Abstract
T regulatory cells that express the transcription factor Foxp3 (Foxp3+ Treg) promote tissue homeostasis in several settings. We now report that symbiotic members of the human gut microbiota induce a distinct Treg population in the mouse colon, which constrains immuno-inflammatory responses. This induction, which we find to map to a broad, but specific, array of individual bacterial species, requires the transcription factor Rorγ, paradoxically in that Rorγ is thought to antagonize FoxP3 and promote T helper 17 (Th17) cell differentiation. Rorγ's transcriptional footprint differs in colonic Tregs and Th17 cells, controlling important effector molecules. Rorγ, and the Tregs that express it, contribute substantially to regulating colonic Th1/Th17 inflammation. Thus, the marked context-specificity of Rorγ results in very different outcomes even in closely related cell-types.
FoxP3 regulatory T (Treg) cells are essential regulators of immunologic homeostasis and responses (1). Beyond their well-described role in regulating the activity of other immunocytes, Tregs located in parenchymal tissues control other, non-immunological, processes. These “tissue Tregs” include those that reside in visceral adipose tissue and regulate metabolic parameters (2, 3), or those that help channel inflammatory and regenerative events in injured muscle (4). The activities, transcriptomes and T cell receptor (TCR) repertoires of these tissue Tregs are distinct from their counterparts in secondary lymphoid organs.
Another essential and quite specific population of tissue Tregs resides in the lamina propria (LP) of the digestive tract, in particular in the colon, where they modulate responses to commensal microbes (reviewed in (5)). Colonic Tregs are an unusual population, which has provoked some contradictory observations. TCRs expressed by colonic Tregs show marked reactivity against microbial antigens which seem important drivers of their differentiation and/or expansion (6, 7). Many of them appear to arise by conversion from FoxP3- conventional CD4+ T cells (Tconv) (6, 7). although arguments for a thymic origin have been made (7). Many colonic Tregs express marker profiles (Nrp1−, Helios−) that differ from Tregs of thymic origin (reviewed in (8)), although the significance of these markers has been questioned ((5, 8)). Accordingly, most studies have found a decreased abundance of colonic Tregs in germ-free (GF) mice (reviewed in (5)), and colonization of GF mice by pools of microbes (Schadler's flora (9), Clostridia combinations (10, 11)) elicited the differentiation or expansion of Helios−Nrp1− colonic Tregs. The ability of single microbes to induce colonic Tregs has been more controversial, and the need for complex combinations (10, 11)) has been questioned (12).
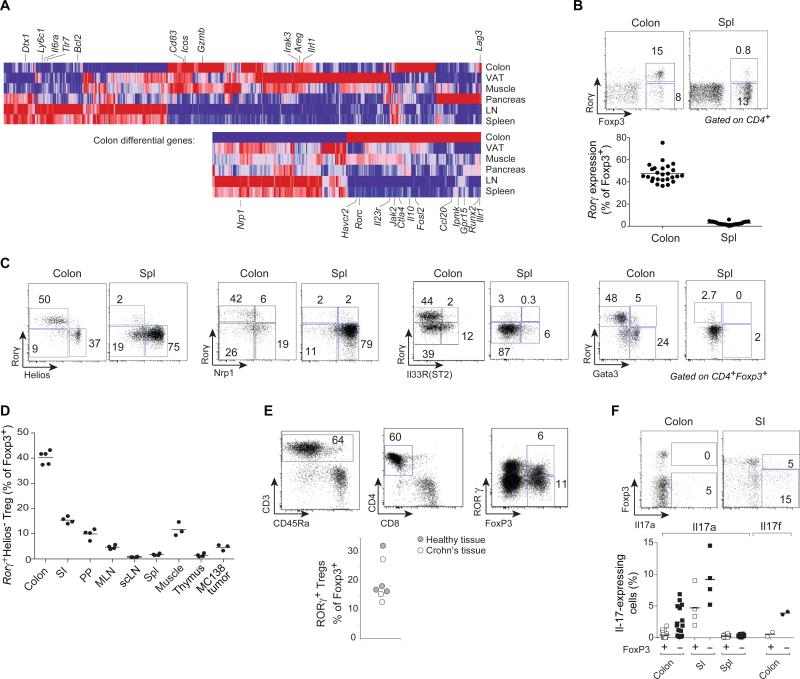
The transcriptomes of tissue-resident Tregs adapt to their location, most strikingly in terms of transcription factors (13), and we searched for such elements in colonic Tregs. Comparison of transcriptomes of highly purified CD4+FoxP3+ Tregs (from Foxp3ires-gfp reporter mice (14)) from colon or spleen uncovered 933 differential transcripts (at a FoldChange>2 and FDR<0.1; Fig. 1A (top), Fig. S1A, Table S1). These encompassed important signaling and effector pathways (Icos, Gzmb, Lag3, Areg, Il1rl1; Fig 1A (top), Table S1), shared in patchwork manner by other tissue Tregs. Yet ~39% (at a colon-specific bias>1.5 fold) had preferential expression in colonic Tregs (including Il10, Ctla4, Havcr2, Ccl20, Jak2, Fosl2; Fig.1A (bottom), Table S2). GeneOntology analysis revealed no enriched function or pathway, except for a high proportion of TFs, including Ahr, Epas1, Hey1, Bcl6, Npas2, Nr1d1, and Maf. Surprisingly, the most differential of these TFs proved to be Rorc (encodes Rorγ; Fig. S1B). Rorγ controls many aspects of immunocyte differentiation (15), but is perhaps best known as the key regulator of interleukin (IL)-17-producing CD4+ T cells (Th17), and as a reciprocal antagonist of FoxP3 during in vitro differentiation in which iTreg and Th17 represent alternative cell fates (reviewed in (16)).
Figure 1. Rorγ, encoded by Rorc, is preferentially expressed in colonic Tregs.
Gene expression profiles from purified Treg cells of various origins. A: Transcripts that are enriched in tissue and colonic Tregs. Top: transcripts differentially represented in tissue vs. splenic Tregs (at a FoldChange>2). Bottom: transcripts that are most biased in colonic Tregs (FoldChange>1.5 vs any other tissue Treg). Means of at least 2 duplicates. B: Transcription factors overrepresented in colonic Tregs vs other tissue and lymphoid organ Tregs. C: Representative flow cytometry plots of CD4+ T cells, and compilation of frequencies (right) of Rorγ+Helios− Tregs within the FoxP3+CD4+TCRβ+ population. Each point is an individual mouse. Data representative of > 3 independent experiments. D: Representative Rorγ vs Helios, Nrp1, Il33R or Gata3 plots for colon or spleen Foxp3+CD4+TCRβ+ Tregs (see Fig. S2 for quantification). Each point is an individual mouse. Data representative of ≥ 3 independent experiments. E: Frequencies of Rorγ+Helios− Tregs among FoxP3+CD4+TCRβ+ cells of different tissues (SI: small intestinal lamina propria; PP: Peyer's patches; MLN: mesenteric lymph nodes; scLN: subcutaneous lymph nodes). Each point is an individual mouse. Data pooled from at least 2 independent experiments. F: Flow cytometry analysis of human colon biopsies and frequencies of human RORγ+ Tregs within the FOXP3+CD4+CD8−CD3+CD45+ population. Healthy tissue were endoscopically-determined normal areas from chronic constipation or irritable bowel syndrome patients; inflamed tissue from Crohn's lesions. Each point is an individual patient. Data pooled from 5 independent experiments). G: Il17a (PMA+ionomycin activation, intracellular staining) or Il17f (reporter in Il17frfp mice) expression among Foxp3+ Treg or FoxP3− Tconv Each point is an individual mouse. Data representative of independent 3 experiments.
Cytometry confirmed that many colonic CD4+FoxP3+ Tregs express Rorγ (40-60% in B6 or other inbred strains – Fig. 1B, S2A), a phenotype largely absent in spleen or lymph node (LN) and among FoxP3+ cells induced in vitro. Helios and Nrp1, described as markers of thymus-derived Tregs (reviewed in (8)), were absent on colonic Rorγ+ Tregs (Fig. 1C), demarcating three distinct subsets of colonic Tregs; Rorγ+ representing the majority of Helios− cells (Fig. 1C, S2B-C). Consistent with the RNA data, Rorγ+ Tregs were also detected in low proportions in the small intestine and the regenerating muscle, (Fig. 1D, S2D). In keeping with a recent report (17), Rorγ+ Tregs were distinct from those expressing the Il33 receptor, most of which were Helios+ (Figs. 1D, S2B-C,E-F), and from Gata3hi Tregs (18), which also belong to the Helios+ Treg subset.
We asked whether RORγ is also expressed by colonic Tregs in humans by staining cells from healthy or inflamed (Crohn's) colon biopsies. Rorγ+ Tregs were indeed detected at comparable levels in both contexts (Fig. 1E)
Rare Tregs expressing Il17 and Rorγ have been observed during chronic inflammation or cancer, usually being Helioshi (reviewed in (19)). We tested Il17 production in colonic Rorγ+ Tregs. While Il17-expressing Tregs could be detected in the small intestine LP, colonic Rorγ+ Tregs did not secrete detectable Il17a or f (Fig. 1F).
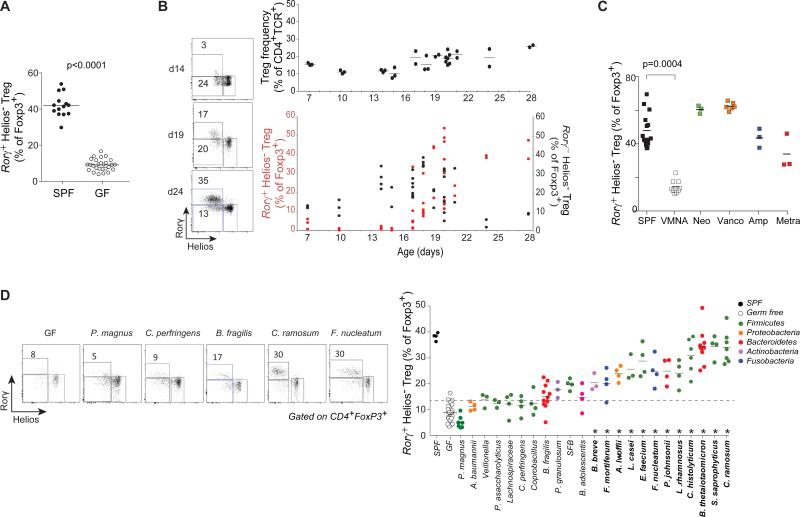
The properties of this dominant colonic Helios−Rorγ+ Treg population suggested a link to the gut microbiota. Indeed, GF mice had a lower proportion of Rorγ+ Tregs than their conventionally-raised specific-pathogen-free (SPF) counterparts (Fig. 2A). During the normal maturation, Rorγ+ Tregs appeared between 15 and 25 days of age (Fig. 2B), coincident with the changes in the gut microbiota that accompany the transition to solid food. Interestingly, Rorγ+ Tregs appeared a few days after Rorγ−Helios− Tregs. Antibiotic treatment strongly affected Rorγ+ Tregs (Fig. 2C), a large reduction following a broad-spectrum antibiotic combination, while individual antibiotics had less or no effect, suggesting the contribution of several microbes. As the reported impacts of various microbial species on total colonic Tregs have differed (10, 12), we took advantage of a panel of mice generated in a large-scale screen in which GF mice were colonized with a single species from a panel of 22 bacterial species from the human gastrointestinal tract (Table S3). A number of microbes elicited colonic Rorγ+ Tregs, with a gradient of responses, and for some at frequencies comparable with those of SPF mice (Fig. 2D). This restoration of Rorγ+ Tregs was independent of bacterial load and not accompanied by inflammation (Fig. S3). Bacteria able to induce Rorγ+ Treg (and colonic FoxP3+ Tregs more generally) belonged to several phyla and genera, and were not restricted to Clostridiae (10, 11). Segmented Filamentous Bacteria (SFB), classic inducers of Rorγ-dependent Th17 cells (20) and which elicit IL17-producing Tregs in the small intestine (SI) (21), were only mediocre inducers of colonic Rorγ+ Tregs, reinforcing the distinction between the cell populations. We noticed diversity within the Bacteroides genus, and assessed a wider Bacteroides panel (Fig. S4A, Table S3). Here again, a range of colonic Rorγ+ Tregs was observed. This distribution did not relate to the Bacteroides phylogeny for these strains with no unique correlation between Treg inducing ability and gene content (Fig. S4B). Colonic Rorγ+ Tregs did not appear immediately after GF colonization, but only after a few days, again following Rorγ−Helios− cells (Fig. S4C).
Figure 2. Rorγ+Helios− Tregs can be induced by several bacterial species.
A: Frequency of Rorγ+Helios− within colon FoxP3+CD4+TCRβ+ Tregs of SPF and germ free mice, p < 0.0001 as determined by Student's t test. Each point is an individual mouse. Data pooled from >3 experiments. B: Induction of Rorγ in colonic Tregs during post-natal development in SPF mice; left: Representative FACS plots; right: frequencies across ages of FoxP3+ Tregs within CD4+TCRβ+ cells, and Rorγ+Helios− (red) and Rorγ−Helios− (black) cells within Tregs. Each point is an individual mouse. Data pooled from ≥ 4 experiments. C; SPF mice were treated with single (Neomycin, Vancomycin, Ampicillin, Metronidazole) or all four (VMNA) antibiotics for 4 weeks. Frequency of colonic Rorγ+Helios− Tregs within the FoxP3+CD4+TCRβ+ population. p=0.0004, Bonferroni-corrected Student's t test. Each point is an individual mouse. Data pooled from 2 experiments. D: GF mice were colonized with single bacterial species and colonic Tregs analyzed after 2 weeks. Representative plots and frequencies of Rorγ+Helios− within FoxP3+CD4+TCRβ+ Tregs, color-coded per phyla Each point is an individual mouse. Data representative 1-3 experiments for each microbe. *: Different from GF at an FDR<0.05.
Several reports have suggested that short-chain fatty acids (SCFA) promote increased colonic Tregs (22-24). To test their relevance to Rorγ+ Tregs, SCFAs were quantitated by LCMS in cecal content of monocolonized mice. No significant correlation between any SCFA and Rorγ+ Treg frequency, or to other Treg parameters, was observed (Fig S5A-B, Table S4). In addition, we could not reproduce previously reported effects of oral or rectal SCFA administration (Fig. S5C-D). Although SCFA combinatorial effects, or inter-colony variation cannot be ruled out, SCFA cannot alone explain microbial impact on colonic Tregs observed here.
To integrate our observations with intercellular pathways that influence intestinal T cells, we measured the relative abundance of Rorγ+ Tregs in mice lacking receptors for key cytokines and alarmins. Signaling through Il23, Il1, or Il33 receptors was not required to sustain Rorγ+ Tregs, as was Il10 (Fig. S6A-D). In fact, only the Helios+ population expanded after Il33 administration (Fig. S6E).
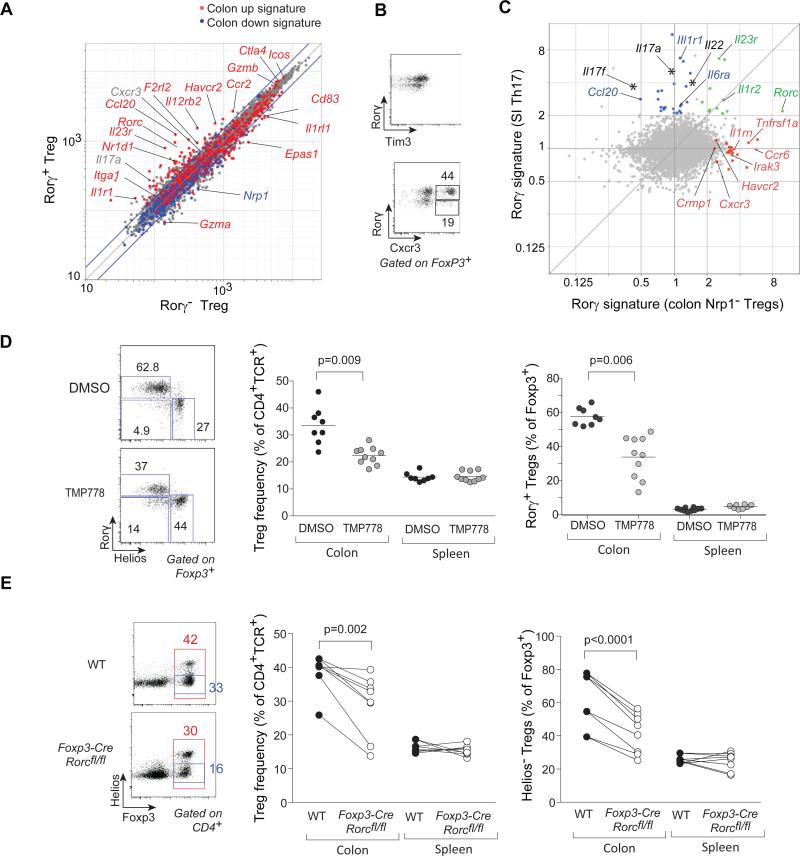
We then asked what transcripts Rorγ controls in Rorγ+ Tregs, and whether Rorγ is necessary to specify this particular Treg lineage. We compared transcriptomes of Rorγ+ and Rorγ− colonic Tregs (sorted from Foxp3Thy1.1 x Rorcgfp intercrossed mice). Rorγ+ cells were enriched in some, but not all, transcripts of the colonic Treg signature, notably Il23r, Cxcr3, Tbx21 and Havcr2 (Fig 3A), as validated at the protein level, including the unexpected CXCR3 (Fig. 3B). Conversely, Il1rl1 (encodes Il33R), Nrp1 and Ikzf2 were underrepresented in Rorγ+ Tregs.
Figure 3. Rorγ determines a specific signature and function in colonic Tregs.
A: Rorγ+ or Rorγ− Tregs were sorted from the colon of Foxp3thy1.1x Rorcgfp intercross male mice, and gene expression profiles determined. Expression values (triplicate averaged) are compared, highlighted according to the colon Treg signature of Fig. S1. B: Flow cytometric validation of some of the Rorγ+/Rorγ− Tregs differential genes (Havcr2 which encodes TIM3, Cxcr3) (represent 2 experiments). C: Comparison by gene expression profiling of the Rorγ signature in different contexts (all mean of triplicates). X-axis: FoldChange between colonic Nrp1− Tregs from WT or Foxp3-cre x Rorcfl/fl mice; Y-axis: FoldChange between SI CD4+ T cells sorted from GF monocolonized with Th17-inducing SFB or from unmanipulated GF. Shared or specific signature genes are color-coded. D: SPF mice were treated with Rorγ antagonist TMP778 or control DMSO for 3 weeks. Representative cytometry plots of colonic Tregs (left) or compiled frequencies of FoxP3+ Tregs (middle) and of Rorγ+Helios− (right) within FoxP3+CD4+TCRβ+ Tregs (right); ); p=0.009 as determined by Student's t test . Each point is an individual mouse. Data representative of ≥ 2 independent experiments.
E: Analysis of Rorγ–deficient Tregs from Foxp3-cre x Rorcfl/fl mice or control (Foxp3-creRorc+/+) littermates. Cytometry plots of colonic Tregs (left) or compiled frequencies of FoxP3+ Tregs (middle) and of Rorγ+Helios− (right) within FoxP3+CD4+TCRβ+ Tregs (right); p=0.002 (middle) and p< 0.0001 (right) as determined by paired Student's t test Each point is an individual mouse. Data representative of > 3 independent experiments.
To further delineate the transcriptional signature of Rorγ in Treg cells, RNAseq profiles were generated from Nrp1− cells of Foxp3-cre.Rorcfl/fl mice, which have a Treg-selective deletion of Rorc (Fig. S7A), or paired WT littermates. Differentially expressed genes were related to the Rorγ-dependent signature in conventional Th17 cells (defined from a comparison of SI CD4+ T cells of mice colonized, or not, with SFB; Fig. 3C, Table S5). Part of the classic Th17 signature was unrelated to Rorγ in colonic Tregs (blue in Fig. 3C;. Il1r1 or the canonical Th17 cytokines Il17a/f and Il22); some were shared (Rorc itself, Il23r); and a third segment was controlled by Rorγ in Nrp1- colonic Tregs but not in Th17 cells (Havrc2, Irak3, Il1rn). Thus, the transcriptional footprint of Rorγ is context-dependent in different T cells.
Next, we explored whether Rorγ contributes to colonic Treg homeostasis. First, mice were treated for 3 weeks with a pharmacologic Rorγ antagonist (25), which reduces SI Th17 levels. This treatment partially decreased both the total frequency of colonic FoxP3+ cells and their Rorγ+ component (Fig. 3D). Second, Foxp3-cre.Rorcfl/fl mice, which have no systemic Treg deficiency or scurfy-like pathology, nor any change in FoxP3 intensity, showed a reduced frequency of colonic Tregs, and more specifically of Helios− Tregs; the proportion of Helios+Gata3+ Tregs was correspondingly increased (Fig. 3E, S7B).
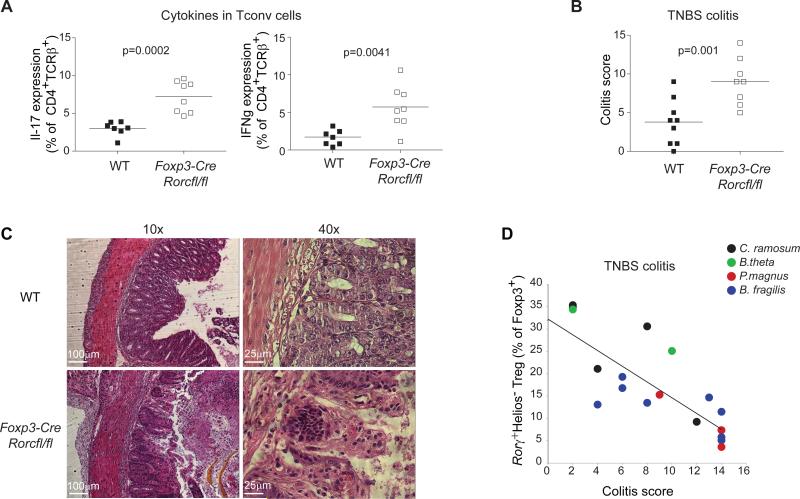
We noted that the loss of Rorγ+ Tregs in Foxp3-cre.Rorcfl/fl mice led to increased production of Il17 and IFNγ, but not Th2 cytokines like IL5 or IL13, by Tconv cells in colons of otherwise unchallenged mice (Fig. 4A), suggesting a decreased ability of colonic Tregs lacking Rorγ to regulate inflammatory responses. We thus assessed Foxp3-cre.Rorcfl/fl mice in the Trinitrobenzenesulfonic acid- (TNBS) induced colitis model, and found an exacerbation of disease severity, in colitis score and histopathology (Figs. 4B/C). Secondly, after TNBS challenge of GF mice monocolonized with different microbes, the frequency of Rorγ+ Tregs correlated with the colitis score (Fig. 4D). These results imply a non-redundant role for Rorγ and Rorγ+ Tregs in colonic homeostasis.
Figure 4. Rorγ+ Tregs control gut inflammation.
A: Frequency of Il17a and IFNγ expression in Foxp3−CD4+ Tconv cells from Foxp3-cre x Rorcfl/fl mice and control Foxp3-cre x Rorc+/+ littermates at steady state; p=0.0002 and p=0.0041, paired Student's t test . Each point is an individual mouse. Data representative of ≥ 3 independent experiments.
B/C: Colitis score (B) and histology (C) of Foxp3-cre x Rorcfl/fl mice and control Foxp3-cre x Rorc+/+ littermates challenged with TNBS, calculated based on weight loss, histologic score and other physical parameters; p=0.001 as determined by paired Student's t test . Each point is an individual mouse. Data representative of >3 independent experiments. D: Correlation between TNBS-colitis score (x-axis) with frequency of Rorγ+Helios− within colonic Tregs in GF mice monocolonized for 2 weeks with bacteria that elicit different levels of Rorγ+Helios− Tregs prior to TNBS colitis induction. Pearson r= 0.82, p<0.0001. Each point is an individual mouse. Data pooled from 4 experiments.
Thus, Rorγ contributes unexpectedly but importantly to the Treg response to commensal microbes. This role contrasts with the accepted dichotomy between FoxP3 and Rorγ, a notion stemming mainly from their antagonism in vitro (14, 26-28), perhaps over-interpreted. There had been indications that the two TFs are not incompatible (19), but these data suggest a collaborative transcriptional impact, consistent with the overlap between their chromatin-binding sites (29). The context-specificity of Rorγ's transcriptional footprint is in line with its broad involvement in many immunological and non-immunological processes (organogenesis, circadian rhythm, lipid metabolism) (15, 30). Rorγ-dependent Il23r expression in Tregs also raises the intriguing speculation that human IL23R genetic variants associated with inflammatory bowel disease (31) might involve balancing effects in effector and regulatory T cells.
Rorγ+ Tregs form the majority of the Helios− Tregs that differentiate locally in response to antigens of commensal microbes in the gut (6), and do not respond to the alarmin Il33, in contrast to Gata3+Helios+ cells that expand during tissue damage (17, 18). Mutually exclusive expression of Gata3 and Rorγ in colonic Tregs suggests that they may distinguish Treg responses to symbiotic (Rorγ) vs aggressive (Gata3) microbes. Contrary to expectations, many individual microbes proved able to elicit Rorγ+ and Helios− Tregs, a property not restricted to Clostridiae (10). The graded range suggests that several mechanisms may be involved. The molecular mediator of Rorγ+ Treg induction remains elusive, but is unlikely to be SCFA alone. Rorγ+ induction must follow different routes in Th17 vs colonic Tregs, since the best Rorγ+ Treg inducers do not affect SI Th17, and vice-versa.
In conclusion, these studies show Rorγ as a uniquely microbe-responsive factor induced in two different cellular contexts, in response to different microbes, with distinct transcriptional consequences, and with diametrically opposite functional outcomes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. A. Onderdonk, C. Dong, A. Rudensky, R. Lee, V. Kuchroo and L. Bry for microbial and mouse strains, S. Edwards, A. T. Sherpa, K. Hattori, K. Rothamel and R. Cruse for help with mice or profiling. TMP778 is available from GSK under a material transfer agreement. The data are tabulated in supplementary materials and deposited at the NCBI GEO (GSE68009). ES/NGZ/DK/DM/CB and HMS have filed a provisional patent application. This work was supported by National Institutes of Health R01-AI51530 and R56-AI110630 and the JPB Foundation (DM/CB); a Sponsored Research Agreement from UCB (DM/CB/DK/AE); the Helmsley Charitable Trust and the Wolpow Family Chair in IBD Treatment and Research (SPS). ES and DZ were supported by fellowships from the Boehringer Ingelheim Fonds, NGZ by HFSP and EMBO (ALTF 251-2011) fellowships and the Weizmann - National Postdoctoral Award for Advancing Women in Science.
Footnotes
Supplementary Materials
Materials and Methods
Supplementary figure legends S1-S6
Supplementary figures S1-S6
Supplementary table legends S1-S5
Supplementary tables S1-S5
References 32-58
REFERENCES
- 1.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 2012;30:531. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cipolletta D, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ai TL, Solomon BD, Hsieh CS. T-cell selection and intestinal homeostasis. Immunol. Rev. 2014;259:60. doi: 10.1111/imr.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cebula A, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilate AM, Lafaille JJ. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu. Rev. Immunol. 2012;30:733. doi: 10.1146/annurev-immunol-020711-075043. [DOI] [PubMed] [Google Scholar]
- 9.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 10.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 12.Faith JJ, et al. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl. Med. 2014;6:220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nat Immunol. 2013;14:1007. doi: 10.1038/ni.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 15.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 2010;10:479. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 16.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu. Rev. Immunol. 2009;27:485. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 17.Schiering C, et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513:564. doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wohlfert EA, et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J. Clin. Invest. 2011;121:4503. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du R, Zhao H, Yan F, Li H. IL-17+Foxp3+ T cells: an intermediate differentiation stage between Th17 cells and regulatory T cells. J. Leukoc. Biol. 2014;96:39. doi: 10.1189/jlb.1RU0114-010RR. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lochner M, et al. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J Immunol. 2011;186:1531. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- 22.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 24.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skepner J, et al. Pharmacologic inhibition of RORgammat regulates Th17 signature gene expression and suppresses cutaneous inflammation in vivo. J. Immunol. 2014;192:2564. doi: 10.4049/jimmunol.1302190. [DOI] [PubMed] [Google Scholar]
- 26.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang XO, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mucida D, et al. Reciprocal Th-17 and Regulatory T Cell Differentiation Mediated by Retinoic Acid. Science. 2007 doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 29.Xiao S, et al. Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40:477. doi: 10.1016/j.immuni.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abraham C, Cho JH. IL-23 and autoimmunity: new insights into the pathogenesis of inflammatory bowel disease. Annu. Rev. Med. 2009;60:97. doi: 10.1146/annurev.med.60.051407.123757. [DOI] [PubMed] [Google Scholar]
- 32.Eberl G, et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 2004;5:64. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 33.Glaccum MB, et al. Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. J. Immunol. 1997;159:3364. [PubMed] [Google Scholar]
- 34.Gonzalez A, et al. Damage control, rather than unresponsiveness, effected by protective DX5+ T cells in autoimmune diabetes. Nat. Immunol. 2001;2:1117. doi: 10.1038/ni738. [DOI] [PubMed] [Google Scholar]
- 35.Townsend MJ, et al. T1/ST2-deficient mice demonstrate the importance of T1/ST2 in developing primary T helper cell type 2 responses. J Exp Med. 2000;191:1069. doi: 10.1084/jem.191.6.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liston A, et al. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc Natl. Acad Sci U S. A. 2008;105:11903. doi: 10.1073/pnas.0801506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 38.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 39.Awasthi A, et al. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J. Immunol. 2009;182:5904. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dillies MA, et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013;14:671. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 44.Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38:500. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 45.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids. 1997;25:955. doi: 10.1093/nar/25.5.955. 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research. 2007;35:3100. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Research. 2008;36:281. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Research. 2001;29:41. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 1999;27:29. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 52.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 53.Tian W, Arakaki AK, Skolnick J. EFICAz: a comprehensive approach for accurate genome-scale enzyme function inference. Nucleic Acids Research. 2004;32:6226. doi: 10.1093/nar/gkh956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 55.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 56.Wapinski I, Pfeffer A, Friedman N, Regev A. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics. 2007;23:549. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- 57.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 58.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.