Abstract
It has been estimated that the sister plant pathogenic fungal species Heterobasidion irregulare and Heterobasidion annosum may have been allopatrically isolated for 34–41 Myr. They are now sympatric due to the introduction of the first species from North America into Italy, where they freely hybridize. We used a comparative genomic approach to 1) confirm that the two species are distinct at the genomic level; 2) determine which gene groups have diverged the most and the least between species; 3) show that their overall genomic structures are similar, as predicted by the viability of hybrids, and identify genomic regions that instead are incongruent; and 4) test the previously formulated hypothesis that genes involved in pathogenicity may be less divergent between the two species than genes involved in saprobic decay and sporulation. Results based on the sequencing of three genomes per species identified a high level of interspecific similarity, but clearly confirmed the status of the two as distinct taxa. Genes involved in pathogenicity were more conserved between species than genes involved in saprobic growth and sporulation, corroborating at the genomic level that invasiveness may be determined by the two latter traits, as documented by field and inoculation studies. Additionally, the majority of genes under positive selection and the majority of genes bearing interspecific structural variations were involved either in transcriptional or in mitochondrial functions. This study provides genomic-level evidence that invasiveness of pathogenic microbes can be attained without the high levels of pathogenicity presumed to exist for pathogens challenging naïve hosts.
Keywords: plant pathogenic fungi, Heterobasidion, structural variations, dN/dS, allopatric speciation
Introduction
Theories predict that drift and differential selection will increase interspecific genetic variation in species diverging allopatrically (Turelli et al. 2001). However, in the absence of strong differential selective pressure, ancestral polymorphisms may be retained. For instance, ancestral polymorphisms may be retained due to the necessity of maintaining a vital biological function or due to the presence of strong disruptive selection pressures. A classical example is that of interfertility: it has been shown that although sympatric and genetically diverging protospecies may experience a reinforcement of genetic isolation through newly developed intersterility, there is little selection pressure to develop intersterility when two divergent populations become geographically isolated (Kohn 2005). In the Kingdom Fungi, it has been reported that differences in genomic architecture mirror the interplay of different mutational processes with constraints provided by their characteristic population biology (Stukenbrock and Croll 2014). Different types of genomic rearrangements leading to changes in genomic architecture and gene expression, as well as amino acid substitutions and duplication events, may play a crucial role in promoting adaptive divergence across species. A genomic analysis of two allopatrically diverged sister taxa may thus reveal substantial structural genomic variations in all regions subject to differential selective pressure. In the case of plant pathogens, it has been commonly assumed that differential adaptive selection on allopatric species leads to divergent evolution of genes involved in plant–pathogen interactions, resulting in greater virulence of pathogens against noncoevolved plant hosts (Parker and Gilbert 2004). Although this appears to be the case for several pathogens, to our knowledge no research has fully addressed adaptive divergent evolution in plant pathogens involving traits related to general fitness and in particular to transmission. The narrow focus on pathogenicity has stifled a broader understanding of divergent evolutionary mechanisms for pathogens (Gonthier and Garbelotto 2013).
The Heterobasidion annosum species complex includes five species of plant pathogens hypothesized to have progressively diverged either in sympatry or in allopatry, depending on the species pair, starting 60 Ma and ending approximately 14 Ma (Dalman et al. 2010). Heterobasidion species can be both saprotrophic decomposers and necrotrophic pathogens. Although this is not uncommon for wood inhabiting microorganisms (Aguilar-Trigueros et al. 2014), Heterobasidion spp. differ from other fungi because the saprotrophic phase can occur both before and after the necrotrophic one. It is the saprotrophic phase preceding the necrotrophic one that is quite unique (Olson et al. 2012; Garbelotto and Gonthier 2013). In fact, it has been repeatedly demonstrated that for Heterobasidion species primarily associated with the genus Pinus, the majority of primary infection occurs as fungal genotypes saprobically colonize pine stumps. Colonized stumps subsequently become the major source of infection for adjacent trees through direct contagion along interconnected root systems.
In nature, only two pairs of species have been documented to hybridize: the sympatric Heterobasidion irregulare and Heterobasidion occidentale in western North America (Garbelotto et al. 1996; Lockman et al. 2014), and the allopatrically diverged H. irregulare and H. annosum in Italy (Gonthier et al. 2007; Gonthier and Garbelotto 2011). Divergent speciation between H. irregulare and H. occidentale has been estimated to have occurred 30–40 Ma, and the two species are known to have substantially different host preference (Dalman et al. 2010; Otrosina and Garbelotto 2010; Garbelotto and Gonthier 2013) and limited interspecific interfertility (Harrington et al. 1989). Thus, it is not surprising that the frequency of hybridization in nature appears to be extremely low and that genomic abnormalities have been reported to be associated with these hybrids (Garbelotto et al. 2004). Conversely, H. irregulare and H. annosum have become sympatric only recently when H. irregulare was transported from North America to Italy, presumably in 1944 (Gonthier et al. 2004). In spite of a history of allopatry lasting 34–41 Ma, the two species have maintained a comparable host preference for the genus Pinus and are known to be highly interfertile (Garbelotto and Gonthier 2013). Massive hybridization has resulted in the generation of hybrid swarms (Gonthier and Garbelotto 2011), a phenomenon known to occur for plants and animals, but so far unreported for the fungi (Brasier 2000). Studies on the fitness of hybrids have highlighted a significant role apparently played by the mitochondrial genome (Olson and Stenlid 2001; Garbelotto et al. 2007), but the mechanisms of this role still remain elusive. Nonetheless, fitness regulated by mitochondrial factors has been reported for a variety of organisms (Jiang et al. 2008; Shen et al. 2010; Greiner and Bock 2013).
The movement of H. irregulare from North America into Italy has resulted not only in the creation of hybrid H. irregulare × H. annosum swarms but also in an ongoing invasion process by the exotic species which is seemingly outcompeting the native one (Gonthier et al. 2007, 2012). Surprisingly, reciprocal inoculation studies and field monitoring efforts have demonstrated that the two species have comparable pathogenicity on Eurasian and North American pine species, but differ significantly in saprobic and sporulation potential (Garbelotto et al. 2010; Giordano et al. 2014; Gonthier et al. 2014).
Despite advances in the understanding of genomic evolution due to hybridization events among species that have long diverged (Clarke et al. 2002; Rieseberg et al. 2003), only scant information is available on the evolution of genomes of fungal pathogens undergoing hybridization (Brasier and Kirk 2010). A comparative genomic analysis would help 1) elucidating the genomic structure of sibling taxa that despite undergoing speciation in allopatry, have maintained a similar biology and host preference; 2) determining the mechanisms underlying the current massive hybridization between the two sibling species, H. irregulare and H. annosum; and 3) the identification of genomic regions providing the advantage the invasive species H. irregulare has over the native H. annosum. Understanding which genomic traits may confer an advantage to an invasive species may be pivotal to identify factors explaining the invasiveness of plant pathogens (Gonthier and Garbelotto 2013). Such understanding might also help to discover genes that may lead to significant adaptive changes if introgressed between species.
In this study, we report on the genome sequencing of three H. irregulare and three H. annosum genotypes, their reciprocal relationship, and their comparison to the reference genome of H. irregulare isolate TC 32-1 sequenced in 2012 (Olson et al. 2012). By using a comparative genomics approach, our specific goals were 1) to confirm that H. irregulare and H. annosum are clearly distinct species in spite of their high morphological and ecological similarities (Otrosina and Garbelotto 2010); 2) to show that in spite of high genetic divergence, the structure of the two genomes has remained extremely similar, as predicted by the ease at which the two species generate fertile hybrids (Gonthier and Garbelotto 2011); 3) to identify the main sources of variation in the genomes of the two sibling species; 4) to identify highly conserved gene families and genomic regions involved in the successful adaptation of both species in similar forest habitats located in two different continents; and 5) to test whether genes involved in saprobic wood decay and fruiting/sporulation may be significantly different between the two species, as expected based on published experimental evidence (Garbelotto et al. 2010; Giordano et al. 2014), thus providing a possible explanation on which traits may confer H. irregulare with an establishment advantage over H. annosum.
Materials and Methods
Biological Materials and DNA Extraction
A total of six haploid genotypes, three of H. irregulare and three of H. annosum, from four Italian sites were used in this study (table 1). Genotypes were deposited at the Mycotheca Universitatis Taurinensis (MUT) (table 1). Based on a previous analysis (Gonthier and Garbelotto 2011), none of the genotypes showed significant genetic admixing. All genotypes derived from single spores collected on woody spore traps and were grown in 2% malt extract liquid medium at 25 °C for 10 days before being harvested. Fungal tissues (200 mg) were collected using a vacuum pump, freeze dried over night, and ground using glass beads (diameter 0.2 and 0.4 mm) in a FastPrepTM Cell Disrupter (FP220-Qbiogene). Total DNA extraction was performed using DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA), following manufacturer instructions. DNA quantification was carried out by using the ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). DNA quality was assessed both by electophoresis of extracted DNA on a 1% agarose gel and by using a chip-based microcapillary electrophoresis system (Experion, Bio-Rad Laboratories, Hemel Hempstead, UK).
Table 1.
Summary of Heterobasidion Genotypes Sequenced
| ID Code | Species | Geographic Origin | MUT Accession No. |
|---|---|---|---|
| 9OA | Heterobasidion irregulare | Castelfusano Pinewood Urban Park; Rome (RM) | MUT00003629 |
| 48NB | Heterobasidion irregulare | Castelfusano Pinewood Urban Park; Rome (RM) | MUT00003627 |
| 49SA | Heterobasidion irregulare | Castelfusano Pinewood Urban Park; Rome (RM) | MUT00003628 |
| 137OC | Heterobasidion annosum | Circeo National Park/the forest of Sabaudia; Sabaudia (LT) | MUT00003656 |
| BM42NG | Heterobasidion annosum | Mesola Forest; Mesola (FE) | MUT00003543 |
| 109SA | Heterobasidion annosum | Feniglia Pinewood; Grosseto (GR) | MUT00003538 |
Genome Sequencing, Mapping, and De Novo Assembly
Paired-end (PE) 100-bp DNA libraries were prepared for each genotype and sequenced using an Illumina HiSeq 2000 platform at the Functional Genomics Laboratory of the University of California, Berkeley. The software FASTQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/, Last accessed November 16, 2015) was used to check read quality. Low-quality 100-bp reads (Q score < 22) were discarded. Raw reads of each genotype were aligned to the reference H. irregulare TC 32-1 genome (Olson et al. 2012) available in the JGI database (Heterobasidion annosum v2.0: Project: 16080, Heterobasidion_annosum.allmasked.fasta) using the Burrows-Wheeler Aligner MEM (BWA-MEM) optimized for 100-bp PE reads (Li and Durbin 2009). The output was used to generate mapping files in SAM format, which were then converted in BAM format, ordered, and indexed. Unmapped reads of H. annosum genotypes were assembled in contigs with Velvet 1.2.08 (Zerbino and Birney 2008) by setting k-mer = 39. Open-reading frame prediction on contigs produced by assembly of unmapped reads was performed by using Augustus v2.5.5 (Stanke and Morgenstern 2005) with a training set from Coprinus species and the following parameters: gene model = partial; protein, introns, start, stop, cds, coding seq, gff3, and UTR = on. The predicted amino acid sequences were identified by similarity search in the NCBI (National Center for Biotechnology Information) nonredundant protein sequence (NR) database (http://www.ncbi.nlm.nih.gov, Last accessed November 16, 2015) and were subsequently characterized by using BLAST2GO (Conesa et al. 2005).
De novo assembly of reads was performed using Velvet 1.2.08 (Zerbino and Birney 2008) with k-mer = 39 bp, chosen after optimization tests using k-mer sizes 37, 39, 41, and 43. Draft assemblies were refined and gaps were minimized using IMAGE2 (Swain et al. 2012) set at ten iterations and taking as inputs both unassembled reads and contigs in FASTA format generated by the Velvet assembly process. The final ordering of contigs and scaffolding process were performed with CONTIGuator (Galardini et al. 2011), by using the genome of H. irregulare TC 32-1 as a backbone (Heterobasidion annosum v2.0: Project: 16080, Heterobasidion_annosum.AssembledScaffolds.fasta). De novo assembly drafts were compared by using progressiveMauve (Darling et al. 2010). Sequences of raw reads, alignments (BAM files), and de novo assembled genomes were submitted to the EMBL database ENA—European Nucleotide Archive as a project under the accession number PRJEB8921.
The alignment of reads to the reference genome was used to identify putative H. annosum structural variations (SVs), copy number variations (CNVs), and single nucleotide polymorphisms (SNPs)/InDels (insertions/deletions). Phylogenomic analysis was also performed on the alignment of reads to the reference genome carried out with bowtie2 (Langmead et al. 2009), which is integrated in the phylogenetic software REALPHY (Bertels et al. 2014). De novo assembly drafts were compared to confirm SVs detected and were used to perform the simple sequence repeat (SSR) survey. Details on the abovementioned analysis are provided in the next paragraphs.
Identification of Putative SVs
SVs among genomes (i.e., large insertions, large deletions, inversions, intratranslocations, and intertranslocations) were predicted by using SVDetect v. r0.7 m (Zeitouni et al. 2010) on filtered SAM files produced by the alignment of reads to the reference genome containing both unmapped and abnormally mapped reads for each genotype. The SAMtools package (Li et al. 2009) was used to filter SAM files. Average insert sizes (µ) and standard deviations (σ) were calculated for each genotype and were used to determine the minimum threshold required for the detection of putative insertions and deletions. Identified SVs were filtered by quality based on the number of reads supporting the predicted variation, that is, SVs supported by less than 20 reads were discarded. SVs shared among H. annosum genotypes but absent among H. irregulare genotypes were putatively considered to be species-specific. Putative SVs, that is, inversions and translocations, were confirmed through comparative genomic analyses among de novo assembly drafts performed by using progressiveMauve (Darling et al. 2010). SVs were then manually scored and compared with those identified by SVDetect. Putative SVs in each scaffold and read depth for two representative genotypes were visualized as circular plots drawn with the Circos software v. 0.64 (Krzywinski et al. 2009). A chi-square test was used to determine whether SVs occurred more frequently in some scaffolds. This test was performed using absolute frequencies of SVs per scaffold and assuming an equal probability of occurrence for each scaffold.
Genes in regions containing inversions and translocations were detected using the BEDtools package “intersect” (Quinlan and Hall 2010). Transcripts were identified by ID and their corresponding encoded proteins were analyzed with BLAST2GO (Conesa et al. 2005) to search for homologs and to determine their Gene Ontology (GO). Significant differences in frequencies of GO terms compared with all H. irregulare gene models were assessed by using a Fisher’s exact test (Blüthgen et al. 2005) with the false discovery rate (FDR) control, which takes into account multiple comparisons (Al-Shahrour et al. 2004).
Identification of Putative CNVs
The CNV analysis was performed with CNV-seq (Xie and Tammi 2009) on alignment files after read-count normalization using a 10−6 P-value threshold. The purpose of this analysis was to identify differences in sequence coverage, that is, to identify variations in copy numbers of mapped sequences using a 2-kb sliding window between the consensus reads data sets of the two species. Results were plotted and drawn using the cnv package and a procedure in R language customized in house. As for inverted and translocated genomic regions, a BLAST2GO analysis was performed on genes detected in regions affected by significant CNV events (P value < 0.05, log2 > 5). Significant differences in frequencies of GO terms compared with all H. irregulare gene models were assessed by using a Fisher’s exact test (Blüthgen et al. 2005) with FDR control.
Identification of Putative SNPs/InDels
A pileup procedure on SAMtools v. 0.1.18 (Li et al. 2009) followed by quality filtering on VCFtools v 0.1.12 a (Danecek et al. 2011) was used to identify SNPs/InDels, with “–minQ” set to 20 and “–min-meanDP” set to 5. By using the tool vcf-isec, the SNPs/InDels consensus data set for H. annosum genotypes was compared with that for H. irregulare genotypes to identify sequence variants present exclusively in H. annosum. The resulting SNPs/InDels panel was annotated with the snpEff package (Cingolani et al. 2012) using the gene model catalog database available for the reference genome (Hannosum_v2.FilteredModels1.gff.gz). The snpEff package was used to determine the genomic position of SNPs/InDels (intergenic/exon/intron) and to identify synonymous and nonsynonymous changes. To confirm putative H. annosum SNPs/InDels, four loci involved in sabrobic growth, four involved in sporulation, and two related to pathogenicity (Olson et al. 2012) were randomly selected to be amplified, sequenced and compared on five additional H. irregulare genotypes and five H. annosum genotypes (listed in supplementary table S1, Supplementary Material online). Primers targeting the selected loci were designed by using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi, Last accessed November 16, 2015). Primer sequences and their respective annealing temperatures are listed in supplementary table S2, Supplementary Material online. DNA extraction was performed by using the Dneasy Plant Mini Kit (QIAGEN, Valencia, CA) on lyophilized fungal cultures that were grown at room temperature for 10 days in a 2% (w/v) malt extract liquid medium (AppliChem GmbH, Darmstadt, Germany). Polymerase chain reaction (PCR) assays were performed in a 25 μl volume containing 5× PCR buffer, 1.5 mM of MgCl2, 0.2 mM of dNTPs mix, 0.5 μM each of the primers, 0.025 U of GoTaq polymerase (Promega, Madison, WI), and 0.1 ng of genomic DNA. PCR reactions were conducted as follows: an initial cycle with a 96 °C denaturation step of 5 min, followed by 35 cycles, with each cycle consisting of a 96 °C denaturation step of 30 s, an annealing step of 30 s and a 72 °C extension step of 45 s, and one final cycle with a 72 °C extension step of 10 min. Amplicons were digested by ExoSAP-IT (Affymetrix, Santa Clara, CA) at 37 °C for 15 min and then at 80 °C for 15 min. The PCR products were sequenced in both directions by the Functional Genomics Laboratory of the University of Berkeley (CA). Sequences were aligned using the algorithm ClustalW in MEGA v.6.0 (Tamura et al. 2013) with default parameters.
Identification of Highly Conserved and Highly Divergent Gene Families in Genomic Regions Shared between the Two Species
The snpEff analysis generated a tab-separated file in which putative H. annosum SNPs/InDels were identified for each annotated gene model of H. irregulare. After gene length normalization, genes harboring less than ten SNPs/InDels per kilobase (nucleotide identity of more than 99%) putatively associated with H. annosum were considered strongly conserved, whereas genes with SNP density higher than 40 SNPs per kilobase (nucleotide identity of less than 96%) were considered as highly divergent and species-specific. A BLAST2GO analysis was used to characterize both conserved and species-specific genes. The GOSSIP program in BLAST2GO was used to perform Fisher’s exact tests with FDR control in order to compare GO distributions of conserved and species-specific genes with distributions predicted by all H. irregulare gene models.
Genes influenced by speciation events (i.e., selection) were identified calculating dN and dS substitution rates for each gene affected by nonsynonymous SNPs/InDels. The dN/dS ratio has been widely used to determine the mode of selection. Ratio of dN/dS >1 is interpreted as a sign of positive selection, that is, natural selection promotes changes in the protein sequence; whereas dN/dS <1 is generally accepted as a sign of purifying selection, where some replacement substitutions have been purified by natural selection, presumably because of their deleterious effects (Hurst 2002). The panel of detected putative H. annosum SNPs was annotated in homologous H. irregulare gene models by using snpEff. This software listed the number of detected synonymous (Sd) and nonsynonymous SNPs (Nd) for each gene, compared pairwise. The program KaKs_calculator (https://code.google.com/p/kaks-calculator/downloads/list, Last accessed November 16, 2015) was used to calculate the number of expected synonymous (S) and nonsynonymous (N) sites for each gene with parameters set according to the Nei–Gojobori method.
The proportions of nonsynonymous and synonymous differences between the two species for each gene compared pairwise (pN and pS, respectively) were calculated with the following formulas (Nei and Gojobori 1986):
The dN/dS ratio for each gene pair was then calculated with the following formulas:
The dN/dS ratios of genes related to saprobic growth, genes related to fruiting body formation, and genes related to pathogenicity (Olson et al. 2012) were compared with the mean dN/dS ratio of the whole gene data set through a permutation t-test (9,999 permutation replicates).
Phylogenomic Analysis
Phylogenomic analysis was carried out with the software REALPHY (reference sequence alignment-based phylogeny builder) (Bertels et al. 2014) using raw reads of the six sequenced genotypes and sequence data from the H. irregulare TC 32-1 genome. REALPHY inferred phylogenetic trees from whole-genome sequence data based on all provided sequences mapped to the reference genome through bowtie2 (Langmead et al. 2009). Phylogenetic trees were inferred on PhyML (phylogenetic estimation using maximum likelihood) (Guindon et al. 2010), and a bootstrap consensus tree was created (1,000 bootstrap replicates, initial tree BioNJ, model of nucleotides substitution GTR [general time reversible]). The phylogenetic analysis was carried out using the alignment of sequenced reads of the six sequenced genotypes on the coding sequences (CDSs) of the reference H. irregulare TC 32-1.
SSR Analysis
An analysis of SSRs was performed on de novo assembly drafts using the MISA mode of SciRoko software (Kofler et al. 2007), with the minimum number of scored repeats set at 14, 7, 5, 4, 4 and 4 for mono-, di-, tri-, tetra-, penta- and hexanucleotides, respectively.
Results
Genome Sequencing, Mapping, and De Novo Assembly
The number of sequenced reads for each genotype was approximately 8 million. The percentages of PE reads that aligned to the reference genome ranged from 87.59 to 92.22 for H. irregulare genotypes (9OA, 48NB, and 49SA) and from 75.90 to 79.61 for H. annosum genotypes (137OC, BM42NG, and 109SA). Sequence coverage ranged between 17× and 22×. Only about 85% of H. irregulare and 70% of H. annosum PE reads were properly mapped. The remaining aligned reads showed an abnormal insert size, highlighting inter- and intraspecific variations in genome architecture (table 2). Unmapped reads of H. annosum genotypes were assembled in 1,104 contigs. AUGUSTUS predicted 249 putative CDSs. After the BLAST2GO analysis, 73 sequences were annotated as related to gag-pol polyproteins, retroelements, retrotransposon nucleocapsid proteins and reverse transcriptases, whereas 176 showed no BLAST (Basic Local Alignment Search Tool) hits.
Table 2.
Statistics of Reads Mapping of the Three Heterobasidion irregulare and the Three Heterobasidion annosum Genotypes to Reference Genome
|
Heterobasidion irregulare Genotypes |
Heterobasidion annosum Genotypes |
|||||
|---|---|---|---|---|---|---|
| 9OA | 48NB | 49SA | 137OC | BM42NG | 109SA | |
| PE reads sequenced | 8,036,722 | 8,025,191 | 8,028,750 | 8,018,594 | 8,018,514 | 8,020,180 |
| Mapped reads | 7,039,599 (87.59) | 7,313,645 (91.13) | 7,404,292 (92.22) | 6,148,803 (76.68) | 6,383,225 (79.61) | 6,086,921 (75.90) |
| Properly mapped Reads | 6,618,004 (82.35) | 6,955,953 (86.68) | 7,051,281 (87.83) | 5,679,959 (70.83) | 5,906,955 (73.67) | 5,650,323 (70.45) |
| Singletons | 63,510 (0.79) | 57,368 (0.71) | 62,502 (0.78) | 206,180 (2.57) | 207,745 (2.59) | 188,937 (2.36) |
| Coverage (×) | 20.52 | 21.78 | 21.61 | 16.97 | 17.66 | 16.86 |
Note.—Values in parentheses are given in percentage.
De novo assembly of reads resulted in 5,262 contigs for genotype 137OC, 4,686 for BM42NG, 4,994 for 109SA, 6,034 for 9OA, 6,992 for 48NB, and 6,119 for 49SA. N50 values ranged from 12.9 to 24 kb. A total of 14 scaffolds for each genome were reconstructed. Sizes of final assemblies ranged from 26.3 to 27.9 Mb. De novo assemblies were smaller than those from the reference genome (33.6 Mb) (table 3).
Table 3.
Statistics of De Novo Assemblies
|
Heterobasidion irregulare Genotypes |
Heterobasidion annosum Genotypes |
|||||
|---|---|---|---|---|---|---|
| 9OA | 48NB | 49SA | 137OC | BM42NG | 109SA | |
| Contigs (n) | 6,034 | 6,992 | 6,119 | 5,262 | 4,686 | 4,994 |
| N50 | 24,007 | 12,945 | 14,781 | 22,892 | 24,566 | 22,448 |
| Max contig length (bp) | 139,331 | 194,148 | 129,618 | 145,724 | 220,315 | 175,157 |
| Total length (bp) | 26,374,808 | 26,374,274 | 26,844,330 | 27,696,343 | 27,988,318 | 27,530,324 |
| Unassembled contigs (n) | 1,694 | 1,984 | 1,551 | 1,578 | 1,385 | 1,492 |
Identification of Putative SVs
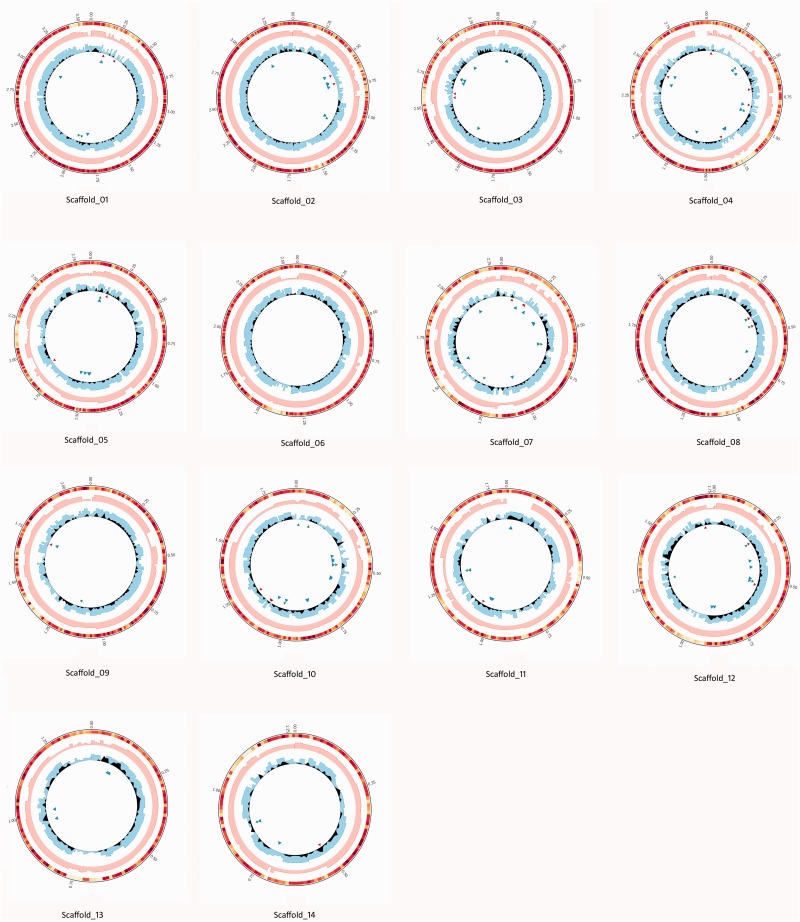
The number of SVs between H. irregulare and H. annosum that were shared by all three H. annosum genotypes varied between 1 and 49, depending on scaffold (table 4). The distribution of large deletions and insertions was not significantly different among scaffolds (P value > 0.05), whereas the distribution of inversions and translocations varied significantly among scaffolds (P value < 0.05). Scaffold_04, Scaffold_07, and Scaffold_10 harbored the highest number of inversions and translocations. Moreover, a single intrascaffold translocation was found in Scaffold_10. Putative SVs detected in H. annosum and their position with respect to reference genome are visualized in figures 1 and 2 and listed in supplementary table S3, Supplementary Material online.
Table 4.
Number of Putative Heterobasidion annosum SVs Divided in Intra- and Interchromosomal for Each Scaffold
| Intra |
Inter |
||||
|---|---|---|---|---|---|
| Large Insertions (>2 kb) | Large Deletions (>2 kb) | Inversions | Trans locations | Trans locations | |
| Scaffold_01 | 2 | 2 | 3 | 0 | 1 |
| Scaffold_02 | 1 | 3 | 5 | 0 | 3 |
| Scaffold_03 | 4 | 2 | 5 | 0 | 1 |
| Scaffold_04 | 3 | 5 | 15 | 0 | 49 |
| Scaffold_05 | 2 | 2 | 10 | 0 | 5 |
| Scaffold_06 | 0 | 0 | 0 | 0 | 4 |
| Scaffold_07 | 2 | 3 | 24 | 0 | 7 |
| Scaffold_08 | 2 | 4 | 9 | 0 | 10 |
| Scaffold_09 | 1 | 1 | 1 | 0 | 1 |
| Scaffold_10 | 4 | 4 | 23 | 1 | 31 |
| Scaffold_11 | 2 | 2 | 12 | 0 | 3 |
| Scaffold_12 | 3 | 3 | 9 | 0 | 19 |
| Scaffold_13 | 0 | 0 | 5 | 0 | 15 |
| Scaffold_14 | 1 | 2 | 5 | 0 | 16 |
Note.—Scaffold number is referred to the reference genome of Heterobasidion irregulare.
Fig. 1.—
Visualization of putative H. annosum SVs and SNPs density for each scaffold. Each of the 14 scaffolds corresponding to chromosomes of the reference H. irregulare TC32-1 is circled and defined. Starting from the outside, circles represent gene density in a window size of 10 kb (color coded from yellow to dark red, with deeper red region representing high gene density), read depth of the representative H. irregulare genotype 49SA (pink histogram), read depth of the representative H. annosum genotype 137OC (cyan histogram). SNP density of H. annosum estimated as density of consensus SNPs of three H. annosum genotypes different from SNPs panel of H. irregulare genotypes (black line plot). Putative H. annosum SVs are represented as triangles and color coded as follows: red for large deletions (>2 kb), green for large insertions (>2 kb), and blue for inversions.
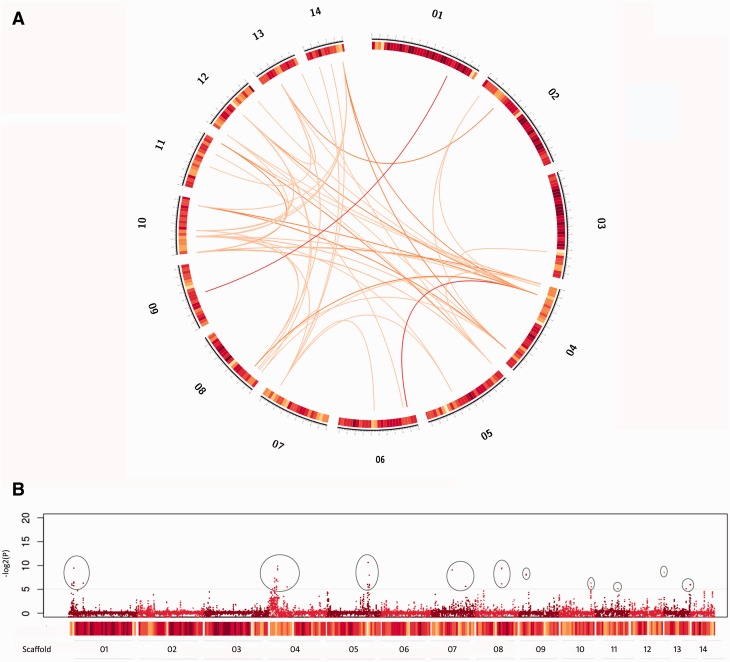
Fig. 2.—
Visualization of putative H. annosum interchromosomal translocations and CNV along scaffolds. Heatmap representing gene density in a window size of 100 kb is color coded from yellow to dark red, with deeper red representing high gene density. (A) Putative interchromosomal translocations are represented as orange lines in the central part of the circle. Red lines are translocations supported by more than 30 reads. (B) CNVs were counted and plotted in a sliding window size of 2 kb and log2 ratios were calculated for all mapped hits. Only significant values (P value < 0.05) are visualized and peaks on y axis represent a possible CNV event. Consistent CNVs (log2 > 5) are circled.
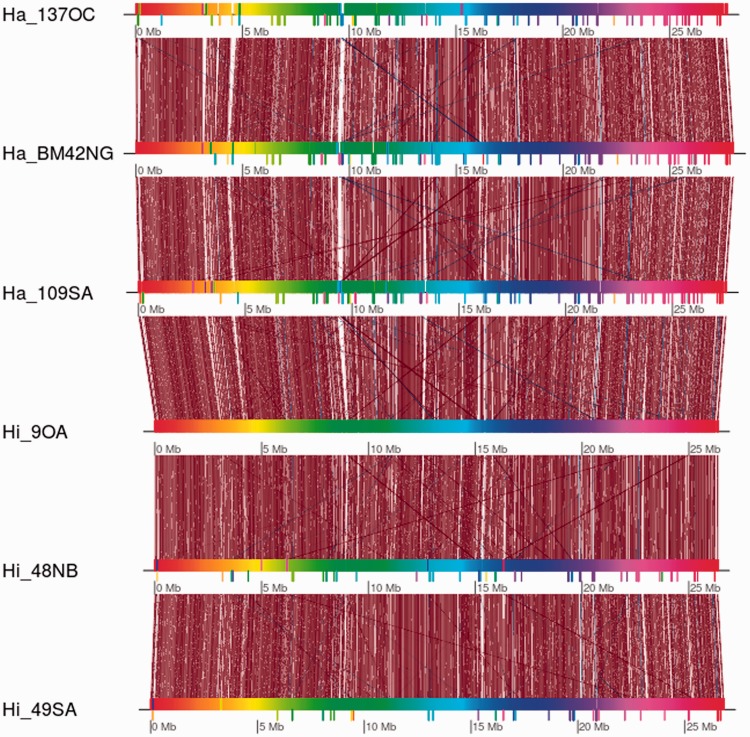
Comparative analyses among de novo assembly drafts by progressiveMauve confirmed the presence of intraspecific variability (fig. 3). Large shared genomic regions as well as inverted blocks among genotypes were detected. Scaffold_07, Scaffold_04, Scaffold_11 and Scaffold_13 showed the largest differences between H. irregulare and H. annosum, possibly suggesting that these scaffolds might contain species-specific genomic regions.
Fig. 3.—
Results of comparative analysis of de novo assembly drafts. The six genomes were compared using progressiveMauve (Darling et al. 2010) and visualized using genoPlotR (Guy et al. 2010). Shared blocks are linked by dark red lines, whereas translocated blocks are linked by blue lines. Inverted blocks are visualized as blocks below the colored bars representing the genomes.
Cumulatively, annotated genes inside inversions were 223, whereas genes detected in translocations were 63. Among these 63 genes, 14 were described as containing zinc-finger-like domains and Gag domains related to retroviruses and retrotransposons. Fisher’s exact test for enrichment analyses on genes in H. annosum-inverted regions showed that GO terms related to mitochondrial factors were overrepresented (FDR-corrected P value < 0.05) (supplementary table S4, Supplementary Material online). In translocated regions, GO terms GO:0008270 (zinc ion binding) and GO:0003676 (nucleic acid binding) were found to be overrepresented (for details, see supplementary table S5, Supplementary Material online).
Identification of Putative CNVs
Putative CNVs were more frequently identified at the beginning of Scaffold_01, Scaffold_04, Scaffold_10, in Scaffold_13 (both telomeric regions), and in Scaffold_05 (about 100 kb from one end). Additionally, CNVs were present but less frequent in Scaffold_07 and Scaffold_08 (fig. 2B). In genomic regions affected by significant CNV events (P value < 0.05, log2 > 5), 36 genes were identified. Fisher’s exact test on these 36 genes showed that only the term GO:0003676 (nucleic acid binding) was significantly overrepresented compared with all H. irregulare gene models (for details, see supplementary table S6, Supplementary Material online).
Identification of Putative SNPs and InDels
The number of consensus SNPs/InDels shared by all H. annosum genotypes and absent in all H. irregulare genotypes was estimated at 674,015 (about 20 SNPs/kb) (table 5). More than 50% of SNPs were distributed in intergenic regions, whereas 36% were in exons, and 14% in introns. In CDSs, the number of SNPs/InDels, normalized for sequence length, resulted as high as 21.05 (SD 9.23). In addition, 10,637 small insertions and 7,903 small deletions were predicted. SNP/InDel annotation showed that about 68% of SNPs/InDels within exons were silent, whereas the remaining 32% caused changes in coding due to frameshift mutations. A total of 146,809 SNPs differentiated the three H. irregulare genotypes naturalized in Italy from the North American H. irregulare reference genome (about four SNPs per kilobase).
Table 5.
Classification of Heterobasidion annosum Annotated Putative SNPs/InDels
| Putative Heterobasidion annosum SNPs/InDels | |
|---|---|
| Number | 674,015 |
| SNPs/InDels distribution | |
| Exons | 217,606 |
| Introns | 97,306 |
| Intergenic regions | 359,103 |
| SNP type | |
| Transitions | |
| A/G | 148,866 |
| C/T | 117,640 |
| G/A | 117,697 |
| T/C | 149,031 |
| Transversions | |
| A/C | 20,795 |
| A/T | 15,384 |
| C/G | 18,847 |
| G/T | 17,355 |
| C/A | 17,677 |
| T/A | 15,248 |
| InDel type | |
| Small insertions | 10,637 |
| Small deletions | 7,903 |
| Number of effects by functional class (%) | |
| Missense | 67,303 (31.175) |
| Nonsense | 381 (0.176) |
| Silent | 148,206 (68.649) |
Alignment of partial sequences of ten loci in additional Heterobasidion genotypes allowed for the validation of the putative H. annosum SNPs/InDels detected in the six whole-genome surveys. In these loci, 159 SNPs/InDels differentiating the two species were detected. All putative H. annosum SNPs/InDels detected by comparative sequence analysis of the six genomes were confirmed in the genotypes for which individual loci were sequenced (see supplementary alignment S1, Supplementary Material online).
In detail, 9 H. annosum SNPs were detected in locus 104812, 26 in locus 105463, 5 in locus 124417, 8 in locus 148906, 20 in locus 150620, 6 in locus 153627, 10 in locus 174687, 18 in locus 43914, 29 in locus 56987, and 19 in locus 63659. In addition, one deletion of 6 bp was confirmed in locus 105463, one deletion of 5 bp in locus 124417, and one deletion of 3 bp in locus 56987. A small insertion of 2 bp was also confirmed for locus 105463. Normalization for sequence length of the number of putative H. annosum SNPs/InDels detected in the ten loci analyzed resulted in an SNP density of 22.20 SNPs/InDels per kilobase. In particular, the density of polymorphism differentiating the two species was 31.35 SNPs/InDels for loci related to saprobic growth, 21.25 SNPs/InDels per kilobase for loci associated with sporulation, and 20.43 SNPs/InDels per kilobase for loci associated with pathogenicity.
Identification of Highly Conserved and Highly Divergent Gene Families in Genomic Regions Shared between the Two Species
The number of conserved genes showing more than 99% of nucleotide identity between the two species was 690. Conversely, 707 H. annosum genes were found to harbor more than 40 putative SNPs/InDels per kilobase (nucleotide identity of less than 96%) and may be regarded as species-specific alleles. Main GO terms (GO terms assigned to more than ten sequences) related to Molecular Function in conserved genes and species-specific alleles are summarized in supplementary figure S1, Supplementary Material online. The comparison between GO terms assigned to both conserved genes and species-specific alleles showed a significant overrepresentation of terms GO:0003676 (oxidoreductase activity) and GO:0020037 (heme binding) in species-specific alleles. On the other hand, GO terms related to ribosomal, cellular metabolic processes, and signal transduction were significantly overrepresented in the data set of conserved genes when compared with species-specific alleles (supplementary table S7, Supplementary Material online).
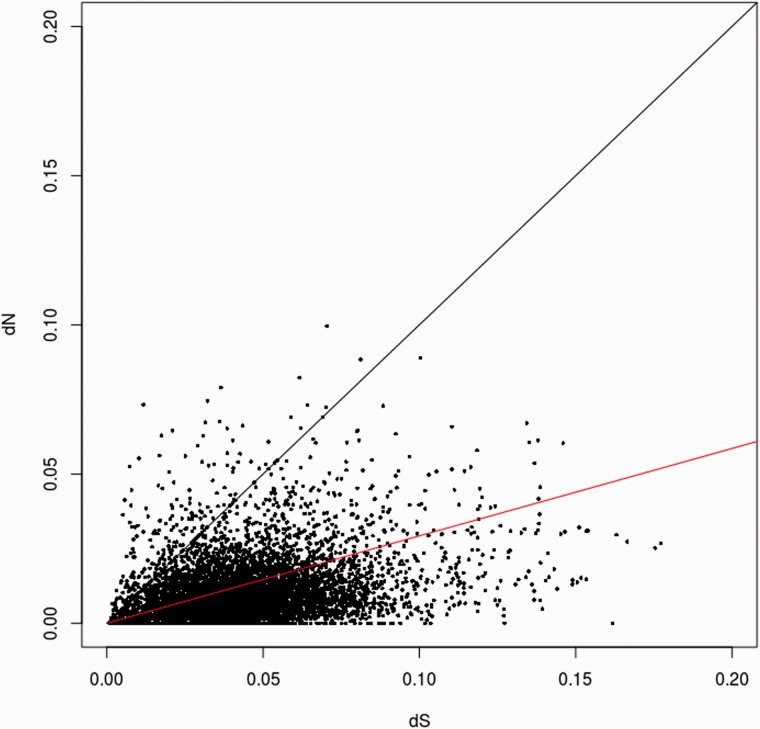
By analyzing SNPs in CDSs, 6,636 gene models were found to contain putative nonsynonymous H. annosum SNPs. After gene length normalization, an average of 10.25 nonsynonymous SNPs per gene was found (SD 9.64). The average ratio of the number of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site (dN/dS ratio) in H. annosum genome compared with H. irregulare was 0.29 (SD 0.39) (fig. 4). Genes involved in saprobic growth (i.e., genes significantly upregulated during growth on wood; Olson et al. 2012) were 61 and had a dN/dS ratio of 0.24 (SD 0.32), whereas genes associated with fruiting body formation (i.e., genes significantly upregulated in fruiting body compared with mycelium growth in liquid medium; Olson et al. 2012) were 81 and had an average dN/dS ratio of 0.21 (SD 0.23). Genes related to pathogenesis (i.e., genes significantly upregulated in necrotic bark tissue; Olson et al. 2012) were 42 and had an average dN/dS ratio of 0.16 (SD 0.19). A permutation t-test comparing each category to the ratio of whole genome determined that only the dN/dS of genes related to pathogenesis was significantly different from the average dN/dS value (difference between means = 0.16, t = −2.6, P = 0.009). Genes with a dN/dS ratio >1 were 153, and were characterized using BLAST2GO (supplementary table S8, Supplementary Material online). Fisher’s exact test on these sequences showed a significant (FDR-corrected P value < 0.05) overrepresentation of several GO terms related to mitochondrial components, mitochondrial functions, transcriptional functions, and metal homeostasis (supplementary table S9, Supplementary Material online).
Fig. 4.—
The dN/dS distribution among homolog pairs of H. irregulare and H. annosum. The mean dN/dS value was 0.29 and is indicated by a red line. Black line indicates the neutral expectation where dN = dS. In total, 153 sequences fall above the black line.
Phylogenomic Analysis
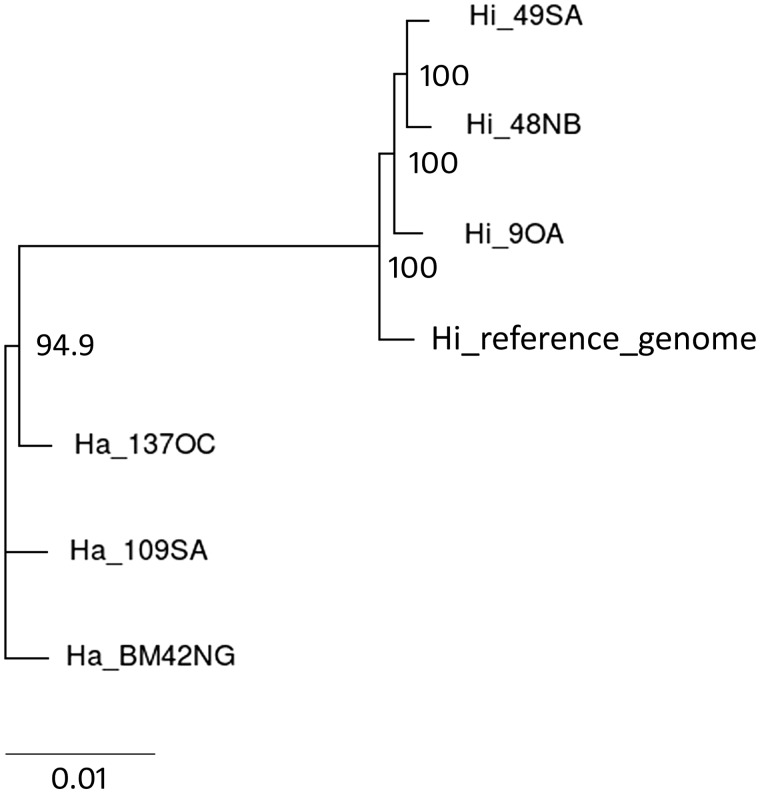
Whole-genome phylogenetic analyses performed by using REALPHY (Bertels et al. 2014) resulted in a consensus tree clearly showing two distinct clades. Genotypes of H. annosum clustered together in one clade, whereas H. irregulare genotypes grouped separately and in the same clade as the reference genome (fig. 5).
Fig. 5.—
Phylogenetic relationships among sequenced genotypes and H. irregulare reference genome as inferred by REALPHY through the alignment of reads on CDSs of reference genome. Only bootstrap values higher than 50% are shown.
SSR Analysis
Results of SSR analysis are summarized in table 6. The raw number of SSRs in H. annosum ranged between 58.27 and 62.88 SSRs per megabase depending on genotype, whereas that in H. irregulare ranged between 70.98 and 74.17 per megabase, again depending on genotype. Although the number of mononucleotide SSRs in H. irregulare genotypes was almost twice as higher than that of H. annosum, differences were not significant (permutation t-test, P = 0.089).
Table 6.
Distribution of SSRs in the Six Analyzed Genomes
| Genotype | Motif | Counts | Average Length | Counts/Mb | |
|---|---|---|---|---|---|
| 137OC | |||||
| No. SSRs | 2,003 | Mononucleotide | 50 | 21.26 | 1.45 |
| Average length | 20.92 | Dinucleotide | 175 | 17.58 | 5.09 |
| Average SD | 5.79 | Trinucleotide | 880 | 19.05 | 25.60 |
| Counts/Mb (whole genome) | 58.27 | Tetranucleotide | 474 | 20.16 | 13.79 |
| Pentanucleotide | 171 | 23.98 | 4.97 | ||
| Hexanucleotide | 253 | 29.02 | 7.36 | ||
| BM42NG | |||||
| No. SSRs | 2,168 | Mononucleotide | 62 | 20.32 | 1.80 |
| Average length | 20.98 | Dinucleotide | 187 | 17.67 | 5.42 |
| Average SD | 5.88 | Trinucleotide | 964 | 19.01 | 27.96 |
| Counts/Mb (whole genome) | 62.88 | Tetranucleotide | 494 | 20.23 | 14.33 |
| Pentanucleotide | 169 | 24.46 | 4.90 | ||
| Hexanucleotide | 292 | 29.03 | 8.47 | ||
| 109SA | |||||
| No. SSRs | 2,080 | Mononucleotide | 46 | 20.11 | 1.34 |
| Average length | 20.83 | Dinucleotide | 194 | 17.27 | 5.65 |
| Average SD | 5.84 | Trinucleotide | 904 | 18.96 | 26.31 |
| Counts/Mb (whole genome) | 60.54 | Tetranucleotide | 505 | 20.27 | 14.70 |
| Pentanucleotide | 167 | 23.89 | 4.86 | ||
| Hexanucleotide | 264 | 29.12 | 7.68 | ||
| 9OA | |||||
| No. SSRs | 2,560 | Mononucleotide | 150 | 21.43 | 4.35 |
| Average length | 21.54 | Dinucleotide | 206 | 17.95 | 5.97 |
| Average SD | 6.17 | Trinucleotide | 1,030 | 19.27 | 29.84 |
| Counts/Mb (whole genome) | 74.17 | Tetranucleotide | 584 | 20.50 | 16.92 |
| Pentanucleotide | 200 | 25.00 | 5.79 | ||
| Hexanucleotide | 390 | 29.30 | 11.30 | ||
| 48NB | |||||
| No. SSRs | 1,872 | Mononucleotide | 88 | 19.41 | 3.34 |
| Average length | 20.90 | Dinucleotide | 149 | 17.38 | 5.65 |
| Average SD | 5.53 | Trinucleotide | 809 | 19.05 | 30.67 |
| Counts/Mb (whole genome) | 70.98 | Tetranucleotide | 415 | 20.05 | 15.74 |
| Pentanucleotide | 146 | 24.31 | 5.54 | ||
| Hexanucleotide | 265 | 28.47 | 10.05 | ||
| 49SA | |||||
| No. SSRs | 2,469 | Mononucleotide | 116 | 20.06 | 3.35 |
| Average length | 21.08 | Dinucleotide | 200 | 17.32 | 5.77 |
| Average SD | 5.62 | Trinucleotide | 1,029 | 19.00 | 29.69 |
| Counts/Mb (whole genome) | 71.24 | Tetranucleotide | 571 | 20.27 | 16.48 |
| Pentanucleotide | 171 | 24.56 | 4.93 | ||
| Hexanucleotide | 382 | 28.62 | 11.02 |
Discussion
Heterobasidion irregulare and H. annosum Are Clearly Distinct Species
Despite the high morphological similarity between H. irregulare and H. annosum (Otrosina and Garbelotto 2010), evidence based on isozyme and phylogenetic analyses limited to a few loci (Otrosina et al. 1993; Linzer et al. 2008; Dalman et al. 2010; Gonthier and Garbelotto 2011) has shown that the two taxa have attained reciprocal monophyly and are distinct based on the phylogenetic species concept (Taylor et al. 2000). However, definitive evidence on their status as distinct species was still lacking. Our phylogenetic analysis on whole-genome sequences proved without any doubt that H. irregulare is clearly distinct from H. annosum.
The Main Structure of the Genomes of the Two Species Has Remained Similar
In spite of the long time since divergence and long allopatric history, the two genomes appeared to be largely syntenic and congruous. The macrosynteny between the two species described in this study may be somewhat spurious due to the fact that a reference genome of H. annosum is not available, and the six newly sequenced genomes representing the two species necessarily had to be referenced to the only annotated and publicly available H. irregulare genome. Nonetheless, the alignment of assembly drafts (fig. 3) showed high number of shared genomic blocks among genomes. As mentioned above, we recognize that this approach may not properly allow to exhaustively determine whether genomes of the two are truly congruent or not, as it is biased by low coverage and quality of the produced contigs. However, taking into consideration the high rate of hybridization between the two species (Garbelotto and Gonthier 2013), these results can be regarded at least as suggestive of significant levels of macrosynteny.
High genomic similarity between the two species is corroborated by the fact that the entire genome of H. irregulare was covered by 75% of H. annosum reads, and that nucleotide interspecific homology reached 98% in several mapped genomic regions. Unmapped H. annosum reads were found to harbor sequences related to retroelements, thus suggesting that abundance of these elements may have contributed to shape those genomic regions that were found to be most divergent between the two species. In spite of the presence of regions that were clearly different between the two species, overall genome sizes, determined by comparing assembly drafts, were similar (26.3–27.9 Mb), with H. annosum genomes slightly larger than H. irregulare genomes.
Although size of the three newly sequenced H. irregulare genomes appeared to be different from that of the reference genome, it should be noted that distinct sequencing approaches may generate artifacts. In this study, the abovementioned intraspecific difference in size should be regarded as a result of variability between traditional Sangerian and Illumina-based new generation sequencing methods. In addition, the difference in length between the new assemblies and the reference genome could be affected by the presence of repetitive sequences, that is, transposable elements (TEs), viruses, and rDNA repeats. In fact, TEs in the genome of H. irregulare were estimated to be as high as 16.2% of the whole genome. This number was also supported by the identification of TEs in the set of unmapped reads of H. annosum. TEs may also have negatively affected the assembly processes, as reported for the de novo assembly draft of H. occidentale (Lind et al. 2012), further explaining some of the differences reported here.
Inversions, Translocations, and CNVs Have Differentiated the Two Genomes
In our survey, putative H. annosum SVs were detected. Genomic SVs, including deletions, insertions, CNVs, inversions and translocations, are generally referred to as genomic rearrangements bigger than 1–2 kb (Alkan et al. 2011). The alignment of H. irregulare and H. annosum reads on the reference genome allowed to identify putative SVs between the two fungal species, removing the bias due to intraspecific variability. The comparison of de novo assembly drafts was used to confirm the presence of SVs. Based on our conservative comparative approach, SVs not shared by all isolates of one species were discarded. We also noticed that some SVs, that is, inversions and interchromosomal translocations, appeared not to be equally distributed across scaffolds: for example, H. annosum Scaffolds 04, 07, 11, and 13 were distinguishable from matching H. irregulare scaffolds due to the presence of inversions and translocations.
Several telomeric and subtelomeric regions of scaffolds of the H. annosum genome were affected by CNV events. It has been reported that fungal pathogens may show relevant genome plasticity due to redundancy related to virulence traits occurring in telomeric regions (Chuma et al. 2011; Raffaele and Kamoun 2012; Stukenbrock 2013) and adaptation to new environments (Denayrolles et al. 1997; Chow et al. 2012). SSR analyses also showed that mononucleotide repeats were more abundant in H. irregulare than in H. annosum genomes, although not significantly. As SSR analysis was performed on de novo assembly drafts, it should not have been biased by the use of H. irregulare as a reference. Given that H. annosum is presumed to be ancestral to H. irregulare (Otrosina et al. 1993; Linzer et al. 2008; Dalman et al. 2010), mononucleotide repeats might be correlated to divergent evolution.
Species-Specific Alleles Were Related to Heme Binding Proteins and to Oxidoreductases
Alignment of reads of the six genomes in relation to the reference genome allowed to identify putative H. annosum SNPs/InDels absent in H. irregulare genotypes. At the intraspecific level, SNPs density detected within H. irregulare genotypes (about four SNPs per kilobase) was consistent with those reported for other basidiomycetes species, such as Lentinula edodes (4.6 SNPs per kilobase; Au et al. 2013) and Melamspora larici-populina (about six SNPs per kilobase; Persoons et al. 2014). On the other hand, interspecific sequence polymorphisms were estimated as high as about 20 SNPs per kilobase. A set of 159 SNPs/InDels were verified and confirmed by the sequencing of ten additional loci in ten isolates, equally representing the two Heterobasidion species.
Conserved genomic regions shared between the two species were assumed to harbor genes related to primary metabolism and to survival, whereas genes in less conserved regions might be related to other pathways (e.g., secondary metabolism). When H. annosum genotypes were compared with H. irregulare genotypes, 690 genes were regarded as highly conserved and had more than 99% of nucleotide identity. On the contrary, 707 alleles harbored high SNPs density resulting in less than 96% of nucleotide identity between the two fungi. The most overrepresented terms among species-specific alleles were related to oxidation-reduction processes (66 of 707 genes with this term) and heme binding proteins (18 of 707 genes). It has been reported that the H. irregulare genome contains a versatile set of oxidoreductases putatively involved in lignin oxidation and conversion, including glyoxal oxidases, quinone-oxidoreductases, and aryl-alcohol oxidases (Olson et al. 2012). In addition, wood decay fungi as Heterobasidion are known to produce large amounts of heme-containing proteins, such as manganese and lignin peroxidases (MacDonald et al. 2012). Lignin peroxidases are among the main enzymes involved in lignin degradation. In a transcriptomics study using H. annosum as a model system, five manganese peroxidases were found to be specifically induced only during saprobic growth (Raffaello et al. 2014). One possible interpretation of the abundance of SNPs/InDels in genes encoding these classes of enzymes could be that they have accumulated mutations during the different adaptive evolution of the two fungal species.
Genes Showing High Values of dN/dS Ratio Were Mainly Related to Transcriptional Activity and Mitochondrial Factors
Nonsynonymous mutations are generally predicted to contribute to phenotypic evolution more than synonymous mutations. In fact, nonsynonymous mutations directly alter the amino acid sequence and are likely to affect protein stability and activity. In our comparisons, only 32% of SNPs/InDels putatively associated with H. annosum coding regions lead to changes in proteins. In particular, approximately 6,600 genes were found to harbor nonsynonymous SNPs between the two species. To evaluate evolutionary pressure acting on genes affected by SNPs/InDels, we estimated the dN/dS ratio between orthologous sequences. Results showed that a majority of sequence pairs (about 95%) had a dN/dS ratio <1, suggesting that these genes covering a wide array of functions evolved under purifying selection without altering the encoded amino acid sequence during the speciation. Loci affected by dN/dS ratios >1 were instead found to be mostly related to transcriptional activity. Transcriptional regulation may trigger several biological processes in a cell or in an organism, both in primary metabolic and physiological balance, and in responses to the environment (Riechmann et al. 2000). In fungi, the comparison of the transcriptional regulatory networks of several ascomycetes suggested that regulatory elements of metabolic pathways have dramatically changed between species and seem extremely plastic (Carroll 2000; Tuch et al. 2008; Lavoie et al. 2009). It could be hypothesized that both large fractions of SNPs on noncoding regions which might represent cis-regulatory elements, that is, enhancers, promoters, 5′-untranslated regions (UTRs), 3′UTRs, introns, as well as nonsynonymous mutations in transcriptional factors, possibly leading to a rewiring of gene expression regulation, may have been involved in the divergence between H. irregulare and H. annosum. The extensive proliferation of nonsynonymous SNPs in genes related to transcriptional activities may reflect the diversification of the regulatory pathways of these two pathogens. According to this scenario, differences in transcription regulation between these two sibling species may be responsible for rapid adaptive evolution. Interestingly, a genome-wide association study performed by Dalman et al. (2013) showed that one of the 14 mutations significantly associated with virulence traits in H. annosum is a synonymous SNP into an SWI5 transcription factor encoding gene, suggesting that this phenotypic trait could also be influenced by changes in transcriptional network. The homologous of SWI5 in the yeast Candida albicans was shown to affect virulence and morphogenesis (Kelly et al. 2004).
Fisher’s exact test for enrichment of GO terms on genes identified as affected by positive selection showed also an enrichment of functional categories related to mitochondrial functions. Most of the mitochondrial functions are encoded by nuclear genes. Mitochondrial bioenergetics has been recently proposed as a potential major force in adaptive evolution and even speciation processes through coevolution of mitochondrion and nuclear-encoded mitochondrial genes (Gershoni et al. 2009). In several fungal pathogens including Heterobasidion, it has been shown that mitochondria affect virulence (Olson and Stenlid 2001; Garbelotto et al. 2007; Shingu-Vazquez and Traven 2011).
SVs Harbored Genes Related to Mitochondrial Functions, TEs, and Nucleic Acid Binding
The presence of SVs disrupting microsynteny between the genomes of the two species might have lead to the creation of small genomic islands harboring ecologically important genes with higher rates of evolution, as it has been documented for genes involved in adaptation to temperature on Neurospora species (Ellison et al. 2011). The gene models detected in inversions, interchromosomal translocations, and regions affected by CNV events were characterized in silico by using BLAST2GO. In inverted regions, the main overrepresented GO terms were related to processes involved in protein targeting to mitochondrion and mitochondrial inner membrane peptidase complex, indicating that these SVs contain genes related to protein involved in mitochondrial factors. Among genes in regions affected by interchromosomal translocations events, 14 of 63 were annotated as encoding for Gag or capsid-related retrotransposon-related proteins. TEs promote chromosomal rearrangements more efficiently than other cellular processes, and TEs have been proposed as drivers of plant pathogen genome evolution due to the fact that they may mediate chromosomal rearrangements by ectopic recombination (Schmidt and Panstruga 2011). The documented proliferation of mobile elements in the genome of H. irregulare (Olson et al. 2012) and the presence of sequences related to retrotransposons in putative interchromosomal translocations indicated that the activity of TEs may have played a role in the differentiation of the H. irregulare genome from that of H. annosum. In addition, the overrepresented GO terms in interchromosomal translocations indicate that these SVs also contain genes related to mitochondrial functions.
Overrepresentation of genes related to nucleic acid binding proteins in regions affected by CNV events in H. annosum suggests that they might be involved in regulation of gene expression, acting as enhancers thanks to their redundancy. It could be inferred that H. irregulare may have lost this type of redundancy present in a common ancestor, or alternatively that H. annosum may have evolved subtelomeric and telomeric redundancy as a result of selective pressure events in its Eurasian ecological niche, in which it coexists with other competitive Heterobasidion species (Garbelotto and Gonthier 2013).
Genes Related to Pathogenicity Are More Conserved than Genes Involved in Saprobic Processes and Fruiting Body Formation
Olson et al. (2012) have shown a clear trade-off between H. irregulare gene families expressed during pathogenesis, saprobic wood decay, and fruiting. Thus, the higher saprobic growth and fruiting potential, demonstrated for H. irregulare compared with H. annosum (Garbelotto et al. 2010; Giordano et al. 2014), should be confirmed at the genomic level by the presence of substantial interspecific differences in genes involved in these two last functions. The presence of nonsynonymous SNPs, of SVs, and the dN/dS ratio in regions harboring genes related to pathogenesis, saprobic ability, and fruiting are in agreement with the documented phenotypic observations. In fact, the significant difference between dN/dS values of pathogenesis genes versus the entire gene data set suggests that these genes have undergone purifying selection and are conserved in both species. In contrast, genes related to saprobic ability and fruiting body formation appeared to have evolved at rates comparable to those of the entire gene data set. A more “relaxed” purifying selection pressure could be inferred for these genes that appear to have tolerated higher densities of nonsynonymous mutations compared with pathogenesis-related genes.
Conclusions
In conclusion, our comparative analyses provide insights on genomic differences between two closely related plant pathogens. SVs including inversions, translocations, and CNVs have played a prominent role in the creation of genomic islands leading to diversification of the two species. In addition, adaptive evolution might have remodeled their transcriptional and post-transcriptional machinery, their mitochondrial-related pathways, and their pattern of mobile elements. Genes encoding heme binding proteins and oxidoreductases exclusively found in regions showing high level of sequence variation might also have been influenced by adaptive processes. Finally, genes involved in sporulation and especially in saprobic growth appeared to be more variable between the two species, compared with genes involved in pathogenicity. This result provides genomic evidence that differences in fitness between the two species are likely to involve saprobic growth and sporulation, two functions positively affecting transmission but not directly involved in pathogenesis.
Further approaches, including postgenomic studies and phenotypic experiments, will allow to understand how specific molecular mechanisms may have evolved during the allopatric speciation of the two sister species. Transcriptomic approaches (i.e., RNA-sequencing) will be pivotal to verify whether transcriptional responses are species specific, thus confirming that allopatric speciation may have influenced the transcriptional and post-transcriptional activities of conserved genes between the two species.
Supplementary Material
Supplementary alignment S1, figure S1, and tables S1–S9 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Guglielmo Gianni Lione for help in statistical analyses and R scripting and to the anonymous reviewers for their helpful suggestions. This work was supported by the Italian Ministry of Education, University and Research, within the FIRB program (grant number RBFR128ONN).
Literature Cited
- Aguilar-Trigueros CA, Powell JR, Anderson IC, Antonovics J, Rillig MC. 2014. Ecological understanding of root-infecting fungi using trait-based approaches. Trends Plant Sci. 19:432–438. [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F, Díaz-Uriarte R, Dopazo J. 2004. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20:578–580. [DOI] [PubMed] [Google Scholar]
- Alkan C, Coe BP, Eichler EE. 2011. Genome structural variation discovery and genotyping. Nat Rev Genet. 12:363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au CH, et al. 2013. Rapid genotyping by low-coverage resequencing to construct genetic linkage maps of fungi: a case study in Lentinula edodes. BMC Res Notes. 6:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertels F, Silander OK, Pachkov M, Rainey PB, Nimwegen E. 2014. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol. 31:1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüthgen N, et al. 2005. Biological profiling of gene groups utilizing Gene Ontology. Genome Inform. 16:106–115. [PubMed] [Google Scholar]
- Brasier C. 2000. Plant pathology: the rise of the hybrid fungi. Nature 405:134–135. [DOI] [PubMed] [Google Scholar]
- Brasier CM, Kirk SA. 2010. Rapid emergence of hybrids between the two subspecies of Ophiostoma novo-ulmi with a high level of pathogenic fitness. Plant Pathol. 59:186–199. [Google Scholar]
- Carroll SB. 2000. Endless forms: the evolution of gene regulation and morphological diversity. Cell 101:577–580. [DOI] [PubMed] [Google Scholar]
- Chow EWL, Morrow CA, Djordjevic JT, Wood IA, Fraser JA. 2012. Microevolution of Cryptococcus neoformans driven by massive tandem gene amplification. Mol Biol Evol. 29:1987–2000. [DOI] [PubMed] [Google Scholar]
- Chuma I, et al. 2011. Multiple translocation of the AVR-pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 7:e1002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KE, Rinderer TE, Franck P, Quezada-Euán JG, Oldroyd BP. 2002. The africanization of honeybees (Apis mellifera L.) of the Yucatan: a study of a massive hybridization event across time. Evolution 56:1462–1474. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Dalman K, et al. 2013. A genome-wide association study identifies genomic regions for virulence in the non-model organism Heterobasidion annosum s.s. PLoS One 8:e53525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalman K, Olson Å, Stenlid J. 2010. Evolutionary history of the conifer root rot fungus Heterobasidion annosum sensu lato. Mol Ecol. 19:4979–4993. [DOI] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayrolles M, de Villechenon EP, Lonvaud-Funel A, Aigle M. 1997. Incidence of SUC-RTM telomeric repeated genes in brewing and wild wine strains of Saccharomyces. Curr Genet. 31:457–461. [DOI] [PubMed] [Google Scholar]
- Ellison CE, et al. 2011. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci U S A. 108:2831–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galardini M, Biondi EG, Bazzicalupo M, Mengoni A. 2011. CONTIGuator: a bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol Med. 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbelotto M, Gonthier P. 2013. Biology, epidemiology, and control of Heterobasidion species worldwide. Annu Rev Phytopathol. 51:39–59. [DOI] [PubMed] [Google Scholar]
- Garbelotto M, Gonthier P, Linzer R, Nicolotti G, Otrosina W. 2004. A shift in nuclear state as the result of natural interspecific hybridization between two North American taxa of the basidiomycete complex Heterobasidion. Fungal Genet Biol. 41:1046–1051. [DOI] [PubMed] [Google Scholar]
- Garbelotto M, Gonthier P, Nicolotti G. 2007. Ecological constraints limit the fitness of fungal hybrids in the Heterobasidion annosum species complex. Appl Environ Microbiol. 73:6106–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbelotto M, Linzer R, Nicolotti G, Gonthier P. 2010. Comparing the influences of ecological and evolutionary factors on the successful invasion of a fungal forest pathogen. Biol Invasions. 12:943–957. [Google Scholar]
- Garbelotto M, Ratcliff A, Bruns TD, Cobb FW, Otrosina WJ. 1996. Use of taxon-specific competitive-priming PCR to study host specificity, hybridization, and intergroup gene flow in intersterility groups of Heterobasidion annosum. Phytopathology 86:543–551. [Google Scholar]
- Gershoni M, Templeton AR, Mishmar D. 2009. Mitochondrial bioenergetics as a major motive force of speciation. Bioessays 31:642–650. [DOI] [PubMed] [Google Scholar]
- Giordano L, Gonthier P, Lione G, Capretti P, Garbelotto M. 2014. The saprobic and fruiting abilities of the exotic forest pathogen Heterobasidion irregulare may explain its invasiveness. Biol Invasions. 16:803–814. [Google Scholar]
- Gonthier P, et al. 2014. An integrated approach to control the introduced forest pathogen Heterobasidion irregulare in Europe. Forestry 87:471–481. [Google Scholar]
- Gonthier P, Garbelotto M. 2011. Amplified fragment length polymorphism and sequence analyses reveal massive gene introgression from the European fungal pathogen Heterobasidion annosum into its introduced congener H. irregulare . Mol Ecol. 20:2756–2770. [DOI] [PubMed] [Google Scholar]
- Gonthier P, Garbelotto M. 2013. Reducing the threat of emerging infectious diseases of forest trees—Mini Review. CAB Rev. 8:1–2. [Google Scholar]
- Gonthier P, Lione G, Giordano L, Garbelotto M. 2012. The American forest pathogen Heterobasidion irregulare colonizes unexpected habitats after its introduction in Italy. Ecol Appl. 22:2135–2143. [DOI] [PubMed] [Google Scholar]
- Gonthier P, Nicolotti G, Linzer R, Guglielmo F, Garbelotto M. 2007. Invasion of European pine stands by a North American forest pathogen and its hybridization with a native interfertile taxon. Mol Ecol. 16:1389–1400. [DOI] [PubMed] [Google Scholar]
- Gonthier P, Warner R, Nicolotti G, Mazzaglia A, Garbelotto MM. 2004. Pathogen introduction as a collateral effect of military activity. Mycol Res. 108:468–470. [DOI] [PubMed] [Google Scholar]
- Greiner S, Bock R. 2013. Tuning a ménage à trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. Bioessays 35:354–365. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PHYML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Guy L, Kultima JR, Andersson SGE. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26:2334–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington TC, Worrall JJ, Rizzo DM. 1989. Compatibility among host-specialized isolates of Heterobasidion annosum from western North America. Phytopathology 79:290–296. [Google Scholar]
- Hurst LD. 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486–487. [DOI] [PubMed] [Google Scholar]
- Jiang H, Guan W, Pinney D, Wang W, Gu Z. 2008. Relaxation of yeast mitochondrial functions after whole-genome duplication. Genome Res. 18:1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MT, et al. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 53:969–983. [DOI] [PubMed] [Google Scholar]
- Kofler R, Schlötterer C, Lelley T. 2007. SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23:1683–1685. [DOI] [PubMed] [Google Scholar]
- Kohn LM. 2005. Mechanisms of fungal speciation. Annu Rev Phytopathol. 43:279–308. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Hogues H, Whiteway M. 2009. Rearrangements of the transcriptional regulatory networks of metabolic pathways in fungi. Curr Opin Microbiol. 12:655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind M, van der Nest M, Olson Å, Brandström-Durling M, Stenlid J. 2012. A 2nd generation linkage map of Heterobasidion annosum s.l. based on in silico anchoring of AFLP markers. PLoS One 7:e48347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzer RE, et al. 2008. Inferences on the phylogeography of the fungal pathogen Heterobasidion annosum, including evidence of interspecific horizontal genetic transfer and of human-mediated, long-range dispersal. Mol Phylogenet Evol. 46:844–862. [DOI] [PubMed] [Google Scholar]
- Lockman B, Mascheretti S, Schechter S, Garbelotto M. 2014. A first generation Heterobasidion hybrid discovered in Larix lyalli in Montana. Plant Dis. 98:1003–1003. [DOI] [PubMed] [Google Scholar]
- MacDonald J1, Suzuki H, Master ER. 2012. Expression and regulation of genes encoding lignocellulose-degrading activity in the genus Phanerochaete. Appl Microbiol Biotechnol. 94(2):339–351. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 3:418–426. [DOI] [PubMed] [Google Scholar]
- Olson Å, et al. 2012. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 194:1001–1013. [DOI] [PubMed] [Google Scholar]
- Olson Å, Stenlid J. 2001. Plant pathogens: mitochondrial control of fungal hybrid virulence. Nature 411:438–438. [DOI] [PubMed] [Google Scholar]
- Otrosina WJ, Chase TE, Cobb FW, Jr, Korhonen K. 1993. Population structure of Heterobasidion annosum from North America and Europe. Can J Bot. 71:1064–1071. [Google Scholar]
- Otrosina WJ, Garbelotto M. 2010. Heterobasidion occidentale sp. nov. and Heterobasidion irregulare nom. nov.: a disposition of North American Heterobasidion biological species. Fungal Biol. 114:16–25. [DOI] [PubMed] [Google Scholar]
- Parker IM, Gilbert GS. 2004. The evolutionary ecology of novel plant-pathogen interactions. Annu Rev Ecol Syst. 35:675–700. [Google Scholar]
- Persoons A, et al. 2014. Patterns of genomic variation in the poplar rust fungus Melampsora larici-populina identify pathogenesis-related factors. Front Plant Sci. 5:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Kamoun S. 2012. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol. 10:417–430. [DOI] [PubMed] [Google Scholar]
- Raffaello T, Chen H, Kohler A, Asiegbu FO. 2014. Transcriptomic profiles of Heterobasidion annosum under abiotic stresses and during saprotrophic growth in bark, sapwood and heartwood. Environ Microbiol. 16:1654–1667. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, et al. 2000. Arabidopsis transcription factors: genome-wide comparative analysis among Eukaryotes. Science 290:2105–2110. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, et al. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216. [DOI] [PubMed] [Google Scholar]
- Schmidt SM, Panstruga R. 2011. Pathogenomics of fungal plant parasites: what have we learnt about pathogenesis? Curr Opin Plant Biol.. 14:392–399. [DOI] [PubMed] [Google Scholar]
- Shen Y-Y, et al. 2010. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc Natl Acad Sci U S A. 107:8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingu-Vazquez M, Traven A. 2011. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell. 10:1376–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33:W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH. 2013. Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. New Phytol. 199:895–907. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH, Croll D. 2014. The evolving fungal genome. Fungal Biol Rev. 28:1–12. [Google Scholar]
- Swain MT, et al. 2012. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nat Protoc. 7:1260–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, et al. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 31:21–32. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Li H, Johnson AD. 2008. Evolution of eukaryotic transcription circuits. Science 319:1797–1799. [DOI] [PubMed] [Google Scholar]
- Turelli M, Barton NH, Coyne JA. 2001. Theory and speciation. Trends Ecol Evol. 16:330–343. [DOI] [PubMed] [Google Scholar]
- Xie C, Tammi MT. 2009. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitouni B, et al. 2010. SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics 26:1895–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.