Summary
Bacillus subtilis and its closest relatives have multiple rap-phr quorum sensing gene pairs that coordinate a variety of physiological processes with population density. Extra-chromosomal rap-phr genes are also present on mobile genetic elements, yet relatively little is known about their function. In this work, we demonstrate that Rap60-Phr60 from plasmid pTA1060 coordinates a variety of biological processes with population density including sporulation, cannibalism, biofilm formation and genetic competence. Similar to other Rap proteins that control sporulation, Rap60 modulates phosphorylation of the transcription factor Spo0A by acting as a phosphatase of Spo0F~P, an intermediate of the sporulation phosphorelay system. Additionally, Rap60 plays a noncanonical role in regulating the autophosphorylation of the sporulation-specific kinase KinA, a novel activity for Rap proteins. In contrast, Rap proteins that modulate genetic competence interfere with DNA binding by the transcription factor ComA. Rap60 regulates the activity of ComA in a unique manner by forming a Rap60–ComA–DNA ternary complex that inhibits transcription of target genes. Taken together, this work provides new insight into two novel mechanisms of regulating Spo0A and ComA by Rap60 and expands our general understanding of how plasmid-encoded quorum sensing pairs regulate important biological processes.
Introduction
Quorum sensing or diffusion monitoring is a process many bacteria use to coordinate biological processes with population density (Redfield, 2002; Winans and Bassler, 2008). Individual cells within a population communicate with one another using extracellular cell–cell signaling molecules. In Gram-negative bacteria, these signals are derived from acylated homoserine lactones. In contrast, Gram-positive bacteria predominately use peptides. Many biological processes are regulated by the quorum response including biofilm formation, cellular motility, the secretion of degradative enzymes, horizontal gene transfer, the production of virulence factors and antibiotics, bioluminescence, sporulation, and genetic competence (see reviews by Waters and Bassler, 2005; Auchtung and Grossman, 2008).
In the soil bacterium Bacillus subtilis, ComA is the master regulator of early competence genes (Roggiani and Dubnau, 1993). The activity of ComA is regulated, in part, by the histidine kinase, ComP and an extracellular cell–cell signaling peptide, ComX, that function in combination to coordinate the phosphorylation of ComA with population density (Weinrauch et al., 1990; Magnuson et al., 1994; Lazazzera et al., 1999 and Fig. 1). Once activated by phosphorylation, ComA binds to a tripartite DNA binding site located within the promoter of target genes (Griffith and Grossman, 2008) and directly activates transcription of 89 genes involved in competence development, antibiotic production and the secretion of degradative enzymes (Ogura et al., 2001; Comella and Grossman, 2005). One of the early competence genes is comS, which encodes an anti-adaptor protein responsible for stabilizing ComK from proteolysis by ClpCP. ComK is the master regulator of late competence genes whose gene products comprise the DNA translocation machinery (Fig. 1). At least five transcription factors (Rok, CodY, DegU, Spo0A and AbrB) converge to directly regulate the transcription of comK. In addition, ComK modulates its own transcription via a positive auto-activation loop (see review by Hamoen et al., 2003). The complex regulation of ComK results in a bimodal distribution of ComK-dependent gene expression across a population of cells ensuring that only a small subset of cells (as high as 10–20% in some laboratory strains) enters into the competent state (Maamar and Dubnau, 2005; Smits et al., 2005).
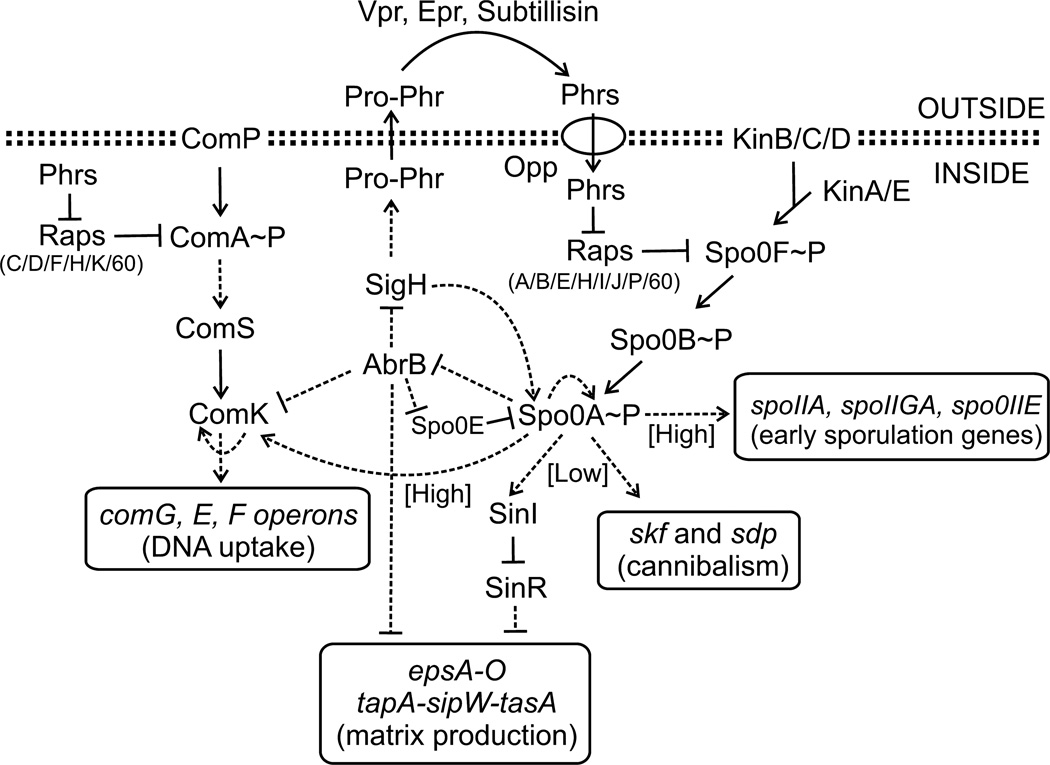
Fig. 1.
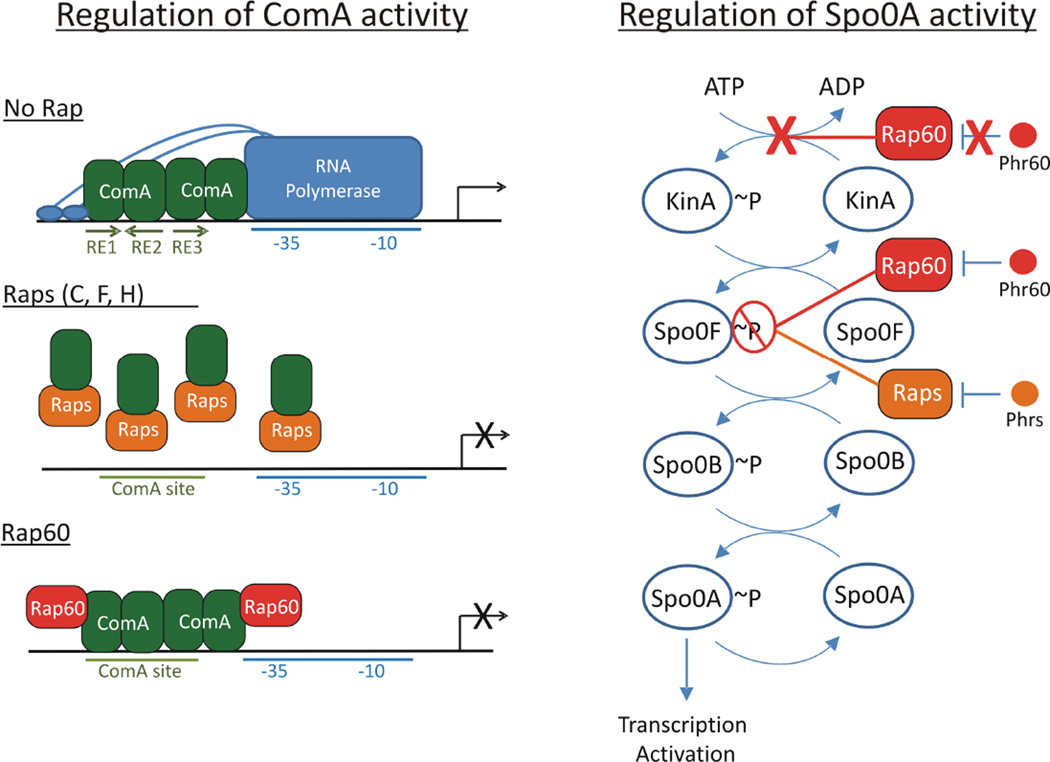
Regulation of sporulation, biofilm formation, cannibalism and genetic competence in B. subtilis. The phosphorylation state of Spo0A and its activity is regulated by the sporulation phosphorelay system that includes kinases KinA-E and two intermediate proteins Spo0F and Spo0B. High levels of Spo0A~P are required for transcription activation of early sporulation and competence genes. In contrast, low levels of Spo0A~P activate genes involved in cannibalism and biofilm formation. Rap proteins inhibit Spo0A by acting as phosphatases of Spo0F~P, whereas competence-specific Rap proteins inhibit ComA binding to DNA. In the case of Rap60, ComA activity is inhibited without perturbing ComA binding to DNA. Phr signaling peptides coordinate the physiological responses with population density. Pro-Phr peptides are exported into the environment where they are processed into mature signaling peptides by extracellular proteases Vpr, Epr and Subtilisin. Mature Phr peptides enter the cell through the oligopeptide permease Opp where they bind to and antagonize the activity of their cognate Rap proteins. Phr peptide production is regulated by Spo0A via the AbrB-SigH pathway. Arrows indicate positive regulation, and perpendicular lines indicate negative control. Solid lines represent regulation by protein–protein interactions, and dashed lines represent transcriptional responses.
Sporulation is also regulated, in part, by the quorum response. During sporulation, a subpopulation of cells undergoes a transformation into an endospore that protects the cell against temperature extremes, desiccation and ionizing radiation (Piggot and Hilbert, 2004). Spo0A is the master regulator of early sporulation genes (Hoch, 1993). The activity of Spo0A is regulated by phosphorylation via kinases (Kin A–E) and a series of phosphorelay proteins that collectively make up the sporulation phosphorelay system (Fig. 1). During conditions that facilitate sporulation, e.g. nutrient limitation, KinA and KinB phosphorylate the intermediate response regulator Spo0F. The phosphoryl group is transferred from Spo0F~P to the histidine phosphotransferase Spo0B, and subsequently, to the response regulator Spo0A (see review by Higgins and Dworkin, 2012). Once activated by phosphorylation, Spo0A~P binds to the ‘0A box’ located in the promoter region of target genes and directly regulates transcription of 121 genes leading to the initiation of sporulation (Fawcett et al., 2000; Molle et al., 2003). Similar to ComK, Spo0A~P regulates its own transcription in a positive auto-activation loop (Hoch, 1991). In addition, spo0A transcription is also modulated by a double-repression system involving the transition state repressor AbrB and the stationary phase sigma factor SigH (Strauch et al., 1990; Predich et al., 1992 and Fig. 1). The complex regulation of Spo0A functions as a bistable switch and ensures that a subpopulation of cells (up to 60%) develops into a mature endospore (Chung et al., 1994).
In addition to sporulation, Spo0A also regulates genetic competence, biofilm formation and cannibalism (Fig. 1). The concentration of phosphorylated Spo0A influences the timing and the developmental fate of individual cells within a population (Fugita et al., 2005). High concentrations of Spo0A~P are required to activate transcription of genes involved in sporulation and genetic competence (Fig. 1). Spo0A~P influences competence by regulating the transcription of comK directly and indirectly through AbrB (Fugita et al., 2005; Mirouze et al., 2012). In contrast, low concentrations of Spo0A~P activate transcription of genes involved in biofilm production and cannibalism (Hamon and Lazazzera, 2001; Branda et al., 2004; Fugita et al., 2005). During biofilm formation, a subset of cells within a population produces a protective extracellular matrix comprised of exopolysaccharide and the amyloid-like protein TasA, encoded by the epsA-O and tapA(yqxM)-sipW-tasA operons respectively. In addition, the biofilm-surface layer protein, encoded by blsA(yuaB), associates with the extracellular matrix and is required for biofilm formation (Stover and Driks, 1999; Branda et al., 2001; Romero et al., 2011; Kobayashi and Iwano, 2012). Transcription of biofilm genes is directly regulated by Spo0A (e.g., bslA) and the AbrB and antirepressor-repressor pair SinI-SinR (e.g., epsA and tasA) (Hamon et al., 2004; Kearns et al., 2005; and Fig. 1). Expression of biofilm and cannibalism genes is coordinated within the same subpopulation of cells (Lopez et al., 2009). It is proposed that cannibalism provides a temporal window for cells to reassess the availability of nutrients prior to committing to sporulation (Gonzalez-Pastor et al., 2003). During cannibalism, cells produce the toxins sporulation-killing factor (SKF) and sporulation-delaying protein, in addition to immunity proteins that protect against the toxins. Sibling cells that lack immunity are lysed by the toxins, and their nutrients were released into the environment. Cannibals feed on the available nutrients and the onset of sporulation is delayed (Gonzalez-Pastor et al., 2003; Ellermeier et al., 2006).
The activities of Spo0A and ComA are regulated, in part, through rap-phr quorum sensing gene pairs that coordinate biological processes with population density (Fig. 1). Rap proteins, originally named for their function as regulators of aspartate phosphatases (Perego et al., 1994), influence Spo0A activity by modulating the sporulation phosphorelay system. Specifically, the chromosomally encoded Rap proteins (RapA, B, E, H, I and J) act as phosphatases of Spo0F~P (Fig. 1). Dephosphorylation of Spo0F~P by the Rap proteins leads to decreased concentrations of Spo0A~P (Perego et al., 1994; Jiang et al., 2000a; Smits et al., 2007; Parashar et al., 2011). Additional phosphatases act directly on Spo0A~P including Spo0E, and homologues YisI and YnzD (Perego, 2001 and Fig. 1). Like Spo0A, the activity of ComA is also regulated by multiple Rap proteins, albeit by a different mechanism. Instead of modulating the phosphorylation state of ComA, five chromosomally encoded Rap proteins (RapC, D, F, H and K) act as ‘anti-activators’ of ComA, presumably by a common mechanism of inhibiting ComA binding to DNA and preventing transcription activation of target genes (Core and Perego, 2003; Bongiorni et al., 2005; Auchtung et al., 2006; Smits et al., 2007; Baker and Neiditch, 2011; and Fig. 1).
Rap protein activity is coordinated with population density via extracellular Phr signaling peptides (Fig. 1). Phr peptides are synthesized as precursor proteins, Pro-Phr, containing an N-terminal signal sequence that is required for export into the extracellular environment by the SecA pathway (Pottathil and Lazazzera, 2003; Stephenson et al., 2003). Once outside the cell, extracellular proteases Subtilisin, Vpr and Epr, process the Phr precursor into the mature signaling peptide (Lanigan-Gerdes et al., 2007). Mature Phr peptides reenter cells through the oligopeptide permease Opp (LeDeaux et al., 1997), and once inside the cell, Phr peptides bind to and inhibit the activity of their cognate Rap proteins (Fig. 1). Rap-Phr quorum sensing pairs function in concert to coordinate biological processes with population density. At low density when the concentration of Phr peptide is low and Rap protein activity is high, Spo0A and ComA are predominately inactive. At high cell density when the concentration of Phr peptide is high and the activity of Rap protein is low, Spo0A~P and ComA~P activate transcription of target genes.
Genes encoding rap-phr pairs are also present on mobile genetic elements including plasmids, bacteriophages and conjugative elements. A rap-phr pair, encoded on the virulence plasmid pX01, regulates sporulation in B. anthracis (Bongiorni et al., 2005). In B. subtilis, RapI-PhrI controls the excision and mating of the conjugative element ICEBs1 (Auchtung et al., 2005). RapLS20 coordinates conjugation of plasmid pLS20 with population density (Singh et al., 2013). Plasmid pBS32, present in the undomesticated B. subtilis strain NCIB3610, encodes RapP-PhrP that regulates biofilm formation, sporulation, motility and genetic competence (Parashar et al., 2013). Rolling circle plasmids pTA1015, pTA1040, pTA1050 and pTA1060 have been isolated from B. subtilis and, in the case of pTA1060, also from B. amyloliquefaciens. Many of these plasmids contain a rap-phr gene pair (see review by Meijer et al., 1998). Koetje et al. (2003) demonstrated that over-expression of Rap60 from pTA1060 regulates the production of extracellular proteases via the Spo0A-AbrB regulatory cascade.
In this work, we expand on the role of Rap60-Phr60 from plasmid pTA1060 and demonstrate its importance in regulating biological processes including sporulation, cannibalism, biofilm formation and genetic competence in B. subtilis. Rap60 regulates these processes by inhibiting the activities of the transcriptional activators Spo0A and ComA. Similar to other Rap proteins that modulate sporulation, Rap60 regulates the activity of Spo0Aby acting as a phosphatase of Spo0F~P. In addition, Rap60 has a non-canonical role in regulating sporulation by interfering with the autophosphorylation of KinA. We also identified a novel mechanism of regulating ComA activity by Rap proteins. Specifically, Rap60 binds to ComA and inhibits its activity without perturbing ComA binding to DNA. Finally, we propose models for these newly identified mechanisms of regulating the activities of transcription factors by Rap60.
Results
Rap60 inhibits sporulation
Koetje et al. (2003) demonstrated that over-expression of Rap60, isolated from plasmid pTA1060, causes a decrease in the production of extracellular proteases in B. subtilis. Impaired protease production by Rap60 is controlled by the Spo0A-AbrB regulatory cascade. Specifically, Rap60 inhibits Spo0A activity resulting in the accumulation of AbrB repressor and direct repression of genes encoding extracellular proteases (Ferrari et al., 1988; Koetje et al., 2003). Interestingly, Koetje et al. (2003) showed that Rap60 inhibits Spo0Aactivity but had no effect on sporulation. This finding is inconsistent with prior knowledge about the regulation of sporulation by Spo0A and warrants further investigation.
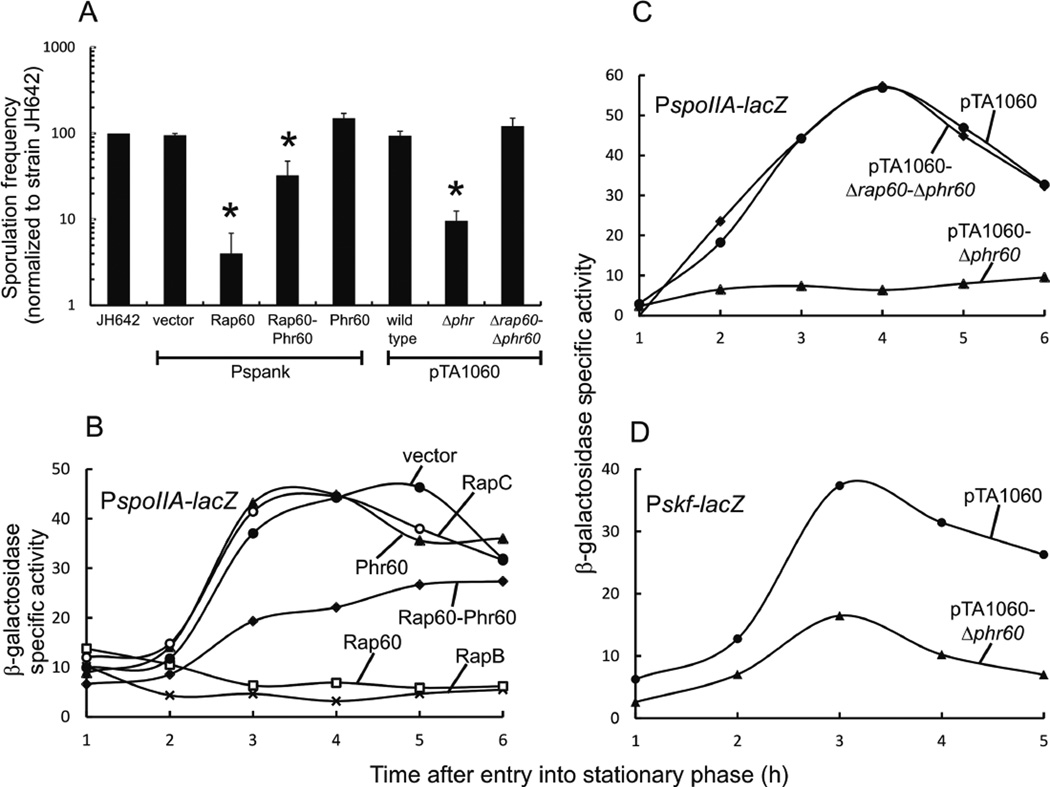
To determine the effect of Rap60 on sporulation, we measured the sporulation frequencies of strains expressing rap60, phr60 and rap60-phr60 from an ectopic Isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter (Pspank) in the absence of plasmid pTA1060 (see Experimental procedures). Strain KB19 containing an empty Pspank vector and wild-type strain JH642, treated with IPTG or left untreated, exhibited similar sporulation frequencies ranging from 20% to 40%, depending on the experiment. Rap60 inhibited sporulation 70% (Fig. 2A). Interestingly, co-expression of Rap60 and Phr60 signaling peptide partially suppressed the effects of Rap60, restoring the sporulation frequency to ~30% of wild type (Fig. 2A). Phr60 had little effect on the sporulation frequency compared with wild type (Fig. 2A). This suggests that Phr60 signaling peptide is specific for Rap60 and does not significantly influence the activity of other Rap proteins that modulate sporulation.
Fig. 2.
Effect of Rap60-Phr60 on sporulation and transcription of early sporulation genes.
A. Sporulation frequencies were measured for each strain and normalized to wild type strain JH642. Strains over-expressing Rap60 (KB26), Phr60 (KB28), Rap60-Phr60 (KB27) and empty vector (KB19) were treated with 0.1 mM IPTG. Strains containing pTA1060 (KG1780), pTA1060-Δphr60 (KG1782) and pTA1060-Δrap60-Δphr60 (KG1781) were grown in the presence of erythromycin to select for plasmid maintenance. Sporulation frequencies ranged from 20% to 40% for strain JH642. Experiments were performed in triplicate and the standard error is shown. Asterisks represent P-values < 0.01.
B-D. Cultures containing promoter fusions to lacZ were grown in DSM. Aliquots were removed at the specified time, and β-galactosidase activity was determined. Experiments were repeated in triplicate with similar results. A single representative experiment is shown.
B. Over-expression of Rap protein and Phr peptide from Pspank in strains containing PspoIIA-lacZ: KB45 empty vector (filled circle); KB63 RapC (open circle); KB48 Phr60 (filled triangle); KB47 Rap60-Phr60 (filled diamond); KB46 Rap60 (open square); and KB138 RapB (X).
C. Stains containing PspoIIA-lacZ and derivatives of plasmid pTA1060: KG1766 pTA1060 (filled circles); KG1772 pTA1060-Δphr60 (filled triangles); and KG1771 pTA1060-Δrap60-Δphr60 (filled diamond).
D. Strains containing Pskf-lacZ and derivatives of plasmid pTA1060: KG1911 pTA1060 (filled circles) and KG1912 pTA1060-Δphr60 (filled triangles).
We sought to determine the effect on sporulation of expressing Rap60 at physiological levels from plasmid pTA1060. Because Phr peptides antagonize the activity of Rap proteins, phr60 was deleted from pTA1060 in order to unmask its effects on Rap60 activity (see Experimental procedures). Strain KG1782 containing pTA1060-Δphr60 had a reduced sporulation frequency, similar to overexpressing Rap60 from the Pspank promoter (Fig. 2A). The deficiency in sporulation is specific to Rap60 and not another gene encoded on pTA1060 as deleting both rap60 and phr60 (pTA1060-Δrap60-Δphr60) restored the sporulation frequency to wild-type levels (Fig. 2A).
Rap60 inhibits Spo0A activity
Rap proteins that modulate sporulation inhibit the activity of Spo0A resulting in decreased transcription of early sporulation genes, e.g., spoIIA, spoIIGA and spoIIIE (Molle et al., 2003; Fugita et al., 2005; and Fig. 1). A spoIIA promoter fusion to lacZ was constructed and used to monitor Spo0A activity (see Experimental procedures). A density-dependent increase in β-galactosidase activity was observed in strain KB45 (empty vector) with maximal activity occurring ~3–4 h after entry into stationary phase (Fig. 2B, filled circles). Consistent with previously published results by Koetje et al. (2003), expression of Rap60 from the Pspank promoter severely inhibited the activity of Spo0A resulting in low β-galactosidase activity (Fig. 2B, open squares). Co-expression of Rap60 and Phr60 partially suppressed the response by Rap60 and restored the density-dependent regulation of PspoIIA-lacZ to an intermediate level compared with strain KB45 (Fig. 2B, filled diamonds). Phr60 had no effect on the transcription of spoIIA (Fig. 2B, closed triangles). This suggests that Phr60 is specific for Rap60 and has no significant effect on the activity of other sporulation-specific Rap proteins. Expression of Rap60 from pTA1060-Δphr60 also inhibited Spo0A activity (Fig. 2C, filled triangles) to comparable levels as Pspank-rap60 (Fig. 2B, open squares). Rap60 is directly responsible for the decreased Spo0A activity as deleting both rap60 and phr60 from pTA1060 restored the β-galactosidase activity to wild-type levels (Fig. 2C, filled diamonds). RapB and RapC are known regulators of Spo0A and ComA respectively (Perego et al., 1994; Core and Perego, 2003). As a control for Rap specificity in these experiments, we found that over-expression of RapB inhibited Spo0A activity, whereas over-expression of RapC had no effect on the activity of Spo0A (Fig. 2B, ‘X’ and open circles).
During sporulation conditions, a subpopulation of cells will initiate cannibalistic behavior in response to low levels of Spo0A~P (Branda et al., 2001; Fugita et al., 2005). To determine the role of Rap60 in cannibalism, β-galactosidase activity was measured in a reporter strain containing a skf (sporulation-killing factor) promoter fusion to lacZ (kind gift from R. Losick). Expression of Rap60 from pTA1060-Δphr60 had a modest inhibitory effect on the transcription of Pskf-lacZ resulting in decreased β-galactosidase activity compared with strain KG1911 containing wild-type pTA1060 (Fig. 2D). Thus, it appears that the major role of Rap60 during sporulation conditions involves inhibiting Spo0A-dependent transcription of early sporulation genes, whereas Rap60 plays a minor role in inhibiting the transcription of cannibalism genes.
Rap60 alters complex colony architecture and biofilms
Undomesticated strains of B. subtilis (e.g., NCIB3610) form complex, structured biofilm communities on solid surfaces and a thick pellicle at the liquid–air interface when grown statically in liquid medium (Branda et al., 2001). In contrast, many common laboratory strains, including strain JH642 used in these studies, have acquired mutations during the process of domestication that inhibit biofilm formation. As a result, many domesticated laboratory strains produce flat and featureless colonies on solid medium and form smooth, fragile pellicles in static liquid cultures (Branda et al., 2001). RapP, present on plasmid pBS32, was recently shown to be important for complex colony architecture in the undomesticated strain 3610. Specifically, deletion of rapP or curing the strain of plasmid pBS32 produced colonies with a hyper-rugose appearance (Konkol et al., 2013).
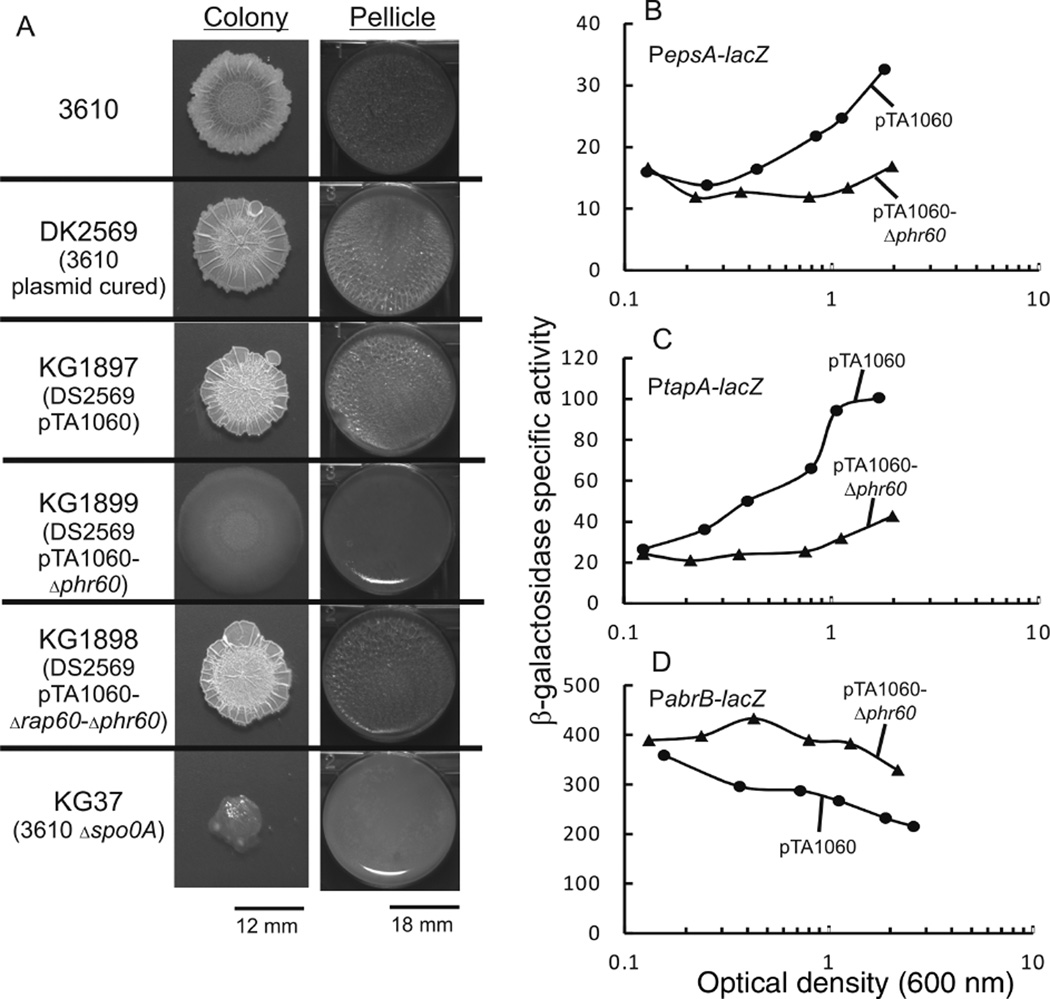
We sought to determine the effect of pTA1060 on colony architecture and pellicle formation in strain DS2569, a plasmid-cured derivative of strain 3610 (kind gift from D. Kearns). Consistent with previously published results, strain DS2569 had a hyper-rugose morphology when grown on solid medium compared with wild-type strain 3610 that formed elaborate colonies with an extensive network of protruding ridges. Despite these differences in colony morphology, strains DS2569 and 3610 both formed robust pellicles at the liquid–air interface (Fig. 3A and Konkol et al., 2013). Expression of Rap60 from pTA1060-Δphr60 in strain KG1899 had a dramatic effect on colony morphology resulting in smooth, flat colonies that lacked the hyper-rugose appearance of strain DS2569 (Fig. 3A). In addition, strain KG1899 formed fragile, paper thin pellicles when grown in static liquid culture, similar to strain KG37, a derivative of 3610 containing a spo0A deletion (Fig. 3A). The effects on colony morphology and pellicle formation are due to Rap60 and not another gene present on pTA1060 as deletion of both rap60 and phr60 (strain KG1898) suppressed the smooth colony morphology and restored the robust pellicle (Fig. 3A).
Fig. 3.
Effect of Rap60-Phr60 on colony morphology, pellicle formation and transcription of biofilm genes.
A. Colony morphology was determined by inoculating 10 µl of an overnight culture onto MSgg solid medium and incubating plates at 24°C for 3 days. Pellicle formation was determined by inoculating 10 µl of an overnight culture into 3 ml of liquid MSgg medium in 6-well microtiter plates, followed by incubation at 24°C for 3 days.
B-D. Cultures containing promoter fusions to lacZ and derivatives of pTA1060 were grown in MSgg medium. Aliquots were removed at the specified time and β-galactosidase activity was determined. Experiments were repeated in triplicate with similar results. A single representative experiment is shown.
B. Strains containing PespA-lacZ: KG1928 pTA1060 (filled circles) and KG1929 pTA1060-Δphr60 (filled triangles).
C. Strains containing PtapA-lacZ: KG1930 pTA1060 (filled circles) and KG1931 pTA1060-Δphr60 (filled triangles).
D. Strains containing PabrB-lacZ: KG1903 pTA1060 (filled circles) and KG1905 pTA1060-Δphr60 (filled triangles).
The epsA-O and tapA(yqxM)-sipW-tasA operons are required for matrix production and biofilm formation (Stover and Driks, 1999; Branda et al., 2001). Promoter-lacZ fusions to epsA and tapA were used to determine the effect of pTA1060-Δphr60 on the transcription of biofilm genes (kind gift from R. Losick). A small decrease in β-galactosidase activity was observed for both reporter strains when Rap60 was expressed from pTA1060-Δphr60 (Fig. 3B and C, solid triangles). The presence of Phr60 from wild-type plasmid pTA1060 suppressed the inhibitory effects of Rap60 resulting in increased β-galactosidase activity from both reporters (Fig. 3B and C, solid circles). Regulation of matrix producing genes is mediated indirectly by Spo0A through the AbrB and SinI-SinR regulatory cascades (Hamon et al., 2004; Kearns et al., 2005; Chu et al., 2006; and Fig. 1).To monitor the activity of Spo0A in the presence of pTA1060, we obtained an abrB promoter fusion to lacZ (kind gift from R. Losick). Strain KG1905, containing pTA1060-Δphr60 and PabrB-lacZ, had increased β-galactosidase activity compared with the reporter strain KG1903 with wild-type pTA1060 (Fig. 3D). Taken together, these results indicate that Rap60 indirectly regulates the transcription of biofilm genes by modulating the activity of Spo0A.
Rap60 inhibits genetic competence
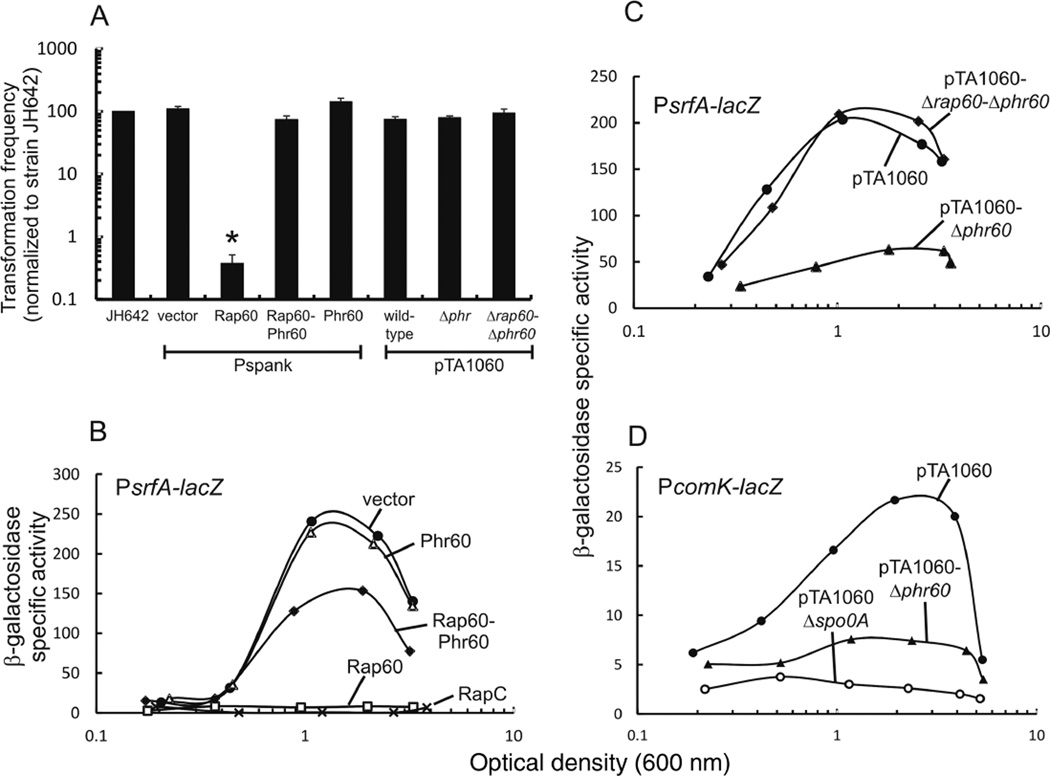
RapH inhibits both sporulation and genetic competence in B. subtilis (Smits et al., 2007). To determine the role of Rap60 in regulating genetic competence, the transformation efficiency was measured in strains expressing Rap60 and Phr60. Strain KB19, containing Pspank vector, had a transformation efficiency of ~0.01%, similar to wild-type strain JH642. Expression of Rap60 inhibited genetic competence reducing the transformation efficiency > 100-fold (Fig. 4A). Co-expression of Rap60 and Phr60 suppressed the effects of Rap60, restoring the transformation efficiency to levels near wild type (Fig. 4A). Phr60 alone had no effect on competence (Fig. 4A), suggesting that Phr60 is specific to Rap60 and has no appreciable effect on the activities of other Rap proteins that regulate genetic competence. Interestingly, although over-expression of Rap60 from the Pspank promoter dramatically reduced the transformation efficiency, expression of Rap60 from pTA1060-Δphr60 had no effect on genetic competence (Fig. 4A).
Fig. 4.
Effect of Rap60-Phr60 on transformation efficiency and transcription of competence genes.
A. Transformation frequency was measured for each strain and normalized to wild-type JH642. Strains over-expressing Rap60 (KB26), Phr60 (KB28), Rap60-Phr60 (KB27) and empty vector (KB19) were treated with 0.1 mM IPTG. Strains containing pTA1060 (KG1780), pTA1060-Δphr60 (KG1782) and pTA1060-Δrap60-Δphr60 (KG1781) were grown in the presence of erythromycin to select for plasmid maintenance. Depending on the experiment, transformation frequencies ranged from 0.01% to 0.07% for strain JH642. Experiments were performed in triplicate, and the standard error is shown. The asterisk represents a P-value < 0.01.
B-D. Cultures containing promoter fusions to lacZ were grown in minimal medium, aliquots removed at the specified times and β-galactosidase activity was determined. Experiments were repeated in triplicate with similar results. A single representative experiment is shown.
B. Over-expression of Rap protein and Phr peptide from Pspank in a PsrfA-lacZ reporter strain. KB22 empty vector (filled circle); KB25 Phr60 (open triangle); KB24 Rap60-Phr60 (filled diamond); KB23 Rap60 (open square); and KB137 RapC (X).
C. Plasmid pTA1060 derivatives in strains containing PsrfA-lacZ. KG1759 pTA1060 (filled circle); KG1764 pTA1060-Δrap60-Δphr60 (filled diamond); and KG1765 pTA1060-Δphr60 (filled triangle).
D. Plasmid pTA1060 derivatives in strains containing PcomK-lacZ. KG1793 pTA1060 (filled circle); KG1796 pTA1060 Δspo0A (open circle); and KG1795 pTA1060-Δphr60 (filled triangle).
Rap60 inhibits ComA activity
Raps C, F and H inhibit competence development by functioning as ‘anti-activators’ of ComA (Core and Perego, 2003; Bongiorni et al., 2005; Smits et al., 2007; Baker and Neiditch, 2011; and Fig. 1). We sought to determine whether Rap60 was also inhibiting competence by regulating ComA. Located within the srfA operon is the gene encoding ComS, an anti-adaptor protein that inhibits degradation of ComK by the ClpCP protease (Hamoen et al., 2003). A srfA promoter fusion to lacZ was constructed and used to monitor ComA activity (see Experimental procedures). Consistent with previously published results, ComA activity was regulated in a density-dependent manner. Specifically, at low population density, low β-galactosidase activity was observed with a 12-fold increase in activity occurring at OD600 ~1–2 as the culture enters stationary phase (Fig. 4B, filled circles and Griffith and Grossman, 2008). Expression of Rap60 abolished the density-dependent regulation resulting in low, basal β-galactosidase activity throughout the growth cycle, similar to over-expressing the competence-specific RapC protein (Fig. 4B, open squares vs. ‘X’ and Core and Perego, 2003). Phr60 partially suppressed the activity of Rap60 (Fig. 4B, filled diamonds). Expression of Phr60 alone had no effect on the activity of ComA (Fig. 4B, open triangles), suggesting that Phr60 is specific for Rap60.
To more accurately determine the role of pTA1060-Δphr60 in regulating competence genes, we used a PsrfA-lacZ and PcomK-lacZ reporter to measure ComA and ComK activity respectively (see Experimental procedures). Deletion of phr60 inhibited the activities of ComA and ComK as judged by decreased β-galactosidase activities from both reporter strains (Fig. 4C and D, filled triangles). Plasmid-encoded Rap60 from pTA1060-Δphr60 had an intermediate effect on ComA activity (Fig. 4C, filled triangles), compared with over-expressing Rap60 from the Pspank promoter that completely abolished all of the density-dependent regulation (Fig. 4B, open squares). Similarly, the presence of pTA1060-Δphr60 reduced comK transcription (Fig. 4D, filled triangles), but to a lesser extent than in strain KG1796 containing a spo0A deletion (Fig. 4D, open circles). The inhibitory effects of Rap60 extend to late competence genes whose products make up the DNAtranslocation machinery. Specifically, we observe a similar reduction in transcription of comGA in the presence of pTA1060-Δphr60 (data not shown). Taken together, expression of Rap60 from pTA1060-Δphr60 clearly inhibits the transcription of competence genes, but to a lesser extent required to affect the transformation efficiency.
Synthetic Phr60 peptides inhibit Rap60
Pro-Phr signaling peptides are secreted into the environment where they are processed into mature signaling peptides by extracellular proteases (Pottathil and Lazazzera, 2003; Stephenson et al., 2003; Lanigan-Gerdes et al., 2007). For the majority of Phr peptides that have been characterized in B. subtilis, the mature signaling peptide is comprised of the carboxyl terminal 5–6 amino acids. Two exceptions are PhrE and PhrH whose mature peptides are internal (Jiang et al., 2000a; Mirouze et al., 2011). Recent work from the Neiditch laboratory demonstrated that synthetic peptides corresponding to the last six amino acids of PhrH and PhrI have enhanced activity over peptides comprised of the last five amino acids, suggesting that hexapeptides might be the biologically relevant cell– cell signaling molecules in B. subtilis (Mirouze et al., 2011).
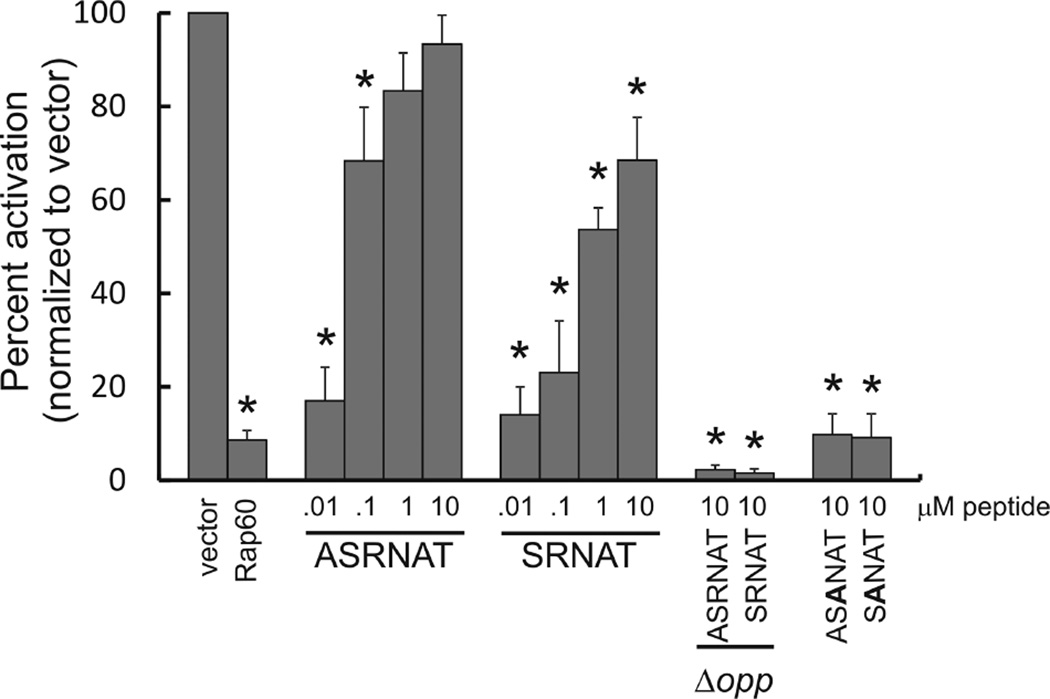
Koetje et al. (2003) demonstrated that a synthetic pentapeptide corresponding to the last five amino acids of Pro-Phr60 (NH2-SRNAT-COOH) inhibited Rap60 activity in vivo. To determine whether the last six amino acids of Pro-Phr60 had an effect on Rap60 activity, synthetic peptides corresponding to the last five (SRNAT) and six (ASRNAT) amino acids of Phr60 were synthesized and added to the growth medium of the reporter strain KB23 containing PsrfA-lacZ and expressing Rap60 from the Pspank promoter (see Experimental procedures). Both the penta- and hexapeptides inhibited Rap60 activity as shown by an increase in β-galactosidase activity from PsrfA-lacZ (Fig. 5). Depending on the concentration, the hexapeptide was 30–70% more potent in inhibiting Rap60 activity compared with the pentapeptide (Fig. 5). The activity of Phr60 was sequence specific as replacing the arginine at position 3 with alanine ((A)SANAT) abolished Phr60 activity even at concentrations as high as 10 µM (Fig. 5). Consistent with previously published results, inhibition of Rap60 activity by the synthetic Phr60 peptides was dependent on the oligopeptide permease Opp (Fig. 5 and Koetje et al., 2003). Specifically, a deletion of the spo0KA operon (encoding Opp) completely abolished the antagonistic effects of the synthetic Phr60 hexapeptide even at concentrations as high as 10 µM (Fig. 5). Interestingly, the reporter strain KB58 containing Pspank-rap60 and the spo0K mutant had significantly reduced β-galactosidase activity compared with strain KB54 containing only Pspank-rap60. This is likely due to unchecked regulation of Rap proteins as the chromosomally encoded Phr peptides (C, F and H) fail to enter the cell and antagonize their cognate Rap proteins.
Fig. 5.
Regulation of Rap60 by synthetic Phr60 peptides. Reporter strain KB23 containing PsrfA-lacZ and Pspank-rap60 was grown with 0.1 mM IPTG in the presence of synthetic peptides corresponding to the carboxyl terminal penta- (SRNAT) and hexapeptides (ASRNAT) of Pro-Phr60. Peptides containing an alanine substitution in the conserved arginine of Phr60 ((A)SANAT) served as controls. Strain KB58 containing PsrfA-lacZ and Pspank-rap60 with an Opp (spo0KA) deletion was used to determine the effects of Opp and synthetic peptides on Rap60 activity. The maximal activity of each culture (typically around OD600 ~1–2) was normalized to strain KB22 containing Pspank empty vector. Experiments were performed in triplicate and the percent standard error is shown. Asterisks represent P-values < 0.01.
Rap60 inhibits multiple steps of the sporulation phosphorelay
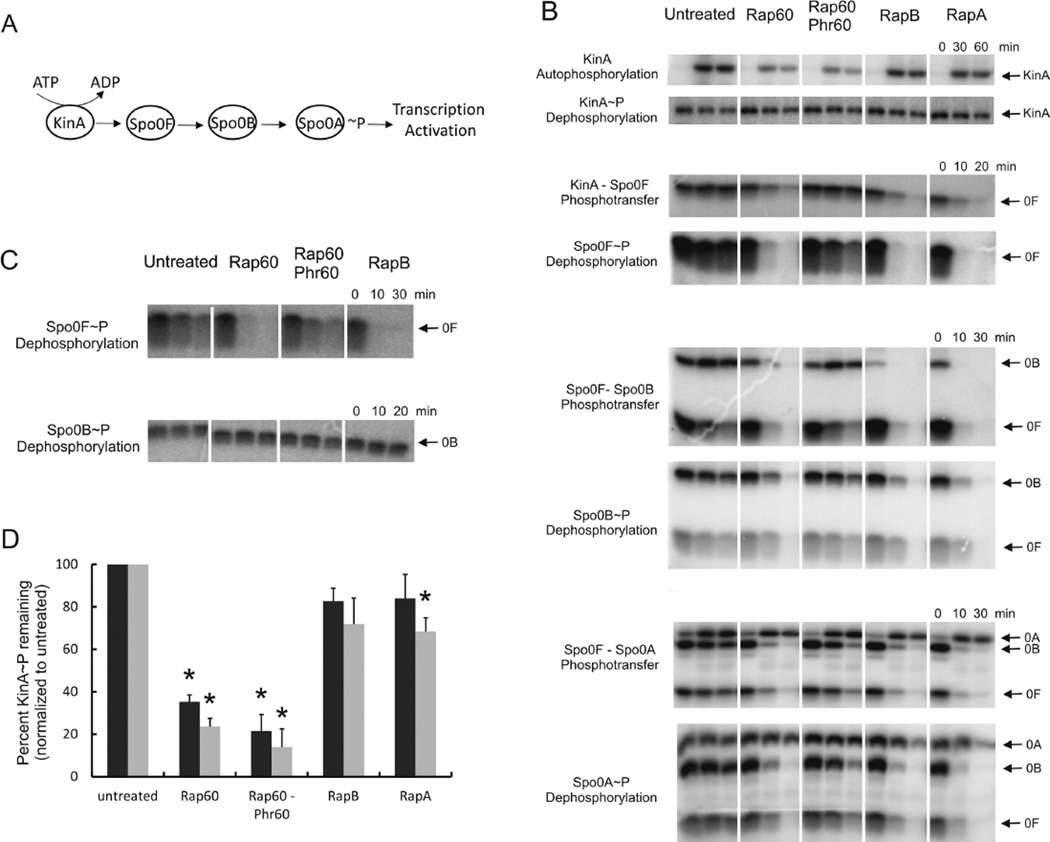
Rap60 controls sporulation, cannibalism and biofilm formation by a common mechanism of inhibiting the activity of Spo0A (Figs 2–4). Spo0A phosphorylation and its activity as a transcriptional activator is regulated by multiple kinases and the sporulation phosphorelay system (see review by Higgins and Dworkin, 2012 and Fig. 6A). To determine which step(s) is affected by Rap60, we purified components of the phosphorelay and developed an assay to monitor protein phosphorylation (see Experimental procedures). Because signaling through the phosphorelay depends both on phosphorylation and dephosphorylation, the effect of Rap60 was systematically determined for both the capacity of a protein to be phosphorylated (phosphotransfer) and the stability of the phosphoryl group (dephosphorylation).
Fig. 6.
Effects of Rap60 on the phosphorylation state of sporulation phosphorelay proteins. Phosphorylation assays were performed using 3 µM his6-KinA, 10 µM his6-Rap, 300 µM Phr60 hexapeptide and 5 µM each of his6-tagged or native Spo0F, Spo0B and Spo0A, where appropriate, except for KinA autophosphorylation and KinA~P dephosphorylation experiments that used 5 µM his6-KinA and 5 µM his6-Rap. Purified proteins were incubated in the reaction buffer with γ32P-ATP for 1 h at 24°C, prior to termination with the addition of 10 mM ATP. Aliquots were removed at the specified times and subjected to SDS-PAGE. Experiments were performed in triplicate and representative gels are shown.
A. Schematic depicting the flow of phosphate through the sporulation phosphorelay system.
B. Phosphotransfer and dephosphorylation of sporulation phosphorelay proteins. Reactions were terminated with 10 mM ATP, aliquots removed at the specified times and subjected to SDS-PAGE. The order of protein addition is described in Experimental procedures.
C. Effects of Rap proteins on dephosphorylation of native Spo0F~P and native Spo0B~P in the absence of other phosphorelay proteins. Spo0F and Spo0B was radiolabeled and purified away from his6-tagged phosphorelay proteins as described in Experimental procedures. The effects on Spo0F~P and Spo0B~P dephosphorylation were determined for Rap60 and RapB.
D. Quantitation of KinA autophosphorylation experiments. The amount of KinA~P remaining after treatment with Rap proteins for 30 min (black bars) and 60 min (gray bars) was normalized to untreated samples. Experiments were repeated in triplicate and percent standard error is shown. Asterisks represent P-values < 0.01.
Spo0F phosphorylation
To measure phosphotransfer from KinA to Spo0F, KinA was first radiolabeled with γ-32P-ATP. The reaction was terminated with the addition of cold ATP. Rap proteins were next added to the mixture, and the transfer of the radiolabeled phosphoryl group from KinA to Spo0F was monitored. To measure Spo0F~P dephosphorylation, KinA and Spo0F were first incubated with γ-32P-ATP to radiolabel Spo0F. The reaction was terminated with cold ATP. Rap60 was then added to the mixture and the disappearance of radiolabeled Spo0F~P was monitored.
Rap60 had a dramatic effect on the phosphorylation state of Spo0F (Fig. 6B). Synthetic Phr60 peptide antagonized Rap60 activity resulting in increased Spo0F~P compared to treatment with Rap60 alone (Fig. 6B). To rule out any potential effects of KinA, Spo0F~P dephosphorylation was monitored in the absence of KinA. Briefly, native Spo0F was first radiolabeled with γ-32P-ATP in the presence of his6-KinA. Next, his6-KinA was removed using Ni-affinity chromatography (see Experimental procedures). Purified his6-Rap60 significantly accelerated the rate of native Spo0F~P dephosphorylation compared with buffer alone, whereas Phr60 peptide inhibited the phosphatase activity of Rap60 (Fig. 6C). Consistent with previously published results, purified his6-RapB also dephosphorylates Spo0F~P (Fig. 6C and Jiang et al., 2000b). Thus, we conclude that Rap60 acts as a bona fide phosphatase of Spo0F~P, and Phr60 peptide antagonizes this phosphatase activity.
Phosphorylation of Spo0B and Spo0A
In addition to decreasing the levels of Spo0F~P, the presence of Rap proteins also decreased the amount of radiolabeled Spo0B~P and Spo0A~P (Fig. 6B). Phosphotransfer between Spo0F and Spo0B is highly reversible (Perego et al., 1994). Thus, a decrease in Spo0F~P, resulting from dephosphorylation by Rap60, could have repercussive effects on the entire phosphorelay system by acting as a molecular sink to switch the equilibrium and reverse the flow of phosphate through the phosphorelay proteins. Alternatively, it is possible that Rap60 directly affects the phosphorylation of Spo0B and Spo0A by blocking phosphotransfer or by acting as a phosphatase. To distinguish between these possibilities, we examined the effects of Rap60 on Spo0B~P dephosphorylation after removing all of the other phosphorelay proteins (see Experimental procedures). We chose Spo0B for our analysis because the effects of Rap60 were greater for Spo0B than Spo0A (Fig. 6B). Neither his6-tagged Rap60 nor RapB had an effect on the dephosphorylation of Spo0B~P (Fig. 6C). Similar results for RapA and RapB were observed by Perego et al. (1994). We conclude that Rap60 does not act as a phosphatase of Spo0B~P. The reduction in Spo0F~P by Rap proteins reverses the flow of phosphate through the phosphorelay system away from Spo0B and Spo0A.
KinA phosphorylation
Surprisingly, Rap60 inhibited KinA autophosphorylation, reducing the amount of radiolabeled KinA~P by 65% and 75% within 30 min and 60 min, respectively, compared with no treatment (Fig. 6B and D). Synthetic Phr60 peptide had no effect on Rap60 inhibition of KinA autophosphorylation (Fig. 6B and D). RapA and RapB had no effect on KinA phosphorylation compared with no treatment, the exception being the 60 min time point for RapA that was reduced ~30% (Fig. 6D and Perego et al., 1994). Because Rap60 acts as a phosphatase of Spo0F~P (Fig. 6B), we sought to determine whether Rap60 could also dephosphorylate KinA~P. The stability of the phosphoryl group on KinA~P was unaffected by any of the Rap proteins (Fig. 6B and Perego et al., 1994). We conclude that the decrease in KinA~P by Rap60 is due to inhibition of KinA autophosphorylation and not dephosphorylation of KinA~P .
Rap60 has no effect on ComA phosphorylation
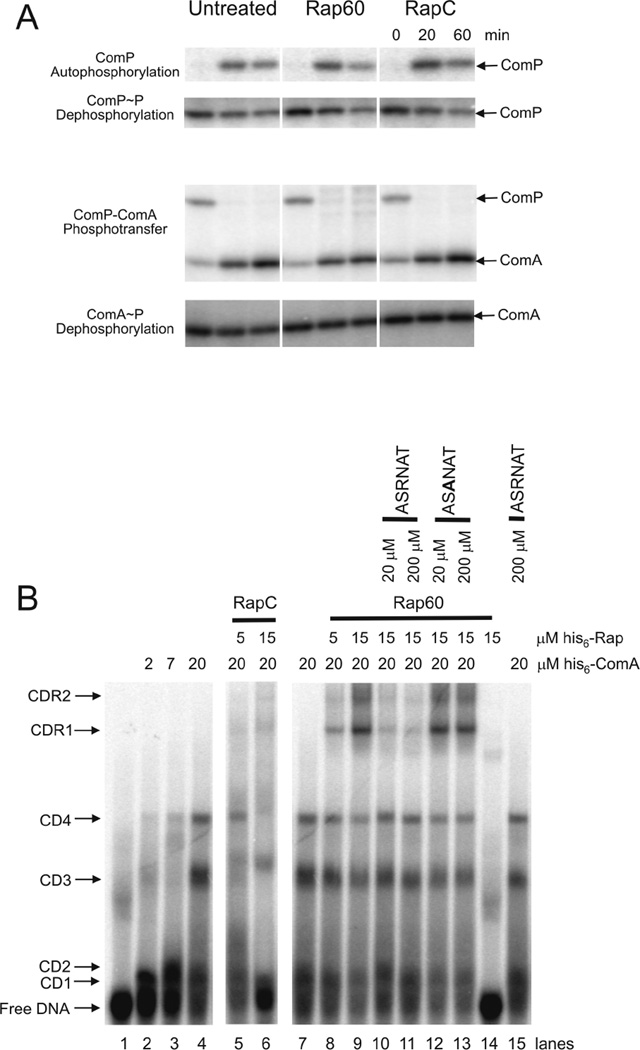
It is clear from the experiments described above that Rap proteins inhibit sporulation by modulating the phosphorylation state of the phosphorelay proteins. During competence development, the response regulator ComA is also activated by phosphorylation from ComP (Weinrauch et al., 1990). Because Rap60 influences the phosphorylation state of KinA in vitro, we sought to determine whether Rap60 was also modulating ComP and ComA phosphorylation. The cytoplasmic portion of ComP (his6-ComP(CD)) was purified and used to phosphorylate his6-ComA (see Experimental procedures). Rap60 had no effect on the phosphorylation of his6-ComP(CD) or his6-ComA, including ComP autophosphorylation, ComP~P dephosphorylation, phosphotransfer from ComP to ComA and ComA~P dephosphorylation (Fig. 7A). The ability of Rap60 to inhibit the autophosphorylation of a kinase appears to be specific to KinA as Rap60 had no effect on ComP phosphorylation.
Fig. 7.
Regulation of ComA activity by Rap60-Phr60.
A. Phosphorylation assays were performed using 3 µM his6-ComP(CD), 10 µM his6-Rap, 300 µM Phr60 hexapeptide and 5 µM his6-ComA, except for ComP autophosphorylation and ComP~P dephosphorylation experiments that used 5 µM his6-Comp(CD) and 5 µM his6-Rap proteins. Reactions were prepared as described in Experimental procedures. Aliquots were removed at the specified times and subjected to SDS-PAGE. Experiments were performed in triplicate and representative gels are shown.
B. Gel mobility shift assays were conducted with purified ComA and Rap proteins using a DNA template containing a minimal ComA binding sequence (tcaTTGCGGcatcCCG CAAgaaactTTGCGGtc). Purified his6-tagged ComA and Rap were incubated with 5 nM 32P-labeled double stranded DNA and 300 µM Phr hexapeptide, where appropriate, in reaction buffer (see Experimental procedures). Proteins were allowed to equilibrate at 24°C for 30 min prior to separation on 10% native acrylamide gels. The numbers to the left indicate different ComA-DNA (CD1–4) and ComA-DNA-Rap60 (CDR1–2) complexes. A representative gel is shown.
Rap60 inhibits ComA without interfering with ComA binding to DNA
The results described above are not surprising as Rap proteins that modulate competence development (RapC, F and H) have no effect on the phosphorylation state of ComA but regulate ComA activity by inhibiting ComA binding to DNA (Core and Perego, 2003; Bongiorni et al., 2005; Smits et al., 2007; Baker and Neiditch, 2011). In contrast, we found that Rap60 does not inhibit ComA binding to DNA. Instead, Rap60 forms a complex with ComA that has no effect on ComA binding to DNA (Fig. 7B). In previous work, we identified a minimal recognition sequence that is required for ComA binding to DNA and transcription activation of target genes (Griffith and Grossman, 2008). To determine the effects of Rap60 on ComA binding to DNA, gel mobility shift assays were performed using a minimal DNA template containing the consensus ComA binding sequence along with purified his6-ComA and his6-Rap60 (see Experimental procedures). The addition of purified his6-ComA (2–20 µM) to a fixed concentration of radiolabeled DNA (5 nM) resulted in the formation of four distinct his6-ComA-DNA binary complexes (CD1–4). The abrupt transitions to slower migrating complexes with increasing concentrations of ComA indicates to us that multiple molecules of his6-ComA probably bind cooperatively to DNA (Fig. 7B, lanes 2–4). However, the stoichiometry of ComA to DNA has not been determined.
Consistent with previously published results, ComA binding to DNA was inhibited with increasing concentrations of his6-RapC (5 µM and 15 µM) resulting in the disappearance of ComA–DNA complexes (CD1–4) and an increase in the amount of free DNA (Fig. 7B, lanes 5–6 and Core and Perego, 2003). In stark contrast to RapC, Rap60 had no effect on ComA binding to DNA. Instead, Rap60 forms two distinct ComA-DNA-Rap60 ternary complexes (CDR1–2) with increasing concentrations of Rap60 (Fig. 7B, lanes 8–9). The super-shifted complexes are a result of interactions between Rap60 and ComA, as purified his6-Rap60 had little effect on the migration of free DNA in the absence of his6-ComA (Fig. 7B, lane 14). It should be noted that in some instances, we observe very faint, slow migrating bands in the presence of RapC, ComA and DNA that migrate to the same position in the gel as the ComA–DNA–Rap60 ternary complexes (Fig. 7B, lanes 5–6). The intensity of these bands is far less than the intensity of the bands corresponding to the ternary complexes formed with Rap60 (Fig. 7B, lanes 8–9). Perhaps, to some small extent, RapC is able to form a higher order complex with ComA and DNA. If so, the significance of these complexes, if any, in regulating the activity of ComA is unknown at this time.
Phr60 hexapeptide (ASRNAT) antagonizes the activity of Rap60 in vivo (Fig. 5). The addition of Phr60 hexapeptide to the reaction mixture containing his6-ComA, radiolabeled DNA and his6-Rap60 absolved the two super-shifted ComA–DNA–Rap60 ternary complexes (CDR1–2) but had no effect on ComA binding to DNA (Fig. 7B, lanes 10–11). A mutant Phr60 peptide (ASANAT), containing an alanine substitution at the conserved arginine, failed to antagonize Rap60 activity in vivo (Fig. 5). Similarly, the mutant Phr60 peptide had no effect on the formation of ComA–DNA binary complexes or ComA–DNA–Rap60 ternary complexes (Fig. 7B, lanes 12–13). Wild-type Phr60 peptide had no effect on ComA binding to DNA (Fig. 7B, lane 15). Taken together, Rap60 is able to form a ternary complex with ComA and DNA that inhibits the activity of ComA. Phr60 peptide disrupts the ternary complex without perturbing ComA binding to DNA.
The DNA binding experiments, described above, were performed using nonphosphorylated ComA. For many response regulators, including ComA, phosphorylation stimulates DNA binding and transcription activation of target genes (Roggiani and Dubnau, 1993; Stock et al., 1995). Attempts were made to determine the effect of Rap60 on ComA~P-DNA using mobility shift assays. Unfortunately, the radiolabeled phosphoryl group present on ComA~P was readily hydrolyzed in the time required for gel electrophoresis, preventing us from accurately measuring Rap60 binding to ComA~P-DNA (data not shown). Because phosphorylation of ComA improves binding to DNA, rather than being obligate for binding (Roggiani and Dubnau, 1993; Griffith and Grossman, 2008), we believe our binding studies using nonphosphorylated ComA still reflects the relevant biology of Rap60.
Identification of amino acids important for Rap60 activity
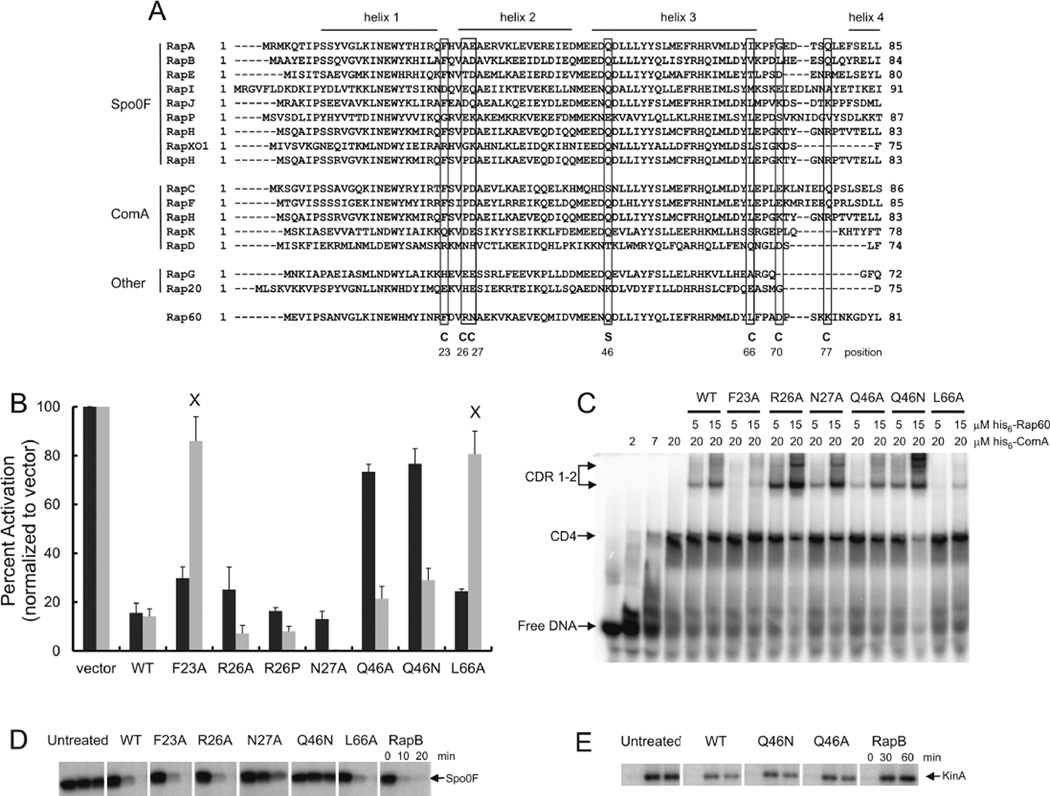
Structural and biochemical studies from the Neiditch laboratory provide insight into how Rap proteins interact with ComA and Spo0F to regulate their activities. Specifically, the RapH-Spo0F co-crystal structure revealed amino acids present in the α-helix 3 and the carboxyl terminal tetratricopeptide repeat sequences of RapH important for interacting with Spo0F (Parashar et al., 2011 and Fig. 8A). Similarly, the co-crystal structure of RapF in complex with the DNA binding domain of ComA identified several amino acids present in the amino terminus of RapF important for interacting with ComA (Baker and Neiditch, 2011 and Fig. 8A). Not surprisingly, these amino acids are highly conserved among Rap proteins that regulate genetic competence and sporulation (Singh et al., 2013 and Fig. 8A). To determine if Rap60 utilizes similar residues, single alanine substitutions were tested for their effects on the activities of Spo0A and ComA using PspoIIA-lacZ and PsrfA-lacZ fusions respectively. In addition, mutants of Rap60 were purified and tested for Spo0F~P phosphatase activity and binding to ComA.
Fig. 8.
Rap60 mutants differentially regulate Spo0F and ComA.
A. Alignment of the N-terminal region of Rap proteins from B. subtilis. Regions corresponding to the α-helices in RapH are indicated on top. Amino acids that comprise the ComA interface in the RapF-ComA crystal structure (Baker and Neiditch, 2011) are indicated with a ‘C’. The catalytic Gln47 of RapH, important for dephosphorylation of Spo0F~P, is shown with an ‘S’.
B. Cultures containing PsrfA-lacZ or PspoIIA-lacZ fusions along with mutations of Pspank-rap60 were grown in the appropriate medium with 0.1 mM IPTG. Aliquots were removed at the specified time and β-galactosidase activity was determined. The maximal activity of each culture (typically around OD600 ~1–2) was normalized to reporter strains containing empty vector. PspoIIA-lacZ (black bars) and PsrfA-lacZ (gray bars). Experiments were performed in triplicate, and the percent standard error is shown. The transcriptional effects of all the Rap60 mutants were statistically different from vector (P-value < 0.01) with the exception of Rap60(F23A) and Rap60(L66A) for srfA transcription (denoted by an ‘X’).
C. Gel mobility shift assay with DNA containing the ComA binding sequence, purified ComA and mutants of Rap60. Binding conditions are as described in Experimental procedures. ComA-DNA binary complexes (CD) and ComA-DNA-Rap ternary complexes (CDR).
D. Spo0F~P dephosphorylation by Rap60 mutants.
E. KinA autophosphorylation in the presence of Rap60(Q46N) and Rap60(Q46A).
A highly conserved glutamine, present in helix 3, constitutes the catalytic residue of RapH responsible for dephosphorylating Spo0F~P (Parashar et al., 2011 and Fig. 8A, marked as S). Not surprisingly, substitutions to alanine or asparagine at the equivalent position 46 of Rap60 abolished Spo0F~P phosphatase activity (Fig. 8D and data not shown) resulting in increased transcription of spoIIA (Fig. 8B, black bars). Interestingly, the catalytically defective phosphatase mutants still inhibited KinA autophosphorylation similar to wild-type Rap60 (Fig. 8E). This might explain why we observe a 20% decrease in β-galactosidase activity from cultures over-expressing Rap60(Q46A) or Rap60(Q46N) compared with vector (Fig. 8B, black bars). Inhibition of KinA autophosphorylation would result in less Spo0A~P in the cell and a decrease in transcription of early sporulation genes. Rap60(Q46N) inhibited ComA activity as judged by ternary complex formation (Fig. 8C) and the regulation of srfA transcription (Fig. 8B, gray bars). This suggests that the overall structure of Rap60 is not affected by these substitutions. Taken together, these data indicate that, like RapH, Rap60 utilizes two very distinct regions of the protein to regulate the activities of Spo0F and ComA. Moreover, different regions of Rap60 appear to be important for regulating the phosphorylation of Spo0F and KinA.
Six amino acids present in RapF comprise the ComA binding interface (Baker and Neiditch, 2011 and Fig. 8A, marked as C). Alanine substitutions at four out of the six equivalent positions were constructed in Rap60, and their effects on Rap60 activity were determined. Position D70 of Rap60 was omitted from our mutational analysis as a glutamic acid to alanine substitution at the equivalent position in RapF (E71A) had no effect on RapF binding to ComA (Baker and Neiditch, 2011). Similarly, Q78 of the RapF-ComA binding pocket was also omitted from our analysis as it is difficult to discern the equivalent amino acid in Rap60 due to gaps in the alignment and a general lack of conservation among different Rap proteins in this region of the protein (Fig. 8A). Instead, we focused our mutagenesis on positions 23, 26, 27 and 66 of Rap60.
The proline at position 27 of RapF (equivalent to R26 in Rap60) is particularly important because it redirects α-helix 2 to form the ComA binding pocket (Baker and Neiditch, 2011). Interestingly, a substitution to alanine or proline at the equivalent position 26 of Rap60 had little effect on Rap60 binding to ComA (Fig. 8C) and the regulation of srfA transcription (Fig. 8B, gray bars). An alanine substitution at position 27 of Rap60 significantly enhanced the activity of Rap60 resulting in a 40-fold reduction in β-galactosidase activity compared with wild-type Rap60 (Fig. 8B, gray bar). Although purified his6-Rap60(N27A) exhibited a modest increase in binding to ComA (Fig. 8C), the magnitude of binding is inconsistent with the 40-fold reduction in ComA activity observed in vivo. This discrepancy is likely attributed to partial activity of purified his6-Rap60(N27A) used in the in vitro studies due to extreme insolubility of the mutant protein during the purification process (data not shown). Consistent with this idea, purified his6-Rap60(N27A) also exhibited decreased Spo0F~P phosphatase activity compared with wild-type Rap60 (Fig. 8D), despite inhibiting spoIIA transcription to wild-type levels (Fig. 8B, black bars).
Single alanine substitutions at positions 23 (F to A) and 66 (L to A) of Rap60 disrupted ternary complex formation (Fig. 8C) resulting in increased β-galactosidase from PsrfA-lacZ, similar to strain KB22 containing an empty vector (Fig. 8B, gray bars annotated with an ‘X’). These amino acids likely make direct contacts with ComA without adversely affecting the overall structure of the protein as Rap60(F23A) and Rap60(L66A) both retain the Spo0F~P phosphatase activity (Fig. 8D) and inhibit spoIIA transcription (Fig. 8B, black bars). Taken together, Rap60 appears to utilize a small subset of the equivalent amino acids that comprise the RapF–ComA interface to regulate the activity of ComA.
Discussion
We identified novel functions for the plasmid-encoded Rap60-Phr60 quorum sensing regulatory pairs in controlling sporulation, cannibalism, biofilms and genetic competence. Similar to other Rap proteins that modulate sporulation, Rap60 controls Spo0A phosphorylation by acting as a phosphatase of Spo0F~P. In contrast, Rap60 plays a noncanonical role in modulating Spo0A phosphorylation by inhibiting the autophosphorylation of KinA. Rap60 also controls the activity of ComA by a novel mechanism. Specifically, Rap60 forms a complex with ComA and DNA that inhibits its activity as a transcriptional activator. We expand on existing models of ComA and Spo0A regulation by Rap proteins to include these novel functions of Rap60.
Rap regulation of genetic competence
In previous work, we demonstrated that ComA binds to a tripartite binding site, comprised of three recognition elements (RE1–3), located within the promoter of target genes. We propose that two dimers of ComA bind to DNA, with one dimer occupying RE1 and RE2 and a second dimer occupying RE3 and nonspecific sequence downstream. Protein– protein contacts between the two dimers of ComA probably help stabilize the complex, allowing for the recruitment of RNA polymerase to the promoter and transcription activation of target genes (Griffith and Grossman, 2008 and Fig. 9).
Fig. 9.
Model of ComA and Spo0A regulation by Rap proteins.
A. Two proposed mechanisms of ComA anti-activation by Rap proteins. In the absence of Rap proteins, ComA binds to a tripartite binding sequence located in the promoter region of target genes and recruits RNA polymerase to activate transcription of target genes. Raps C, F and H bind to the DNA binding domain of ComA, inducing a comformational change that disrupts ComA dimerization and interferes with ComA binding to DNA. In contrast, Rap60 forms a ternary complex with ComA and DNA that inhibits the activity of ComA without perturbing binding to DNA. We speculate that Rap60 binds to a region of ComA that prevents ComA access to the transcriptional machinery. Phr peptides antagonize their cognate Rap proteins allowing ComA-dependent transcription activation of target genes.
B. Rap proteins regulate Spo0A by modulating the sporulation phosphorelay system. Rap proteins, including Rap60, act as phosphatases of Spo0F~P. Dephosphorylation of Spo0F~P prevents the phosphorylation of Spo0A and reverses the flow of phosphate through the phosphorelay system resulting in decreased levels of Spo0B~P and Spo0A~P. Phr peptide antagonizes Rap phosphatase activity of Spo0F~P resulting in the accumulation of Spo0A~P and transcription activation of early sporulation genes. Rap60 has an additional, noncanonical role in regulating Spo0A activity by inhibiting the autophosphorylation of KinA. Phr60 has no effect on this activity by Rap60.
There exists at least two models for ComA anti-activation by Rap proteins based on previously published work from other laboratories, and the work described herein for Rap60 (Core and Perego, 2003; Bongiorni et al., 2005; Smits et al., 2007; Baker and Neiditch, 2011). Rap proteins (C, F, and H) bind to the DNA binding region of ComA, disrupt ComA dimerization and inhibit ComA binding to promoter DNA (Fig. 9). In contrast, Rap60 interacts with ComA and inhibits its activity without perturbing ComA binding to DNA (Figs 7B and 9). Results from the gel mobility shift assays support this mechanism of anti-activation by Rap60. Specifically, the formation of two distinct ComA–DNA–Rap60 complexes suggests that two molecules of Rap60 likely form a ternary complex with ComA and DNA (Figs 7B and 9). The twofold dyad symmetry and the location of the ComA binding site relative to the −35 element position ComA in close proximity to interact with RNA polymerase (Griffith and Grossman, 2008 and Fig. 9). The simplest model is that Rap60 binding to ComA sterically inhibits ComA access to the transcriptional machinery by blocking an overlapping region(s) of ComA important for interacting with RNA polymerase (Fig. 9). Alternatively, instead of limiting RNA polymerase access to ComA, Rap60 could inhibit RNA polymerase access to DNA. Irrespective of the mechanism, the ComA-Rap60-DNA ternary complex inhibits the activity of ComA (Figs 7B and 9). Phr60 peptide disrupts the ternary complex without perturbing ComA binding to DNA, allowing ComA to activate transcription of target genes (Figs 1–5, 7B and 9).
The two models of anti-activation make very different predictions about how Rap proteins interact with ComA. Previously published results indicate that RapF interacts with the DNA-binding domain of ComA (Baker and Neiditch, 2011). In contrast, Rap60 interacts with a different surface of ComA, one that is not required for binding to DNA (Fig. 7B). Amino acids P27 and D28 in RapF comprise part of the ComA binding pocket (Baker and Neiditch, 2011). The equivalent residues are not conserved in Rap60 (R26 and N27) and single alanine substitutions at these positions have no adverse effects on the activity of Rap60 (Fig. 8B – D). Moreover, two additional positions of the RapF–ComA interface (the equivalent amino acids D70 and K77 in Rap60) are also not conserved in Rap60, suggesting they are not likely involved in contacting ComA. In contrast, F23 and L66 of Rap60 are the only two equivalent amino acids of the RapF–ComA interface that are conserved in Rap60 and both are required for Rap60 regulation of ComA (Fig. 8B – D). Taken together, Rap60 utilizes a different region of the protein than RapF to regulate the activity of ComA. This provides additional support for an alternate mechanism of ComA anti-activation by Rap60.
Over-expression of RapD or RapK also inhibits srfA transcription (Auchtung et al., 2006; Ogura and Fugita, 2007). However, biochemical evidence is needed to determine if RapD and RapK act directly on ComA. Interestingly, none of the equivalent amino acids that comprise the RapF–ComA interface are conserved in RapK or RapD (Fig. 8A). Moreover, the amino acids equivalent to positions 23 and 66 of Rap60, which were shown to be important for Rap60 binding to ComA, are also not conserved in RapK or RapD. Assuming RapK and RapD directly regulate the activity of ComA, neither protein is likely to function by one of the two mechanisms of anti-activation described above (Fig. 9). Perhaps a third mechanism of anti-activation exists. Alternatively, RapD and RapK might function similarly to the plasmid-encoded RapP protein that modulates srfA transcription indirectly via the Spo0A-AbrB-SigH regulatory loop (Parashar et al., 2013).
Rap regulation of sporulation
All of the Rap proteins that modulate sporulation, including Rap60, influence the activity of Spo0A by a common mechanism of dephosphorylating Spo0F~P (Perego et al., 1994; Jiang et al., 2000a; Smits et al., 2007; Parashar et al., 2011; and Fig. 6). Rap-mediated dephosphorylation of Spo0F~P prevents Spo0A phosphorylation and transcription activation of Spo0A-dependent genes. Moreover, a decrease in the concentration of Spo0F~P reverses the flow of phosphate through the phosphorelay system leading to the dephosphorylation of preexisting Spo0A~P in vitro (Fig. 6B and Perego et al., 1994). Assuming this also occurs in vivo, it provides an effective mechanism to regulate the activity of Spo0A and inactivate the response even after it has already initiated, a role analogous to the phosphatase Spo0E (Perego, 2001 and Fig. 1).
Structural and biochemical analyses identified the RapH-Spo0F interface and the catalytic site of RapH (glutamine at position 47) required for dephosphorylation of Spo0F~P (Parashar et al., 2011 and Fig. 8A). These amino acids are highly conserved in Rap60 and other Rap proteins that regulate sporulation (Parashar et al., 2011; Singh et al., 2013). Not surprisingly, mutations in the active site of Rap60 (Q46A or Q46N) were defective for Spo0F~P phosphatase activity resulting in increased transcription of early sporulation genes (Fig. 8B, black bars). Rap60 appears to play a noncanonical role in modulating the activity of Spo0A by inhibiting KinA autophosphorylation (Fig. 6B and D). This activity is specific for KinA as Rap60 had no effect on the phosphorylation state of a second histidine kinase, ComP (Fig. 7A).
It is conceivable that inhibition of KinA autophosphorylation by Rap60 is an artifact of the in vitro system, possibly due to a contaminant that co-purified with Rap60. However, we do not favor this explanation, and results from two experiments support this noncanonical role of Rap60 in regulating KinA phosphorylation. First, mutants Rap60(Q46A) and Rap60(Q46N) are catalytically defective for Spo0F~P dephosphorylation (Fig. 8D and data not shown) but retain the ability to inhibit KinA autophosphorylation (Fig. 8E). Over-expression of these mutants inhibits transcription of spoIIA ~20% compared with empty vector (Fig. 8B, black bars). We presume this reduced Spo0A activity is due to a decrease in Spo0A~P resulting from inhibition of KinA autophosphorylation. Second, Phr60 peptide has no effect on KinA autophosphorylation by Rap60. Constitutive inhibition of KinA phosphorylation by Rap60 explains why Phr60 only partially suppressed the activity of Rap60 in vivo (Fig. 2B). Typically, rap-phr gene pairs are transcriptionally coupled and many phr genes have additional promoters (SigA- and SigH-dependent) that function to increase Phr peptide production and inactivate Rap proteins as the culture density increases (McQuade et al., 2001). We speculate that expression of Rap60 from the Pspank promoter is sufficient to saturate all of the Rap60 binding partners, e.g., Spo0F, ComA, KinA and other if they exist. Co-expression of Rap60 and Phr60 is expected to inhibit the SpooF~P phosphatase activity of Rap60 but have no effect on the inhibition of KinA autophosphorylation by Rap60 (Fig. 6B–D). This unchecked activity of Rap60 will suppress the levels of KinA~P in the cell causing a decrease in transcription of early sporulation genes (Fig. 2B). In addition, a decrease in KinA~P might also explain why over-expression of Rap60-Phr60 partially suppressed ComA-dependent transcription of srfA (Fig. 4B). Decreased Spo0A~P, caused by Rap60 inhibition of KinA phosphorylation, leads to decreased Phr production via the AbrB-SigH pathway resulting in enhanced Rap activity and a reduction in the transcription of ComA-dependent genes (Fig. 4B). Taken together, these results support a noncanonical role of Rap60 in regulating KinA phosphorylation. However, additional experiments are required to fully understand the regulation of KinA by Rap60.
The biological significance of the regulation of KinA phosphorylation by Rap60 is less pronounced in a wild-type background. Specifically, this activity is only observed when Rap60 is over-expressed from the Pspank promoter and is not present when Rap60 is expressed at physiological concentrations from plasmid pTA1060 (Figs 2 and 4). This makes sense given that unchecked regulation of Rap60 could prove deleterious to the cell, especially because many important biological processes are regulated by Spo0A. From a mechanistic standpoint, however, it will be interesting to determine how Rap60 inhibits KinA autophosphorylation and whether Rap60 influences the phosphorylation of other kinases involved in sporulation and biofilm formation including KinB-E.
Experimental procedures
Growth media
Liquid cultures of B. subtilis were grown in Luria Broth (LB), Difco nutrient broth sporulation medium (Harwood and Cutting, 1990), MSgg medium (Branda et al., 2001) or S7 defined minimal medium salts (Vasantha and Freese, 1980) containing 50 mM 4-morpholinepropanesulfonic acid instead of 100 mM (S750) and supplemented with 1% glucose, 0.1% glutamate, tryptophan (40 µg ml−1), phenylalanine (40 µg ml −1) and threonine (200 µg ml−1), where appropriate. B. subtilis was grown on solid medium plates containing Spizizen salts (Harwood and Cutting, 1990) supplemented with 1% glucose, 0.1% glutamate and amino acids, where appropriate. LB agar plates were used for routine cloning and growth of B. subtilis and Escherichia coli. The following concentrations of antibiotics were used: ampicillin (100 µg ml−1), neomycin (2.5 µg ml−1), tetracycline (8–10 µg ml−1), phleomycin (5 µg ml−1), chloramphenicol (5 µg ml−1), spectinomycin (100 µg ml−1), and erythromycin (0.5 µg ml−1) and lincomycin (12.5 µg ml−1) together to select for macrolidelincosamide-streptogramin B (MLS) resistance.
Plasmids, strains and alleles
Escherichia coli strains DH5α and AG1111 (a MC1061 derivative with F′(lacIq) lacZM15 Tn10) were used for routine cloning. B. subtilis strains in Table 1 were derived from the parental strain JH642 (trpC2 pheA1) (Perego et al., 1988) or strain DS2569, which is cured of the plasmid pBS32 and is a prototrophic derivative of strain NCIB3610 (Konkol et al., 2013).
Table 1.
Plasmids and strains used in this study.
| Plasmid or straina | Relevant genotype |
|---|---|
| Sporulation, competence, and colony architecture | |
| KB19 | amyE::Pspank-(empty vector) spc |
| KB26 | amyE::Pspank-rap60 spc |
| KB27 | amyE::Pspank-rap60-phr60 spc |
| KB28 | amyE::Pspank-phr60 spc |
| KG1780 | pTA1060 |
| KG1782 | pTA1060-Δphr60 |
| KG1781 | pTA1060-Δrap60Δphr60 |
| KG1897 | DS2569 pTA1060 |
| KG1899 | DS2569 pTA1060-Δphr60 |
| KG1898 | DS2569 pTA1060-Δrap60Δphr60 |
| KG37 | 3610 Δspo0A::cm |
| Sporulation and cannabilism gene promoter-lacZ fusions | |
| KB45 | lacA:PspoIIA-lacZ tet amyE:: Pspank-(empty vector) spc |
| KB46 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rap60 spc |
| KB47 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rap60-phr60 spc |
| KB48 | lacA::PspoIIA-lacZ tet amyE:: Pspank-phr60 spc |
| KB63 | lacA:PspoIIA-lacZ tet amyE:: Pspank-rapC spc |
| KB138 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rapB spc |
| KG1766 | amyE::PspoIIA-lacZ neo pTA1060 |
| KG1771 | amyE::PspoIIA-lacZ neo pTA1060-Δrap60Δphr60 |
| KG1772 | amyE::PspoIIA-lacZ neo pTA1060-Δphr60 |
| KG1911 | amyE::Pskf-lacZ spc pTA1060 |
| KG1912 | amyE::Pskf-lacZ spc pTA1060-Δphr60 |
| PH62 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rap60(F23A) spc |
| PH28 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rap60(R26A) spc |
| PH63 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rap60(R26P) spc |
| PH64 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rap60(N27A) spc |
| PH65 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rap60(Q46A) spc |
| PH66 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rap60(Q46N) spc |
| PH67 | lacA::PspoIIA-lacZ tet amyE:: Pspank-rap60(L66A) spc |
| Matrix gene promoter-lacZ fusions | DS2569 amyE::PepsA-lacZ cat pTA1060 |
| KG1928 | DS2569 amyE::PepsA-lacZ cat pTA1060-Δphr60 |
| KG1929 | DS2569 amyE::PtapA-lacZ cat pTA1060 |
| KG1930 | DS2569 amyE::PtapA-lacZ cat pTA1060-Δphr60 |
| KG1931 | DS2569 amyE::PabrB-lacZ spc pTA1060 |
| KG1903 | DS2569 amyE::PabrB-lacZ spc pTA1060-Δphr60 |
| KG1905 | |
| Competence gene promoter-lacZ fusions | |
| KB22 | thrC::PsrfA-lacZ erm amyE:: Pspank-(empty vector) spc |
| KB23 | thrC::PsrfA-lacZ erm amyE:: Pspank-rap60 spc |
| KB24 | thrC::PsrfA-lacZ erm amyE:: Pspank-rap60-phr60 spc |
| KB25 | thrC::PsrfA-lacZ erm amyE:: Pspank-phr60 spc |
| KB137 | thrC::PsrfA-lacZ erm amyE:: Pspank-rapC spc |
| KB53 | lacA::PsrfA-lacZ tet amyE::Pspank-(empty vector) spc |
| KB54 | lacA::PsrfA-lacZ tet amyE::Pspank-rap60 spc |
| KB58 | lacA::PsrfA-lacZ tet amyE::Pspank-rap60 spcΔspoOK::erm |
| KG1759 | amyE::PsrfA-lacZ neo pTA1060 |
| KG1764 | amyE::PsrfA-lacZ neo pTA1060-Δrap60Δphr60 |
| KG1765 | amyE::PsrfA-lacZ neo pTA1060-Δphr60 |
| KG1793 | amyE::PcomK-lacZ neo pTA1060 |
| KG1796 | amyE::PcomK-lacZ neo pTA1060 Δspo0A::cat |
| KG1795 | amyE::PcomK-lacZ neo pTA1060-Δphr60 |
| PH55 | lacA::PsrfA-lacZ tet amyE:: Pspank-rap60(F23A) spc |
| PH9 | lacA::PsrfA-lacZ tet amyE:: Pspank-rap60(R26A) spc |
| PH56 | lacA::PsrfA-lacZ tet amyE:: Pspank-rap60(R26P) spc |
| PH57 | lacA::PsrfA-lacZ tet amyE:: Pspank-rap60(N27A) spc |
| PH58 | lacA::PsrfA-lacZ tet amyE:: Pspank-rap60(Q46A) spc |
| PH59 | lacA::PsrfA-lacZ tet amyE:: Pspank-rap60(Q46N) spc |
| PH60 | lacA::PsrfA-lacZ tet amyE:: Pspank-rap60(L66A) spc |
| Protein purificationb | |
| KB98 | pBADAp18-Nhis6-KinA |
| KG1878 | pBADAp18-Nhis6-Spo0F |
| KG1879 | pBADCm33-Nhis6-Spo0B |
| KG1880 | pBADCm33-Nhis6-Spo0A |
| KB95 | pBADCm33-Nhis6-Rap60 |
| PH537 | pBADCm33-Nhis6-Rap60(F23A) |
| PH538 | pBADCm33-Nhis6-Rap60(R26A) |
| PH539 | pBADCm33-Nhis6-Rap60(R26P) |
| PH540 | pBADCm33-Nhis6-Rap60(N27A) |
| PH541 | pBADCm33-Nhis6-Rap60(Q46A) |
| PH542 | pBADCm33-Nhis6-Rap60(Q46N) |
| PH543 | pBADCm33-Nhis6-Rap60(L66A) |
| KG1881 | pBADCm33-Nhis6-RapA |
| KG1882 | pBADCm33-Nhis6-RapB |
| KG1883 | pBADAp18-Nhis6-RapC |
| KG1958 | pBADAp18-Nhis6-ComP(CD) |
| KG1959 | pBADAp18-Nhis6-ComA |
| KG1884 | pET21a–Nhis6-SUMO-Spo0F |
| KG1885 | pET21a–Nhis6-SUMO-Spo0B |
| KG1960 | pET21a–Nhis6-SUMO-ComA |
All B. subtilis strains are derivatives of JH642 and contain mutations in trpC and pheA (not shown) unless otherwise noted. Strain DS2569 is a pBS32 plasmid-cured derivative of the prototrophic strain NCIB3610.
Plasmids used for protein purification are in strain DH5α.
Promoter-lacZ fusions
Promoter fusions to lacZ were constructed and integrated into the B. subtilis chromosome. Primers were used to amplify, by the Polymerase Chain Reaction (PCR), fragments of the promoter regions of srfA (−434 to +30), spo0IIA (−562 to +30) and comK (−271 to +30). A termination codon (TAA) was engineered after the first 10 amino acids of each coding sequence. EcoRI and BamHI restriction enzyme recognition sites were incorporated into the forward and reverse primers respectively. PCR products were digested with EcoRI and BamHI restriction enzymes (NEB) and ligated with T4 DNA Ligase (NEB) into the following plasmids that were digested with the same two enzymes: pKS2 for integration into the amyE locus (Magnuson et al., 1994), pCAL215 for integration into the thrC locus (Auchtung et al., 2007) and pKG1877 for integration into the lacA locus (this study). Ligation reactions were transformed into strain DH5α and plated on LB solid medium with ampicillin. Clones were verified by DNA sequencing (UMASS Core Facility). Plasmid DNA was linearized by digestion with NcoI restriction enzyme, transformed into B. subtilis strain JH642 and plated on LB solid medium with neomycin (pKS2), MLS (pCAL215) or tetracycline (pKG1877).
Pspank-rap and phr
Constructs were engineered to express Rap60 and Phr60 from the inducible Pspank promoter and integrated into the B. subtilis chromosome at the amyE locus. Primers were used to amplify by PCR the gene encoding rap60, phr60 and rap60-phr60 from pTA1060 plasmid DNA. Each forward primer contains the sequence 5′-GCT TAGTGAAGCTTAAGGAGGTATACATATG-3′ with a HindIII restriction site underlined, an optimal ribosome binding sequence in bold, the initiation codon in italics and 15–17 bp of gene-specific sequence immediately downstream of the ATG beginning with amino acid 2 of each gene. The reverse primers have the sequence 5′-TGCTACGAGCATGCTTA-3′ with a SphI restriction site underlined, the termination codon in bold and 15–17 bp of gene-specific sequence preceding the termination codon. PCR products were digested with HindIII and SphI restriction enzymes and ligated into pDR110 (kind gift from D. Rudner), which was also digested with the same two restriction enzymes. Ligation reactions were transformed into strain DH5α and plated on LB solid medium with ampicillin. Correct clones were verified by DNA sequencing. Plasmid DNAwas linearized by digestion with NcoI restriction enzyme, transformed into B. subtilis strain JH642 and plated on LB solid medium with spectinomycin. Single amino acid substitutions were created in Pspank-rap60 using the Polymerase Chain Reaction Splicing by Overlap Extension (PCR SOE) (Horton et al., 1990), by the same procedure as described above.
Modified pTA1060
Plasmid pTA1060 DNA was isolated from B. amyloliquefaciens strain IFO3022 using the alkaline lysis method according to the manufacturer (Qiagen) with a single modification. Cell pellets were resuspended in P1 buffer with the addition of lysozyme (3 mg ml−1) and allowed to incubate for 20 min at 37°C. Several modifications were made to plasmid pTA1060 to make it more suitable to genetic manipulation. First, a medium copy pBR322 origin of replication and a gene encoding ampicillin resistance were cloned into pTA1060 for selection and plasmid maintenance in E. coli. Primers were used to amplify a 2.8 KB region of plasmid pBR322 DNA containing the origin of replication and the gene encoding ampicillin resistance. Unique restriction endonuclease sites were engineered into each primer flanking the pBR322 cassette. The PCR product and plasmid pTA1060 were digested with NcoI restriction enzyme and ligated with T4 ligase. The ligation mixture was transformed into strain DH5α and plated onto LB solid medium supplemented with ampicillin. Plasmid DNA was isolated and used in a second step to introduce an erythromycin resistance gene for selection of pTA1060 in B. subtilis. Primers were used to amplify a 1.2 KB fragment containing the erythromycin resistance gene from plasmid pHP13. The restriction enzyme sites XhoI and SpeI were engineered into the primers for directional cloning into the modified pTA1060 plasmid. An erythromycin resistant colony was selected, and the plasmid was confirmed by restriction digests. These modifications were introduced into the intergenic region of pTA1060 between orf7 and the mob gene flanked by transcriptional terminators to prevent read through transcription into surrounding plasmid genes. The modified plasmid was named pTA1060::erm but is referred to in the text simply as pTA1060.
Plasmid pTA1060-Δrap60-Δphr60 was created by digesting the modified pTA1060 plasmid with NaeI restriction enzyme. NaeI recognition sites are present in the middle of rap60 and ~250 bp downstream of the phr60 coding sequence. The fragment corresponding to pTA1060 was gel purified from the rap60-phr60 fragment and re-ligated with T4 ligase. Plasmid pTA1060-Δphr60 was created by introducing two tandem termination codons early in the coding sequence of phr60 by add-on PCR.
pBAD-his6-tagged proteins
Constructs to over-express N-terminal hexa-histidine fusion proteins were created by PCR amplification of the genes of interest from B. subtilis genomic DNA (Table 1). Rap60 was amplified from the modified pTA1060 plasmid. Each forward primer contains the sequence 5′-GCTTAGTGGGTACCAAGGAGATATACAT ATGcatcaccatcaccatcac-3′ with a KpnI restriction site underlined, an optimal ribosome binding sequence in bold, the initiation codon in italics, the his6-tag in lowercase and 15–17 bp of gene-specific sequence immediately downstream of the his6-tag beginning with amino acid 2 of each gene. The reverse primers contain the sequence 5′-TGCTACGAGCATGCTTA-3′ with a SphI restriction site underlined, the termination codon in bold, and 15–17 bp of gene-specific sequence preceding the termination codon. PCR products were digested with KpnI and SphI restriction enzymes and ligated into pBAD-Cm33 or pBAD-Ap18, which was also digested with the same two restriction enzymes. Ligations were transformed into strain DH5α and plated on LB solid medium with the appropriate antibiotic. Correct clones were confirmed by DNA sequencing. Amino acid substitutions were created in pBADCm33-Nhis6-Rap60 by PCR SOE, using the same procedure as described above.
pET-his6-SUMO-taggedproteins
Constructs to over-express Nhis6-SUMO fusion proteins were created by amplifying spo0F, spo0B, and comA from B. subtilis genomic DNA. Each forward primer contains the sequence 5′-GCTTAGTGACCGGTGGT-3′ where the AgeI restriction site is underlined, two codons corresponding to glycine are in bold (required for efficient cleavage by his6-Ulp1) and 15–17 bp of gene-specific sequence immediately downstream of the last glycine beginning with amino acid 2 of each gene. The downstream primer contains the sequence 5′-TGCTACGAGCGGCCGCTTA with a NotI restriction site underlined, the termination codon in bold and 15–17 bp of gene-specific sequence preceding the termination codon. PCR products were digested with AgeI and NotI restriction enzymes and ligated into pET-his6-SUMO (Lee et al., 2008) that was digested with the same two enzymes. Ligations were transformed into strain DH5α and plated on LB solid medium with ampicillin. The correct clones were confirmed by DNA sequencing.
Oligonucleotides and peptides
All oligonucleotides used in this study were synthesized and desalted by Integrated DNA Technologies. Sequences are available upon request. Peptides used in this study were synthesized using solid phase peptide synthesis and purified using reverse phase HPLC by AnaSpec. Peptide concentrations were determined by the manufacturer and found to be >95% pure for full-length protein using mass spectrometry (AnaSpec).
Computational analyses
Statistical analyses were performed using Microsoft Excel and the suite of statistical tools from http://www.in-silico.net. Single sample Student’s t-tests were performed, and a P-value of 0.01 was used as a cut-off for statistical significance. Alignments of the primary amino acid sequences of the Rap proteins were made using the ClustalW program from the Galaxy Project (http://www.usegalaxy.org). Standard default program settings were used.
Sporulation assays
Overnight cultures were inoculated into Difco nutrient broth sporulation medium (DSM) with 0.1 mM IPTG or 0.25 µg ml −1 erythromycin, where appropriate. Cultures were grown at 37°C with vigorous aeration for ~20 h after entry into stationary phase and heated to 80°C for 20 min. Serial dilutions were prepared with each culture using Spizizen’s salts (Harwood and Cutting, 1990), and 100 µl was spread on LB plates. The number of viable colonies was determined after incubation overnight at 37°C. The sporulation frequency was determined as the number of heat-resistant colonies per milliliter divided by the total number of colony forming units (before heat treatment) per milliliter and was normalized to wild-type strain JH642.
Colony morphology and pellicle formation
Overnight cultures were inoculated in LB at 30°C. A 10 µl aliquot was spotted onto MSgg plates that were poured fresh the day before and placed in a laminar flow hood for 20 min prior to inoculating with bacteria. Inoculated plates were left at 24°C for 3 days prior to being photographed with a Hamatsu camera using a UVP gel documentation system. To assess pellicle formation, 10 µl of fresh overnight cultures were added to 3 ml of liquid MSgg medium in 6 well microtiter plates (Corning). The microtiter plates were left undisturbed at 24°C for 3 days prior to being photographed as described above.
Competence assays
A two-step procedure was used to make cells competent for DNA uptake. A single colony from an LB plate was used to inoculate LB liquid medium containing 0.1 mM IPTG or 0.25 µg ml −1 erythromycin, where appropriate. Cultures were grown at 37°C with vigorous aeration until OD600 ~1, and diluted 1:20 into MD medium pH 7.5 (61 mM K2HPO4, 44 mM KH2PO4, 3.4 mM trisodium citrate, 2% glucose, 50 µg ml −1 tryptophan, 50 µg ml −1 phenylalanine, 11 µg ml −1 ammonium iron(III) citrate and 3 mM MgSO4). The cultures were grown at 37°C with constant aeration for 4 h. Genomic DNA (500 ng) from the prototrophic, undomesticated strain NCIB3610 was added to 100 µl of competent cell culture and incubated at 37°C for 45 min. Serial dilutions were prepared in Spizizen’s salts (Harwood and Cutting, 1990), and 100 µl was spread onto solid medium plates containing LB and plates containing Spizizen minimal medium with either tryptophan (40 µg ml−1) or phenyalanine (40 µg ml−1) to select for integration of the trp or phe alleles, respectively. The transformation frequency was determined as the number of transformants (Trp + or Phe + colonies) that grew on minimal medium plates per milliliter divided by the total number of colony forming units that grew on LB per milliliter and normalized to wild-type strain JH642. Similar transformation efficiencies were observed when selecting for Trp + or Phe + transformants (data not shown).
Growth conditions and β-galactosidase assays
Spo0A activity was monitored in reporter strains by inoculating single colonies from an LB plate into shaker flasks containing DSM medium to measure sporulation genes. To monitor ComA activity and biofilm genes, overnight cultures of reporter strains were grown as light lawns on Spizizen minimal medium plates and used to inoculate shaker flasks containing S750 minimal medium (ComA activity) or MSgg medium (biofilm genes) at a final OD600 ~0.02 as described in Griffith and Grossman (2008).
Liquid cultures were grown in shaker flasks at 37°C with vigorous aeration. One milliliter aliquots were removed and placed in a 2.2 ml 96-well polypropylene block, which was stored at −20°C until time to assay β-galactosidase activity. A second aliquot was taken to determine OD600. β-galactosidase assays were performed as previously described (Griffith and Grossman, 2008). Briefly, cells were prepared by thawing to room temperature, adding 20 µl of toluene to each well and permeabilizing cells directly in the block by vigorous pipetting up and down using a multi-channel pipettor. A0.2 ml aliquot of the permeabilized cell suspension was transferred to a second polypropylene block containing 0.8 ml Z-buffer (Miller, 1972).
A 100 µl aliquot of the cell suspension in Z-buffer was transferred to a microtiter plate. The assay was initiated with the addition of 20 µl freshly prepared Ortho-nitrophenyl-β-D-galactopyranoside (4 mg ml−1) and terminated with the addition of 40 µl 1M Na2CO3. Cell debris was pelleted in a microtiter plate by centrifugation at 3000 g for 10 min. The supernatant was transferred to a new plate using a multichannel pipettor. A420 was determined using a SpectraMax plate reader (Molecular Dynamics), and data analysis was performed using Microsoft Excel. β-galactosidase specific activity was calculated as follows: 1000 × [(ΔA420 min−1 ml−1)/OD600 of culture].
Protein expression and purification
Afresh overnight culture of strain DH5α containing the appropriate plasmid (Table 1) was diluted 1:100 into LB (Fisher) containing the appropriate antibiotic. Cultures were grown to OD600 ~0.5 at 37°C with vigorous aeration. L-arabinose (Sigma) was added to a final concentration of 0.2% to induce expression from pBAD plasmids and 1 mM IPTG was used to induce expression from pET vectors. Cultures over-expressing his6-tagged KinAand Rap60 were induced for 48 h at 25°C and 15°C respectively. All other cultures containing his6-tagged constructs were induced for 5–6 h at 37°C. Cells were harvested after induction by centrifugation at 5000 g for 10 min at 4°C and cell pellets were stored at −20°C until further use.
His6-tagged proteins were purified by standard Ni-NTA chromatography as previously described (Griffith and Grossman, 2008). The cell pellet from 1 l of culture was thawed on ice, resuspended in 10 ml sonication buffer (10 mM Tris pH 8, 0.3 M NaCl, 5% glycerol, 5 mM imidazole, 5 mM β-mercaptoethanol and 5 mM MgCl2), and cells were lysed by sonication using a Branson sonifier (10 cycles of 20 s on and 40 s off, at setting 6). The culture was cleared by centrifugation at 12 000 rpm for 30 min at 4°C, and the cell extract was passed over 2 ml of Ni-NTA (Qiagen). After 10 washes with 25 ml of sonication buffer, his6-tagged proteins were eluted from the column in 10 ml sonication buffer with increasing concentrations of imidazole (25 mM, 50 mM, 120 mM, 200 mM and 300 mM). For his6-Rap60 and his6-KinA, two additional washes in sonication buffer containing 25 mM and 50 mM imidazole were performed prior to elution in buffer containing 120 mM, 200 mM and 300 mM imidazole. Fractions were analyzed for purity by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie staining. Fractions with the greatest purity were pooled and dialyzed at 4°C against three buffer changes of 2 l dialysis buffer (10 mM Tris pH 8, 0.3 M NaCl, 5% glycerol, 10 mM β-mercaptoethanol and 5 mM MgCl2). Dialyzed proteins were concentrated to 5–15 mg ml−1 using a Centricon-10 (Amicon), with the exception of Nhis6-Rap60, which remained soluble ≤ 1 mg ml−1. His6-Ulp1 and his6-RapB were stored at −20°C in dialysis buffer. His6-tagged proteins Rap60, KinA and RapC were stored 4°C in dialysis buffer and remained stable for several months without significant loss of activity (data not shown). The remaining his6-tagged proteins were stored at −20°C in dialysis buffer containing 40% glycerol. Protein concentrations were determined by Bradford assay using bovine serum albumin as protein standard. Protein preparations were estimated to be > 95% pure as determined by SDS-PAGE followed by Coomassie staining (data not shown).
In vitro cleavage with Nhis6-Ulp1
Spo0F, Spo0B and ComA were purified as N-terminal his6-SUMO fusion proteins, similar to the methods described above. His6-Ulp1 protease was also purified by Ni-NTA as previously described (Lee et al., 2008). Purified his6-Ulp1 was added to the pooled his6-SUMO-tagged protein fractions in a 1:10 ratio (Ulp1 to his6-SUMO-tagged proteins) and dialyzed against three buffer changes of 1 l dialysis buffer at 4°C (see above). Ni-affinity chromatography was used to remove the his6-tagged proteins away from the native proteins. Briefly, the imidazole concentration of each sample was adjusted to 10 mM prior to incubation with 0.3 ml Ni-NTA (Qiagen) at 4°C for 1 h. The flow through contained only native Spo0F, Spo0B or ComA, and no his6-tagged proteins were present as shown by SDS-PAGE followed by Coomassie staining (data not shown). Native proteins were concentrated using a Centricon-10 device (Amicon). Protein concentrations were determined by Bradford assay.
Gel mobility shift assays
DNA corresponding to the minimal optimal ComA binding sequence was prepared by annealing two complementary oligonucleotides containing the following sequence: 5′-tcaTTGCGGcatcCCGCAAgaaactTTGCGGtc-3′, where the bases in uppercase represent Recognition Elements 1–3. DNA templates contain bases 5′-TCA preceding the ComA binding sequence and bases TC-3′ following it and are underlined. One of the oligonucleotides from each pair was labeled on its 5′ end using γ-32P-ATP (Perkin Elmer) and T4 polynucleotide kinase (NEB). The kinase reaction was terminated by incubation at 70°C for 20 min. A 1.3-fold molar excess of the complimentary oligonucleotide was added to the mixture and heated to 95°C for 5 min, followed by slow cooling to room temperature to facilitate annealing of the oligonucleotides. Duplex DNA was purified away from the unincorporated label using a G-25 Centrispin 10 column (Princeton Separations).
In vitro binding reactions contained 13 mM Tris pH 8, 50 mM 4-(2-Hydroxyethyl)piperazine-1-propanesulfonic acid (EPPS) pH 8.5, 20 mM MgCl2, 0.1 mM Ethylenediaminetet-raacetic acid (EDTA), 100 mM KCl, 3 mM Dithiothreitol (DTT) and 10% glycerol in a 20 µl final volume. Radiolabeled DNA (5 nM) was added to the binding reaction along with the appropriate amount of his6-tagged Rap and ComA, and 300 µM purified hexapeptide, where appropriate. Protein–DNA complexes were allowed to equilibrate at 24°C for 30 min, prior to the addition of 5 µl of 5X agarose gel loading dye. Samples were loaded into the wells of a 10% polyacrylamide gel containing 5% glycerol and electrophoresed into the gel at 300 V. Once the loading dye had entered the gel, the voltage was reduced to 120 V, and gels were run for 5–6 h at 4°C. Gels were dried and analyzed using a Typhoon PhosphorImager (Molecular Dynamics). The amount of free DNA and DNA in complex with ComA and Rap60 was determined using ImageQuant software (Molecular Dynamics) and Microsoft Excel.
In vitro phosphorylation assays
Sporulation phosphorylation assays were conducted using 3 µM his6-KinA, 10 µM his6-Rap and 5 µM each of his6-tagged or native Spo0F, Spo0B and Spo0A, where appropriate. Similar amounts of protein were used to measure ComA and ComP phosphorylation. Specifically, 3 µM his6-ComP(CD), 10 µM his6-Rap and 5 µM his6-ComA was used to monitor ComA dephosphorylation and phosphotransfer from ComP to ComA. Atotal of 5 µM of his6-tagged kinase and 5 µM his6-Rap was used to monitor KinA and ComP(CD) autophosphorylation and dephosphorylation. The following reaction conditions were used: 13 mM Tris pH 8, 50 mM EPPS pH 8.5, 20 mM MgCl2, 0.1 mM EDTA, 100 mM KCl, 3 mM DTT, 10% glycerol, 0.5 mM Adenosine triphosphate (ATP), and 5 µCi γ-32P-ATP. Reactions were assembled and allowed to equilibrate for 1 h at 24°C, prior to termination with the addition of a final concentration of 10 mM ATP to the reaction mixture. Alternatively, reactions were terminated by removing all ATP from the reaction mixture using a G-25 Centrispin 10 column (Princeton Separations). Similar results were obtained with both methods. However, a ~50% loss of Spo0F and his6-Spo0A occurred with the G-25 columns (data not shown). To compensate for the poor yield from the G-25 columns, the concentration of his6-Rap proteins was adjusted in the reaction mixture to maintain a 1:2 molar ratio of phosphorelay protein to his6-Rap. Aliquots were removed at the specified times and subjected to SDS-PAGE. Gels were dried and analyzed using a Typhoon PhosphorImager (Molecular Dynamics). The amount of radiolabeled protein was quantitated using ImageQuant software (Molecular Dynamics) and Microsoft Excel.
Each step of the phosphorelay was monitored for phosphotransfer and dephosphorylation. As an example, to monitor phosphotransfer from Spo0B to Spo0A, Spo0B was first phosphorylated in the presence of KinA, Spo0F and γ-32P-ATP at 24°C for 1 h. The reaction was terminated as described above using a large excess of cold ATP or by removing ATP using a G-25 column. Spo0A was then added to the mixture, and the transfer of radiolabeled phosphate from Spo0B~P to Spo0A was monitored in the presence and absence of Rap proteins. To monitor dephosphorylation of Spo0A~P, Spo0A was first radiolabeled in the presence of KinA, Spo0F, Spo0B and γ-32P-ATP at 24°C for 1 h. The reaction was terminated, as described above, and the disappearance of radiolabeled Spo0A~P was monitored in the presence and absence of Rap proteins. Similar order of addition was used to monitor phosphorylation of KinA, Spo0F, Spo0B, Spo0A, ComA and ComP(CD).
Dephosphorylation of native Spo0F~P and Spo0B~P
To determine if Rap proteins act as phosphatases of Spo0F and Spo0B in the absence of other phosphorelay proteins, native Spo0F or Spo0B was first radiolabeled with γ-32P-ATP in the presence of his6-tagged phophorelay proteins for 1 hr at 24°C. The his6-tagged proteins were purified away from the native proteins using Ni-NTA chromatography. Briefly, 20 mM imidazole was added to the reaction mixture prior to incubation with 100 µl Ni-NTA (Qiagen). The tube was gently agitated for 30 s. Native Spo0F and Spo0B were each recovered after centrifugation at 500 rpm for 30 s to pellet the his6-tagged proteins bound to the Ni-NTA resin. Approximately 50% of native Spo0B and 10% of native Spo0F was recovered from the chromatography step, and no his6-tagged proteins were recovered as shown by SDS-PAGE followed by autoradiography (data not shown). The concentration of his6-Rap was adjusted to maintain a 1:2 molar ratio of Spo0F or Spo0B to his6-tagged Rap protein. Aliquots were removed at the specified times and subjected to SDS-PAGE. Gels were analyzed as described above.
Acknowledgements
We would like to thank Drs. Peter Chien and Steve Sandler for stimulating discussions and for critical reading of this manuscript and Drs. Richard Losick and Daniel Kearns for sharing strains. This work was supported by a grant from the Healey Endowment Foundation awarded to K.L.G., a summer fellowship from the Howard Hughes Medical Institute awarded to K.M.B., and funding from the Microbiology Department and the College of Natural Sciences at the University of Massachusetts-Amherst.
References
- Auchtung JM, Grossman AD. Extracellular peptide signaling and quorum responses in development, self-recognition, and horizontal gene transfer in Bacillus subtilis. In: Winans SC, Bassler BL, editors. Chemical Communication Among Microbes. Washington, DC: ASM Press; 2008. pp. 13–30. [Google Scholar]
- Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci USA. 2005;102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Grossman AD. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol. 2006;188:5273–5285. doi: 10.1128/JB.00300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Garrison KL, Grossman AD. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis . Mol Microbiol. 2007;64:1515–1528. doi: 10.1111/j.1365-2958.2007.05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MD, Neiditch MB. Structural basis of response regulator inhibition by a bacterial anti-activator protein. PLoS Biol. 2011;9:1–15. doi: 10.1371/journal.pbio.1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiorni C, Ishikawa S, Stephenson S, Ogasawara N, Perego M. Syngeristic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. 2005;187:4353–4361. doi: 10.1128/JB.187.13.4353-4361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis . PNAS. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Dervyn JE, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis . J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis . Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- Chung JD, Stephanopoulos G, Ireton K, Grossman AD. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J Bacteriol. 1994;176:1977–1984. doi: 10.1128/jb.176.7.1977-1984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comella N, Grossman AD. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum sensing transcription factor ComA in Bacillus subtilis. Mol Micriobiol. 2005;57:1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- Core L, Perego M. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis . Mol Microbiol. 2003;49:1509–1522. doi: 10.1046/j.1365-2958.2003.03659.x. [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis . Proc Natl Acad Sci USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E, Henner DJ, Perego M, Hoch JA. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol. 1988;170:289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugita M, Gonzalez-Pastor JE, Losick R. High- and Low-threshold genes in the Spo0A regulon of Bacillus subtilis . J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garti-Levi S, Eswara A, Smith Y, Fujita M, Ben-Yehuda S. Novel modulators controlling entry into sporulation in Bacillus subtilis . J Bacteriol. 2013;195:1475–1483. doi: 10.1128/JB.02160-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Griffith KL, Grossman AD. A degenerate tripartite DNA binding site required for activation of ComA-dependent quorum response gene expression in Bacillus subtilis . J Mol Biol. 2008;381:261–275. doi: 10.1016/j.jmb.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis . Mol Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. Identification ofAbrB-regulated genes involved in biofilm formation by Bacillus subtilis . Mol Microbiol. 2004;52:847–860. doi: 10.1111/j.1365-2958.2004.04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Chichester, England: John Wiley & Sons; 1990. [Google Scholar]
- Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. Microbiol Rev. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JA. Bacillus subtilis and other gran-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: ASM Press; 1991. spo0A genes, the phosphorelay, and the initiation of sporulation; pp. 747–755. [Google Scholar]
- Hoch JA. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis . Ann Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Jiang M, Grau R, Perego M. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis . J Bacteriol. 2000a;182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis . Mol Microbiol. 2000b;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis . Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- Koetje EJ, Hajdo-Milasinovic A, Kiewiet R, Bron S, Tjalsma H. A plasmid-borne Rap-Phr system of Bacillus subtilis can mediate cell-density controlled production of extracellular proteases. Microbiology. 2003;149:19–28. doi: 10.1099/mic.0.25737-0. [DOI] [PubMed] [Google Scholar]
- Konkol MA, Blair KM, Kearns DB. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis . J Bacteriol. 2013;195:4085–4093. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanigan-Gerdes S, Dooley AN, Faull KF, Lazazzera BA. Identification of subtilisin, Epr, and Vpr as enzymes that produce CSF, an extracellular signaling peptide of Bacillus subtilis . Mol Microbiol. 2007;65:1321–1333. doi: 10.1111/j.1365-2958.2007.05869.x. [DOI] [PubMed] [Google Scholar]
- Lazazzera BA, Kurtser IG, McQuade RS, Grossman AD. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis . J Bacteriol. 1999;181:5193–5200. doi: 10.1128/jb.181.17.5193-5200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDeaux JR, Solomon JM, Grossman AD. Analysis of non-polar deletion mutations in the genes of the spo0K (opp) operon of Bacillus subtilis . FEMS Microbiol Lett. 1997;153:63–69. doi: 10.1111/j.1574-6968.1997.tb10464.x. [DOI] [PubMed] [Google Scholar]
- Lee CD, Sun HC, Hu SM, Chiu CF, Homhaun A, Liang SM, et al. An improved SUMO fusion protein system for effective production of native proteins. Protein Sci. 2008;17:1241–1248. doi: 10.1110/ps.035188.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis . Mol Microbiol. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Dubnau DA. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade RS, Comella N, Grossman AD. Control of a family of phosphatase regulatory genes (phr) by the alternate sigma factor sigma-H of Bacillus subtilis . J Bacteriol. 2001;183:4905–4909. doi: 10.1128/JB.183.16.4905-4909.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis . Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- Meijer WJ, Wisman GB, Terpstra P, Thorsted PB, Thomas CM, Holsappel S, et al. Rolling-circle plasmids from Bacillus subtilis: complete nucleotide sequences and analyses of genes of pTA1015, pTA1040, pTA1050, and pTA1060, and comparisons with related plasmids from Gram-positive bacteria. FEMS Microbiol Rev. 1998;21:337–368. doi: 10.1111/j.1574-6976.1998.tb00357.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1972. [Google Scholar]
- Mirouze N, Parashar V, Baker MD, Dubnau DA, Neiditch MB. An atypical Phr peptide regulates the developmental switch protein RapH. pJ. Bacteriology. 2011;193:6197–6206. doi: 10.1128/JB.05860-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze N, Desai Y, Raj A, Dubnau DA. Spo0A~P imposes a temporal gat for the bimodal expression of competence in Bacillussubtilis . PLoS Genet. 2012;8:1–15. doi: 10.1371/journal.pgen.1002586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. The Spo0A regulon of Bacillus subtilis . Mol Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- Ogura M, Fugita M. Bacillus subtilis rapD, a direct target of transcription repression by RghR, negatively regulates srfA expression. FEMS Microbiol Lett. 2007;268:73–80. doi: 10.1111/j.1574-6968.2006.00559.x. [DOI] [PubMed] [Google Scholar]
- Ogura M, Yamaguchi H, Yoshida K, Fujita Y, Tanaka T. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B.subtilis two-component regulatory systems. Nucleic Acids Res. 2001;29:3804–3813. doi: 10.1093/nar/29.18.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar V, Mirouze N, Dubnau DA, Neiditch MB. Structural basis of response regulator dephosphorylation by Rap phosphatases. PLoS Biol. 2011;9:1–18. doi: 10.1371/journal.pbio.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar V, Konkol MA, Kearns DB, Neiditch MB. Aplasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. J Bacteriol. 2013;195:2437–2448. doi: 10.1128/JB.02030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis . Mol Microbiol. 2001;42:133–143. doi: 10.1046/j.1365-2958.2001.02611.x. [DOI] [PubMed] [Google Scholar]
- Perego M, Spiegelman GB, Hoch JA. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis . Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Perego M, Hanstein C, Welsh KM, Djavakhishvili T, Glasser P, Hoch JA. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis . Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis . Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Pottathil M, Lazazzera BA. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis . Front Biosci. 2003;8:32–45. doi: 10.2741/913. [DOI] [PubMed] [Google Scholar]
- Predich M, Nair G, Smith I. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing σH . J Bacteriol. 1992;174:2771–2778. doi: 10.1128/jb.174.9.2771-2778.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- Roggiani M, Dubnau D. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA . J Bacteriol. 1993;175:3182–3187. doi: 10.1128/jb.175.10.3182-3187.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D, Vlamakis H, Losick R, Kolter R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol. 2011;80:1155–1168. doi: 10.1111/j.1365-2958.2011.07653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Ramachandran G, Ramos-Ruiz R, Peiro-Pastor R, Abia D, Wu LJ, Meijer WJJ. Mobility of the native Bacillus subtilis conjugative plasmid pLS20 is regulated by intercellular signaling. PLoS Genet. 2013;9:1–14. doi: 10.1371/journal.pgen.1003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Eschevins CC, Susanna KA, Bron S, Kuipers OP, Hamoen LW. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol. 2005;56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- Smits WK, Bongiorni C, Veening JW, Hamoen LW, Kuipers OP, Perego M. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis . Mol Microbiol. 2007;65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- Stephenson S, Mueller C, Jiang M, Perego M. Molecular analysis of Phr peptide processing in Bacillus subtilis . J Bacteriol. 2003;185:4861–4871. doi: 10.1128/JB.185.16.4861-4871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JB, Surette MG, Levit M, Park P, editors. Two-Component Signal Transduction Systems: Structure-Function Relationships and Mechanisms of Catalysis. Washington, DC: ASM Press; 1995. [Google Scholar]
- Stover AG, Driks A. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J Bacteriol. 1999;181:1664–1672. doi: 10.1128/jb.181.5.1664-1672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch MA, Webb V, Spiegelman G, Hoch JA. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci USA. 1990;87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantha N, Freese E. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J Bacteriol. 1980;144:1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Weinrauch Y, Penchev R, Dubnau E, Smith I, Dubnau D. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 1990;4:860–872. doi: 10.1101/gad.4.5.860. [DOI] [PubMed] [Google Scholar]
- Winans SC, Bassler BL. Chemical Communication among Bacteria. Washington, D.C: ASM Press; 2008. [Google Scholar]