Abstract
Leptospirosis is the most widespread zoonotic disease in the world. It is caused by pathogenic spirochetes of the genus Leptospira spp. and is maintained in nature through chronic renal infection of carrier animals. Rodents and other small mammals are the main reservoirs. Information on leptospirosis in marine mammals is scarce; however, cases of leptospirosis have been documented in pinniped populations from the Pacific coast of North America from southern California to British Columbia. We report the isolation of a Leptospira spp. strain, here named Manara, from a kidney sample obtained from a Southern Right Whale (Eubalaena australis) calf, which stranded dead in Playa Manara, Península Valdés, Argentina. This strain showed motility and morphology typical of the genus Leptospira spp. under dark-field microscopy; and grew in Ellinghausen-McCullough-Johnson-Harris (EMJH) medium and Fletcher medium after 90 days of incubation at 28°C. Considering the source of this bacterium, we tested its ability to grow in Fletcher medium diluted with seawater at different percentages (1%, 3%, 5%, 7% and 10% v/v). Bacterial growth was detected 48 h after inoculation of Fletcher medium supplemented with 5% sea water, demonstrating the halophilic nature of the strain Manara. Phylogenetic analysis of 16S rRNA gene sequences placed this novel strain within the radiation of the pathogenic species of the genus Leptospira spp., with sequence similarities within the range 97–100%, and closely related to L. interrogans. Two different PCR protocols targeting genus-specific pathogenic genes (G1-G2, B64I-B64II and LigB) gave positive results, which indicates that the strain Manara is likely pathogenic. Further studies are needed to confirm this possibility as well as determine its serogroup. These results could modify our understanding of the epidemiology of this zoonosis. Until now, the resistance and ability to grow in seawater for long periods of time had been proven for the strain Muggia of L. biflexa, a saprophytic species. To the best of our knowledge, this is the first isolation of a Leptospira sp. from cetaceans. Our phenotypic data indicate that strain Manara represents a novel species of the genus Leptospira, for which the name Leptospira brihuegai sp. nov. is proposed.
Introduction
Leptospirosis is a neglected zoonotic disease, endemic in most tropical and subtropical regions of the world. The causative agents of this zoonosis are pathogenic strains belonging to the order Spirochaetales, family Leptospiracea and genus Leptospira spp. Leptospirosis is maintained in nature through chronic renal infection of carrier animals, with rodents and other small mammals as the most important reservoirs [1–3].
In marine animals, seropositivity has been reported in North America, in California sea lions (Zalophus californianus) [4], Northern fur seals (Callorhinus ursinus) [5], Northern elephant seals (Mirounga angustirostiris) [6], and harbor seals (Phoca vitulina) [7,8]. Furthermore, Leptospira spp. can cause disease in pinnipeds [9–12], periodic large scale stranding and mortality events, every three to four years, of California sea lions along the pacific coast of North America from southern California to British Columbia have been attributed to L. interrogans serovar Pomona infections [4, 10, 11, 12, 13,14]. Seroprevalence in marine mammals has been reported also in the Pacific coast of South America, in Peruvian Amazon manatees (Trichechus inunguis) [15] and in Chilean South American sea lions (Otaria byronia) [16].
Cameron et al. [10], describe clinical signs of Leptospirosis in sea lions as renal failure, dehydration, polydipsia, vomiting, and depression. Using species-specific primer pairs Cameron et al. [10] revealed host specificity of L. interrogans for sea lions and of L. kirschneri for elephant seals.
The genus Leptospira spp. displays a great genomic plasticity and several schemes for the genotyping of pathogenic species have been developed during the last years around the world [17–19]. Bacterial isolation followed by molecular characterization has recently been successfully applied to genotype pathogenic Leptospira spp. strains in Argentina. A considerable number of pathogenic Leptospira spp. from wildlife [20–22], domestic animals [23–25] and water samples [26] were thus characterized.
Environmental factors are key determinants in leptospiral distribution, as water is the vehicle by which leptospires travel and disseminate into ecosystems. To date, only one strain of Leptospira spp., L. biflexa strain Muggia, has been isolated from seawater in a region near Trieste, Italy [27, 28]. Growth of other Leptospira spp., including L. interrogans, in the presence of diluted seawater has always yielded negative results [2, 3, 29, 30]. However, Saito et al. [31] isolated two leptospiral strains after a typhoon in Philippines in soil samples, and commented that both isolated strains could live in seawater for only three and four days respectively.
The present work describes the characterization of a Leptospira sp. strain, isolated from the kidney of a stranded Southern Right Whale in Patagonia, Argentina, that grows optimally in the presence of seawater. This is the second halophilic Leptospira spp. reported to date, and to the best of our knowledge, the first one ever isolated from a cetacean.
Materials and Methods
Sample collection
A total of 27 kidney samples from 27 dead stranded Southern Right Whales (Eubalaena australis) Península Valdés, Argentina, were collected by the Southern Right Whale Health Monitoring Program (SRWHMP) between 2009 and 2010. Field permits for this work were issued annually by the Department of Wildlife of the Subsecretary of Tourism and Protected Areas, Chubut Province, Argentina. Necropsies were performed using a Right Whale necropsy protocol developed by the SRWHMP (Chirife et al., 2014 unpublished) based on the methods of McCellan et al. [32], F. Gulland (pers. comm.), A. Carribero (unpublished) and Geraci & Lounsbury [33]. Carcass decomposition was graded subjectively on a scale from condition code 2 to 5 (2 = fresh, 3 = decomposed but tissues largely intact, 4 = advanced decomposition, 5 = mummified or skeletonized) [33]. Samples were placed in Whirlpack® bags, stored in liquid nitrogen at the site, and later transferred on dry ice to the laboratory for diagnosis. Samples were stored at -70°C until processing.
Isolation and maintenance of cultures
All kidney samples were cultured in leptospire-specific Ellinghausen-McCullough-Johnson-Harris (EMJH) and Fletcher media (Difco Laboratories). Cultures were incubated at 28°C for 90 days and observed every 15 days under dark-filed microscopy to evaluate possible bacterial growth. Once the culture was positive, subcultures were conducted to maintain the isolated strain alive.
Growth assay in the presence of seawater
After weak growth in both EMJH and Fletcher media and considering the source of isolation, we tested the ability of these bacteria to grow in each of these media after the addition of seawater at different percentages (1%, 3%, 5%, 7% and 10%, v/v) [30,31]. Seawater was collected from Puerto Madryn, Chubut, Argentina, and sterilized by filtration through 0.22 μm Millipore filters. Cultures were incubated at 28°C for up to 12 days. A control experiment was carried out, where filter-sterilized seawater was cultured with both EMJH and Fletcher Media. Immunofluorescence staining of the strain was performed with a multivalent FA, LEP-FAC (Seasinglab, Argentina) conjugate specific for Leptospira spp.
Molecular characterization
Two different PCR protocols were tested to indicate if the isolated strain was pathogenic, amplifying genus-specific genes that are considered pathogenicity determinants. DNA templates were obtained after nucleic acid purification using the Chelex-100 resin (Bio Rad) protocol [22].
Multiplex PCR
The combined primers G1-G2 and B64I-B64II previously described in Gravekamp et al., [34] were used in this study. The PCR reaction was carried out in a final volume of 50 μl, which contained 2 μl purified DNA template, and the PCR mixture and the cycling program used was the same as indicated in Gravekamp et al. [34]. PCR was carried out in a My Cycler TM thermocycler (Bio Rad). Amplification products were analyzed by electrophoresis in ethidium bromide stained 2% agarose gels, followed by exposure to UV light (Uvi Tec transiluminator BTS-20.M). Amplicon sizes were estimated using a 100 bp ladder (Embiotech).
PCR for LigB
A 1 kb sequence of the adhesin Lig B gene, which is present in all pathogenic Leptospira spp. strains, was amplified by PCR using primers LigBpetF and LigBpetR [35]. The PCR mixture and cycling was carried out as indicated in Martínez et al. [35]. PCR was carried out in a My Cycler TM thermocycler (BIO RAD) and amplified samples were analyzed by electrophoresis in 1% agarose gels stained with Sybr® Safe (Invitrogen) and visualized under UV light.
16S rRNA sequencing
A PCR targeting the 16S rRNA gene was carried out for bacterial identification after sequencing. The following primers were used: 5′-GGCGGCGCGTCTTAAACATG-3′ and 5′-GTCCGCCTACGCACCCTTTACG-3′ [36]. These primers have the ability to amplify all pathogenic and nonpathogenic species of Leptospira spp. PCR was performed as indicated in Djadid et al. [36]. After verification of the amplicon by electrophoresis in an ethidium bromide-containing 2% agarose gel and visualization upon UV light exposure, PCR products were purified using a commercial kit (EMBIOTECH). The samples were sequenced at the Institute of Biotechnology, National Institute of Agricultural Technology using a 3130xl Genetic Analyzer (Applied Biosystems). For alignment and construction of the phylogeny, the program MEGA version 6.06 [37] was used. The dendrogram was constructed using Neighbor-joining with a bootstrap of 100, partial sequences of the 16S rRNA gene were used.
Results and Discussion
Isolation of Leptospira sp. and maintenance of the cultures
One positive culture of Leptospira sp. was obtained from one Southern Right Whale, a newborn female calf (ID 092410PV-Ea23), 5.05 meters length. The calf stranded dead on September 24th 2010 at Playa Manara in Golfo Nuevo, Península Valdés (42° 40’ 18.9”S 64° 59’ 22.9”W), and was necropsied the following day. Carcass condition at necropsy was 3.
The isolated strain was designated strain Manara. This strain showed typical motility and morphology of the genus Leptospira spp. under dark-field microscopy and grew in EMJH and Fletcher media after 90 days of incubation at 28°C. When seawater (1%, 3%, 5%, 7%, 9% and 10%, v/v) was added, no growth was observed in EMJH medium during the first 6 days. At day 7, positive growth based on the turbidity of the culture was detected at 1% seawater (Fig 1). A control experiment, where only seawater was incubated in the same media did not show any leptospiral growth. On the other hand, growth of strain Manara was observed at 48 h in Fletcher medium, with a typical dinger ring formation in the culture. After 72 h, growth was achieved at percentages of 1% and 3%, and after 96 h, all cultures were positive (Table 1). At day 8 of this assay, new subcultures were started in Fletcher medium supplemented with the same seawater dilutions as before (Table 2). Importantly, the same growth behavior was observed. These experiments confirmed the ability of strain Manara to grow in vitro in the presence of diluted seawater demonstrating its halophilic nature.
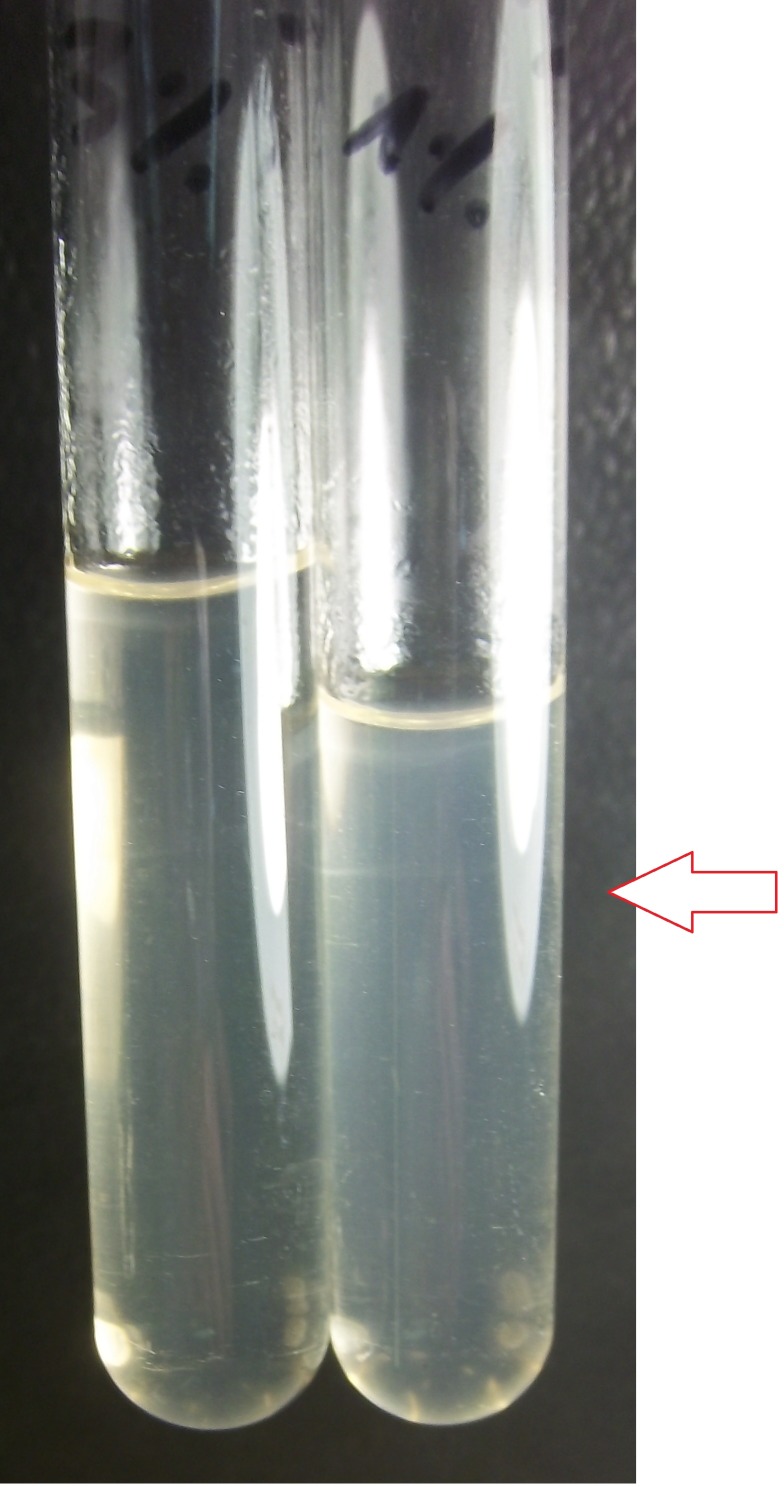
Fig 1. Picture of two cultures of Leptospira sp. strain Manara.
Two cultures of Leptospira sp. strain Manara in Fletcher medium supplemented with 3% (left tube) and 1% (right tube) seawater after 48 h of incubation at 28°C. The dinger rings corresponding to Leptospira sp. growth are indicated with red arrows.
Table 1. Growth results of strain Manara in EMJH and Fletcher media diluted with seawater.
| EMJH Medium | Fletcher Medium | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| day | 1% | 3% | 5% | 7% | 10% | 1% | 3% | 5% | 7% | 10% |
| 1 | - | - | - | - | - | - | - | - | - | - |
| 2 | - | - | - | - | - | - | - | + | - | - |
| 3 | - | - | - | - | - | + | + | + | - | - |
| 4 | - | - | - | - | - | + | + | + | + | + |
| 5 | - | - | - | - | - | + | + | + | + | + |
| 6 | - | - | - | - | - | + | + | + | + | + |
| 7 | + | - | - | - | - | + | + | + | + | + |
| 8 | + | - | - | - | - | +* | +* | +* | +* | +* |
| 9 | + | - | - | - | - | + | + | + | + | + |
| 10 | + | - | - | - | - | + | + | + | + | + |
| 11 | + | - | - | - | - | + | + | + | + | + |
| 12 | + | - | - | - | - | + | + | + | + | + |
Growth results of strain Manara in EMJH and Fletcher media diluted with seawater at percentages 1%, 3%, 5%, 7% and 10% (v/v). + and–correspond to detection and no detection of growth, respectively; Subcultures (marked with an asterisk) under the same conditions were started at day 8.
Table 2. Growth results of subcultures of strain Manara in Fletcher media diluted with seawater after 8 days.
| Subculture from Fletcher Medium after 8 days | |||||
|---|---|---|---|---|---|
| day | 1% | 3% | 5% | 7% | 10% |
| 1 | - | - | - | - | - |
| 2 | - | - | - | - | - |
| 3 | + | + | + | + | + |
| 4 | + | + | + | + | + |
| 5 | + | + | + | + | + |
Growth results of subcultures strain Manara started at day 8 of the first culture assay, in Fletcher media diluted with seawater at percentages 1%, 3%, 5%, 7% and 10% (v/v). + and–correspond to detection and no detection of growth, respectively.
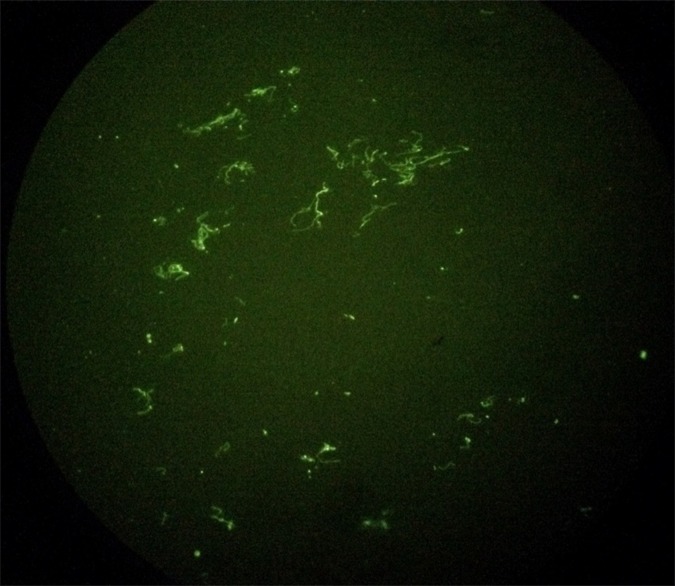
Immunofluorescence detection was positive and leptospires could be microscopically observed. (Fig 2).
Fig 2. Immunofluorescence of strain Manara.
Immunofluorescence of strain Manara isolated from a kidney sample of a Southern right whale (Eubalaena australis) 40X.
To the best of our knowledge, there are no recognized halophilic pathogenic leptospiral strains. Previous studies have demonstrated that Leptospira spp. can grow in seawater [27, 28] but the identification of serogroup and serovar was not provided and the authors indicated that this unique strain Muggia could belong to the species L. biflexa. In the study of Cinco et al. [28], the isolated halophilic strain grew in Korthof-Babudieri seawater medium for about 16 to 18 h, and a maximal concentration of 2 x 108 cells/ml was reached after 10 days of incubation at 30°C. In their study, the strain was considered as halodependent to specific NaCl requirements for growth. On the other hand, Saito et al. [31], could isolate two leptospiral strains from soil samples and tested the ability to grow in seawater, one strain called MS422 could grew in seawater for four days and the second strain MS432 only for three days. Both strains grew also in media with PBS for six days. These studies did not extend for a longer time period. In the same study [31], both strains were sequenced using the 16S rRNA gene and both showed similarity to L. kmetyi.
In our study, the strain Manara was capable of growing in the presence of different concentrations of seawater (Table 1). In media that was not supplemented with seawater, growth of Manara strain was considerably slower and could not be detected until 30 days (results not shown), whereas growth could be detected as soon as 48 h after inoculation when cultured in media with 5% seawater in Fletcher media. Further studies are necessary to characterize the biological features of this novel strain.
Molecular characterization
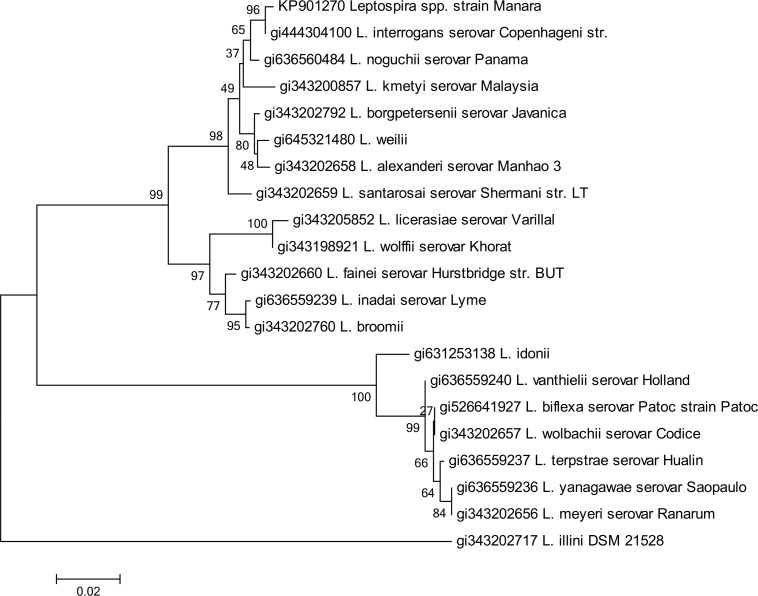
Two different PCR protocols were used to explore the possible pathogenicity of the Manara strain, amplifying the genus-specific and virulence-associated genes G1-G2, B64I-B64II and LigB, both with positive results. Phylogenetic analysis of 16S rRNA gene sequences placed this novel strain within the radiation of the pathogenic species of the genus Leptospira spp., with sequence similarities within the range 97–100% (results not shown), and closely related to L. interrogans (Fig 3). Further molecular characterization must be pursued to confirm this possibly new species, such as sequencing secY gene. The sequence obtained is available in GenBank under Accession number KP901270.
Fig 3. Neighbor-joining analysis of the sequence 16S rRNA of strain Manara.
Phylogenetic analysis based of 16S rRNA including 19 representative species of Leptospira spp. The corresponding sequence of Leptonema illini strain DSM 21528 was used as outgroup. The dendrogram was constructed using Neighbor-joining. Bootstrap values are displayed as percentages.
Detection of new strains in the environment and carrier animals is vital to the understanding of enzootic and epizootic leptospirosis in marine mammals. Since pathogenic Leptospira spp. serovars are extremely difficult to cultivate in vitro [29,30], the mode of transmission of this organism in marine species is not understood [10–13]. Detection by PCR of leptospiral DNA in sand was observed by Cameron et al. [10], however this study could not determine if Leptospira sp. was dead or alive in sand. In this study, no samples were taken at site, where the Southern Right Whale stranded dead, but the possibility exists that leptospiral DNA could be also in sand. Potential environmental sources of pathogen exposure in the marine environment could increase the zoonotic potential of Leptospirosis in marine mammals, however the way of transmission is yet unclear [5,7,8,10,11,12,14].
Confirmation that this isolate contained the gene LigB, a gene associated with virulence and present in all pathogenic Leptospira spp., suggests this strain is pathogenic and is consistent with our sequencing results which place strain Manara within the radiation of other pathogenic Leptospira spp. Further studies in animal models must be conducted to confirm this possibility. In addition, determination of the serogroup and serovar of this strain are the object of on-going research.
Due to the phenotypic findings in this study we propose that strain Manara belongs to a new species of Leptospira spp., phylogenetically closely related to L. interrogans, but with the particular features of being able to survive during a long period of time in seawater, and to colonize unreported hosts, as the Southern right whale.
Ethiology of Leptospira brihuegai sp. Nov
The proposed name for strain Manara is Leptospira brihuegai, after professor Bibiana Brihuega, an Argentinean veterinarian and bacteriologist who has made significant contributions to the study of Leptospirosis.
Conclusion
This study reports the first isolation of a Leptospira sp. from a cetacean. Phylogenetic analysis of a sequence obtained by targeting the 16S rRNA as well as its host specificity and growth requirements identified this as a novel halophilic Leptospira sp. strain These findings contribute highly relevant information to current knowledge on leptospirosis epidemiology and ecology
New findings concerning the biology of leptospires is a challenge and more extensive studies are required to monitor the presence of Leptospira spp. in the environment and different mammalian hosts. Our study contributes valuable information to our knowledge on leptospires overall, and on marine mammals in particular.
Acknowledgments
We thank members, researchers and collaborators of the SRWHMP: M. Sironi, V. Rowntree, La Sala, L. M. Pozzi, L. Musmeci, N. Mohamed, J. E. Sala, J. Andrejuk, L. Bandieri, A. Chirife, M. Di Martino, L. Beltramino, D. Taboada, R. Schteinbarg, L. Valenzuela, M. Ricciardi, P. de Diego, C. Marón, and the many volunteers who make fieldwork possible. The Instituto de Conservación de Ballenas, Wildlife Conservation Society, Ocean Alliance, Fundación Patagonia Natural, Ecocentro Puerto Madryn, Armada Argentina, Prefectura Naval Argentina and Aluar also provided invaluable support of various kinds. Research permits for this work were issued annually by the Dirección de Fauna y Flora Silvestre and the Subsecretaría de Turismo y Áreas Protegidas of Chubut Province, Argentina.
Data Availability
All relevant data are within the paper.
Funding Statement
These authors have no support or funding to report.
References
- 1. Sterling CR, Thiermann AB. Urban rats as chronic carriers of leptospirosis: an ultrastructural investigation. 1981Vet Pathol, 18:628–637. [DOI] [PubMed] [Google Scholar]
- 2. Faine S. Guidelines for the control of leptospirosis, WHO offset publication N° 67. 1982. World Health Organization, Geneva, 161 pg. [PubMed] [Google Scholar]
- 3. Levett PN. Leptospirosis. 2001Clin Microbiol Rev, 14:296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gulland FMD, Koski M, Lowenstine LJ, Colagross A, Morgan L, Spraker T. Leptospirosis in California sea lions (Zalophus californianus) stranded along the central California coast, 1981–1994. 1996. J. Wildl. Dis. 32; 572–80. [DOI] [PubMed] [Google Scholar]
- 5. Smith AW, Brown RJ, Skilling DE, Bray HL, Keyes MC. Naturally occurring Leptospirosis in Northern fur seals (Callorhinus ursinus). 1977J. Wildl. Dis. 13;144–8. [DOI] [PubMed] [Google Scholar]
- 6. Colegrove KM, Lowenstine JL, Gulland FMD. Leptospirosis in northern elephant seals (Mirounga angustirostris) stranded along the California coast. 2005. J. Wildl. Dis.41; 426–30. [DOI] [PubMed] [Google Scholar]
- 7. Stamper MA, Gulland FMD, Spraker T. Leptospirosis in rehabilitated Pacific harbor seals from California. 1998. J. Wildl. Dis. 34; 407–10. [DOI] [PubMed] [Google Scholar]
- 8. Greig DJ, Gulland FM, Smith WA, Conrad PA, Field CL, Fleetwood M, et al. Surveillance for zoonotic and selected pathogens in harbor seals Phoca vitulina from central California. 2014. Dis Aquat Org 111;93–106. 10.3354/dao02762 [DOI] [PubMed] [Google Scholar]
- 9. Colagross-Schouthern A, Mazet JA, Gulland FMD, Miller MA, Hietala S. Diagnosis and seroprevalence of leptospirosis in California sea lions from coastal California. 2002. Journal of Wildlife Diseases 38(1);7–17. [DOI] [PubMed] [Google Scholar]
- 10. Cameron C, Zuerner RL, Raverty S, Colegrove KM, Norman S, Lambourn DM, et al. Detection of Pathogenic Leptospira bacteria in Pinniped Population via PCR and Identification of a source of Transmission for zoonotic leptospirosis in the Marine Environment. 2008. Journal of Clinical Microbiology, 46(5);1728–33 10.1128/JCM.02022-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zuerner RL, Cameron CE, Raverty S, Robinson J, Colegrove KM, Norman SA, et al. Geographical dissemination of Leptospira interrogans serovar Pomona during seasonal migration of California sea lions.2009Vet. Microbiol.137; 105–10. 10.1016/j.vetmic.2008.12.017 [DOI] [PubMed] [Google Scholar]
- 12. Waltzek TB, Cortés-Hinojosa G, Wellehan JFX Jr, Gray GC. Marine Mammal Zoonoses: A review of Disease Manifestations. 2012. Zoonoses and Public Health Dec 59(8); 521–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Norman SA, DiGiacomo RF, Gulland FMD, Meschke JS, Lowry MS. Risk factors for an outbreak of leptospirosis in California sea lions (Zalophus californianus) in California, 2004.2008J. Wildl. Dis. 44; 837–44. [DOI] [PubMed] [Google Scholar]
- 14. Lloyd-Smith J, Greig DJ, Hietala S, Ghneim G, Palmer L, Leger J, et al. Cyclical changes in seroprevalence of leptospirosis in California sea lions: endemic and epidemic disease in one host species? 2007. BMF Infectious Diseases 7; 125–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delgado PM, Sanchez Perea N, Biffi Garcia C, Garcia Davila CR. Detection of infection with Leptospira spp. in manatees (Trichechus inunguis) of the Peruvian Amazon. 2015. Latin American Journal of Aquatic Mammals 10(1); 58–61. [Google Scholar]
- 16. Sepulveda MA, Seguel M, Alvarado-Rybak M, Verdugo C, Muñoz-Zanzi C, Tamayo R. Postmortem Findings in Four South American Sea Lions (Otaria byronia) from a Urban Colony in Valdivia, Chile. Journal of Wildlife Disease 51(1);279–282. [DOI] [PubMed] [Google Scholar]
- 17. Cerqueria GM, Picardeau M. A century of Leptospira strain typing. 2009. Infect Genet Evol 9 (5); 760–8. 10.1016/j.meegid.2009.06.009 [DOI] [PubMed] [Google Scholar]
- 18. Cerqueira GM, McBride AJ, Picardeau M, Ribeiro SG, Moreira AN, Morel V, et al. Distribution of the leptospiral immunoglobin-like (lig) genes in pathogenic Leptospira species and application of ligB to typing leptospiral isolates. 2009. Journal of Medical Microbiology 58,1173–1181. 10.1099/jmm.0.009175-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nalam L, Ahmed A, Majulata S, Francalacci P, Baig M, Sechi L, et al. Genetic Affinities within a Large Global Collection of Pathogenic Leptospira: Implications for Strain Identification and Molecular Epidemiology. 2010. PLOS ONE 5(8):e12637 10.1371/journal.pone.0012637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brihuega B, Pavan M, Cairo F, Auteri C, Funes D, Romero G, et al. Leptospira patógena en riñón de Didelphis albiventris (comadreja). 2007. Rev Arg Microb; 39:19. [PubMed] [Google Scholar]
- 21. Grune Loffler S, Pavan ME, Vanasco B, Samartino L, Suarez O, Auteri C. et al. Pathogenic Leptospira spp. from urban and periurban rodents in Argentina. 2014. Memorias do Instituto Oswaldo Cruz. Rio de Janeiro: Fundaco Oswaldo Cruz (2);1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grune S. Aislamiento y caracterización genotípica de leptospiras provenientes de animales silvestres en tres ecoregiones argentinas mediante la técnica del Múltiple-Locus Variable-number tandem repeat Análisis (MLVA): Coincidencia con genotipos provenientes de animales de producción. 2014. PhD Thesis. Universidad de Buenos Aires, 135pg.
- 23. Pavan M, Cairo F, Brihuega B, Samartino L. Múltiple-locus variable-number tandem repeats analysis (MLVA) of Leptospira interrogans serovar Pomona from Argentina reveals four new genotypes. 2008. Comp Immun Microbiol & Infec Diseases, 31 (1); 37–45. [DOI] [PubMed] [Google Scholar]
- 24. Pavan ME, Cairó F, Pettinari MJ, Samartino L, Brihuega B. Genotyping of Leptospira interrogans strains from Argentina by Multiple-Locus Variable-number tandem repeat Analysis (MLVA). 2011. Comp Immunol Microbiol Infect Dis. 34(2); 135–41. 10.1016/j.cimid.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 25. Grune Loffler S, Passaro D, Samartino L, Soncini A, Romero G, Brihuega B. Genotypes of Leptospira spp. strains isolated from dogs in Buenos Aires, Argentina. 2014. Rev Argent Microbiol. 46(3);1–13. [DOI] [PubMed] [Google Scholar]
- 26. François S, Brihuega B, Grune S, Gattarello V, Correa D, Petrakovsky J, et al. Aislamiento de Leptospira borgpetersenii de fuentes de agua en Argentina. 2013. Revista Cubana de Medicina Tropical. Nr. 2. [Google Scholar]
- 27. Cinco M, Tamaro M, Cociancich L. Taxonomical, cultural and metabolic characteristics of halophilic leptospirae. Zentralblatt für Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. 1975. Erste Abteilung Originale Reihe A: Medizinische Mikrobiologie und Parasitologie 233(3);400–57. [PubMed] [Google Scholar]
- 28. Cinco M, Tamaro M, Rottini GD, Monti-Bragadin C. Comparative Serological Studies Between a newly Isolated Halophilic Leptospira and two other Leptospiras isolated from Brackish Water. 1974. International Journal of Systematic Bacteriology 24(1); 131–135. [Google Scholar]
- 29. Khairani-Bejo S, Bahaman AR, Zamri-Saad M, Mutalib AR. The survival of Leptospira interrogans serovar Hardjo in the Malaysian environment. 2004. J. Anim. Vet. Adv. 3;123–129 [Google Scholar]
- 30. Trueba G, Zapata S, Madrid K, Cullen P, Haake D. Cell aggregation: a mechanism of pathogenic Leptospira to survive in fresh water. 2004. International microbiology 7; 35–40. [PubMed] [Google Scholar]
- 31. Saito M, Miyahara S, Villanueva SYAM, Aramaki N, Ikejiri M, Kobayashi Y, et al. PCR and Culture Identification of Pathogenic Leptospira spp. from Coastal Soil in Leyte, Philippines, after a Storm Surge during Super Typhoon Haiyan (Yolanda). 2014. Appl. Enviorn. Microbiol 80(22);6926–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McClellan WA, Rommel SA, Moore MJ, Pabst DA. Right whale necropsy protocol. 2004. Contract report to NOAA Fisheries. [Google Scholar]
- 33. Geraci JR, Lounsbury VJ. Marine mammals ashore: a field guide for strandings (Geraci JR and Lounsbury VJ, Eds.2005. 2nd edn. National Aquarium of Baltimore, Inc., Baltimore, MD.344pg. [Google Scholar]
- 34. Gravekamp C, Van de Kemp H, Franzen M, Carrington D, Schoone GJ, Van Eys GJ, et al. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. 1993. J. Gen. Microbiol. 139(8);1691–700 [DOI] [PubMed] [Google Scholar]
- 35. Martínez M, Grune Loffler S, Samartino L, Romero G, De la Fuente I, Brihuega B. Detección de leptospiras patógenas mediante PCR usando los iniciadores LigBF y LigBR. 2014. Revista Argentina de Zoonosis y Enfermedades Infecciosas Emergentes 9(2);41. [Google Scholar]
- 36. Djadid ND, Ganji ZF, Gouya MM, Rezvani M, Zakeri S. A simple and rapid nested polymerase chain reaction-restriction fragment length polymorphism technique for differentiation of pathogenic and nonpathogenic Leptospira spp. 2009. Diagn. Microbiol. Infect Dis. 63(3); 251–6. 10.1016/j.diagmicrobio.2008.10.017 [DOI] [PubMed] [Google Scholar]
- 37. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA 5: Molecular Evolutionary Genetics Analysis using maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. 2011. Molecular Biology and Evolution 28; 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.