Summary
The transporter of phosphatidylcholine Mdr2/MDR3 not only plays an essential role for bile formation but also is involved in the maintenance of lipid homeostasis. Deficiency of Mdr2 leads to accumulation of ROS, cell transformation and susceptibility to intestinal carcinogenesis.
Abstract
Multidrug resistance 2 (Mdr2), also called adenosine triphosphate-binding cassette B4 (ABCB4), is the transporter of phosphatidylcholine (PC) at the canalicular membrane of mouse hepatocytes, which plays an essential role for bile formation. Mutations in human homologue MDR3 are associated with several liver diseases. Knockout of Mdr2 results in hepatic inflammation, liver fibrosis and hepatocellular carcinoma (HCC). Whereas the pathogenesis in Mdr2 −/− mice has been largely attributed to the toxicity of bile acids due to the absence of PC in the bile, the question of whether Mdr2 deficiency per se perturbs biological functions in the cell has been poorly addressed. As Mdr2 is expressed in many cell types, we used mouse embryonic fibroblasts (MEF) derived from Mdr2 −/− embryos to show that deficiency of Mdr2 increases reactive oxygen species accumulation, lipid peroxidation and DNA damage. We found that Mdr2 −/− MEFs undergo spontaneous transformation and that Mdr2 −/− mice are more susceptible to chemical carcinogen-induced intestinal tumorigenesis. Microarray analysis in Mdr2−/− MEFs and cap analysis of gene expression in Mdr2 −/− HCCs revealed extensively deregulated genes involved in oxidation reduction, fatty acid metabolism and lipid biosynthesis. Our findings imply a close link between Mdr2 −/−-associated tumorigenesis and perturbation of these biological processes and suggest potential extrahepatic functions of Mdr2/MDR3.
Introduction
Multidrug resistance P-glycoproteins, Mdr2 in mouse and MDR3 in human, are the choline-containing phospholipid phosphatidylcholine (PC) transporters that belong to the subfamily B4 of ATP-binding cassette transporters (ABCB4) (1). In the liver, Mdr2/MDR3 are involved in the translocation of PC from the inner leaflet to the outer leaflet of the canalicular membrane of hepatocytes for direct extraction by bile acids (BA). BA and PC are essential components in the bile that form BA micelles to reduce the toxic detergent activity of BA for hepatocytes and cholangiocytes and to establish physiological bile flow. Mutations in the ABCB4 gene in human result in deficiency of PC in the bile, causing diseases including progressive familial intrahepatic cholestasis type 3 (PFIC3), intrahepatic cholestasis of pregnancy and low-phospholipid-associated cholelithiasis (2). A constitutive Mdr2 knockout mouse has been generated, which successively develops liver inflammation, hepatic fibrosis and hepatocellular carcinoma (HCC) at late ages (3). The pathogenesis in Mdr2 −/− mice is commonly considered to be caused by toxic BA in the biliary canaliculus, which damages hepatocytes and cholangiocytes and provokes liver inflammation. However, the contribution of ABCB4 deficiency to the development of the pathologies has been largely unknown.
The ABC transporters are integral membrane proteins. The plasma membrane displays an asymmetric arrangement of the various phospholipids, with PC and sphingomyelin enriched in the exoplasmic leaflet and the aminophospholipids phosphatidylserine and phosphatidylethanolamine in the cytosolic leaflet (4). MDR3 can influence lipid components at the plasma membrane, as it has been shown that exogeneous MDR3 expression in HEK293 cells increased sphingomyelin and cholesterol enrichment in lipid rafts (5). Moreover, expression of MDR3 alters lipid composition on the envelop of HIV vector, thus reducing viral infectivity (6). MDR3-mediated transport of PC at the cell membrane results in the formation of lipid vesicles, which induces cholesterol diffusion from cell membrane (7). These observations suggest that Mdr2/MDR3 are not only the PC transporters for bile formation, but also play a role in the maintenance of lipid homeostasis. Indeed, a previous report showed association of alterations in lipid metabolism and liver pathogenesis in Mdr2 −/− mice (8).
Although hepatocytes are the major cell type in which Mdr2/MDR3 are expressed, Mdr2/MDR3 mRNAs have been detected in many tissues such as adrenal glands, muscle, tonsil and spleen and various cell types including T and B lymphocytes, epithelial cells and fibroblasts (9,10). In this study, we used mouse embryonic fibroblasts (MEF) isolated from Mdr2 −/− embryos to investigate if Mdr2 deficiency impacts on other cell functions than bile formation. We found accumulated reactive oxygen species (ROS), increased lipid peroxidation and DNA damages in Mdr2 −/− MEFs. These findings were corroborated by observations in Mdr2 −/− liver. Our results imply that Mdr2 deficiency can contribute to liver diseases through perturbation of ROS homeostasis and underscore possible extrahepatic functions of Mdr2.
Materials and methods
Animals
Mdr2 −/− (FVB.129P2-Abcb4 tm1Bor/J) and WT FVB/NCrl mice were obtained from The Jackson Laboratory. Mdr2 −/− mice were first mated to WT mice to generate Mdr2 −/+ animals. Intercrossing Mdr2 −/+ animals gave rise to Mdr2 −/− and Mdr2 +/+ littermates. All the experiments were performed with age- and sex-matched Mdr2 −/− and Mdr2 +/+ littermates. For azoxymethane (AOM) and dextran sodium sulfate (DSS)-induced intestinal tumorigenesis, female mice were used. Experimental procedures were approved by Ethic Committee Paris Center and South in accordance with the French government regulations. Mice were bred in a pathogen-free environment at the Institut Pasteur animal facility in accordance with welfare criteria outlined in the ‘Guide for the Care and Use of Laboratory Animals’.
Cell culture
MEF and derivatives, established and extensively characterized by us at the Institut Pasteur during this study, were derived from Mdr2 −/−, Mdr2 −/+ and Mdr2 +/+ littermate embryos on day 13.5 generated by intercrossing Mdr2 −/+ mice. Spontaneously immortalized cell lines were established using 3T3 protocol as described (11). Three independent MEFs preparations were performed. The earliest frozen stocks of all cell lines have been stored in nitrogen containers at the Institut Pasteur. We have propagated cells from frozen stocks of the original vials that were authenticated by qPCR on the expression of Mdr2. The validation of the identity of cells at the earliest stocks or grown >20 passages was tested on regular basis. MEFs were cultured in Dulbecco’s modified Eagle medium (DMEM (1×)-glutaMAXTM-I supplemented with 10% fetal bovine serum, in 95% air and 5% CO2 atmosphere and humidified incubator at 37°C. To measure cell proliferation, MEFs were plated at a density of 2×104 cells into six-well plates in triplicate. Cell numbers were determined using a Z1 Coulter Counter® (Beckman Coulter). To examine resistance to doxorubicin (Dox)-induced apoptosis, 2×103 primary and immortalized MEFs were seeded into 96-well plates and treated with increasing doses of Dox for 2h. Cell viability was measured with MTT assay.
Mdr2 cDNA was synthesized and cloned in pCMV6 by OriGene and stably transfected into Mdr2 −/− MEFs by Lipofectamine 2000 (Life Technologies) followed by G418 selection.
RNA analysis
For qPCR, total RNA was isolated from cells, mouse livers and mouse intestines with RNeasy minikit (Qiagen). 1 μg of RNA was reverse-transcribed using random primers and the cDNA Sythesis Kit (Life Technologies). The mean copy number of each gene from a triplicate determination was normalized to the mean copy number of hypoxanthine-guanine phosphoribosyltransferase. The sequences of primers used for qPCR are: mouse Mdr2 5′-GAGCAAAGTCCAGGTCTGC-3′, 5′-TGCTGCCTAGTTCAAAGTCG-3′; mouse Mdr1a 5′-CAACATCCACCAGTTCATCG-3′, 5′-AATGTGAGGCTGT CTGACGA-3′; mouse Mdr1b 5′-tgggtgcatacaaccagtgt-3′, 5′-tcagccccataaccagaaag-3′. The sequences of hypoxanthine-guanine phosphoribosyltransferase were published previously (12).
For microarray experiment, 50ng of total RNA from MEFs was reverse transcribed and amplified using the Ovation Pico WTA System v2 (Nugen Technologies, Inc. #3302–12), cDNA was biotin labeled before hybridization on Affymetrix Genechip Mouse Gene 2.0 ST Array. Two independent WT and three independent Mdr2 −/− primary or immortalized MEF clones were profiled via microarrays according to the ‘Target Preparation for Affymetrix GeneChip® Eukaryotic Array Analysis’ protocol. Microarray data are available through the Gene Expression Omnibus (GEO) database under accession number GSE72615.
CAGE libraries (for Cap Analysis of Gene Expression) with total RNA extracted from WT or Mdr2 −/− livers were constructed following the protocol reported previously (13). About 5 μg of total RNA was used to synthesize cDNA with random primers. Full-length cDNAs were biotinylated and captured by streptavidin-coated magnetic beads. The cDNAs were released from cap-trapped RNAs, ligated to 5′ linkers including barcode sequences, and digested with EcoP15I. CAGE libraries were sequenced with single-end reads of 50bp on the Illumina HiSeq 2000 platform. CAGE data are available through GEO database under accession number GSE60982.
Microarray and CAGE data analyses
After quality assessment and normalization of the arrays, gene-level expression values were derived from the CEL file probe-level hybridization intensities using the model-based Robust Multichip Average algorithm. The estimated false discovery rate (FDR) of the local pooled error test was calculated using the BH approach. The list of deregulated genes in biological processes analyzed using the Gene Ontology is available upon request.
For CAGE data analysis, differentially expressed genes between tumor versus normal were identified using a Bayesian T-test (14) on the normalized data. For each P value, the Benjamini–Hochberg procedure was used to calculate the FDR to avoid the problem of multiple testing (15). The significant genes were selected using 0.05 as threshold for the FDR.
For each pair of genes we computed the Spearman Correlation Coefficient (SCC) obtaining a correlation coefficient matrix. We estimated the SCC significance for each pair of genes by computing t-statistics of each SCC value and then using Student’s t-test distribution to estimate the P value. To control the number of false positives owing to the multiple hypotheses test problem, we estimated the degrees of freedom of the t-test distribution from the data by fitting the parameters of a Student’s t-location-scale distribution to the t statistics computed for all the genes pairs. We estimated the parameters by minimizing the squared error between the theoretical and the empirical distribution. We then generated a network by connecting two genes if the associated SCC was significant (corrected P < 0.05). From the total network, we extracted the subnetwork of the significant differentially expressed genes and the groups of densely interconnected genes (i.e. communities) were detected via the Walktrap Algorithm by Pons and Latapy (16). This algorithm uses a hierarchical agglomerative method and the distance between two nodes is defined in terms of random walks. The idea is that shorts random walks tend to stay in the same communities. We assessed that 40% of the identified communities are enriched for a specific biological function by Gene Ontology analysis performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Functional Annotation Clustering tool (17). The list of Gene Ontology enrichment for each of the community is available upon request.
Protein analysis
1.5×105 MEFs were seeded into six-well plates and synchronized by incubation in 0.5% fetal bovine serum in DMEM for 72h. Synchronized cells were stimulated by 10% fetal bovine serum in DMEM. Cellular extracts were prepared at indicated times post-stimulation and analyzed by immunoblotting. The following primary antibodies were used: cyclin D1 (sc-450), cyclin A (sc-751), β-actin (Sigma-Aldrich), caspase-3 (Cell Signaling), HP1α (Euromedex), Akt and phospho-Akt (Cell Signaling). Mitotic cells were analyzed by immunofluorescence with antibodies against phosphorylated H2AX on Ser139 and 53BP1 (Cell Signaling). Immunohistochemistry (IHC) with antibodies against phosphorylated H2AX on Ser139 and 53BP1 was performed with 4-month-old liver as described previously (18).
Measurement of ROS and malondialdehyde (MDA)
Intracellular ROS contents were measured by flow cytometry analysis using 2,7-dichlorodihydrofluorescein diacetate dye (H2DCFDA, Life Technologies) as described previously (19). Briefly, MEFs were treated for 1h with 0.1mM H2O2, then allowed to recover in fresh media and harvested after 1 and 6h. Then cells were incubated in pre-warmed Hanks’ balanced salt solution in the presence of 10 µM H2DCFDA and incubated 1h at room temperature. Dye loading buffer was then removed and replaced by pre-warmed complete growth media. Cells were incubated for 30min at 37°C to allow H2DCFDA oxidation generating 2,7-dichlorofluorescein (DCF), a fluorescent compound. ROS accumulation was analyzed using a MACSQuant® (Miltenyi Biotec) flow cytometer and FlowJo software.
The end product of lipid peroxidation MDA was measured using Lipid Peroxidation Assay Kit (Abcam). Cells or liver tissues were homogenized in MDA lysis buffer. Insoluble materials were removed by centrifugation. Supernatant was used for the Colorimetric Assay to quantify MDA amounts according to the Manufacturer’s Protocol.
Cell transformation assays and tumorigenesis
For focus assay, MEFs were seeded at 150, 300 or 600 cells/well in six-well plates. After 2 weeks, cells were stained with crystal violet and colonies more than 2mm in diameter were counted under a Nikon-Eclipse TS100 light microscope. The experiments were repeated twice with two independent MEFs preparations. For soft agar assay, cells were resuspended in 0.3% agarose in DMEM at a density of 5×104 cells/well in six-well plates and plated in triplicate onto solidified 0.6% agarose-containing medium. Cells grew for 4 weeks and were supplemented with fresh media two times a week. Colonies were stained by 0.005% crystal violet and counted using a light microscope. The experiments were repeated twice with different MEFs preparations. For in vivo tumorigenesis assays, 5×106 cells were implanted s.c. into each flank of five 8-week-old female athymic BALB/c mice (Charles River Laboratories). Tumor growth was monitored with a caliper for 3 weeks. No tumor developed in control group during the period of observation of 10 weeks. The experiment was repeated once.
Intestinal tumor induction
Intestinal tumors were induced by AOM and DSS according to a previous report (20). 6- to 8-week-old female mice (Mdr2 +/+ and Mdr2 −/− littermates on FVB background) were injected intraperitoneally with 10mg/kg body weight AOM (Sigma-Aldrich) and exposed seven days later to three cycles of 2.5% of Dextran Sulfate 40 Sodium (A3261, AppliChem, average molecular weight 40000g/mol) administrated in the drinking water for 7 days, followed by 14 days of regular water. Mice were killed after the last cycle. Intestines were removed from mice and fixed as ‘Swiss rolls’ in 4% paraformaldehyde. Tumor scoring and histopathological analysis were described previously (21).
Results
Deficiency of Mdr2 in MEFs increases cell proliferation and resistance to Dox-induced apoptosis
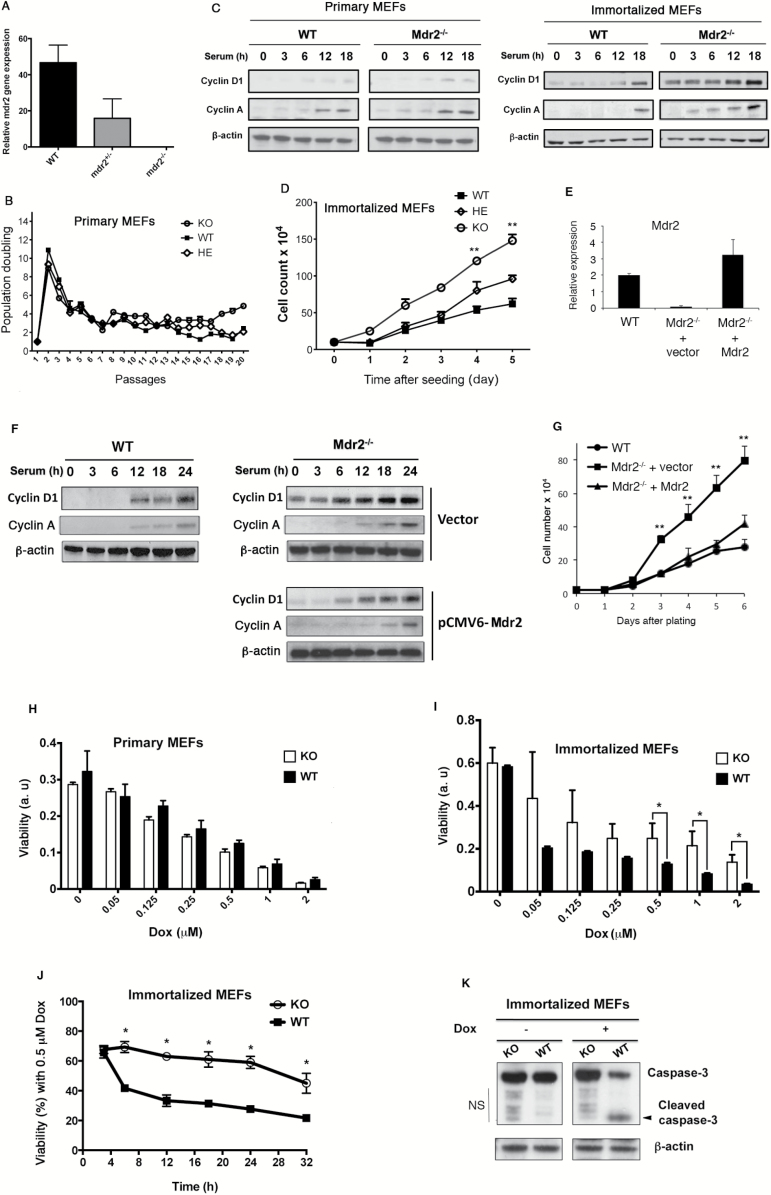
We isolated WT and Mdr2 −/− MEFs derived form Mdr2 +/+ and Mdr2 −/− littermate embryos. After confirming expression of Mdr2 mRNA by qPCR in WT MEFs (Figure 1A), primary WT and Mdr2 −/− cells were subjected to passages using the 3T3 protocol to obtain spontaneously immortalized cell lines. No difference in growth and senescence kinetics was observed between primary Mdr2 +/+, Mdr2 +/− and Mdr2 −/− cells (Figure 1B). When primary WT and Mdr2 -/- cells were subjected to serum starvation for 72h followed by serum addition, expression of the cell cycle regulators cyclin D1 and cyclin A displayed similar intensity and kinetics in response to serum stimulation (Figure 1C, left panel). In sharp contrast, immortalized Mdr2 −/− cells showed distinct proliferation profiles. They grew significantly faster than Mdr2 +/+ MEFs, whereas Mdr2 +/− cells exhibited an intermediate growth rate (Figure 1D). Accordingly, in Mdr2 −/− cells, the kinetics of induction of cylcin D1 and cyclin A was drastically advanced in response to serum stimulation, and the levels of the induction were greatly augmented (see Figure 1C, right panel).
Figure 1.
Loss of Mdr2 increases cell proliferation and resistance to apoptosis. (A) Mdr2 mRNA expression in Mdr2 +/+, Mdr2 −/+ and Mdr2 −/− MEFs at passage 3 by qPCR. (B) Primary Mdr2 +/+, Mdr2 −/+ and Mdr2 −/− cell proliferation on a 3T3 protocol. Data shown are representative of cell proliferation of three independent MEF preparations from littermate embryos. (C) Primary and immortalized WT and Mdr2 −/− cells were synchronized by serum starvation and stimulated by 10% serum. Cell lysates were prepared at the indicated time after stimulation and analyzed by immunoblotting. Similar results were obtained with two other independent clones. (D) Increased cell proliferation in immortalized Mdr2 −/− cells. Equal numbers of spontaneously immortalized WT and Mdr2 −/− MEFs were plated in triplicate. Cell numbers were determined during 5 days. Data presented are the mean values ± standard deviation obtained from three independent cell cultures. The experiments were repeated with two other independent clones. The asterisk indicates significant difference between WT and KO. **P < 0.005, Student’s test. KO: Mdr2 knockout, HE: Mdr2 heterozygote. (E) Ectopic expression of Mdr2 in Mdr2 −/− MEFs. Mdr2 mRNA expression in WT MEFs, Mdr2 −/− MEFs transfected with empty vector and Mdr2 −/− MEFs transfected with vector expressing Mdr2 by qPCR. (F) Reduced cyclin D1 and cyclin A expression in Mdr2 −/− cells after introduction of Mdr2 was detected by immunoblotting with anti-cyclin D1 and anti-cyclin A antibodies. (G) Growth curve of immortalized vector-transfected Mdr2 −/− MEFs, Mdr2-transfected Mdr2 −/− MEFs and WT MEFs. Equal numbers of cells were plated in triplicate at the beginning of the experiments. Cell numbers were determined every day for a total of 6 days. Data presented are the mean values ± standard deviation obtained from three independent cell cultures. The experiments were repeated with two other independent Mdr2-transfected Mdr2 −/− pools. **P < 0.005, Student’s test. Primary (H) and immortalized (I) WT and Mdr2 −/− MEFs were treated with increasing doses of Dox. Cell viability was analyzed by MTT assay. Data presented are the mean values ± standard deviation obtained from three independent experiments. The experiments were repeated with two other independent preparations of MEFs. a.u.: arbitrary unit. (J) Immortalized MEFs were treated with 0.5 μM Dox for different times. Cell viability was examined at indicated time points by MTT assay. *P < 0.05, Student’s test. KO: Mdr2 knockout cells. (K) Proteins were extracted from immortalized MEFs without treatment or with treatment of 0.5 μM Dox for 12h and analyzed by immunoblotting with anti-caspase-3 antibody. NS: non-specific.
To confirm that Mdr2 deficiency was directly involved in the phenotype, we re-expressed Mdr2 in primary Mdr2 −/− MEFs at early passages (Figure 1E) and assessed the effect of exogenous Mdr2 expression on cyclin D1, cylcin A and cell growth. Continuous passage of Mdr2-restored MEFs readily gave rise to established 3T3 cell lines. Restoration of Mdr2 in Mdr2 −/− MEFs delayed cyclin D1 and cyclin A induction and decreased their levels of expression (Figure 1F). In addition, exogenous expression of Mdr2 significantly decelerated cell growth (Figure 1G).
As deregulation of cell proliferation is frequently associated with deregulation of cell death, we next examined the effects of Mdr2 deficiency on apoptosis. Treatment with Dox, an apoptosis inducer with a broad-spectrum antibiotic activity, caused a decrease of cell viability in both primary WT and Mdr2 −/− MEFs in a dose-dependent manner with similar kinetics (Figure 1H). In contrast, immortalized Mdr2 −/− MEFs were more resistant to Dox-induced apoptosis, showing significant higher viability than WT counterparts when treated with 0.5 μM Dox or higher doses (Figure 1I and J). While analysis by immunoblotting readily revealed cleaved caspase-3 in WT cells treated with Dox, indicative of the activation of this critical executioner of apoptosis, the cleaved form of caspase-3 was barely detectable in Mdr2 −/− MEFs (Figure 1K). As a previous report showed that moderate increase in mRNA levels of Mdr1a and Mdr1b in murine breast cancer can cause Dox resistance (22), we assessed expression of Mdr1a and Mdr1b in immortalized Mdr2 −/− MEFs by qPCR. There was a trend but not significant increase in Mdr1a and Mdr1b expression in Mdr2 −/− cells (Supplementary Figure 1, available at Carcinogenesis Online). Together, these results demonstrate that Mdr2 deficiency can increase cell proliferation as well as resistance to Dox-induced apoptosis.
Accumulation of ROS and increased lipid peroxidation in immortalized Mdr2 −/− MEFS
To gain insight into the impact of Mdr2 deficiency on gene expression at the whole genome level, we performed cDNA microarray of primary and immortalized Mdr2 −/− and WT cells to identify genes differentially expressed. As expected, primary Mdr2 −/− and WT cells exhibited undistinguishable expression profile. In contrast, 827 probe sets were revealed differentially expressed between WT and immortalized Mdr2 −/− cells with FDR < 0.1% as the criterion for differential expression. Pathway analysis using the Gene Ontology evidenced deregulation of genes involved in different biological processes. These include regulation of cell proliferation, positive regulation of biological processes, lipid metabolic process, regulation of cell differentiation, defense response, lipid biosynthetic process, oxidation reduction and fatty acid metabolism (Supplementary Tables 1 and 2, available at Carcinogenesis Online), which have been previously identified in Mdr2 −/− liver (8,23,24). Furthermore, in agreement with Katzenellenbogen et al. (23), deregulation of expression of Jun, MCM4, MCM7 and Pparg involved in DNA replication was also observed in Mdr2 −/− MEFs (Supplementary Table 3, available at Carcinogenesis Online).
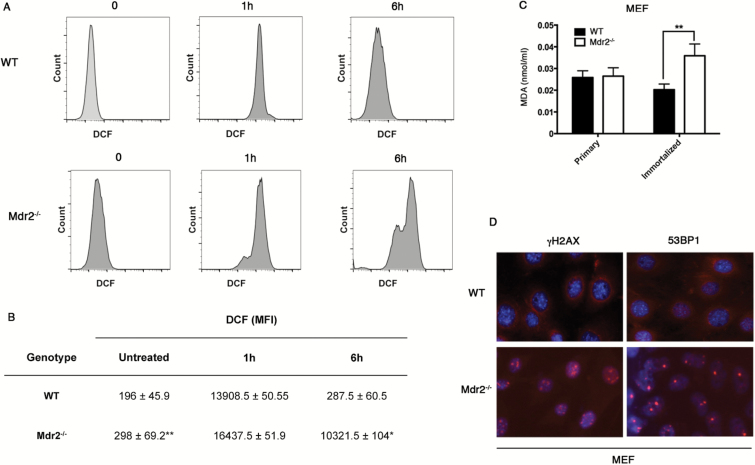
Since ROS acts as critical signaling molecules in cell proliferation and survival, we set up to measure ROS production in immortalized MEFs. Cells were incubated with the probe H2DCFDA whose intracellular oxidation into the green fluorescein derivative 2,7-dichlorofluorescein (DCF) served as indicator of ROS formation (25). Analysis of the percentage of ROS-positive cells was carried out by flow cytometry. As shown in Figure 2A and B, immortalized Mdr2 −/− cells displayed significantly higher levels of ROS than WT cells. We then treated cells with H2O2 for 1h and measured ROS-positive cells 1 and 6h post-treatment. Both WT and Mdr2 −/− cells showed high levels in ROS contents at 1h point because of H2O2 addition (Figure 2A and B). Whereas WT cells almost eliminated all exogenous H2O2 6h after treatment, large amount of exogenous H2O2 still remained in Mdr2 −/− cells (Figure 2A and B), suggesting dysfunction of the antioxidant systems (26). These findings are correlated with microarray data showing deregulation of gene expression in biological process of oxidative reduction (see Supplementary Table 2, available at Carcinogenesis Online).
Figure 2.
Reactive oxygen species (ROS) accumulation, increased lipid peroxidation and DNA damage in immortalized Mdr2 −/− MEFs. (A, B) Cells were treated for 1h with 0.1mM H2O2. ROS accumulation was analyzed before treatment or 1 and 6h after treatment by flow cytometry. MFI: mean fluorescence intensity. *P < 0.05, **P < 0.005, Student’s test. (C) Primary and immortalized MEF extract was quantified for malondialdehyde (MDA) using Colorimetric Assay. **P < 0.005, Student’s test. (D) Analysis by immunofluorescence of immortalized WT and Mdr2 −/− MEFs with antibodies against γH2AX and 53BP1.
Oxidative stress occurs when the production of ROS exceeds their catabolism through either an increase in ROS levels or a decrease in the cellular antioxidant capacity. Oxidative stress results in direct or indirect ROS-mediated damage of lipids, proteins and nucleic acids. ROS can react with polyunsaturated fatty acids of lipid membranes and induces lipid peroxidation (27). We thus measured the end product of lipid peroxidation MDA using colorimetric assay. As shown in Figure 2C, while no significant difference in the production of MDA was detected between primary WT and Mdr2 −/− cells, immortalized Mdr2 −/− cells produced significantly higher levels of MDA than WT counterparts, attesting increased lipid peroxidation. ROS can cause double-strand DNA breaks, damaging DNA. The DNA damage-induced foci, marked by the histone variant H2AX phosphorylated on Ser139 (γH2AX) and the recruitment of chromatin-associated genome caretakers such as p53-binding protein 1 (53BP1), represent sites of DNA breaks. Analysis by immunofluorescence detected numerous positive foci of γH2AX and 53BP1 in the nucleus of Mdr2 −/− cells in contrast to few foci in WT cells (Figure 2D), indicative of DNA damage in Mdr2 −/− cells.
Increased lipid peroxidation and DNA damage in Mdr2 −/− liver
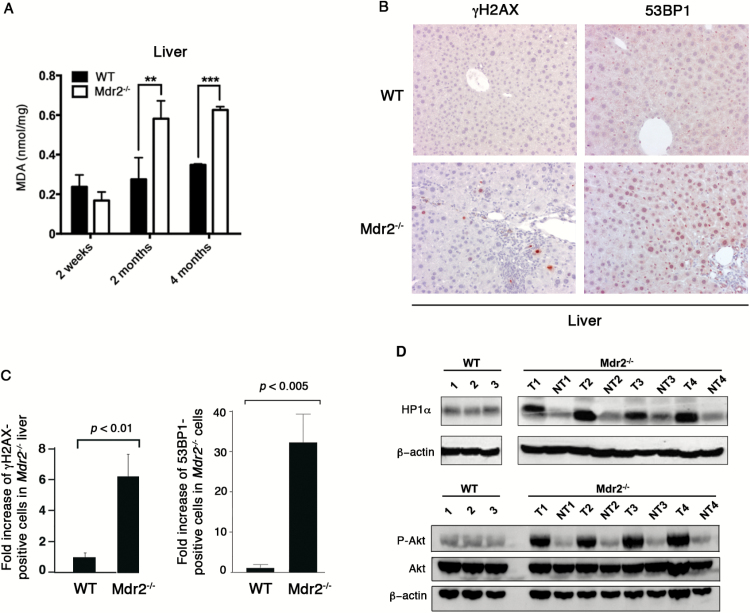
We next wanted to investigate how Mdr2 deficiency affected lipid peroxidation in vivo. To address it, liver tissues were dissected from Mdr2 −/− and WT mice at different ages and were subjected to quantification of MDA. As shown in Figure 3A, no difference was observed in MDA production between Mdr2 −/− and WT livers from 2-week-old mice in which inflammation is not found yet (3). However, at 2-month-old and 4-month-old when inflammation is present, Mdr2 −/− livers generated significant higher levels of MDA than WT counterparts (Figure 3A). Moreover, analysis by IHC in the 4-month-old liver revealed six-fold increase of γH2AX-positive cells and 30-fold increase of 53BP1-positive cells in Mdr2 −/− mice, showing significant difference compared to the WT liver (Figure 3B and C). This result is in agreement with previous report (28). DNA damage leads either to cell death or aberrant cell proliferation, which is one of the characteristics of cancer cells. In Mdr2 −/− HCCs, we found dramatic increases in heterochromatin protein 1 isoform α (HP1α) abundance and Akt phosphorylation (Figure 3D). HP1α is overexpressed in many types of cancer and its high level expression is associated with rapid cell proliferation (29). Activation of the PI3K/Akt pathway results in a disturbance of control of cell growth and survival, which contributes to a competitive growth advantage.
Figure 3.
Increased lipid peroxidation and DNA damage in the Mdr2 −/− liver. (A) Liver extract was prepared from 2-week-old, 2-month-old and 4-month-old mice and quantified for malondialdehyde (MDA) by Colorimetric Assay. Three mice were used for each genotype. The data are presented as the MDA quantity per mg of liver weight. **P < 0.005, ***P < 0.0005, Student’s test. (B) Immunostaining with antibodies against γH2AX and 53BP1 on liver sections of WT and Mdr2 −/− mice (n = 4). (C) Fold increase of γH2AX- and 53BP1-positive cells in Mdr2 −/− mice over WT mice. (D) Immunoblotting analysis of expression of HP1α and Akt and phosphorylation of Akt in three WT livers and paired tumorous (T) and non-tumorous (NT) liver tissues from four Mdr2 −/− mice.
To gain insight into alterations of gene expression at the whole-genome level in Mdr2 −/− liver, CAGE was performed using liver samples from 6 WT mice and 31 Mdr2 −/− mice (30). We analyzed CAGE data using a recently developed network-based approach (31). Briefly, we first identified differentially expressed genes (DEGs) between WT and Mdr2 −/− mice for a total of 5844 genes with FDR < 1% and 8653 with FDR < 5%. Due to the large number of significant genes, we used a co-expression network approach to identify significantly dysregulated pathways in Mdr2 −/− mice. To this end, we first generated a coexpression network from all the 37 WT and Mdr2 −/− CAGE-seq expression profiles where genes are connected to each other if they are significantly coexpressed across the samples according to a correlation measure. We then extracted from the subnetwork containing only the significant DEGs and identified ‘communities’ of densely interconnected genes. Finally, we performed Gene Ontology enrichment analysis for each of the community. The top enriched Biological Process terms include ‘oxidation reduction’, ‘lipid biosynthetic process’ and ‘fatty acid metabolic process’ (Table 1), which is in accordance with previous report showing alterations of gene expression in lipid metabolism in the liver of Mdr2 −/− mice (8,24). This network-based approach has the advantage to identify enriched biological processes with differentially expressed connected genes, even though the genes involved in the same biological process are not necessarily the same when they are from different communities (Supplementary File 1, available at Carcinogenesis Online).
Table 1.
Representation of communities of the Mdr2 −/− CAGE
| ID | N1 | N2 | Gene Ontology | Biological process |
|---|---|---|---|---|
| 1 | 259 | 19 | GO:0055114 | Oxidation reduction |
| 1 | 259 | 12 | GO:0008610 | Lipid biosynthetic process |
| 1 | 259 | 13 | GO:0006631 | Fatty acid metabolic process |
| 1 | 259 | 4 | GO:0006695 | Cholesterol biosynthesis process |
| 2 | 122 | 17 | GO:0055114 | Oxidation reduction |
| 2 | 122 | 7 | GO:0008202 | Steroid metabolic process |
| 3 | 200 | 32 | GO:0055114 | Oxidation reduction |
| 3 | 200 | 7 | GO:0009309 | Amine biosynthetic process |
| 4 | 127 | 11 | GO:0010604 | Positive regulation of macromolecule metabolic process |
| 5 | 27 | 7 | GO:0051301 | Cell division |
| 5 | 27 | 7 | GO:0007049 | Cell cycle |
ID: the community identification number, N1: total number of genes in each community, N2: number of genes involved in the specified biological process, GO: Gene Ontology number.
Collectively, ablation of Mdr2 results in increased levels of lipid peroxidation and DNA damage in the liver.
Spontaneous transformation of Mdr2 -/- MEFs
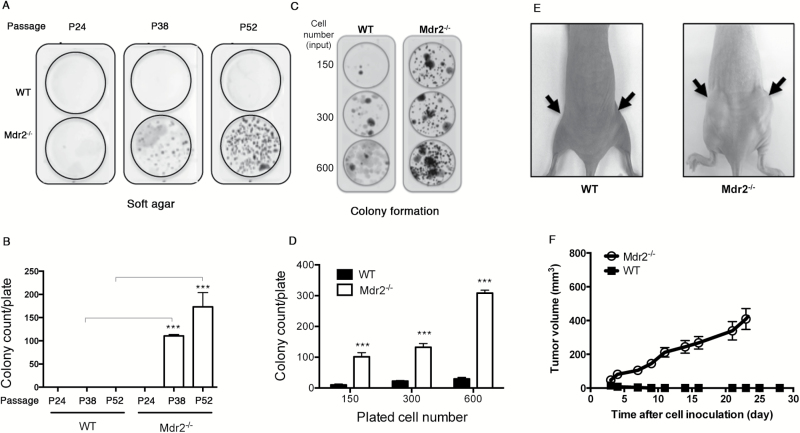
To investigate direct impact of Mdr2 deficiency on cell transformation, we tested anchorage-independent growth of Mdr2 −/− MEFs at different passages on 3T3 protocol by soft agar assays. As shown in Figure 4A and B, no colony developed in neither WT nor Mdr2 −/− cells at passage 24, but strikingly, numerous colonies were detected at passage 38 in Mdr2 −/− cells but not in WT cells. At passage 52, colonies were even more abundant in Mdr2 −/− cells (Figure 4A). Quantification showed that about 100 and 170 colonies were formed in a plate in Mdr2 −/− cells at P38 and P52, respectively (Figure 4B). Next, cells at passage 38 were analyzed for the loss of contact inhibition by focus formation assays. Loss of Mdr2 markedly augmented colony formation (Figure 4C). The number of colonies per plate developed about 100 times more in Mdr2 −/− cells than WT cells (Figure 4D). When injected s.c. into BALB/c nude mice, transformed Mdr2 −/− MEFs induced tumor formation in 10/10 mice, while no tumor developed in 10/10 mice injected with WT MEFs (Figure 4E and F). Together, these results indicate that deficiency of Mdr2 drives cell transformation.
Figure 4.
Spontaneous transformation in Mdr2 −/− MEFs. (A, B) Soft agar assay of WT and Mdr2 −/− MEFs at different passages. Data presented are the mean values ± standard deviation obtained from three independent cell cultures. The experiments were repeated with two other clones from independent MEF preparations. The asterisk indicates significant difference between Mdr2 −/− and WT cells at the same passage. ***P < 0.0005, Student’s test. (C, D) Focus formation assay of WT and Mdr2 −/− MEFs at passage 38. Data presented are the mean values ± standard deviation obtained from three independent cell cultures. The experiments were repeated with two other clones from independent MEF preparations. The asterisk indicates significant difference between Mdr2 −/− and WT cells when seeded with the same number of cells. ***P < 0.0005, Student’s test. (E, F) Tumorigenicity of transformed Mdr2 −/− MEFs in athymic BALB/c mice. Data presented are the mean values ± standard deviation obtained from two flanks of five mice. The experiments were repeated once.
Deletion of Mdr2 increases intestinal polyposis in a mouse model of colitis-associated cancer
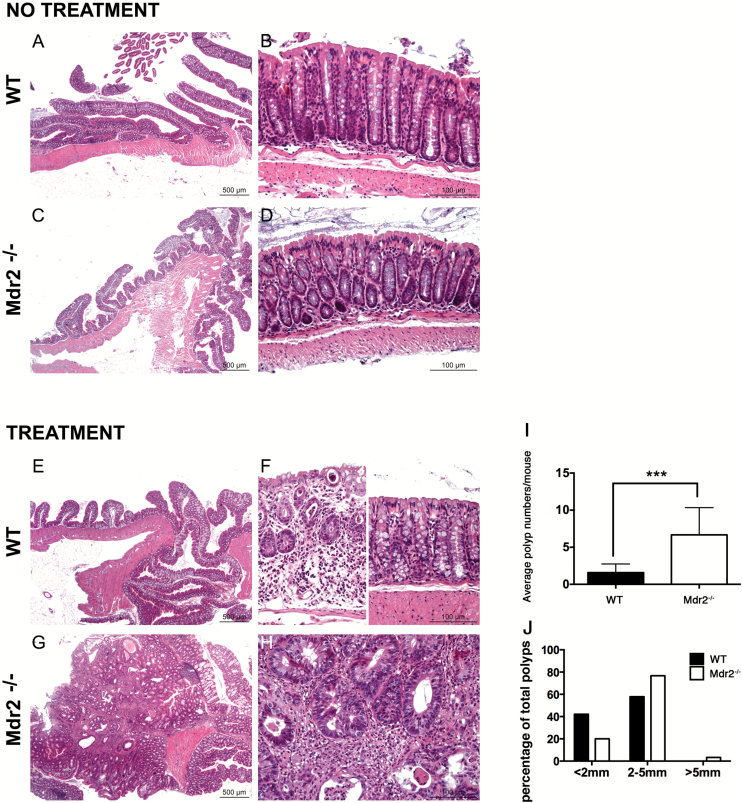
To determine the in vivo effects of Mdr2 deficiency on cell transformation in other tissues than the liver, we took advantage of the mouse model of AOM/DSS-induced intestinal tumorigenesis to investigate how Mdr2 affects the phenotype associated with AOM/DSS-induced cancer (32). The protocol of AOM/DSS administration was applied to Mdr2 −/− mice and their Mdr2 +/+ littermate control mice. Expression of Mdr2 mRNA in WT intestine was detectable by qPCR (data not shown). Because of enterohepatic circulation of bile, we first examined if the lack of phospholipids in the bile had impact on the intestine in Mdr2 −/− mice. As shown in Figure 5A–D, deletion of Mdr2 did not alter crypt-villus architecture of the small intestine. Regardless of genotype, all mice treated with AOM plus DSS developed tumors, which were mostly adenomas with high-grade dysplasia (Figure 5E and H). However, we observed a seven-fold increase in tumor incidence in the Mdr2 −/− group (Figure 5I). We measured the sizes of polyps and showed that tumor sizes in Mdr2 −/− group were larger than in the Mdr2 +/+ group (Figure 5J). Precisely, there were proportionally more tumors in Mdr2 −/− mice with sizes superior to 5mm than in WT mice (Figure 5J). Together, these results suggest that Mdr2 is important in both tumor initiation and tumor growth.
Figure 5.
Deletion of Mdr2 increases intestinal tumor incidence. (A–H) Histological analysis of untreated and AOM/DSS treated intestines in WT and Mdr2 −/− mice. (I) Total number of intestinal polyps was counted in 12 WT and 12 Mdr2 −/− mice after AOM/DSS treatment. Compared to WT littermates, significant increase in poly number was observed in Mdr2 −/− mice. ***P < 0.0005, Student’s test. (J) Mdr2 deficiency enhances tumor growth. The percentage of total number of polyps in 12 WT and 12 Mdr2 −/− mice with size inferior to 2mm, between 2 and 5mm and superior to 5mm is presented.
Discussion
Ablation of Mdr2 abolishes the PC transporter function causing bile defect-associated diseases including liver inflammation, hepatic fibrosis and HCC. It could also produce other pathogenic properties that contribute to cell proliferation, survival and tumorigenesis. Here, we explored the phenotypic effects of Mdr2 deficiency in MEFs and in chemical carcinogen-induced intestinal tumor and showed that Mdr2 is necessary to maintain ROS homeostasis. Mdr2 −/− MEFs produce high levels of ROS leading to increased lipid peroxidation and DNA damage. These cells display enhanced proliferation, resistance to Dox-induced apoptosis and spontaneously undergo transformation at late passages. The phenotypes in MEFs are correlated with the findings in Mdr2 −/− mice in which were observed enhanced levels of lipid peroxidation and DNA damage in the liver and increased susceptibility of carcinogen-induced tumor development in the intestine. These results contribute to the understanding of Mdr2 actions in pathogenesis and raise questions of potential extrahepatic behaviors of MDR3 mutations in human.
Most of our understanding of how deletion of Mdr2 mediates its pathogenic activity has been derived from exploring the consequences of the bile defect in the liver. Whereas hepatic inflammation resulted from hepatocyte and cholangiocyte injury by toxic BAs in Mdr2 −/− mice promotes production of ROS, oxidative DNA damage and reduced DNA repair (28), the contribution of Mdr2 deficiency to pathogenesis is poorly understood. Here, we show that deletion of Mdr2 enhances ROS production in MEFs, fostering lipid peroxidation and DNA damage. Questions remain as to how the transporter of PC acts mechanistically in the regulation of ROS. Previous report showed that alterations in lipid metabolism and disruption of lipid homeostasis contribute to the pathogenesis and progression of cholestatic liver disease in Mdr2 −/− mice (8). We hypothesize that disruption of lipid homeostasis owing to Mdr2 deletion may represent the major event with consequences throughout pathways involved in ROS production and catabolism and ROS-mediated damage repair (26). This view is supported by gene expression profiling in MEFs by microarray and in Mdr2 −/− HCCs by CAGE that revealed significant expression changes in genes involved in oxidative reduction, lipid biosynthesis and fatty acid metabolism. Furthermore, our preliminary results suggest increased levels of lipids in Mdr2 −/− cells even though we have not been able to identify which specific species of lipids were changed.
It has long been known that lipid can serve as second message for signal transductions and mediate an array of cellular response including inflammation, cell proliferation and apoptosis (33). Recently, specific species of PC have been identified as ligands for the nuclear receptors peroxisome proliferator-activated receptor a (PPARa) and the liver receptor homolog 1 (LRH-1) (34,35), which not only are the key modulators of metabolism of glucose, lipid and BAs but also play important roles in carcinogenesis. Signaling through PPARa contributes to the upregulation of peroxisomal enzymes, such as acyl CoA oxidase, that produce the ROS H2O2 increasing oxidative stress. Besides, PPARa activation also leads to elevated hepatocellular proliferation and inhibition of apoptosis (36). Double knockout of Mdr2 and PPARa protected animals from chronic liver injury, resulting in reduced bile duct proliferation and less fibrosis, whereas administration of PPARa activator fenofibrate in Mdr2 −/− mice not only aggravated liver injury and fibrosis but also increased inflammation (8), demonstrating a close link between PPARa-mediated signaling and the pathogenesis in Mdr2 −/− mice. LRH-1 is a driving factor in development and progression of cancer, especially colon, pancreatic and breast cancers, by promoting cell growth and proliferation (37–39). Thus nuclear receptor activation by PC ligands may constitute a mechanism for lipid second message signaling functions in carcinogenesis. Moreover, endonuclear PC biosynthesis supports cell proliferation (40). Further study is needed to investigate the quantity, composition and cellular localization of PC species in Mdr2 −/− cells and their impact on signal transductions.
Given the predominant and restricted functions of Mdr2 in bile formation, most of the pathologic properties of Mdr2 deficiency are thought to reflect toxic effects of BAs in the bile devoid of phospholipids. Indeed, previous work on the role of partial hepatectomy in HCC initiation and progression in Mdr2 −/− mice suggests an accumulation of DNA double-strand breaks in the hepatocytes of the Mdr2 −/− mice consequential to chronic inflammation (28). Though inflammation is a crucial force to promote increase in ROS accumulation in Mdr2 −/− liver, we identified additional effects of Mdr2 deficiency that also contribute to oxidative stress. However, the fact that only immortalized but not primary Mdr2 −/− MEFs showed increased proliferation, ROS accumulation and alterations in gene expression profile suggests that a ‘second hit’ event is necessary to sustain Mdr2 deficiency-mediated oxidative stress. Our data provide evidence of broader functional roles of Mdr2, which is in line with previous report showing that Mdr2 is a modulator of glucose homeostasis (41). Although we cannot exclude effects of abnormal bile composition in the increased propensity of Mdr2 −/− mice to AOM/DSS-induced intestinal tumorigenesis, AOM treatment alone showed development of few tumors in the intestine of Mdr2 −/− mice similar to WT animals (data not shown), suggesting that bile components may not be a determinant factor of intestinal tumor development in this setting.
The identification of broader functions of the PC transporters Mdr2/MDR3 may have impact on the appreciation of human diseases associated with MDR3 mutations. Although extrahepatic phenotypes have never been reported in patients harboring homozygous mutations in the MDR3 gene (42), analysis of potential effects of MDR3 defects in other tissues than the liver may provide full insight into disease pathogenesis. In particular, the question of whether the functions of immune cells are affected by MDR3 mutations may be worth to be addressed, as these cells express the ABCB4 gene (10) and are key factors in mediating inflammatory response in the diseased liver caused by MDR3 mutations.
Supplementary material
Supplementary Tables 1–3 and Figure 1 can be found at http://carcin.oxfordjournals.org/
Funding
European Union’s Seventh Framework Program (FP7) under grant agreement No. 259743 (MODHEP consortium) to the Institut Pasteur (A.T., F.L., Y.W.), RIKEN Center (K.H., A.M.S., P.C.) and Telethon Institute of Genetics and Medicine (D.B.).
Supplementary Material
Acknowledgements
We are grateful to Agnese Collino and Paola Nicoli in Gioacchino Natoli’s laboratory at the European Institute of Oncology (IEO), Milan, for sample preparation for CAGE experiments, Estelle Mignot and Laura Garnier for technical help, and Justine Bertrand-Michel at MetaToul-Lipidomique Inserm U1048, Toulouse, for lipidomic analysis. We thank Marie-Annick Buendia at Inserm U785, Villejuif, Anne-Marie Cassard-Doulcier at Inserm UMR_S996, Clamart, Edith Heard, Jean-Pierre Quivy and Geneviève Almouzni at Institut Curie, Paris, France, for insightful discussion.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AOM
azoxymethane
- BA
bile acid
- CAGE
Cap Analysis of Gene Expression
- DSS
dextran sodium sulfate
- Dox
doxorubicin
- FDR
false discovery rate
- HCC
hepatocellular carcinoma
- MDA
malondialdehyde
- MEF
mouse embryonic fibroblasts
- Mdr2
multidrug resistance 2
- PC
phosphatidylcholine
- PPARa
peroxisome proliferator-activated receptor a
- ROS
reactive oxygen species
- SCC
Spearman Correlation Coefficient
References
- 1. Borst P., et al. (2002) Mammalian ABC transporters in health and disease. Annu. Rev. Biochem., 71, 537–592. [DOI] [PubMed] [Google Scholar]
- 2. Jacquemin E., et al. (2001) The wide spectrum of multidrug resistance 3 deficiency: from neonatal cholestasis to cirrhosis of adulthood. Gastroenterology, 120, 1448–1458. [DOI] [PubMed] [Google Scholar]
- 3. Smit J.J., et al. (1993) Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell, 75, 451–462. [DOI] [PubMed] [Google Scholar]
- 4. Simons K., et al. (1997) Functional rafts in cell membranes. Nature, 387, 569–572. [DOI] [PubMed] [Google Scholar]
- 5. Morita S.Y., et al. (2013) Bile salt-stimulated phospholipid efflux mediated by ABCB4 localized in nonraft membranes. J. Lipid Res., 54, 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Til N.P., et al. (2008) Alteration of viral lipid composition by expression of the phospholipid floppase ABCB4 reduces HIV vector infectivity. Retrovirology, 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morita S.Y., et al. (2007) Bile salt-dependent efflux of cellular phospholipids mediated by ATP binding cassette protein B4. Hepatology, 46, 188–199. [DOI] [PubMed] [Google Scholar]
- 8. Moustafa T., et al. (2012) Alterations in lipid metabolism mediate inflammation, fibrosis, and proliferation in a mouse model of chronic cholestatic liver injury. Gastroenterology, 142, 140–151.e12. [DOI] [PubMed] [Google Scholar]
- 9. Smit J.J., et al. (1994) Tissue distribution of the human MDR3 P-glycoprotein. Lab. Invest., 71, 638–649. [PubMed] [Google Scholar]
- 10. Chin J.E., et al. (1989) Structure and expression of the human MDR (P-glycoprotein) gene family. Mol. Cell. Biol., 9, 3808–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Labalette C., et al. (2008) The LIM-only protein FHL2 regulates cyclin D1 expression and cell proliferation. J. Biol. Chem., 283, 15201–15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dahan J., et al. (2013) LIM-only protein FHL2 activates NF-κB signaling in the control of liver regeneration and hepatocarcinogenesis. Mol. Cell. Biol., 33, 3299–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takahashi H., et al. (2012) 5’ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc., 7, 542–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baldi P., et al. (2001) A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics, 17, 509–519. [DOI] [PubMed] [Google Scholar]
- 15. Benjamini Y., et al. (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B (Methodological), 57, 289–300. [Google Scholar]
- 16. Pons P., et al. (2005) Computing communities in large networks using random walks. http://arxivorg/abs/physics/0512106.
- 17. Huang da W., et al. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 18. Nouët Y., et al. (2012) The four and a half LIM-only protein 2 regulates liver homeostasis and contributes to carcinogenesis. J. Hepatol., 57, 1029–1036. [DOI] [PubMed] [Google Scholar]
- 19. Tebbi A., et al. (2011) TAp73 induction by nitric oxide: regulation by checkpoint kinase 1 (CHK1) and protection against apoptosis. J. Biol. Chem., 286, 7873–7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neufert C., et al. (2007) An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc., 2, 1998–2004. [DOI] [PubMed] [Google Scholar]
- 21. Labalette C., et al. (2010) Deficiency of the LIM-only protein FHL2 reduces intestinal tumorigenesis in Apc mutant mice. PLoS One, 5, e10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pajic M., et al. (2009) Moderate increase in Mdr1a/1b expression causes in vivo resistance to doxorubicin in a mouse model for hereditary breast cancer. Cancer Res., 69, 6396–6404. [DOI] [PubMed] [Google Scholar]
- 23. Katzenellenbogen M., et al. (2006) Multiple adaptive mechanisms to chronic liver disease revealed at early stages of liver carcinogenesis in the Mdr2-knockout mice. Cancer Res., 66, 4001–4010. [DOI] [PubMed] [Google Scholar]
- 24. Potikha T., et al. (2013) Interstrain differences in chronic hepatitis and tumor development in a murine model of inflammation-mediated hepatocarcinogenesis. Hepatology, 58, 192–204. [DOI] [PubMed] [Google Scholar]
- 25. LeBel C.P., et al. (1992) Evaluation of the probe 2’,7’-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress. Chem. Res. Toxicol., 5, 227–231. [DOI] [PubMed] [Google Scholar]
- 26. Nathan C., et al. (2013) Beyond oxidative stress: an immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol., 13, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janero D.R. (1990) Malondialdehyde and thiobarbituric acid-reactivity as diagnostic indices of lipid peroxidation and peroxidative tissue injury. Free Radic. Biol. Med., 9, 515–540. [DOI] [PubMed] [Google Scholar]
- 28. Barash H., et al. (2010) Accelerated carcinogenesis following liver regeneration is associated with chronic inflammation-induced double-strand DNA breaks. Proc. Natl. Acad. Sci. USA, 107, 2207–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De Koning L., et al. (2009) Heterochromatin protein 1alpha: a hallmark of cell proliferation relevant to clinical oncology. EMBO Mol. Med., 1, 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashimoto, K. et al. (2015) CAGE profiling of ncRNAs in hepatocellular carcinoma reveals a strong activation of retroviral LTR promoters in virus-induced tumors. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 31. Amato R., et al. (2014) A network-based approach to dissect the cilia/centrosome complex interactome. BMC Genomics, 15, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Greten F.R., et al. (2004) IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell, 118, 285–296. [DOI] [PubMed] [Google Scholar]
- 33. Spiegel S., et al. (1996) Signal transduction through lipid second messengers. Curr. Opin. Cell Biol., 8, 159–167. [DOI] [PubMed] [Google Scholar]
- 34. Chakravarthy M.V., et al. (2009) Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell, 138, 476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee J.M., et al. (2011) A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature, 474, 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tachibana K., et al. (2008) The Role of PPARs in Cancer. PPAR Res., 2008, 102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee Y.K., et al. (2008) Liver receptor homolog-1, an emerging metabolic modulator. Front. Biosci., 13, 5950–5958. [DOI] [PubMed] [Google Scholar]
- 38. Chand A.L., et al. (2012) The orphan nuclear receptor LRH-1 and ERα activate GREB1 expression to induce breast cancer cell proliferation. PLoS One, 7, e31593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benod C., et al. (2011) Nuclear receptor liver receptor homologue 1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proc. Natl. Acad. Sci. USA, 108, 16927–16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hunt A.N. (2006) Completing the cycles; the dynamics of endonuclear lipidomics. Biochim. Biophys. Acta, 1761, 577–587. [DOI] [PubMed] [Google Scholar]
- 41. Hochrath K., et al. (2012) The hepatic phosphatidylcholine transporter ABCB4 as modulator of glucose homeostasis. FASEB J., 26, 5081–5091. [DOI] [PubMed] [Google Scholar]
- 42. Jacquemin E. (2012) Progressive familial intrahepatic cholestasis. Clin. Res. Hepatol. Gastroenterol., 36 (Suppl 1), S26–S35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.