Abstract
Glyceridae (Annelida) are a group of venomous annelids distributed worldwide from intertidal to abyssal depths. To trace the evolutionary history and complexity of glycerid venom cocktails, a solid backbone phylogeny of this group is essential. We therefore aimed to reconstruct the phylogenetic relationships of these annelids using Illumina sequencing technology. We constructed whole-genome shotgun libraries for 19 glycerid specimens and 1 outgroup species (Glycinde armigera). The chosen target genes comprise 13 mitochondrial proteins, 2 ribosomal mitochondrial genes, and 4 nuclear loci (18SrRNA, 28SrRNA, ITS1, and ITS2). Based on partitioned maximum likelihood as well as Bayesian analyses of the resulting supermatrix, we were finally able to resolve a robust glycerid phylogeny and identified three clades comprising the majority of taxa. Furthermore, we detected group II introns inside the cox1 gene of two analyzed glycerid specimens, with two different insertions in one of these species. Moreover, we generated reduced data sets comprising 10 million, 4 million, and 1 million reads from the original data sets to test the influence of the sequencing depth on assembling complete mitochondrial genomes from low coverage genome data. We estimated the coverage of mitochondrial genome sequences in each data set size by mapping the filtered Illumina reads against the respective mitochondrial contigs. By comparing the contig coverage calculated in all data set sizes, we got a hint for the scalability of our genome skimming approach. This allows estimating more precisely the number of reads that are at least necessary to reconstruct complete mitochondrial genomes in Glyceridae and probably non-model organisms in general.
Keywords: Glyceridae, venomous annelids, mitogenomics, whole-genome shotgun sequencing, sequencing coverage, group II introns
Introduction
Glyceridae Grube, 1850 (Annelida) are a group of venomous annelids that possess an eversible pharynx bearing four cross arranged teeth that are connected to venom glands (Ehlers 1868; Fauchald and Rouse 1997; Wolf 1977). Venom systems evolved several times independently in the animal kingdom and serve predominantly for predation, defense, and competition (Fry et al. 2009; Casewell et al. 2013; von Reumont, Campbell, and Jenner 2014). Earlier studies indicated that the venom of Glycera tridactyla includes an unusual neurotoxin, namely α-Glycerotoxin (GLTx), which is able to upregulate presynaptic Cav2.2 channels (N-type Ca2+ channels) (Meunier et al. 2002) and that venom of Glycera dibranchiata comprises components able to induce ion-permeable pores in lipid bilayers (Kagan et al. 1982). A recent computational study elucidated a complex mixture of transcripts representing known toxin classes as well as Glycera-specific ones by analyzing venom gland transcriptomes of three different glycerid species (von Reumont, Campbell, Richter, et al. 2014). To allow an extended investigation and understanding of the venom evolution in this group, a solid backbone phylogeny of Glyceridae is needed.
At present, Glyceridae comprises 46 valid species (Böggemann 2014). It is a group of worldwide distributed annelids, consisting of the three genera Glycera, Glycerella, and Hemipodia, distinguishable by concise genus-specific morphological details as reviewed in Böggemann (2002). So far, only few phylogenetic studies exist which aim to investigate the relationships within Glyceridae and most of them are solely based on morphological characters (e.g., Böggemann 2002, 2006). These analyses propose the monophyly of Glyceridae and support furthermore a sister group relationship between Glyceridae Grube, 1850, and Goniadidae Kinberg, 1866, which are unified as Glyceriformia Fauchald, 1977 (Pleijel 2001). Unfortunately, glycerid species share a rather uniform morphology, thereby hampering the ability of morphological studies to distinguish different morphological characters resulting mostly in low node support (Böggemann 2002). We aim to overcome this problem by reconstructing the phylogeny of Glyceridae on a molecular level using mitochondrial (and nuclear) target genes, which had already been proven informative in other phylogenetic (e.g., Botero-Castro et al. 2013; Gillett et al. 2014; Williams et al. 2014) and phylogeographic studies (e.g., Morin et al. 2010). Especially the genes of the mitochondrial genome, known to harbor higher substitution rates compared with the slower evolving nuclear genes (Curole and Kocher 1999), are well-suited for resolving phylogenies of different taxonomic levels and young radiations, respectively.
Traditional approaches of complete mitochondrial genome sequencing usually followed protocols involving random sequencing of clones resulting from fragmentation of mitochondrial DNA and blunt-end cloning (Burger et al. 2007) or long-range polymerase chain reactions (PCRs) and subsequent sequencing through primer walking (e.g., Helfenbein et al. 2001; Dreyer and Steiner 2004; Bleidorn et al. 2006). The generation of primers requires prior sequence information and was thus a comparatively time- and also cost-intensive strategy to reveal complete mitochondrial genomes, especially of distantly related species. The advent of next-generation sequencing (NGS) techniques leveraged the sequencing of mitochondrial genomes. An improvement concerning the sequencing depth of mitochondrial target genes was reached by sequencing longer amplicons through NGS (e.g., Lloyd et al. 2012) or the inclusion of an enrichment step for mitochondrial DNA in the case of low-quality DNA (e.g., Maricic et al. 2010; Horn et al. 2011; Winkelmann et al. 2013). The enrichment step was conducted through generated baits (laboratory baits) whose generation is dependent once again, upon knowledge of closely related reference genomes. Nowadays, enrichment techniques are no longer restricted to mitochondrial genes, but rather expanded to numerous other loci of interest for their implementation in phylogenetic analyses (e.g., Lemmon et al. 2012; Peñalba et al. 2014). In contrast, bait sequences (bioinformatic baits) serve to identify complete mitochondrial genomes in a mixed pool of untagged samples that were sequenced and assembled together (e.g., Rubinstein et al. 2013; Gillett et al. 2014). However, these approaches usually need prior knowledge of reference sequences. Particularly when working with non-model organisms, the access to a priori sequence information can be difficult and time-intensive. Thus, more and more approaches/pipelines were developed to recover complete mitochondrial genomes directly from whole-genome shotgun (WGS) sequencing data (Botero-Castro et al. 2013; Hahn et al. 2013; Lavrov et al. 2013; Williams et al. 2014; Li et al. 2015) using reference-independent assembly strategies and species-specific tagging of samples.
Here, we are following such an approach to reconstruct a robust glycerid backbone phylogeny. We generated WGS data for 19 glycerid specimens and 1 outgroup species. The phylogenetic analyses are based on multigene analyses of different data set sizes targeting 15 mitochondrial genes (atp6, atp8, cox1, cox2, cox3, cytb, nad1, nad2, nad3, nad4, nad4l, nad5, nad6, sRNA, and lRNA) and 4 loci from the nuclear ribosomal cluster (18SrRNA, 28SrRNA, ITS1, and ITS2). To test the influence of the sequencing depth on the mitochondrial target genes, we generated reduced data sets (10 million, 4 million, and 1 million reads) and analyzed the recovery of mitochondrial genomes per data set size. A comparison of the contig coverage elucidated in all data sets gave us an idea for the scalability of our genome skimming approach. Consequently, we will be able to estimate the number of WGS reads necessary for reconstructing complete mitochondrial genomes in Glyceridae and probably also other non-model organisms.
Materials and Methods
Library Reconstruction, Illumina Sequencing, and Processing
WGS libraries were constructed for 19 glycerid specimens and 1 outgroup species (Glycinde armigera, FS17) (table 1). Genomic DNA was sheared through sonication for 130 s with the focused-ultrasonicator Covaris S2 (Covaris, Inc., Woburn, MA) to generate fragments with a predominant length of 250 bp. To evaluate length distributions as well as the amount of sheared DNA fragments, the samples were run on a High Sensitivity DNA Chip using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) (Panaro et al. 2000). Beginning with blunt-end repair, the Illumina libraries were processed according to the Illumina multiplex protocol of Meyer and Kircher (2010). Afterwards, double-indexed libraries (Kircher et al. 2012) were pooled and sequenced on one lane of the HiSeq 2500 (Illumina, San Diego, CA) at the Max Planck Institute for Evolutionary Anthropology (Leipzig, Germany). The generated paired-end reads (96 bp) were sorted according their indices, adapters were clipped, and base calling was conducted using freeIbis (Renaud et al. 2013). Overlapping paired-end reads were trimmed and merged to a single sequence (Renaud et al. 2014), hereinafter referred to as single reads. The library of G. tridactyla (Glytri) was processed with the same Illumina multiplex protocol (Meyer and Kircher 2010), but solely consists of single reads (75 bp).
Table 1.
List of Glycerid Species (Glyceridae, Annelida) and One Outgroup Species Glycinde armigera (FS17) for Which WGS Libraries Were Constructed
| Species | Origin of Species | Labcode | Accession |
|---|---|---|---|
| Glycera americana | Drakes Bay, 45 m depth, CA, coll. April 2003 | FS01 | |
| Glycera americana | Barnstable Harbor, MA, coll. September 2001 | FS12 | KT989321 |
| Glycera americana | Tampa Bay, FL, coll. March 2013 | FS23 | KT989330 |
| Glycera capitata | Bamfield, Vancouver Island, BC, Canada, coll. March 2008 | FS10 | KT989319 |
| Glycera capitata | White Sea Biological Station, Russia, coll. July 2010 | FS11 | KT989320 |
| Glycera cf. capitata | Antarctica, Long 2°59.33′ W, Lat 62°0.64′ S, coll. December 2007 | FS09 | |
| Glycera dibranchiata | Wellfleet/Loagy Bay, MA, coll. September 2001 | FS05 | KT989318 |
| Glycera fallax | Roscoff, France, coll. April 2011 | FS14 | KT989323 |
| Glycera lapidum | Baie de Morlaix, France, coll. June 2012 | FS06 | |
| Glycera lapidum | Baie de Morlaix, France, coll. June 2012 | FS07 | |
| Glycera nicobarica | Asamushi, Japan, coll. August 2012 | FS22 | |
| Glycera oxycephala? | Monterey Bay, CA, coll. March 2003 | FS21 | KT989329 |
| Glycera sp. | Antarctica, Long 0°01.12′ W, Lat 52°01.98′ S, coll. December 2007 | FS08 | |
| Glycera tesselata | Banyuls-sur-Mer, France, coll. November 2003 | FS18 | KT989326 |
| Glycera tridactyla | Roscoff, France, coll. April 2010 | Glytri | KT989331 |
| Glycera cf. tridactyla | Eilat, Israel, coll. March 2011 | FS19 | KT989327 |
| Glycera cf. tridactyla | Saint-Efflam, France, coll. April 2011 | FS20 | KT989328 |
| Glycera unicornis | Banyuls-sur-Mer, France, coll. November 2003 | FS15 | KT989324 |
| Glycinde armigera | Bellingham Bay, WA, coll. August 2002 | FS17 | KT989325 |
| Hemipodia simplex | Bamfield, Vancouver Island, BC, Canada, coll. March 2008 | FS13 | KT989322 |
Note.—The sequences of complete mitochondrial genomes have been deposited at GenBank under the mentioned accession numbers.
Filtering of Sequencing Data
The Illumina reads were filtered with ConDeTri v.2.2 (Smeds and Künstner 2011) to eliminate low-quality reads (supplementary table S1, Supplementary Material online). Thereby, only reads of which 93.75% of the nucleotides have a PHRED score (Ewing et al. 1998; Ewing and Green 1998) above 15 (filter 15) were kept for further analyses.
Generation of Reduced Data Sets
In addition to the original data sets, reduced data sets consisting of 10 million, 4 million, and 1 million reads were analyzed to perform coverage studies (fig. 1). The reduced data sets were constructed using a Python script for “random sampling fastq files” downloaded from hitseq (subsampler.py, http://www.hitseq.com/forum/topic/13, last accessed December 2, 2015). The filtered reads of the original data sets were randomly pruned to data set sizes of 10 million, 4 million, and 1 million reads. Per species and data set size, ten replicates were generated. To allow comparable coverage studies, reduced data sets are constructed for libraries in which the complete mitochondrial genome is represented by a single contig in the original data sets (FS05, FS10, FS11, FS12, FS13, FS14, FS15, FS17, FS18, FS19, FS20, FS21, FS23, Glytri). The original data sets of FS20 and FS21 comprised less than 10 million reads (supplementary table S1, Supplementary Material online), which is why these libraries were only reduced to data set sizes of 4 million and 1 million reads.
Fig. 1.—
Overview of the methodical approaches focused in this study. The studied data sets comprise the original and reduced data sets (10 million, 4 million, and 1 million reads) (boxes marked in green). To resolve the regarding scientific questions (boxes marked in red), several methodical approaches were used (boxes marked in white).
Sequence Assembly
The filtered sequence reads of the original (supplementary table S1, Supplementary Material online) and reduced data sets were assembled de novo using IDBA-UD v.1.1.1 (Peng et al. 2012). IDBA-UD assemblies are constructed using an initial k-mer size of 20, an iteration size of 5, and a maximum k-mer size of 60.
Searching for Target Genes
The assemblies of the original data sets were screened for mitochondrial and nuclear target genes using the Basic Local Alignment Search Tool v.2.2.28+ (Altschul et al. 1997; Zhang et al. 2000). First, a contig comprising all mitochondrial genes had been identified in G. tridactyla by BLAST-searches utilizing the mitochondrial genes of Platynereis dumerilii (GenBank accession NC_000931.1) as query. Subsequently, the mitochondrial genes (atp6, atp8, cox1, cox2, cox3, cytb, nad1, nad2, nad3, nad4, nad4l, nad5, nad6, sRNA, and lRNA) of G. tridactyla (Glytri) were annotated using the MITOS webserver (Bernt et al. 2013) and served as reference for BLAST-searches in all other glycerid databases as well as in Glyci. armigera (FS17). Ribosomal candidate genes (5.8SrRNA, 18SrRNA, 28SrRNA, ITS1, and ITS2) were screened through BLAST in G. tridactyla (Glytri) using published nuclear genes of Glycera americana 28SrRNA (EU418864.1), Proceraea cornuta 5.8SrRNA (AF212165.1), and G. americana 18SrRNA (EU418856.1) as reference. Afterwards, annotated ribosomal candidate genes identified in G. tridactyla (Glytri) were used as query for BLAST-searches in all other glycerid databases as well as in Glyci. armigera. Protein-coding genes were searched with tBLASTx, whereas BLASTn was used for ribosomal mitochondrial genes and the nuclear target loci. All contigs with an e value below 1e−5 were used for phylogenetic reconstructions.
Phylogenetic Analyses
Phylogenetic reconstructions are based on the original data sets of 19 glycerid specimens and Glyci. armigera (FS17) (table 1 and supplementary table S1, Supplementary Material online). To avoid frameshift errors in the nucleotide sequences during alignment step, the deduced amino acid sequences of the protein-coding genes were aligned and subsequently retranslated using ClustalW implemented in BioEdit v.7.1.11 (Hall 1999). However, the nucleotide sequences of the ribosomal mitochondrial genes and the nuclear ribosomal cluster were aligned with the L-INS-i option of Mafft v.7.130b (Katoh et al. 2002) (alignments see supplementary data set S1, Supplementary Material online). Additionally, third codon positions of protein-coding genes were deleted in all alignments using the R package APE (Paradis et al. 2004) to test the influence of this position on our phylogenetic analyses. Phylogenetic analyses of different data sets (see below) were conducted with maximum likelihood (ML) using RAxML v.8.0.5 (Stamatakis 2014) and Bayesian inference (BI) using PhyloBayes MPI v.1.5a (Lartillot et al. 2009; Lartillot et al. 2013). The ML phylogenies (raxmlHPC-PTHREADS-AVX -f b -m GTRGAMMAI -q partition -N1000) represent the best-obtained tree for each data set under a GTR + GAMMA + I substitution model. Bootstrap values were determined from 1,000 pseudoreplicates. For Bayesian inference, 50% majority rule consensus trees were obtained from two independent runs per data set (CAT-GTR; 30,000 generations each, burn-in 5,000 each). The data sets comprise 1) the 13 protein-coding mitochondrial genes (atp6, atp8, cox1, cox2, cox3, cytb, nad1, nad2, nad3, nad4, nad4l, nad5, and nad6) including and excluding the third codon position of the protein-coding genes, 2 ribosomal mitochondrial genes (sRNA and lRNA), and 4 loci from the nuclear ribosomal cluster (18SrRNA, 28SrRNA, ITS1, and ITS2), hereafter referred to as MLall/BIall (with and without third position); 2) the 15 mitochondrial genes including and excluding the third codon position of the protein-coding genes, hereafter referred to as MLmt/BImt (with and without third position); 3) the 13 protein-coding mitochondrial genes including and excluding the third codon position of the protein-coding genes, 2 ribosomal mitochondrial genes (sRNA and lRNA) and 18SrRNA as well as 28SrRNA, hereafter referred to as MLmt + nucl/BImt + nucl (with and without third position); 4) four loci from the nuclear ribosomal cluster (18SrRNA, 28SrRNA, ITS1, and ITS2), hereafter referred to as MLnucl + ITS/BInucl + ITS; and 5) 18SrRNA and 28SrRNA, hereafter referred to as MLnucl/BInucl, respectively (table 2). The data sets were partitioned by gene when analyzing multiple genes from either the mitochondrial or nuclear genome (MLmt/BImt, MLnucl + ITS/BInucl + ITS, and MLnucl/BInucl). In analyses comprising both, mitochondrial and nuclear genes, the data sets were subdivided in only two partitions accordingly (MLall/BIall, MLmt + nucl/BImt + nucl). The uncorrected (p) genetic distances were calculated using DAMBE v.5.6.7 (Xia 2013) for the data set comprising both, the 15 mitochondrial genes and the 4 nuclear loci (MLall/BIall, with third position), and for the data set comprising only the 4 nuclear loci (MLnucl + ITS/BInucl + ITS) (supplementary table S2, Supplementary Material online). The outgroup taxa were Lumbricus terrestris, Nephtys sp., Nephtys incisa, Orbinia latreillii, Orbinia swani, P. dumerilii, Sipunculus nudus, Terebellides stroemii (supplementary table S3, Supplementary Material online), and Glyci. armigera (FS17). Another ML analysis (GTR + GAMMA + I; 1,000 pseudoreplicates) was performed for the MLall (with third position) data set which additionally included 125 cox1 sequences of Glyceridae and Goniadidae published in National Center for Biotechnology Information (NCBI) (alignment see supplementary data set S1, Supplementary Material online). All phylogenetic trees were visualized and edited with iTOL (Letunic and Bork 2007).
Table 2.
Data Sets Used for Maximum likelihood analyses (ML) and Bayesian Inference (BI)
| Data Set | Loci | Third Position | Alignment Positions (bp) |
|---|---|---|---|
| MLall/BIall | mt, 18S, 28S, ITS1, ITS2 | Yes | 19,991 |
| MLall/BIall | mt, 18S, 28S, ITS1, ITS2 | No | 16,274 |
| MLmt + nucl/BImt + nucl | mt, 18S, 28S | Yes | 18,802 |
| MLmt + nucl/BImt + nucl | mt, 18S, 28S | No | 15,085 |
| MLmt/BImt | mt | Yes | 13,270 |
| MLmt/BImt | mt | No | 9,553 |
| MLnucl + ITS/BInucl + ITS | 18S, 28S, ITS1, ITS2 | — | 6,721 |
| MLnucl/BInucl | 18S, 28S | — | 5,532 |
Note.—The third codon positions of the protein-coding genes were either included or excluded from the analyses. mt, mitochondrial genome, comprising the 13 protein-coding mitochondrial genes and the 2 ribosomal mitochondrial genes (sRNA and lRNA).
Determination of the Mitochondrial Gene Order
Gene order annotations were performed for libraries in which all mitochondrial target genes were recovered on a single contig (hereafter referred to as “mitocontig”) (13 × Glyceridae, 1 × Goniadidae, see table 1 for accession numbers). First, it has been tested whether the mitocontigs represent complete mitochondrial genomes (hereafter referred to as “mitogenomes”) by screening for identical sequence parts in the 3′- and 5′-end of each mitocontig which allowed for closing the circularly organized mitogenome. Afterwards, mitocontigs were uploaded to the MITOS webserver (revision 567, 2014-08-25) (Bernt et al. 2013). Mitochondrial genomes had been automatically annotated with default settings using the invertebrate genetic code for mitochondria. Mitochondrial gene orders were visualized using Circos v.0.67 (Krzywinski et al. 2009) (for Circos configuration files, see supplementary data set S2, Supplementary Material online).
Composition of Mitochondrial Genomes
To measure the strand-specific bias of nucleotide composition, the AT and GC skews were calculated according to the formula AT skew = (A − T)/(A + T) and GC skew = (G − C)/(G + C) (Perna and Kocher 1995). Calculations were performed for the complete mitochondrial genome (13 × Glyceridae, 1 × Goniadidae) and separately only for the 13 protein-coding genes, 2 ribosomal RNAs (rRNAs) (sRNA and lRNA), 22 transfer RNAs (tRNAs), and group II introns (see supplementary data set S3, Supplementary Material online). Moreover, codon usage in the 13 protein-encoding genes and the putative secondary structures of the 22 inferred tRNAs were analyzed. Gene annotation was conducted using the MITOS webserver (revision 671, 2015-05-05) with default settings and the invertebrate genetic code for mitochondria.
Phylogenetic Analyses of Group II Introns
Two group II introns of Glycera fallax (FS14) (I1 and I2) and one of Glycera unicornis (FS15) were analyzed together with the alignment of Vallès et al. (2008) which built upon an analysis of Zimmerly et al. (2001). The deduced amino acid sequences were aligned with the L-INS-i option of Mafft v.7.204. Afterwards, uninformative positions were removed using Gblocks v.0.91b (Castresana 2000; Talavera and Castresana 2007) and the best-fitting substitution model (LG + I + G + F) was determined with ProtTest v.3.4 (Guindon and Gascuel 2003; Darriba et al. 2011) (alignment see supplementary data set S1, Supplementary Material online). Finally, an ML analysis was conducted with RAxML v.8.0.5 (raxmlHPC-PTHREADS-AVX -f a -m PROTGAMMAILGF -N1000). Bootstrap support was determined from 1,000 pseudoreplicates.
Coverage Studies
Coverage studies were performed for libraries in which the complete mitochondrial genome was represented by a single contig obtained from the original data sets (FS05, FS10, FS11, FS12, FS13, FS14, FS15, FS17, FS18, FS19, FS20, FS21, FS23, Glytri). Hence, the studied data sets comprise 14 original data sets (13 × Glyceridae, 1 × Goniadidae) and 400 corresponding reduced data sets as ten replicates per data set size (10 million, 4 million, and 1 million reads) were analyzed. The sequencing coverage (C), hereafter referred to as “contig coverage,” has been calculated in the original data sets as well as the corresponding reduced data sets according to the Lander/Waterman equation: C = LN/G (Lander and Waterman 1988). Thereby L describes the read length (96/75 bp) and N the number of reads which mapped to the mitocontig. Mapping was conducted with segemehl v.0.2.0 (Hoffmann et al. 2009; Hoffmann et al. 2014). The number of mapped reads was derived from the output files (SAM-files) using a common bash command (wc -l). The mitocontig length recovered from each of the corresponding original data sets served as reference length (G) in the above-mentioned formula. To further allow comparison of different data set sizes of one specimen, the contig coverage was normalized to 1 million reads (referred to as “cov per million”; supplementary data set S4, Supplementary Material online). To test the influence of the data set size on the recovery of complete mitochondrial genomes, the relative size of the mitochondrial genome (referred to as “relative mitosize”) and the number of contigs representing a mitochondrial genome (referred to as “contig count”) had been determined for each data set (supplementary data set S4, Supplementary Material online). Therefore, for all IDBA-UD assemblies (see “Sequence Assembly” in the Materials and Methods section) BLAST-searches were executed (BLASTn, e value: 1e−50, output format: -outfmt 6) in which the species-specific reference mitocontig recovered from the original data sets served as query. The number of contigs representing a complete or broken mitochondrial genome was ascertained using common bash commands (awk, sort -u, wc -l). To calculate the relative mitosize, the cumulative length of the retained contigs or the length of a single mitocontig was compared with the length of the complete mitochondrial reference genome (equal to 100%) which was determined in each of the original data sets. Based on ten replicates constructed for each specimen per data set size (10 million, 4 million, and 1 million), mean values and standard deviations (SD) were calculated for the following parameters: Coverage per 1 million reads (cov per million), contig coverage, relative mitosize, contig count, and the number of mapped reads (referred to as “read count”) (supplementary data set S4, Supplementary Material online). Calculations were performed with Microsoft Excel 2010.
Results
Genome Sequencing
WGS libraries of 19 glycerid specimens and Glyci. armigera (FS17) were constructed. After filtering (filter 15), the number of Illumina reads obtained varied from 4,179,924 reads (FS01) to mostly about 16 million reads to 35,587,134 reads (Glytri) (supplementary table S1, Supplementary Material online). Four libraries (FS01, FS20, FS21, and FS22) consist of less than 10 million reads. Furthermore, for FS05, FS06, FS07, FS08, and FS09, the quality of DNA available for library construction was low and/or the number of molecules per microliter revealed by quantitative real-time PCR during library preparation was remarkably lower than in the remaining glycerid libraries. These samples contain only 106–107 molecules per microliter in contrast to the remaining samples which contain 109–1010 molecules per microliter. In the low-quality libraries FS06, FS07, FS08, FS09, and also in FS01 and FS22, the mitochondrial genome found by BLAST-searches is broken in several contigs. However, in all other 13 glycerids and the outgroup species Glyci. armigera (FS17) BLAST-searches revealed a single contig comprising almost all expected mitochondrial genes. Finally, we were able to determine the complete mitochondrial nucleotide sequence and gene arrangement of 14 specimens. In eight glycerid specimens, we could close the circular mitochondrial genome (FS05, FS10, FS11, FS13, FS18, FS19, FS20, and Glytri). In the other six specimens (FS12, FS14, FS15, FS17, FS21, and FS23), the mitochondrial genome is broken within a noncoding unknown region (UNK, equivalent to the control region in vertebrates) (cf. supplementary fig. S1, Supplementary Material online). The analyzed mitochondrial genomes are around 15,500 bp in size, except of G. fallax (FS14) and G. unicornis (FS15) which are exhibiting a size of approximately 20,600 bp (supplementary fig. S1, Supplementary Material online).
Among Glyceridae, the mitochondrial genomes show similar values concerning AT content, AT skew, and GC skew. The mean AT content of the complete mitochondrial genome calculated for 13 glycerid taxa is 65.95% (fig. 2A). The AT skew is slightly negative (mean AT skew of −0.021, fig. 2B) and ranges between values of −0.097 (FS19) and 0.057 (FS14) (supplementary data set S3, Supplementary Material online). Different from that, the complete (+)-strand genome sequence is enriched for cytosine as the average GC skew is negative (mean GC skew of −0.246). The GC skew is most prominent at the third codon position (mean GC skew of −0.623) (cf. fig. 2C and supplementary data set S3, Supplementary Material online). Noticeably, the AT skew for group II introns of G. fallax (FS14) and G. unicornis (FS15) is highly positive inferring more A versus T (values of 0.250 and 0.220 in FS14, value of 0.168 in FS15), whereas the GC skew is as negative as in the remaining mitogenome (cf. supplementary data set S3, Supplementary Material online). The mitochondrial genome of the outgroup species Glyci. armigera (FS17) has a higher AT content of 68.54%, an AT skew of −0.105, and a GC skew of −0.182 (cf. fig. 2A–C). Generally, the biased base composition within the mitochondrial genome is congruent with a biased codon usage (cf. supplementary data set S3, Supplementary Material online). In the 13 studied glycerid taxa and the outgroup species Glyci. armigera (FS17), all typical 22 tRNAs were detected (cf. supplementary fig. S2, Supplementary Material online). In two species, one loop of the trnR is modified (FS21) or missing (FS15). Although the tRNAs of the G. americana specimens (FS12 and FS23) and Glycera capitata (FS10 and FS11) each show similar structures, more differences are present for G. tridactyla (Glytri), Glycera cf. tridactyla (Eilat, ISR, FS19), and Glycera cf. tridactyla (Saint-Efflam, FRA, FS20). However, the 22 inferred tRNAs show no clade-specific features which could be putatively phylogenetically informative (cf. supplementary fig. S2, Supplementary Material online).
Fig. 2.—
Nucleotide composition of complete mitochondrial genomes in 13 glycerid taxa (mean values ± standard deviations) and the outgroup species Glycinde armigera (FS17). (A) AT content, (B) AT skew, (C) GC skew. The values are shown for the complete mitochondrial genome (Genome), the 13 protein-coding genes considering all codon positions (all) as well as only the first and second (1 + 2) and the third (3rd) codon position, the 2 rRNAs and 22 tRNAs (tRNA), respectively. Note the disparity in the outgroup species for most of the parameters.
The analyzed taxa show different gene orders, three of them within Glyceridae, which vary in the position of the tRNAs trnL1, trnS2, and trnA (fig. 3A). A further rearrangement in tRNA position could be observed in Glycera tesselata (FS18). In this species, the trnQ, which is located between the genes cox3 and nad6 in all other analyzed glycerid specimens and Glyci. armigera (FS17), is translocated to a position between the genes rrnL and nad1 (fig. 3A). Surprisingly, gene order annotation revealed two group II introns (I1 and I2) inside the cox1 gene of G. fallax (FS14) and one group II intron inside the cox1 gene of G. unicornis (FS15) (fig. 3B). Each group II intron contains the typical starting (GTGCG) and ending sequence (AC). Moreover, Pfam-A searches (Finn et al. 2014) determined one open reading frame (ORF) for a reverse transcriptase and a type II intron maturase per intron. Interestingly, the group II intron (I2) of G. fallax (FS14) and the one of G. unicornis (FS15) start at the exactly same position (directly after position 700) of the coding sequence (CDS) of the cox1 gene (fig. 3B). An ML analysis (RAxML, LG + I + G + F; 1,000 pseudoreplicates) revealed a sister group relationship between the group II intron (I2) of G. fallax (FS14) and a group II intron identified in the annelid species Nephtys sp. (see Vallès et al. 2008), and both occur as sister to the group II intron of G. unicornis (FS15) (supplementary fig. S3, Supplementary Material online). Contrary, the group II intron (I1) of G. fallax (FS14), which starts directly after position 184 of the CDS of the cox1 gene, does not cluster together with the above-mentioned annelid group II introns. Apart from this, gene order annotation revealed an approximately 2,988-bp-long intergenic region between the genes nad3 and nad2 in G. unicornis (FS15) (fig. 3C). The coverage for the group II introns as well as for the intergenic region was as high as for the remaining mitochondrial genome implicating that these regions are indeed part of the mitochondrial genome rather than assembly artefacts of nuclear integrations.
Fig. 3.—
Mitochondrial gene orders and group II introns within Glyceridae. (A) Mitochondrial gene order arrangements of the complete mitochondrial genome of (Δ) the outgroup species Glycinde armigera (FS17), (○) the glycerid species Glycera tesselata (FS18), (•) Glycera dibranchiata (FS05), Glycera tridactyla (Glytri), Glycera cf. tridactyla (FS19), Glycera cf. tridactyla (FS20) (cf. clade 1 in figs. 4 and 5), and (▪) Glycera americana (FS12), G. americana (FS23), Glycera capitata (FS10), G. capitata (FS11), Glycera fallax (FS14), Glycera oxycephala? (FS21), Glycera unicornis (FS15), and Hemipodia simplex (FS13) (cf. clades 2 and 3 in figs. 4 and 5). (B) Group II introns identified inside the cox1 gene of G. fallax (FS14) (I1 and I2) and G. unicornis (FS15). Note that the group II intron (I2) of G. fallax and the group II intron of G. unicornis start at the exactly same position (directly after CDS position 700) of the CDS of the cox1 gene. (C) Intergenic region of approximately 2,988 bp located between the genes nad3 and nad2 in G. unicornis (FS15).
Phylogenetic Reconstructions
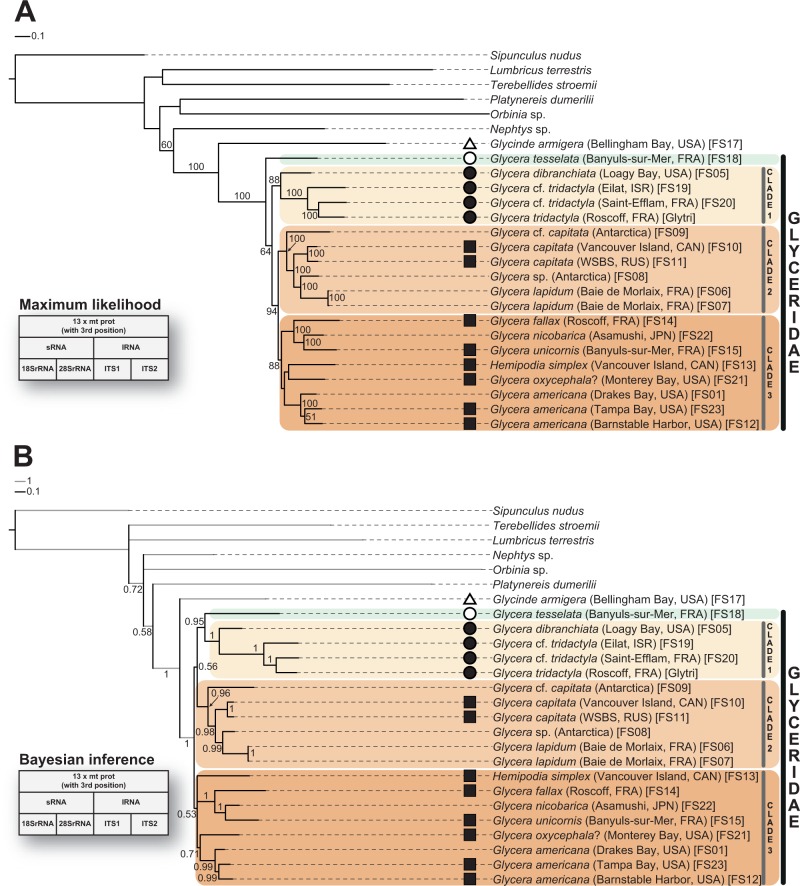
Partitioned ML analyses (RAxML, GTR + GAMMA + I; 1,000 pseudoreplicates) and Bayesian inference (CAT-GTR; 30,000 generations each, burn-in 5,000 each) resolved a quite robust glycerid phylogeny. The alignment, which contains 15 mitochondrial genes (including the third codon position of the protein-coding genes) and 4 nuclear loci (18SrRNA, 28SrRNA, ITS1, and ITS2), comprises approximately 20,000 base positions with almost no missing data. First, ML and Bayesian analyses congruently recover a highly supported sister group relationship of Glyceridae Grube, 1850 and Goniadidae Kinberg, 1866 (figs. 4 and 5). Second, both approaches congruently resolve the maximally supported monophyly of Glyceridae and reveal three monophyletic clades (clades 1–3) within Glyceridae (figs. 4 and 5). In the ML analysis MLall (with third position), clade 1 occurs as sister to clade 2 + clade 3, and G. tesselata (FS18) emerges as earliest branching taxon forming the sister to all other analyzed glycerid specimens (fig. 4A). Clade 1 comprises four glycerid species, namely G. dibranchiata (FS05) and three genetically different OTUs of the G. tridactyla morphotype (FS19, FS20, and Glytri) from different localities. Glycera dibranchiata (FS05) appears as sister to the remaining three glycerid species including the GLTx producing G. tridactyla (Glytri). The monophyly of clade 2 is fully supported and also the nodes within clade 2 are supported by bootstrap values of 100%. This clade consists of genetically different G. capitata morphotypes (FS09, FS10, and FS11) from different localities, an undescribed Glycera sp. (FS08) from Antarctica, and Glycera lapidum (FS06 and FS07). Clade 3 includes G. fallax (FS14), Glycera nicobarica (FS22), G. unicornis (FS15), Hemipodia simplex (FS13), Glycera oxycephala? (FS21) and three genetically different G. americana morphotypes (FS01, FS12, and FS23) from different localities. Within clade 3, G. fallax (FS14), G. nicobarica (FS22), and G. unicornis (FS15) form a maximally supported monophyletic group in which G. fallax emerges as sister to the two other glycerid species (fig. 4A). The mitochondrial gene order of all analyzed glycerid species differs from that of the studied outgroup species Glyci. armigera (FS17). Glycera tesselata (FS18) shows a species-specific mitochondrial gene order, and the species of clade 1 harbor a common gene order. Moreover, the species of clades 2 and 3 share the same gene order arrangements (figs. 3A and 4A) which are identical to the recently published mitochondrial gene order of Goniada japonica (KP867019.1).
Fig. 4.—
Phylogeny of Glyceridae based on ML and Bayesian inference for a data set comprising the 13 protein-coding mitochondrial genes, 2 ribosomal mitochondrial genes, and four loci from the nuclear ribosomal cluster (18SrRNA, 28SrRNA, ITS1, and ITS2). The data set includes the third codon position of the protein-coding genes. Scale bars indicate the number of substitutions per site. (A) The ML phylogeny obtained with RAxML v.8.0.5 represents the best tree under a data set-specific GTR + GAMMA + I substitution model. Bootstrap support values (>50%) from 1,000 pseudoreplicates are given at the nodes. (B) For the Bayesian analysis, the 50% majority rule consensus tree was obtained from two independent runs using PhyloBayes MPI v.1.5a (CAT-GTR; 30,000 generations each, burn-in 5,000 each). Posterior probability values (>0.50) are given at the nodes.
Fig. 5.—
Phylogeny of Glyceridae based on ML and Bayesian inference for a data set comprising the 13 protein-coding mitochondrial genes and the 2 ribosomal mitochondrial genes. The data set includes the third codon position of the protein-coding genes. Scale bars indicate the number of substitutions per site. (A) The ML phylogeny obtained with RAxML v.8.0.5 represents the best tree under a data set-specific GTR + GAMMA + I substitution model. Bootstrap support values (>50%) from 1,000 pseudoreplicates are given at the nodes. (B) For the Bayesian analysis, the 50% majority rule consensus tree was obtained from two independent runs using PhyloBayes MPI v.1.5a (CAT-GTR; 30,000 generations each, burn-in 5,000 each). Posterior probability values (>0.50) are given at the nodes.
In contrast to the ML analysis, the Bayesian analysis BIall (with third position) places G. tesselata (FS18) as sister taxon to clade 1, which together are recovered as being weakly supported (posterior probability value 0.56) as the sister to clade 2 (fig. 4B). Furthermore, the phylogenetic position of H. simplex (FS13) could not be resolved in this Bayesian analysis. A slightly better supported position of H. simplex could be recovered in a Bayesian analysis of the BImt data set (with third position) (fig. 5B). Even if the phylogenetic analyses recover incongruent phylogenetic relationships within clade 3, H. simplex (FS13) remains always nested within the genus Glycera (cf. figs. 4 and 5 and supplementary fig. S4A–D, Supplementary Material online).
In contrast to the above-mentioned topologies, the three monophyletic clades within Glyceridae could not be recovered by ML analyses of the small data sets MLnucl and MLnucl + ITS. Moreover, Bayesian analyses of these data sets, namely BInucl and BInucl + ITS, even failed to recover the monophyly of Glyceridae (cf. supplementary fig. S4E and F, Supplementary Material online).
Coverage Studies to Assess the Scalability of Reconstructing Mitochondrial Genomes
To estimate the number of reads needed to reconstruct complete mitochondrial genomes, coverage studies were performed. Furthermore, the relative size of the mitochondrial genome (relative mitosize) as well as the corresponding number of contigs representing the mitochondrial genome (contig count) had been determined to describe the scalability of genome skimming in respect to recover complete mitochondrial genomes. The data sets studied in this context comprise 14 original data sets (each consisting of around 16 million reads) and 400 reduced data sets, as ten independent subsamples per data set size (10 million, 4 million, and 1 million reads) were constructed for each taxa (fig. 1).
The normalized contig coverage (cov per million) adopts similar values in all data sets of one specimen, regardless the fact that the absolute number of reads is adequate or inadequate for reconstructing complete mitochondrial genomes (cf. supplementary data set S4, Supplementary Material online). When comparing among species, the normalized coverage shows remarkable discrepancies. It varies in the glycerid species studied from a minimum of 0.82 (G. unicornis, FS15) to a maximum of 6.97 (Glycera cf. tridactyla, FS20), and the values even rise to 21.18 for the outgroup species Glyci. armigera (FS17) (cf. supplementary data set S4, Supplementary Material online). In glycerid specimens with a comparatively higher basic coverage drawn by values larger than 5 (cov per million of FS13, FS20, and Glytri), more than 89% of the mitochondrial genome could be reconstructed even based on the smallest tested data set size (1 million reads) (supplementary data set S4, Supplementary Material online). A comparison of all data set sizes among the studied glycerid specimens elucidates that a contig coverage of at least 6–7 × is sufficient to reconstruct more than 95% of a partially fragmented mitochondrial genome (fig. 6A–C and supplementary data set S4, Supplementary Material online). For Glyceridae which have an average mitogenome size of approximately 15.5 kb, this corresponds to approximately 1,000 reads of mitochondrial origin (fig. 6A).
Fig. 6.—
Summarized statistics of the coverage analyses in Glyceridae. (Stacked) Bar graphs (A–C) illustrating the original data sets (marked in orange) of 13 studied glycerid taxa and the corresponding reduced data sets consisting of 10 million reads (marked in black), 4 million reads (marked in dark gray) and 1 million reads (marked in light gray). Absolute numeric values are plotted for the original data sets, mean values are plotted for the reduced data sets. Mean values and standard deviations were calculated from ten subsamples analyzed for each specimen per data set size. (A) Number of reads that mapped to the species-specific mitocontig originated from the original data sets and the resulting contig coverage. The asterisk (*) indicates a multiplication factor of 103. For details on the calculation of the contig coverage, see “Coverage Studies” in the Materials and Methods section. Successful recovery of mitochondrial genomes dependent on data set sizes. (B) To determine the “relative mitosize,” the cumulative length of broken mitocontigs was referred to the length of the corresponding complete mitogenome (equal to 100%) originating from the original data sets. (C) Number of contigs representing the mitochondrial genome for each studied glycerid specimen and data set size (referred to as “contig count”). Boxplots showing the distribution of the relative mitosize (D) and the number of obtained mitocontigs (E) across all subsamples and data set sizes. They are based on the data of ten replicates per data set size (10 million, 4 million, and 1 million reads) per studied glycerid specimen (13 libraries), here plotted as dark gray data points. The interquartile range, comprising the middle 50% of the data points, is highlighted in orange.
As a general tendency, the coverage studies clearly show that the original data set sizes of around 16 million reads (cf. supplementary table S1, Supplementary Material online) are sufficient for reconstructing complete mitochondrial genomes, whereas its reduction to 1 million reads causes in most of the glycerid taxa incomplete and massively broken mitochondrial genomes (fig. 6D and E and supplementary data set S4, Supplementary Material online). Based on data set sizes of 1 million reads, the interquartile range which describes 50% of data, ranges from highly incomplete reconstructed mitogenomes (∼15% of the original size) to more complete mitogenomes (∼85% of the original size). Furthermore, the number of recovered contigs contributing to a broken mitogenome ranges from values of 5 up to 17. The median is drawn by a relative mitogenome size of approximately 47% to which correspond approximately ten short contigs representing the broken mitochondrial genome (fig. 6D and E and supplementary data set S4, Supplementary Material online). In summary, using 1 million reads mitogenomes tend to be incompletely recovered and broken in several contigs. In contrast to this, with data sets sizes of 4 million reads in most cases (except of FS15 and FS18) more than approximately 95% of the mitochondrial genome was recovered (fig. 6B and supplementary data set S4, Supplementary Material online). The average number of contigs representing this mitochondrial genome size ranges from 1.5–3.8 (e.g., FS12, FS14, FS19, FS21, and FS23) to maximally 11 (FS11), and for smaller reconstructed mitosizes to 13.9 (FS18) or 25.6 (FS15) (fig. 6C and supplementary data set S4, Supplementary Material online). Summarizing, 4 million reads already allow the retrieval of nearly complete mitochondrial genomes, which were represented by few long contigs. In data set sizes of 10 million reads, more than 98% of the mitochondrial genome could be always recovered. In four occasions, the complete mitochondrial genome was depicted by a single mitocontig (FS13, FS14, FS19, and Glytri). In the other cases, the complete mitochondrial genome was maximally broken in 2.8 separate contigs at average (fig. 6B and C and supplementary data set S4, Supplementary Material online).
Discussion
Phylogenetic Relationships of Glyceridae and Features of Their Mitochondrial Genomes
Using a genome skimming approach we were able to retrieve 13 complete mitochondrial genomes for glycerids, and the complete mitogenome of Glyci. armigera. The AT content, AT skew, and GC skew describing the composition of the mitochondrial genome are similar among all studied glycerids and different from that of the outgroup species (cf. fig. 2A–C and supplementary data set S3, Supplementary Material online). The mitochondrial genomes of Glyceridae are AT-rich (mean AT content of 65.95%, fig. 2A), the AT skew is only slightly negative (mean AT skew of −0.021, fig. 2B), whereas the GC skew is more negative (mean GC skew of −0.246, fig. 2C). These findings are consistent with the outcome of other annelid studies (e.g., Bleidorn et al. 2006; Mwinyi et al. 2009; Aguado et al. 2015). The highest level of bias in base composition is revealed for the third codon position (cf. fig. 2C) due to its high variability in codon usage (wobble base). Notably, the AT skew of the group II introns identified inside the cox1 gene of G. fallax and G. unicornis is remarkably different from that of the remaining mitochondrial genome (cf. supplementary data set S3, Supplementary Material online) indicating an independent origin.
The phylogenetic reconstructions of Glyceridae are based on ML analyses as well as Bayesian inference of different data sets varying in the number of included genes (table 2). Concerning the number of taxa and genes, our analyses represent the currently most extensive molecular approach resolving glycerid relationships. Although ML analyses reveal a quite robust glycerid phylogeny (figs. 4A and 5A), Bayesian inference using CAT-GTR seems to be unsuitable, especially for shorter alignments consisting of 6,721 aligned positions (BInucl + ITS) or 5,532 alignment positions (BInucl) (cf. table 2 and supplementary fig. S4E and F, Supplementary Material online). As the ribosomal data sets suffer from low levels of phylogenetic signal (cf. supplementary table S2, Supplementary Material online), we assume that these data sets do not yield enough information for fitting the complex nonparametric model used in our Bayesian analyses. Larger data sets seem to be more suitable as the distribution of site-specific effects of the underlying data sets and the substitutional heterogeneity will be better modelled (Lartillot and Philippe 2004; Lartillot et al. 2009).
The results of our phylogenetic analyses strongly support a sister group relationship between Glyceridae Grube, 1850 and Goniadidae Kinberg, 1866, namely Glyceriformia Fauchald, 1977 (Pleijel 2001) (figs. 4 and 5). The monophyletic status of Glyceriformia (Pleijel 2001) was doubted as some molecular analyses based on the nuclear ribosomal markers 18SrRNA and 28SrRNA (Struck et al. 2008; Böggemann 2009) rejected this hypothesis. Other molecular analyses using 16SrRNA (Böggemann 2009) and three combined genes 18SrRNA, 28SrRNA and EF1-alpha (Struck et al. 2007), as well as a comprehensive morphological analysis (Böggemann 2002) supported the monophyly of Glyceriformia, but only with weak support values. Besides the monophyly of Glyceriformia, also the monophyly of Glyceridae was revealed with maximal support values (figs. 4 and 5). This is in line with former studies based on single genes (18SrRNA, 16SrRNA, cox1) (Böggemann 2009) and a morphological approach built up on immense taxon sampling (Böggemann 2002). So far, no sequence data are available for Glycerella, the third described genus within this family. However, the morphology clearly indicates that this taxon will be part of a monophyletic Glyceridae (Böggemann 2002).
Contrary to the morphological study (Böggemann 2002), our actual molecular work could resolve the majority of nodes within Glyceridae with high support values. Our analyses recover three main clades, comprising all analyzed taxa, with exception of G. tesselata whose phylogenetic position could not be firmly resolved. Although this species occurs as sister taxon to clade 1 in the Bayesian analysis (fig. 4B), G. tesselata emerges as sister to all other glycerids in the ML tree (fig. 4A). More data are needed to clarify the position of this taxon. The relationships between the three main clades remain controversial. Although ML analysis gives strong support for a sister group relationship of clades 2 and 3 (fig. 4A), Bayesian analysis recovers clade 2 as sister to clade 1 (fig. 4B). Clade 1 includes the GLTx producing species G. tridactyla and will be discussed in more detail below. Clade 2 includes G. lapidum, G. capitata, and two undescribed Antarctic species. As revealed in a ML analysis comprising all published glycerid cox1 sequences, the Antarctic species belong to a previously discovered species complex. We could show that Glycera cf. capitata (Antarctica, FS09) is identical with Glycera sp. clade III (sensu Schüller 2011), and Glycera sp. (Antarctica, FS08) is a member of Glycera sp. clade I (sensu Schüller 2011) (cf. supplementary fig. S5, Supplementary Material online). These taxa show a spatial and depth-dependent distribution pattern, but have not been formally described yet (Schüller 2011). Within clade 2 our actual topology (fig. 4A) further resolves a sister group relationship of G. capitata and G. lapidum. The relationships recovered within clade 3 (fig. 4A) are congruent with the morphological approach (Böggemann 2002) revealing a sister group relationship of G. nicobarica and G. unicornis, occurring as sister to a clade containing G. fallax, and all together emerge as sister to a clade including G. americana. By sampling multiple individuals of some species, we got some potential hints for the existence of cryptic species within Glyceridae as the G. americana, G. tridactyla, and G. capitata individuals sampled at different locations are genetically distinct (cf. fig. 4A). These findings are congruent with a study of Schüller (2011) observing genetic differences in same morphotypes within the glycerid species Glycera kerguelensis. Our analysis of all available cox1 sequences also recovers more candidates of putatively cryptic species (see supplementary fig. S5, Supplementary Material online). However, as glycerids are difficult to identify several cases of misidentification may be present in the NCBI GenBank data (e.g., some glycerids cluster outside Glyceridae and Goniadidae; see supplementary fig. S5, Supplementary Material online). Future studies using an integrative taxonomic approach and implementing an increased taxon sampling will be crucial to analyze this question in more detail.
An unexpected phylogenetic position was recovered for H. simplex. Apart from the genus Glycera, Glyceridae comprise two additional genera, namely Glycerella and Hemipodia. All three genera had been regarded as monophyletic based on obviously distinct, and genus-specific morphological characters (Böggemann 2002). However, ML and Bayesian analyses revealed for H. simplex always a nested position within the genus Glycera (figs. 4 and 5 and supplementary fig. S4A–D, Supplementary Material online). Future taxonomic revisions should consider transferring H. simplex into the genus Glycera. With respect to the glycerid gene orders, we can show that the gene order of the protein-coding mitochondrial genes as well as of the ribosomal mitochondrial genes is conserved within Glyceridae and harbors gene arrangements identical to the majority of the yet known annelid mitochondrial genomes (Jennings and Halanych 2005; Bleidorn et al. 2006; Bleidorn et al. 2007; Golombek et al. 2013; Li et al. 2015) (fig. 3A). Within Glyceridae, three different gene orders could be distinguished. There are also two different gene orders found in Goniadidae (cf. Glyci. armigera and Go. japonica [see Chen et al. 2015]). Even though Glyci. armigera and Nephtys sp. (NC_010559.1) share the same gene order arrangement, it is not possible to determine the ancestral gene order for Glyceridae and Glyceriformia based on the yet published data, as Go. japonica shows an identical gene order as clades 2 and 3 of Glyceridae in our study. The typical 22 tRNAs could not contribute any clade-specific features (cf. supplementary fig. S2, Supplementary Material online). Nevertheless, in all cases the mitochondrial gene order is consistent with the phylogeny inferred from the sequence data and no convergent changes have to be assumed (cf. figs. 4 and 5).
Morphological cladistic analyses (Böggemann 2002) always recovered G. dibranchiata and the known GLTx producing species G. tridactyla in different groups, whereas our actual phylogenetic analyses indicate a close relationship of these two species within clade 1 (figs. 4 and 5 and supplementary fig. S4A–D, Supplementary Material online). This close relationship is further supported by an identical gene order identified in both species (figs. 3A, 4, and 5). The glycerid species G. tridactyla is known to possess a neurotoxin, namely α-Glycerotoxin, which activates specifically presynaptic Cav2.2 channels (N-type Ca2+ channels) causing increased neurotransmitter releases (Manaranche et al. 1980; Morel et al. 1983; Bon et al. 1985; Meunier et al. 2002). Its effects have been shown to be dose-dependent and reversible (Manaranche et al. 1980; Thieffry et al. 1982). Interestingly, the venom of G. dibranchiata is also able to induce ion-permeable channels in lipid bilayers (Kagan et al. 1982), but this rather seem to be evoked by an arsenal of pore-forming and membrane disrupting toxins (von Reumont, Campbell, Richter, et al. 2014). Even if computational transcriptome analyses revealed in G. dibranchiata venom transcripts coding for putative neurotoxins, like the gigantoxin I-like neurotoxin (von Reumont, Campbell, Richter, et al. 2014), the G. dibranchiata venom does not cause any increase in transmitter releasing effects like described for G. tridactyla venom (Bon et al. 1985). Consequently, it seems that these closely related glycerid species developed differently acting venom cocktails. As the G. tridactyla library analyzed in the study of von Reumont, Campbell, Richter, et al. (2014) was only shallowly sequenced, further RNAseq data are necessary and more species should be analyzed to reach more detailed insights into the complexity of glycerid venoms. The here presented backbone phylogeny will provide the necessary framework to trace the venom evolution in this annelid group.
Group II Introns in Glycerid Mitochondrial Genomes
A surprising finding of our analyses was the detection of group II introns inside the mitochondrial genomes of two glycerids. Group II introns are self-splicing mobile genetic elements that are built-up of a catalytically active intron RNA and an intron-encoded protein (IEP). Furthermore, they are thought to be the evolutionary ancestors of eukaryotic spliceosomal introns and retrotransposons (review Lambowitz and Zimmerly 2011). Although group II introns are not present in eukaryotic nuclear genomes, they are found in organelle genomes of, for example, yeast, fungi, liverwort, green plants and algae, as well as in Bacteria and Archaea (Zimmerly et al. 2001; Dai et al. 2003). In metazoans only for Trichoplax adhaerens (see Dellaporta et al. 2006) and the annelid Nephtys sp. (see Vallès et al. 2008), group II introns are known to be present in their mitochondrial genome, the latter which marked the first occurrence for Bilateria. Furthermore, Zanol et al. (2010) report possible candidate introns within cox1 sequences of eunicid annelids, and Zhong et al. (sensu Bleidorn et al. 2009) in Endomyzostoma scotia, which all still need confirmation. Interestingly, we found two additional annelid species, namely G. fallax and G. unicornis, harboring group II introns inside the cox1 gene. This allows speculating, that the ongoing application of NGS techniques in conjunction with a growing number of genomes possibly lead to recover additional group II introns in Bilateria yet remained undiscovered. The here found glycerid group II introns and the known one from Nephtys sp. share common characteristics as they are all located inside the cox1 gene, exhibit the same typical starting and ending sequence as well as an ORF for a reverse transcriptase and a type II intron maturase per intron (fig. 3B). Moreover, the group II introns of G. unicornis, G. fallax (I2), and Nephtys sp. start at the exact same position inside the gene, directly after position 700 of the cox1 coding sequence (fig. 3B). These introns cluster together in an ML analysis (supplementary fig. S3, Supplementary Material online). A second group II intron (I1) in G. fallax starts after CDS position 184 and does not cluster with the above-mentioned bilaterian group II introns. It is supposed that the initial DNA target site recognition is accomplished by the intron IEP, which recognizes specific bases of the DNA target site through major groove interactions cf. (Singh and Lambowitz 2001; Lambowitz and Zimmerly 2011). As the IEPs are encoded by the intron RNA, they recognize intron-specific DNA target sites (Lambowitz and Zimmerly 2011). We assume that G. fallax possess two different group II introns colonizing the cox1 gene independently. The absence of group II introns in most of the analyzed glycerids as well as its presence in the outgroup species Nephtys sp. shows evidence that the occurrence of group II introns inside the cox1 gene of the here analyzed annelid species results from separate events. An imaginable scenario already hypothesized in a recent study explains group II introns as the result of a horizontal gene transfer from a bacterial or viral vector into the mitochondrial genome of its host (Vallès et al. 2008).
Genome Skimming Allows Unravelling Complete Mitochondrial Genomes
High-throughput sequencing promoted the generation and utilization of mitochondrial genomes to resolve phylogenetic relationships (e.g., Gillett et al. 2014; Williams et al. 2014) as well as phylogeographic questions (e.g., Morin et al. 2010). In our study, we followed a time- and cost-efficient approach to sequence in parallel complete mitochondrial genomes of closely related non-model organisms performing multiplexed Illumina HiSeq sequencing.
The pipeline used here consisting of tagged NGS sequencing, de novo assembly and phylogenetic studies, was used in a similar manner in other recent studies (e.g., Botero-Castro et al. 2013; Hahn et al. 2013; Williams et al. 2014; Li et al. 2015). However, the sufficient sequencing depth remained obscure. Botero-Castro et al. (2013) proposed that based on a higher ratio of mitochondrial DNA over nuclear DNA in most of the tissues, already less than 10 million reads should be mostly sufficient to recover complete mitochondrial genomes by an adequate coverage. To test the scalability of the genome skimming approach in view of reconstructing complete mitochondrial genomes, we performed in silico analyses on 14 original data sets (13 × Glyceridae, 1 × Goniadidae) and 400 corresponding reduced data sets (10 million, 4 million, and 1 million reads) as for each data set size ten subsamples were generated.
Our results clearly show that the percentage of retrieved mitochondrial reads is scalable regarding the total number of sequence reads (cf. cov per million; supplementary data set S4, Supplementary Material online). Consequently, rather than the relative number of reads referring to a mitochondrial genome, the absolute number is crucial to reconstruct complete mitochondrial genomes. Comparing the different analyzed classes of read numbers (10 million, 4 million, and 1 million) we find that using only 1 million reads did in most cases not allow the assembly of complete mitochondrial genomes. BLAST-searches revealed that the recovered genomes are highly incomplete and broken into many contigs (fig. 6B–E). Moreover, the variance between subsets is quite high. This is no surprise, as the estimated coverage in these data sets is relatively low, in many cases below 5 × (fig. 6A). In contrast, the coverage increases in data sets of 4 million reads to 6–7 × and higher which refers to approximately 1,000 mapped reads (fig. 6A and supplementary data set S4, Supplementary Material online). These data set sizes allow assembling at least 95% of the original mitochondrial genome content, but the mitochondrial genomes are broken several times in different contigs (fig. 6B–E). We assume that most de novo assemblers should need at least a 6×to 7× coverage to create long mitochondrial contigs, as found for the here used IDBA-UD. In data set sizes of 10 million reads, each time more than 98% of the glycerid mitochondrial genome was recovered. Thereby, the mitochondrial genomes were recovered as single or in two contigs in most cases (maximally in ∼2.8 contigs at average, cf. supplementary data set S4, Supplementary Material online and fig. 6D and E), which would enable gene order annotations. Therefore, we would propose data set sizes of at least 10 million reads as adequate dimensioned to reconstruct mitochondrial genomes in case of Glyceridae. Consequently, Illumina HiSeq 2500 which produces approximately 500–600 million paired-end reads per lane (Illumina 2015, last accessed December 3, 2015) will enable the parallel processing of 50 glycerid individuals, of 10 million reads each, on a single lane.
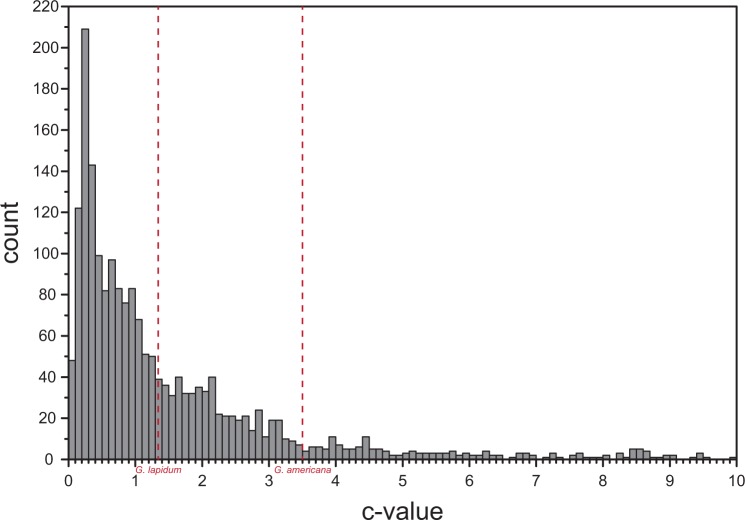
The number of mitochondrial reads in WGS data is not only strongly influenced by the number of mitochondria but also by the genome size of the target organism. Assuming a similar number of mitochondria per cell when compared between two species, the relative number of mitochondrial reads will be lower for species with a larger genome. The genome size can be measured by the c-value which is defined as the haploid nuclear DNA content in picogram (1 pg = 978 Mb) (Doležel et al. 2003; Gregory 2005). Compared with Glyceridae having published genome sizes of c-value = 1.33 (G. lapidum) and c-value = 3.5 (G. americana) (Gregory et al. 2007; Gregory 2014), the majority of invertebrate species harbors considerably smaller genomes sizes (fig. 7). As a consequence, it seems justified to propose that for most invertebrate target species already less than 10 million reads are needed for the reconstruction of complete mitochondrial genomes (but see Tilak et al. [2015] for problems with genome skimming in tunicates). This is likely reflected in the outgroup species Glyci. armigera in which the determined normalized coverage (cov per million) of 21.18 is three times higher than the highest value recovered in all studied glycerids (supplementary data set S4, Supplementary Material online). However, it has to be noted that the number of mitochondria per cell can be highly dependent on the type of tissue and developmental stage (e.g., Robin and Wong 1988; Kawamura et al. 2012). In our study we used body wall and muscle tissue for all analyzed specimens, allowing a valid comparison. Usage of tissue types enriched for mitochondria will have obviously a positive influence on the ration of recovered mitochondrial reads.
Fig. 7.—
Histogram comprising the c-values of 1,985 invertebrate species. The published c-values of two glycerid species are highlighted in red (c-value = 1.33 Glycera lapidum; c-value = 3.5 Glycera americana). Note that 62.42% of the included invertebrate species have comparatively smaller genome sizes than G. lapidum. c-Values above a value of 10 are not shown. The c-values were taken from the Animal Genome Size Database (Gregory 2015, last accessed December 2, 2015).
Genome skimming gained attention for metagenome analyses of insect communities (e.g., Andújar et al. 2015; Crampton-Platt et al. 2015; Linard et al. 2015). For this approach, WGS sequencing was conducted for untagged DNA libraries from preselected insect individuals and it has been shown that it is possible to assemble larger mitochondrial contigs from such mixed sequence data. For example, for mixed DNA samples of around 500 Coleoptera it was possible to retain 107 complete mitochondrial genomes using two Illumina MiSeq runs (∼34 million reads, 250-bp paired-end) (Crampton-Platt et al. 2015). The authors also barcoded all investigated beetles by sequencing a cox1 fragment, which allows to refer the retained mitochondrial genomes to individual specimens. Due to relatively easy protocols coupled with an ever increasing sequencing power it seems obvious that genome skimming approaches will be of huge interest for phylogenetic studies of evolutionary younger groups (as shown here for Glyceridae), as well as for metagenomic studies of animal communities.
In summary, the genome skimming approach offers numerous advantages for assembling mitochondrial data sets of nonmodel taxa: 1) The usage of DNA allows to analyze freshly preserved material as well as longer stored museum specimens; the fact that no prior resources are required allows a direct start of library preparation, which makes it a comparatively 2) fast and 3) cost-efficient method (table 3). Besides genome skimming several other approaches exist to address phylogenomic studies of non-model organisms (table 3). One such class of methods is target enrichment, which usually amplifies selected exons or ultraconserved elements of the genome (e.g., Lemmon et al. 2012; McCormack et al. 2013). These methods enable enrichment for “moderate target sizes (25–100 loci)” (Peñalba et al. 2014) as well as a rapid capturing of hundreds of loci for phylogenetic analyses. However, prior genomic resources are always required for probe construction. Moreover, enrichment of thousands of loci is still very expensive and as such only cost effective, when huge data sets (>96 species) are analyzed. Interestingly, using the unbiased genome skimming approach we were able to support the integration of group II introns within the mitochondrion due to a comparison with the coverage of mitochondrial genes. Such analyses would be highly biased using target enrichment and may lead to the exclusion of such unusual mitochondrial features. RNA-sequencing, which provides potentially phylogenetically informative genes (Wang et al. 2009) and has been successfully used in several deep phylogenomic studies (Kocot et al. 2011; Fernández et al. 2014; Weigert et al. 2014), requires high-quality RNA for library construction (table 3) which was not available for most of the hitherto studied glycerid specimens.
Table 3.
Comparison of Three High-Throughput Sequencing Strategies regarding Their Application, Potential Advantages and Disadvantages, and Technological Issues
| RNA Sequencing | Target Enrichment | Genome Skimming | |
|---|---|---|---|
| Technology | |||
| Principle | High-throughput sequencing | High-throughput sequencing | High-throughput sequencing |
| Material | RNA | DNA | DNA |
| Hints for application | |||
| Prior genomic resources required | No | Yes | No |
| Limitations by starting material | RNA has to be available | DNA | DNA |
| Recommend taxon number | Flexible | Huge number recommended | Flexible |
| Required amount of RNA/DNA | Low | Low | Low |
| Genome size of species | Less relevant | Less relevant | Important |
| Workload | Time intensive | Time intensive | Fast and easy method |
| Application | |||
| Ability to identify single copy genes | Yes | Yes | Maybe |
| Ability to distinguish different isoforms | Yes | No | No |
| Ability to analyze expression levels | Yes | No | No |
| Ability to analyze intron–exon structure | No | Yes (require prior information) | Yes |
Conclusions
We were able to resolve a robust backbone phylogeny for Glyceridae, which will be essential to choose taxa for further venom transcriptome studies aiming to understand venom evolution in this group. Our phylogenetic analyses demonstrate that mitochondrial genome data still represent a valuable marker for phylogenetics in postgenomic times, especially suitable when working on a higher taxonomic level or with young radiations. Furthermore, NGS revolutionized the acquisition of mitochondrial genomes by generating huge amounts of data in a comparatively short time. Consequently, the number of available mitogenomes increases continually and their screening will further contribute to the detection of unusual features, as for example, group II introns. Apart from this, to plan time- and cost-efficient NGS projects and to increase the outcome of such studies, it is obviously advantageous to know the depth of sequencing that should be achieved. Our in silico analyses show that in Glyceridae, low contig coverages of around 6–7× are already adequate to reconstruct more than 95% of the mitochondrial genome. Generally, for species harboring a mitogenome of approximately 15.5 kb coupled with c-values ranging from approximately c-value = 1.33 to c-value = 3.5, about 10 million sequencing reads seemingly adequate for resolving more than 98% of the mitochondrial genome. Nevertheless, genome skimming is one of several approaches as discussed above. The best one should be chosen according to the scientific question, taxon sampling, and/or available starting material.
Supplementary Material
Supplementary figures S1–S5, tables S1–S3, and data sets S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Myriam Schüller (University of Bochum, Germany), Kenneth M. Halanych (Auburn University, USA), Franziska A. Franke (University of Leipzig, Germany), and Sabrina Kaul-Strehlow (University of Vienna, Austria) for collecting and providing specimens. Furthermore, they are thankful to Tara A. Macdonald (Biologica Environmental Services, Victoria, Canada) who supported the collection of glycerids in Bamfield, Canada. Moreover, they thank Lahcen I. Campbell (Natural History Museum, London), the editor Dennis V. Lavrov (Iowa State University, USA), and three unknown reviewers for comments on the manuscript. Furthermore, they thank Anne Weigert (University of Leipzig, Germany) for help in library preparation. They gratefully acknowledge the Max Planck Institute for Evolutionary Anthropology Leipzig for Illumina sequencing opportunities, especially Marie-Theres Gansauge, Birgit Nickel, Matthias Meyer, Gabriel Renaud, Martin Kircher, and Svante Pääbo. Moreover, they thank Michael Gerth (University of Leipzig, Germany) for introductive help in the visualization program Circos. They are thankful to the group of Peter F. Stadler (University Leipzig, Germany), especially Jens Steuck and Stephan H. Bernhart as well as the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig for providing computational resources for bioinformatic analyses. They acknowledge support from the German Research Foundation (DFG) and Universität Leipzig within the program of Open Access Publishing. This work was supported by the German Research Foundation (DFG; grant BL787/7-1) and an EU ASSEMBLE grant (No. 227799; http://www.assemblemarine.org) to C.B.
Literature Cited
- Aguado MT, Glasby CJ, Schroeder PC, Weigert A, Bleidorn C. 2015. The making of a branching annelid: an analysis of complete mitochondrial genome and ribosomal data of Ramisyllis multicaudata. Sci Rep. 5:12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andújar C, et al. 2015. Phylogenetic community ecology of soil biodiversity using mitochondrial metagenomics. Mol Ecol. 24:3603–3617. [DOI] [PubMed] [Google Scholar]
- Bernt M, et al. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Bleidorn C, et al. 2007. Mitochondrial genome and nuclear sequence data support Myzostomida as part of the annelid radiation. Mol Biol Evol. 24:1690–1701. [DOI] [PubMed] [Google Scholar]
- Bleidorn C, Podsiadlowski L, Bartolomaeus T. 2006. The complete mitochondrial genome of the orbiniid polychaete Orbinia latreillii (Annelida, Orbiniidae)–a novel gene order for Annelida and implications for annelid phylogeny. Gene 370:96–103. [DOI] [PubMed] [Google Scholar]
- Bleidorn C, et al. 2009. On the phylogenetic position of Myzostomida: can 77 genes get it wrong? BMC Evol Biol. 9:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böggemann M. 2014. Glyceridae Grube, 1850. In Handbook of Zoology Online. Berlin, Boston: De Gruyter; Available from: http://www.degruyter.com/view/Zoology/bp_029147-6_27. [Google Scholar]
- Böggemann M. 2009. Polychaetes (Annelida) of the abyssal SE Atlantic. Org Divers Evol. 9:251–428. [Google Scholar]
- Böggemann M. 2002. Revision of the Glyceridae Grube 1850 (Annelida: Polychaeta). Stuttgart (Germany): E. Schweizerbart'sche Verlagsbuchhandlung. [Google Scholar]
- Böggemann M. 2006. Worms that might be 300 million years old. Mar Biol Res. 2:130–135. [Google Scholar]
- Bon C, Saliou B, Thieffry M, Manaranche R. 1985. Partial purification of α-glycerotoxin, a presynaptic neurotoxin from the venom glands of the polychaete annelid Glycera convoluta. Neurochem Int. 7:63–75. [DOI] [PubMed] [Google Scholar]
- Botero-Castro F, et al. 2013. Next-generation sequencing and phylogenetic signal of complete mitochondrial genomes for resolving the evolutionary history of leaf-nosed bats (Phyllostomidae). Mol Phylogenet Evol. 69:728–739. [DOI] [PubMed] [Google Scholar]
- Burger G, Lavrov DV, Forget L, Lang BF. 2007. Sequencing complete mitochondrial and plastid genomes. Nat Protoc. 2:603–614. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. 2013. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 28:219–229. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chen X, et al. 2015. The complete mitochondrial genome of the polychaete, Goniada japonica (Phyllodocida, Goniadidae). Mitochondrial DNA. Advance Access published June 29, 2015, doi: 10.3109/19401736.2015.1053124. [DOI] [PubMed] [Google Scholar]
- Crampton-Platt A, et al. 2015. Soup to tree: the phylogeny of beetles inferred by mitochondrial metagenomics of a bornean rainforest sample. Mol Biol Evol. 32:2302–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curole JP, Kocher TD. 1999. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol. 14:394–398. [DOI] [PubMed] [Google Scholar]
- Dai L, Toor N, Olson R, Keeping A, Zimmerly S. 2003. Database for mobile group II introns. Nucleic Acids Res. 31:424–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, et al. 2006. Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum. Proc Natl Acad Sci U S A. 103:8751–8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. 2003. Letter to the editor: nuclear DNA content and genome size of trout and human. Cytometry A 51A:127–128. [DOI] [PubMed] [Google Scholar]
- Dreyer H, Steiner G. 2004. The complete sequence and gene organization of the mitochondrial genome of the gadilid scaphopod Siphonondentalium lobatum (Mollusca). Mol Phylogenet Evol. 31:605–617. [DOI] [PubMed] [Google Scholar]
- Ehlers EH. 1868. Die Borstenwürmer (Annelida Chaetopoda) nach systematischen und anatomischen Untersuchungen. Leipzig (Germany): Verlag von W. Engelmann. [Google Scholar]
- Ewing B, Green P. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175–185. [DOI] [PubMed] [Google Scholar]
- Fauchald K, Rouse G. 1997. Polychaete systematics: past and present. Zool Scr. 26:71–138. [Google Scholar]
- Fernández R, Hormiga G, Giribet G. 2014. Phylogenomic analysis of spiders reveals nonmonophyly of orb weavers. Curr Biol. 24:1772–1777. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, et al. 2009. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 10:483–511. [DOI] [PubMed] [Google Scholar]
- Gillett CPDT, et al. 2014. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol Biol Evol. 31:2223–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golombek A, Tobergte S, Nesnidal MP, Purschke G, Struck TH. 2013. Mitochondrial genomes to the rescue–Diurodrilidae in the myzostomid trap. Mol Phylogenet Evol. 68:312–326. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2014. Animal Genome Size Database. Available from: http://www.genomesize.com.
- Gregory TR, editor. 2005. Genome size evolution in animals. The evolution of the genome. Burlington (MA)/San Diego (CA)/London (United Kingdom): Elsevier Academic Press; p. 3–88. [Google Scholar]
- Gregory TR, et al. 2007. Eukaryotic genome size databases. Nucleic Acids Res. 35:D332–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98. [Google Scholar]
- Helfenbein KG, Brown WM, Boore JL. 2001. The complete mitochondrial genome of the articulate brachiopod Terebratalia transversa. Mol Biol Evol. 18:1734–1744. [DOI] [PubMed] [Google Scholar]
- Hoffmann S, et al. 2014. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 15:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S, et al. 2009. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol. 5:e1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, et al. 2011. Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species. PLoS One 6:e14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina. 2014. HiSeq System Performance Parameters. Available from: http://www.illumina.com/systems/hiseq_2500_1500/performance_specifications.html.
- Jennings RM, Halanych KM. 2005. Mitochondrial genomes of Clymenella torquata (Maldanidae) and Riftia pachyptila (Siboglinidae): evidence for conserved gene order in annelida. Mol Biol Evol. 22:210–222. [DOI] [PubMed] [Google Scholar]
- Kagan BL, Pollard HB, Hanna RB. 1982. Induction of ion-permeable channels by the venom of the fanged bloodworm Glycera dibranchiata. Toxicon 20:887–893. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Kitamura S, Sekida S, Tsuda M, Sunanaga T. 2012. Molecular anatomy of tunicate senescence: reversible function of mitochondrial and nuclear genes associated with budding cycles. Development 139:4083–4093. [DOI] [PubMed] [Google Scholar]
- Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocot KM, et al. 2011. Phylogenomics reveals deep molluscan relationships. Nature 477:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. 2011. Group II Introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 3:a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Waterman MS. 1988. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2:231–239. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21:1095–1109. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI. Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62:611–615. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, et al. 2013. Mitochondrial DNA of Clathrina clathrus (Calcarea, Calcinea): six linear chromosomes, fragmented rRNAs, tRNA editing, and a novel genetic code. Mol Biol Evol. 30:865–880. [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Emme SA, Lemmon EM. 2012. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst Biol. 61:727–744 [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. 2015. Mitogenomics reveals phylogeny and repeated motifs in control regions of the deep-sea family Siboglinidae (Annelida). Mol Phylogenet Evol. 85:221–229. [DOI] [PubMed] [Google Scholar]
- Linard B, Crampton-Platt A, Timmermans MJTN, Vogler AP. 2015. Metagenome skimming of insect specimen pools: potential for comparative genomics. Genome Biol Evol. 7:1474–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RE, Foster PG, Guille M, Littlewood DTJ. 2012. Next generation sequencing and comparative analyses of Xenopus mitogenomes. BMC Genomics 13:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaranche R, Thieffry M, Israel M. 1980. Effect of the venom of Glycera convoluta on the spontaneous quantal release of transmitter. J Cell Biol. 85:446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic T, Whitten M, Pääbo S. 2010. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One 5:e14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JE, et al. 2013. A phylogeny of birds based on over 1,500 loci collected by target enrichment and high-throughput sequencing. PLoS One 8:e54848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier FA, Feng Z-P, Molgó J, Zamponi GW, Schiavo G. 2002. Glycerotoxin from Glycera convoluta stimulates neurosecretion by up-regulating N-type Ca2+ channel activity. EMBO J. 21:6733–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010 (6), doi:10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Morel N, Thieffry M, Manaranche R. 1983. Binding of a Glycera convoluta neurotoxin to cholinergic nerve terminal plasma membranes. J Cell Biol. 97:1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PA, et al. 2010. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res. 20:908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwinyi A, et al. 2009. Mitochondrial genome sequence and gene order of Sipunculus nudus give additional support for an inclusion of Sipuncula into Annelida. BMC Genomics 10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaro NJ, et al. 2000. Evaluation of DNA fragment sizing and quantification by the Agilent 2100 Bioanalyzer. Clin Chem. 46:1851–1853. [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. [DOI] [PubMed] [Google Scholar]
- Peñalba JV, et al. 2014. Sequence capture using PCR-generated probes: a cost-effective method of targeted high-throughput sequencing for non-model organisms. Mol Ecol Resour. 14:1000–1010. [DOI] [PubMed] [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. [DOI] [PubMed] [Google Scholar]
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41:353–358. [DOI] [PubMed] [Google Scholar]
- Pleijel F. 2001. Glyceriformia Fauchald, 1977. In: Rouse GW, Pleijel F, editors. Polychaetes. Oxford: Oxford University Press; p. 111–114. [Google Scholar]
- Renaud G, Kircher M, Stenzel U, Kelso J. 2013. freeIbis: an efficient basecaller with calibrated quality scores for Illumina sequencers. Bioinformatics 29:1208–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud G, Stenzel U, Kelso J. 2014. leeHom: adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res. 42:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin ED, Wong R. 1988. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 136:507–513. [DOI] [PubMed] [Google Scholar]
- Rubinstein ND, et al. 2013. Deep sequencing of mixed total DNA without barcodes allows efficient assembly of highly plastic ascidian mitochondrial genomes. Genome Biol Evol. 5:1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller M. 2011. Evidence for a role of bathymetry and emergence in speciation in the genus Glycera (Glyceridae, Polychaeta) from the deep Eastern Weddell Sea. Polar Biol. 34:549–564. [Google Scholar]
- Singh NN, Lambowitz AM. 2001. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J Mol Biol. 309:361–386. [DOI] [PubMed] [Google Scholar]
- Smeds L, Künstner A. 2011. ConDeTri—a content dependent read trimmer for Illumina data. PLoS One 6:e26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck T, et al. 2007. Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evol Biol. 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck TH, Nesnidal MP, Purschke G, Halanych KM. 2008. Detecting possibly saturated positions in 18S and 28S sequences and their influence on phylogenetic reconstruction of Annelida (Lophotrochozoa). Mol Phylogenet Evol. 48:628–645. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577. [DOI] [PubMed] [Google Scholar]
- Thieffry M, Bon C, Manaranche R, Saliou B, Israël M. 1982. Partial purification of the Glycera convoluta venom components responsible for its presynaptic effects. J Physiol. 78:343–347. [PubMed] [Google Scholar]
- Tilak M-K, et al. 2015. A cost-effective straightforward protocol for shotgun Illumina libraries designed to assemble complete mitogenomes from non-model species. Conserv Genet Resour. 7:37–40. [Google Scholar]
- Vallès Y, Halanych KM, Boore JL. 2008. Group II introns break new boundaries: presence in a bilaterian’s genome. PLoS One 3:e1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Reumont BM, Campbell LI, Jenner RA. 2014. Quo vadis venomics? A roadmap to neglected venomous invertebrates. Toxins 6:3488–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Reumont BM, Campbell LI, Richter S, et al. 2014. A polychaete’s powerful punch: venom gland transcriptomics of Glycera reveals a complex cocktail of toxin homologs. Genome Biol Evol. 6:2406–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert A, et al. 2014. Illuminating the base of the annelid tree using transcriptomics. Mol Biol Evol. 31:1391–1401 [DOI] [PubMed] [Google Scholar]
- Williams ST, Foster PG, Littlewood DTJ. 2014. The complete mitochondrial genome of a turbinid vetigastropod from MiSeq Illumina sequencing of genomic DNA and steps towards a resolved gastropod phylogeny. Gene 533:38–47. [DOI] [PubMed] [Google Scholar]
- Winkelmann I, et al. 2013. Mitochondrial genome diversity and population structure of the giant squid Architeuthis: genetics sheds new light on one of the most enigmatic marine species. Proc R Soc B Biol Sci. 280:20130273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G. 1977. Kieferorgane von Glyceriden (Polychaeta)–ihre Funktion und ihr taxonomischer Wert. Senckenbergiana Marit. 9 (5/6):261–283. [Google Scholar]
- Xia X. 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 30:1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanol J, Halanych KM, Struck TH, Fauchald K. 2010. Phylogeny of the bristle worm family Eunicidae (Eunicida, Annelida) and the phylogenetic utility of noncongruent 16S, COI and 18S in combined analyses. Mol Phylogenet Evol. 55:660–676. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol. 7:203–214. [DOI] [PubMed] [Google Scholar]
- Zimmerly S, Hausner G, Wu X-C. 2001. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 29:1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.