Abstract
For both historical and technical reasons, 16S ribosomal RNA has been the most common molecular marker used to analyze the contents of microbial communities. However, its slow rate of evolution hinders the resolution of closely related bacteria—individual 16S-phylotypes, particularly when clustered at 97% sequence identity, conceal vast amounts of species- and strain-level variation. Protein-coding genes, which evolve more quickly, are useful for differentiating among more recently diverged lineages, but their application is complicated by difficulties in designing low-redundancy primers that amplify homologous regions from distantly related taxa. Given the now-common practice of multiplexing hundreds of samples, adopting new genes usually entails the synthesis of large sets of barcoded primers. To circumvent problems associated with use of protein-coding genes to survey microbial communities, we develop an approach—termed phyloTAGs—that offers an automatic solution for primer design and can be easily adapted to target different taxonomic groups and/or different protein-coding regions. We applied this method to analyze diversity within the gorilla gut microbiome and recovered hundreds of strains that went undetected after deep-sequencing of 16S rDNA amplicons. PhyloTAGs provides a powerful way to recover the fine-level diversity within microbial communities and to study stability and dynamics of bacterial populations.
Keywords: phylotypes, bacterial species, population structure, bacterial strain diversity, microbiome, community profiling
Introduction
Characterization of microbial communities has been transformed through the application of ribosomal RNA profiling methods, which allow the cultivation-independent identification of community constituents based on full or partial small subunit rRNA (hereafter, simply called 16S) sequences (Woese et al. 1990; Pace 1997; DeLong and Pace 2001; Smith et al. 2001). The basic approach involves the use of universally conserved polymerase chain reaction (PCR) primers to amplify segments containing the more variable regions, which are then sequenced through technologies that have become increasingly advanced over the past decades (Bartram et al. 2011; Caporaso et al. 2011, 2012). Due to the read depths currently afforded by next-generation sequencing platforms, we are approaching the point where it will be possible to catalog all of the lineages within a microbial community.
Despite the extraordinary insights that have been gained through 16S profiling analyses, there are several shortcomings with present methodologies, particularly at the shallowest taxonomic levels, which leave numerous questions about the structure and contents of communities unanswered. The slow rate of rRNA evolution serves well for resolving relationships at deep phylogenetic levels, but renders it inadequate for assessing extent of strain- or species-level variation. The practice of clustering OTUs at 97% (or even 99%) 16S sequence identity will group together functionally diverse lineages and disregard any microevolutionary processes that contribute to adaptation. Moreover, the variation in the numbers of rRNA operons possessed by different bacterial species introduces problems when quantifying cell numbers or taxon abundances based on 16S phylotypes (Pei et al. 2010; Sun et al. 2013).
Many studies have ventured beyond the analysis of 16S sequences by targeting coding regions with conserved primers or by extracting coding-gene orthologs from shotgun metagenomic surveys (e.g., Vos et al. 2012; Sunagawa et al. 2013; Schloissnig et al. 2013; Barret et al. 2015). However, there is no broadly applicable, community-profiling method based on protein-coding genes analogous to those available for rRNA. One difficulty in devising such a method stems from the high variability of protein-coding genes, particularly at synonymous codon positions, which thwarts the design of universally conserved primers. Additionally, bacterial lineages vary in their genomic contents, suggesting that different genes might be needed to resolve the diversity within certain taxonomic groups.
In this article, we describe, develop, and apply a new experimental method—termed phyloTAGs—which parallels current 16S-profiling approaches but is based on the characterization of protein-coding genes, which, due to their more rapid evolutionary rates, allow examination of the contents of complex microbial communities at the level of bacterial strains and species. We present a systematic approach for the design of phylogenetically targeted primers that maximize the recovery of sequence variants across a broad range of taxonomic ranks. Additionally, the phyloTAG method integrates a procedure that circumvents the need (and costs) to produce, for each new gene or primer pair, new sets of indexing primers, each with its on barcode, thereby enabling the multiplexing of scores of samples into a single sequencing run. Focusing on the diversity within two bacterial families with this method, we show that 16S phylotypes fail to uncover hundreds, if not thousands, of bacterial strains in the gut microbiota.
Materials and Methods
To develop a portable, flexible, low-cost, high-throughout, ultra-high-coverage system based on protein-coding regions, two issues must be overcome: 1) Designing conserved primers for genes that are highly variable, and 2) reducing the upfront costs associated with introducing barcode identifier sequences into primers so that multiple samples can be multiplexed into a single sequencing run. Below, we describe the procedure and pipeline for obtaining low-degeneracy primers targeted to a selected gene (with emphasis given the taxonomic level that will be assayed by the specific primer pair), and then outline the method, originally reported by Faith et al. (2013), for producing sets of phased, barcoded primers through an annealing/amplification step. A manual describing the application, execution and output of the phyloTAGs pipeline, and all related scripts, is supplied as supplementary file S1, Supplementary Material online, and deposited at GitHub.
DNA Samples
As proof of concept, we outline and apply our approach, termed phyloTAGS, to evaluate the extent of microbial diversity within the fecal microbiome of wild-living, nonhabituated gorillas (Gorilla gorilla and Gorilla beringei). We elected to examine the gut microbiome of gorillas because it has previously been shown, based on 16S analyses, to contain high levels of bacterial diversity for one of bacterial families that we are targeting (Lachnospiraceae) and none for the other (Bacteroidaceae), so we reasoned that this would provide a good test case for assessing diversity at finer taxonomic levels. The source of samples, and procedures for DNA extraction and 16S rDNA amplicon sequencing are described in Moeller et al. (2013).
Gene and Amino Acid Sequences
The phyloTAGs approach targets single-copy protein-coding genes, making it superior to the analysis of 16S for quantifying bacterial taxon abundances because 16S operons may be in multiple copies and polymorphic within a genome. To identify single-copy genes that are conserved among genomes of a designated taxonomic rank, we downloaded the set of fully sequenced bacterial genomes from National Center for Biotechnology Information (as of September 2013, there were 2,639 complete genomes). Our analysis centered on the diversity within two of the major bacterial families within the gut microbiome (Bacteroidaceae and Lachnospiraceae), so we supplemented this data set with 18 Bacteroidaceae and 7 Lachnospiraceae high-coverage draft genomes available from the Broad Institute (www.broadinstitute.org).
Because the magnitude of sequence divergence encompassed by organisms classified at a particular taxonomic rank is highly variable across the tree of life, genomes constituting a given taxonomic rank (in this case, family) are first grouped based on their degree of 16S divergence instead of relying solely on taxonomic nomenclature. Thus, to determine whether the Bacteroidaceae and the Lachnospiraceae span approximately the same phylogenetic depth and range of variation, we examined the maximum difference in 16S genes among the sequenced members of each family. To calculate the percent identity of 16S genes between genomes, 16S rDNA sequences were extracted from the noncoding RNA file for each genome (“.frn”), and the pairwise sequence identities were calculated using USEARCH global alignments (Edgar 2010). Draft genomes that did not have annotated 16S genes were queried for 16S rDNA homologs using Basic Local Alignment Search Tool (BLAST). In these cases, regions greater than 1,400 nt and having greater than 70% identity to a 16S gene present in any of the complete genomes were extracted and annotated as 16S genes. The resulting set of 16S genes was dereplicated by removing identical sequences with USEARCH, and the nonredundant set of sequences was subjected to an all-versus-all comparison to establish the extent of 16S rDNA divergence within a given taxonomic rank. Because the 16S genes within those genomes containing multiple copies can be polymorphic, the degree of 16S divergence between two genomes is taken to be the minimum pairwise difference between 16S gene copies from each genome. Members of the same bacterial family were defined as those pairs of organisms whose 16S genes differ by less than 10%, a value that conforms to the previously reported correspondence between 16S divergence and taxonomic classification (Konstantinidis and Tiedje 2005).
Selection of Target Gene(s)
Groups of genomes within a specified level of 16S divergence were examined to identify protein-coding genes common to all members of the group. The gyrB gene, encoding subunit B of DNA gyrase, was selected as the target gene for both the Lachnospiraceae and the Bacteroidaceae because 1) it was present in only one copy in each of the evaluated genomes; 2) it has a low reported frequency of horizontal gene transfer; 3) it is routinely used for bacterial identification and phylogenetics; and in studies of bacterial diversity (Yamamoto and Harayama 1996; Wang et al. 2007; Caro-Quintero et al. 2011; Deng et al. 2014), 4) it contains at least two highly conserved regions that are suitable for designing low degeneracy primers (see below) and are separated by distance appropriate to the high-throughput sequencing platform (≈500 nt in the case of the Illumina MiSeq).
Primer Design Using the phyloTAGs Bioinformatic Pipeline
Designing low-degeneracy primers that generate amplicons for all genomes within a specified taxonomic group (i.e., level of 16S divergence) requires the alignment, search, and assessment of polymorphic regions within the target gene. To expedite this process, we generated a set of PERL scripts that assist in the selection of primer sequences, and that can be applied to any gene and any designated taxonomic rank. The scripts require the input of two parameters: 1) A FASTA file containing all orthologs of the target gene and 2) the length, in nucleotides, of the window to be analyzed, which is dictated by the favored size of PCR primers (21 is the default value).
The script first conceptually translates the genes using transeq (Rice et al. 2000), and the protein sequences are aligned using the ClustalW algorithm (Thompson et al. 1994). Using the protein sequence alignment as a guide, the corresponding nucleotide sequences are subsequently aligned, codon by codon, using pal2nal (Suyama et al. 2006). The resulting alignment is then searched in short blocks according to the specified window size, using a sliding window of one nucleotide, to identify regions of the gene suitable for designing low degeneracy primers. To locate such regions, a consensus nucleotide is assigned to each nucleotide position based on the following criteria: Where one of the four nucleobases is present in at least 80% of the aligned sequences (this stringency threshold can be modified), we assigned that nucleobase as the consensus, for sites with higher levels of polymorphism, we combined all nucleobases occurring at frequencies over 20% and assigned a single-letter nucleotide in accordance with the standard IUPAC degeneracy code. For each window of specified length, the script calculates the total number of degeneracies in the sense and antisense DNA strand across the entire gene. All results are saved in a tab-delimited output file “phyloTAGs.txt” (an example of which is presented as supplementary table S3, Supplementary Material online). A second output file displays the degree of nucleotide conservation (% identity) for each sliding window along the reference sequence at different levels of 16S rDNA divergence (supplementary fig. S1, Supplementary Material online), showing the taxonomic level at which low-degeneracy primers will likely anneal.
Primer Selection, Phasing, and Barcoding for Sample Multiplexing
Pairs of 21-nt primers that anneal to highly conserved regions and that span a ≈500-bp variable region of the gyrB gene were synthesized (supplementary table S1, Supplementary Material online). Because the phyloTAG approach employs an amplification method to affix barcodes to primers, it is possible to test many primer pairs and to alter the primer pairs or the targeted region at no additional cost. Thirty pairs of primers were initially tested for the amplification of fragments of the correct length with genomic DNAs purified from representatives of each of the selected bacterial families (Bacteroidacea family: Bacteroides thetaiotaomicron, Bacteroidesvulgatus, Bacteroideseggerthii [obtained from Andrew Goodman, Yale University]; Lachnospiraceae family: strains 6_1_63FAA, ACC2 and 7_1_58FAA [obtained from BEI Resources, NIAID and NIH, as part of the Human Microbiome Project]).
Primer pairs yielding the most robust results were resynthesized to incorporate adaptor sequences at their 5′-ends. Adaptor sequences contain a phase and a linker region, which are used to produce sets of primers that generate amplicons suitable for barcoding and sample multiplexing (Faith et al. 2013). The phase region is a 1–7 nt sequence that offsets the start position of sequencing, thereby generating a more even distribution of the four nucleobases at each sequencing position when the samples are pooled. An even distribution of nucleobases is required by the Illumina software for successful sequencing and is usually accomplished through the addition of phiX DNA to the sequencing library. By introducing sequence complexity through phasing, the entire Illumina flow cell can be devoted to the resolution of phyloTAG sequences. The linker region is a ≈30-nt sequence that provides the template for annealing the primers containing the sample-identifying barcode sequence, added to the amplicons during a second round of PCR amplification. Different linker sequences can be added to the forward and reverse primers to allow the dual barcoding of amplicons, which vastly increases the numbers of uniquely coded samples that can be multiplexed into a single sequencing run.
Barcode primers possess three features: A sequence that anneals to the linker, a 10-nt barcode, and the Illumina flow-cell capture sequence. We synthesized 30 unique barcode primers for the forward linker sequence and 30 for the reverse linker sequence, which together produce 900 combinations that can be used to index samples for multiplexing in a single Illumina run. Because these barcodes can be introduced into any primer that possesses a corresponding linker sequence, after the initial cost of synthesizing a set of barcode primers, the phyloTAG approach can be applied to any selected region. The sequences of the adaptors (phasing and linker regions) incorporated into primers used to target a specific genic region and the list of primers used for barcoding are provided as supplementary tables S4–S6, Supplementary Material online.
Amplification and Sequencing of phyloTAGs
After testing primer pairs designed to target the gyrB gene, we selected the most proficient to generate amplicons suitable for Illumina sequencing. Barcoded amplicons were produced by two consecutive PCR amplifications, following the procedure described in the previous section. The primers used in the first PCR amplified a region of the gyrB (600 bp in Bacteroidacea; 500 bp in Lachnospiraceae) from DNA samples extracted from gorilla feces, and added the appropriate phase and linker sequences to the amplified fragment. The amplicons from each sample were subsequently subjected to a second PCR using different combinations of barcode primers for each sample.
The first PCR, which targeted the specified portion of gyrB, was performed in triplicate and carried out in 20 µl reaction volumes containing 8 µl of 5-Prime HotMasterMix 2.5×, 1 µl (100 ng) of sample DNA, 1 µl (10 µM) of each of the forward and reverse primers containing adaptors sequences, and 9 µl of UltraPure Distilled Water (Invitrogen). Because the primers used in the first PCR possess a long adaptor that is not complementary to the DNA template, it is sometimes necessary to add 5 µM of each of the corresponding primers that do not contain the adaptor sequence in order to produce additional templates for the primers with adaptors.
The PCR starts with denaturation at 95 °C for 120 s, followed by 30 cycles of at 95 °C for 45 s, 50 °C for 60 s, and 72 °C for 90 s, and a final extension at 72 °C for 10 min. Amplifications were verified on agarose gels, replicates combined, and reaction products purified with AMPure XP beads. The second PCR using the amplicons from each sample as template is carried out by adding 5 µl of purified product, 1 µl (10 µM) of each of the forward and reverse barcoding primers, 8 µl of the 5-Prime HotMasterMix 2.5×, and 5 µl of UltraPure Distilled Water (Invitrogen). PCR was performed using the same reaction conditions as above but allowed to proceed for only 12 cycles. Products of the second PCR were purified with AMPure XP beads, and DNA concentrations were quantified on a Qubit 2.0 flourometer (Invitrogen). Samples were normalized to contain equal concentrations of amplicons, combined and pair-end sequenced (250-nt reads) on the Illumina MiSeq by the Genomic Sequencing and Analysis Facility at the University of Texas at Austin.
Trimming and Merging of phyloTAGs
The trimming and filtering of sequencing reads were performed using the FASTX-toolkit (hannonlab.cshl.edu/fastx_toolkit). Initially, reads are end-trimmed using the fastq_quality_trimmer script, with the following parameters “-t 20 -l 75.” Trimmed reads were then filtered for overall quality with the fastq_quality_filter script, with the following parameters “-q 25 -p 90,” and reads that did not pass this filter were removed. Reads were converted to FASTA format, and paired reads, whose ends overlapped greater than 10% of their total lengths and were greater than 99% identical, were merged into a single sequence. Primer and phasing sequences were trimmed from sequences, and both merged-pair reads and individual unmerged reads were used in analyses.
The Diversity and Community Structure of Recovered by the phyloTAGs
Community descriptors (e.g., diversity, richness, and coverage) were estimated for operational taxonomic units (OTUs) clustered at different degrees of sequence identity. Rarefaction curves, for comparing across OTUs and phylogenetic markers, were generated with 100 bootstrap replicates, using a sample size of twice that of the smaller data set (Chao and Jost 2012). All parameters and curves were obtained using the iNEXT R package (glimmer.rstudio.com/tchsieh/inext/).
Reconstruction of the community structure recovered by phyloTAGs was done by the progressive clustering of reads obtained for each of the two bacterial families, as follows: Clustering begins at the highest level of sequence identity (e.g., 99%), then a representative sequence central to the cluster was extracted and used as input for clustering at the next highest level. Subsequent rounds of clustering followed by the extraction of representative sequences are conducted until the lowest selected identity value is reached or until all sequences are grouped in a single cluster. In this study, DNA sequences were progressively clustered at 99–88% identity, at 1% intervals, and amino acid sequences were clustered at 95–55% identity, at 5% intervals. Clustering of DNA and amino acid sequences was performed with USEARCH. The affiliation of reads to the clusters generated at each identity level was depicted as a cladogram-like structure in Cytoscape (Smoot et al. 2011).
Results
Bacterial Diversity Resolved by phyloTAGs
Using the phyloTAG approach, we amplified a region of the gyrB gene from members of two bacterial families, Bacteroidacea and Lachnospiraceae, present in the fecal microbiome of gorilla. Using primer pairs F_La_334_354, R_La_816_836 for amplification of Lachnospiraceae and the primer pairs F_Bt_330_350, R_Bt_918_938, for amplification of Bacteroidaceae, all sequence reads could be assigned to one of these two bacterial families. When assessing the bacterial diversity in these same samples using universal primers targeted to the V4 region of 16S rDNA, only about 0.3% of the tens of thousands of reads generated for each sample were assigned to Lachnospiraceae, and not a single read was assigned to the Bacteroidacea (table 1). The ability to target-specific bacterial families allowed the phyloTAGs to produce a significantly higher number reads and to resolve much higher levels of diversity within both of these bacterial families than that obtained by the analogous methods based on 16S rDNA variable regions.
Table 1.
Comparison of Diversity Recovered by 16S iTAGs and gyrB phyloTAGs
| Gorilla Sample | Family | Marker | Reads Total | Reads Assigned | 100 (%) | 99 (%) | 98 (%) | 97 (%) | 88 (%) |
|---|---|---|---|---|---|---|---|---|---|
| 5248 | Lachnospiraceae | 16S rDNA | 45,699 | 113 | 56 | 31 | 26 | 22 | — |
| 5249 | Lachnospiraceae | 16S rDNA | 36,604 | 116 | 63 | 42 | 34 | 31 | — |
| 5274 | Lachnospiraceae | 16S rDNA | 46,947 | 196 | 81 | 43 | 36 | 29 | — |
| All | Lachnospiraceae | 16S rDNA | 129,250 | 425 | 155 | 73 | 57 | 48 | — |
| 5248 | Lachnospiraceae | gyrB | 4,395 | 4,395 | 3,591 | 1,170 | 444 | 218 | 98 |
| 5249 | Lachnospiraceae | gyrB | 2,033 | 2,033 | 1,691 | 872 | 242 | 171 | 88 |
| 5274 | Lachnospiraceae | gyrB | 2,013 | 2,013 | 1,719 | 929 | 277 | 176 | 89 |
| All | Lachnospiraceae | gyrB | 8,441 | 8,441 | 6,920 | 3,502 | 866 | 366 | 149 |
| 5248 | Bacteroidaceae | 16S rDNA | — | — | — | — | — | — | — |
| 5249 | Bacteroidaceae | 16S rDNA | — | — | — | — | — | — | — |
| 5274 | Bacteroidaceae | 16S rDNA | — | — | — | — | — | — | — |
| All | Bacteroidaceae | 16S rDNA | — | — | — | — | — | — | — |
| 5248 | Bacteroidaceae | gyrB | 376 | 376 | 85 | 70 | 50 | 38 | 22 |
| 5249 | Bacteroidaceae | gyrB | 446 | 446 | 120 | 110 | 70 | 44 | 31 |
| 5274 | Bacteroidaceae | gyrB | 668 | 668 | 283 | 224 | 116 | 65 | 35 |
| All | Bacteroidaceae | gyrB | 1,490 | 1,490 | 488 | 383 | 207 | 109 | 53 |
Numbers of OTUs were obtained by clustering reads based on their degree of sequence identity (table 1). Sequence tags containing the variable regions of 16S are often clustered at 99% or 97% sequence identity (hereafter termed 99%-OTUs and 97%-OTUs, respectively). Applying these thresholds, samples averaged 39 99%-OTUs and 27 97%-OTUs ofLachnospiraceae based on the 16S rDNA V4 region. In contrast, there were nearly 1,000 99%-OTUs of Lachnospiraceae per sample based on a similarly sized region of the gyrB gene. However, the absolute numbers of Lachnospiraceae OTUs recovered by 16S rDNA iTAGs and gyrB phyloTAGs are not directly comparable because of the differences in the sampling depth (i.e., number of reads representing each family). Applying a statistic that imparts the level of diversity at finer taxonomic scales and is independent of sampling depth is the ratio of 99%-OTUs to 97%-OTUs: For 16S assigned to Lachnospiraceae in these samples, this ratio is approximately 2:1, but for gyrB in Lachnospiraceae, it is nearly 10:1. In contrast, this ratio is only 3:1 for the gyrB phyloTAGS in Bacteroidacea, indicating that the variation within this family assorts at higher taxonomic ranks.
Detecting Strain- and Species-Level Diversity with phyloTAGs
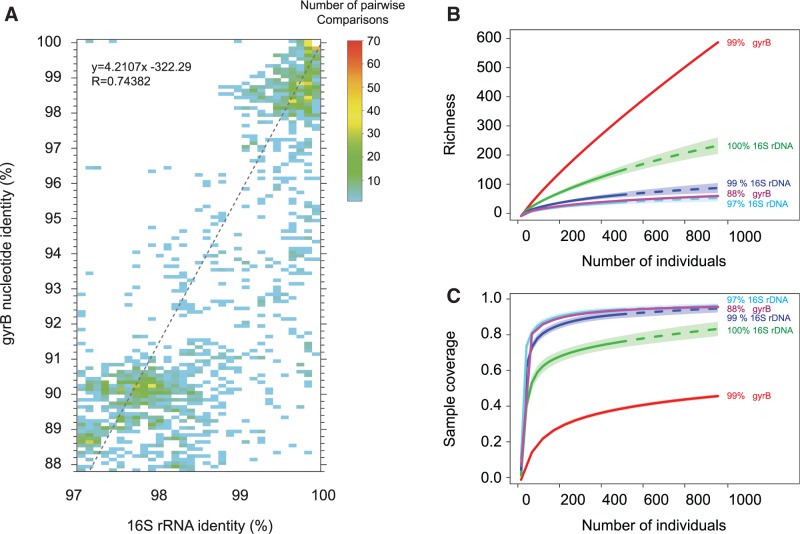
To estimate how much resolution is gained by using coding-gene phyloTAGs instead of 16S rDNA sequences, we plotted the average pairwise identities of the targeted gyrB region against the average pairwise identities of full-length 16S sequences for organisms classified at the same taxonomic rank (fig. 1A). Using this regression, we find that organisms belonging to a given bacterial species, conventionally defined as organisms with ≥97% overall 16S rDNA identity, have agyrB nucleotide identity ≥88%. Thus, comparisons of this short region of the gyrB gene yield four times the amount of polymorphism than provided by the entire 16S rDNA gene. [Note that the extent of sequence identity in the V4 region of 16S rDNA is representative of the 16S molecule as a whole (supplementary fig. S2, Supplementary Material online), justifying the application of a 97% identity threshold to delineate species using this restricted region.]
Fig. 1.—
Correspondence between levels of sequence divergence and estimates of OTU richness for gyrB and 16S rDNA. (A) Association between the degree of sequence identity of 16S rDNA and the gyrB gene for pairs of genomes assigned to the same species. Note that 1) a 16S rDNA sequence identity value of 97%, which is conventionally used to delineate bacterial species, corresponds to 88% nucleotide sequence identity for gyrB, and 2) a 16S rDNA sequence identity value of 99%, which has been used to delineate strains within a designated bacterial species, corresponds to 96% nucleotide sequence identity for gyrB. A total of 604 genomes were examined. (B) Richness of the Lachnospiraceae family within all samples, as estimated by the Chao1 index for gyrB phyloTAGs and the 16S iTAGs at several values of OTU clustering. Estimation of parameters based on subsampling the data sets for each marker gene to the same depth with 100 bootstraps replicates. Shaded zones around rarefaction curves represent the 95% confidence intervals. Dashes show read numbers obtained after extrapolation to sample sizes larger than the actual total number of reads for the 16S data set. (C) Rarefaction analysis of sample coverage for data sets analyzed in panel (B), using identical subsampling parameters. As in (B), shaded zones around the rarefaction curves represent 95% confidence intervals, and dashed lines indicating trends after extrapolation to sample sizes larger than the actual total number of reads.
Applying the sequence identity threshold described above, there was a total of 149 species of Lachnospiraceae (89–98 species per sample) and 53 species of Bacteroidaceae (22–35 species per sample) based on the gyrB phyloTAGs. The numbers of species detected by 16S rDNA iTAGs are much lower, but as mentioned above, species numbers obtained by the two methods are not directly comparable due to differences in sampling depths of the taxa in question. To accommodate these differences, we subsampled the Lachnospiraceae data sets using rarefaction analysis. Using the same sample sizes and 100 bootstraps, we found a striking correspondence between the number of estimated species richness and sample coverage for the Lachnospiraceae gyrB phyloTAGs and 16S rDNA iTAGs, 70 and 63 species, respectively (fig. 1B and C).
Within species (i.e., strain-level) variation can be assessed by examining the numbers of unique OTUs. But because sequencing errors can produce sequence variants that are not actually present in the community, we tested the phyloTAGs approach on cultivable strains of known sequence in order to determine the extent of artifacts. We found that clustering reads at 99% sequence identity subsumed all artifacts caused by sequencing errors—therefore, bacterial strains were discriminated as unique 99%-OTUs. (Analogously, 99%-OTUs based on 16S sequences are often view as bacterial strains or subspecies.) Applying these criteria, the gyrB phyloTAGs targeted to the Lachnospiraceae family contained a total of 3,502 strains representing 149 species, the 16S rDNAiTAGs identified 73 strains typed to 48 species ofLachnospiraceae, and the gyrB phyloTAGs targeted to the Bacteroidaceae family contained 383 strains representing 52 species (remembering that no 16S sequences were assigned to the Bacteroidaceae). Rarefaction analysis for 99%-OTUs showed that gyrB phyloTAGs provide substantially higher resolution, recovering up to six times more Lachnospiraceae strains than 16S-profiling by iTAGs (fig. 1B).
Assessing Community Contents and Structure
An added advantage of using protein-coding genes for analyzing the contents of microbial communities is that their nucleotide sequences are suitable for resolving the relationships among the closely related constituents of communities whereas their translated amino acid sequences are as useful as 16S rDNA sequences for establishing more distant relationships [as evident by the strong linear association between divergence in gyrB amino acid sequences and 16S sequences (supplementary fig. S3, Supplementary Material online)].
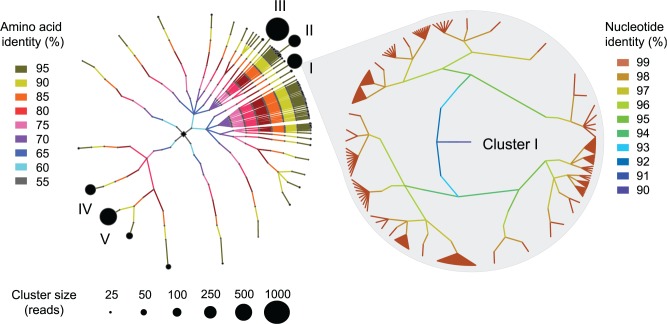
To illustrate the apportionment of community diversity at different taxonomic levels, we clustered phyloTAG sequences at progressively decreasing levels of sequence identity: DNA sequences were clustered from 99% to 88% identity at 1% intervals to assess the intraspecies population structure, whereas the amino acid sequences were clustered from 95% to 55% identity at 5% intervals to assess the community structure from the species to the family rank. Note that the clusters of gyrB phyloTAGs based on 95% amino acid identity or 88% nucleotide identity correspond to the 97% 16S sequence identity threshold for species (fig. 1A and supplementary fig. S3, Supplementary Material online). This progressive clustering procedure links sequences into progressively higher taxonomic groups (fig. 2A) and provides a comprehensive picture of the contents, diversity and relationships within the community.
Fig. 2.—
Community structure and OTU diversity recovered by gyrB phyloTAGs. Progressive clustering of gyrB phyloTAG sequences at decreasing levels of amino acid identity (based on conceptually translated nucleotide sequences) reconstructs the diversity and number of taxa within the Lachnospiraceae at different phylogenetic depths. Branches in cladogram are colored according to level of identity at which sequences are clustered. Four major branches (i.e., lineages) occur when clustered at 55% amino acid identity (black), 5 at 60% identity (turquoise), 12 at 65% identity (blue), and so on, with amount of diversification at lower taxonomic scales being highly variable among lineages. Black circles at the terminal end of each branch are sized according to the number of sequences affiliated to a cluster at levels >95% amino acid identity, and the five largest clusters are labeled with roman numerals (I–V). The inset shows the fine-level resolution of OTU variation in Lachnospiraceae cluster I. In this case, progressive clustering of gyrB phyloTAGs was performed on nucleotide sequences and revealed that most of the sequence variation assorts into 99% OTUs, that is, at the level of closely related strains within species.
The clustering of amino acid sequence targeting the Lachnospiraceae family revealed the existence of 127 sequence clusters (>95% amino acid identity OTUs) that might each be considered species. The relationships and representation of these sequence clusters are highly variable. For the most part, the cladogram based on amino acid sequence identities is skeletal (fig. 2A), a pattern produced by the occurrence of many deep-branching lineages that only sporadically diversified. For example, in two cases, a sequence cluster is represented by a single strain that is very distantly related (<55% amino acid identity) to any other strain. In contrast, there are three clades that display a fan-like structure in which there is a burst of diversification into a large number of sequence clusters at 70% amino acid identity, followed by little or no lineage splitting into sequence clusters at higher identity levels.
Despite the many deep-branching sequence clusters represented by a single strain, several contain large numbers of closely related strains and were found to correspond to several known clades (species groups) within the Lachnospiraceae. For example, Cluster I representing 704 sequences was related to the Lachnospiraceae bacterium COE1, Cluster II representing 562 sequences was related to Lachnospiraceae 6 1 63 FAA, Cluster III representing 1,146 sequences was related to Blautia sp CAG:237, Cluster VI representing 474 sequences was most closely related to Lachnospiraceae 2 1 46FAA, and Cluster V representing 821 sequences was related to Dorea spp. The nucleotide sequence information derived from phyloTAGs allows examination of the structure and contents of these highly populated clusters in detail. The cladogram in figure 2B presents a fine-grained view of the distribution of lineages in Cluster I and shows that much of the variation is partitioned into 99%-OTUs, such that certain species are represented by multiple closely related strains.
Discussion
There is extensive species- and strain-level variation present in many microbial communities, but in most assessments of community diversity, its scope remains unobserved and unknown. The application of deep-sequencing technologies, including 454 (Sogin et al. 2006; Huber et al. 2007; Liu et al. 2007) and Illumina (Claesson et al. 2010; Gloor et al. 2010; Caporaso et al. 2011; Zhou et al. 2011; Degnan and Ochman 2012), to the analysis of microbial communities has led to the rapid and in-depth characterization of the microbes inhabiting a broad array of habitats. Despite increases in read lengths and sequencing depths, these methods still provide little resolution of the fine-grained diversity at lower taxonomic ranks.
Both for historical and technical reasons, the majority of studies rely on analysis of small subunit ribosomal RNA sequences to survey bacterial community diversity (Woese and Fox 1977; Pace 1997; Hugenholtz 2002; Tringe and Hugenholtz 2008). Given the state of current databases, the partial 16S rRNA sequences generated by high-throughput, deep-sequencing technologies will usually classify organisms to the level of bacterial genus. However, a single 16S OTU or phylotype can potentially encompass vast amounts of species- and strain-level variation, which remains largely unexplored in studies that are confined to even the most highly variable regions of 16S rRNA.
There have been attempts to further resolve taxa either through the use of more complete rRNA sequences, through molecular methods that control for errors (Faith et al. 2013), or by examining nucleotide positions that distinguish among very closely related taxa (Eren et al. 2014). However, even organisms having identical 16S sequences can be genetically and ecologically distinct (Jaspers and Overmann 2004; Hahn and Pockl 2005; Caro-Quintero et al. 2011), which indicates a need for alternative strategies to study the composition of microbial communities at a finer taxonomic scales. Such information is useful for understanding the temporal stability or replacement of strains, the coevolution of bacteria and hosts, and whether broad taxonomic ranks comprise a single or multiple lineages.
The use of protein-coding regions to assess the diversity with microbial communities offers several advantages. Foremost among these is the fact that sequences from protein-coding genes provide a fine-grained view of variation at lower taxonomic levels and enable the resolution of individual strains within a bacterial species. Although protein-coding genes are regularly assayed for epidemiological and population genetic studies (Maiden et al. 1998; Brettar et al. 2001; Hill et al. 2002; Santos and Ochman 2004; Thompson et al. 2005), they have only rarely been used to examine noncultivable bacteria or in high-throughput studies (Hou et al. 2008; Vos et al. 2012). As no protein-coding genes, even those that are universally distributed, are highly conserved in its DNA sequence, the design of low-redundancy primers that allow amplification of homologous regions from divergent taxa has been the major obstacle in their application for studying microbiomes. Our pipeline addresses this issue by offering a systematic and automatic approach for primer design that can be modified according to the specific gene and particular taxonomic groups in question.
In the application of phyloTAGs described in this article, we targeted a single gene, gyrB, in two of the dominant bacterial families within the gut microbiome and recovered, for the Lachnospiraceae, more than six times the resolution provided by the 16S iTAGs, and for the Bacteroidaceae, higher taxonomic groups that went completely undetected by 16S sequence analysis. The phyloTags method provides a more complete resolution of the strain-level diversity within microbial communities than that provided by 16S tagging approaches, but how these strains assort into species is a matter beyond what can be established with a single gene. If bacteria were completely clonal, classification and assignment to a bacterial species could be based on some prescribed genomic signature or sequence thresholds, as computed from the variation present in currently designated taxa (Konstantinidis and Tiedje 2005; Thompson et al. 2013). The occurrence of homologous exchange and gene transfer among strains presents the possibility that bacteria might also be classified into biological species in a manner analogous to that applied to sexual organisms. We note, however, that phyloTAGs can be used to reconstruct deep-branching as well as strain-level relationships as analyses of diversity can be based on either gene or protein sequences. In this way, it is possible to link the diversity at multiple taxonomic levels, thereby providing a more comprehensive view of community structure.
The large number of reads offered by current sequencing platforms facilitates the multiplexing of hundreds (soon to be thousands) of samples into a single sequencing lane. In order to identify each of the samples that are mixed together and sequenced together, unique identifying sequences (aka barcodes) are added to the amplification primers used for each sample. The incorporation of these barcodes usually involves the synthesis of large sets of amplification primers for each targeted region—a costly endeavor when one is assaying a new gene. In order to circumvent the need of synthesizing large sets of primers, we have adopted the method of Faith et al. (2012) that uses separate sets of primers for gene amplification and barcoding, allows the use of the same set of barcode primers with any gene of interest.
Several methods that do not rely upon the amplification of specific target-genes have been used to study the fine level diversity within microbial communities. Early attempts to extract single-copy protein-coding genes assembled from deep metagenomic sequencing (Venter et al. 2004; Roux et al. 2011; Sunagawa et al. 2013) usually produced consensus sequences representing the most abundant variants and did not directly assess strain diversity. More recently, a study mapped shotgun metagenomic reads to sequenced bacterial genomes in order to quantify the relative abundance of individual strains in the human microbiome (Kraal et al. 2014), and an assessment of the variation in human microbiomes, again surveyed by shotgun metagenomics, indicated that individual hosts each harbor many unique strains of the commonly occurring microbial taxa (Schloissnig et al. 2013). But due to the costs and analytical procedures associated with shotgun-metagenomic sequencing, such approaches are often not feasible unless the genomic contents and complexity of the community are already well-established.
Despite the general utility of phyloTAGs for assessing the species- and strain-level diversity within microbial community, its application has limitations. Second, because rates of evolution can differ across and among genes, phyloTAGs from different single-copy protein genes (or even different portions of the same gene) might recover different numbers of variants. Second, and as observed in studies that target 16S sequences (Suzuki and Giovannoni 1996; Acinas et al. 2005; Pinto and Raskin 2012), there can be amplification biases that will affect the interpretation of strain abundances. Third, insufficient representation of the taxonomic group of interest in the databases may have consequences on the design and specificity of primers. To evaluate the effect of this last factor, we tested the extent to which the primer sequences designed for our study recruited raw reads from two large gut microbiome metagenomic libraries. We found that both primer pairs aligned to the recruited metagenomics reads and would, in principle, amplify the corresponding regions from the original samples (supplementary fig. S4, Supplementary Material online). Therefore, by targeting primers to well-conserved regions from diverse genomes, phyloTAGs will capture the variation that is present in the microbial communities at large.
Supplementary Material
Supplementary file S1, figures S1–S4, and tables S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Andrew Goodman for providing bacterial strains and for help with experimental protocols, and Kim Hammond for assistance with the preparation of figures. This work was supported by the National Institute of Health grant number R01GM101209 to H.O.
Literature Cited
- Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. 2005. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microbiol. 71:8966–8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret M, et al. 2015. Emergence shapes the structure of the seed-microbiota. Appl Environ Microbiol. 81:1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. 2011. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol. 77:3846–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettar I, Moore ER, Hofle MG. 2001. Phylogeny and abundance of novel denitrifying bacteria isolated from the water column of the central Baltic Sea. Microb Ecol 42:295–305. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, et al. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 108:4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 6:1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Quintero A, et al. 2011. Unprecedented levels of horizontal gene transfer among spatially co-occurring Shewanella bacteria from the Baltic Sea. Isme J. 5:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Jost L. 2012. Coverage-based rarefaction and extrapolation: standardizing samples by completeness rather than size. Ecology 93:2533–2547. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, et al. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Ochman H. 2012. Illumina-based analysis of microbial community diversity. Isme J. 6:183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Pace NR. 2001. Environmental diversity of bacteria and archaea. Syst Biol. 50:470–478. [PubMed] [Google Scholar]
- Deng J, et al. 2014. Stability, genotypic and phenotypic diversity of Shewanella baltica in the redox transition zone of the Baltic Sea. Environ Microbiol. 16:1854–1866. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Eren AM, Borisy GG, Huse SM, Mark Welch JL. 2014. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci U S A. 111:E2875–E2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, et al. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, et al. 2010. Functionally compensating coevolving positions are neither homoplasic nor conserved in clades. Mol Biol Evol. 27:1181–1191. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Pockl M. 2005. Ecotypes of planktonic actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Appl Environ Microbiol. 71:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, et al. 2002. Extensive profiling of a complex microbial community by high-throughput sequencing. Appl Environ Microbiol. 68:3055–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XL, Cao QY, Jia HY, Chen Z. 2008. Pyrosequencing analysis of the gyrB gene to differentiate bacteria responsible for diarrheal diseases. Eur J Clin Microbiol Infect Dis 27:587–596. [DOI] [PubMed] [Google Scholar]
- Huber JA, et al. 2007. Microbial population structures in the deep marine biosphere. Science 318:97–100. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3:REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers E, Overmann J. 2004. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl Environ Microbiol. 70:4831–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraal L, Abubucker S, Kota K, Fischbach MA, Mitreva M. 2014. The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS One 9:e97279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MC, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 95:3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, et al. 2013. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 23:1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740. [DOI] [PubMed] [Google Scholar]
- Pei AY, et al. 2010. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol. 76:3886–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AJ, Raskin L. 2012. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One 7:e43093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Roux S, Enault F, Bronner G, Debroas D. 2011. Comparison of 16S rRNA and protein-coding genes as molecular markers for assessing microbial diversity (Bacteria and Archaea) in ecosystems. FEMS Microbiol Ecol 78:617–628. [DOI] [PubMed] [Google Scholar]
- Santos SR, Ochman H. 2004. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ Microbiol. 6:754–759. [DOI] [PubMed] [Google Scholar]
- Schloissnig S, et al. 2013. Genomic variation landscape of the human gut microbiome. Nature 493:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z, McCaig AE, Stephen JR, Embley TM, Prosser JI. 2001. Species diversity of uncultured and cultured populations of soil and marine ammonia-oxidizing bacteria. Microb Ecol 42:228–237. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc Natl Acad Sci U S A. 103:12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun DL, Jiang X, Wu QL, Zhou NY. 2013. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol. 79:5962–5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, et al. 2013. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods 10:1196–1199. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 62:625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson FL, et al. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol. 71:5107–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, Hugenholtz P. 2008. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol. 11:442–446. [DOI] [PubMed] [Google Scholar]
- Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. [DOI] [PubMed] [Google Scholar]
- Vos M, Quince C, Pijl AS, de Hollander M, Kowalchuk GA. 2012. A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity. PLoS One 7:e30600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LT, Lee FL, Tai CJ, Kasai H. 2007. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA-DNA hybridization in the Bacillus subtilis group. Int J Syst Evol Microbiol. 57:1846–1850. [DOI] [PubMed] [Google Scholar]
- Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 74:5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 87:4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Harayama S. 1996. Phylogenetic analysis of Acinetobacter strains based on the nucleotide sequences of gyrB genes and on the amino acid sequences of their products. Int J Syst Bacteriol 46:506–511. [DOI] [PubMed] [Google Scholar]
- Zhou HW, et al. 2011. BIPES, a cost-effective high-throughput method for assessing microbial diversity. Isme J. 5:741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.