Abstract
Previous studies focused on psychrophilic adaptation generally have demonstrated that multiple mechanisms work together to increase protein flexibility and activity, as well as to decrease the thermostability of proteins. However, the relationship between high and low temperature adaptations remains unclear. To investigate this issue, we collected the available predicted whole proteome sequences of species with different optimal growth temperatures, and analyzed amino acid variations and substitutional asymmetry in pairs of homologous proteins from related species. We found that changes in amino acid composition associated with low temperature adaptation did not exhibit a coherent opposite trend when compared with changes in amino acid composition associated with high temperature adaptation. This result indicates that during their evolutionary histories the proteome-scale evolutionary patterns associated with prokaryotes exposed to low temperature environments were distinct from the proteome-scale evolutionary patterns associated with prokaryotes exposed to high temperature environments in terms of changes in amino acid composition of the proteins.
Keywords: low temperature adaptation, amino acid composition, asymmetric substitution, homologous protein
Introduction
Thermophilic adaptation in prokaryotes has been a hot research topic in microbial ecology and evolutionary biology. Understanding what allows proteins from thermophiles to function at high temperatures, not only will be beneficial to protein engineering (Wijma et al. 2013), but also will help to answer evolutionary questions (Boussau et al. 2008; Puigbò et al. 2008; Hobbs et al. 2012). Temperature exerts strong selective pressure on organisms and is one of the most important drivers of the composition of microbial communities (Sharp et al. 2014). The mechanisms in microorganisms that respond to high temperatures have been shown to be multidimensional, involving DNA-binding proteins, specific topoisomerases, repair mechanisms that protect against DNA damage (Forterre 2002; Brochier-Armanet and Forterre 2007), more rigid membranes (Koga 2012), etc. Besides these physiological strategies to cope with high temperature, on the aspect of protein structure, an important thermal adaptation in a wide range of proteins is the inclusion of more hydrophobic amino acid residues and residues with altered charge or polarity in their sequences (López-García et al. 2015).
Until now, there have been more reports on high temperature adaptation than on low temperature adaptation (Metpally and Reddy 2009; Paredes et al. 2011; Struvay and Feller 2012); however, some studies have reported psychrophilic adaptations of individual enzymes (Pulido et al. 2007; Parvizpour et al. 2015). Psychrophilic proteins are generally characterized by a higher degree of structural flexibility, lower thermostability, and higher specific activity at low temperatures compared with their mesophilic counterparts (de Maayer et al. 2014). For instance, comparative genome analysis indicated that there is reduced use of Pro, Arg, and acidic amino acids, in a significant portion of the Psychrobacter arcticus proteome (Ayala-del-Rio et al. 2010). Similar results were also found in the psychrophilic strains of Shewanella and gamma-proteobacteria (Zhao et al. 2010). For the cold-adapted Archaea, proteins are characterized by a higher content of noncharged polar amino acids, particularly Gln and Thr and a lower content of hydrophobic amino acids, particularly Leu (Saunders et al. 2003). Therefore, it has been considered that low temperature adaptation may involve mechanisms opposite to those of high temperature adaptation, based on some single protein comparisons (Nakashima et al. 2003; Siglioccolo et al. 2010); for example, more flexible proteins with reduced hydrophobic cores, less charged surfaces, and more glycine residues (Feller 2010; Reed et al. 2013).
However, structural analyses of some individual psychrophilic enzymes have shown that amino acid substitutions at some crucial sites could effectively change the flexibility of proteins (Xie et al. 2009; Parvizpour et al. 2015), and different low temperature adaptation strategies have been reported in different organisms (Casanueva et al. 2010). If targeted amino acid substitutions are a common phenomenon, then amino acid frequencies in whole proteomes should not change drastically in the opposite direction for high temperature adaptation. Decade ago, Methé et al. (2015) had found that there are more significant differences in amino acid composition between the thermophiles and either mesophiles or psychrophiles than between mesophiles and psychrophiles, by comparing the amino acid compositions of 22 predicted proteomes. Thus, low temperature adaptation may not be the opposite process of high temperature adaptation, at least in terms of the extent of changes in the amino acid composition of their proteomes.
Many studies have attempted to correlate protein thermal stability with sequence-derived (Zeldovich et al. 2007; de Vendittis et al. 2008) or structure-derived (Maugini et al. 2009; Ma et al. 2010) features. So far, two major strategies have been used to investigate whether some amino acids are more favored than others in thermophilic proteins. One strategy is to compare the overall proportions of amino acid residues in protein sequences from microorganisms that live at different temperatures (Fukuchi and Nishikawa 2001; Singer and Hickey 2003; Berezovsky et al. 2007), and then to calculate the correlation between the proportions of amino acid residues with the optimal growth temperature (OGT) (Suhre and Claverie 2003; Zeldovich et al. 2007; Wang et al. 2008). This approach suffered from the phylogenetic dependences of the analyzed samples, as well as the effect of genome-wide GC content bias (McDonald 2010). A second strategy is to compare the abundances of amino acid residues from mesophilic and thermophilic proteins in pairs (Glyakina et al. 2007), and/or to evaluate the substitutional asymmetry based on the homologous proteins of species pairs (Haney et al. 1999; McDonald et al. 1999; McDonald 2001, 2010). The main purpose of pairing was to reduce the effects of natural evolution and GC content bias when the species were phylogenetically close. However, because of the limited amount of genomic data that were publicly available, it was usually difficult to collect enough data that matched the conditions.
The main goal of this study is to test the hypothesis that low temperature adaptation might not be an opposite trend of high temperature adaptation in terms of changes in the amino acid composition, and to determine the relationship between high and low temperatures adaptations. We analyzed variations in the amino acid residues of homologous proteins between psychrophiles and mesophiles and compared our results with the results obtained from a data set for high temperature adaptation based on statistically enough species pairs. Our results confirmed the original hypothesis and enriched our knowledge of the mechanisms of temperature adaptation of proteins.
Materials and Methods
Complete predicted proteome sequences of 2,806 prokaryotes were retrieved from the NCBI FTP server (ftp://ftp.ncbi.nih.gov/genomes/archive/old_genbank/Bacteria/; last accessed April 4, 2015). The OGT information was extracted from the DSMZ (http://www.dsmz.de; details shown in supplementary table S1, Supplementary Material online). We obtained OGT information for 915 of the 2,806 prokaryotes and tried to pair each of them with another species. Paired species were required to share taxonomic assignment at least to the family level (the genus level was preferential), have enough of an OGT difference (≥10 °C), and have no more than 5% difference in genomic GC content. When there were more than one candidate meeting the same requirements about taxonomic assignment and GC content, the one having the maximum OGT difference with target species was selected; if the maximum OGT difference appeared between target species and several candidates, the program selected one at random. Then, according to the phylogenetic analysis and the OGTs of the members of each pair, the species pairs were divided into two categories: lower temperature adaptation and higher temperature adaptation (details shown in table 1). The OGT ranges of psychrophiles and thermophiles were defined as ≤ 15 °C and ≥50 °C, respectively. The pairs containing merely species with OGTs from 15 to 50 °C were removed. Moreover, if the species appeared in two pairs, and then another species paired with it belonged to the same genera, only pairs with maximum OGT difference were kept.
Table 1.
Information about the Species Pairs Selected for the Analyses of Amino Acid Variations in Homologous Protein Pairs
| Types of Adaptations | The Member of Species Pair with Relative Lower OGTs |
The Member of Species Pair with Relative Higher OGTs |
Difference in OGT | Difference in GC Content (%) | Number of Orthologous Proteins | Number of Orthologous Transmembrane Proteins | ||
|---|---|---|---|---|---|---|---|---|
| Organism Name | OGT | Organism Name | OGT | |||||
| Lower temperature adaptation | Aequorivita sublithincola | 4 | Flavobacterium psychrophilum | 20 | 16 | 3.7 | 1,032 | 149 |
| Octadecabacter arcticus | 4 | Roseobacter litoralis | 20 | 16 | 1.8 | 1,472 | 247 | |
| Aequorivita sublithincola | 4 | Gramella forsetii | 25 | 21 | 0.4 | 1,384 | 264 | |
| Photobacterium profundum | 10 | Vibrio parahaemolyticus | 37 | 27 | 3.7 | 1,939 | 407 | |
| Shewanella halifaxensis | 15 | Shewanella putrefaciens | 25 | 10 | 0.4 | 1,981 | 415 | |
| Cellulophaga algicola | 15 | Cellulophaga lytica | 25 | 10 | 1.6 | 1,784 | 371 | |
| Marinomonas sp. MWYL1 | 15 | Marinomonas mediterranea | 25 | 10 | 1.6 | 1,830 | 353 | |
| Flavobacterium columnare | 15 | Flavobacterium indicum | 30 | 15 | 0.3 | 1,247 | 237 | |
| Renibacterium salmoninarum | 15 | Rothia dentocariosa | 37 | 22 | 1.8 | 775 | 100 | |
| Higher temperature adaptation | Amycolatopsis mediterranei | 28 | Saccharomonospora viridis | 50 | 22 | 4.1 | 1,969 | 389 |
| Halorubrum lacusprofundi | 30 | Natrinema sp. J7 | 50 | 20 | 0.2 | 1,392 | 187 | |
| Natrinema pellirubrum | 37 | Natrinema sp. J7 | 50 | 13 | 0.1 | 2,139 | 431 | |
| Bacillus selenitireducens | 26 | Geobacillus thermoglucosidasius | 55 | 29 | 4.6 | 855 | 109 | |
| Amycolatopsis mediterranei | 28 | Thermobispora bispora | 55 | 27 | 0.7 | 1,090 | 110 | |
| Desulfotomaculum gibsoniae | 35 | Desulfotomaculum carboxydivorans | 55 | 20 | 0.7 | 1,021 | 150 | |
| Aminobacterium colombiense | 37 | Thermovirga lienii | 55 | 18 | 1.7 | 605 | 92 | |
| Methanococcus voltae | 30 | Methanothermococcus okinawensis | 65 | 35 | 0.0 | 918 | 91 | |
| Mesotoga prima | 37 | Kosmotoga olearia | 65 | 28 | 4.2 | 863 | 151 | |
| Tepidanaerobacter acetatoxydans | 37 | Thermoanaerobacter pseudethanolicus | 65 | 28 | 3.7 | 619 | 82 | |
| Salinibacter ruber | 37 | Rhodothermus marinus | 65 | 28 | 1.5 | 1,082 | 186 | |
| Meiothermus ruber | 50 | Thermus scotoductus | 65 | 15 | 0.9 | 1,210 | 226 | |
| Thermoanaerobacter sp. X514 | 55 | Thermoanaerobacter pseudethanolicus | 65 | 10 | 0.0 | 1,618 | 336 | |
| Meiothermus ruber | 50 | Marinithermus hydrothermalis | 70 | 20 | 4.3 | 1,179 | 216 | |
| Thermus scotoductus | 65 | Thermus thermophilus | 75 | 10 | 4.5 | 1,532 | 296 | |
| Caldicellulosiruptor hydrothermalis | 65 | Caldicellulosiruptor bescii | 75 | 10 | 0.9 | 1,676 | 361 | |
| Hydrogenobacter thermophilus | 70 | Thermocrinis albus | 80 | 10 | 2.9 | 1,073 | 166 | |
| Thermococcus sibiricus | 78 | Pyrococcus horikoshii | 95 | 17 | 1.5 | 1,005 | 134 | |
The homologous proteins of members of species pair were identified using OrthoMCL version 2.0 (Li et al. 2003), and the transmembrane proteins among them were identified by using TMHMM 2.0 (Krogh et al. 2001). To estimate the contribution of amino acids to protein stability, the scale developed by Takano and Yutani (2001) was used to calculate quantificationally changes of stability in the homologous proteins caused by the amino acid substitutions. In addition, because the contributions of amino acids to protein stability depend on their positions in the secondary structure, so we predicted the secondary structures of homologous proteins using PSIPRED 3.0 (Jones 1999).
For each pair of species, all orthologous proteins were aligned using ClustalW (Larkin et al. 2007), and only orthologous proteins with distances less than 0.5 were used in the following calculations, in order to reduce the number of unreliable alignments caused by proteins that were too divergent. The distance of orthologous protein was defined as the ratio of the amount of variable sites (including sites with gaps) to the whole alignment length. To calculate the amino acid frequencies, whole homologous sites with different amino acids were extracted from the protein sequences. The substitution patterns were also recorded for the asymmetric substitution analyses. Then, the changes of protein stability were evaluated by summing the difference of the scales for the side-chain contribution to protein stability caused by the amino acid substitutions. Different scales were adopted where amino acids appeared in different secondary structure. If the summation was greater than zero, it indicated the substitutions make the proteins more stable.
The related calculations were executed using custom scripts in Python. The paired Student’s t-test was used to test the significance of changes in amino acid frequencies between the low and high OGTs groups. For each pair of amino acids, one-sample Student’s t-test was used to test the significance of the deviation of substitutions in two directions (i.e., A->B and B->A) from the expected zero, to determine the substitutional asymmetry. All statistical analyses were implemented using the R project software (R Core Team 2014).
Results and Discussion
The great increase of genome data has also made more available genomic data from psychrophiles (see supplementary table S1, Supplementary Material online). To investigate whether low temperature adaptation is an opposite process of high temperature adaptation, we screened paired species from the predicted proteomes downloaded from the NCBI Microbial Genomes database. The OGT information was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ; http://www.dsmz.de) resource. Paired species from the same genus (or at least the same family) were chosen. Differences in the genomic GC content of the paired organisms were restricted to 5%. Further, only the relative temperature difference was controlled in this study. Our approach allowed the observed variations of amino acid frequencies to be correlated with the relative OGT differences, as well as with mesophile-thermophile evolution. There are two advantages to this approach: one, the sample size to be analyzed was increased; and two, species pairs were not limited to mesophile-thermophile pairs, but two thermophile pairs were also compared.
Based on above-mentioned criterions, we obtained 915 prokaryotic predicted proteomes with OGT information from the NCBI and DSMZ databases and then further selected 47 species pairs consisting of 79 genomes with pairwise OGT differences from 10 to 35 °C. Furthermore, for discriminating the psychrophiles and thermophiles more strictly, we defined the OGTs ranges of psychrophiles and thermophiles as ≤15 °C and ≥50 °C respectively. Of 47 species pairs, 17 pairs including merely species with OGTs from 15 to 50 °C were removed. Then, for taking into account the number of samples and the violation of sample repetition to the statistical tests, three pairs having same species that were already appeared in other pairs were removed (pairs with greater OGT differences were kept). Notably, there were still six species appeared in two pairs, but the compared species with them in different pairs were from different genera. Finally 27 species pairs consisting of 48 genomes were screened for the following analyses. The details about the species pairs are shown in table 1. The average of GC content difference is 1.92%; there are 5/9 psychrophiles having higher GC content; and there are 10/18 thermophiles having higher GC content. These 27 species pairs were divided into two categories according to the OGTs of the members of the species pairs; that is, those with lower temperature adaptations (≤15 °C; 9 species pairs) and those with higher temperature adaptations (18 species pairs).
Our study has two caveats. First, the partitioning of the data sets to represent lower and higher temperature adaptations was arbitrary. Besides the actual OGTs, a more reasonable way of determining whether a psychrophilic species had undergone lower temperature adaptation would be one based on exhaustive phylogenetic analysis and an investigation of their ecology. However, because information about the close neighbors of these psychrophiles was limited, so we estimated their temperature adaptation simply according to the actual OGTs and phylogenetic relationships presented in supplementary fig. S1, Supplementary Material online. Second, most of the species pairs (18/27) were from the same families. Therefore, the complexities of evolutionary scenarios may have resulted in the loss of some predictive sensitivity, due to the potential long evolutionary distance between the members of these species pairs. Unfortunately, the trade-off between accuracy and comprehensiveness was entirely dependent on the data that are currently available.
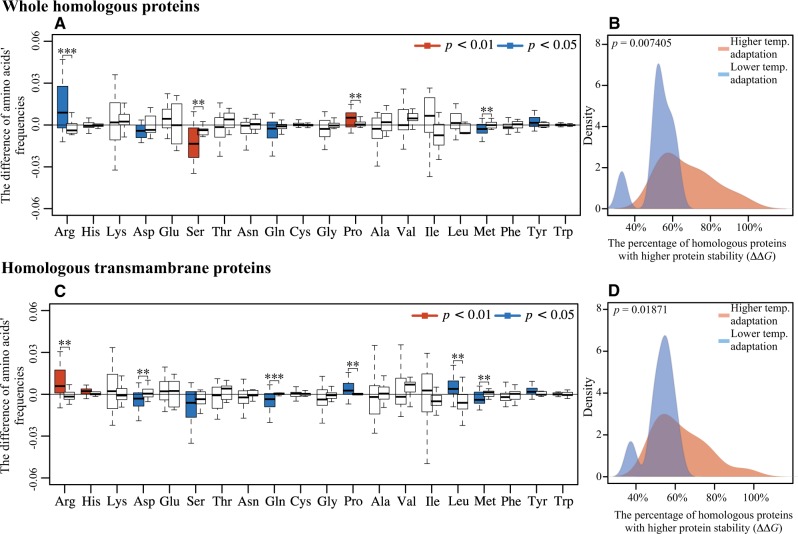
We calculated the frequencies of amino acids at variable sites in whole homologous proteins to determine the difference in their frequencies between the groups with lower and higher OGTs (supplementary table S2, Supplementary Material online; the amino acids frequencies at all aligned sites are shown in supplementary table S3, Supplementary Material online). For each amino acid, two boxes are shown (fig. 1A): the left boxes show the amino acid changes in a subset of data that represented higher temperature adaptations and the right boxes show the amino acid changes in a subset of data that represented lower temperature adaptations. For higher temperature adaptations, of the polar uncharged amino acids, only the frequencies of Ser (P = 7.74 × 10−4) and Gln (P = 0.0384) decreased significantly; of the charged amino acids, the frequency of Arg increased (P = 0.0114) and the frequency of Asp decreased significantly (P = 0.0331); and of the eight hydrophobic amino acids, only the frequencies of Tyr (P = 0.0221) increased significantly. Four of the other seven hydrophobic amino acids had increased frequencies, although they were not significant statistically (P > 0.05). Another important variation was the increased frequency of Pro (P = 0.0024) (Goihberg et al. 2007). These results related to higher temperature adaptations were generally consistent with previous reports (Sterner and Liebl 2001; Singer and Hickey 2003; Sadeghi et al. 2006), except that some of the previously reported variations were not observed in this study, perhaps because of the data collection strategy that we used. For lower temperature adaptations, clearly the species (OGTs from 20 to 37 °C; table 1) that were compared with psychrophiles (OGTs ≤15 °C) did not contain the same pattern of changes in amino acid frequencies. Among the 20 amino acids, there is no any amino acid showed significant (P < 0.05) change in frequency, based on the present genomes data set. Overall, four amino acids including Arg, Ser, Pro, and Met showed different patterns between the subsets of data for higher and lower temperature adaptations.
Fig. 1.—
Changes of amino acid frequencies at homologous sites and protein stability between low and high OGT groups. (A and C) Boxplots showing changes of amino acid frequencies in homologous sites in the all the proteins tested and in a subset of transmembrane proteins, respectively. Two boxes are shown for each amino acid: the left box shows the amino acid changes in a subset of data that represented high temperature adaptations and the right box show the amino acid changes in a subset of data that represented low temperature adaptations. Boxes that are not colored indicate changes that were not significant in the Student’s t-test. (C and D) Kernel density plots showing the percentages of homologous proteins with higher stability in the all the proteins tested and in a subset of transmembrane proteins, respectively.
To quantify the amino acid variations in the proteins of microorganisms faced with temperature changes, we calculated the protein stability (ΔΔGaa, see supplementary table S4, Supplementary Material online) of all the homologous proteins (fig. 1B). The stabilities of more than half of homologous proteins were enhanced through amino acid substitutions in the members of species pairs with higher OGTs (thermophiles, OGTs ≥50 °C) in the subset of data for higher temperature adaptation. On the contrary, the kernel density based on the subset of data for lower temperature adaptation was different (P < 0.01). It indicates that the percentage of homologous proteins with enhanced stabilities, in the species (OGTs from 20 to 37 °C) compared with psychrophiles, is significant lower. In other words, lower temperature did not make psychrophiles to decrease the protein stabilities extensively. Thus, the psychrophiles also contain a high proportion of homologous proteins with higher stability values indicating the protein stability. These results indicate that the psychrophiles did not extensively decrease the stability of proteins through amino acids substitutions. This supports the idea that only minor structural modifications are needed to engineer small increases or decreases of thermostability in psychrophilic enzymes, with an emphasis on local rather than global flexibility (Pasi et al. 2009; Xie et al. 2009; Casanueva et al. 2010).
Transmembrane proteins often use different strategies from intracellular proteins to adapt to high temperatures (Trivedi et al. 2006); therefore we analyzed them separately from the whole homologous proteins. Although some of the details are different (fig. 1C and D), the transmembrane proteins also showed that lower temperature adaptation was not the opposite process of higher temperature adaptation. Surprisingly, the frequency of Leu in proteins from psychrophiles was higher than in proteins from the other species in the pairs (most of them are mesophiles). This finding is inconsistent with a previous report (Metpally and Reddy 2009).
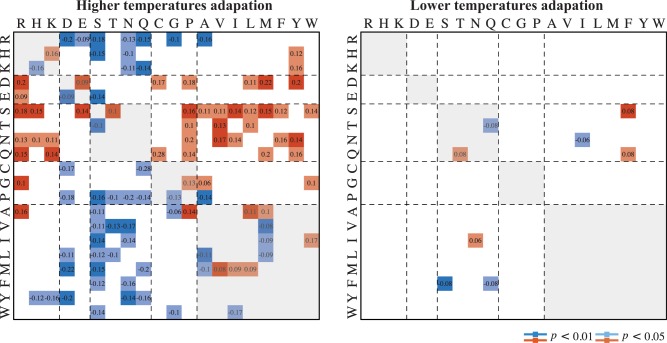
The impetus underlying the amino acid frequency changes may be correlated directly with asymmetric substitutions (McDonald et al. 1999); therefore, we analyzed the patterns of amino acid substitution (fig. 2; the detail data are shown in supplementary table S5, Supplementary Material online). For the higher temperature adaptation, the observed increase of Arg may be explained by asymmetric substitutions of Asp, Glu, Ser, Asn, Gln, Gly, and Ala, whereas the observed increase of Pro could be explained mainly by asymmetric substitutions of Asp, Ser, Thr, Asn, Gln, Gly, and Ala. Thus, the asymmetric substitutions from polar uncharged amino acids to hydrophobic ones contributed to an increased number of hydrophobic amino acids in the proteins. Positive charged amino acids were more favored than negative charged ones. Remarkably, seven amino acids, including His, Lys, Glu, Thr, Ala, Ile, and Met, were both favored and unfavored at the same time. Although this may explain why there were different kinds of asymmetric substitutions, these substitutions did not give rise to the significant changes of amino acid frequencies that were detected. In the subset of data for lower temperatures adaptation, of the eight hydrophobic amino acids, only Phe were favored by the paired species compared with psychrophiles. The polar uncharged amino acids trended to be substituted by Phe. The increase of Pro was found to be an important feature for high temperature adaptation (Goihberg et al. 2007); however, Pro was also found to be favored at certain homologous sites in proteins from psychrophiles. For verifying the difference of the patterns of asymmetric substitutions related to higher and lower temperature adaptations, respectively, we repeated the analysis based on three subdata sets: one was generated by removing all species pairs with OGTs difference under 15 °C; second subdata set was generated by removing all species pairs belonging to domain Archaea; third subdata set was generated by removing all species including halophiles. All these results demonstrate that the patterns of amino acid substitutions under higher and lower temperatures are apparent different (supplementary fig. S2, Supplementary Material online).
Fig. 2.—
Asymmetric substitution matrix showing the patterns of amino acid substitutions in homologous sites in lower and higher OGT groups. Only statistically significant asymmetric substitutions are shown (P < 0.05). Decreases in amino acid frequencies are shown in red; increases in amino acid frequencies are shown in blue. For example, the frequency difference between Asp→Arg substitution and Arg→Asp substitution was equal 0.2 (larger than zero significantly, P < 0.01), in the subset of data for higher temperature adaptation, indicating that the substitution from Asp to Arg is more frequent than the substitution from Arg replaced Asp. The depth of the color indicates a different degree of significance.
In summary, the evidence based on the variations of amino acids frequencies and substitutional asymmetry demonstrated that lower temperature adaptation was not the opposite process of higher temperature adaptation. This result is consistent with previous biochemical studies (Pulido et al. 2007; Fields et al. 2015; Parvizpour et al. 2015). The evolutionary issue about the origin of the last universal common ancestor has been a focus of evolutionary biology (Galtier et al. 1999; Gribaldo and Brochier-Armanet 2006). Through uncovering and analyzing the imprints left by the effects of temperature on ancestral organisms, the bacterial ancestor was predicted to be thermophilic and subsequently adapted to lower temperatures (Gaucher et al. 2008). Similarly, the archaeal ancestor was also thought to have been thermophilic (Gribaldo and Brochier-Armanet 2006). Although there might be two environmental temperature-related phases during the evolutionary history of the tree of life (Boussau et al. 2008), high temperature is a crucial environmental factor for prokaryotic early evolution. Nevertheless, at present, more than three-quarter of the earth’s surface is occupied by cold ecosystems (Feller and Gerday 2003), which implies that microorganisms that have experienced lower temperatures adaptation should be more extensive than microorganisms that experienced higher temperature adaptation during evolution. Our conclusion will provide new insight into the adaptive evolution of prokaryotes in response to changes in temperature. Recently, López-García et al. (2015) hypothesized that horizontal gene transfer might have been crucial for the adaptation of archaea to mesophilic lifestyles after their thermophilic origin. This hypothesis implies that the proteomes of extant mesophilic archaea retained some of the features of their thermophilic ancestors. Certainly, further and exhaustive analyses of mesophilic archaea and bacteria are required to test whether the features that were inherited from their thermophilic ancestors will still be needed in the future.
Supplementary Material
Supplementary figures S1 and S2 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant number 31560309 and 31160003) and the opening project of the State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences (grant number SKLMR-20140601).
Literature Cited
- Ayala-del-Rio HL, et al. 2010. The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl Environ Microbiol. 76:2304–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovsky IN, Zeldovich KB, Shakhnovich EI. 2007. Positive and negative design in stability and thermal adaptation of natural proteins. PLoS Comput Biol. 3:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M. 2008. Parallel adaptations to high temperatures in the Archaean eon. Nature 456:942–945. [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Forterre P. 2007. Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers. Archaea 2:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanueva A, Tuffin M, Cary C, Cowan DA. 2010. Molecular adaptations to psychrophily: the impact of 'omic' technologies. Trends Microbiol. 18:374–381. [DOI] [PubMed] [Google Scholar]
- De Maayer P, Anderson D, Cary C, Cowan DA. 2014. Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep 15:508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vendittis E, et al. 2008. Adaptation of model proteins from cold to hot environments involves continuous and small adjustments of average parameters related to amino acid composition. J Theor Biol. 250:156–171. [DOI] [PubMed] [Google Scholar]
- Feller G. 2010. Protein stability and enzyme activity at extreme biological temperatures. J Phys Condens Matter 22:323101. [DOI] [PubMed] [Google Scholar]
- Feller G, Gerday C. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol. 1:200–208. [DOI] [PubMed] [Google Scholar]
- Fields PA, Dong Y, Meng X, Somero GN. 2015. Adaptations of protein structure and function to temperature: there is more than one way to 'skin a cat'. J Exp Biol. 218:1801–1811. [DOI] [PubMed] [Google Scholar]
- Forterre P. 2002. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 18:236–237. [DOI] [PubMed] [Google Scholar]
- Fukuchi S, Nishikawa K. 2001. Protein surface amino acid compositions distinctively differ between thermophilic and mesophilic bacteria. J Mol Biol. 309:835–843. [DOI] [PubMed] [Google Scholar]
- Galtier N, Tourasse N, Gouy M. 1999. A nonhyperthermophilic common ancestor to extant life forms. Science 283:220–221. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Govindara Jan S, Ganesh OK. 2008. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451:704–707. [DOI] [PubMed] [Google Scholar]
- Glyakina AV, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV. 2007. Different packing of external residues can explain differences in the thermostability of proteins from thermophilic and mesophilic organisms. Bioinformatics 23:2231–2238. [DOI] [PubMed] [Google Scholar]
- Goihberg E, et al. 2007. A single proline substitution is critical for the thermostabilization of Clostridium beijerinckii alcohol dehydrogenase. Proteins 66:196–204. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Brochier-Armanet C. 2006. The origin and evolution of archaea: a state of the art. Philos Trans R Soc Lond B Biol Sci. 361:1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney PJ, et al. 1999. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc Natl Acad Sci U S A. 96:3578–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs JK, et al. 2012. On the origin and evolution of thermophily: reconstruction of functional precambrian enzymes from ancestors of Bacillus. Mol Biol Evol. 29:825–835. [DOI] [PubMed] [Google Scholar]
- Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 292:195–202. [DOI] [PubMed] [Google Scholar]
- Koga Y. 2012. Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea 2012:789652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a Hidden Markov Model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Zivanovic Y, Deschamps P, Moreira D. 2015. Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol. 13:447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma BG, Goncearenco A, Berezovsky IN. 2010. Thermophilic adaptation of protein complexes inferred from proteomic homology modeling. Structure 18:819–828. [DOI] [PubMed] [Google Scholar]
- Maugini E, Tronelli D, Bossa F, Pascarella S. 2009. Structural adaptation of the subunit interface of oligomeric thermophilic and hyperthermophilic enzymes. Comput Biol Chem. 33:137–148. [DOI] [PubMed] [Google Scholar]
- McDonald JH. 2001. Patterns of temperature adaptation in proteins from the bacteria Deinococcus radiodurans and Thermus thermophilus. Mol Biol Evol. 18:741–749. [DOI] [PubMed] [Google Scholar]
- McDonald JH. 2010. Temperature adaptation at homologous sites in proteins from nine thermophile-mesophile species pairs. Genome Biol Evol. 2:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methé BA, et al. 2005. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci U S A. 102:10913–10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metpally RP, Reddy BV. 2009. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: insights into the molecular basis of cold adaptation of proteins. BMC Genomics 10:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima H, Fukuchi S, Nishikawa K. 2003. Compositional changes in RNA, DNA and proteins for bacterial adaptation to higher and lower temperatures. J Biochem. 133:507–513. [DOI] [PubMed] [Google Scholar]
- Paredes DI, Watters K, Pitman DJ, Bystroff C, Dordick JS. 2011. Comparative void-volume analysis of psychrophilic and mesophilic enzymes: structural bioinformatics of psychrophilic enzymes reveals sources of core flexibility. BMC Struct Biol. 11:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvizpour S, Razmara J, Jomah AF, Shamsir MS, Illias RM. 2015. Structural prediction of a novel laminarinase from the psychrophilic Glaciozyma antarctica PI12 and its temperature adaptation analysis. J Mol Model 21:63. [DOI] [PubMed] [Google Scholar]
- Pasi M, Riccardi L, Fantucci P, De Gioia L, Papaleo E. 2009. Dynamic properties of a psychrophilic alpha-amylase in comparison with a mesophilic homologue. J Phys Chem B. 113:13585–13595. [DOI] [PubMed] [Google Scholar]
- Puigbò P, Pasamontes A, Garcia-Vallve S. 2008. Gaining and losing the thermophilic adaptation in prokaryotes. Trends Genet. 24:10–14. [DOI] [PubMed] [Google Scholar]
- Pulido MA, Koga Y, Takano K, Kanaya S. 2007. Directed evolution of Tk-subtilisin from a hyperthermophilic archaeon: identification of a single amino acid substitution responsible for low-temperature adaptation. Protein Eng Des Sel 20:143–153. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; Available from: http://www.R-project.org/. [Google Scholar]
- Reed CJ, Lewis H, Trejo E, Winston V, Evilia C. 2013. Protein adaptations in archaeal extremophiles. Archaea 2013:373275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi M, Naderi-Manesh H, Zarrabi M, Ranjbar B. 2006. Effective factors in thermostability of thermophilic proteins. Biophys Chem. 119:256–270. [DOI] [PubMed] [Google Scholar]
- Saunders NF, et al. 2003. Mechanisms of thermal adaptation revealed from the genomes of the Antarctic archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 13:1580–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp CE, et al. 2014. Humboldt's spa: microbial diversity is controlled by temperature in geothermal environments. Isme J. 8:1166–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siglioccolo A, Bossa F, Pascarella S. 2010. Structural adaptation of serine hydroxymethyltransferase to low temperatures. Int J Biol Macromol 46:37–46. [DOI] [PubMed] [Google Scholar]
- Singer GAC, Hickey DA. 2003. Thermophilic prokaryotes have characteristic patterns of codon usage, amino acid composition and nucleotide content. Gene 317:39–47. [DOI] [PubMed] [Google Scholar]
- Sterner R, Liebl W. 2001. Thermophilic adaptation of proteins. Crit Rev Biochem Mol Biol. 36:39–106. [DOI] [PubMed] [Google Scholar]
- Struvay C, Feller G. 2012. Optimization to low temperature activity in psychrophilic enzymes. Int J Mol Sci. 13:11643–11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre H, Claverie JM. 2003. Genomic correlates of hyperthermostability, an update. J Biol Chem. 278:17198–17202. [DOI] [PubMed] [Google Scholar]
- Takano K, Yutani K. 2001. A new scale for side-chain contribution to protein stability based on the empirical stability analysis of mutant protines. Protein Eng 14:535–538. [DOI] [PubMed] [Google Scholar]
- Trivedi S, Gehlot HS, Rao SR. 2006. Protein thermostability in archaea and eubacteria. Genet Mol Res. 5:816–827. [PubMed] [Google Scholar]
- Wang J, Ma BG, Zhang HY, Chen LL, Zhang SC. 2008. How does gene expression level contribute to thermophilic adaptation of prokaryotes? An exploration based on predictors. Gene 421:32–36. [DOI] [PubMed] [Google Scholar]
- Wijma HJ, Floor RJ, Janssen DB. 2013. Structure-and sequence-analysis inspired engineering of proteins for enhanced thermostability. Curr Opin Struct Biol. 23:588–594. [DOI] [PubMed] [Google Scholar]
- Xie BB, et al. 2009. Cold adaptation of zinc metalloproteases in the thermolysin family from deep sea and arctic sea ice bacteria revealed by catalytic and structural properties and molecular dynamics: new insights into relationship between conformational flexibility and hydrogen bonding. J Biol Chem. 284:9257–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldovich KB, Berezovsky IN, Shakhnovich EI. 2007. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JS, Deng Y, Manno D, Hawari J. 2010. Shewanella spp. genomic evolution for a cold marine lifestyle and in-situ explosive biodegradation. PLoS One 5:e9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.