Abstract
The sunflower (Helianthus annuus) genome contains a very large proportion of transposable elements, especially long terminal repeat retrotransposons. However, knowledge on the retrotransposon-related variability within this species is still limited. We used next-generation sequencing (NGS) technologies to perform a quantitative and qualitative survey of intraspecific variation of the retrotransposon fraction of the genome across 15 genotypes—7 wild accessions and 8 cultivars—of H. annuus. By mapping the Illumina reads of the 15 genotypes onto a library of sunflower long terminal repeat retrotransposons, we observed considerable variability in redundancy among genotypes, at both superfamily and family levels. In another analysis, we mapped Illumina paired reads to two sets of sequences, that is, long terminal repeat retrotransposons and protein-encoding sequences, and evaluated the extent of retrotransposon proximity to genes in the sunflower genome by counting the number of paired reads in which one read mapped to a retrotransposon and the other to a gene. Large variability among genotypes was also ascertained for retrotransposon proximity to genes. Both long terminal repeat retrotransposon redundancy and proximity to genes varied among retrotransposon families and also between cultivated and wild genotypes. Such differences are discussed in relation to the possible role of long terminal repeat retrotransposons in the domestication of sunflower.
Keywords: Helianthus annuus, long terminal repeat retrotransposons, plant domestication, repetitive DNA, retrotransposon redundancy
Introduction
Transposable elements (TEs) are mobile DNA sequences, which are able to change their chromosomal location (transposition). TEs are present in the nuclear genomes of all eukaryotes, with the potential to replicate faster than the host (Naito et al. 2009; Belyayev et al. 2010). Based on their transposition mechanisms, TEs can be classified into two groups, retrotransposons or Class I elements, and DNA transposons or Class II elements (Wicker et al. 2007).
Retrotransposons move through an RNA intermediate that is reverse transcribed into a DNA copy that can insert elsewhere in the genome (Kumar and Bennetzen 1999). In contrast, DNA transposons move without creating a new copy of the elements, using a DNA-based enzymatic method for excision and transposition of the parent copy itself (Wicker et al. 2007); consequently, Class II TEs are generally less abundant than retrotransposons.
The elements belonging to Class I can be classified into five taxonomic orders (Wicker et al. 2007). The most abundant and diverse order in plants, the long terminal repeat retrotransposons (LTR-RTs), are composed of a coding portion flanked by two direct repeats, LTRs, and can be primarily attributed to two superfamilies, Ty1/Copia and Ty3/Gypsy (Wicker et al. 2007), which differ in the position of the integrase domain within the encoded polyprotein (Kumar and Bennetzen 1999). LTR-RTs vary in size from a few hundred base pairs to over 10 kb, with LTRs that usually contain the promoter and RNA processing signals starting with “TG” and terminating with “CA” (Kumar and Bennetzen 1999). In addition to the two identical LTRs, a typical intact element contains the primer-binding site and the polypurine tract, which provide the signals for reverse transcription of retrotransposon transcripts into the cDNA that will be reintegrated into the genome. These two sequence sites flank a region that contains Open Reading Frame (ORFs) for Gag, a structural protein of the virus-like particles, and for Pol. Pol encodes a polyprotein with protease, reverse transcriptase, RNaseH, and integrase enzyme domains, which are required for the replication and the integration of the elements in the host chromosomes (Kumar and Bennetzen 1999).
Now that much genomic data are available, it has been shown that LTR-RTs comprise a large portion of plant genomes. The relative proportions of LTR-RTs may vary between species (Hua-Van et al. 2011). For example, retrotransposon sequences compose about 39.5% of the rice genome, 50.3% of the soybean genome, and 84.2% of the maize genome (Vitte et al. 2014). It has been suggested that variation in the relative proportion of these repetitive elements in a genome could either be the result of different insertion site preferences (Peterson-Burch et al. 2004; Gao et al. 2008) or be due to differences in the host-encoded mechanisms that limit TE proliferation (Du et al. 2010).
Superfamilies like Copia and Gypsy can be also classified into different families, the members of which share sequence similarity. Six major evolutionary Copia and six Gypsy families have been identified (Wicker and Keller 2007, Llorens et al. 2011) across different plant species. Among species, DNA sequence similarity within a family is minimal and limited to those coding regions which exhibit a high level of conservation (Wicker et al. 2007). Generally, the bulk of the repetitive fraction in a genome is composed of a few families, whose relative proportions may differ among species. For example, the Angela family of Copia elements is predominant in wheat (Wicker et al. 2001), but Gypsy-like Ogre elements predominate in some Pisum and Vicia species (Neumann et al. 2003).
Despite the differences in transposition mechanism and genomic abundance, both retrotransposons and DNA transposons are capable of introducing genetic variation, and some of these variations may have important effects on the course of plant evolution (Lisch 2013). TEs are not only able to cause genetic mutations, but they also play a role in the epigenetic settings of the genome, regulate chromatin organization in the nucleus, and act as control elements for the expression of genes (van Driel et al. 2003; Song et al. 2004). For example, TEs are associated with reduced gene expression and also with gene expression differences between orthologs in Arabidopsis species (Hollister and Gaut 2009; Hollister et al. 2011).
In addition to the effects on gene function, LTR-RTs are a major driver of genome size increase, resulting in variation in the composition of repetitive DNA. For example, in Oryza australiensis, a wild relative of rice, the genome size doubled by the amplification of only three LTR-RT families within the last 3 million years (Piegu et al. 2006).
Although several papers have investigated the role of TEs in changing the structure and function of plant genomes, only a few studies focused on intraspecific variability of the repetitive component of plant genomes. Moreover, these studies have been limited to a few model species, especially maize and Arabidopsis (Springer et al. 2009; Albert et al. 2010; Hollister et al. 2011). Hence, we decided to investigate the contribution of LTR-RTs to genome structure in different genotypes of a crop that exhibits wide morphological diversity, the sunflower (Helianthus annuus L., Asteraceae).
The sunflower is the most important crop belonging to the genus Helianthus. The genus Helianthus originated relatively recently, ranging between 4.75 and 22.7 Ma (Schilling 1997), likely in Mexico, with subsequent migration from North America (Schilling et al. 1998). Sunflower domestication probably occurred in the eastern regions of North America. A molecular genetic study has shown that modern sunflower cultivars, collected primarily in the United States, are most close genetically to wild sunflower populations of the Midwestern United States (Harter et al. 2004). Another study argued for an earlier domestication event in Mexico, that is, an independent domestication event in this area (Lentz et al. 2008), but molecular genetic studies showed that Mexican cultivars also cluster with wild sunflower populations from the Midwestern United States (Blackman, Scascitelli, et al. 2011). Thus, it is clear that cultivated sunflower arose from a single domestication event in eastern North America.
Although a genome sequence of H. annuus became publicly available only recently (http://www.sunflowergenome.org, last accessed December 1, 2015), it has been evident for more than a decade that the sunflower genome contains many thousands of TEs (Santini et al. 2002; Natali et al. 2006, 2013; Staton et al. 2012). Mobilization and consequent amplification of retrotransposons have been reported during Helianthus speciation, even in relatively recent times (Ungerer et al. 2009). Specific sunflower LTR-RTs have been shown to be transcribed regularly and, at small rates, reinserted into the genome (Vukich, Giordani, et al. 2009).
Overall characterization of the repetitive fraction of the sunflower genome was first obtained using a Sanger-sequenced small insert library. Combining sequence analysis with slot blot hybridization and fluorescent in situ hybridization, the fraction of sequences that can be classified as repetitive amounted to 62% in total (Cavallini et al. 2010).
Later, a sequencing strategy that combined whole-genome shotgun sequencing (Solexa and 454 platforms) with high-density genetic and physical maps estimated that 78% of the sunflower genome consists of repetitive sequences (Kane et al. 2011). With the further improvement in NGS technologies, a great effort has been made to characterize the repetitive component of the sunflower genome. In a recent study, a large set of whole-genome shotgun sequence reads representing approximately 25% of the sunflower genome was analyzed; the results suggest that the sunflower genome is composed of more than 81% TEs, 77% of which are LTR-RTs, especially of Gypsy superfamily and Chromovirus family (Staton et al. 2012).
In another recent experiment, Natali et al. (2013) used NGS technologies to produce sunflower sequences and create a database of sunflower repetitive sequences (SUNREP). The results confirmed that LTR-RTs are by far the most abundant class of sequences in the sunflower genome, accounting for at least 79.53% of the reads matching the contigs. Among LTR-RTs, sequences belonging to the Gypsy superfamily were 2.3-fold more represented than those belonging to the Copia superfamilies. The larger abundance of Gypsy elements compared with Copia can be explained by two hypotheses: Gypsy elements have been more active during sunflower evolution and/or they have been active more recently, so that they are more easily recognizable by similarity searches, having been subject to fewer mutations.
Now that a characterization of the repetitive component of sunflower has been achieved, it is important to analyze variation in the relative proportion of the LTR-RTs among genotypes, cultivars, and wild accessions of the genus Helianthus. In fact, thanks to the wide variety of wild accessions, which occupy habitats ranging from open plains to sand dunes and salt marshes (Heiser et al. 1969), Helianthus lends itself to be an exemplar genus for the study of genetic variation in the wild.
Here we present a comparative analysis of the LTR-RT component of the genome among different genotypes of H. annuus. This study focuses on eight worldwide sunflower cultivars and seven wild accessions from North America, in order to assess genomic differences and similarities due to these repetitive sequences, concerning especially the differences between wild and domesticated genotypes.
Materials and Methods
Plant Materials and DNA Isolation
The sunflower (H. annuus) genotypes used in these experiments are listed in supplementary material S1, Supplementary Material online. Wild accessions and cultivars were obtained from United States Department of Agriculture (USDA), Agricultural Research Service (ARS), National Genetic Resources Program, USA. Further data on analyzed wild and cultivated genotypes can be found at the National Germplasm Resources Laboratory homepage (http://www.ars-grin.gov/npgs/searchgrin.html, last accessed December 1, 2015) and in a previous work (Vukich, Schulman, et al. 2009).
Seeds were germinated in moistened paper in Petri dishes and then plantlets were grown in pots in the greenhouse. Leaf tissue was sampled from single individuals of each genotype and total genomic DNA was extracted using a Cetyl TrimethylAmmonium Bromide (CTAB) procedure (Doyle JJ and Doyle JL 1989).
Illumina and 454 Sequencing
DNAs were randomly (mechanically) sheared into fragments for sequencing. Paired-end libraries were prepared as recommended by Illumina (Illumina Inc., San Diego, CA) with minor modifications. Illumina reads were preprocessed to remove Illumina adapters by using CLC-BIO Genomic Workbench 7.0.4 (CLC-BIO, Aarhus, Denmark). This tool was also used for quality trimming with default setting and to define the length of the reads at 75 nt.
For some experiments, reads obtained by 454 sequencing (454 Life Science, Branford, CT) of genomic DNA of the highly inbred sunflower line HA412-HO were used. Also, these reads were trimmed for quality with default setting, checked for adapters and cut at 400 nt in length using CLC-BIO Genomic Workbench 7.0.4.
For both 454 and Illumina sequences, all reads containing organellar DNA sequences were removed using CLC-BIO Genomic Workbench 7.0.4, by mapping to an in-house developed library of chloroplast and mitochondrial sequences of sunflower and other dicotyledons.
Graph-based Clustering of Sequences of a Homozygous Line
In order to identify putative repeat families, graph-based clustering (using RepeatExplorer; Novák et al. 2010) was performed on a random set of cleaned genomic 454 reads (790,742 reads for a total coverage of 0.1X) of the highly inbred sunflower line HA412-HO. The output of RepeatExplorer contained both annotated and nonannotated clusters. To increase the number of annotated clusters, similarity searches on the remaining unknown clusters were performed by BLASTN and TBLASTX search against a library of repetitive sequences of sunflower, SUNREP (Natali et al. 2013), and against a library composed of 18 full-length LTR-RTs, 6 incomplete LTR-RTs, and 2 nonautonomous LTR-RTs (Buti et al. 2011). All annotated clusters were collected to prepare an in-house reference library of sunflower LTR-RT-related sequences.
Finally, pairwise clustering between a random set of Illumina reads of the line HA214-HO and a random set of reads for each of the analyzed genotypes were performed using RepeatExplorer, in order to verify that no supplementary repeats occurred in those genotypes compared with the HA214-HO line.
Redundancy Estimation of Clusters
Relative redundancy of each LTR-RT-related cluster was estimated by mapping Illumina reads of each of the 15 genotypes to the reference library of LTR-RTs. Mapping was performed using CLC-BIO Workbench 7.0.4, with the following parameters: Mismatch cost = 1, deletion cost = 1, insertion cost = 1, similarity = 0.9, and length fraction = 0.9.
In this analysis, multireads (i.e., those reads that matched multiple distinct sequences) were distributed randomly, and hence the number of mapped reads to a single sequence would be only an indication of its redundancy. On the other hand, if all sequences of a sequence class are taken together, the total number of mapped reads (in respect to total genomic reads) reveals the effective redundancy of that class. Each redundancy value was reported as total number of mapped reads per million reads used for mapping.
Analysis of Proximity of LTR-RTs to Genes
For every genotype a set of Illumina paired-end reads (trimmed for quality and adapters but not at a specific length) were mapped onto a library, obtained joining the set of LTR-RT clusters assembled by RepeatExplorer and a set of protein-encoding genes representing the whole sunflower transcriptome (Rowe and Rieseberg 2013).
Mapping was carried on using BWA (Li and Durbin 2009) version 0.7.10-r789 with the following parameters: aln -t 4 -l 12 -n 4 -k 2 -o 3 -e 3 -M 2 -O 6 -E 3. The resulting paired-end mappings were resolved via the “sampe” module of BWA, and the output was converted into a “bam” file using SAMtools (Li et al. 2009) version 0.1.19. SAMtools was used to extract the reads mapping in pair with the function “view,” option -F 12.
Statistical Analyses
Redundancy variations of retrotransposons among genotypes were investigated with a principal component analysis (PCA) and a nonparametric multivariate analysis, namely the permutational Multivariate ANalysis Of VAriance (MANOVA) (Anderson 2001). For each cluster, the redundancy data on 15 genotypes were used to build a Euclidean distance matrix. The PCA was performed using the implementation of the R package FactoMineR version 1.26 (Lê et al. 2008); the permutational MANOVA was carried out using the implementation provided by the R package vegan version 2.0-10 (Oksanen et al. 2013). An in-house R script was used for building the distance matrices and performing statistical tests for all the clusters.
The R package pvclust version 1.3-2 (Suzuki and Shimodaira 2006) was used to build a dendrogram on the redundancy data by assessing the uncertainty in hierarchical cluster analysis via multiscale bootstrap resampling with 1,000 bootstrap replications.
To define the extent of variation related to random sampling, concerning the redundancy and the number of mapped paired reads (MPR) that match onto a gene and an LTR-RT, we randomly sampled the Illumina read set of the wild accession North Dakota into six subsets of 6 million reads each, with reads trimmed at 75 nt, for the analysis of redundancy, and into six subsets of 7 million reads each, with reads of variable length, to study the proximity to genes. Each subset was mapped with the parameters reported above. The maximum percent difference measured between subsets was used as a threshold to establish the percentage of random variation: Differences between genotypes higher than the threshold values were considered as relevant, that is, not related to random sampling of the reads.
Results
Characterization of the LTR-Retrotransposons of the Inbred HA412-HO
The repetitive component of the sunflower genome (line HA412-HO) was initially investigated in a random sample of 454 reads, corresponding to a total coverage of 0.1X, using RepeatExplorer (Novak et al. 2010).
This tool discovers and characterizes repetitive sequences in eukaryotic genomes, allowing de novo repeat identification, based on finding and quantifying similarities between individual sequence reads. This approach produced separate and automatically annotated clusters of frequently connected reads that represented individual families of repetitive elements. Overall, 601,190 of the 790,742 reads were grouped into 46,563 clusters, representing about 76% of the genome. In our experiment, top clusters, that is, all those clusters representing more than 0.01% of the genome, amounted to 288. The other clusters represent low-copy repeat families and were not considered in our analyses. It is presumable that variations of low-copy retrotransposons can have a role in determining phenotypic differences between individuals; however, low-copy retrotransposon variation analysis requires the availability of a reference genome sequence and of extensive resequencing of genotypes, at least at loci carrying such elements.
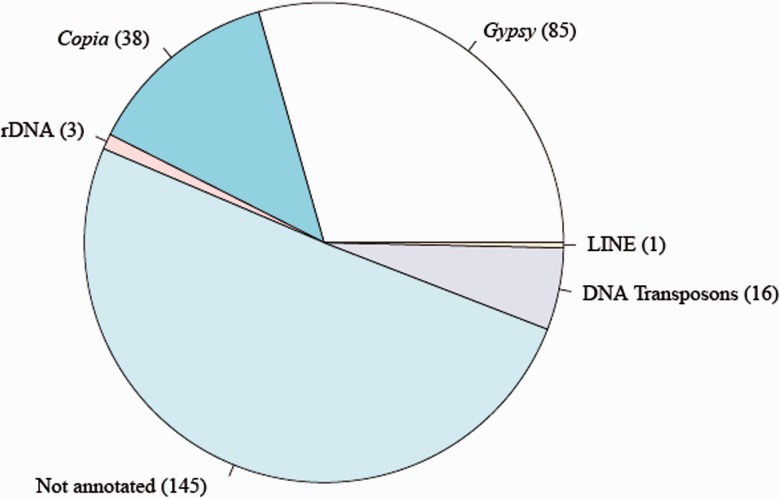
The number of singletons, that is, reads that were not assembled by RepeatExplorer, was 189,544, which corresponds to 24% of the total nuclear reads. Hence, we estimate that 24% of the genome belongs to the low-copy fraction. A representation of the abundance of the clusters produced by RepeatExplorer is presented in figure 1.
Fig. 1.—
The repeat class distribution of the 288 top (most frequent) clusters obtained assembling a random set of 454 reads (0.1X coverage) using RepeatExplorer. The amount of clusters composing each repeat class is reported inside brackets.
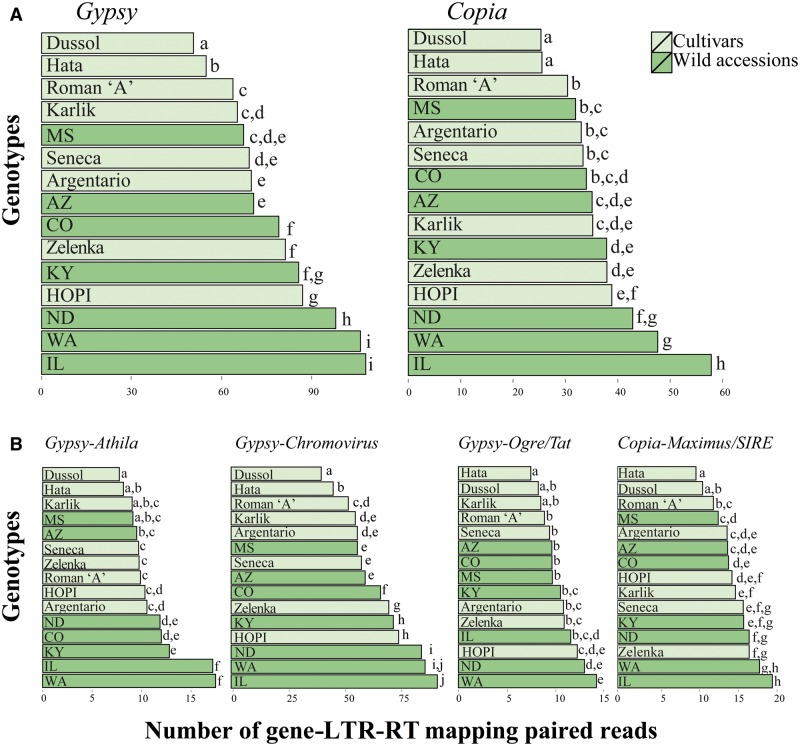
The majority of top clusters identified by RepeatExplorer were not annotated. Among the annotated clusters, 123 were identified as similar to LTR-RTs of Gypsy (85) and Copia (38) superfamilies. This analysis is consistent with previous results that found that Gypsy LTR-RTs are largely more redundant than Copia in the sunflower genome (Cavallini et al. 2010; Staton et al. 2012; Natali et al. 2013).
Each cluster is a group of sequences which share sequence similarity and hence a common progenitor. The identification of protein-encoding domains in LTR-RT clusters allowed us to establish that clusters belong to different families within the Gypsy and the Copia superfamilies. Hence, different clusters belonging to the same LTR-RT family can be considered as LTR-RT subfamilies. For the Copia superfamily, seven families were identified (table 1), the most redundant being Maximus/SIRE (11 clusters, corresponding to 5.5% of the genome) and AleII elements (7 clusters, 4.3%). Only three families were identified within the Gypsy superfamily, in which the Chromovirus family was largely the most redundant, with 58 clusters, accounting for more than 23% of the genome (table 1).
Table 1.
Description of the 123 Clusters Obtained using RepeatExplorer and Annotated as LTR-Retrotransposons, and Their Genome Proportion in the Inbred Sunflower Line HA412-HO
| Superfamily | Family | Number of Clusters | Genome Proportion (%) |
|---|---|---|---|
| Copia | AleI/Retrofit | 2 | 0.688 |
| AleII | 7 | 4.269 | |
| Angela | 5 | 0.610 | |
| Bianca | 3 | 0.126 | |
| Ivana/Oryco | 2 | 0.076 | |
| Maximus/SIRE | 11 | 5.523 | |
| TAR/Tork | 5 | 0.509 | |
| Unknown | 3 | 0.060 | |
| Total | 38 | 11.861 | |
| Gypsy | Athila | 13 | 3.182 |
| Chromovirus | 58 | 23.097 | |
| Ogre/Tat | 14 | 4.241 | |
| Total | 85 | 30.520 |
All sequences included in the 123 clusters annotated as LTR-RTs were collected to produce a library that was used as a reference for the subsequent analyses of LTR-RT-related intraspecific variability of H. annuus.
Previous results indicated that, in this inbred line, retrotransposons accounted for around 80% of the genome (Natali et al. 2013). Mapping the reference library with the same set of reads as in Natali et al. (2013), we could estimate that sequences included in the library accounted for about 40% of the sunflower genome and hence more than 50% of its repetitive component.
LTR-Retrotransposon Redundancy Variability among Sunflower Genotypes
Assuming that Illumina reads in our experiments are sampled with uniform biases, if any, for particular sequence types, we estimated the frequency of each LTR-retrotransposon-related cluster in each genotype by counting the total number of reads (per million) of that genotype, that mapped to the sequences of the cluster. This method has already been used in many plant species (Swaminathan et al. 2007; Tenaillon et al. 2011; Barghini, Natali, Cossu, et al. 2014; Barghini, Natali, Giordani, et al. 2014) and also in sunflower (Natali et al. 2013).
We analyzed LTR-RTs in 15 genotypes of H. annuus: 8 wild accessions and 7 cultivars. The wild accessions represented widespread provenances in North America; the cultivars were randomly chosen from different countries in which sunflower is a major crop, representing a broad sample of genetic diversity in the domesticated materials of this species. The occurrence of large genetic variability among these sunflower cultivars was already shown by Inter-Retrotransposons-Amplification-Polymorphism (Kalendar et al. 1999) analysis (Vukich, Schulman, et al. 2009).
To avoid the exclusion of genotype-specific LTR-RT families from the reference library, a read sample of the HA412-HO line was used for pairwise clustering against read samples of each of the 15 genotypes used (see Materials and Methods). No clusters specific to the analyzed genotypes and absent in the HA412-HO line were found.
The Illumina reads of the 15 genotypes were mapped onto the reference library of 123 clusters (made up by 11,456 contigs) allowing the evaluation of differences in the overall redundancy of this set of retrotransposons in cultivars and wild genotypes.
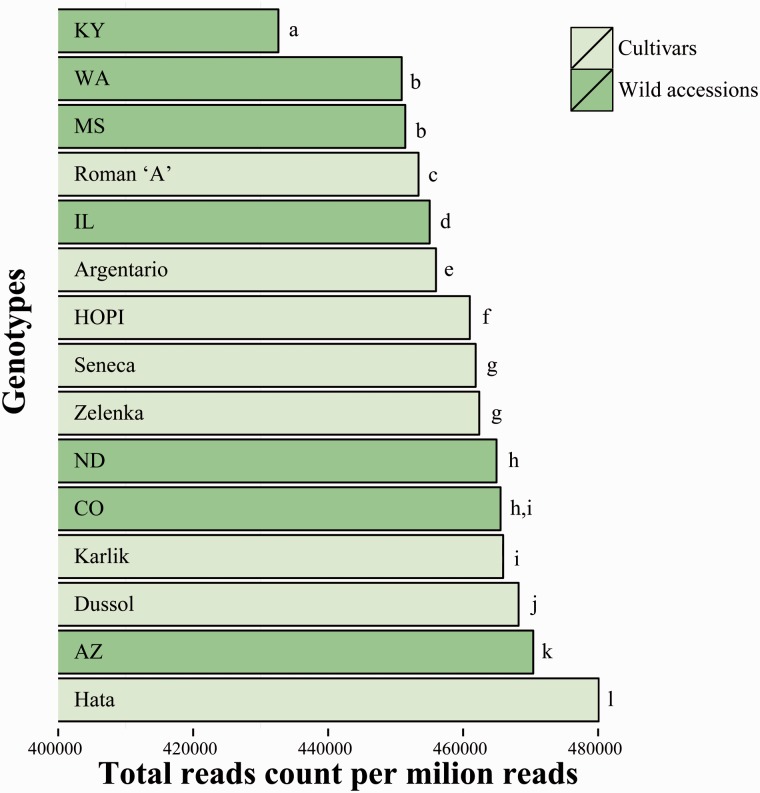
The redundancy of the whole set of analyzed LTR-RTs is reported in figure 2. The extent of redundancy ranges from 433,000 (43.26%, for the Kentucky accession) to 480,000 mapped reads per million (48.00%, for the cv. Hata).
Fig. 2.—
LTR-RT redundancy in 15 Helianthus annuus genotypes, measured by counting the number of reads (per million) mapping the set of LTR-RTs included in the reference library. Bars not sharing the same letter are to be considered as different according to a threshold indicating the extent of differences related to random sampling of reads (see Materials and Methods).
Variations in redundancy data between genotypes, obtained by mapping with Illumina reads, could be related to the stochasticity in read packages used for mapping, rather than to real differences in redundancy. To determine the extent of redundancy variation attributable to random sampling of reads, we produced six random subsets of Illumina reads from one wild accession (North Dakota), and then we mapped these reads to the reference library and counted the number of mapped reads. The maximum percent difference in the total number of mapped reads between subsets amounted to 0.14% (supplementary material S2, Supplementary Material online). Hence, we assumed 0.14% as a threshold value to compare LTR-RT frequency among the different genotypes: When two genotypes differed in LTR-RT frequency for more than the threshold, then their difference in LTR-RT redundancy was considered relevant. Differences between genotypes were generally larger than the threshold, indicating that differences in redundancy were biologically meaningful, not related to random sampling (fig. 2).
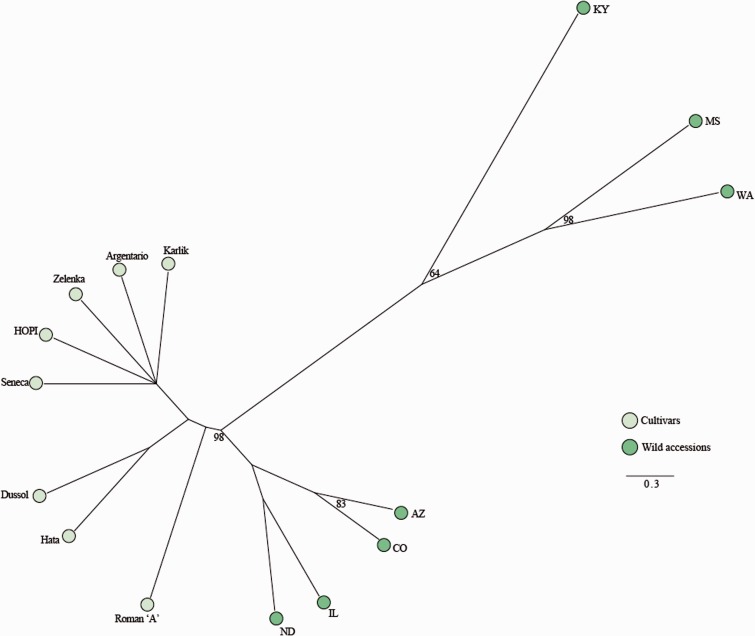
Interestingly, wild accessions have generally lower levels of LTR-RT redundancy compared with cultivars (fig. 2). In fact, 3 of the 4 genotypes with the lowest LTR-RT frequencies are wild accessions, and 3 of the 4 genotypes with the highest LTR-RT frequencies are cultivars. Furthermore, a dendrogram based on LTR-RT redundancy values separated significantly cultivated genotypes from wild accessions (fig. 3).
Fig. 3.—
Unweighted pair group method with arithmetic mean dendrogram produced by a distance matrix based on LTR-RT redundancy in 15 sunflower genotypes (7 wild accessions and 8 cultivars). Numbers indicate multiscale bootstrap resampling (only values > 60% are given). The bar represents the genetic distance.
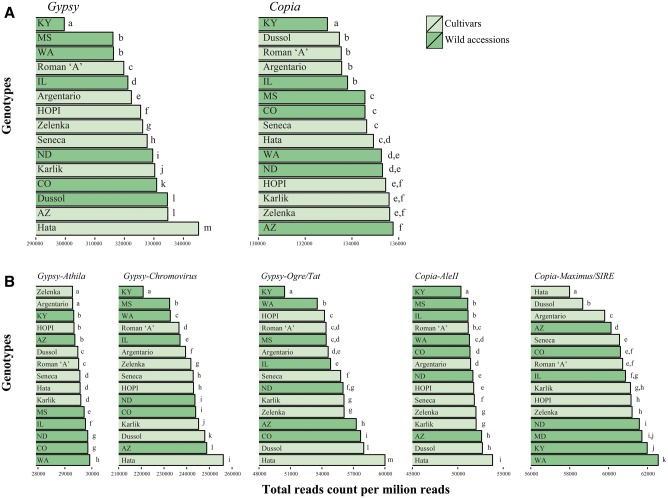
The number of mapped reads per million was also counted in all genotypes keeping the Gypsy- and Copia-related clusters separate and, at a family level, Chromovirus, Ogre/Tat, Athila, Maximus/SIRE, and AleII LTR-RTs, that is, the most redundant families. First, the maximum percentage of variation was calculated among the six randomly produced read packages of the North Dakota accession, as above, for each superfamily or family (supplementary material S2, Supplementary Material online); then, these values were used as threshold to compare frequencies of different superfamilies and families. It is to be noted that the largest maximum percent variation due to random sampling was for the Ogre/Tat family and corresponded to less than 0.5%.
This allowed us to define the level of redundancy of the different LTR-RT superfamilies and families (fig. 4), and even different clusters, collected in the library, in all the genotypes studied.
Fig. 4.—
Redundancy of Gypsy and Copia superfamilies (A) and families (B) in the 15 genotypes analyzed. Bars not sharing the same letter are to be considered as different according to a threshold indicating the extent of differences related to random sampling of reads (see Materials and Methods). The respective genome proportion of the superfamily or family is reported inside brackets.
The same pattern of redundancy variation was found in both Copia and Gypsy superfamilies, that is, wild accessions generally showed lower levels of LTR-RT redundancy compared with cultivars. A similar pattern was observed for individual families, although there were exceptions as well. For example, the Gypsy–Chromovirus LTR-RTs and the Gypsy–Ogre/Tat LTR-RTs, which represent 23.097% and 4.241% of the genome of line HA412-HO, respectively (table 1), were more abundant in cultivars than in wild accessions, but Gypsy–Athila LTR-RTs (3.182% of the genome;1 table 1) were more abundant in wild accessions compared with cultivars. Within the Copia superfamily, the redundancy levels of families that have a genome representation >1% (table 1) resulted higher in cultivars than in wild accessions for the AleII family and the opposite trend was observed for the Maximus/SIRE family.
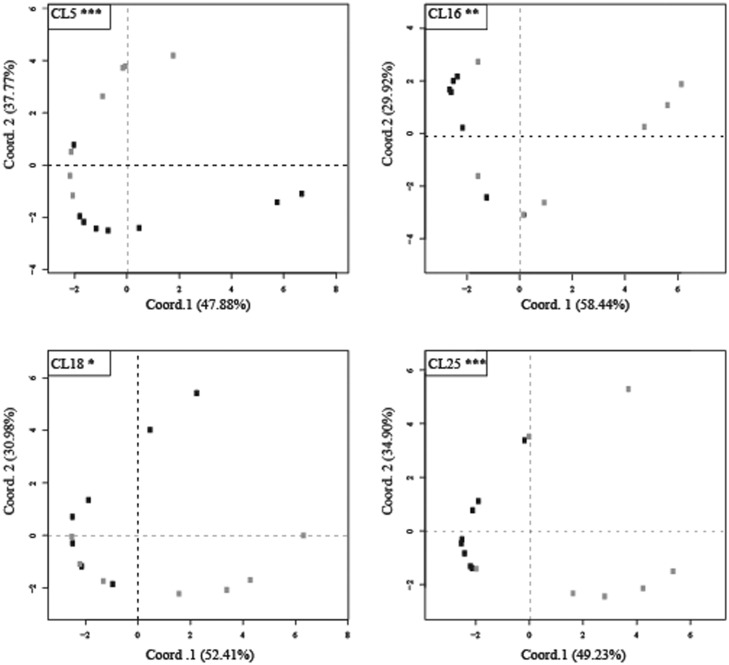
At cluster (subfamily) level, PCA of the intraspecific relative redundancy (of which four examples are reported in fig. 5) was performed. Keeping wild and cultivated genotypes separate, the mean redundancy was significantly (P < 0.05) higher or lower in cultivars compared with wild accessions for 27 of the 123 clusters (supplementary material S3, Supplementary Material online).
Fig. 5.—
PCA plots of redundancy values of four clusters (CL), produced by RepeatExplorer, in cultivars (dark dots) and wild accessions (light dots) of Helianthus annuus. The percentage of variation accounted by each axis is reported. Asterisks mark permutational MANOVA significance with the following significance codes: ***0.001; **0.01; *0.05.
Of these clusters, 8 showed higher redundancy values in cultivars compared with wild accessions (7 Gypsy–Chromovirus, 1 Copia–TAR/Tork) and 19 showed lower redundancy values (3 Gypsy–Athila, 11 Gypsy–Chromovirus, 1 Gypsy–Ogre/Tat, 1 Copia–AleII, 1 Copia–Angela, 2 Copia–Maximum/SIRE).
Proximity of Retrotransposons to Genes
Retrotransposons increase their frequency by inserting retrotranscribed copies in loci widespread in the genome. An important phenotypic effect of TE mobility derives from insertion of elements in proximity to or within genes, which consequently loose or change their function (Butelli et al. 2012; Falchi et al. 2013). To infer the potential impact of TEs on gene function, we analyzed the association between LTR-RTs and protein-encoding genes in the sunflower genome.
The proximity of LTR-RTs to genes in the 15 selected H. annuus genotypes was studied by mapping Illumina paired-ends reads to both the reference library of LTR-RTs and a set of protein-encoding genes representing the whole sunflower transcriptome (Rowe and Rieseberg 2013). The analysis was performed with a set of Illumina paired-end reads from every accession. Different patterns of paired-ends mapped can be obtained (supplementary material S4, Supplementary Material online).
Table 1 reports the number of mapping paired reads (MPR) of which at least one mapping onto an LTR-RT and the number of paired reads of which one mapped onto an LTR-RT and the other onto a gene (hereafter called gene-RT MPR) in the analyzed genotypes.
Table 2.
Total Number of Illumina Paired Reads of which at least One Mapped to an LTR-RT and Number of Illumina Paired Reads of which One Mapped to an LTR-RT and the Other onto a Gene
| Genotype | Total Number of Paired Reads of which at least One Mapped onto an LTR-RT | No. of Gene-RT MPR | No. of Gene-RT MPR × Million of Paired Reads of which at least One Mapped onto an LTR-RT |
|---|---|---|---|
| Hata | 9,520,320 | 2,604 | 273.52 |
| Dussol | 7,471,766 | 1,961 | 262.45 |
| Argentario | 2,849,970 | 1,120 | 392.99 |
| Karlik | 6,608,140 | 2,386 | 361.07 |
| Zelenka | 2,503,434 | 1,075 | 429.41 |
| Roman “A” | 5,200,227 | 1,847 | 355.18 |
| Hopi | 4,267,411 | 1,982 | 464.45 |
| Seneca | 3,819,193 | 1,439 | 376.78 |
| Mean (cultivars) | 364.48 | ||
| AZ | 4,036,773 | 1,549 | 383.72 |
| CO | 6,274,800 | 2,642 | 421.05 |
| IL | 4,753,115 | 3,063 | 644.42 |
| KY | 3,541,359 | 1,851 | 522.68 |
| MS | 5,771,509 | 2,253 | 390.37 |
| ND | 1,330,2214 | 7,137 | 536.53 |
| WA | 1,680,349 | 1,061 | 631.42 |
| Mean (wild accessions) | 504.31 |
Because the coverage used for this analysis was relatively low, it was necessary to establish the extent of variation in the number of gene-RT MPR (i.e., the extent of proximity of LTR-RTs to genes) determined by the stochasticity in read packages used for mapping. Hence, we analyzed the frequency of gene-RT MPR in six subpackages of Illumina paired-end reads of the same genotype (North Dakota accession). In supplementary material S2, Supplementary Material online, the maximum percent difference in these subpackages is reported for the whole set of retrotransposons, for the two superfamilies, and for the most redundant LTR-RT families. Such values were assumed as thresholds to compare the gene-RT MPR frequency among the different genotypes: If two genotypes differed in gene-RT MPR frequency for more than the threshold, then it was assumed that in one genotype more retrotransposons lie close to genes than in the other genotype. It can be observed that, for Copia families (except Maximus/SIRE), the maximum percent variation due to stochasticity can be high (exceeding 25%), presumably because of the low frequency of these elements in the sunflower genome. In these cases, the analysis of gene-RT MPR was not taken into consideration. In contrast, the stochastic variation for Gypsy LTR-RT families was always lower than 20%, and we therefore focused on these families for the following analyses.
The occurrence of gene-RT MPR is reported in figure 6 for the Gypsy superfamily. The frequency of gene-RT MPR, indicating the number of sites in which a Gypsy LTR-RT lies close to a gene, was generally higher in wild accessions than in cultivars. This trend was also confirmed in each of the three Gypsy families to which individual clusters that make up the library of LTR-RTs belonged (fig. 6). PCA confirmed that Gypsy, as a whole superfamily, and in specific Athila and Chromovirus LTR-RTs, differed in the proximity of these elements to genes between cultivars and wild accessions (supplementary material S5, Supplementary Material online); the elements of these families were found to be close to more genes in wild than in cultivated genotypes.
Fig. 6.—
Frequency of gene-RT mapping paired reads in the 15 genotypes analyzed in all Gypsy and Copia LTR-RTs found (A) and in Athila, Chromovirus, Ogre/Tat, and Maximus/SIRE LTR-RTs (B). Bars not sharing the same letter are to be considered as different according to a threshold indicating the extent of differences related to random sampling of reads (see Materials and Methods).
The number of gene-RT MPR × million paired reads of which at least one mapped onto an LTR-RT was also calculated separately for each LTR-RT family with the aim of establishing if some LTR-RT families are more prone than others to insert in proximity of genes. This analysis was performed only in the North Dakota wild accession, for which the largest number of reads were available. The results of such analysis are reported in table 3. The number of gene-RT MPR per million was different among families, ranging from 0.33 for the Ogre/Tat family to 2.27 for the Ivana/Oryco family.
Table 3.
Total Number of Illumina Paired Reads of North Dakota Wild Accession of which at least One Mapped onto an LTR-RT of a Certain Family and Number of Paired Reads of which One Mapped onto an LTR-RT of the Same Family and the Other onto a Gene
| LTR-RT Family | Total Number of Paired Reads of which at least One Mapped onto an LTR-RT | No. of Gene-RT MPR | No. of Gene-RT MPR × Million of Paired Reads of which at least One Mapped onto an LTR-RT |
|---|---|---|---|
| Copia-AleI/Retrofit | 176,212 | 122 | 692.35 |
| Copia-AleII | 1,320,690 | 697 | 527.75 |
| Copia-Angela | 221,598 | 224 | 1,010.84 |
| Copia-Bianca | 47,430 | 37 | 780.10 |
| Copia-Ivana/Oryco | 19,394 | 44 | 2,268.74 |
| Copia-Maximus/SIRE | 1,760,696 | 834 | 473.68 |
| Copia-TAR/Tork | 193,224 | 146 | 755.60 |
| Copia-Unknown | 17,356 | 20 | 1,152.34 |
| Gypsy-Athila | 899,879 | 586 | 651.20 |
| Gypsy-Chromovirus | 6697673 | 3,780 | 564.38 |
| Gypsy-Ogre/Tat | 1,948,062 | 647 | 332.12 |
Concerning the genes lying in proximity to LTR-RTs, we focused on those belonging to large gene families (represented by more than 100 sequences in the sunflower transcriptome) because of the relatively small number of paired-end reads used in this analysis. The number of gene-RT MPR × million paired reads for each analyzed gene family is reported in table 4. Overall, larger values of gene-RT MPR × million paired reads were found in wild than in cultivated genotypes. For three large gene families (encoding Leucine-Rich-Repeat containing proteins, Pentatricopeptide-Repeat containing proteins, and Sodium Transporters) differences between cultivars and wild accessions resulted significant.
Table 4.
Mean Number (and standard error, S.E.) of Illumina Paired Reads (per million) of 8 Cultivars and 7 Wild Genotypes of Sunflower, of which at least One Mapped onto an LTR-RT and the Other onto a Gene Belonging to a Gene Family Represented by at least 100 Sequences in the Sunflower Transcriptome (Rowe and Rieseberg 2013)
| Encoded Protein | No. of Sequences | Cultivars |
Wild Accessions |
|||
|---|---|---|---|---|---|---|
| Mean No. of Gene-RT MPR | S.E. | Mean No. of Gene-RT MPR | S.E. | |||
| Leucine-Rich-Repeat-related | 1,448 | 3.09 | 0.19 | 3.72 | 0.27 | * |
| ABC-Transporter | 425 | 0.84 | 0.11 | 0.94 | 0.17 | ns |
| DNAJ Homologous | 343 | 0.92 | 0.09 | 1.07 | 0.14 | ns |
| Pentatricopeptide-Repeat | 269 | 0.89 | 0.12 | 1.22 | 0.10 | * |
| S-locus-related | 243 | 0.53 | 0.07 | 0.66 | 0.11 | ns |
| Heat Shock Responsive | 222 | 0.61 | 0.12 | 0.76 | 0.07 | ns |
| NAC Transcription Factor | 179 | 0.70 | 0.04 | 0.70 | 0.08 | ns |
| Terpenoid Cyclases | 176 | 0.35 | 0.02 | 0.29 | 0.06 | ns |
| Clathrin | 163 | 0.26 | 0.05 | 0.35 | 0.06 | ns |
| Alcohol Dehydrogenase | 161 | 0.36 | 0.05 | 0.38 | 0.03 | ns |
| Sodium Transporter | 151 | 0.21 | 0.03 | 0.37 | 0.03 | ** |
| Histone | 146 | 0.18 | 0.02 | 0.20 | 0.03 | ns |
| Cellulose Synthase | 124 | 0.13 | 0.03 | 0.13 | 0.02 | ns |
| Aquaporin | 117 | 0.23 | 0.03 | 0.26 | 0.06 | ns |
| Lipid Transfer Protein | 101 | 0,41 | 0.07 | 0.38 | 0.08 | Ns |
Note.—Each gene family is identified by the encoded protein and for each gene family the number of sequences in the transcriptome is indicated. The significance of differences between cultivars and wild accessions is reported. ns: not significant.
*P < 0.05; **P < 0.01.
Discussion
Retrotransposon-related Intraspecific Variability in Helianthus annuus
The occurrence of large variation in plant genome size has been ascertained both within and among species by means of cytophotometric and biochemical analysis and has been attributed to variation in the proportion of repetitive DNA (Flavell 1986).
The development of DNA sequencing techniques has greatly improved the knowledge of sequences underlying genome size variation and that of the mechanisms that produce such variation.
In plants, most of the variation relates to the retrotransposon component of the genome that is subject to rapid turnover (Ma and Bennetzen 2004; Wang and Dooner 2006). Retrotransposons can proliferate in a relatively short time span, based on the capacity of some of them to escape epigenetic control by the host genome (Vitte and Bennetzen 2006). TEs can also be rapidly removed through unequal homologous and illegitimate recombination (Devos et al. 2002; Vitte and Panaud 2003).
Retrotransposon proliferation and DNA loss can determine the production of haplotypes with large differences in the occurrence of retrotransposons at specific loci, as observed comparing large orthologous regions through Bacterial Artificial Chromosome (BAC) sequencing in maize and rice (Brunner et al. 2005; He et al. 2006). Even in sunflower, a huge amount of retrotransposon insertion site variation has been reported (Vukich, Schulman, et al. 2009). Retrotransposon proliferation has been documented in the genus Helianthus, including H. annuus (Ungerer et al. 2009; Vukich, Giordani, et al. 2009; Buti et al. 2011). If LTR-RT proliferation and/or loss have occurred at different frequency in different haplotypes, then the number of LTR-RTs in the genome could be further subject to variation by the random combination of LTR-RT-rich haplotypes.
Such processes have been studied primarily in model species. At the intraspecific level, a well-studied case of variation in the repetitive fraction of the genome is maize (Springer et al. 2009; Albert et al. 2010). However, even in nonmodel species, which lack a reference genome sequence, novel methods that employ NGS and bioinformatics analysis can be conveniently used to explore the repetitive component of the genome (Swaminathan et al. 2007). These new approaches enable large, genome-wide comparative characterization and profiling of DNA repeats indifferent to genotypes of one and the same species.
In sunflower, we employed NGS techniques to produce a library of retrotransposon sequences. The composition of this library reflects the structure of sunflower genome reported in previous studies (Staton et al. 2012; Natali et al. 2013; reviewed in Giordani et al. 2014). Gypsy elements represented the majority of sequences in the library. Both Gypsy- and Copia-related clusters have been further characterized identifying the LTR-RT families to which they putatively belong. Gypsy elements belonged to three families, with a large prevalence of Chromovirus-related LTR-RTs, an aspect previously described in this species (Staton et al. 2012). Six Copia families were identified in the library.
The library of retrotransposon-related sequences was then used to perform a quantitative and qualitative survey of intraspecific variation of the redundancy of this set of LTR-RTs among 7 wild and 8 cultivated genotypes of H. annuus.
Analyzing the whole library, considerable variation in redundancy was observed among genotypes. Such variation was found for both Gypsy and Copia LTR-RTs and even for each LTR-RT family. Within each superfamily, different families showed different trends: For example, Athila and Maximus/SIRE LTR-RTs were more redundant in wild than in cultivated genotypes, while the opposite trend was found for Chromovirus and AleII LTR-RTs.
The effects of retrotransposon mobility on the plant phenotype are related to their insertion in proximity to genes, of which they may affect the expression rate (Butelli et al. 2012; Falchi et al. 2013). Another mechanism by which retrotransposon mobility affects the phenotype of the host is related to the inactivation by methylation of the region into which the retrotransposon is inserted: If such region includes a gene, this gene may be inactivated (Hollister and Gaut 2009).
The proximity of LTR-RTs to genes was evaluated counting the number of gene-RT MPR. Analyzing the different families separately, the number of paired reads of which one mapped onto an LTR-RT and the other onto a gene was the highest for the Ivana/Oryco family and the lowest for the Ogre/Tat family.
In this sense, according to Venner et al. (2009), it is confirmed that LTR-RTs are a community of different organisms in the genome, with superfamilies, which can be described as species, and with “subspecies,” i.e., the families, characterized by different protein sequences, activity, and evolution history.
For the whole set of analyzed LTR-RTs, the number of gene-RT MPR per million of paired reads of which at least one mapped onto an LTR-RT ranged from 262.45 to 644.42 among the analyzed genotypes. Although such values were small, they are apparently not related to random sampling of the read pairs, hence they should be considered as meaningful. On the other hand, even a very low number of insertions close to genes can have dramatic effects on the phenotype of the plant. For example, the occurrence of different number of repeats around 60 kbp upstream of the tb1 gene heavily affects the maize phenotype with regard to apical dominance (Doebley et al. 1997). Hence the observed trend that LTR-RTs are more close to genes in wild than in cultivated sunflower genotypes could have phenotypic consequences.
A Role for LTR-Retrotransposons in the Sunflower Domestication?
It is generally suggested that only a few loci contribute to the process of domestication of plants and animals from their wild progenitors (Wang et al. 1999; Gepts and Papa 2002; Olsen and Purugganan 2002; Doebley 2004), hence the divergence at the molecular level might be relatively small. However, domestication is the direct effect of artificial selection, which can determine extensive molecular divergence on characters that are naturally selected in the wild but neutral in cultivation (Innan and Kim 2004). The reduction in the effective population size during artificial selection can also contribute to increase in divergence between domesticated and wild genotypes (Eyre-Walker et al. 1998).
Many of the mutations that accumulate during domestication might be even deleterious, as reported by Lu et al. (2006) in rice. Normally, the accumulation of deleterious mutations in sexually reproducing species is infrequent because recombination enables these mutations to be removed (Carvalho 2003). However, the practice of inbreeding during domestication can reduce the effectiveness of crossover in breaking up regions of low recombination.
Among mutations accumulated during domestication, those related to amplification or loss of transposons might have a prominent role. The involvement of transposons in the domestication of plant species has been reported in a few cases. For example, a striking amplification of the mPing miniature-inverted-repeat-TE has been reported in domesticated rice (Naito et al. 2006). In our experiments, 27 of the 123 sunflower LTR-RT-related clusters showed significant (according to permutational MANOVA) differences in redundancy between cultivated and wild genotypes. The different LTR-RT redundancy between wild and cultivated genotypes can be explained by different hypotheses: 1) such variation might be related to the random sampling of the genotypes selected for this study, although the number of variable clusters is apparently too high (27 of 123, that is, 22.9%); 2) the different LTR-RT redundancies between cultivars and wild accessions might be related to the origin of cultivated sunflowers from relatively few genotypes initially selected by native Americans and then bred by European explorers (Harter et al. 2004); if differences in LTR-RT redundancy did not result in advantages to those genotypes, then the smaller or higher (depending on the LTR-RT family) LTR-RT redundancies in cultivars than in wild accessions could be a consequence of genetic drift; 3) on the other hand, if the occurrence of a lower number of certain LTR-RTs in cultivars conferred advantages for those genotypes, one could deduce that low or high redundancies of certain LTR-RTs could have been unconsciously selected by the first breeders.
The effect—if any—of different redundancy of certain LTR-RTs on the phenotype would likely be related to the genomic regions in which such variants occurred. In fact, retrotransposon insertion usually determines both structural and functional chromatin modifications, which in turn is related to alterations in the function of neighboring genes. Counting the number of gene-RT MPR in the different genotypes allowed us to establish that, for some LTR-RT families, there is a trend to be inserted in proximity to genes with higher frequency in cultivars than in wild accessions. Focusing on gene families represented by at least 100 sequences in the sunflower transcriptome, three showed significant differences between wild and cultivated genotypes. Interestingly, one gene family encodes Leucine-Rich Repeat (LRR) domain-containing proteins, that is, one of the largest plant gene families, involved in plant defense (McHale et al. 2006). Another gene family encodes pentatricopeptide-repeat containing proteins, a very heterogeneous class of proteins, often targeted to mitochondria or chloroplasts, and involved in RNA editing, with effects on many characters, for example, plant development and environmental adaptation (Barkan and Small 2014).
Such differences in the proximity of retrotransposons to genes could contribute—at least in part—to the striking phenotypic differences between wild and cultivated sunflowers. During recent years, more than 100 genes have been shown to be involved in the sunflower domestication, for example, genes involved in plant architecture (i.e., branching), flowering time, and fatty acid synthesis (Blackman, Rasmussen, et al. 2011; Chapman and Burke 2012; Mandel et al. 2013, 2014; Baute et al. 2015); their involvement was shown by their extremely low sequence variability in cultivated materials and/or extreme genetic differentiation between wild and cultivated accession. Many of these so-called domestication genes likely affect the phenotype through variation in their expression level, which in turn can be affected by a different retrotransposon landscape in the neighboring chromosomal regions between cultivated and wild haplotypes.
The achievement of a complete reference genome sequence for sunflower, that has finally been made publicly available, will represent a primary tool for resequencing of specific loci in different genotypes and discovering the effects of retrotransposon variability on phenotype and, consequently, also on sunflower domestication.
Supplementary Material
Supplementary materials S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This research work was supported by PRIN-MIUR, Italy, Project “SUNREP: caratterizzazione molecolare della componente ripetitiva del genoma di girasole”. Thanks are due to Prof. Brandon Gaut for his critical reading of the manuscript.
Literature Cited
- Albert PS, Gao Z, Danilova TV, Birchler JA. 2010. Diversity of chromosomal karyotypes in maize and its relatives. Cytogenet Genome Res. 129:6–16. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. 2001. A new method for non‐parametric multivariate analysis of variance. Austral Ecol. 26:32–46. [Google Scholar]
- Barghini E, Natali L, Cossu RM, et al. 2014. The peculiar landscape of repetitive sequences in the olive (Olea europaea L.) genome. Genome Biol Evol. 6:776–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barghini E, Natali L, Giordani T, et al. 2014. LTR retrotransposon dynamics in the evolution of the olive (Olea europaea) genome. DNA Res. 22:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 65:415–442. [DOI] [PubMed] [Google Scholar]
- Baute GJ, Kane NC, Grassa C, Lai Z, Rieseberg LH. 2015. Genome scans reveal candidate domestication and improvement genes in cultivated sunflower, as well as post-domestication introgression with wild relatives. New Phytol. 206:830–838. [DOI] [PubMed] [Google Scholar]
- Belyayev A, et al. 2010. Transposable elements in a marginal plant population: temporal fluctuations provide new insights into genome evolution of wild diploid wheat. Mob DNA. 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman BK, Rasmussen DA, et al. 2011. Contributions of flowering time genes to sunflower domestication and improvement. Genetics 187:271–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman BK, Scascitelli M, et al. 2011. Sunflower domestication alleles support single domestication center in eastern North America. Proc Natl Acad Sci U S A. 108:14360–14365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner S, Pea G, Rafalski A. 2005. Origins, genetic organization and transcription of a family of non-autonomous helitron elements in maize. Plant J. 43:799–810. [DOI] [PubMed] [Google Scholar]
- Butelli E, et al. 2012. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24:1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buti M, et al. 2011. Temporal dynamics in the evolution of the sunflower genome as revealed by sequencing and annotation of three large genomic regions. Theor Appl Genet. 123:779–791. [DOI] [PubMed] [Google Scholar]
- Carvalho AB. 2003. The advantages of recombination. Nat Genet. 34:128–129. [DOI] [PubMed] [Google Scholar]
- Cavallini A, et al. 2010. Analysis of transposons and repeat composition of the sunflower (Helianthus annuus L.) genome. Theor Appl Genet. 120:491–508. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Burke JM. 2012. Evidence of selection on fatty acid biosynthetic genes during the evolution of cultivated sunflower. Theor Appl Genet. 125:897–907. [DOI] [PubMed] [Google Scholar]
- Devos KM, Brown JK, Bennetzen JL. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12:1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. 2004. The genetics of maize evolution. Annu Rev Genet. 38:37–59. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. 1997. The evolution of apical dominance in maize. Nature 386:485–488. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1989. Isolation of plant DNA from fresh tissue. Focus 12:13–15. [Google Scholar]
- Du J, et al. 2010. Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J. 63:584–598. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS. 1998. Investigation of the bottleneck leading to the domestication of maize. Proc Natl Acad Sci U S A. 95:4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchi R, et al. 2013. Three distinct mutational mechanisms acting on a single gene underpin the origin of yellow flesh in peach. Plant J. 76:175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell RB. 1986. Repetitive DNA and chromosome evolution in plants. Philos Trans R Soc Lond B Biol Sci. 312:227–242. [DOI] [PubMed] [Google Scholar]
- Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. 2008. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 18:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P, Papa R. 2002. Evolution during domestication. In: Encyclopedia of life sciences. London (United Kingdom): Nature Publishing Group. [Google Scholar]
- Giordani T, Cavallini A, Natali L. 2014. The repetitive component of the sunflower genome. Curr Plant Biol. 1:45–54. [Google Scholar]
- Harter AV, et al. 2004. Origin of extant domesticated sunflowers in eastern North America. Nature 430:201–205. [DOI] [PubMed] [Google Scholar]
- He G, et al. 2006. Haplotype variation in structure and expression of a gene cluster associated with a quantitative trait locus for improved yield in rice. Genome Res. 16:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser CB, Smith DM, Clevenger SB, Martin WC. 1969. The North American sunflowers (Helianthus). Torrey Bot Club Mem. 22:1–218. [Google Scholar]
- Hollister JD, Gaut BS. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19:1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, et al. 2011. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci U S A. 108:2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A, Le Rouzic A, Boutin TS, Filée J, Capy P. 2011. The struggle for life of the genome’s selfish architects. Biol Direct. 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kim Y. 2004. Pattern of polymorphism after strong artificial selection in a domestication event. Proc Natl Acad Sci U S A. 101:10667–10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalendar R, Grob T, Regina M, Suoniemi A, Schulman AH. 1999. IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet. 98:704–711. [Google Scholar]
- Kane NC, et al. 2011. Progress towards a reference genome for sunflower. Botany 89:429–437. [Google Scholar]
- Kumar A, Bennetzen JL. 1999. Plant retrotransposons. Annu Rev Genet. 33:479–532. [DOI] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. J Stat Softw. 25:1–18. [Google Scholar]
- Lentz DL, Pohl MD, Alvarado JL, Tarighat S, Bye R. 2008. Sunflower (Helianthus annuus L.) as a pre-Columbian domesticate in Mexico. Proc Natl Acad Sci U S A. 105:6232–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. 2013. How important are transposons for plant evolution? Nat Rev Genet. 14:49–61. [DOI] [PubMed] [Google Scholar]
- Llorens C, et al. 2011. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res. 39:D70–D74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, et al. 2006. The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet. 22:126–131. [DOI] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 101:12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel JR, McAssey EV, Nambeesan S, Garcia-Navarro E, Burke JM. 2014. Molecular evolution of candidate genes for crop-related traits in sunflower (Helianthus annuus L.). PLoS One 9:e99620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel JR, et al. 2013. Association mapping and the genomic consequences of selection in sunflower. PLoS Genet. 9:e1003378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW. 2006. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 7:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, et al. 2006. Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci U S A. 103:17620–17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, et al. 2009. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461:1130–1134. [DOI] [PubMed] [Google Scholar]
- Natali L, et al. 2006. Distribution of Ty3-gypsy- and Ty1-copia-like DNA sequences in the genus Helianthus and other Asteraceae. Genome 49:64–72. [DOI] [PubMed] [Google Scholar]
- Natali L, et al. 2013. The repetitive component of the sunflower genome as shown by different procedures for assembling next generation sequencing reads. BMC Genomics 14:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Požárková D, Macas J. 2003. Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol Biol. 53:399–410. [DOI] [PubMed] [Google Scholar]
- Novák P, Neumann P, Macas J. 2010. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, et al. 2013. Package ‘vegan’, Community ecology package. R package, version 2.0–10. Available from: http://CRAN.R-project.org/package=vegan. [Google Scholar]
- Olsen KM, Purugganan MD. 2002. Molecular evidence on the origin and evolution of glutinous rice. Genetics 162:941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson-Burch BD, Nettleton D, Voytas DF. 2004. Genomic neighborhoods for Arabidopsis retrotransposons: a role for targeted integration in the distribution of the Metaviridae. Genome Biol. 5:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piegu B, et al. 2006. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16:1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Rieseberg LH. 2013. Genome-scale transcriptional analyses of first-generation interspecific sunflower hybrids reveals broad regulatory compatibility. BMC Genomics 14:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini S, et al. 2002. Ty1/copia-and Ty3/gypsy-like DNA sequences in Helianthus species. Chromosoma 111:192–200. [DOI] [PubMed] [Google Scholar]
- Schilling EE. 1997. Phylogenetic analysis of Helianthus (Asteraceae) based on chloroplast DNA restriction site data. Theor Appl Genet. 94:925–933. [Google Scholar]
- Schilling EE, Linder CR, Noyes RD, Rieseberg LH. 1998. Phylogenetic relationships in Helianthus (Asteraceae) based on nuclear ribosomal DNA internal transcribed spacer region sequence data. Syst Bot. 23:177–187. [Google Scholar]
- Song X, Sui A, Garen A. 2004. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: effects on steroidogenesis and oncogenesis. Proc Natl Acad Sci U S A. 101:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, et al. 2009. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5:e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staton SE, et al. 2012. The sunflower (Helianthus annuus L.) genome reflects a recent history of biased accumulation of transposable elements. Plant J. 72:142–153. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Varala K, Hudson ME. 2007. Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey. BMC Genomics 8:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Hufford MB, Gaut BS, Ross-Ibarra J. 2011. Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome Biol Evol. 3:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Strakosh SC, Stimpson KM. 2009. Proliferation of Ty3/Gypsy-like retrotransposons in hybrid sunflower taxa inferred from phylogenetic data. BMC Biol. 7:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel R, Fransz PF, Verschure PJ. 2003. The eukaryotic genome: a system regulated at different hierarchical levels. J Cell Sci. 116:4067–4075. [DOI] [PubMed] [Google Scholar]
- Venner S, Feschotte C, Biémont C. 2009. Dynamics of transposable elements: towards a community ecology of the genome. Trends Genet. 25:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Bennetzen JL. 2006. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci U S A. 103:17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Fustier MA, Alix K, Tenaillon MI. 2014. The bright side of transposons in crop evolution. Brief Funct Genomics. 13:276–295. [DOI] [PubMed] [Google Scholar]
- Vitte C, Panaud O. 2003. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice (Oryza sativa L.). Mol Biol Evol. 20:528–540. [DOI] [PubMed] [Google Scholar]
- Vukich M, Giordani T, Natali L, Cavallini A. 2009. Copia and Gypsy retrotransposons activity in sunflower (Helianthus annuus L.). BMC Plant Biol. 9:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukich M, Schulman AH, et al. 2009. Genetic variability in sunflower (Helianthus annuus L.) and in the Helianthus genus as assessed by retrotransposon-based molecular markers. Theor Appl Genet. 119:1027–1038. [DOI] [PubMed] [Google Scholar]
- Wang Q, Dooner HK. 2006. Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc Natl Acad Sci U S A. 103:17644–17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Stec A, Hey J, Lukens L, Doebley J. 1999. The limits of selection during maize domestication. Nature 398:236–239. [DOI] [PubMed] [Google Scholar]
- Wicker T, Keller B. 2007. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 17:1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. 2001. Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 26:307–316. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8:973–982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.