Abstract
Movement of transposons causes insertions, deletions, and chromosomal rearrangements potentially leading to premature lethality in Drosophila melanogaster. To repress these elements and combat genomic instability, eukaryotes have evolved several small RNA-mediated defense mechanisms. Specifically, in Drosophila somatic cells, endogenous small interfering (esi)RNAs suppress retrotransposon mobility. EsiRNAs are produced by Dicer-2 processing of double-stranded RNA precursors, yet the origins of these precursors are unknown. We show that most transposon families are transcribed in both the sense (S) and antisense (AS) direction in Dmel-2 cells. LTR retrotransposons Dm297, mdg1, and blood, and non-LTR retrotransposons juan and jockey transcripts, are generated from intraelement transcription start sites with canonical RNA polymerase II promoters. We also determined that retrotransposon antisense transcripts are less polyadenylated than sense. RNA-seq and small RNA-seq revealed that Dicer-2 RNA interference (RNAi) depletion causes a decrease in the number of esiRNAs mapping to retrotransposons and an increase in expression of both S and AS retrotransposon transcripts. These data support a model in which double-stranded RNA precursors are derived from convergent transcription and processed by Dicer-2 into esiRNAs that silence both sense and antisense retrotransposon transcripts. Reduction of sense retrotransposon transcripts potentially lowers element-specific protein levels to prevent transposition. This mechanism preserves genomic integrity and is especially important for Drosophila fitness because mobile genetic elements are highly active.
Keywords: antisense, double-stranded RNA, Drosophila, convergent transcription, Dicer-2
MOBILE genetic elements are one source of genetic alterations that drive evolution, but can also lead to catastrophic genomic instability. Thus, maintaining an appropriate balance between the potential harm and benefit of transposons (Tns) is vital. If active Tns are not adequately controlled by their hosts, mutations produced by their movement can be detrimental (Lee and Marx 2013). Specifically in Drosophila, genetic rearrangements that cause hybrid digenesis syndrome (Kidwell et al. 1977; Picard et al. 1978) are linked to transposon movement (Bingham et al. 1982; Rubin et al. 1982).
Since the discovery of Tns by Barbara McClintock more than 60 years ago (McClintock 1950), researchers have elucidated key mechanisms describing how Tns incorporate into genomes and how hosts combat these potentially toxic genomic perturbations. However, many aspects of Tn biology remain elusive. While ∼44% of the human genome is composed of Tns (Cordaux and Batzer 2009), there is little diversity in active transposons (Mills et al. 2007); only autonomous LINE-1 and nonautonomous Alu and SVA retrotransposons are currently mobile (Brouha et al. 2003; Cordaux and Batzer 2009; Deininger 2011). While the Drosophila genome is only ∼22% transposons, many (∼30%) of these elements are full length and thought to be active (Kaminker et al. 2002; Lerat et al. 2003; Kofler et al. 2015). Having active transposons from all three major classes of mobile elements to investigate offers a unique opportunity to understand silencing mechanisms in eukaryotic organisms.
Tns are defined by their approach to mobility. Terminal inverted repeat (TIR) Tns encode a Transposase that binds Tn inverted repeats (in most cases), creates double-strand breaks at the ends of the Tn, and integrates the Tn into a new genomic location. This mechanism can create genomic rearrangements such as insertions, deletions, and inversions. Unlike TIR Tns, retrotransposons (retroTns) include an RNA intermediate in their movement mechanism and therefore encode a reverse transcriptase (RT). RetroTns are divided into long terminal repeat (LTR) and non-LTR retroTns. LTR retroTns are similar to retroviruses and contain several hundred nucleotide terminal repeats at both the 5′ and 3′ ends (Figure 1A). While some Drosophila LTR retroTns have gag and env genes homologous to retroviruses, others have more divergent ORFs that function in retroTn mobility (Figure 1). Non-LTR retroTns lack these terminal repeats and sequences homologous to the env gene (Figure 2A), but have conserved RTs (Figure 2). Both LTR and non-LTR retroTns often have an internal promoter located in the 5′ untranslated region (UTR) and a 3′ UTR containing a polyadenylation signal (Gogvadze and Buzdin 2009 and this work). The initial transposition step for all retroTns is RNA polymerase II (RNAPII)-dependent transcription of the entire element followed by translation of each independent ORF in different reading frames from this single, polygenic messenger RNA (mRNA).
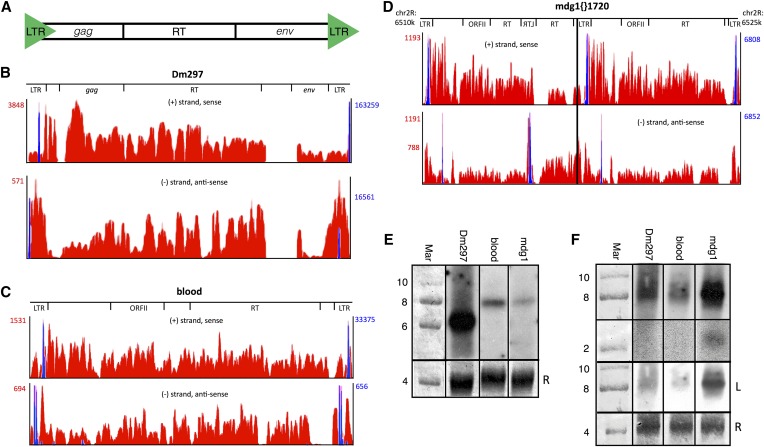
Figure 1.
LTR retroTns Dm297, blood, and mdg1{}1720 produce AS transcripts from intraelement tss in or near the LTRs. (A) Schematic of Drosophila LTR retroTns. (B–D) Bedgraphs representing S (top) and AS (bottom) nonunique RNA-seq reads mapping to each LTR retroTn are shown in red. Peak reads per million (RPM) are listed to the left (red numbers). For mdg1, two AS RPM values are listed; the top is the RPM for mdg1{}1720 and the bottom is the RPM for only the downstream canonical mdg1 element (right of the black line). Only the chromosome location of mdg1{}1720 is shown as Dm297 and blood bedgraphs are representative examples. Relative locations of specific ORFs are shown above the bedgraphs. Nonunique small-capped RNA-seq reads representing tss are overlaid in blue and RPM values are listed to the right (blue numbers). (E) Representative Northern blots of S LTR retroTn transcripts. The probe used for each blot is indicated above. The first lane is methylene blue-stained RNA marker; the sizes of bands are shown to the left of the blots. Methylene blue-stained 28S rRNA is used as a loading control (bottom) and is marked with an “R.” (F) Representative Northern blots of AS LTR retroTn transcripts. The top two panels are from the same longer exposure film while the third panel (“L”) is a lighter exposure. Other details are as in E.
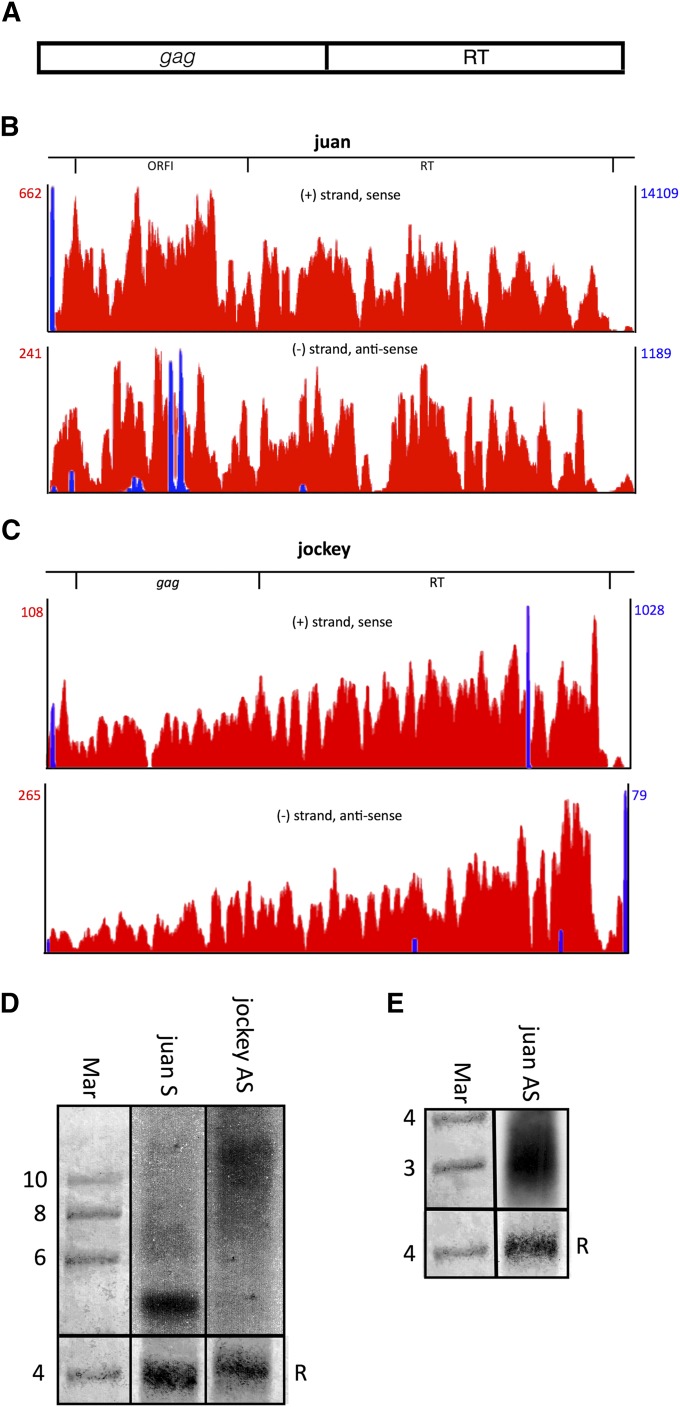
Figure 2.
Non-LTR retroTns juan and jockey produce AS transcripts from intraelement tss. (A) Schematic of non-LTR retroTns in Drosophila. (B and C) Bedgraphs representing S (top) and AS (bottom) nonunique RNA-seq reads mapping to each non-LTR retroTn are shown in red. Other details are as described in Figure 1. (D and E) Representative Northern blots of juan S and jockey AS (D), and juan AS (E) transcripts are shown. Details are as in Figure 1E.
Eukaryotic cells have evolved several noncoding RNA-mediated mechanisms to control further genomic spread of retroTns. In humans, mobility of the LINE-1 (L1) retroTn is regulated by both canonical RNA interference (RNAi) (Yang and Kazazian 2006) and endogenous small interfering (esi)RNA-mediated chromatin modifications (Chen et al. 2012). Similarly, in Drosophila, two distinct RNAi-like processes for silencing Tns have been elucidated. In the germline, the Piwi-interacting RNA (piRNA) pathway generates small RNAs that suppress Tns by inducing heterochromatin formation (Vagin et al. 2006; Aravin et al. 2007; Brennecke et al. 2007; Sentmanat and Elgin 2012; Le Thomas et al. 2013). In somatic cells, esiRNAs silence retroTns via a Dicer 2 (Dcr-2)/Argonaute 2 (Ago2)-dependent mechanism (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Saito and Siomi 2010; Xie et al. 2013). Global analysis of small RNA libraries generated from embryo-derived Drosophila somatic cells (S2) (Schneider 1972) showed that 86% of esiRNAs mapped to Tns; esiRNAs mapping to LTR retroTns were highly enriched (Ghildiyal et al. 2008). Dcr-2 is required for generation of esiRNAs (Czech et al. 2008; Okamura et al. 2008a,b) and retroTn expression increases following RNAi depletion of Dcr-2 (Ghildiyal et al. 2008; Marques et al. 2010).
The production of esiRNAs by Dcr-2 requires a double-stranded RNA (dsRNA) precursor (Tomari et al. 2007; Ghildiyal et al. 2008; Marques et al. 2010). While dsRNAs generated by hybridization of natural antisense transcripts and their sense counterparts are substrates for Dcr-2 in Drosophila (Czech et al. 2008; Okamura et al. 2008a), retroTn dsRNA precursors have not been systematically investigated. As Drosophila does not encode an RNA-dependent RNA polymerase to generate a complementary strand, the origin of the antisense (AS) transcript necessary to form the dsRNA retroTn precursor is unknown. Here, we provide evidence that both non-LTR and LTR retroTns produce sense (S) and AS transcripts from intraelement transcription start sites (tss) with canonical Drosophila promoters. We then use a novel polyA+/− fractionation followed by strand-specific RT-qPCR technique to show that most S and AS retroTn transcripts are not enriched for polyadenylation. Finally, increases in AS retroTn transcript levels in Dmel-2 cells RNAi depleted of Dcr-2 indicate that AS and S transcripts are substrates for Dcr-2.
Materials and Methods
Large and small RNA preparation/PolyA+ RNA selection
Total RNA from 8 × 106 Drosophila Dmel-2 tissue culture cells was isolated using QIAzol Lysis Reagent (Qiagen). Total RNA was fractionated into large (>200 nt) and small (<200 nt) fractions using RNeasy Mini spin columns and RNeasy MinElute spin columns, respectively (Qiagen). DNA was removed from the large fraction by on-column DNase digestion (Qiagen). Fractionation and DNA removal were verified by RT-qPCR. RNA integrity and size fractionation were confirmed using small RNA and RNA 6000 Pico Bioanalyzer chips (Agilent). Total RNA was then fractionated into polyA+ and polyA− fractions using the MicroPoly(A) Purist Kit (Ambion AM1919). Fractionation was verified by RT-qPCR.
Ribosomal RNA depletion
The 28S, 18S, and 5S ribosomal RNAs (rRNAs) were depleted from 5 µg of each large RNA fraction using the Ribo-Zero Magnetic Kit (Epicentre). While this kit was designed for human/mouse/rat, it performs adequately for Drosophila. rRNA depletion was confirmed by RT-qPCR and validated using RNA 6000 Pico Bioanalyzer chips (Agilent).
The 2S rRNA was depleted from the small RNA fraction according to Seitz et al. (2008) with the following modifications: 0.1 nM 2S rRNA complementary oligo was bound to 500 µg streptavidin beads in 1 ml 0.5× SSC for 1 hr at 4°. The beads were then washed five times in 0.5× SSC followed by a 5-min incubation at 65° to remove secondary structure. A total of 2 µg of the small RNA fraction was diluted to 12.5 ng/µl and 160 µl was added to the bead slurry. The remaining steps of the protocol were as described (Seitz et al. 2008). Following rRNA depletion from both small and large RNA fractions, RNA integrity, and rRNA depletion were validated on a Bioanalyzer.
Library preparation/next-generation sequencing
RNA-seq libraries were prepared in triplicate from 35 ng of the rRNA-depleted large RNA fraction using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB). Small RNA sequencing (smRNA-seq) libraries were prepared in triplicate from ∼475 ng of the 2S rRNA-depleted small RNA fraction using the NEBNext Small RNA Library Prep Set for Illumina (NEB). Each small interfering RNA (siRNA)- and RNA-seq library was amplified with a primer having a unique barcode. The appropriate size of each library was validated on a Bioanalyzer using a high sensitivity DNA chip (Agilent) and quantitated using the Qubit dsDNA BR Assay Kit (Molecular Probes) according to the manufacturer’s instructions. All siRNA-seq libraries were multiplexed and sequenced in one flow cell using a MiSeq and MiSeq Reagent Kit v2 (50-cycle) (Illumina). RNA-seq libraries were multiplexed and sequenced in two HiSeq lanes by the Genome Access Technology Center (GATC) at Washington University.
Next-generation sequencing data analyses
All adapter sequences were trimmed and the libraries cleaned using Cutadapt (Martin 2011). We aggressively trimmed the siRNA reads to 25 nt from the 5′ end following adapter removal to filter remaining rRNAs, small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), and transfer RNAs (tRNAs) out of the dataset before mapping. All datasets were mapped to the Drosophila melanogaster genome and transcriptome using the RNA-seq Unified Mapper (RUM) (Grant et al. 2011). The NEB kit used to prepare the RNA-seq samples produces libraries with high directionality and RUM utilized this feature to strand specifically map the RNA-seq reads. RUM separated unique and nonuniquely mapping sequences into separate output files that could be further analyzed. Detailed mapping statistics can be found in Supporting Information, Table S1.
The University of California Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu, Dm6 assembly, August 2014) was used to visualize nonunique and unique bedgraph output files (Kent et al. 2002; dos Santos et al. 2015). The genome browser displays a peak normalized read count (reads per million, RPM) on the y-axis for the visualized genomic location. Nonunique RPM were used to calculate the average S and AS reads for Tn families shown in Table 1, and S and AS transcription of individual Tns having three or more full-length elements (Table S3). For individual Tns, a “+” system was devised to represent relative transcription among the Tns (Table S3). Unique RPM were used to measure S and AS expression levels of protein coding genes (Table S2).
Table 1. Drosophila Tn transcripts.
| Class | Family | No. Tns | %S | %AS | Avg S RPM | Avg AS RPM | S/AS |
|---|---|---|---|---|---|---|---|
| Non-LTR | Jockey | 5 | 80.0 | 80.0 | 302.0 | 104.8 | 2.88 |
| I | 1 | 0.0 | 0.0 | 0.0 | 0.0 | — | |
| R1 | 2 | 0.0 | 50.0 | 0.0 | 32.0 | — | |
| LTR | Gypsy | 18 | 66.7 | 72.2 | 871.3 | 261.1 | 3.34 |
| Pao | 3 | 100.0 | 100.0 | 1046.7 | 153.7 | 6.81 | |
| Copia | 1 | 100.0 | 100.0 | 64258.0 | 486.0 | 132.22 | |
| TIR | ProtoP | 1 | 100.0 | 100.0 | 80.0 | 31.0 | 2.58 |
| Tc1 | 5 | 0.0 | 40.0 | 0.0 | 24.5 | — | |
| Transib | 1 | 100.0 | 100.0 | 48.0 | 21.0 | 2.29 | |
| Pogo | 1 | 100.0 | 100.0 | 785.0 | 28.0 | 28.0 |
Transposons sorted by class and family were analyzed. No. Tns, the number of individual Tns within a family having three or more full-length elements; %S or %AS, the percentage of Tns included in column 3 with S or AS nonuniquely mapping RNA-seq reads; AVG S or AVG AS, the average normalized nonunique S or AS read count (RPM) for each Tn family; and S/AS, the ratio of S RPM to AS RPM.
Small-capped RNA-seq datasets (SRA: SRP001584, SRR032457, and SRR032458) were obtained from the Gene Expression Omnibus (GEO) accession number GSE18643 (Nechaev et al. 2010). FASTQ files were mapped strand specifically using RUM to obtain nonuniquely mapping reads. The UCSC genome browser (www.genome.ucsc.edu) was used to visualize the bedgraph output files. For presentation in Figure 1 and Figure 2, screen captures of nonunique S and AS reads and tss mapping to full-length, representative (for Dm297, blood, juan, and jockey) or individual (for mdg1{}1720) Tns were taken and overlaid to scale.
cDNA synthesis
A total of 50 ng of total Dmel-2 RNA isolated using the RNeasy MinElute Cleanup Kit (Qiagen) were reverse transcribed with RevertAid reverse transcriptase (Thermo Scientific) and a strand-specific, gene-specific reverse transcription primer (RT sense or antisense primer). This primer contains a unique nucleic acid tag 5′ of the complementary sequence that does not map to the Drosophila transcriptome (Table S5). The RT reaction contained 5× reaction buffer (no random hexamers or oligo dT), 1 µl Ribolock (40 units/µl), 1 mM dNTPs, 100 nM RT primer, and 2 µl of RevertAid (200 units/µl) in a total volume of 20 µl. The reaction incubated at 50° for 1 hr was heat inactivated at 85° for 5 min and was then diluted 1:10 with nuclease free water.
Quantitative PCR
Quantitative PCR (qPCR) was optimized and performed on a Bio-Rad CFX96 Real-Time system using SYBR Green detection chemistry (Bio-Rad SsoAdvanced Universal SYBR green). Briefly, 4 µl of diluted cDNA (10-fold) was mixed with 5 µl 2× SYBR green (Bio-Rad) and 0.5 µl of forward and 0.5 µl of reverse primer (500 nM final concentration). Initial denaturation was carried out at 95° for 3 min followed by a 30-sec denature step and a 30-sec annealing step (40×). Gene-specific primers for strand-specific qPCR are provided in Table S5. All RT-qPCR experiments were conducted in technical triplicates.
Northern blot analysis
A total of 50 µg of total RNA was separated on a 1% agarose denaturing gel at 100 V for ∼4.5 hr. RNA was transferred to hybond+ nitrocellulose membrane, UV cross-linked, and rRNA was stained with methylene blue. Following prehybridization, blots were probed with 32P-end labeled ∼50-nt probes (Table S7). Blots were grouped based on predicted transcript levels (group no. 1-S Dm297, S blood and S mdg1; group no. 2-AS Dm297, AS blood and AS mdg1; group no. 3-S and -AS juan, and S and AS jockey) and exposed to film. This treatment ensured that qualitative levels of transcripts within a group could be assessed.
RNAi, nuclear extract preparation, and antibodies
RNAi was performed as described (Sullivan et al. 2009). For the LacZ control, dsRNA targeting E. coli β-galactosidase was added to Dmel-2 cells to initiate the RNAi pathway without targeting Drosophila mRNAs. Comparing experimental knockdowns to this control ensures that molecular phenotypes observed upon RNAi depletion result from knockdown of specific mRNAs and not simply induction of RNAi. On day 5, the cells were counted with a Nexcelom cell counter and harvested. Nuclear extracts (NEs) were prepared as described (Sullivan et al. 2009) from 1 × 108 cells and then flash frozen in liquid N2 and stored at −80°. Commercial α-Actin (Abcam ab8224) and α-Dcr-2 (Abcam ab4732) antibodies were used at 1:5000 and 1:1000, respectively, for Western blot.
Data availability
Gene expression data are available at GEO with the accession number: GSE67725
Results
To investigate Tn AS transcription, we performed strand-specific high throughput sequencing (HTS) of rRNA-depleted RNA (>200 nt) from control Dmel-2 cells (LacZ). These libraries were prepared in triplicate and sequenced on an Illumina HiSeq, resulting in an average read depth of 101.5× and >98% of reads mapping to the Drosophila genome (Table S1). Surprisingly, 41.9% of the reads mapped nonuniquely (Table S1), indicating that a large percentage of transcripts are derived from non-rRNA repetitive sequences.
To compare abundance of S and AS Tn transcripts, we visualized nonunique RNA-seq reads using the UCSC genome browser. Because Tns are highly conserved and multicopy, RNA-seq reads corresponding to S and AS Tn transcripts map to more than one genomic location. Therefore, the normalized reads per million value identified for each Tn generally represents total cellular S or AS transcription of all copies of that element. Only Tns having more than three full-length annotated elements in the Drosophila genome were investigated.
LTR retroTns generate the majority of AS Tn transcripts
This analysis revealed S and AS nonunique reads for the majority of Tns examined (Table 1), while little to no AS transcription of non-Tn genes is evident (Table S2). These observations are consistent with previous data that Drosophila does not exhibit AS transcription upstream of mRNA genes (Lapidot and Pilpel 2006; Nechaev et al. 2010; Core et al. 2012). Tn nonunique RPM show that the most abundant and active Drosophila Tn class, LTR retroTns, is highly expressed in the S and AS direction (Kaminker et al. 2002; Kofler et al. 2015) (Table 1). Non-LTR retroTn and TIR DNA Tns are generally transcribed at lower levels (Table 1). These data are consistent with previous analyses showing that LTR retroTn-derived esiRNAs are more prevalent in S2 cells than esiRNAs originating from non-LTR retroTn or TIR DNA Tns (Ghildiyal et al. 2008).
While AS transcription is observed for all but the “I” family of Tns, the ratio of S to AS nonunique reads differs dramatically when S and/or AS RPM are >100. Non-LTR retroTns in the Jockey family and LTR retroTns in the Gypsy family have generally low S/AS ratios (2.88 and 3.34, respectively) while LTR retroTns in the Pao and copia families and TIR DNA Tns in the Pogo family have much higher S/AS ratios (6.81, 132.22, and 28.04, respectively) (Table 1). LTR retroTns generating the most esiRNAs in S2 cells all belong to the Gypsy family of retroTns (Chung et al. 2008; Czech et al. 2008; Kawamura et al. 2008). Because Gypsy retroTns Dm297, blood, and mdg1 generate abundant esiRNAs and produce ample AS transcripts (Table S3), these retroTns were chosen for further investigation. Additionally, only Tns in the Jockey family of non-LTR retroTns generate both S and AS transcripts (Table 1) and esiRNAs (Kawamura et al. 2008). Jockey family members jockey and juan produce the highest levels of AS RNAs (Table S3). Therefore, to explore the importance of LTR sequences in retroTn AS transcription, juan and jockey were chosen for further study. TIR DNA Tns were not further investigated.
LTR retroTn AS transcription initiates from within or near LTRs
Bedgraphs of nonunique, strand-specific RNA-seq reads mapping to Dm297 (Figure 1B), blood (Figure 1C), and mdg1 (Figure 1D, right half of mdg1{}1720) representative full-length elements are shown. These data indicate that both S and AS transcripts are distributed across the elements including the three ORFs (Figure 1, B–D). Dm297, blood, and mdg1 AS reads tend to be concentrated in the LTRs, while S RPM are higher in the ORFs (Figure 1, B–D). In all three cases, total S transcript levels were higher than AS (Figure 1, B–D, red numbers). Some sequences were removed by splicing (data not shown).
To identify S and potential AS transcription start sites (tss), we remapped publicly available short-capped RNA high-throughput sequencing datasets (Nechaev et al. 2010; Henriques et al. 2013) and filtered to isolate only nonunique reads. Potential S and AS tss were observed for all three LTR retroTns (Figure 1, B–D, blue). RPM for tss on the S strand were higher than RPM for AS tss for Dm297 and blood (Figure 1, B and C, blue numbers). These data correlate with S and AS transcription levels for each retroTn. For Dm297 and blood, S and AS transcription could begin near the 3′ end of the LTR (Figure 1, B and C, top, blue) and transcription initiating from this location could result in S and AS RNAs spanning the entire element (Figure 1, B and C, blue).
In contrast, no LTR AS tss were observed for full-length mdg1 elements. An AS tss is visible (Figure 1D, right, bottom, blue); however, transcription from this tss would only produce AS RNAs of the 5′ LTR. One mdg1 element, mdg1{}1720, was identified that could produce the observed AS transcripts. Mdg1{}1720 consists of two tandemly repeated mdg1 elements with an inverted and centralized LTR in the RT ORF of the first mdg1 retroTn (Figure 1D). Transcription initiating from the non-LTR tss in the downstream mdg1 element could result in AS transcription of the first mdg1 retroTn. While only nonuniquely mapping AS reads are shown in Figure 1D, AS reads corresponding to unique sequences in the upstream mdg1 repeat are observed (data not shown), indicating that this specific retroTn, mdg1{}1720, is transcribed.
We next performed strand-specific RT-qPCR (Purcell et al. 2006; Vashist et al. 2012) to confirm S and AS transcription of Dm297, blood, and mdg1. Each potential transcript was reverse transcribed with a strand-specific, gene-specific primer having a unique nucleic acid tag (Table S5). The unique tag provided a primer binding site for the downstream qPCR reaction to ensure detection of only the transcript of interest. Random priming was evaluated in the absence of an RT primer and no target transcripts were detected (data not shown). For Dm297, blood, and mdg1, we detected both S and AS transcription using several primer sets spanning the coding sequence (Table S5). We calculated the difference between S and AS Cts (ΔCt(S − AS)) for Dm297, blood, and mdg1 (Table S6). AS transcripts were less abundant in all cases and the differences between S and AS transcription correlated with RNA-seq data (Figure 1, B–D).
Lastly, we performed Northern blot analysis to detect S and AS transcripts (Table S7). RNA from Dmel-2 cells was transferred to a membrane and probed with radioactively labeled Dm297, blood, and mdg1 complementary S and AS probes. S (Figure 1E) and AS (Figure 1F) transcripts were observed for Dm297, blood, and mdg1. S Dm297 (6995 nt) appears slightly smaller than its predicted size while S blood (7410 nt) and mdg1 (7480 nt) are slightly larger (Figure 1E). No other prominent bands were detected. The Dm297 S transcript is most abundant while mdg1 and blood S transcripts are much less prevalent. Each lane contained equal amounts of 28S rRNA (Figure 1E, bottom). These data correlate with LTR retroTn S transcript levels observed in the RNA-seq data (Figure 1, B–D). We observe multiple Dm297 and blood AS transcripts resulting in a smear between 8 and 10 kb, while a single AS mdg1 transcript is present at ∼8 kb (Figure 1F). An ∼2 kb mdg1 AS transcript is also visible (Figure 1F). The sizes of these RNAs support AS transcription of full-length Dm297 and blood LTR retroTns from the bioinformatically identified tss (Figure 1, B–D). These data also indicate that the mdg1 AS tss (Figure 1D, first and third blue peaks) is functional, producing the predicted ∼8 kb and ∼2 kb RNAs, while the inverted LTR S tss is not active in this context. Multiple Dm297 AS transcripts may result from inefficient RNAPII termination. A lighter exposure (Figure 1F, bottom “L”) of the AS transcripts reveals that the ∼8 kb AS mdg1 transcript is most abundant, while Dm297 and blood AS RNAs are less prevalent, mirroring the RNA-seq data (Figure 1, B–D). S2 culture cells have amplified Tn content (Potter et al. 1979; Junakovic et al. 1988; Wen et al. 2014) and RNA-seq reads cannot be mapped to the S2-specific mdg1 copies as the S2 genome has not been sequenced. Regardless, the data presented here support AS transcription of mdg1{}1720 from non-LTR tss.
To gain a more complete view of AS retroTn transcription, we considered the genomic context of each annotated full-length Dm297, blood, and mdg1 element (Table S4). Individual retroTns were found both intergenically and within introns. Intronic Dm297 and mdg1 retroTns were more often AS to their host coding gene, while blood elements were in the S orientation. These data indicate that transcription of intronic LTR retroTns, together with RNAs produced from intraelement tss, could contribute to AS transcript abundance.
We also investigated S and AS transcription of individual Dm297, blood, and mdg1 elements (Table S4). If unique intraelement RNA-seq reads and RNA-seq reads corresponding to the retroTn-intergenic/intronic sequence junction could be identified, we concluded that the individual Dm297, blood, or mdg1 element was transcribed. S transcription was confirmed for 9/18 (50%) of Dm297, 2/15 (13%) of mdg1, and 9/22 (41%) of blood retroTns (Table S4). AS transcription was verified for 4/18 (22%) of Dm297, 1/15 (7%) of mdg1, and 3/22 (14%) of blood elements (Table S4). Of all mdg1 retroTns, only AS transcription of mdg1{}1720 could be confirmed using this analysis. These numbers of transcribed individual elements are probably an underestimate of the total as not all elements have mutations allowing observation of unique internal reads. Also, RPM for unique reads were low, reflecting less RNA from one individual element as compared to nonunique RPM corresponding to total transcription of all individuals of a retroTn type. Collectively, the RNA-seq analyses, strand-specific RT-qPCR, and Northern blots indicate that individual LTR retroTns in Dmel-2 cells undergo convergent S and AS transcription and that transcription can initiate within or near Dm297, blood, and mdg1 LTRs.
Non-LTR retroTns juan and jockey produce AS transcripts
To investigate the role of LTRs in AS transcription, we examined non-LTR retroTns juan and jockey. Similar to LTR elements, strand-specific nonunique RNA-seq reads map the entire length of jockey and juan (Figure 2, B and C, red); however, non-LTR S and AS transcripts are less abundant than corresponding LTR retroTn RNAs. Additionally, jockey is the only retroTn investigated for which the AS transcript is more highly expressed than the S transcript.
A juan S tss is observed at the 5′ end of the retroTn (Figure 2B, top, blue) and initiation of transcription from this location could result in a single complete element S transcript. Several AS tss were also observed; however, these tss cannot be responsible for reads mapping to the 3′ half of juan (Figure 2B, bottom, blue). The source of these AS RNA-seq reads is unclear, although unique AS reads were identified for both intergenic and intragenic juan elements (Table S4), indicating AS transcription of individual elements (see previous section). Two S tss are observed for jockey. Transcription beginning at these tss could produce a S transcript the entire length of the element (Figure 2C, top, blue). For jockey, a potential AS tss is observed at the 5′ end, but the number of reads mapping to this tss do not correlate with higher AS RPM (Figure 2C, bottom, blue). Collectively, these data indicate that non-LTR retroTns are transcribed in both S and AS directions, albeit at lower levels than LTR retroTns.
To verify S and AS transcription, we performed strand-specific qPCR of non-LTR retroTns juan and jockey as described in the previous section (Table S5). S and AS transcription were detected for juan and jockey and the ΔCt(S − AS) mirrored the ratio of S to AS RPM of each ORF investigated (Table S6). Additionally, Northern blot analysis revealed juan S and AS, and jockey AS transcription (Figure 2, D and E); however, the S jockey transcript was not visible presumably because of its low abundance. Previously, S jockey transcripts initiating from an internal promoter were identified in Drosophila cell culture (Mizrokhi et al. 1988). A probe to the S juan transcript (4236 nt) reveals one band ∼5 kb, while AS jockey (5020 nt) and AS juan probes show smears indicating multiple transcripts (Figure 2, D and E). Juan AS RNAs range from ∼2 kb to ∼4 kb (Figure 2E). If juan AS transcription initiates from bioinformatically identified tss (Figure 2B) and RNAPII termination is inefficient, multiple ∼2 to ∼4 kb AS transcripts could be produced. Jockey AS RNAs range from ∼7 kb to greater than 10 kb (Figure 2D). We hypothesize that smaller transcripts in this range could be produced from the bioinformatically identified tss (Figure 2C). Additionally, one jockey retroTn (jockey{}1630), has a LTR retroTn, roo (9092 nt), inserted in the gag ORF making this element 14,127 nt. Transcripts originating from an observed tss (data not shown) at the 5′ end of jockey{}1630 could result in jockey AS RNAs greater than 10 kb. These data suggest that non-LTR retroTns juan and jockey are transcribed in both the S and AS directions from intraelement tss.
S and AS tss have canonical Drosophila promoter elements
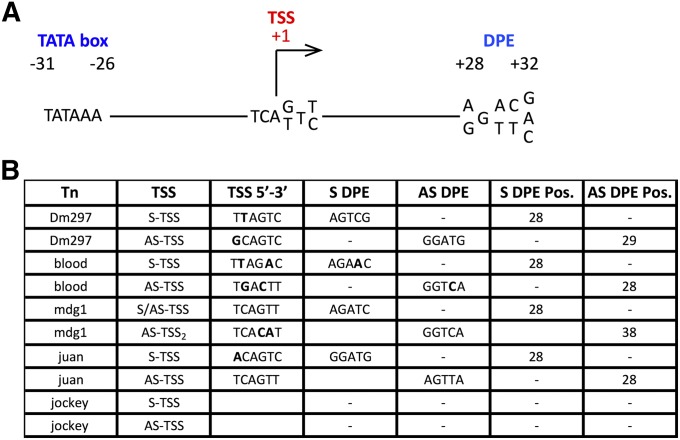
We next wanted to determine if the observed LTR and non-LTR retroTn tss were flanked by traditional Drosophila promoter elements. Drosophila transcription initiates at T-C-A+1-G/T-T-T/C (where A+1 is the tss) within a promoter composed of a TATA box (−31 to −26) and/or a downstream promoter element (DPE) located between +28 to +32 (Butler and Kadonaga 2002) (Figure 3A). The TATA box or DPE occur in core promoters 29% and 26% of the time, respectively, while 14% contain both a TATA box and a DPE (Butler and Kadonaga 2002). Further evaluation of Dm297 revealed near-canonical tss and DPEs in appropriate locations for both S and AS promoters (+28 and +29, respectively, Figure 3B). Blood S and AS transcripts initiate from tss having two noncanonical bases but also have canonical DPEs +28 from S and AS tss (Figure 3B). The mdg1 LTR has a canonical tss with a perfect DPE +28 downstream, while the AS tss not located in the LTR has two nonideal bases and an inappropriately spaced DPE (Figure 3B). These data support previous characterization of the mdg1 S promoter (Arkhipova and Ilyin 1991). The non-LTR retroTn juan has a near canonical S tss and a canonical AS tss. Both tss have canonical DPEs +28 bases downstream of the tss (Figure 3B). These data support bonafide S tss for all three LTR-retroTns and non-LTR retroTn juan. Finally, promoter analysis revealed no canonical initiation site or DPE for either S or AS jockey tss. We hypothesize that this promoter is unique compared to more canonical core Drosophila promoters.
Figure 3.
S and AS tss have canonical Drosophila RNAPII promoter elements. (A) A schematic representing canonical Drosophila promoter elements is shown. (B) HTS analysis at nucleotide resolution of LTR and non-LTR retroTns tss is depicted. The tss, tss sequence, S DPE, AS DPE, and position of each DPE are shown for each retroTn. Bold nucleotides represent divergence from canonical nucleotide/s shown in A.
LTR and non-LTR retroTn AS transcripts lack strong polyadenylation
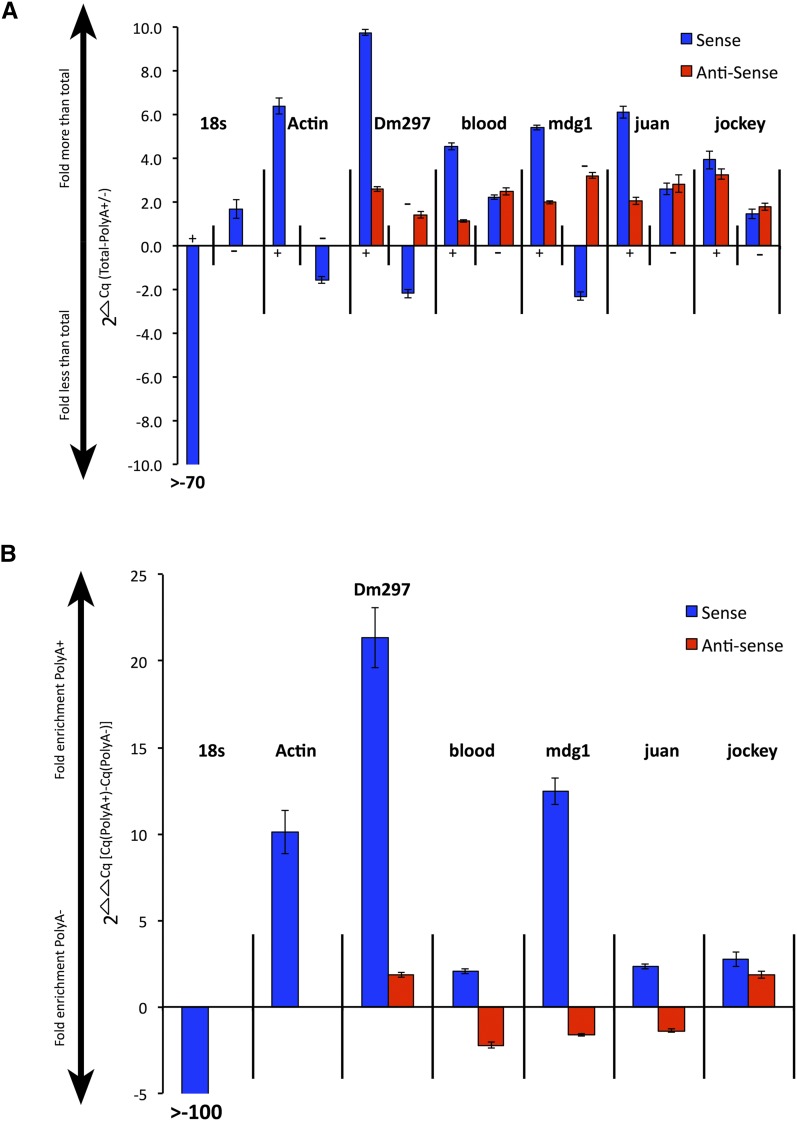
Our data suggest that S and AS LTR and non-LTR retroTns are convergently transcribed from canonical Drosophila promoters. As these RNAs are likely RNAPII transcripts (Gogvadze and Buzdin 2009), we wanted to determine their polyadenylation status. S retroTn transcripts have canonical polyadenylation signals (Gogvadze and Buzdin 2009; this work) and polyadenylation of these RNAs has previously been reported (Gogvadze and Buzdin 2009). We first fractionated total RNA using an oligo d(T) column and then performed strand-specific qPCR to the RT ORF of each retroTn on total RNA, polyA+ RNA and polyA− RNA (Table S5). We used the amount of transcript in total RNA to normalize polyA+ and polyA− fractions by subtracting polyA+ and polyA− Cq values from those of total RNA (ΔCq = total-polyA +/−) (Figure 4A). We then determined a fold enrichment of polyadenylation by calculating the difference between these ΔCq values (ΔΔCq = [(total-polyA +)-(total-polyA −)]) (Figure 4B). 18s rRNA (polyA−) and Actin (polyA+) were used as controls. Total RNA was efficiently separated into polyA+ and polyA− fractions as 18s S transcripts were >70-fold less in the polyA+ fraction than in total RNA (Figure 4A, 18s +) and actin S transcripts were ∼6-fold increased in the polyA+ fraction as compared to total RNA (Figure 4A, actin +). 18s S RNAs were more than 100-fold depleted in polyA+ transcripts, while Actin S RNAs were approximately 10-fold enriched in polyA+ transcripts (Figure 4B), indicating that our assay to assess polyadenylation was working properly.
Figure 4.
LTR and non-LTR retroTn AS transcripts lack strong polyadenylation. (A) Graph of S and AS transcript fold differences in polyA+ or polyA− fractions compared to total RNA. 2ΔCq is the y-axis and represents (total RNA – polyA+ or polyA−). PolyA+ or polyA− fractions are indicated along the x-axis as + or – signs. S transcripts are blue bars and AS transcripts are red bars. The name of each retroTn or control is listed above the appropriate group. Error bars represent standard deviation of strand-specific qPCR technical triplicates. (B) Graph of direct comparison of polyA+ to polyA− levels for each retroTn S/AS transcript pair. Fold enrichment values for polyA+ or polyA− fractions are shown along the y-axis (2ΔΔCq) where the ΔΔCq equals [(total-polyA +)-(total-polyA −)]). S and AS transcripts are shown along the x-axis; S bars are blue and AS bars are red. Error bars represent standard deviation of strand-specific qPCR technical triplicates.
S Dm297 and mdg1 transcripts were ∼10- and ∼5-fold more, respectively, in the polyA+ fraction than in total RNA and ∼2-fold less in the polyA− fraction than in total RNA (Figure 4A, Dm297 and mdg1, blue). These S transcripts are enriched for polyadenylation at least as much (mdg1) if not more (Dm297) than the polyadenylated Actin control (Figure 4B). Dm297 and mdg1 AS transcripts, and blood, juan, and jockey S and AS transcripts were between ∼1.5- and ∼3-fold more in both polyA+ and polyA− fractions than in total RNA, indicating a mixture of both polyA+ and polyA− transcripts (Figure 4A). ΔΔCq calculations suggest that blood, juan, and jockey S transcripts are enriched for polyadenylation although much less than Dm297 and mdg1 (Figure 4B). None of the AS transcripts are highly enriched with polyA+ transcripts. Blood, mdg1, and juan are slightly enriched in polyA− RNAs (Figure 4B). These data suggest that while all retroTn S transcripts have polyadenylation signals, only Dm297 and mdg1 S transcripts are polyadenylated. Bioinformatic assessment did not reveal strong polyadenylation sites for any of the AS transcripts examined (data not shown). Collectively, these data support a hypothesis that retroTn AS transcripts are not strongly polyadenylated.
Dcr-2 depletion decreases retroTn-derived esiRNA levels
Previous studies show that esiRNAs, many of which map to retroTns, are cleaved from long dsRNA precursors by Dcr-2 (Tomari et al. 2007; Ghildiyal et al. 2008; Chung et al. 2008; Kawamura et al. 2008; Siomi et al. 2008). Knockdown of Dcr-2 results in decreased esiRNA levels and a corresponding increase in precursor RNAs (Chung et al. 2008; Ghildiyal et al. 2008). Small RNA (<200 nt) high-throughput sequencing (HTS) libraries were constructed in triplicate from Dcr-2-depleted and control (LacZ) cells (Figure 5A). Greater than 99% of reads from all six libraries mapped to the Drosophila genome (Table S1). Dcr-2 knockdown resulted in a statistically significant (P = 0.00154) decrease in nonunique reads (22.7%) compared to the LacZ control (34.4%) (Table S1), indicating that Dcr-2 is required for global production of nonuniquely mapping esiRNAs.
Figure 5.
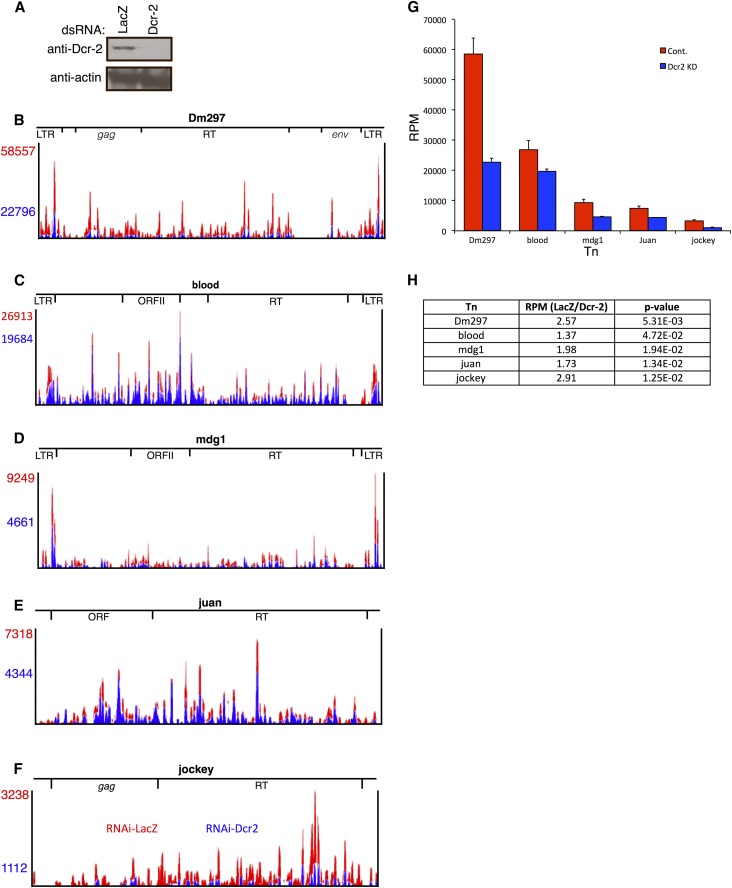
Dcr-2 depletion decreases retroTn-derived esiRNA levels. (A) Representative Western blot of Dcr-2 depletion. The antibody used is indicated to the left of blots and dsRNA for RNAi is labeled above blots. (B–F) Bedgraphs of esiRNAs mapping to retroTns in control and Dcr-2-depleted Dmel-2 cells. The control is red and the Dcr-2 knockdown is in blue. RPM values are listed to the left of bedgraphs and are color coordinated. Relative locations of specific ORFs are shown above the bedgraphs. (G) Graphs of esiRNA levels in control and Dcr-2-depleted Dmel-2 cells (control, red; Dcr-2 knockdown, blue). RPM values are on the y-axis and retroTns are indicated along the x-axis. Error bars represent standard deviation of technical triplicates. (H) A table reporting the ratio of esiRNA levels (RPM) between the control and Dcr-2 knockdown for each retroTn (middle column) is shown. RetroTn is indicated in the left column and P-value (unpaired t-test) is indicated in the right column.
Nonunique siRNA-seq reads map across Dm297, blood, mdg1, juan, and jockey for both the LacZ control and the Dcr-2-depleted samples (Figure 5, B–F) and esiRNA patterns are generally similar for both the control and the Dcr-2 knockdown. RPM vary considerably among the five retroTns with the most esiRNAs mapping to Dm297 and blood, and fewer mapping to mdg1, juan, and jockey (Figure 5, B–F, red numbers). Both Dm297 and mdg1 have a higher concentration of reads mapping to LTRs, as previously described (Chung et al. 2008; Ghildiyal et al. 2008) (Figure 5, B and D). RPM for esiRNAs mapping to all five retro Tns were decreased in Dcr-2-depleted Dmel-2 cells (Figure 5, B–F). Average RPM calculated from triplicate sequencing experiments for each retroTn in both control and Dcr-2-depleted samples (Figure 5G) were used to determine the ratio of esiRNAs in the LacZ control as compared to the Dcr-2 knockdown (Figure 5H). Dcr-2 depletion led to statistically significant reduction of the number of esiRNAs mapping to Dm297 (2.6-fold), jockey (2.9-fold), mdg1 (2.0-fold), juan (1.7-fold), and blood (1.4-fold) (Figure 5H). These data indicate that depletion of Dcr-2 causes a decrease in retroTn-derived esiRNA levels without changing the specific esiRNAs produced. These data strengthen the previous hypothesis that Dcr-2 produces retroTn-derived esiRNAs in Dmel-2 cells.
Sense and antisense retroTn transcript levels increase with Dcr-2 knockdown
Previous RT-qPCR studies suggest that some retroTn transcript levels increase after knockdown of Dcr-2 in S2 cells (Chung et al. 2008; Ghildiyal et al. 2008). To determine S and AS retroTn RNA levels globally, we performed strand-specific RNA-seq on Dcr-2-depleted, large RNA (>200 nt) resulting in an average read depth of 78.3× and ≥98% of reads mapping to the Drosophila genome (Table S1). Dcr-2 depletion resulted in a lower read depth as compared to the control (Table S1). The percentage of nonuniquely mapping reads was significantly increased in the Dcr-2 knockdown (46.7%) compared to the LacZ knockdown (41.9%) (P = 9.6 × 10−5, Table S1).
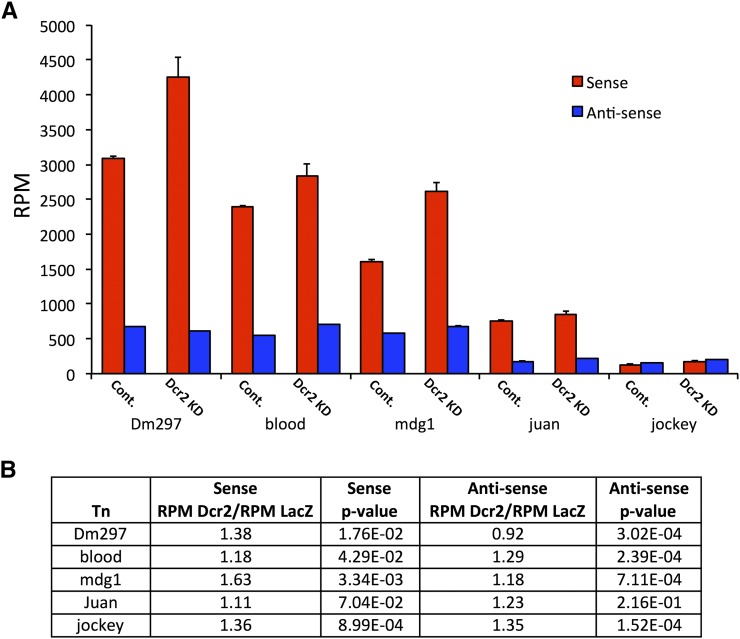
We compared Dcr-2 knockdown and LacZ control RPM for Dm297, blood, mdg1, juan, and jockey (Figure 6A). Generally, S retroTn transcript levels were increased in the Dcr-2-depleted samples, as previously reported (Figure 6, A, red, and B) (Chung et al. 2008; Ghildiyal et al. 2008). We observed a similar trend for AS retroTn transcripts except for Dm297, which showed a slight reduction in AS transcript levels following Dcr-2 knockdown (Figure 6, A, blue, and B). These results suggest that Dcr-2 generates esiRNAs from dsRNA precursors consisting of S and AS retroTn transcripts.
Figure 6.
S and AS retroTn transcript levels increase with Dcr-2 knockdown. (A) A graph of retroTn transcript levels upon RNAi depletion of Dcr-2 is shown (sense, red; antisense, blue). RPM values are shown on the y-axis and retroTn (control vs. Dcr-2 knockdown) is indicated on the x-axis. Error bars represent standard deviation of technical triplicates. (B) A table reporting the ratio of retroTn S and AS transcript levels (RPM) between the control and Dcr-2 knockdown for each retroTn is shown. RetroTn is indicated in the first column. P-values (unpaired t-test) are reported for these comparisons.
Discussion
Understanding the mechanisms that balance retroTn amplification and repression in eukaryotes is critical, as misregulation can lead to detrimental genomic damage. Many retroTns are active in Drosophila (Kofler et al. 2015), providing a unique opportunity to understand molecular mechanisms of retroTn repression. In Drosophila somatic cells, silencing of retroTns requires a dsRNA precursor that is processed into esiRNAs by Dcr-2 (Tomari and Zamore 2005; Ghildiyal et al. 2008; Marques et al. 2010). To better understand the origin of this retroTn-derived dsRNA precursor, we performed RNA-seq, strand-specific qPCR, and Northern blotting of control Dmel-2 cells. Most Tns produce S and AS transcripts, although S and AS expression are highest for LTR retroTns (Table 1). Bioinformatic analysis of representative LTR retroTns Dm297 and blood, a specific mdg1 element (mdg1{}1720), and representative non-LTR retroTns juan and jockey showed S and AS transcripts originating from intraelement transcription start sites for all elements investigated (Figure 1 and Figure 2). These initiation sites are generally canonical RNAPII transcription start sites with conserved DPEs (Figure 3). Collectively, these data suggest that AS retroTn RNAs are convergently transcribed from these start sites. Interestingly, we also observed that AS transcripts derived from retroTns are not strongly polyadenylated (Figure 4). By sequencing small RNAs, we determined that esiRNAs are globally derived from locations of retroTn S/AS convergent transcription and that Dcr-2 knockdown decreases esiRNA levels (Figure 5). Consistently, we showed that both S and AS retroTn transcript levels increase when Dcr-2 is knocked down (Figure 6). Taken together, these data support a model in which AS retroTn transcripts hybridize to their S counterparts forming dsRNAs that are substrates for esiRNAs production by Dcr-2 (Figure 7).
Figure 7.
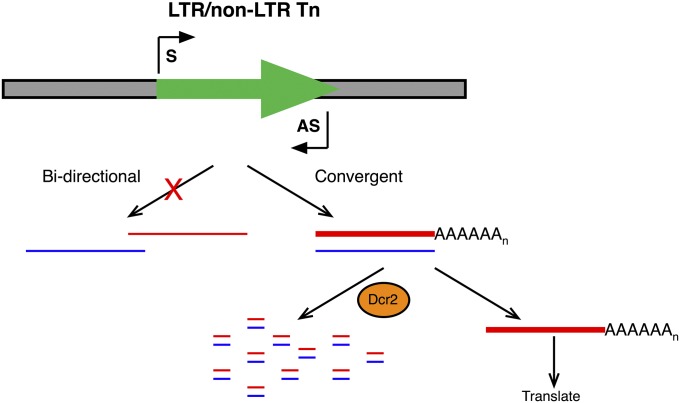
Dcr-2 generates esiRNAs from dsRNAs derived from convergent S and AS transcription of retroTns. Shown is a model depicting convergent S and AS transcription (arrows, black, “S” and “AS,” respectively) of retroTns (arrow, green) in Drosophila. S transcripts (red) are polyadenylated and more abundant (thick line) compared to AS transcripts (blue, thin line). AS transcripts act as a molecular sponge isolating a portion of S transcripts resulting in the formation of dsRNA Dcr-2 substrates. Some S transcripts are translated promoting mobility of retroTns.
Drosophila retroTns are convergently transcribed from independent, canonical S and AS tss
Unlike in mammals, Drosophila RNAPII transcription does not initiate bidirectionally from promoters to generate AS transcripts (Lapidot and Pilpel 2006; Nechaev et al. 2010; Core et al. 2012). Also, the >100 predicted overlapping cis-natural pairs in Drosophila are most often complementary ORF 3′ UTRs (Okamura et al. 2008a), not S transcript-noncoding AS RNA pairs as in other organisms (Pelechano and Steinmetz 2013). Because protein coding genes do not produce AS transcripts in Drosophila, mechanisms of AS transcription and downstream regulatory functions have not been fully elucidated. AS retroTn transcription of Drosophila telomere LTR retroTns has been observed previously (Danilevskaya et al. 1999). Herein, we provide the first evidence of global AS transcription of LTR and non-LTR retroTns, and TIR DNA Tn families in Drosophila (Table 1 and Table S3). Additionally, we identify and quantitate AS transcription of individual Tns and specific elements (Figure 1, Figure 2, and Table S4).
Bioinformatically identified AS transcription start sites and promoter analysis provide the first clues about how retroTn AS transcripts are produced (Figure 3). Interestingly, Dm297, blood, and mdg1 AS transcription start sites and promoter elements are located within the retroTn and AS transcripts initiating from these locations could explain all observed AS RNA-seq reads, suggesting that external sequences are not required for LTR retroTn AS transcription. Evidence to support this hypothesis comes from identifying several individual, intergenic LTR retroTns that are transcribed in the AS direction (Table S4). It seems unlikely that multiple intergenic LTR retroTns simultaneously evolved external promoters in different genomic locations and is more plausible that the observed internal Dm297, blood, and mdg1 AS transcription start sites are functional. AS transcription start sites are observed for non-LTR retroTn juan, but these cannot be responsible for transcription of the 5′ end of the element (Figure 2). The AS transcription start site identified for jockey does not have adequate normalized read counts to account for the amount of AS transcription (Figure 2). We hypothesize that the additional AS jockey and 5′ juan transcripts originate from intragenic elements oppositely oriented to mRNA S transcripts (Table S4). Collectively, these data suggest that LTR retroTns are transcribed from intraelement transcription start sites, while some non-LTR retroTns RNAs are generated indirectly by transcription of protein coding genes.
AS transcripts can arise from bidirectional transcription at RNAPII promoters (Core et al. 2008; Guil and Esteller 2012) or convergent transcription from strand-specific promoters (Gullerova and Proudfoot 2012). Bidirectional transcription initiates in both directions from one promoter, while convergent transcription requires independent transcription start sites. Bidirectional transcription of a retroTn from a single promoter would not result in a full-length dsRNA (Figure 7). Therefore, our results suggest that double-stranded retroTn RNAs are derived from convergent transcription of S and AS RNAs from independent transcription start sites (Figure 7). Transcriptional gene silencing mediated by convergent transcription is highly efficient in both fission yeast and mammalian cells (Gullerova and Proudfoot 2012). Our data support a model in which formation of dsRNAs by convergent transcription of retroTns is the first step in Drosophila somatic cell retroTn silencing.
Production of dsRNAs by convergent transcription is a novel retroTn regulatory mechanism
A well-studied mammalian non-LTR retroTn, L1, initiates AS transcription in the S RNA 5′ UTR in humans (Speek 2001; Nigumann et al. 2002) and the S RNA ORF1 in mice (Li et al. 2014). Most full-length intragenic L1 elements are oriented AS to protein coding genes (Szak et al. 2002). Therefore, AS transcription from the identified transcription start sites proceeds into neighboring mRNAs forming fusion transcripts that regulate expression of numerous genes (Speek 2001; Nigumann et al. 2002; Mätlik et al. 2006; Cruickshanks and Tufarelli 2009) and affect mobility of L1 elements (Li et al. 2014). The L1 retroTn is closely related to Drosophila non-LTR retroTns jockey and juan (Mizrokhi et al. 1988; Speek 2001). No AS transcription start sites were observed in jockey analogous to L1 AS transcription start sites (Figure 2C). Juan AS transcription start sites were located in the S RNA ORFI (Figure 2C), but only two full-length juan elements (of seven total) are intragenic (Table S4) limiting the impact of a potential L1-like AS fusion transcript regulatory mechanism. Additionally, juan AS transcripts were identified upstream of the observed transcription start sites. Together, these data indicate that the mechanism of Drosophila non-LTR retroTn AS transcript initiation and the functions of these AS RNAs may differ from their mammalian L1 counterparts.
In fission yeast and the Drosophila germline, movement of repetitive sequences and retroTns, respectively, are repressed by a transcriptional gene silencing mechanism wherein siRNAs induce heterochromatin formation (Huisinga and Elgin 2009; Wang and Elgin 2011; Sienski et al. 2012; Huang et al. 2013; Le Thomas et al. 2013; Rozhkov et al. 2013). In Schizosaccharomyces pombe, siRNAs are produced from RNA-dependent RNA polymerase generated dsRNA precursors by Dicer 1 (Volpe et al. 2002; Yu et al. 2014; Holoch and Moazed 2015). In Drosophila, retroTns are silenced in the germline by piRNAs cleaved from single-stranded substrates and amplified via a mechanism that does not include long dsRNAs (Saito et al. 2006; Aravin et al. 2007; Gunawardane et al. 2007). Therefore, our proposed model that esiRNAs are generated from hybridized convergently transcribed S and AS retroTn transcripts, is novel as other mechanisms do not require dsRNA substrates, utilize an RNA-dependent RNA polymerase to produce dsRNA substrates, or use AS transcripts to regulate gene expression in a way that does not require siRNAs.
Lack of AS retroTn polyadenylation may lead to nuclear retention of dsRNAs
Efficient polyadenylation of transcripts can promote export to the cytoplasm and removal of polyA signals may cause nuclear retention of RNAs (Dower et al. 2004). We propose that convergently transcribed S and AS retroTn transcripts hybridize in the nucleus forming dsRNAs. Because only Dm297 and mdg1 S transcripts are polyadenylated (Figure 4), all double-stranded retroTn RNAs investigated would contain at least one polyA− component, encouraging nuclear retention of these dsRNAs. As the number of retroTn AS transcripts is often significantly less than the number of S transcripts (Figure 1 and Figure 2), unhybridized S RNAs are exported to the cytoplasm for translation, leading to a balance of repression and expansion of retroTns. A nuclear pool of Dcr-2 (Cernilogar et al. 2011, and data not shown) may use nuclear-retained retroTn dsRNA as substrates for esiRNAs biogenesis.
Dcr-2 generates esiRNAs from dsRNAs derived from convergent S and AS transcription of retroTns
LTR and non-LTR retroTns produce esiRNAs from dsRNA precursors through Dcr-2-dependent mechanisms in Drosophila somatic cells (Ghildiyal et al. 2008; Kawamura et al. 2008; Siomi et al. 2008). Here, we show that expression of both S and AS retroTn transcripts is regulated by Dcr-2. Specifically, depletion of Dcr-2 leads to reduction in esiRNA levels (Figure 5) and a corresponding increase in both S and AS retroTn transcript levels (Figure 6). The mechanism of Dcr-2 mediated AS silencing is likely similar to S silencing as S and AS esiRNAs are often equally abundant (Ghildiyal et al. 2008).
Others have hypothesized that esiRNAs are processed from double-stranded LTR hairpins because of higher concentrations of small RNA-seq reads from LTRs (Chung et al. 2008; Ghildiyal et al. 2008). Generally, our data do not support this model. EsiRNA reads from retroTns span the entire element, suggesting that LTR hairpins cannot be the only dsRNA substrates (Figure 5). Additionally, S and AS RNA-seq reads map to all regions of retroTns, indicating that the entire element has the potential to form a dsRNA precursor. Also, we observe convergent transcription of non-LTR retroTns and Dcr-2-regulated esiRNAs mapping to these retroTns (Figure 5). Thus, an LTR is not required for dsRNA formation and subsequent siRNA biogenesis.
Mechanisms of AS transcription and esiRNA biogenesis are conserved in tissue culture and Drosophila
The data presented here were collected in Dmel-2 cells, a derivative of Schneider 2 (S2) cells, a somatic cell line derived from Drosophila embryos (Schneider 1972). Previous parallel investigation of esiRNA biogenesis in S2 cells and Drosophila heads indicated no mechanistic differences between these two tissues (Ghildiyal et al. 2008). Most importantly, esiRNAs were equally derived from S and AS retroTn strands and mapped evenly across retroTn precursors (Ghildiyal et al. 2008), indicating that a full-element dsRNA precursor is required to generate the observed esiRNAs in both fly tissues and cell lines. These data are consistent with our results that retroTns in S2 cells produce AS transcripts (Figure 1 and Figure 2).
Previous studies show that Drosophila tissue culture cells have amplified Tn content (Potter et al. 1979; Tchurikov et al. 1981; Maisonhaute et al. 2007; Wen et al. 2014) and hypothesize that this amplification is necessary for creating immortal cell lines (Junakovic et al. 1988). Once established, Tn location and number appear stable in Drosophila Kc and S2 cell lines (Junakovic et al. 1988). This amplification is reflected as a greater portion of retroTn-derived esiRNAs mapping to Tns in S2 cells than in Drosophila heads (Ghildiyal et al. 2008). While having more Tn copies in tissue culture potentially increases the absolute levels of S and AS retroTn transcripts (and esiRNAs generated from dsRNA precursors) the molecular mechanisms required to produce AS transcripts and generate esiRNAs appear conserved between flies and culture cells (Ghildiyal et al. 2008). Additionally, a higher concentration of esiRNAs and dsRNA precursors is a tremendous advantage of the S2 cell system.
In conclusion we show, for the first time, that Drosophila retroTns are transcribed in the AS direction from intraelement transcription start sites. We observed convergent transcription of S and AS transcripts in all retroTns investigated, suggesting that this is a global dsRNA formation mechanism in Drosophila. The experiments described here will provide the basis for future mechanistic studies of retroTn AS transcription and allow determination of the role of convergent transcription in retroTn gene silencing.
Acknowledgments
We thank Joshua Daugherty, Michael McKain, and Michael Hughes for technical assistance; Daniel Michalski for helpful discussions; and Ambrose R. Kidd, III and Lon Chubiz for critical review of the manuscript.
Footnotes
Communicating editor: J. Birchler
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177196/-/DC1.
Literature Cited
- Aravin A. A., Hannon G. J., Brennecke J., 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764. [DOI] [PubMed] [Google Scholar]
- Arkhipova I. R., Ilyin Y. V., 1991. Properties of promoter regions of mdg1 Drosophila retrotransposon indicate that it belongs to a specific class of promoters. EMBO J. 10: 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham P. M., Kidwell M. G., Rubin G. M., 1982. The molecular basis of P-M hybrid dysgenesis: the role of the P element, a P-strain-specific transposon family. Cell 29: 995–1004. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., et al. , 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Brouha B., Schustak J., Badge R. M., Lutz-Prigge S., Farley A. H., et al. , 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 100: 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. E. F., Kadonaga J. T., 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16: 2583–2592. [DOI] [PubMed] [Google Scholar]
- Cernilogar F. M., Onorati M. C., Kothe G. O., Burroughs A. M., Parsi K. M., et al. , 2011. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Dahlstrom J. E., Lee S.-H., Rangasamy D., 2012. Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. Epigenetics 7: 758–771. [DOI] [PubMed] [Google Scholar]
- Chung W.-J., Okamura K., Martin R., Lai E. C., 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 18: 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R., Batzer M. A., 2009. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J., Lis J. T., 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J., Gilchrist D. A., Fargo D. C., Kwak H., et al. , 2012. Defining the status of RNA polymerase at promoters. Cell Reports 2: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshanks H. A., Tufarelli C., 2009. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics 94: 397–406. [DOI] [PubMed] [Google Scholar]
- Czech B., Malone C. D., Zhou R., Stark A., Schlingeheyde C., et al. , 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O. N., Traverse K. L., Hogan N. C., DeBaryshe P. G., Pardue M. L., 1999. The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol. Cell. Biol. 19: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P., 2011. Alu elements: know the SINEs. Genome Biol. 12: 236–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos G., A. J. Schroeder, J. L. Goodman, V. B. Strelets, M. A. Crosby et al, 2015 FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43: D690–D697. [DOI] [PMC free article] [PubMed]
- Dower K., Kuperwasser N., Merrikh H., Rosbash M., 2004. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA 10: 1888–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M., Seitz H., Horwich M. D., Li C., Du T., et al. , 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogvadze E., Buzdin A., 2009. Retroelements and their impact on genome evolution and functioning. Cell. Mol. Life Sci. 66: 3727–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant G. R., Farkas M. H., Pizarro A. D., Lahens N. F., Schug J., et al. , 2011. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics 27: 2518–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S., Esteller M., 2012. Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 19: 1068–1075. [DOI] [PubMed] [Google Scholar]
- Gullerova M., Proudfoot N. J., 2012. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat. Struct. Mol. Biol. 19: 1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L. S., Saito K., Nishida K. M., Miyoshi K., Kawamura Y., et al. , 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Henriques T., Gilchrist D. A., Nechaev S., Bern M., Muse G. W., et al. , 2013. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol. Cell 52: 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D., Moazed D., 2015. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. A., Yin H., Sweeney S., Raha D., Snyder M., et al. , 2013. A major epigenetic programming mechanism guided by piRNAs. Dev. Cell 24: 502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K. L., Elgin S. C. R., 2009 Small RNA directed heterochromatin formation in the context of development: what flies might learn from fission yeast. Biochimica et Biophysica Acta. 1789: 3–16. [DOI] [PMC free article] [PubMed]
- Junakovic N., Di Franco C., Best-Belpomme M., Echalier G., 1988. On the transposition of copia-like nomadic elements in cultured Drosophila cells. Chromosoma 97: 212–218. [DOI] [PubMed] [Google Scholar]
- Kaminker J. S., Bergman C. M., Kronmiller B., Carlson J., Svirskas R., et al. , 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y., Saito K., Kin T., Ono Y., Asai K., et al. , 2008. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453: 793–797. [DOI] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., et al. , 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M. G., Kidwell J. F., Sved J. A., 1977. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86: 813–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Nolte V., Schlötterer C., 2015. Tempo and mode of transposable element activity in Drosophila. PLoS Genet. 11: e1005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M., Pilpel Y., 2006. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 7: 1216–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A., Rogers A. K., Webster A., Marinov G. K., Liao S. E., et al. , 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 27: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-C., Marx C. J., 2013. Synchronous waves of failed soft sweeps in the laboratory: remarkably rampant clonal interference of alleles at a single locus. Genetics 193: 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E., Rizzon C., Biémont C., 2003. Sequence divergence within transposable element families in the Drosophila melanogaster genome. Genome Res. 13: 1889–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Kannan M., Trivett A. L., Liao H., Wu X., et al. , 2014. An antisense promoter in mouse L1 retrotransposon open reading frame-1 initiates expression of diverse fusion transcripts and limits retrotransposition. Nucleic Acids Res. 42: 4546–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonhaute C., Ogereau D., Hua-Van A., Capy P., 2007. Amplification of the 1731 LTR retrotransposon in Drosophila melanogaster cultured cells: origin of neocopies and impact on the genome. Gene 393: 116–126. [DOI] [PubMed] [Google Scholar]
- Marques J. T., Kim K., Wu P.-H., Alleyne T. M., Jafari N., et al. , 2010. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 17: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., 2011 Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17: 10–12.
- Mätlik K., Redik K., Speek M., 2006. L1 antisense promoter drives tissue-specific transcription of human genes. J. Biomed. Biotechnol. 2006: 71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1950. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 36: 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R. E., Bennett E. A., Iskow R. C., Devine S. E., 2007. Which transposable elements are active in the human genome? Trends Genet. 23: 183–191. [DOI] [PubMed] [Google Scholar]
- Mizrokhi L. J., Georgieva S. G., Ilyin Y. V., 1988. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell 54: 685–691. [DOI] [PubMed] [Google Scholar]
- Nechaev S., Fargo D. C., G. dos Santos, L. Liu, Y. Gao et al 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigumann P., Redik K., Mätlik K., Speek M., 2002. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79: 628–634. [DOI] [PubMed] [Google Scholar]
- Okamura K., Balla S., Martin R., Liu N., Lai E. C., 2008a Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat. Struct. Mol. Biol. 15: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Chung W.-J., Ruby J. G., Guo H., Bartel D. P., et al. , 2008b The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Steinmetz L. M., 2013. Gene regulation by antisense transcription. Nat. Rev. Genet. 14: 880–893. [DOI] [PubMed] [Google Scholar]
- Picard G., Bregliano J. C., Bucheton A., Lavige J. M., Pélisson A., et al. , 1978. Non-mendelian female sterility and hybrid dysgenesis in Drosophila melanogaster. Genet. Res. 32: 275–287. [DOI] [PubMed] [Google Scholar]
- Potter S. S., Brorein W. J., Dunsmuir P., Rubin G. M., 1979. Transposition of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell 17: 415–427. [DOI] [PubMed] [Google Scholar]
- Purcell M. K., Hart S. A., Kurath G., Winton J. R., 2006. Strand-specific, real-time RT-PCR assays for quantification of genomic and positive-sense RNAs of the fish rhabdovirus, Infectious hematopoietic necrosis virus. J. Virol. Methods 132: 18–24. [DOI] [PubMed] [Google Scholar]
- Rozhkov N. V., Hammell M., Hannon G. J., 2013. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 27: 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Kidwell M. G., Bingham P. M., 1982. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell 29: 987–994. [DOI] [PubMed] [Google Scholar]
- Saito K., Siomi M. C., 2010. Small RNA-mediated quiescence of transposable elements in animals. Dev. Cell 19: 687–697. [DOI] [PubMed] [Google Scholar]
- Saito K., Nishida K. M., Mori T., Kawamura Y., Miyoshi K., et al. , 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20: 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I., 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27: 353–365. [PubMed] [Google Scholar]
- Seitz H., Ghildiyal M., Zamore P. D., 2008. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA* strands in flies. Curr. Biol. 18: 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentmanat M. F., Elgin S. C. R., 2012. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proc. Natl. Acad. Sci. USA 109: 14104–14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G., Dönertas D., Brennecke J., 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M. C., Saito K., Siomi H., 2008. How selfish retrotransposons are silenced in Drosophila germline and somatic cells. FEBS Lett. 582: 2473–2478. [DOI] [PubMed] [Google Scholar]
- Speek M., 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21: 1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. D., Steiniger M., Marzluff W. F., 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell 34: 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak S. T., Pickeral O. K., Makalowski W., Boguski M. S., Landsman D., et al. , 2002. Molecular archeology of L1 insertions in the human genome. Genome Biol. 3: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchurikov N. A., Ilyin Y. V., Skryabin K. G., Ananiev E. V., Bayev A. A., et al. , 1981. General properties of mobile dispersed genetic elements in Drosophila melanogaster. Cold Spring Harb. Symp. Quant. Biol. 45(Pt 2): 655–665. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Zamore P. D., 2005. Perspective: machines for RNAi. Genes Dev. 19: 517–529. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Du T., Zamore P. D., 2007. Sorting of Drosophila small silencing RNAs. Cell 130: 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V. V., Sigova A., Li C., Seitz H., Gvozdev V., et al. , 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. [DOI] [PubMed] [Google Scholar]
- Vashist S., Urena L., Goodfellow I., 2012. Development of a strand specific real-time RT-qPCR assay for the detection and quantitation of murine norovirus RNA. J. Virol. Methods 184: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I. S., et al. , 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wang S. H., Elgin S. C. R., 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. USA 108: 21164–21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Mohammed J., Bortolamiol-Becet D., Tsai H., Robine N., et al. , 2014. Diversity of miRNAs, siRNAs, and piRNAs across 25 Drosophila cell lines. Genome Res. 24: 1236–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Donohue R. C., Birchler J. A., 2013. Quantitatively increased somatic transposition of transposable elements in Drosophila strains compromised for RNAi. PLoS One 8: e72163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Kazazian H. H., 2006. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13: 763–771. [DOI] [PubMed] [Google Scholar]
- Yu R., Jih G., Iglesias N., Moazed D., 2014. Determinants of heterochromatic siRNA biogenesis and function. Mol. Cell 53: 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Gene expression data are available at GEO with the accession number: GSE67725