Summary
There is an urgent need for new drugs to treat malaria, with broad therapeutic potential and novel modes of action, to widen the scope of treatment and to overcome emerging drug resistance. We describe the discovery of DDD107498, a compound with a potent and novel spectrum of antimalarial activity against multiple life-cycle stages of the parasite, with good pharmacokinetic properties, and an acceptable safety profile. DDD107498 demonstrates potential to address a variety of clinical needs, including single dose treatment, transmission blocking and chemoprotection. DDD107498 was developed from a screening programme against blood stage malaria parasites; its molecular target has been identified as translation elongation factor 2 (eEF2), which is responsible for the GTP-dependent translocation of the ribosome along mRNA, and is essential for protein synthesis. This discovery of eEF2 as a viable antimalarial drug target opens up new possibilities for drug discovery.
Introduction
The WHO estimates there were approximately 200 million clinical cases and 584,000 deaths from malaria in 2013, predominantly amongst children and pregnant women in sub-Saharan Africa1. The malaria parasite has developed resistance to many of the current drugs, including emerging resistance to the core artemisinin component of artemisinin-based combination therapies that comprise current first-line therapies2. To support the current treatment and eradication agenda3, there are a number of requirements for new antimalarials: novel modes of action with no cross-resistance to current drugs; single dose cures; activity against both the asexual blood stages that cause disease and gametocytes responsible for transmission; compounds which prevent infection (chemoprotective agents); and compounds which clear P. vivax hypnozoites from the liver (anti-relapse agents)4.
Discovery of a novel antimalarial
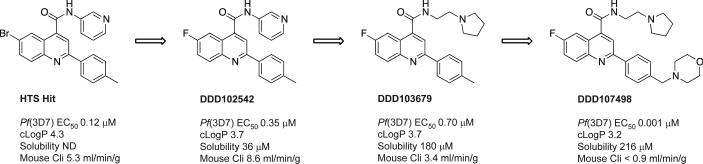
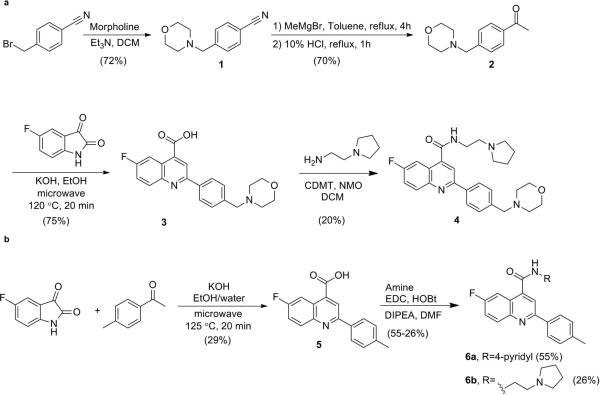
A phenotypic screen of the Dundee protein kinase scaffold library5 (then 4731 compounds) was performed against the blood stage of the multi-drug sensitive P. falciparum 3D7 strain. A compound series from this screen, based on a 2,6-disubstituted quinoline-4-carboxamide scaffold, had sub-micromolar potency against the parasites, but suffered from poor physicochemical properties. Chemical optimisation (Fig. 1 and Extended Data Fig. 1) led to DDD107498 with improved physicochemical properties (Supplementary Methods Tables S1 and S2) and a 100-fold increase in potency. The key stages involved were: replacing the bromine with a fluorine atom to reduce molecular weight and lipophilicity; replacing the 3-pyridyl substituent with an ethylpyrrolidine group, and addition of a morpholine group via a methylene spacer. Initial cost of goods estimates together with likely human dose projections suggest a low cost (approximately US$1 per treatment), which is important, given most of the patient population is living in poverty.
Figure 1. Chemical evolution of DDD107498 from the phenotypic hit.
Cli = intrinsic clearance in mouse liver microsomes.
Blood-stage activity and developability
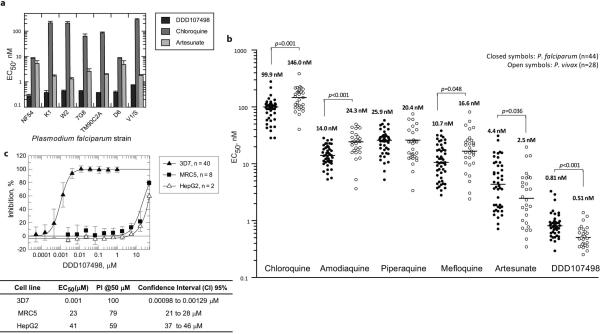
DDD107498 showed excellent activity against 3D7 parasites: EC50 =1.0 nM (95% Confidence Interval (CI) 0.8-1.2 nM); EC90 = 2.4 nM (95% CI 2.0-2.9 nM); EC99 = 5.9 nM (95% CI 4.5-7.6 nM), (n=39). It was also almost equally active against a number of drug-resistant strains (Extended Data Fig. 2a)6. Furthermore, DDD107498 was more potent than artesunate in ex vivo assays against a range of clinical isolates of both P. falciparum (median EC50 = 0.81 [Range 0.29-3.29] nM, n=44) and P. vivax (median EC50 = 0.51 [Range 0.25-1.39] nM, n=28), collected from patients with malaria from Southern Papua, Indonesia, a region where high-grade multidrug-resistant malaria is endemic for both species (Extended Data Fig. 2b)7,8. In contrast the compound was not toxic to human cells (MRC5 and Hep-G2 cells) at much higher concentrations (> 20,000 fold selectivity, Extended Data Fig. 2c).
DDD107498 showed good drug-like properties: metabolic stability when incubated with hepatic microsomes or hepatocytes from several species; good solubility in a range of different media; and low protein binding (Supplementary Methods, Tables S1 and S2). DDD107498 displayed excellent pharmacokinetic properties in preclinical species, including good oral bioavailability, an important pre-requisite for use in resource-poor settings, and long plasma half-life, important for single dose treatment and chemoprotection (Extended Data Table 1a).
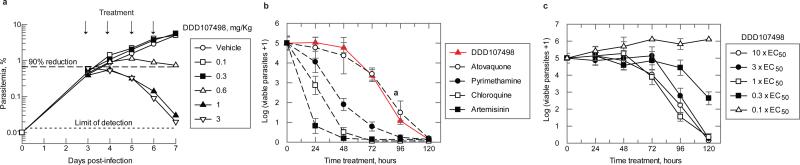
DDD107498 was very active in several mouse models of malaria, with comparable or greater efficacy than current antimalarials (Extended Data Table 1b). DDD107498 had an ED90 (90% reduction in parasitaemia) of 0.57 mg/kg after a single oral dose in mice infected with the rodent parasite P. berghei. Efficacy was also tested in NOD-scid IL-2R_null mice engrafted with human erythrocytes and infected with P. falciparum strain 3D70087/N9 (Fig. 2a)9. When dosed orally daily for 4 days, the ED90 on day 7 after infection was 0.95 mg/kg per day. Blood sampling from the infected SCID mice suggested a minimum parasiticidal concentration (MPC) for DDD107498 of 10-13 ng/mL for asexual blood stage infections.
Figure 2. Efficacy studies and parasite killing rate.
a. In vivo activity against P. falciparum in NOD-scid IL-2R_null mice. Six mice were used, each serially sampled. The variability of cytometry for repeated acquisitions of a sample is < 2-3 %. The percentage of parasitemia was calculated by acquiring a minimum number of 500 parasitized erythrocytes. An independent experiment with 3 further mice was performed to confirm the ED90.
b. Determination of the in vitro killing rate of DDD107498. The in vitro PRR assay was used to determine onset of action and rate of killing as previously described10. P. falciparum was exposed to DDD107498 at a concentration corresponding to 10 × EC50. The number of viable parasites at each time point was determined as described10. Four independent serial dilutions were done with each sample to correct for experimental variation and the error bars shown are the standard deviation. Previous results reported on standard antimalarials tested at 10 × EC50 using the same conditions are shown for comparison10.
c. The in vitro PRR assay was used to determine the minimal concentration of compound needed for achieving maximal killing effects. Parasites were exposed to DDD107498 at concentrations of 0.1, 0.3, 1, 3 and 10 × EC50 using conditions described above. Error bars shown are the standard deviation. Concentrations of DDD107498 ≥ 1 × EC50 are sufficient to produce maximal killing effects on treated parasites.
The effects of DDD107498 on circulating parasites in the SCID mouse model could be observed in one replication cycle (48 h) and led to trophozoites with condensed cytoplasm (Extended Data Fig. 3). Stage specificity studies using synchronized cultures showed that at a concentration of 4nM for 24 h, DDD107498 led to: (1) from the ring stage, formation of abnormal trophozoites; (2) from the trophozoite stage, prevention of schizont formation with a 50% reduction in parasites, indicative of cidal activity; (3) from the schizont stage, prevention of ring formation with a 98% reduction in parasites, indicative of cidal activity (Extended Data Fig. 3b,c).
DDD107498 showed a similar parasite killing profile both in vitro (Fig. 2b,c) and in vivo (Fig. 2a) which is supportive of a common mode of action in cellular and animal models of disease. Using a Parasite Reduction Rate (PRR) assay10 there was a lag of about 24-48 h during which time the effects of the compounds were reversible following wash-out. Rapid killing occurred after parasites had been exposed to DDD107498 for more than 48 h (Figs. 2b,c).
All these experiments suggest for the blood stage form that treatment with DDD107498 prevented development of trophozoites and schizonts and at least in the case of schizonts caused rapid killing. Any ring stage parasites only developed as far as abnormal trophozoites, which appeared to survive for about 48 h under drug pressure, but then were killed.
In safety studies, DDD107498 showed no significant inhibition of any of the major human cytochrome P450 (CYP) isoforms and CYP450 induction risk was low (Supplementary Methods, Table S3), indicating a low risk of clinical drug-drug interactions. DDD107498 is non-mutagenic and has very weak inhibitory potencies on IKr (hERG) and other ion channels indicating a very low risk for adverse cardiovascular activity. However, given its potency, long half-life observed in preclinical species, and safety margins from a rat 7-day toxicology study, DDD107498 has potential for both single dose treatment and once weekly chemoprotection11.
Activity against other life-cycle stages
Activity against intra-hepatocytic parasites (liver schizont stages)
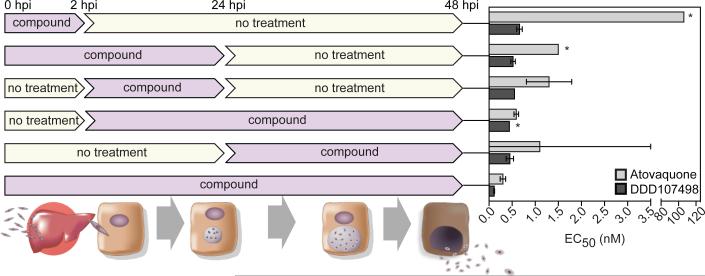
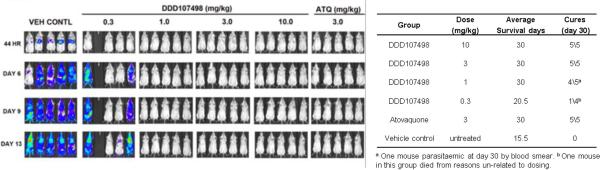
This is the first stage of human infection after injection of sporozoites by anopheline mosquitoes, where the parasites invade and multiply in the liver. Compounds active against this stage have potential for use in chemoprotection. DDD107498 showed an EC50 ~ 1 nM against the liver schizont forms of P. berghei and P. yoelii12. DDD107498 was active when dosed for only 2 h during the initial infection (hepatocyte invasion) of the liver cells (Fig. 3). In contrast, atovaquone (clinically used for chemoprotection in combination with proguanil) had a significantly reduced activity during this period (EC50 ~ 106 nM versus 0.3 nM for continuous treatment). Further, DDD107498 showed equivalent potency against the P. berghei liver stage when dosed for a short period of time after initial infection had been established (Fig. 3). This suggests that intermittent treatment may be sufficient for chemoprotection. To assess chemoprotective potential in vivo, mice were treated with DDD107498 2 h before being infected with luciferase-expressing P. berghei sporozoites (Extended Data Fig. 4). At a dose of 3 mg/kg, DDD107498 was fully curative with no sign of parasitaemia after 30 days. Thus, DDD107498 demonstrates potent chemoprotection using in vitro and in vivo models, where blood sampling from the mice during the experiment suggests a MPC of 15-20 ng/mL.
Figure 3. In vitro activity of DDD107498 against P. berghei liver stages.
hpi is hours post-infection. Each time point was the average of 4 technical replicates. 95% Confidence limits are shown. * more than one curve fitting possible.
Transmission Blocking
The parasite erythrocytic form differentiates into the asymptomatic male and female gametocytes (stages I-V) within the human host. Mature stage V gametocytes are infective to mosquitoes, but are not eliminated by the majority of current antimalarial agents, and remain circulating in the human bloodstream for up to 3 weeks, long after the disappearance of clinical symptoms of malaria13,14. After ingestion by the mosquito, gametocytes differentiate into gametes. DDD107498 potently inhibited both male and female gamete formation from the gametocyte stage at similar concentrations (1.8nM [95% CI 1.6-2.1nM] and 1.2nM [95% CI 0.8-1.6nM] respectively, Extended Data Fig. 5), indicating that it is an extremely potent inhibitor of the functional viability of both male and female mature gametocytes15. In line with this, DDD107498 blocked transmission as determined by the Standard Membrane Feeding Assay (SMFA). In this assay, parasite cultures containing P. falciparum stage V gametocytes were exposed to compound for 24 h prior to mosquito feeding. DDD107498 blocked subsequent oocyst development in the mosquito (measured after 7 days) with an EC50 of 1.8 nM. At a baseline oocyst intensity of 27 oocysts per mosquito in the DMSO controls, prevalence of infection was inhibited with an EC50 of 3.7 nM as measured by the number of infected mosquitoes. Repeating the SMFA in which DDD107498 was added at the moment of mosquito feeding gave an EC50 of 10 nM, indicating potent activity against the parasite sexual stages that develop in the mosquito midgut (Extended Data Fig. 5)16.
A P. berghei mouse-to-mouse17,18 model was additionally used to examine transmission-blockade. Mice were infected with P. berghei (PbGFPCON507)19, and then treated orally with compound 24 h before mosquitoes took a direct blood meal17. After a 3 mg/kg dose of DDD107498, a 90.7% reduction in infected mosquitoes and a 98.8% reduction in oocysts per midgut was observed at day 10 in comparison with mosquitoes fed on untreated mice (Extended Data Fig. 6). A corresponding reduction in sporozoite intensity and prevalence was observed (93.8% and 88.6% respectively). Mosquitoes previously fed on infected, drug treated mice were allowed to feed on uninfected mice17. Over multiple mosquito biting rates, a 89.5% (95% Cls 71.4-100) reduction in the number of mice that developed blood stage infection was observed in comparison with mice bitten by mosquitoes that had fed on non drug-treated infected mice. The overall effectiveness of an intervention over a round of transmission (from mouse to mosquito to mouse) can be quantified by estimating its ability to reduce the basic reproductive number (R0). This has been termed the “effect size” of an intervention. By fitting data from the mouse-to-mouse assay to a chain binomial model we can estimate the effect size of the intervention17, assessing the ability of DDD107498 usage to reduce the basic reproductive number Ro (assuming 100% coverage). Our results estimate an effect size of 90.5% (95% Cls 78.3-94.2), suggesting that DDD107498 is capable of acting as a potent transmission-blocking drug over multiple transmission settings within a field context18. The combination of these key in vitro and in vivo assays demonstrate the very strong potential of DDD107498 for blocking transmission; importantly, the required doses are likely to be similar to those required for treatment of blood-stage malaria.
DDD107498 targets PfeEF2
To determine the molecular target for DDD107498, asexual blood stage P. falciparum were cultured in the presence of DDD107498 at 5 × EC50, until parasites became resistant (Extended Data Table 2)20. Resistance was obtained in the 3D7 (drug sensitive) and 7G8 and Dd2 (multi-drug resistant) strains, with minimum inocula of 107, 107 and 106 respectively. Genomic DNA was extracted from resistant lines and whole genome sequencing of 10 drug-resistant lines identified shared mutations in one gene, which were not present in the parental lines: Pf3D7_1451100 (Supplementary Data 1). This gene encodes P. falciparum translation elongation factor 2 (PfeEF2). Three lines had two single nucleotide polymorphisms (SNPs) in PfeEF2, with a mixture of wildtype (WT) and mutant reads at each position (Supplementary Data 2), suggesting that these lines were mixtures of two clones, each with independent mutations in PfeEF2. SNPs were confirmed by Sanger sequencing and nine validated mutations in PfeEF2 clustered in three regions of the encoded protein. The fact that resistance to DDD107498 can be associated with multiple independent mutations is in keeping with observations from both artemisinin and compounds in development21-23. In two cases identical SNPs were identified in two independent lines, indicating mutations in functionally significant residues that were acquired independently in separate selection experiments.
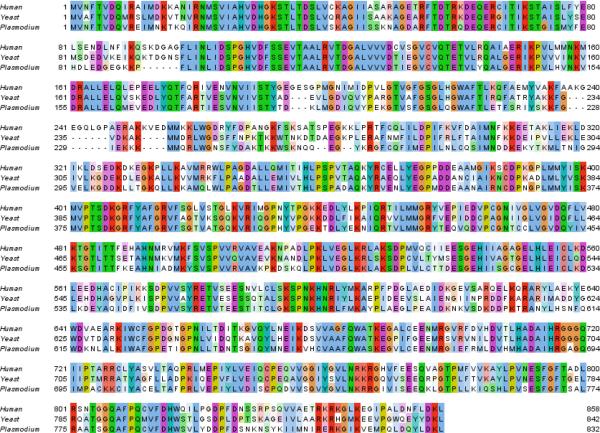
eEF2 is one of several essential elongation factors required in eukaryotic protein synthesis, by mediating GTP-dependent translocation of the ribosome along mRNA (Fig. 4a)24. Yeast eEF2 is the target of the antifungal compound sordarin25,26. Sordarin is selective for the Saccharomyces cerevisiae eEF2 over mammalian eEF2 in vitro, demonstrating that although protein synthesis in eukaryotes is conserved, it is possible to obtain selective inhibitors, despite the relatively high homology (67.2% identity) between the yeast and human eEF2 (Extended Data Fig. 7)25,27. In keeping with the potential for selectivity, DDD107498 is not toxic to mammalian cells (Extended Data Fig 2c). The PfeEF2 mutations associated with resistance mapped to several areas on the surface of a homology model of the protein (based on the apo structure of S. cerevisiae eEF228), with the mutations giving the highest degree of resistance clustering together (Fig. 4b). A DDD107498 binding pocket could not be elucidated through these modeling studies.
Figure 4. eFF2.
a. eEF2 promotes the GTP-dependent translocation of the ribosome along mRNA during protein synthesis.
b. Homology model of Plasmodium falciparum eEF2. The mapped mutations from each strain are colour coded by EC50 fold (red high, amber moderate, green low).
c. Live cell imaging of P. falciparum expressing an extra copy of eEF2 (WT) fused to GFP. The image is representative of >50 parasites visualized on two independent occasions.
d. Protein and DNA/RNA synthesis were evaluated by measuring the incorporation of [35S]-labelled methionine and cysteine ([35S]-Met/Cys) (upper panel) and [3H]-labelled hypoxanthine (lower panel) into asynchronous 3D7 wild-type (○) and 3D7 DDD107498-resistant line (eEF2-E134A/P754A) (●) after 40 min incubation with DDD107498, cycloheximide or actinomycin D. Radiolabeled incorporation, measured as cpm, was normalised as % of incorporation against inhibitor concentration (means ± s.d.; n=3 independent experiments each run in duplicate).
e. The EC50 values for transfectants against DDD107498 (means ± s.d.; n=4-7 independent experiments, each run in duplicate). Statistical significance was determined by the Mann-Whitney U test: *P<0.05; **P<0.01.
f. DDD107498-resistant line (eEF2-Y186N) transfected episomally with plasmids expressing either WT-eEF2 or eEF2-Y186N (means ± s.d.; n=3 independent experiments each run in duplicate).
We confirmed that brief pre-incubation with DDD107498 specifically inhibited P. falciparum protein synthesis, by measuring short-term incorporation of [35S]-Met/Cys in WT and DDD107498-resistant 3D7 P. falciparum parasites (Fig. 4d)21. Cycloheximide (a protein synthesis inhibitor) and DDD107498 both prevented [35S]-Met/Cys incorporation into WT P. falciparum 3D7, whereas actinomycin D (an inhibitor of RNA transcription) had no effect. Notably, DDD107498 was 100-fold less effective in DDD107498-resistant 3D7 compared to WT parasites, whereas cycloheximide prevented [35S]-Met/Cys incorporation equally in resistant and WT lines. DDD107498 and cycloheximide had minimal effects on DNA/RNA biosynthesis in both sensitive and drug-resistant parasites (measured by incorporation of [3H]-hypoxanthine), demonstrating specificity. In contrast actinomycin D caused a significant dose-dependent reduction in [3H]-incorporation.
To confirm that PfeEF2 is the target for DDD107498, we integrated transgenes expressing either WT PfeEF2 or resistance-associated alleles of PfeEf2 (Y186N, observed in mutant Dd2 and P754S, observed in mutant 3D7) using attPxattB integrase-mediated recombination21,29. These transfectants also express endogenous PfeEF2. Imaging of GFP-fusions of PfeEF2 showed cytoplasmic localisation (Fig. 4c), indicating that DDD107498 inhibits protein synthesis in the cytoplasm as opposed to the apicoplast30, the site of action of tetracycline and azithromycin.
Dose-response assays with DDD107498 showed a similar inhibition profile between PfeEF2 transgene-expressing lines and the WT Dd2 strain, indicating that the endogenous WT-PfeEF2 has a dominant effect in these experiments (Fig. 4e). This may be due to stable complex formation between the ribosome, WT-PfeEF2 and DDD107498, resulting in ribosome stalling, which would explain why in mixed populations the WT-PfeEf2 is dominant. For example in bacteria, fusidic acid binds to the complex between EF-G and the ribosome, preventing dissociation and blocking protein translation31.
To determine whether WT-PfeEF2 was dominant in poisoning translation, we introduced episomal plasmids encoding WT or Y186N mutant PfeEF2 into the resistant PfeEF2 Y186N line (Fig. 4f). Plasmid-borne PfeEF2-Y186N had no effect on sensitivity to DDD107498 (EC50 ~ 3100 nM), whereas WT-PfeEF2 restored sensitivity (EC50 = 2 nM). This demonstrated a dominant effect of WT-PfeEF2 on parasite susceptibility and confirmed that PfeEF2 is the primary molecular target of the compound. We note that the shallow slope observed for the WT-PfeEF2 transfected line is likely a result of heterogeneous episomal plasmid copy number seen with episomally transformed parasite lines29. Structural studies will be required to define precisely how DDD107498 interacts with eEF2 and the ribosome.
Resistance has been reported for all clinical antimalarials, including artemisinins2. Whilst the correlation between the rate of resistance generation in laboratory and clinical settings for antimalarials is not fully understood, it is important to evaluate the risk of all new antimalarials in both preclinical and clinical studies. In our studies, the minimum inoculum for generating resistance to DDD107498 is within acceptable limits32. Furthermore, selected resistant Dd2 lines revealed impaired growth rates in the absence of drug pressure compared to WT-Dd2 (Extended Data Fig. 8); moreover the higher the resistance, the lower the growth fitness. Importantly genome sequences from 1685 clinical isolates of P. falciparum from 17 countries23,33 reveal a high degree of PfeEF2 sequence conservation in the field. The sole non-synonymous SNP (T16S) identified was unique to West Africa (allele frequency of 0.002) and is in a PfeEF2 domain distinct from mutations associated with in vitro resistance to DDD107498.
Conclusion
DDD107498 represents a promising prospect for development as an anti-malarial agent, with both a potent activity profile against multiple life-cycle stages (sub-10 nM), a novel mode of action and excellent drug-like properties. It has potential for single dose treatment, which has major implications for ensuring patient compliance and practical deployment. Its complementary activity on the sexual stages of the parasites has potential to reduce transmission and its action on the liver stage suggests a possible role in chemoprevention. Chemoprotection and transmission blocking properties are fundamental to the goal of elimination and eradication of malaria, for which the high potency and long half-life of DDD107498 are well suited.
Due to general concerns about the emergence of resistance, all antimalarials are developed as combination therapies, a strategy shown to improve efficacy and reduce the development of drug resistance. In terms of treatment of the erythrocytic stage, DDD107498 fulfils the criteria as a long duration partner to complete the clearance of blood stage parasites11. Therefore, it should be combined with a fast acting compound, ideally with a pharmacological duration of action as close to DDD107498 as possible. This would reduce the initial level of infection, with the prolonged activity of DDD107498 eliminating the remaining parasites11.
Inhibition of protein synthesis by DDD107498 through PfeEF2, which is expressed in multiple life cycle stages34, provides mechanistic support for the observed broad-spectrum profile. This highlights PfeEF2 as a novel drug target in malaria, and also implies that inhibition of protein synthesis is an effective intervention for achieving multi-stage activity in Plasmodium. DDD107498 has now been progressed into advanced nonclinical development, with the aim of entering into human clinical trials.
Extended Data
Extended Data Figure 1. Synthetic Methodology.
a. Synthesis of DDD107498 (4). b. Synthesis of DDD102542 (6a) and DDD103679 (6b).
Extended Data Figure 2. In vitro Activity.
a. In vitro activity against a panel of resistant and sensitive strains of P. falciparum. Abbreviations: chloroquine (CQ), pyrimethamine (PYR), cycloguanil (CYC), quinine (QUI), sulfadoxine (SUL) and/or mefloquine (MFQ). Resistance as follows: K1 (CQ, SUL, PYR, CYC); W2 (CQ, SUL, PYR, CYC); 7G8 (CQ, PYR, CYC); TM90C2A (CQ, PYR, MFQ, CYC); D6 (MFQ); V1/S (CQ, SUL, PYR, CYC). Data are the means ± s.d. of n=3 independent [3H]hypoxanthine incorporation experiments (each run in duplicate). b. Ex-vivo activity against P. falciparum and P. vivax clinical isolates from Papua (Indonesia). c. Effect of DDD107498 on P. falciparum 3D7, HepG2 and MRC5 cells. Data are the means ± s.d. of n reported independent experiments.
Extended Data Figure 3. Effect of DDD107498 on parasite morphology.
a. Phenotype of P. falciparum in peripheral blood of NOD-scid IL-2R_null mice engrafted with human erythrocytes. Blood samples were taken at day 5 and 7 of the assay (1 and 2 asexual cycles, respectively) after the start of treatment with vehicle or DDD107498 at day 3. The bidimensional flow cytometry plots measure the murine (Ter-119-PE+) and human (Ter-119-PE−) erythrocytes, and the presence of nucleic acids (infected SYTO-16+ events). The blue circles indicate the region of infected erythrocytes. Vehicle-treated mice showed a characteristic pattern of staining with SYTO-1635, which correlated with the presence of healthy rings, trophozoites and schizonts in blood smears. Conversely, mice treated with DDD107498 at 50 mg/kg showed only trophozoites with condensed cytoplasm and some pyknotic cells at day 5 (red circle in flow cytometry plot and corresponding blood smears). By day 7, few infected erythrocytes were detected by flow cytometry and blood smears revealed parasites with a similar morphology to those at day 5. This suggests that trophozoites are the most sensitive population since the cycle is interrupted at this stage. The images displayed are taken from a mouse with high levels of parasitemia. At least 50 parasites were counted per sample screened in the microscope. Of these, 4 photos of representative parasite phenotype were selected to represent the morphology of the most prevalent phenotype. Thus, this is a qualitative assessment. b. Stage specificity assays using synchronised cultures. For morphological analysis of antimalarial drug action, thin blood smears were prepared, fixed and stained with Giemsa followed by examination with an upright microscope using an oil-immersion lens (100×). For parasitemia determination, a total number of 1000 red blood cells (corresponding to 5 microscopic fields) were counted. R to T. Abnormal trophozoites observed after 24h exposure of synchronized rings to DDD107498. T to S. Trophozoites do not develop into schizonts after 24h exposure to DDD107498. S-R. No ring stages are observed 24h after treatment of schizonts with DDD107498. c. Percentage parasitemia in the red blood cells. R = ring stage, T = trophozoite, S = schizont.
Extended Data Figure 4. Prophylactic activity of DDD107498 against sporozoite challenge.
P. berghei (luciferase) sporozoite in vivo mouse model of chemoprotection. A dose of 3mg/kg was fully protective. Data are the mean of n=5.
Extended Data Figure 5. DDD107498 in vitro activity on the different life cycle stages of Plasmodium spp.
For comparator data with known antimalarials see Figure 5 in Delves et al.6 DDD107498 also showed potent activity in vitro against P. berghei ookinetes16, (5.0nM [CI 4.4-5.7nM]), confirming that if DDD107498 was taken up during a blood meal, it could continue to kill parasites within the mosquito.
Extended Data Figure 6. In vivo P. berghei mouse-to-mouse assay.
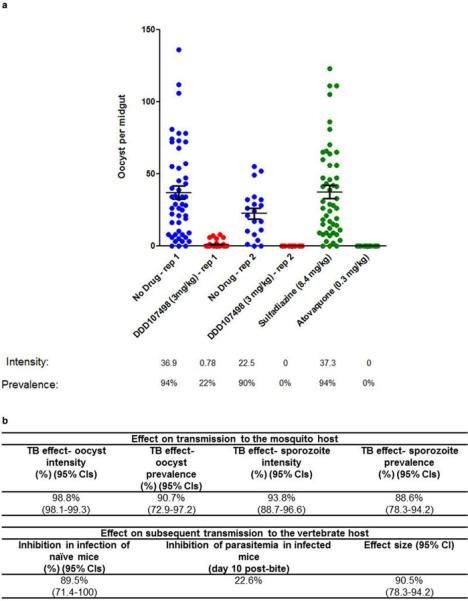
a. Impact of DDD107498 treatment on in vivo mosquito infection. Mice infected with P. berghei (PbGFPCON507 clone 1)19 were dosed orally with either DDD107489 at 3 mg/kg, atovaquone at 0.3 mg/kg, sulphadiazine at 8.4 mg/kg, or were not drug treated (negative control). After 24 h, populations of treated mice (n=5) were exposed to 500 female A. stephensi mosquitoes, and oocyst intensity and infection prevalence in the mosquito midgut was measured 10 days post-feeding. Individual data points represent the number of oocysts found in individual mosquitoes. Two replicates were performed. Sulphadiazine (8.4 mg/kg, i.p.) and atovaquone (0.3 mg/kg, i.p.) were used as negative and positive transmission-blocking drug controls, respectively. Horizontal bars indicate mean intensity of infection, whilst error bars indicate S.E.M. within individual mosquito populations. b. Impact of treatment with DDD107498 over a complete transmission cycle in vivo. Following drug treatment of infected mice and mosquito feeds, surviving potentially infectious mosquitoes were allowed to blood-feed on naive mice at a range of transmission settings (biting rates of 2,5 and 10 bites per naive mouse) to assess ability of drug treatment to reduce the number of new malarial cases post mosquito bite. Efficacy is expressed as: i) impact on the mosquito population – expressed as reduction in both oocyst and sporozoite intensity and prevalence; ii) impact on subsequent transmission to new naïve vertebrate hosts – expressed as reduction in infection of naïve mice (reduction in number of new malarial cases post-mosquito bite) and inhibition of subsequent parasitema (day 10 post-bite) in mice that do become infected; iii) Effect size generated – by fitting the data achieved within this assay to a chain-binomial model we can assess the ability of DDD107498 to reduce Ro (assuming 100% coverage). If Ro is reduced to <1, transmission is unsustainable and elimination will occur. 95% Cls are shown in brackets. Efficacy is calculated in comparison with no-drug controls. TB is transmission blocking
Extended Data Figure 7. ClustalWS alignment of eEF2 sequences from Human (Hs), Yeast (Sc) and Plasmodium falciparum (Pf).
Alignment made using Jalview ClustalX default colouring.
Extended Data Figure 8. Fitness phenotypes of DDD107498-resistant parasite lines.
Unmarked Dd2 and DDD107498-selected parasites with various levels of resistance were assessed for growth in a competition assay, relative to a Dd2-GFP reference line. a. Equal numbers of unmarked test lines were mixed with the Dd2-GFP reference, in triplicate wells, and the ratio of non-fluorescent and fluorescent cells assessed by flow cytometry over time. At day 0, all lines had a 1:1 ratio with the Dd2-GFP reference. Increased growth of the test line over the Dd2-GFP reference, which has a slower growth rate than unmarked WT Dd2, would result in an increased ratio of Test:Dd2-GFP. b. Growth assay of 4 different test lines: i) WT Dd2, ii) EF2-E134D, iii) EF2-L775F, and iv) EF2-Y186N, relative to Dd2-GFP. A faster growth rate of WT Dd2 (DDD107498 IC50 0.14 nM) relative to the fluorescent Dd2-GFP line is reflected in an increased ratio over time. The low-level resistant line EF2-E134D (IC50 5.8 nM) did not attain a WT growth rate, and the high-level resistant lines EF2-L775F (IC50 660 nM) and EF2-Y186N (3100 nM) were further impaired. Means ± s.d.; n=4 independent experiments each run in triplicate.
Extended Data Table 1. Pharmacokinetics and Rodent Efficacy of DDD107498.
a. In vivo pharmacokinetic parameters in preclinical species (n=3, all species). b. Oral in vivo single dose efficacy of antimalarial drugs and DDD107498 in the murine P. berghei model.
| a | |||
|---|---|---|---|
| Mouse | Rat | Dog | |
| Intravenous | 1 mg/kg | 1 mg/kg | 1 mg/kg |
| CI (ml/min/g) | 12 | 18 | 30 |
| Vdss (L/kg) | 15 | 15 | 30 |
| T½ (h) | 16 | 10 | 13 |
| Oral | 3 mg/kg | 5 mg/kg | 3 mg/kg |
|---|---|---|---|
| Cmax (ng/ml) | 90 | 180 | 27 |
| Tmax (h) | 1 | 4 | 4 |
| T½ (h) | 19 | 18 | 20 |
| F% | 74 | 84 | 46 |
| b | ||||||
|---|---|---|---|---|---|---|
| 30 mg/kg | 10 mg/kg | 3 mg/kg | ||||
| Compound | Activity % | Survival days | Activity % | Survival days | Activity % | Survival days |
| Chloroquine | §99.9 | 10 | 99.5 | 7 | 83 | 7 |
| Mefloquine | §99.6 | 22 | 95 | 16 | <40 | 7 |
| Artemether | 98 | 7 | 89 | 6 | 59 | 6 |
| Dihydroartemsinin | 99 | 7 | 97 | 6 | 61 | 7 |
| Artesunate | §92 | 9 | 75 | 6 | <40 | 6 |
| DDD107498 | 99.1 | 23 | 99.0 | 14 | 99 | 9 |
Data are the means of n=3 for DDD107498 and n≥5 for the reference compounds.
Data from Charman et al.36.
Extended Data Table 2.
Summary of resistance selection experiments with DDD107498
| Strain | Pressure (nM) | Pressure (x IC90) | Inoculum* Number of positive cultures (day of recrudescence) |
MIR | EC50 shift (inoculum)† | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 104 | 105 | 106 | 107 | 108 | 109 | |||||
| Dd2 | 1.25 | 2.7 | ND‡ | ND | 2 (13, 41) | 3 (13,13,15) | 3 (13,13,16) | 3 (29/29/29) | ND | 2315 (109) |
| 7 (108) | ||||||||||
| 4810 (106) | ||||||||||
| Dd2 | 1.26 | 27 | 0 | 0 | 0 | 3 (20,22,22) | ND | ND | 107 | 13 (107) |
| Dd2 | 1.25 | 2.7 | ND | 0 | 1 (39) | ND | ND | ND | 106 | 5567 (106) |
| 7G8 | 1.25 | 3.1 | ND | ND | 0 | 3 (25, 25, 47) | 1 (25) | 3 (15,15,15) | 107 | 70 (109) |
| 60 (108) | ||||||||||
| 82 (107) | ||||||||||
| 3D7 | 1.5 | 3.3 | ND | ND | 0 | 2 (38. 38) | 2 (28.28) | 3 (20.20.20) | 107 | 19 (109) |
| 99 (108) | ||||||||||
| 14(107) | ||||||||||
Number of positive cultures (n=3) and day of recrudescence for each inoculum.
EC50 fold change was determined on bulk cultures.
Not determined. MIR is the Minimum Inoculum of Resistance.
Supplementary Material
Acknowledgements
This work was supported by grants from Medicines for Malaria Venture, the Wellcome Trust (100476 (IHG, AHF), 091625 (RNP) and 098051 (JCR, WP, TDO)), the Bill and Melinda Gates Foundation (OPP1043501 (MD, RS), the NIH (R01 AI103058 to EAW and DAF) and the European Union (EVIMalaR (TDO)). DDU infrastructure was supported by the European Regional Development Fund 2007-2013 and UK Research Partnership Investment Fund awards to Michael Ferguson, who we also thank for continued support. We would like to thank Prof Carol Sibley, University of Washington for helpful discussions. We acknowledge the East Scotland Blood Transfusion Service, Ninewells Hospital, Dundee for erythrocyte supply to Dundee. We thank L. D. Shultz and The Jackson Laboratory for providing access to nonobese diabetic scid IL2Rγc null mice through their collaboration with GSK Tres Cantos Medicines Development Campus. The following are acknowledged for technical assistance: all members of the DDU (Dundee), Matthew Berriman, Jolanda Kamber, Enny Kenangalem, Alexis LaCrue, Oliver Montagnat, Jeanne Rini Poespoprodjo, Mandy Sanders, Sibylle Sax, Christian Scheurer, Leily Trianty, Mark Tunnicliff (detailed in Supplementary Information).
Footnotes
Author Contributions The author contributions are detailed in the Supplementary Information.
Author Information A patent relating to this work has been filed (PCT/GB2009/002084). Koen Dechering and Robert Sauerwein have shares in TropIQ Health Sciences.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.WHO World Malaria Report 2014. 2014 ISBN 978 992 974 156483 156480. [Google Scholar]
- 2.Ariey F, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso PL, et al. A research agenda for malaria eradication: Drugs. PLoS Med. 2011;8:e1000402. doi: 10.1371/journal.pmed.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wells TNC, Gutteridge WE. Neglected Diseases and Drug Discovery. 2012:1–32. RSC. [Google Scholar]
- 5.Brenk R, et al. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem. 2008;3:435–444. doi: 10.1002/cmdc.200700139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delves M, et al. The activities of current antimalarial drugs on the life cycle stages of Plasmodium: A comparative study with human and rodent parasites. PLoS Med. 2012;9:e1001169. doi: 10.1371/journal.pmed.1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell B, et al. Determinants of in vitro drug susceptibility testing of Plasmodium vivax. Antimicrob. Agents Chemother. 2008;52:1040–1045. doi: 10.1128/AAC.01334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karyana M, et al. Malaria morbidity in Papua Indonesia, an area with multidrug resistant Plasmodium vivax and Plasmodium falciparum. Malaria J. 2008;7:148. doi: 10.1186/1475-2875-7-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angulo-Barturen I, et al. A murine model of falciparum-malaria by in vivo selection of competent strains in non-myelodepleted mice engrafted with human erythrocytes. PLoS One. 2008;3:e2252. doi: 10.1371/journal.pone.0002252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanz LM, et al. P. falciparum in vitro killing rates allow to discriminate between different antimalarial mode-of-action. PLoS One. 2012;7:e30949. doi: 10.1371/journal.pone.0030949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burrows JN, van Huijsduijnen RH, Mohrle JJ, Oeuvray C, Wells TN. Designing the next generation of medicines for malaria control and eradication. Malar. J. 2013;12:187. doi: 10.1186/1475-2875-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meister S, et al. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science. 2011;334:1372–1377. doi: 10.1126/science.1211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adjalley SH, et al. Quantitative assessment of Plasmodium falciparum sexual development reveals potent transmission-blocking activity by methylene blue. Proc. Natl. Acad. Sci. U S A. 2011;108:E1214–1223. doi: 10.1073/pnas.1112037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bousema T, Drakeley C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin. Microbiol. Rev. 2011;24:377–410. doi: 10.1128/CMR.00051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delves MJ, et al. Male and female Plasmodium falciparum mature gametocytes show different responses to antimalarial drugs. Antimicrob. Agents Chemother. 2013;57:3268–3274. doi: 10.1128/AAC.00325-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delves MJ, et al. A high-throughput assay for the identification of malarial transmission-blocking drugs and vaccines. Int. J. Parasitol. 2012;42:999–1006. doi: 10.1016/j.ijpara.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Blagborough AM, et al. Transmission-blocking interventions eliminate malaria from laboratory populations. Nat. Commun. 2013;4:1812. doi: 10.1038/ncomms2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Upton LM, et al. Lead clinical and preclinical antimalarial drugs can significantly reduce sporozoite transmission to vertebrate populations. Antimicrob. Agents Chemother. 2015;59:490–497. doi: 10.1128/AAC.03942-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janse CJ, et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol. Biochem. Parasitol. 2006;145:60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Flannery EL, Fidock DA, Winzeler EA. Using genetic methods to define the targets of compounds with antimalarial activity. J. Med. Chem. 2013;56:7761–7771. doi: 10.1021/jm400325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rottman M, et al. Spiroindolones, a potent compound class for the treatment of malaria. Science. 2010;329:1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNamara CW, et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature. 2013;504:248–253. doi: 10.1038/nature12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miotto O, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen R, Merrill AR, Andersen GR. The life and death of translation elongation factor 2. Biochem. Soc. Trans. 2006;34:1–6. doi: 10.1042/BST20060001. [DOI] [PubMed] [Google Scholar]
- 25.Justice MC, et al. Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 1998;273:3148–3151. doi: 10.1074/jbc.273.6.3148. [DOI] [PubMed] [Google Scholar]
- 26.Capa L, Mendoza A, Lavandera JL, de las Heras FG, Garcia-Bustos JF. Translation elongation factor 2 is part of the target for a new family of antifungals. Antimicrob. Agents Chemother. 1998;42:2694–2699. doi: 10.1128/aac.42.10.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shastry M, et al. Species-specific inhibition of fungal protein synthesis by sordarin: identification of a sordarin-specificity region in eukaryotic elongation factor 2. Microbiology. 2001;147:383–390. doi: 10.1099/00221287-147-2-383. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen R, et al. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Biol. 2003;10:379–385. doi: 10.1038/nsb923. [DOI] [PubMed] [Google Scholar]
- 29.Nkrumah LJ, et al. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat. Methods. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biswas S, et al. Interaction of apicoplast-encoded elongation factor (EF) EF-Tu with nuclear-encoded EF-Ts mediates translation in the Plasmodium falciparum plastid. Int. J. Parasitol. 2011;41:417–427. doi: 10.1016/j.ijpara.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Cox G, et al. Ribosome clearance by FusB-type proteins mediates resistance to the antibiotic fusidic acid. Proc. Natl. Acad. Sci. U S A. 2012;109:2102–2107. doi: 10.1073/pnas.1117275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding XC, Ubben D, Wells TN. A framework for assessing the risk of resistance for anti-malarials in development. Malar. J. 2012;11:292. doi: 10.1186/1475-2875-11-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Florens L, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez-Diaz MB, et al. Quantitative measurement of Plasmodium-infected erythrocytes in murine models of malaria by flow cytometry using bidimensional assessment of SYTO-16 fluorescence. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2009;75:225–235. doi: 10.1002/cyto.a.20647. [DOI] [PubMed] [Google Scholar]
- 36.Charman SA, et al. Synthetic ozonide drug candidate OZ439 offers new hope for a single-dose cure of uncomplicated malaria. Proc. Natl. Acad. Sci. U S A. 2011;108:4400–4405. doi: 10.1073/pnas.1015762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.