. 2012;51:5466–5469. doi: 10.1002/anie.201109004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Chang S.-c., Fedeles BI, Wu J, Delaney JC, Li D, Zhao L, Christov PP, Yau E, Singh V, Jost M, Drennan CL, Marnett LJ, Rizzo CJ, Levine SS, Guengerich FP, Essigmann JM. Next-generation sequencing reveals the biological significance of the N2,3-ethenoguanine lesion in vivo. Nucleic Acids Res. 2015;43:5489–5500. doi: 10.1093/nar/gkv243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4 - Analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J. Biol. Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- (50).Chowdhury G, Guengerich FP. Current Protocols in Nucleic Acid Chemistry. Suppl. 47. John Wiley & Sons, Inc.; New York: 2011. Liquid Chromatography-Mass Spectrometry Analysis of DNA Polymerase Reaction Products. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- (52).Yang Z, Price NE, Johnson KM, Gates KS. Characterization of interstrand DNA–DNA cross-links derived from abasic sites using bacteriophage ϕ29 DNA polymerase. Biochemistry. 2015;54:4259–4266. doi: 10.1021/acs.biochem.5b00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol. Cell. Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase η-mediated error-prone removal of mitomycin C interstrand cross-links. Mol. Cell. Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Jansen JG, Temviriyanukul P, Wit N, Delbos F, Reynaud C-A, Jacobs H, de Wind N. Redundancy of mammalian Y family DNA polymerases in cellular responses to genomic DNA lesions induced by ultraviolet light. Nucleic Acids Res. 2014;42:11071–11082. doi: 10.1093/nar/gku779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor J-S, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase κ in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Choi J-Y, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J. Biol. Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- (59).Takeiri A, Wada NA, Motoyama S, Matsuzaki K, Tateishi H, Matsumoto K, Niimi N, Sassa A, Gruz P, Masumura K, Yamada M, Mishima M, Jishage K, Nohmi T. In vivo evidence that DNA polymerase kappa is responsible for error-free bypass across DNA cross-links induced by mitomycin C. DNA Repair. 2014;24:113–121. doi: 10.1016/j.dnarep.2014.09.002. [DOI] [PubMed] [Google Scholar]
- (60).Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase iota incorporates dCTP opposite template G via a G.C + Hoogsteen base pair. Structure. 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- (61).Takata K.-i., Tomida J, Reh S, Swanhart LM, Takata M, Hukriede NA, Wood RD. Conserved overlapping gene arrangement, restricted expression and biochemical activities of DNA polymerase ν (POLN) J. Biol. Chem. 2015;290:24278. doi: 10.1074/jbc.M115.677419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48:11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Moldovan G-L, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D’Andrea AD. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol. Cell. Biol. 2010;30:1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Sharma S, Helchowski CM, Canman CE. The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2013;743–744:97–110. doi: 10.1016/j.mrfmmm.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Guèranger Q, Glover TW, Canman CE. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol. Cell. Biol. 2010;30:1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik NS, van Gent DC, Takata M. Multiple roles of Rev3, the catalytic subunit of polζ in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Wu F, Lin X, Okuda T, Howell SB. DNA polymerase ζ regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 2004;64:8029–8035. doi: 10.1158/0008-5472.CAN-03-3942. [DOI] [PubMed] [Google Scholar]
- (68).Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, Kohzaki M, Takeda S. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol. Cell. Biol. 2005;25:6103–6111. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol ζ) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- (71).Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Schörer OD. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 2011;39:7455–7464. doi: 10.1093/nar/gkr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Räschle M, Smeenk G, Hansen RK, Temu T, Oka Y, Hein MY, Nagaraj N, Long DT, Walter JC, Hofmann K, Storchova Z, Cox J, Bekker-Jensen S, Mailand N, Mann M. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science. 2015;348:1253671. doi: 10.1126/science.1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.