Abstract
Purpose
MR fingerprinting (MRF) is a technique for quantitative tissue mapping using pseudorandom measurements. To estimate tissue properties such as T1, T2, proton density, and B0, the rapidly acquired data are compared against a large dictionary of Bloch simulations. This matching process can be a very computationally demanding portion of MRF reconstruction.
Theory and Methods
We introduce a fast group matching algorithm (GRM) that exploits inherent correlation within MRF dictionaries to create highly clustered groupings of the elements. During matching, a group specific signature is first used to remove poor matching possibilities. Group principal component analysis (PCA) is used to evaluate all remaining tissue types. In vivo 3 Tesla brain data were used to validate the accuracy of our approach.
Results
For a trueFISP sequence with over 196,000 dictionary elements, 1000 MRF samples, and image matrix of 128 × 128, GRM was able to map MR parameters within 2s using standard vendor computational resources. This is an order of magnitude faster than global PCA and nearly two orders of magnitude faster than direct matching, with comparable accuracy (1–2% relative error).
Conclusion
The proposed GRM method is a highly efficient model reduction technique for MRF matching and should enable clinically relevant reconstruction accuracy and time on standard vendor computational resources.
Keywords: MR fingerprinting, clustering, matching, pruning
INTRODUCTION
NMR based techniques are commonly used in both clinical and medical research settings to investigate tissue properties related to disease (1–4). Many of the most commonly used imaging and spectroscopy techniques provide weighted measurements or images from which quantitative tissue relationships are estimated. There have been several attempts at efficient quantitative or multiparameter acquisitions in MR (5–8). A promising new approached called MR fingerprinting (MRF) has been introduced (9) as an acquisition and reconstruction strategy to estimate multiple tissue properties (such as T1, T2, and off-resonance B0). MRF experiments involve the creation of a time series evolution, which is later compared or “matched” against simulation. This time series of measurements is created by changing acquisition parameters such as flip angle and TR in a pseudorandom manner. The optimization of the acquisition parameters is an active research topic and the feasibility of schemes can be constrained by limitations in signal to noise and reconstruction artifacts. Bloch simulations across a range of physical parameters are completed a priori and used to assign multiple quantitative values to the observed signals. In this way, MRF builds upon compressed sensing (10–15) principles that allow for the fitting of underdetermined data through prior assumptions.
A bottleneck for the clinical usefulness of such approaches is often the computational complexity of the image reconstruction or matching of patient observations against precomputed tissue models. In the case of MRF, clinically acceptable resolutions for the tissue properties will translate into hundreds of thousands of dictionary elements. This creates a demanding computational problem as correlation between these dictionary elements, each with thousands of time points, will need to be compared against thousands of voxel time courses per image. For example, the direct MRF matching corresponding to a clinically relevant protocol can take several minutes for a single slice (9,16). This is approaching the total desired acquisition time for a whole brain accelerated clinical scan.
In this work, we introduce a fast group based matching algorithm (GRM) that exploits inherent clustering properties of the Bloch simulation dictionary used for MRF (9,17). Unlike drawing from random distributions that can produce uncorrelated time series, MRF signals are tied to the underlying physics of the tissue types. Similar tissue responses can require a large number of samples to differentiate accurately. In GRM, tissues that produce similar time courses are automatically grouped and assigned a unique signature that will lead to early identification in the matching process. Group principal component analysis (PCA) is then used to efficiently evaluate only meaningful dictionary comparisons. GRM allows for the accurate reconstruction of T1, T2, and B0 quantities orders of magnitude faster than direct matching. We will demonstrate that GRM removes a major computational bottleneck associated with MRF acquisitions and enables clinically relevant reconstruction times on standard vendor’s computational hardware.
THEORY
There is a rich history of algorithms for combinatorial problems that focus on iteratively reducing optimization search spaces to achieve good solution quality in practical time. Some of the most classic are mixed integer programming solutions focusing on branching choices for optimization variables and pruning/cutting of the solution space (18–20). In this work, we leverage these principles through the use of a compact MRF matching method. We will demonstrate that pruning techniques can be quickly applied based upon a compact MRF dictionary, enabling a 99% reduction in the total number of comparisons required to match. Finally, group PCA can be used to quickly evaluate the dictionary candidates still under consideration. The efficiency of the proposed GRM technique is a result of the inherent clustering properties found in Bloch simulations of standard tissue types.
Grouping of MRF Dictionaries
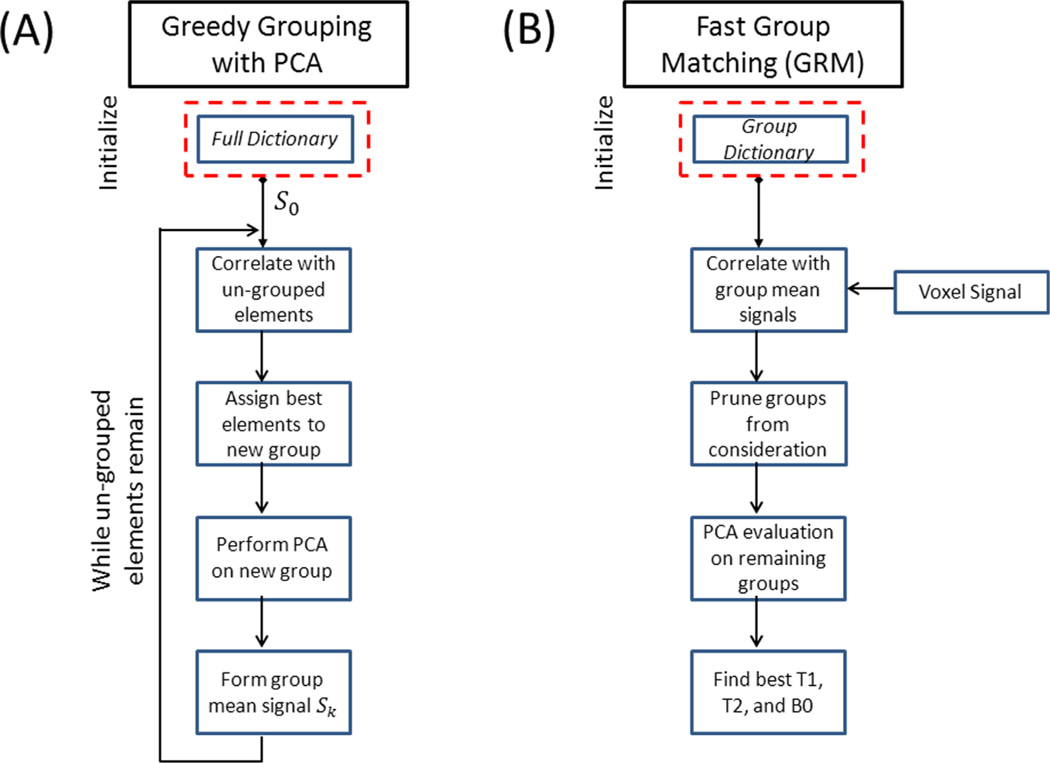
There are many approaches for the clustering of elements based upon strength of correlation. Sparse methods such as K-way partitioning (21) repetitively solve minimum cut problems to sub-divide elements in a hierarchical manner. K-way partitioning has many advantages but can be computationally demanding for large problems. In this work, we apply a highly scalable greedy grouping scheme toward the MRF dictionary elements. Figure 1A shows the flow diagram of the grouping process. For simplicity of illustration, we will assume the normalized MRF dictionary with M dictionary elements can be evenly spread across N groups.
FIG. 1.
Greedy grouping process for GRM is illustrated in (A). The MRF dictionary elements are iteratively assigned to groups based upon inherent correlation between Bloch simulations. The GRM process is shown in (B). An early pruning stage removes unnecessary comparisons and group PCA enables fast evaluation of dictionary elements that remain under consideration.
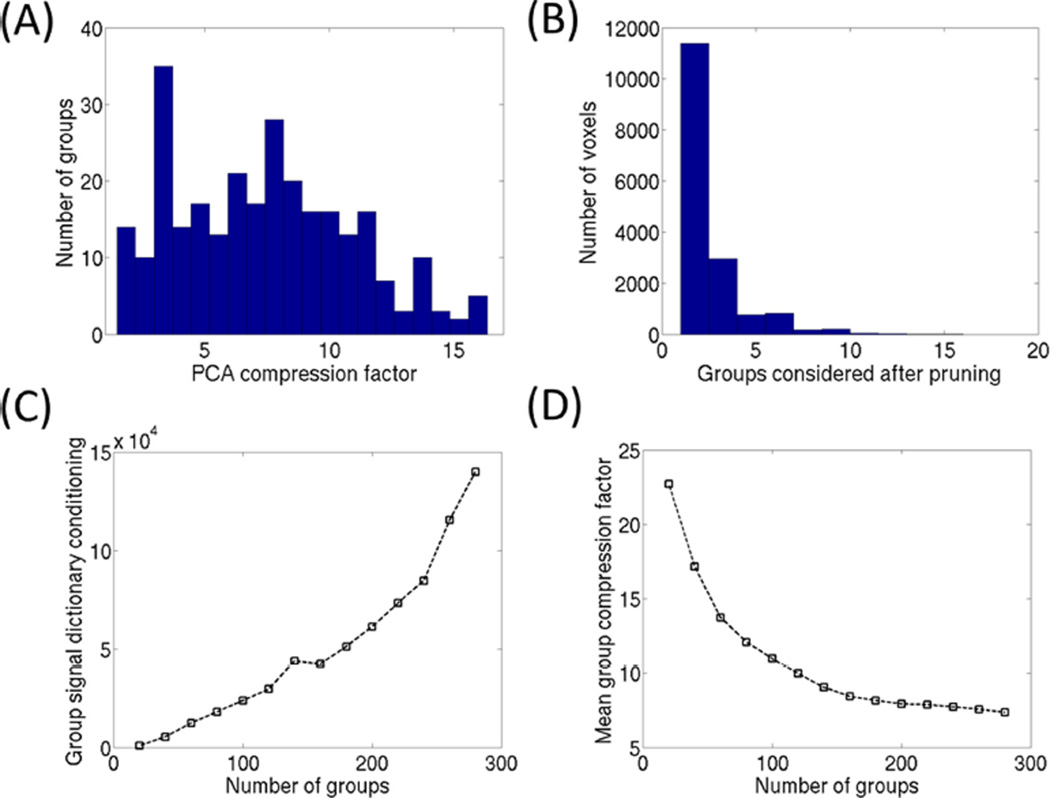
The GRM process begins with the choice of an initial signal S0. There are many choices available for this signal, e.g., a random dictionary element, the mean gray/white matter signals, etc. As there have been no dictionary elements assigned to groups, the signal S0 is compared against all other dictionary elements. Based upon a predetermined group size (M/N), the top correlations to the signal S0 are assigned to the first group. A new signal S1 is then created to best represent the time courses contained within the first group. In this work, we define S1 to be the mean signal for the group. This will allow for a quick evaluation of the average correlation of the acquired signal against all elements within a grouping. In a similar manner to the global PCA approach of (22), smaller group level PCA are computed using the singular value decomposition. The group PCA will be later used as part of the matching process. It is important to note that PCA will be highly effective on these group signals as they were chosen based upon strength of correlation or linear dependence. The grouping process repeats until all dictionary elements have been assigned. Figure 2A shows the histogram of compression rates on 280 (nearly evenly sized) groups that contain 196,000 typical dictionary elements considered for a true fast imaging with steady state precession (true-FISP) based MRF sequence. Assuming a conservative 10−5 PCA truncation tolerance, the average compression rate is over 7×. In comparison, if this truncation level was applied to the full dictionary there would be nearly no compression as it would require 999 of 1000 of the singular vectors to be retained.
FIG. 2.
A: Shows the distribution of compression factors for 280 groups through the GRM process. The compression factor is the ratio of the number of dictionary elements in the group against the number of singular vectors retained using a 10−5 relative truncation tolerance. B: Shows the number of groups that each voxel will need to be evaluated against after the GRM pruning stage. The inclusion of groups for matching consideration is based upon a relative truncation level of 5 × 10−3 below the best group mean signal correlation for each voxel. C: Shows the condition number for the group mean signals assuming varying group sizes. As the number of groups increases the linear independence of the group signals is degraded. D: shows the average compression rate across the group PCA, where smaller group sizes are less likely to be highly compressible.
Matching with Grouped MRF Dictionaries
Figure 1B shows the flow diagram for the fast group matching algorithm. After the acquisition of patient data, an initial compact matching is performed against the representative group signals [S1 ⋯ SN]. This allows for efficient pruning of groups from consideration, i.e., if a voxel’s signal cannot reasonably be matched to the representative signal for several groups, the signals within these groups should no longer be considered as candidates. The pruning criteria can be determined through a relative or absolute correlation threshold (relative threshold is used in this work). Finally, the remaining groups under consideration for a voxel are evaluated for quality of fit through PCA projection. The best fit for each voxel is used to assign T1, T2, and B0 tissue properties.
In practice we observed that the relative pruning criteria of 5 × 10−3 below the best group match for the voxel ensured good final matching accuracy. Figure 2B shows a histogram of groups remaining under consideration after the initial compact matching for a representative single-slice trueFISP MRF acquisition, using the 280 group dictionary described above. Across the 128 × 128 voxels in this example, the average number of groups remaining was 2.3 of the 280 total groups. This corresponds to a pruning of over 99% of possible dictionary comparisons.
METHODS
To investigate the efficiency of the proposed GRM method, in vivo data were acquired from a single healthy volunteer subject to institutionally approved protocol consent. The data were acquired on a 3 Tesla (T) Siemens Skyra with the standard Siemens 16-channel head array coil. A trueFISP sequence was used to acquire data at 1000 time points, where each data point was acquired using one of 48 highly undersampled (40×) variable density spiral interleaves (9,23–26). The choice of balanced SSFP was based upon the efficiency of the sequence with respect to SNR. Unlike a spoiled GRE sequence, the temporal dependency between the T1, T2, and B0 parameters in the balanced SSFP sequence allows for a larger variance across the fingerprint dictionary. The number of samples used in this work was determined by comparing the estimated maps to those found through longer acquisitions. A sufficient number of samples were chosen to ensure accuracy.
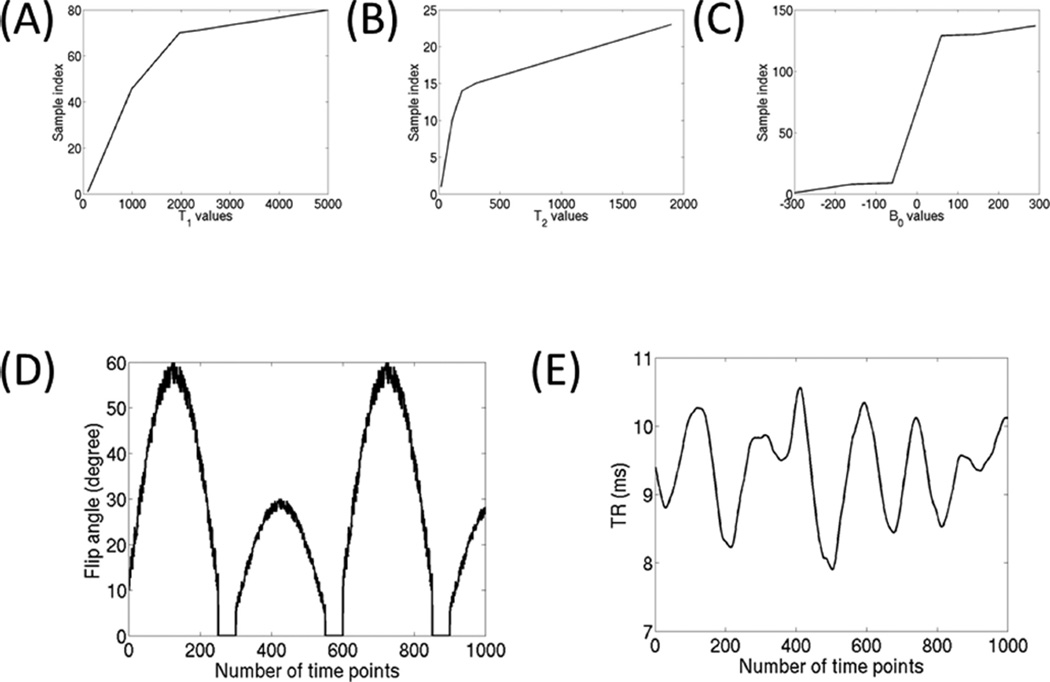
As described in Ma et al (9), gridding was used to reconstruct each time point and channel sensitivity maps were estimated using the average image across the initial 100 reconstructed time points. The MRF experiment was performed with a TR ranging from 7.84 ms to 10.56 ms and the flip angle was restricted to be within 0° to 60° to ensure good quality slice profiles (see Figures 3D,E for illustration of the acquisition strategy used in this work). The choice of flip angle and TR variation was selected based upon robustness with respect to signal to noise and artifact levels. Heavily undersampled spiral readouts were used in this work, leading to aliasing artifacts at each time point. The slow varying flip angle trends allow for us to exploit temporal dependencies to reduce the influence of artifact/noise from the measurements. The regions with flip angle of 0° degrees are inserted to allow for T1 recovery. By combining this strategy with small random fluctuations, we are able to differentiate tissues with similar responses. This trade-off between coherent and random parameter components can change based upon the artifact/noise level in the measurements.
FIG. 3.
A–C: Show the T1, T2, and B0 values considered in the trueFISP dictionary. All possible combinations of these parameters produce the over 196,000 dictionary elements. D,E: Respectively show the flip angle and TR acquisition strategies used in this work.
The field of view was 300 × 300 mm2, with an image matrix of 128 × 128, slice thickness of 5mm, and TI=12.7ms. As was described in Ma et al (9), the initial inversion pulse is included to increase the sensitivity to T1. The MRF dictionary was created using 1435 combinations of T1 and T2 values, each of which are considered against 137 possible B0 values. Varying step sizes were used to cover T1, T2 and B0 values in the ranges of [100, 5000], [20, 1900], and [−300, 290] respectively, see Figures 3A–C. To experimentally tune the GRM algorithm, several computational experiments were performed in MATLAB on a cluster of AMD Opteron 6282 SE 2.6 GHz processors. To demonstrate the trade-off between matching time, MRF reconstruction accuracy, PCA group compression factors, and group mean signal independence we performed a sensitivity analysis for the GRM method. These numeric parameters were then used as part of a multithreaded C++ implementation of the GRM method that has been implemented within Siemens ICE framework and tested on the standard Siemens Skyra computational hardware.
RESULTS
Figure 2C shows the relationship between the numerical condition number for the dictionary of group mean signals [S1 ⋯ SN] as the number of groups N increases. The condition number of the group dictionary is the ratio of the largest to the smallest singular value. A high condition number suggests that there is linear dependency between these signals. The inverse of the condition is related to the PCA drop tolerance used in this work. The number of groups can be determined using the stability of the condition number as the metric, i.e., different grouping levels can be tested until the appropriate condition number is achieved. This process is nondata dependent and can be performed offline. Even when considering 280 groups the condition number remains stable, i.e., there is almost no redundancy in the group signals. This is based upon the reciprocal relationship between condition number and a PCA based truncation threshold (10−5 in our experiments). Figure 2D shows the average PCA compression rate across varying numbers of groups. As the size of a group decreases (number of groups increases), it becomes more unlikely to observe redundancy in the elements within the group. However, the inherent clustering of MRF dictionary elements is still strong when considering 280 groups (see Figure 2B for a finer breakdown of the 7× average compression ratio in this case).
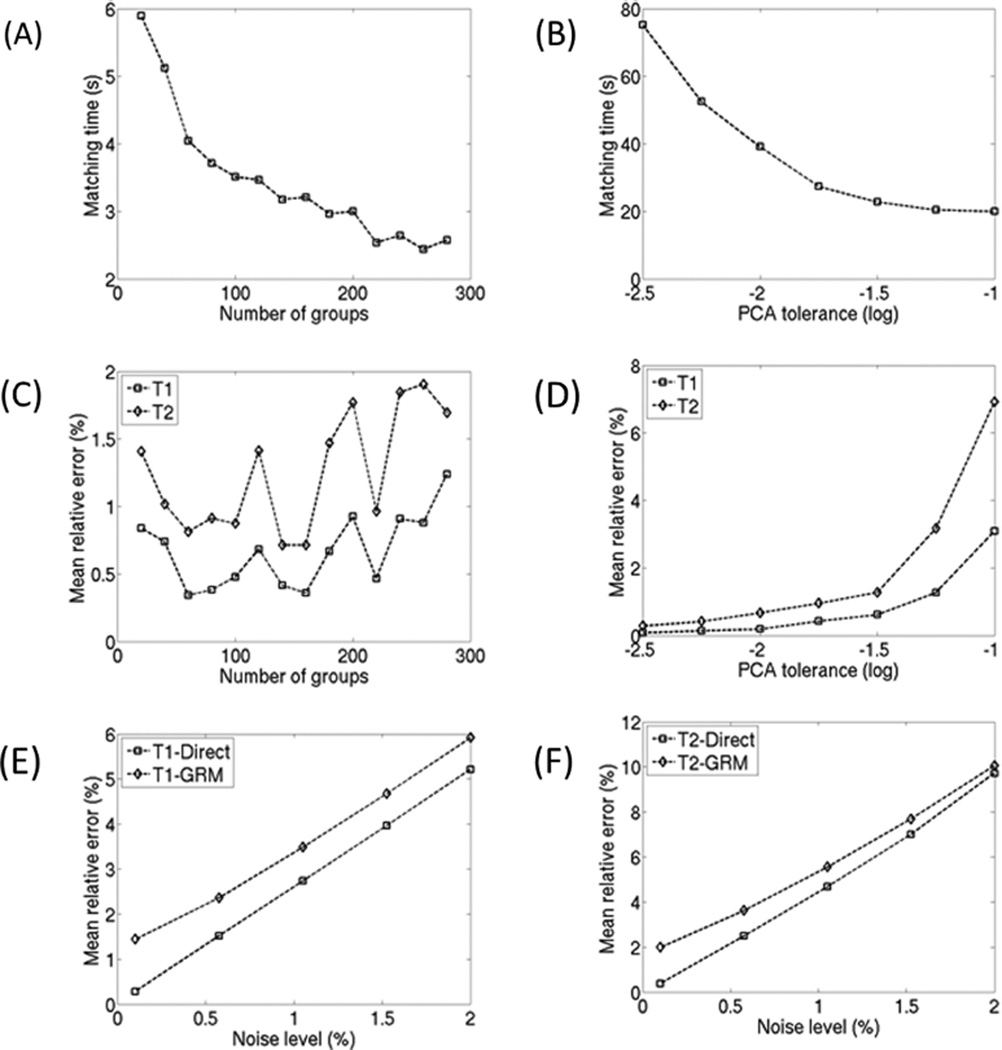
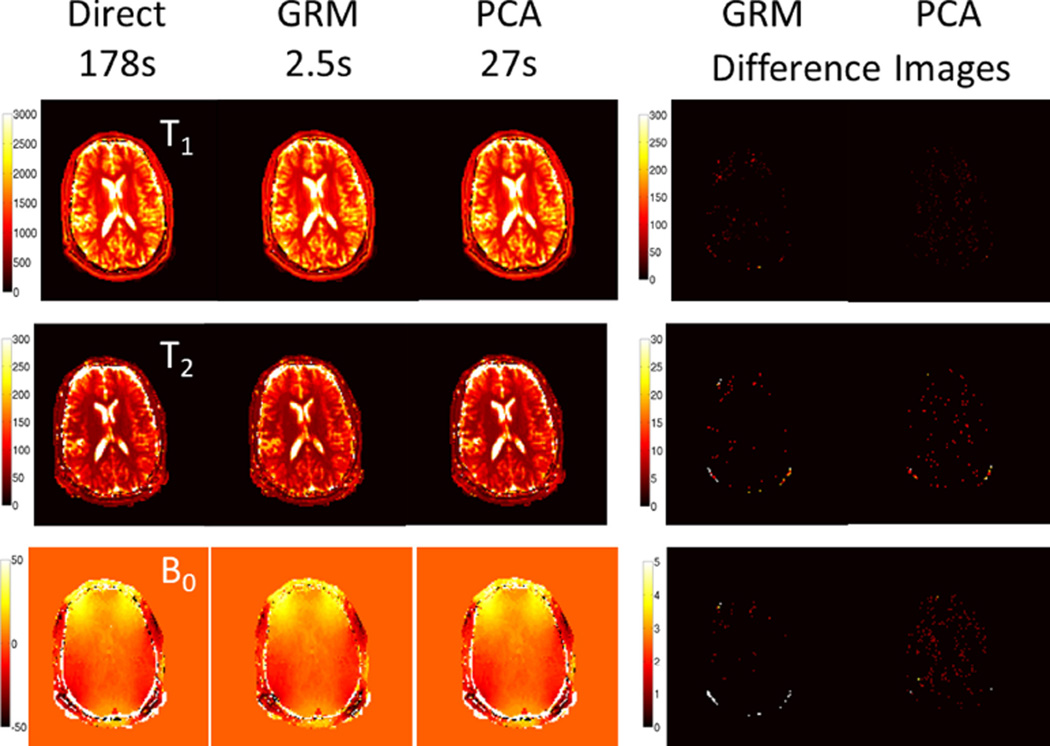
Figures 4A and C illustrate the computation and accuracy trade-off when increasing the number of the groupings. Here, accuracy is measured as the mean relative error across typical white and gray matter regions (T1, T2 under 1300 and 150, respectively). Similarly, Figures 4B and D illustrate the computation and accuracy trade-off when increasing the global PCA drop tolerance. Next, the robustness of the direct matching and 280 group GRM methods for T1 and T2 estimation are evaluated in the presence of increased noise levels. Figures 4E and F show the mean relative error assuming 100 random experiments with complex Gaussian noise included at the illustrated noise level. Figure 5 shows the reconstructed T1, T2, and B0 images for the direct matching, global PCA using a tolerance of 0.02, and GRM with 220 groups. The gray/white matter masked difference images are provided. The direct matching time was over 70× longer than GRM with an average relative error difference of less than 2% (see Figure 4). The global PCA matching time was over 10× longer than GRM with similar error levels.
FIG. 4.
A: Shows the decrease in GRM matching time (MATLAB implementation) as the number of groups is increased. B: Shows the decrease in matching time as the global PCA drop tolerance is increased. C: Shows the accuracy of GRM with respect to the number of groups. D: Shows the accuracy of global PCA with respect to the drop tolerance. E,F: Shows the robustness of the T1 and T2 estimations respectively. Both the direct matching and 280 group GRM algorithms are evaluated with respect to increases in noise level. Each figure point represents mean error assuming 100 random experiments with complex Gaussian noise included at the illustrated noise level.
FIG. 5.
The reconstructed quantitative T1, T2, and B0 images for GRM using 220 groups, global PCA using a tolerance of 0.02, and the direct matching. The difference maps are masked to only consider white and gray matter regions, see Figure 4 for average relative error level.
As a final computational experiment, we have integrated a multithreaded C++ implementation of GRM within Siemens online reconstruction framework. The 280 group GRM dictionary information was loaded into the scanner memory during the data acquisition, and the total time for matching T1, T2, and B0 for the slice was under 2 s using the standard CPU hardware.
DISCUSSION AND CONCLUSIONS
In this work, we propose an efficient group based matching for MRF dictionaries. The GRM method leverages techniques from discrete optimization and numerical linear algebra to match quantitative values within clinically relevant time. GRM is an order of magnitude faster than global PCA techniques (22) and nearly two orders of magnitude faster than direct matching, with comparable accuracy (1–2% relative error).
We have demonstrated the accuracy of the GRM method across a wide range of group sizes. In addition, we have introduced a guiding principal for determining appropriate group sizes. It can be observed from Figure 2 that there is an important balance between the condition number for the group mean signals and the average PCA compression rate within the groups. While it is advantageous to add more groups for early pruning, a lack of compression within the groups will lead to increased time for final evaluation. In addition, if there is redundancy across the mean group signals then the pruning efficiency will be degraded. In our experiments, we observed a condition number for the group mean signals matching the PCA truncation level of 10−5 resulted in good computational performance and reconstruction accuracy. It is important to note that the choice of GRM pruning threshold used in this work may not generalize to all types of MRF dictionaries. However, the threshold can easily be relaxed from an empirical setting to investigate convergence in the map estimation. That is, additional group comparisons (corresponding to a relaxed pruning threshold) can easily be appended to update previously computed GRM solutions.
Finally, the group based model reduction exploited in GRM should be a valuable tool in reducing the complexity of many dictionary reconstruction and optimization problems for MRF. This includes forward modeling of the entire reconstruction/matching process, where the number of optimization variables can be prohibitive. That is, a parallel imaging forward model could be regularized through the addition of a sparsity constraint against the smaller number of group mean signals (as opposed to the full dictionary). The sparsity across the group mean signals is currently being used to guide the pruning stage of GRM, see Figure 2. We also think group based analysis methods similar to GRM are the key to reducing the complexity associated with the difficult optimization of MRF acquisition parameters such as TR and flip angle. For example, with a fixed grouping scheme across the tissue properties, the strength of correlation between the group mean signals could be used to quickly evaluate the quality of acquisition scenarios.
Acknowledgments
Grant sponsor: National Institutes of Health (NIH); Grant numbers: R01EB006847, R00EB012107, R01EB017219, P41RR014075, and U01MH093765.
REFERENCES
- 1.Bartzokis G, Sultzer D, Cummings J, Holt LE, Hance DB, Henderson VW, Mintz J. In vivo evaluation of brain iron in Alzheimer disease using magnetic resonance imaging. Arch Gen Psychiatry. 2000;57:47–53. doi: 10.1001/archpsyc.57.1.47. [DOI] [PubMed] [Google Scholar]
- 2.Larsson HBW, Nordenbo A. Assessment of demyelination, edema, and gliosis by in vivo determination of T1 and T2 in the brain of patients with acute attack of multiple sclerosis. Magn Reson Med. 1989;11:337–348. doi: 10.1002/mrm.1910110308. [DOI] [PubMed] [Google Scholar]
- 3.Pitkanen A, Laakso M, Kiilviainen R, Partanen K, Vainio P. Severity of hippocampal atrophy correlates with the prolongation of MRI T, relaxation time in temporal lobe epilepsy but not in Alzheimer’s disease. Neurology. 1996;46:1724–1730. doi: 10.1212/wnl.46.6.1724. [DOI] [PubMed] [Google Scholar]
- 4.Williamson P. Frontal, temporal, and striatal proton relaxation times in schizophrenic patients and normal comparison subjects. Am J Psychiatry. 1992;149:549–551. doi: 10.1176/ajp.149.4.549. [DOI] [PubMed] [Google Scholar]
- 5.Warntjes JBM, Dahlqvist O, Lundberg P. Novel method for rapid, simultaneous T1, T*2, and proton density quantification. Magn Reson Med. 2007;57:528–537. doi: 10.1002/mrm.21165. [DOI] [PubMed] [Google Scholar]
- 6.Warntjes JBM, Leinhard OD, West J, Lundberg P. Rapid magnetic resonance quantification on the brain: optimization for clinical usage. Magn Reson Med. 2008;60:320–329. doi: 10.1002/mrm.21635. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt P, Griswold MA, Jakob PM, Kotas M, Gulani V, Flentje M, Haase A. Inversion recovery TrueFISP: quantification of T(1), T(2), and spin density. Magn Reson Med. 2004;51:661–667. doi: 10.1002/mrm.20058. [DOI] [PubMed] [Google Scholar]
- 8.Ehses P, Seiberlich N, Ma D, Breuer FA, Jakob PM, Griswold MA, Gulani V. IR TrueFISP with a golden-ratio-based radial readout: fast quantification of T1, T2, and proton density. Magn Reson Med. 2013;69:71–81. doi: 10.1002/mrm.24225. [DOI] [PubMed] [Google Scholar]
- 9.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature. 2013;495:187–192. doi: 10.1038/nature11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein T, Osher S. The Split Bregman method for L1-regularized problems. SIAM J Imaging Sci. 2009;2:323–343. [Google Scholar]
- 12.Bilgic B, Goyal VK, Adalsteinsson E. Multi-contrast reconstruction with Bayesian compressed sensing. Magn Reson Med. 2011;66:1601–1615. doi: 10.1002/mrm.22956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malioutov D, Member S, Çetin M, Willsky AS. A sparse signal reconstruction perspective for source localization with sensor arrays. IEEE Trans Signal Process. 2005;53:3010–3022. [Google Scholar]
- 14.Donoho DL. Compressed sensing. IEEE Trans Inf Theory. 2006;52:1289–1306. [Google Scholar]
- 15.Smith DS, Li X, Gambrell JV, Arlinghaus LR, Quarles CC, Yankeelov TE, Welch EB. Robustness of quantitative compressive sensing MRI: the effect of random undersampling patterns on derived parameters for DCE- and DSC-MRI. IEEE Trans Med Imaging. 2012;31:504–511. doi: 10.1109/TMI.2011.2172216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tropp JA, Gilbert AC. Via orthogonal matching pursuit. IEEE Trans Inform Theory. 2007;53:4655–4666. [Google Scholar]
- 17.Doneva M, Börnert P, Eggers H, Stehning C, Sénégas J, Mertins A. Compressed sensing reconstruction for magnetic resonance parameter mapping. Magn Reson Med. 2010;64:1114–1120. doi: 10.1002/mrm.22483. [DOI] [PubMed] [Google Scholar]
- 18.Wagner HM. Principles of operations research: with applications to managerial decisions. 2nd. Englewood Cliffs, NJ: Prentice-Hall; 1975. p. 1039. [Google Scholar]
- 19.Luenberger DG. Linear and nonlinear programming. 2nd. New York: Springer; 2003. p. 516. [Google Scholar]
- 20.Applegate D, Bixby R, Chvátal V, Cook W. On the solution of traveling salesman problems. Doc Math. 1998:645–656. Extra Volu. [Google Scholar]
- 21.Karypis G, Kumar V. Parallel multilevel k-way partitioning scheme for irregular graphs; Proceedings of the ACM/IEEE Conference on Supercomputing; Pittsburgh, Pennsylvania, USA. 1996. [Google Scholar]
- 22.McGivney D, Ma D, Saybasil H, Jiang Y, Griswold M. Singular value decomposition for magnetic resonance fingerprinting in the time domain; Proceedings of the 22nd Annual Meeting of ISMRM; Milan, Italy. 2014. Abstract 4287. [Google Scholar]
- 23.Schmitt P, Griswold MA, Gulani V, Haase A, Flentje M, Jakob PM. A simple geometrical description of the TrueFISP ideal transient and steady-state signal. Magn Reson Med. 2006;55:177–186. doi: 10.1002/mrm.20738. [DOI] [PubMed] [Google Scholar]
- 24.Lee JH, Hargreaves BA, Hu BS, Nishimura DG. Fast 3D imaging using variable-density spiral trajectories with applications to limb perfusion. Magn Reson Med. 2003;50:1276–1285. doi: 10.1002/mrm.10644. [DOI] [PubMed] [Google Scholar]
- 25.Marseille G, de Beer R, Fuderer M, Mehlkopf A, van Ormondt D. Nonuniform phase-encode distributions for MRI scan time reduction. J Magn Reson B. 1996;111:70–75. doi: 10.1006/jmrb.1996.0061. [DOI] [PubMed] [Google Scholar]
- 26.Tsai CM, Nishimura DG. Reduced aliasing artifacts using variable-density k-space sampling trajectories. Magn Reson Med. 2000;43:452–458. doi: 10.1002/(sici)1522-2594(200003)43:3<452::aid-mrm18>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]