Abstract
Neural tube defects including spina bifida are common and severe congenital disorders. In mice, mutations in more than 200 genes can result in neural tube defects. We hypothesized that this large gene set might include genes whose homologs contribute to morphogenesis in diverse animals. To test this hypothesis, we screened a set of Caenorhabditis elegans homologs for roles in gastrulation, a topologically similar process to vertebrate neural tube closure. Both C. elegans gastrulation and vertebrate neural tube closure involve the internalization of surface cells, requiring tissue-specific gene regulation, actomyosin-driven apical constriction, and establishment and maintenance of adhesions between specific cells. Our screen identified several neural tube defect gene homologs that are required for gastrulation in C. elegans, including the transcription factor sptf-3. Disruption of sptf-3 in C. elegans reduced the expression of early endodermally expressed genes as well as genes expressed in other early cell lineages, establishing sptf-3 as a key contributor to multiple well-studied C. elegans cell fate specification pathways. We also identified members of the actin regulatory WAVE complex (wve-1, gex-2, gex-3, abi-1, and nuo-3a). Disruption of WAVE complex members reduced the narrowing of endodermal cells’ apical surfaces. Although WAVE complex members are expressed broadly in C. elegans, we found that expression of a vertebrate WAVE complex member, nckap1, is enriched in the developing neural tube of Xenopus. We show that nckap1 contributes to neural tube closure in Xenopus. This work identifies in vivo roles for homologs of mammalian neural tube defect genes in two manipulable genetic model systems.
Keywords: Morphogenesis, gastrulation, C. elegans
DURING development, an embryo transitions from a cluster of cells to a well-organized body plan in a process known as morphogenesis. One important morphogenetic process is the internalization of cells and tissues from the surface of an embryo. Cells that give rise to internal tissues, such as the intestine and neural tube, are initially borne on the outside of the embryo and undergo a series of cell shape changes, rearrangements, and migrations that move cells to their final positions within the body.
Although diverse cell types undergo internalization, the mechanisms driving these movements are directed in part by a common set of cell behaviors and molecular mechanisms (Solnica-Krezel and Eaton 2003; Rauzi and Lecuit 2009; Sawyer et al. 2010). For example, during neural tube formation in vertebrates, the surface neuroepithelium becomes internalized and folds into a closed tube (Figure 1). The initial bending of the surface neuroepithelium into the body depends on apical constriction (reviewed in Schoenwolf and Smith 1990). Apical constriction, which has been observed in diverse animal systems, occurs when the contraction of a cell’s apical actomyosin network results in shrinkage of the apical side (reviewed in Sawyer et al. 2010). In vertebrate neuroepithelia, this cell shape change is partially responsible for bending of the neuroepithelium and its resulting invagination into the body. In parallel, cell rearrangements and cell migration act in a coordinated fashion and contribute to formation of the closed neural tube (reviewed in Wallingford et al. 2013).
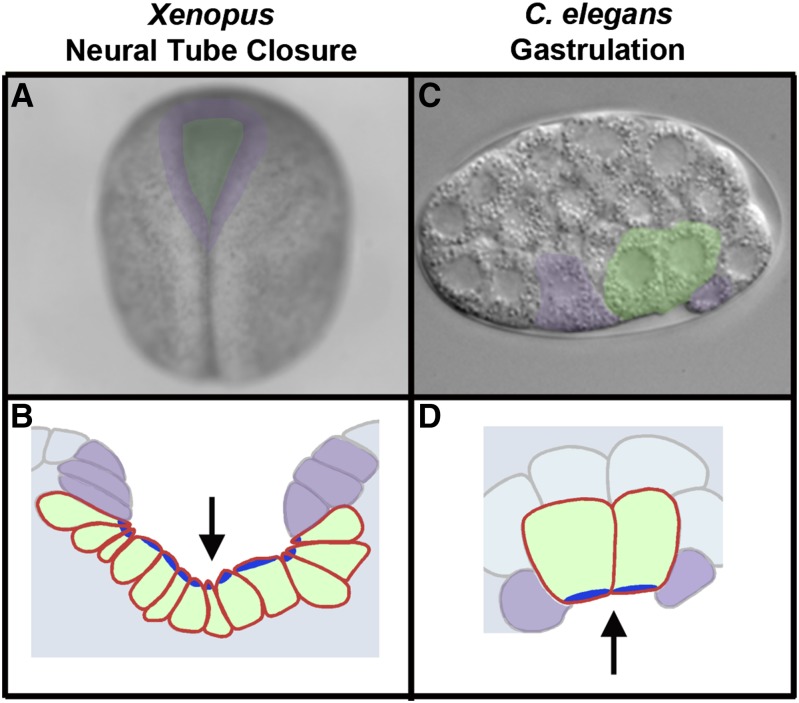
Figure 1.
Tissue internalization during Xenopus neural tube closure and cell internalization during C. elegans gastrulation. (A) Dorsal view of a Xenopus embryo undergoing neural tube closure, with anterior at the top. (B) Tracing of neural tube closure in a cross section (after Lee et al. 2007). Cells are pseudocolored to mark the neuroepithelium (green) and surrounding neural folds (purple); arrow marks the apical side. (C) C. elegans embryo in early gastrulation, lateral view with anterior to the left. (D) Tracing of a C. elegans embryo with cells pseudocolored to mark the internalizing E cells (green) and neighboring cells (purple); arrow marks the apical side. Dark blue marks sites of actomyosin-driven apical constriction [see Sawyer et al. (2010) for review].
Genetic studies in the mouse have identified over 200 genes with roles in neural tube closure; however, the functions of most of these neural tube defect (NTD) genes in neural tube closure are not well understood (Harris and Juriloff 2007, 2010). Notable exceptions include Shroom3, which encodes an actin binding protein that recruits a myosin activator to specific sites in cells (Hildebrand and Soriano 1999; Haigo et al. 2003; Hildebrand 2005; Nishimura and Takeichi 2008), and planar cell polarity genes, which encode proteins with roles in cell rearrangements and apical constriction (reviewed in Muñoz-Soriano et al. 2012; Nishimura et al. 2012; Ossipova et al. 2014).
Given the large number of genes with poorly understood roles in vertebrate neural tube closure, we postulated that some of these genes might have homologs with conserved roles in other model systems. We took a screening approach to identify NTD gene homologs with roles in morphogenesis in other systems. Such an approach has the potential to identify human birth defect genes with in vivo roles that can be studied efficiently in nonmammalian model systems, as well as to identify candidates for core mechanisms of morphogenesis that might be shared by multiple animal systems. We probed NTD gene homologs in the roundworm Caenorhabditis elegans, a powerful model system for genetics and live imaging studies (Hodgkin 2005; Nance et al. 2005). C. elegans does not produce a neural tube, but many cells internalize by apical constriction during embryonic development (Nance et al. 2005; Harrell and Goldstein 2011). In particular, gastrulation—the internalization of endodermal, mesodermal, germline, and sometimes other tissues from an embryo’s surface—involves internalization of scores of cells by apical constriction (Nance et al. 2005; Harrell and Goldstein 2011).
We focused on an early event, internalization of endoderm cells during gastrulation. In C. elegans, two endoderm lineage cells (called Ea and Ep, for the endodermal founder cell’s anterior and posterior daughters, and referred to here for convenience as E cells) are borne on the outside of the embryo at the 26-cell stage (Nance et al. 2005). These two cells undergo apical constriction, much as apical constriction contributes to vertebrate neurulation (Figure 1). As in vertebrate neurulation, apical constriction in the E cells requires factors driving cell polarization, cytoskeletal factors regulating myosin localization and activation, and cell adhesion molecules (Nance and Priess 2002; Severson et al. 2002; Lee and Goldstein 2003; Nance et al. 2003; Anderson et al. 2008; Roh-Johnson and Goldstein 2009; Grana et al. 2010; Roh-Johnson et al. 2012). Once internalized, the E cells undergo multiple divisions, giving rise to the entire intestine.
Here we report the screening of a large set of C. elegans NTD gene homologs for roles in gastrulation using a two-step RNA interference (RNAi) enhancer screen developed previously (Sawyer et al. 2011). Our screen succeeded in identifying four NTD homologs with roles in C. elegans gastrulation. One of these genes, sptf-3, encodes a transcription factor with homology to the Sp family of transcription factors (Ulm et al. 2011; Hirose and Horvitz 2013). We found that sptf-3 is required for normal expression of genes involved in endodermal lineage specification. Our screen also identified two members of the WAVE complex of actin regulators, abi-1/Abi1 and gex-3/Nckap1. The requirement for these genes appears to reflect a requirement for the WAVE complex more generally because we found that all five C. elegans genes encoding members of the WAVE complex (wve-1/Wave1, gex-2/Sra1, gex-3/Nckap1, abi-1/Abi1, and nuo-3a/Brick1) have roles in C. elegans gastrulation. Although WAVE complex members appear to be expressed in all or most tissues in C. elegans embryos (Patel et al. 2008), we show that in the vertebrate Xenopus, nckap1 messenger RNA (mRNA) is enriched in the developing neural tube. We report that as in mouse (Rakeman and Anderson 2006; Yokota et al. 2007), neural tube closure in Xenopus depends on Nckap1/Nap1. Our results identify in vivo roles for homologs of mammalian NTD genes in two model systems, forming a platform for future work on mechanistic in vivo roles for these genes.
Materials and Methods
C. elegans strains and worm maintenance
C. elegans were cultured and handled as described in Brenner (1974). Experiments were performed using the following strains: wild-type Bristol N2, LP77 end-3(ok1448); ced-5(n1812) (Sawyer et al. 2011), JJ1317 zuIs3 [end-1::GFP], JR2274 wIs137 [rol-6;end-3::END-3[P202L]::GFP], MS126 irIs16[tbx-35::NLS::GFP; unc-119(+)], JJ1531 zuIs70 [end-1::GFP::CAAX;unc-119(+)] (Wehman et al. 2011), end-1(ok558), end-3(ok1448), LP54 PH::mCherry; NMY-2::GFP, MT2547 ced-4(n1162), MT2551 ced-4(n1162), dpy-17(e164), MT5287 ced-4(n1894), MT5816 ced-4(n2273), MT5851 ced-4(n2274), WM43 gex-3(zu196), OX232 wve-1(zu469) unc-101(m1)/hin1 (gift from the Soto laboratory), OX309 wve-1(zu469) unc-101(m1)/hin1 (gift from the Soto laboratory), VC1180 gex-2(ok1603)/dpy-9(e12), LP163 unc-119(ed3) III; nuo-3(cp14[nuo-3aΔ + unc-119(+)]) IV/nT1[qIs51], LP418 sptf-3(cp154[mNG^SEC^3xFlag::sptf-3]) I, LP419 spft-3(cp155[mNG^3xFlag::sptf-3]) I, and LP362 gex-3(cp114[mNG^3xFlag::gex-3]) IV (Dickinson et al. 2015).
RNAi screening and quantification of embryonic lethality
Primary screen: RNAi-by-feeding:
RNAi-by-feeding was performed by a protocol outlined previously (Sawyer et al. 2011) with minor modifications. Three to five L4 larvae from a wild-type background or LP77 end-3(ok1448); ced-5(n1812) gastrulation-sensitized background were placed on RNAi plates seeded with double-stranded RNA (dsRNA)–producing bacterial strains (Timmons and Fire 1998; Kamath et al. 2001). Bacterial feeding strains were obtained from a dsRNA feeding library from the Medical Research Council (MRC) Geneservice (Kamath and Ahringer 2003). Bacterial clones were confirmed using the sequencing primer L4440F (5′-GAGTGAGCTGATACCGC-3′). Worms were transferred to fresh plates approximately every 12 hr. After ∼48 hr, worms were removed from the plates. The F1 embryos and larvae were counted at least 24 hr after the embryos were laid, and embryonic lethality (percent of embryos that failed to hatch) was determined. dsRNA-targeting GFP was used to determine embryonic lethality in the wild-type and LP77 end-3(ok1448); ced-5(n1812) gastrulation-sensitized backgrounds. Enhancement of lethality for each gene tested was determined by subtracting the percent embryonic lethality (percent of embryos that failed to hatch) of the LP77 end-3(ok1448); ced-5(n1812) gastrulation-sensitized background from the wild-type background. We used 1 SD above the mean of lethality in the LP77 end-3(ok1448); ced-5(n1812) gastrulation-sensitized background, which was 8.43%, as a threshold for defining enhancers: any gene whose dsRNA produced lethality in the gastrulation-sensitized background of more than 8.43% above the lethality produced by the same dsRNA in the wild-type background was considered an enhancer and was carried forward to the secondary screen. All RNAi-by-feeding experiments were performed at 20°.
Secondary screen: RNAi-by-injection:
PCR templates for in vitro transcription of dsRNA were generated from a plasmid isolated either from the bacterial feeding library or from complementary DNA (cDNA) isolated from embryos (SuperScript III First-Strand Synthesis System, Life Technologies). For plasmid PCR, we used a primer recognizing the T7 promoter sequence; for cDNA PCR, we used a two-step PCR strategy outlined previously (Sawyer et al. 2011). All PCR products were gel purified and used as a template in an in vitro transcription reaction (T7 RiboMAX Express RNAi System, Promega) according to the manufacturer’s instructions. dsRNA integrity was assessed by gel electrophoresis, and dsRNA concentration was determined by spectrophotometry. dsRNA was injected at a concentration of 1 μg/μl into adult hermaphrodites using a Narishige injection apparatus and a Parker Instruments Picospritzer II mounted on a Nikon Eclipse TE300 microscope. dsRNA was stored in one volume of 100% ethanol at −80°.
Genome engineering
The nuo-3 gene produces an mRNA that encodes two distinct polypeptides. The 5′ nuo-3a ORF (Wormbase accession number Y57G11C.12a) encodes a BRICK homolog, while the 3′ nuo-3b ORF (Wormbase accession number Y57G11C.12b) encodes an NADH-ubiquinone oxidoreductase α-subunit. We generated knockouts of nuo-3a alone by Cas9-triggered homologous recombination (Dickinson et al. 2013). We constructed a homologous repair template in which the 727 bp of intergenic sequence between nuo-3 and the next upstream gene, arl-8, was fused directly to the nuo-3b ORF such that the nuo-3a ORF at the 5′ end of the nuo-3 transcript was deleted, but nuo-3b was still under the control of the native nuo-3 promoter. The unc-119(+) selectable marker was inserted upstream of the 727-bp arl-8/nuo-3 intergenic region. To avoid disrupting arl-8 expression, a second copy of the arl-8/nuo-3 intergenic region was placed upstream of unc-119(+). Finally, the repair template plasmid contained 2.0 and 1.7 kb of unmodified sequence for homologous recombination at the 5′ and 3′ ends of the construct, respectively. The complete sequence of this plasmid is available on request. The Cas9 expression plasmid pDD162 (Dickinson et al. 2013) was modified by insertion of the guide sequence 5′-tcctttagttgttgttgaa-3′. The Cas9 plasmid, repair template, and co-injection markers were prepared and injected into young adults of strain HT1593 unc-119(ed3) III as described previously (Dickinson et al. 2013). We isolated four independent alleles with deletions of nuo-3a. All four alleles caused a maternal-effect lethal phenotype as homozygotes, consistent with the known phenotypes of other WAVE complex subunits (Patel et al.2008). To facilitate strain maintenance, these alleles were placed over the GFP-marked balancer nT1[qIs51] (IV; V). Using RT-PCR, we confirmed that nuo-3b mRNA was still expressed in embryos homozygous for the nuo-3a deletion (Supporting Information, Figure S2).
To modify the sptf-3 locus, we inserted mNeonGreen^SEC^3xFlag at the 5′ end of the coding region, following a published procedure (Dickinson et al. 2015). Briefly, we generated a Cas9-sgRNA expression construct by inserting the guide sequence 5′-gctgttgctgttgcatttgt-3′ into pDD162. In parallel, we cloned homology arms into the mNG-SEC vector pDD268. The 5′ homology arm comprised 642 bp upstream of the SPTF-3 start codon, and the 3′ homology arm comprised the first 560 bp of the SPTF-3 coding region (including the first exon and part of the first intron). A silent c→t mutation was introduced at nucleotide residue 33 of the SPTF-3 coding region to prevent cleavage of the repair template by Cas9. The complete sequence of the repair template plasmid is available on request. The Cas9 plasmid and repair template were injected into young adults of strain N2, and knock-in animals were isolated by selecting on Hygromycin. We found that the mNG^SEC^3xFlag::sptf-3 insertion caused a maternal-effect lethal phenotype, so the insertion allele was maintained in heterozygotes. To isolate mNG^SEC^3xFlag::sptf-3 homozygotes (which produce embryos lacking sptf-3 expression), we singled L4 rollers and checked for the presence of viable progeny a day later. Animals that had laid only dead eggs were dissected and their embryos used for RNA sequencing (RNA-seq). To isolate an mNG^3xFlag::sptf-3 protein fusion, the L1/L2 offspring of animals heterozygous for mNG^SEC^3xFlag::sptf-3 were heat shocked at 32° for 5 hr. Five days after heat shock, marker-excised animals were identified by the combination of a wild-type phenotype and mNG fluorescence. We found that two of nine filmed mNG^3xFlag::sptf-3 embryos were gastrulation defective, consistent with partial rescue and suggesting that the fusion protein does not fully rescue function.
Microscopy, image quantification, mRNA quantification
Differential interference contrast (DIC) imaging:
C. elegans embryos were mounted on poly-l-lysine-coated coverslips at the one- to four-cell stage mounted on a 2.5% agarose pad with egg buffer (as in Sawyer et al. 2011). Four-dimensional (4D) DIC video microscopy was performed on a Nikon Eclipse 800 microscope using a Diagnostic Instruments SPOT2 camera. Images were acquired with a 60× oil objective every 60–90 sec at 1-μm optical sections. Images were analyzed with MetaMorph software (MetaMorph, Inc.).
Fluorescence microscopy:
The spinning-disk confocal images of JJ1531 zuIs70; JJ1317 zuIs3 [spe-26(+)/end-1::GFP], JR2274 wIs137 [end-3::END-3(P202L)-GFP; rol-6], MS126 unc-119(ed4) III; irIs16 [tbx-35::NLS::GFP]; and Xenopus membrane (CAAX)::GFP RNA were acquired on a Nikon Eclipse Ti-E microscope equipped with a CSU-X1 Yokogawa head and a Hamamatsu ImagEM EMCCD camera. The Nikon Inverted Eclipse TE2000-U microscrope equipped with a Yokogawa Spinning Disk Confocal Scanner Unit CSU10 and a Spot Insight 2-megapixel camera was used to acquire images of the LP54 PH::mCherry; NMY-2::GFP strain.
Quantification of expression from transgenes:
Embryos from the following backgrounds were analyzed as controls or injected with sptf-3 dsRNA: JJ1317 zuIs3 [spe-26(+)/end-1::GFP], JR2274 wIs137 [end-3::END-3(P202L)-GFP; rol-6], and MS126 unc-119(ed4) III; irIs16 [tbx-35::NLS::GFP]. Embryos at the two- to four-cell stage were mounted on agarose pads 24–30 hr after injection and filmed by DIC microscopy until near the time of P4 cell division. Images were acquired once every 3 min at 4-μm optical sections through 20 μm total with the 60× oil objective. A 488-nm laser was used at 15% power for imaging end-1::GFP and end-3::END-3-GFP embryos and at 25% power for tbx-35::GFP embryos (exposure times 200 msec). Fluorescence intensities were calculated by subtracting the average pixel values of off-embryo background from the average pixel values of E cell nuclei, with pixel values measured using MetaMorph software.
RNA-seq and analysis:
Four replicates of five to six embryos were collected at the 8AB stage for each genotype: N2 and sptf-3(cp155). For each replicate, we weakened eggshells with chitinase (Edgar and Goldstein 2012) and lightly disrupted them mechanically by aspiration. We flash froze these embryos in liquid nitrogen and stored them at −80°. We prepared cDNA (Clontech SMARTer Legacy Kit) and RNA-seq libraries (Nextera XT Kit) according to the manufacturers’ guidelines. We multiplexed these libraries and sequenced them on a single HiSequation 2500 lane for 50 cycles from a single end.
Identical reads were collapsed using fastx_collapser (FASTX-Toolkit v. 0.0.13.2; http://hannonlab.cshl.edu/fastx_toolkit/) and aligned to the ce10 genome using TopHat2 v. 2.0.11 (Kim et al. 2013). Count files were generated using HTseq-count-0.5.3p3 (Anders et al. 2015) and a WBcel235.78.gtf reference file (http://ftp.ensembl.org/pub/release-78/gtf/caenorhabditis_elegans/). To analyze lineage-restricted genes that in our results were robustly expressed without excessive variation between replicates, we included in our analysis all genes whose mRNAs had (1) lineage-restricted expression in Supplementary Table 1 in Hashimshony et al. (2015), (2) a minimal mean of 10 RPKM in our results, and (3) total RPKM values that were greater than total 95% confidence intervals between replicates from our results.
NMY-2::GFP accumulation and apical membrane length analysis:
For NMY-2::GFP accumulation analysis, LP54 PH::mCherry; NMY-2::GFP embryos and gex-3(RNAi); LP54 embryos were analyzed as described previously (Roh-Johnson and Goldstein 2009). Images were acquired once every 60 sec at 1.5-μm optical sections (10 μm total).
For analysis of apical membrane lengths, JJ1531 zuIs70 [pJN152: end-1::gap::caax;unc-119(+)] (provided by J. Nance) embryos and gex-3(RNAi); zuIs70 embryos were mounted on agarose pads at the two- to four-cell stage 30–36 hr after injection and filmed by DIC microscopy until near the time of P4 cell division. Images were acquired once every 90 sec at 2-μm optical sections (10 μm total) with the 60× objective. The 488-nm laser was used at 25% power (exposure time 200 msec). Apical membranes lengths were measured as described previously (Roh-Johnson and Goldstein 2009). Statistical tests were determined using GraphPad Prism software (GraphPad Software, Inc.) employing an unpaired t-test with Welch’s correction. In cases where the Ep apical membranes extended around the posterior boundary of P4 (as was often the case in gex-3(RNAi) embryos), the length of the apical membrane in contact with eggshell was determined.
Movies of membranes blebs were filmed with LP54 PH::mCherry; NMY-2::GFP (uninjected or wve-1(RNAi)) embryos mounted in a ventral position. Images were acquired every 3 sec at 1-μm optical sections (through 5–8 μm total).
Contact length ratios:
P1 cells were isolated from wild-type and WM43 gex-3(zu196) embryos at the two-cell stage using the same method of isolation described in Werts et al. (2011). The cells were mounted in Shelton’s growth medium (Shelton and Bowerman 1996) either as P1 cells or, after one division, as P2-EMS cell pairs. Cells were imaged using 4D DIC video microscopy on a Nikon Eclipse 800 microscope with a Diagnostic Instruments SPOT2 camera. The contact length ratios are represented as contact length/radius, as in Grana et al. (2010). Measurements were taken using Image J.
Xenopus laevis (Xenopus) manipulations
In situ hybridization:
Xenopus embryos were staged according to Nieuwkoop and Faber (1967). Whole-mount in situ hybridization was carried out as described by Harland (1991). The probe for nckap1 in situ hybridization was generated from nckap1; IMAGE clone ID: 4970729.
Xenopus plasma membrane imaging experiments:
For imaging membranes, Xenopus embryos were injected at the one-cell stage with 100 ng of membrane (CAAX)::GFP RNA or with 100 ng of membrane (CAAX)::GFP RNA together with 45 ng of Nckap1 MO1. The pCS2_mem(CAAX)::GFP plasmid, kindly provided by the Wallingford laboratory, was digested with Not1 and used as the template for RNA preparation. RNA was prepared by in vitro transcription of CAAX::GFP using the Ambion mMessage Kit with the Sp6 polymerase. Neurula-stage embryos (stage 16) were mounted in glass-bottom dishes in 2% agarose submerged in 0.1× MBS buffer and oriented using eyebrow hairs glued to toothpicks. Images were acquired once every 2 min at 10-μm optical sections with the 10× objective and a 1.5× magnifying lens using the Nikon Eclipse Ti-E microscope. The 488-nm laser was used at 50% power (exposure time 150 msec).
Morpholino injections:
Two nonoverlapping translation-blocking morpholinos (MOs) were designed against the start site of nckap1 and upstream 5′ UTR region (Gene Tools) (Figure S6 shows MO target sequences). Xenopus embryos were injected with the following range of Nckap1 MO concentrations: control mismatch MO (43–45 ng), MO1 (43–45 ng for NTD phenotype; 25–30 ng for suboptimal dosage), and MO2 (25–30 ng for NTD phenotype; 20 ng for suboptimal dosage). MO injections were performed as described previously (Tandon et al. 2012).
Western blotting:
The 166-bp 5′ UTR of Xenopus nckap1 was cloned together with 123 bp of 5′ cds (41 aa) from stage 25 whole-embryo cDNA using the iProof High Fidelity PCR Kit (Bio-Rad) with 45° annealing temperature and 10-sec extension. This PCR product (289 bp) was purified (QIAquick PCR Purification Kit, Qiagen) and digested with HindIII and BamHI restriction enzymes (NEB) for insertion into the pEGFP-N1 vector (Clontech). This allowed the fusion of eGFP in frame at the 3′ end of the PCR product. This nckap1 5′ UTR-GFP fusion (1023 bp) was then digested from the vector using XhoI and NotI restriction enzymes and ligated into the modified pSP64TxB vector (with β-globin UTR elements). The new vector was linearized (XmaI), purified, and transcribed to make capped mRNA for injections (mMessage mMachine SP6 Transcription Kit, Invitrogen). Fertilized Xenopus embryos were injected with a range of Nckap1 MO1, MO2, and control MO concentrations and then subsequently injected with 1 ng of nckap1 5′ UTR-GFP RNA. Embryos were cultured in 0.1× MBS solution at 16° until stage 11, when they were then resuspended in lysis buffer for protein analysis, as performed previously (Tandon et al. 2012, 2013; Sojka et al. 2014). Western blots were probed with anti-mouse-JL8 GFP (Clontech, 632381, 1:10,000 dilution) to ascertain blocking of GFP-RNA translation by the MO and anti-mouse-SHP2 (BD Transduction, 610622, 1:2500 dilution) as a loading control, followed by anti-mouse HRP (Jackson, 1:10,000) and ECL substrate to develop the blot.
Data availability
Strains are available upon request. Table S1 contains information about the genes we identified as C. elegans homologs to mammalian neural tube defect genes and includes accession numbers.
Results
Identifying NTD gene homologs in C. elegans
Mammalian NTD genes were compiled using a previously generated list of NTD genes (Harris and Juriloff 2007, 2010), which we augmented by additional literature searches for mouse and human NTD genes. We also included a set of folic acid metabolism genes because mutations in some folic acid metabolism genes have been shown to affect neural tube closure in mouse embryos (Harris 2009). C. elegans homologs were identified by BLAST searches using protein sequences. C. elegans BLAST hits with an E-value score below a threshold (1 × 10−9) were considered candidate NTD gene homologs. In addition, four C. elegans genes (ced-4/Apaf-1, brc-1/Brc1, pfn-1/Profilin1, and unc-60/Cofilin1) with higher E-values above the threshold were considered in this study because they have been previously recognized as homologous genes (Table S1) (Zou et al. 1997; Ono and Benian 1998; Severson et al. 2002; Boulton et al. 2004). We performed a reciprocal best BLAST hit analysis using C. elegans protein sequences against the mouse or human genome. We categorized these results according to whether the corresponding genes were the best homolog or one of multiple genes with similar E-values. In some cases, we considered multiple C. elegans genes as potential NTD gene homologs in order to include in our screen genes likely to have diverged recently by gene duplication. In total, we searched 234 candidate mammalian NTD genes by BLAST against the C. elegans genome, including 3 human genes with known SNPs as risk factors for NTDs and 5 folic acid metabolism genes. We identified 191 C. elegans genes with homology to mammalian NTD genes (Table S1).
Identifying genes with roles in C. elegans embryonic development by an RNAi enhancer screen
Our laboratory previously developed a two-step screen as an efficient method to identify genes with roles in C. elegans gastrulation from among candidate genes (Sawyer et al. 2011), and we applied that screen here to the NTD homologs. In brief, we first used an RNAi-by-feeding approach to disrupt gene functions (Kamath et al. 2001; Kamath and Ahringer 2003), recording percent embryonic lethality in the wild-type and gastrulation-sensitized end-3(ok1448); ced-5(n1812) backgrounds. We used these results to identify genes in which the RNAi phenotype resulted in increased embryonic lethality in the gastrulation-sensitized background (Figure 2). For convenience, we refer to these genes as enhancers (see Materials and Methods for details). We next targeted the enhancers in wild-type worms using RNAi-by-injection because this method can induce a stronger loss of function than RNAi-by-feeding, filming embryonic development to directly identify genes that are required for gastrulation. We included in our primary screen 104 of the 191 NTD gene homologs, including genes that were available in an RNAi-by-feeding library and whose feeding clones we could confirm by sequencing (Table S2).
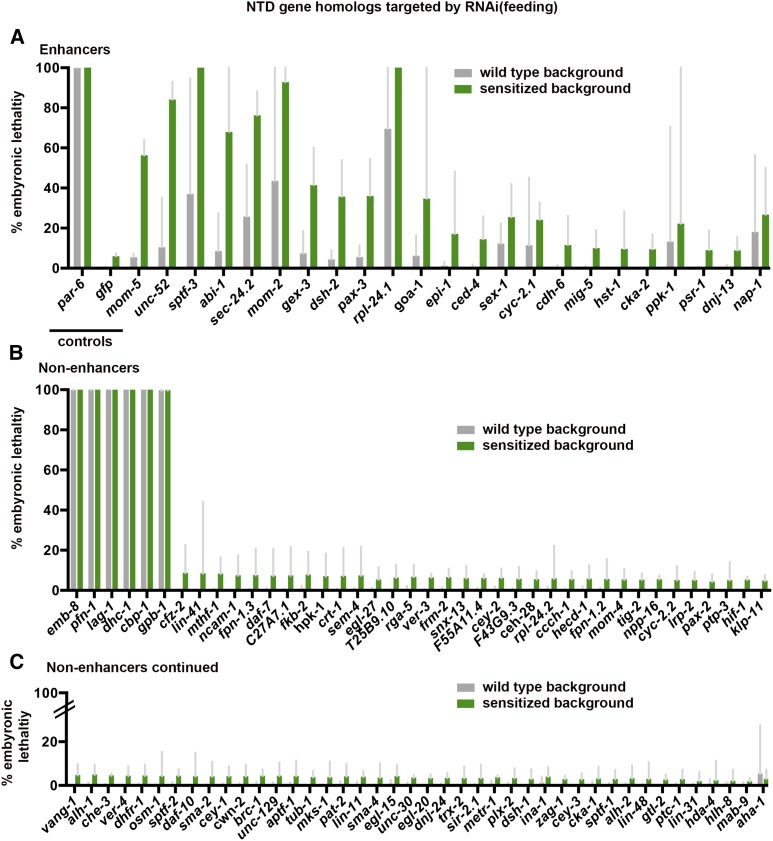
Figure 2.
Primary RNAi screen. dsRNAs targeting 104 NTD gene homologs were fed into wild-type and gastrulation-sensitized backgrounds. Shown are bar graphs indicating percent embryonic lethality in wild-type (gray) and gastrulation-sensitized backgrounds (green), performed in triplicate, ±95% confidence intervals. (A) Putative enhancers. RNAi targeting genes listed in this category resulted in increased embryonic lethality beyond a threshold in the gastrulation-sensitized background (see Materials and Methods). Controls: dsRNA targeting par-6, gfp, and mom-5. (B and C) Nonenhancers. RNAi targeting these genes resulted in 100% embryonic lethality in both backgrounds or below the threshold of enhanced embryonic lethality in the gastrulation-sensitized background.
For each of the 104 genes, RNAi-by-feeding was performed in triplicate in the wild-type and gastrulation-sensitized backgrounds. As controls for RNAi effectiveness in each trial, we used bacterial strains targeting par-6 (used as a positive control for effective RNAi, which gave an average of 99.88 ± 0.39 and 100.00 ± 0.00% embryonic lethality in the wild-type and gastrulation-sensitized backgrounds, respectively) and gfp (negative control, 0.40 ± 0.40 and 5.92 ± 2.5% embryonic lethality in the wild-type and gastrulation-sensitized backgrounds, respectively). We also included mom-5, a homolog of the Wnt receptor Frizzled, as a positive control for the effectiveness of the gastrulation-sensitized background (5.48 ± 3.1 and 56.33 ± 13.49% embryonic lethality in the wild-type and gastrulation-sensitized backgrounds, respectively). mom-5 was previously implicated in gastrulation (Lee et al. 2006), and dsRNAs targeting mom-5 were previously shown to enhance lethality in the gastrulation-sensitized strain (Sawyer et al. 2011).
Screening NTD gene homologs in the wild-type and gastrulation-sensitized backgrounds, we identified 22 NTD gene homologs as enhancers, which we considered as candidate C. elegans gastrulation genes (Figure 2 and Table S2). Among the 22 enhancers, we identified three Wnt pathway genes that were previously known to function in C. elegans gastrulation: mom-2/Wnt, dsh-2/Dsh, and mig-5/Dsh (Lee et al. 2006). We view this result as validating our approach as a method for identifying bona fide gastrulation genes among NTD gene homologs. We also identified a predicted heparan sulfotransferase gene, hst-1, that is predicted to function with Wnt and other signaling pathways (Yan and Lin 2009) and a well-studied G protein α-subunit gene with known embryonic roles, goa-1 (Bastiani and Mendel 2006). The functions of these proteins in gastrulation were not pursued further here.
Secondary screen for genes with roles in C. elegans gastrulation
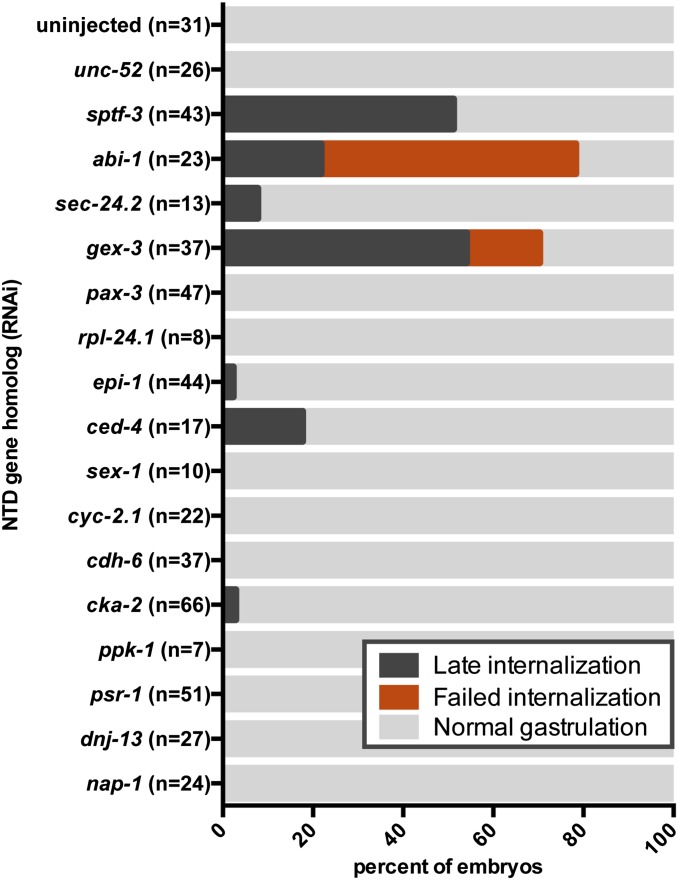
To determine which of the enhancer genes are required for gastrulation in C. elegans, we injected dsRNAs targeting each of the remaining 17 enhancers into wild-type adult worms and monitored the development of embryos produced at least 24 hr after injection. In our experience, injection of dsRNAs often results in penetrant phenotypes, phenocopying null mutants more often than does RNAi-by-feeding (preliminary unpublished observations). 4D DIC microscopy of living embryos was used to observe development at the 26- to 28-cell stage, looking specifically for gastrulation defects. A gastrulation defect was defined by the failure of the E cells to internalize before their division. Embryos were scored as gastrulation defective if one or both the E cells divided on the surface of the embryo (Figure 3 and Figure 4, asterisks). Among gastrulation-defective embryos, we further distinguished two phenotypic classes: late internalization (if Ea and/or Ep failed to internalize, but all four of their daughter cells internalized before dividing) and failed internalization (if some of the four daughter cells failed to internalize before dividing) (Figure 3 and Figure 4, asterisks). We found that RNAi targeting four genes (sptf-3, abi-1, gex-3, and ced-4) resulted in embryos consistently showing gastrulation defects (Figure 4 and File S1, File S2, File S3, File S4, and File S5). We also identified three genes, not considered further here, in which only 1 embryo of 13 or more analyzed showed a gastrulation-defective phenotype after RNAi: sec-24.2 (which also showed cytokinesis defects), epi-1, and cka-2 (Figure 3). Among the four genes consistently showing gastrulation defects, ced-4(RNAi) had the weakest effect on E cell internalization (Figure 3). We analyzed four different mutant alleles of ced-4, including the null allele ced-4(n1162) (Yuan and Horvitz 1992), and found similarly weak but consistent gastrulation defects (Figure S1). Taken together, these results identify four NTD gene homologs—sptf-3, abi-1, gex-3, and ced-4—as having direct or indirect roles in C. elegans gastrulation. Given the low penetrance of gastrulation defects after targeting ced-4, we did not consider it further here. Later we probe the basis for gastrulation defects in sptf-3(RNAi), abi-1(RNAi), and gex-3(RNAi) embryos.
Figure 3.
Secondary RNAi screen. dsRNAs targeting the enhancers identified in the primary screen were injected into wild-type animals. Embryos from each background were filmed by 4D DIC microscopy. Films were analyzed for internalization of the endoderm cells (see Figure 4 for examples of gastrulation defects indicated here).
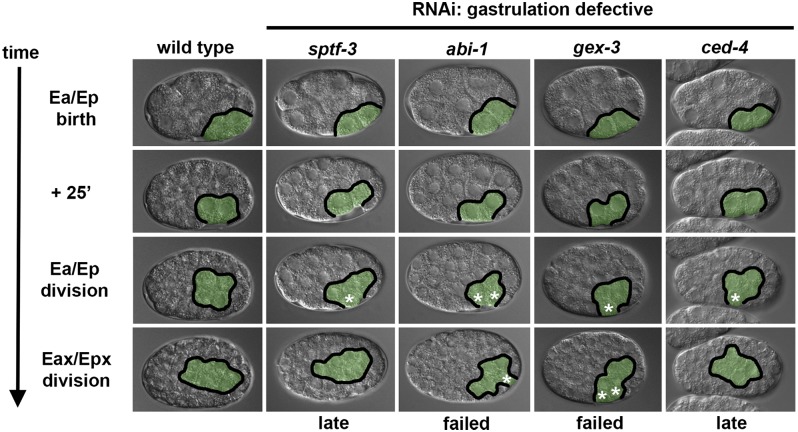
Figure 4.
Gastrulation-defective phenotypes in sptf-3(RNAi), abi-1(RNAi), gex-3(RNAi), and ced-4(RNAi) embryos. The endoderm precursors (pseudocolored in green) were internalized before Ea/Ep division in wild-type embryos and were exposed on the surface in gastrulation-defective embryos. In sptf-3(RNAi) and ced-4(RNAi) embryos, the endoderm cells generally failed to internalize by the time of Ea/Ep division and internalized before the next round of cell division (Eax/Epx division, where x represents both anterior and posterior daughters). In abi-1(RNAi) and gex-3(RNAi) embryos, the endoderm failed to complete internalization by Eax/Epx division in 56.5 and 16.2% of embryos, respectively (see Figure 3). White asterisks mark cells that remain on the embryo’s surface.
sptf-3 affects the length of E cell cycles and endoderm cell fate and encodes a protein that is enriched in nuclei during gastrulation
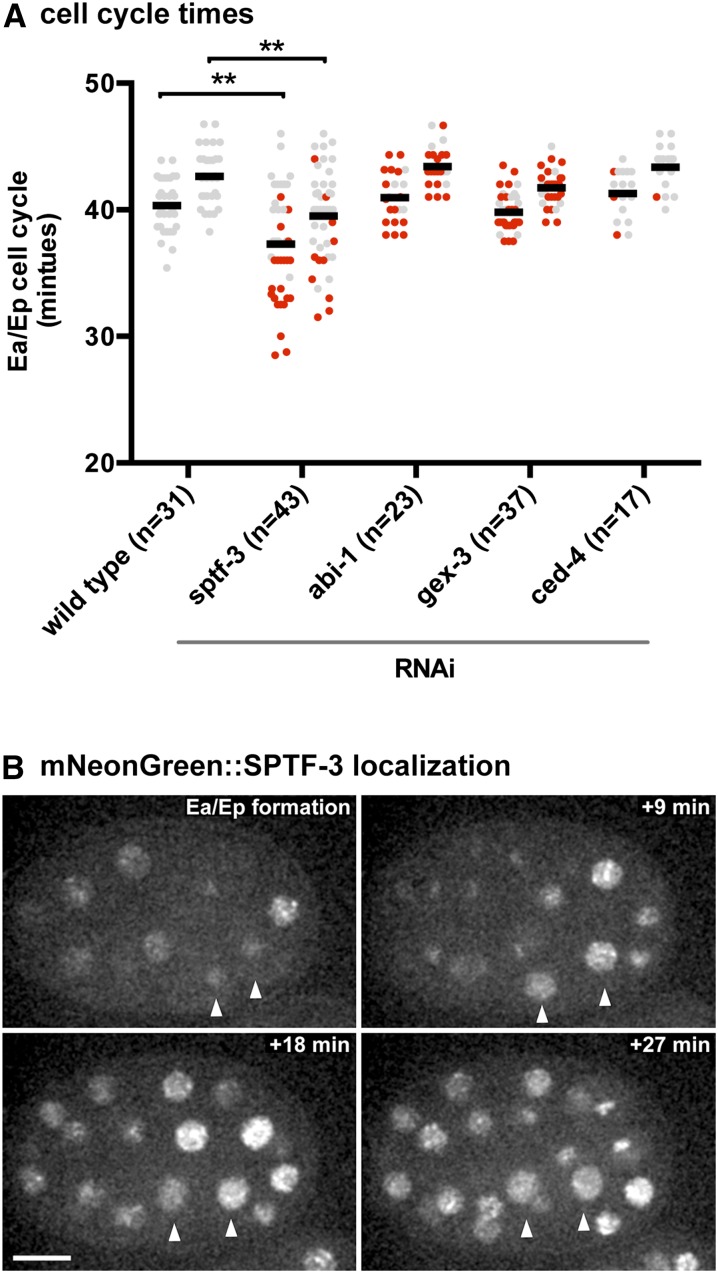
The E cells are the first cells in C. elegans embryonic development to introduce a gap phase to the cell cycle, resulting in an ∼20-min extension compared to their cousin MS cells (Edgar and McGhee 1988). In wild-type embryos, the cell cycle length of the Ea cell (40.33 ± 0.39 min) is slightly but consistently shorter than that of the Ep cell (42.64 ± 0.39 min), as observed previously (Boeck et al. 2011). To determine whether sptf-3, abi-1, or gex-3 regulates E cell cycle lengths, we injected dsRNAs targeting these genes. Targeting abi-1 or gex-3 by RNAi did not affect cell cycle lengths (P > 0.05) (Figure 5A) (ced-4 gave similar results). However, RNAi against sptf-3 reduced E cell cycle lengths significantly (P < 0.01; Ea, 37.28 ± 0.65 min; Ep, 39.51 ± 0.57 min). Most cells that failed to internalize in sptf-3(RNAi) embryos had a shortened cell cycle. However, there were some cells that failed to internalize despite having normal cell cycle timing, indicating that the reduced cell cycle alone does not fully explain the gastrulation defects of sptf-3(RNAi) embryos.
Figure 5.
(A) Ea/Ep cell cycle times are reduced in some sptf-3(RNAi) embryos. In wild-type embryos, the Ea cell cycle (left column) is 40.33 ± 0.39 min, and the Ep cell cycle (right column) is 42.64 ± 0.39 min. In sptf-3(RNAi) embryos, the cell cycle of the Ea and Ep cells is reduced to 37.28 ± 0.65 and 39.51 ± 0.57 min, respectively. Ea and Ep cell cycle timing is not changed in abi-1(RNAi), gex-3(RNAi), and ced-4(RNAi) embryos. **P < 0.01. Excluded is one abi-1(RNAi) embryo that had an anomalous delay outside the range shown here (Ea = 62 min; Ep = 64 min). Cells that failed to internalize are shown in red. Gray indicates cells that internalized. Means are represented by thick black bars. (B) mNeonGreen::SPTF-3 is enriched in all nuclei during gastrulation (nine of nine embryos filmed). Arrowheads mark Ea/Ep nuclei. Bar, 10 µm.
Genes that regulate endoderm cell fate are known to affect the timing of E cell division and E cell internalization during gastrulation. Endodermal cell fate is marked by the expression of two partially redundant GATA transcription factors, end-1 and end-3 (Maduro et al. 2005, 2007; Owraghi et al. 2010; Boeck et al. 2011). When the endoderm cell fate regulators end-1 and end-3 are disrupted, the E cells have a shorter cell cycle and divide prematurely on the outside of the embryo, before internalization (Zhu et al. 1997; Maduro et al. 2005; Boeck et al. 2011). The sptf-3 gene encodes an Sp1 family transcription factor with multiple recognized roles in C. elegans, including vulval development, oocyte production, embryonic development, and specification of apoptotic cell fate (Ulm et al. 2011; Hirose and Horvitz 2013). Given the previously recognized role of sptf-3 in cell fate specification and our data showing that sptf-3 affects cell cycle length in the E cells, we hypothesized that sptf-3 plays a role in specifying E cell fate. Because sptf-3 has diverse functions in different cells in C. elegans, we predicted that sptf-3 also might contribute to other early cell lineages. Consistent with this prediction, we found that a fluorescent SPTF-3 fusion protein resulting from tagging the endogenous locus localized to all nuclei in gastrulation-stage embryos (Figure 5B).
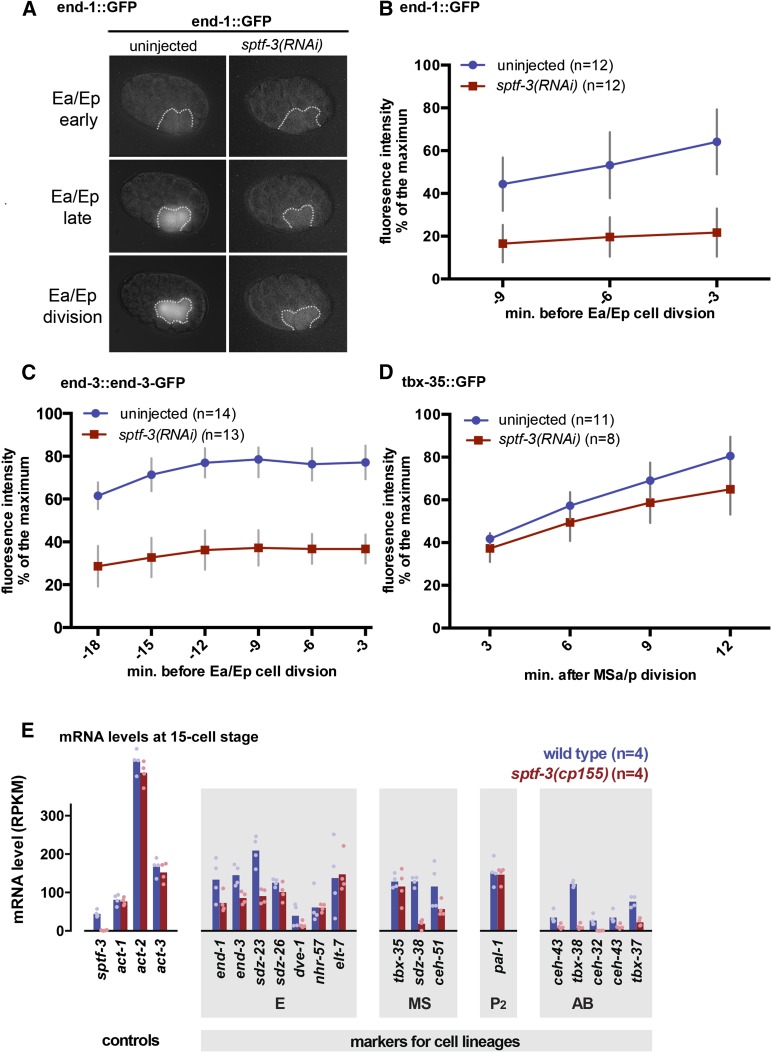
To assess the effects on E cell fate specification, we examined GFP reporters of end-1 and end-3 expression in sptf-3(RNAi) embryos. sptf-3(RNAi) resulted in a significant reduction of end-1::GFP reporter intensities throughout the Ea and Ep cell cycle (Figure 6, A and B). end-3 expression is apparent earlier in the Ea/Ep cell cycle than is end-1 expression (Maduro et al. 2007; Boeck et al. 2011). We found that sptf-3(RNAi) also resulted in decreased levels of end-3::GFP (Figure 6C). In addition to the end-1 and end-3 endodermal markers, we analyzed the fluorescence intensities of an MS cell reporter when sptf-3 was disrupted to determine whether cell fates are affected more generally. The MS lineage gives rise to multiple tissue types, including muscle and neurons but not endoderm cells (Sulston et al. 1983). Unlike the endoderm markers, sptf-3(RNAi) did not significantly decrease the expression of tbx-35::GFP, a marker of MS cell fate (Broitman-Maduro et al. 2006), in mesoderm progenitor cells (P > 0.05) (Figure 6D). To determine whether sptf-3 affects gene expression more broadly beyond the E lineage, we performed transcriptome profiling from wild-type and sptf-3 knockout 15-cell-stage embryos and analyzed mRNA levels of genes whose mRNAs were shown in previous quantitative experiments to be lineage restricted (Hashimshony et al. 2015). sptf-3 knockout resulted in the expected loss of sptf-3 mRNA, little change to three actin mRNAs expressed at distinct levels, and effects on end-1, end-3, and tbx-35 mRNA levels that were consistent with the earlier reporter construct results for each of these genes. Analysis of additional lineage-specific markers demonstrated that sptf-3 knockout decreased levels of specific mRNAs from the MS and AB lineages (Figure 6E). Thus, sptf-3 is likely to affect endodermal internalization at least in part by regulating the expression of genes that determine early endodermal fate and regulate gastrulation, and sptf-3 also has broader roles in regulating expression of genes in multiple cell lineages.
Figure 6.
Endoderm cell fate marker expression was reduced in sptf-3(RNAi) embryos. (A) end-1::GFP DIC overlay from video stills. White dotted line is drawn around the E cells. (B) Fluorescence intensity from the end-1::GFP reporter strain was reduced in sptf-3(RNAi) embryos. (C) Fluorescence intensity of end-3::end-3-GFP was reduced in sptf-3(RNAi) embryos. (D) Fluorescence intensity of the mesodermal MS lineage marker tbx-35::GFP in MSa/p’s daughters was not significantly reduced in sptf-3(RNAi) embryos (P > 0.05). In B, C, and D, 100% represents the maximum fluorescence value measured from a single data point. (E) RNA-seq shows reduced mRNA level for certain markers of multiple lineages in an sptf-3 deletion mutant (bars, mean values; dots, individual replicates).
Although sptf-3(RNAi) had detectable effects on the expression of certain early endoderm cell fate markers earlier, sptf-3 had little effect on expression levels of the endodermally expressed genes nhr-57 or elt-7 (Figure 6E), nor did they affect a marker of terminal differentiation of the endoderm. We found that sptf-3(RNAi) embryos were capable of forming birefringent rhabditin granules, sometimes called gut granules (Table 1) (Babu 1974; Laufer et al. 1980). However, disruption of sptf-3 by RNAi enhanced the disruption of intestinal development in end-3(ok1448) worms (and not in end-1(ok558) worms) (Table 1).
Table 1. Birefringent gut granules failed to form in sptf-3(RNAi); end-3(ok1448) embryos.
| Genotype | Percent gut granules | n |
|---|---|---|
| Wild type | 100 | 24 |
| end-1(ok558) | 88 | 34 |
| end-3(ok1448) | 91 | 34 |
| sptf-3(RNAi) in wild type | 100 | 33 |
| sptf-3(RNAi) in end-1(ok558) | 100 | 19 |
| sptf-3(RNAi) in end-3(ok1448) | 0 | 14 |
dsRNA targeting sptf-3 was injected into wild-type, end-1(ok558), and end-3(ok1448) worms, and percent of embryos with gut granule formation was scored.
Taken together, these results indicate that sptf-3 has critical roles in the expression of early endodermal cell fate genes and the subsequent internalization of the endoderm cells, as well as gene expression in other cell lineages. The results establish sptf-3 as a broad regulator of gene expression that, among other roles, makes important contributions to the well-studied C. elegans endodermal specification pathway.
The WAVE complex is required for gastrulation in C. elegans
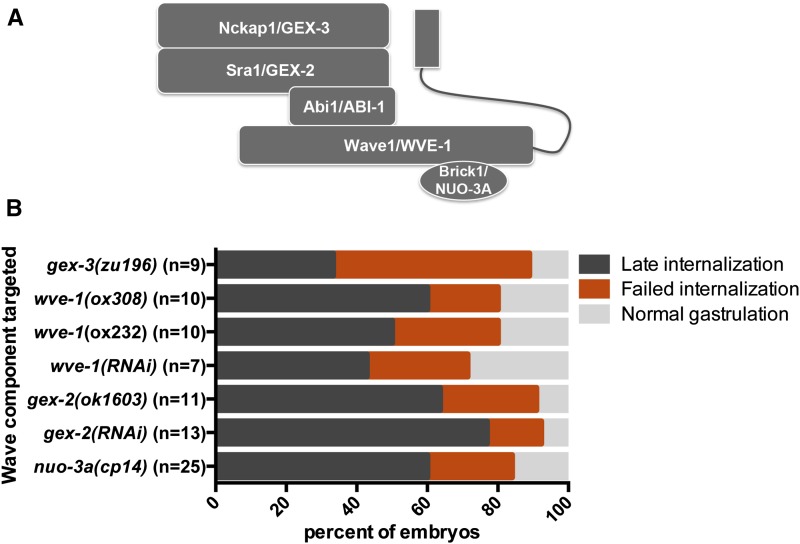
Our secondary screen revealed that genes encoding two members of the WAVE complex of actin regulators, abi-1/Abi1 and gex-3/Nckap1, are required for gastrulation in C. elegans. The WAVE complex is highly conserved, regulating actin-dependent processes in plants, Dictyostelium, yeast, Drosophila, C. elegans, and vertebrate animal systems (reviewed in Takenawa and Suetsugu 2007). Wave proteins were discovered initially by homology to an actin regulatory domain in the Wiskott-Aldrich syndrome proteins (WASPs): WAVE is an abbreviation of WASP family verprolin-homologous (Miki et al. 1998). Activation of the WAVE complex is regulated by multiple stimuli, including Rac GTPases (Miki et al. 1998, Eden et al. 2002), resulting in a tightly controlled, polarized actin assembly mediated via Arp2/3 (Machesky and Insall 1998; Machesky et al. 1999). The WAVE complex is composed of five proteins (Eden et al. 2002), whose C. elegans homologs are GEX-3/Nckap1, GEX-2/Sra1, ABI-1/Abi1, WVE-1/Wave1, and NUO-3A/Brick1 (Figure 7A). In C. elegans, GEX-3, GEX-2, ABI-1, and WVE-1 exist in a protein complex, and as in other organisms, disruption of one component results in degradation of the other subunits (Patel et al. 2008). To determine whether the role we found for ABI-1/Abi1 and GEX-3/Nckap1 in gastrulation reflects a role for the entire WAVE complex in gastrulation, we disrupted gex-2 and wve-1 using RNAi and also analyzed phenotypes in existing null mutants (Soto et al. 2002; Patel et al. 2008). Results from RNAi and mutant analyses indicate that both gex-2 and wve-1 are required for gastrulation (Figure 7B). In addition, a genetic null of gex-3 validated the RNAi gastrulation-defective phenotype (Figure 7B).
Figure 7.
Targeting members of the C. elegans WAVE complex affected gastrulation. (A) Schematic diagram of the WAVE complex: Nckap1/GEX-3, Sra1/GEX-2, Abi1/ABI-1, Wave1/WVE-1, and Brick1/NUO-3A. (B) Embryos from each background were analyzed by 4D DIC microscopy. Internalization of the endoderm precursors was scored.
The smallest member of the WAVE complex, nuo-3a/Brick1, has been difficult to study in C. elegans by traditional RNAi approaches because the nuo-3 gene produces an RNA that encodes two distinct polypeptides: the BRICK homolog (the nuo-3a ORF) and the NADH-ubiquinone oxidoreductase α-subunit (nuo-3b ORF). As a result, dsRNA targeting the nuo-3 transcript should affect the RNA levels of both polypeptides. To overcome this challenge, we used CRISPR/Cas9-triggered homologous recombination (Dickinson et al. 2013) to generate a knockout of nuo-3a/Brick1 while leaving the nuo-3b NADH-ubiquinone oxidoreductase coding sequence intact (Figure S2). We found that like the other members of the WAVE complex, the nuo-3a/Brick1 homolog was required for gastrulation in C. elegans (Figure 7B).
We analyzed the E cell cycle timing of gex-3, wve-1, gex-2, and nuo-3a mutants. We found that E cell cycle lengths were similar in these backgrounds as in wild-type worms, showing only a modest decrease in the Ea cell cycle length in gex-2 mutants and a modest increase in the Ea cell cycle length in nuo-3a mutants (P < 0.05) (Figure S3). We conclude that WAVE complex members are unlikely to affect gastrulation via effects on cell cycle length.
Apical narrowing and cell compaction are affected by gex-3
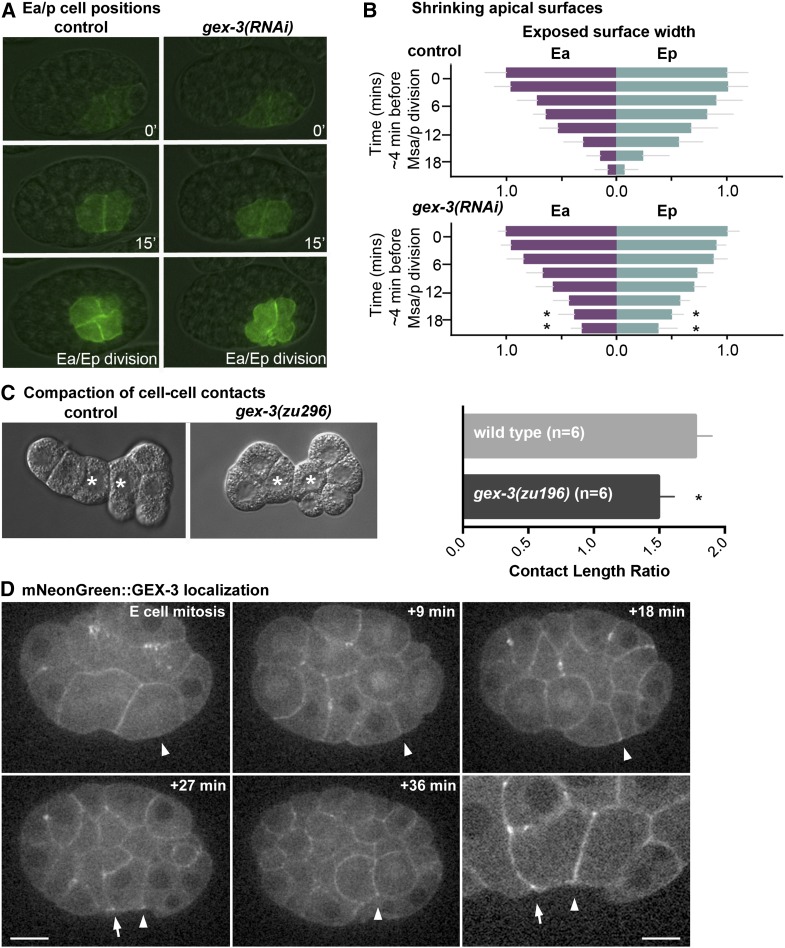
We hypothesized that mutations in gex-3 may affect apical constriction in the E cells. When members of the WAVE complex were disrupted, we noticed that plasma membranes formed blebs (File S6), similar to the phenotype observed previously when members of the Arp2/3 complex, a downstream target of the WAVE complex, were disrupted (Roh-Johnson and Goldstein 2009). Furthermore, previous studies have indicated that Arp2/3 has critical roles in C. elegans gastrulation (Severson et al. 2002; Roh-Johnson and Goldstein 2009). To determine whether members of the WAVE complex affect E cells’ apical constriction, we analyzed the narrowing of Ea/Ep cells’ apical domains over time using an end-1::GFP::CAAX background (Wehman et al. 2011) in which the plasma membranes of the endoderm precursor cells are labeled. To disrupt the WAVE complex, we used gex-3(RNAi) because disruption of gex-3 is known to reduce levels of other members of the WAVE complex (Patel et al. 2008). We measured the maximal anterior-to-posterior length of the exposed apical surfaces over time during the Ea/Ep cell cycle. We found that the Ea and Ep apical lengths in gex-3(RNAi) embryos began to narrow similarly to control embryos, but the final stages of endoderm internalization were affected, with more apical surface left exposed in gex-3(RNAi) embryos late in the cell cycle than in wild-type embryos (P < 0.05) (Figure 8, A and B). These results suggest that while initiation of apical constriction appeared normal, later stages were disrupted, similar to what was observed in Arp2/3 knockdown embryos (Roh-Johnson and Goldstein 2009). Although shrinkage of apical cell surfaces was affected, we found that myosin became apically enriched in gex-3(RNAi) embryos to a similar level as in control embryos (Figure S4), much as found in Arp2/3(RNAi) embryos previously (Roh-Johnson and Goldstein 2009). We conclude that gex-3 is likely to affect apical constriction without strong apparent effects on myosin distribution.
Figure 8.
Ea/Ep cell apical surface shrinking and cell compaction are affected by gex-3(RNAi) and mNeonGreen::GEX-3 localization. (A) E cell positions over time; images are from end-1::CAAX-GFP uninjected control and gex-3(RNAi) embryos. (B) Lengths of exposed E cell apical surfaces were measured in end-1::CAAX-GFP uninjected control and gex-3(RNAi) embryos over time. The exposed surfaces began to narrow in gex-3(RNAi) embryos, but at later time points the membranes remained exposed on the embryo surface. *P < 0.05. (C) Cell isolation experiments with control and gex-3(zu196) cells. The contact length ratio (cell-cell contact length/cell diameter) is used as a metric for cell compaction (see Materials and Methods for details). gex-3(zu196) cells have a small but statistically significant reduction in cell contact length ratios. *P < 0.05. White asterisks mark the Ea/Ep cells. (D) mNeonGreen::GEX-3 localization in an embryo during gastrulation. Arrowheads mark the single E cell in the first image and the apical contact between its daughter cells, Ea/p, in other images. Arrows mark an Ea/MS granddaughter apical border, at which mNG::GEX-3 appeared enriched in 10 of 14 embryos. Bar = 10 µm except for lower-right frame, which is a magnified view of the +27-min time point in another focal plane, bar, 5 µm.
Because the WAVE complex has been shown to affect the formation of cell junctions in C. elegans (Bernadskaya et al. 2011), and because we observed blebbing after targeting the WAVE complex (File S6), we predicted that the WAVE complex affects cell shapes. To examine cell shapes, we removed eggshells from two-cell-stage embryos, separated the cells, and followed development of these isolates. C. elegans embryos with eggshells removed are known to undergo compaction, expanding cell contacts and resulting in an increasingly flattened appearance at the embryo surface where cells meet (Grana et al. 2010). Previous studies have shown that when proteins involved in cell adhesion (SAX-7/L1CAM and HMR-1/cadherin) were depleted, compaction of the Ea/Ep cells was reduced, as measured by a decreased length of cell contact between the Ea and Ep cells (Grana et al. 2010). We performed similar experiments in control and gex-3 mutant embryos and observed a modest but statistically significant decrease in contact length ratios in gex-3 mutant embryos (Figure 8C), suggesting that the WAVE complex has a modest effect on cell shapes in vivo. A functional mNG-tagged form of GEX-3 (Dickinson et al. 2015) showed cortical localization with some enrichment at apical borders between Ep and MS granddaughters (Figure 8D), a known site of Arp2/3-dependent transient F-actin enrichment (Roh-Johnson and Goldstein 2009). The findings from our in vivo studies—that the WAVE complex affects apical narrowing, cell compaction, and gastrulation in C. elegans without strong effects on cell cycle timing or myosin localization—are consistent with the known effects of the WAVE complex, which affects F-actin architecture generally and cell junctions of other cells in C. elegans specifically (Takenawa and Suetsugu 2007; Bernadskaya et al. 2011).
Nckap1 contributes to neural tube closure in Xenopus
We hypothesized that genes implicated in mammalian neural tube closure and C. elegans morphogenesis might have broadly conserved roles in morphogenesis. To test whether genes we found by screening in C. elegans have conserved cellular functions, we turned to Xenopus embryos, which allow for imaging neural tube closure at the level of individual cells. Xenopus is a valuable vertebrate model in NTD research (see, e.g., Wallingford and Harland 2002; Haigo et al. 2003; Gray et al. 2009; Liu et al. 2011; Ossipova et al. 2014) owing in part to its external development, which facilitates in vivo imaging of neural tube closure (Kieserman et al. 2010). To determine whether Xenopus might serve as a model for probing the WAVE complex’s roles in neural tube formation, we tested whether a homolog of gex-3, nckap1, affects neural tube closure in Xenopus. We first examined whether nckap1 was expressed in embryos during neural tube closure at stage 18. Using RNA in situ hybridization, we found that nckap1 mRNA was expressed at neural tube closure stages and that the mRNA was enriched in the neural tube and surrounding neural tissues (Figure S5).
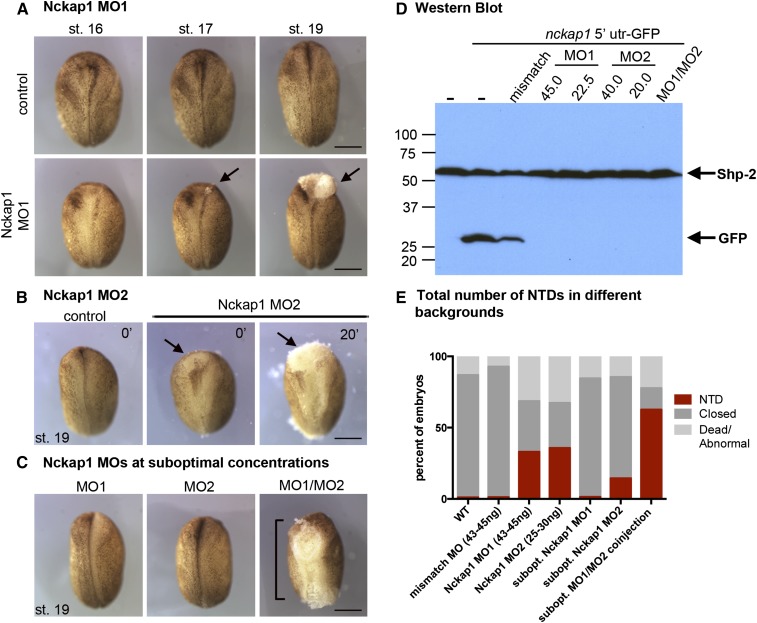
To test whether Nckap1 has a role in neural tube closure, we designed two nonoverlapping MOs to the 5′ UTR of nckap1 (MO1 and MO2; for MO design, see Figure S6) to disrupt Nckap1 expression. Both MOs disrupted neural tube closure (Figure 9, A and B). Co-injection of Nckap1 MO1 and MO2 together at suboptimal concentrations (concentrations at which each MO alone had little or no effect) resulted in an increased incidence of NTDs (χ2 test, P < 0.01), suggesting that the MOs are specific to Nckap1 (Figure 9, C and E). Disruption of Nckap1 was confirmed by co-injecting GFP-tagged nckap1-5′ UTR RNA with various concentrations of Nckap1 MOs (Figure 9D). Using a Western blot, we were unable to observe a GFP signal from MO1- and MO2-injected embryos. Further validating the specificity of the MOs, we showed that injection of a 5-bp mismatch MO resulted in reduced but not absent GFP signal (Figure 9D), and embryos injected with 45 ng of the 5-bp mismatch MO did not develop NTDs (data not shown).
Figure 9.
Targeting nckap1 affected neural tube closure in Xenopus. (A) Nckap1 MO1 (Figure S5 shows MO target sites) resulted in a NTD and cell dissociation in the neural tube (arrows). (B) A second, nonoverlapping Nckap1 MO (MO2) also resulted in a NTD (arrows). (C and E) Suboptimal concentrations of MO1 and MO2 did not result a NTD; however, co-injecting MO1/MO2 at suboptimal concentrations resulted in NTDs (brackets). (D) Western blot from protein lysates of embryos injected with Nckap1 5′ UTR-GFP RNA, probed with anti-GFP. Nckap1 5′ UTR-GFP levels were decreased in the mismatch control (fold change 0.614), while levels were undetectable in the MO1 and MO2 samples. Shp-2 was used as a loading control. All figures are dorsal views, anterior up. Staging of MO embryos was based from stage of control embryos. Bars, 0.5 mm.
Injection of the MO1 and MO2 at 43–45 and 25–30 ng, respectively, resulted in embryos that appeared superficially normal prior to neural tube closure stages. However, a delay in neural tube closure was evident between stages 16 and 19 (Figure 9, A and B). At various points during neural tube closure, lesions or small tears formed in the ectoderm, often beginning as a small lesion (Figure 9A, arrow, stage 17) that expanded in the neural tube (Figure 9A, arrow, stage 19). These lesions were observed at various sites along the neural tube in different embryos, often in the cranial and posterior regions. At later stages, the neural tube remained open, and additional lesions often were seen in the ventral surface (Figure S7). To examine the morphology of individual cells in Nckap1 MO-injected embryos, we injected embryos with GFP-CAAX mRNA to mark plasma membranes and filmed neural tube closure. At times when the neural folds converge in control embryos, cells were seen to round up and then become detached from the neuroepithelium in the Nckap1 MO-injected embryos at stage 16 (File S7, File S8, and Figure S8). Cells were seen to dissociate from the lesions, suggesting loss of cell adhesion. Although the loss of adhesion observed in Xenopus and the more moderate effect on compaction in C. elegans appear superficially similar, we note that either defect could be a primary effect of loss of a WAVE complex member or secondary consequences of unknown additional defects.
Discussion
Using a two-step enhancer screen, we discovered NTD gene homologs as newly recognized contributors to C. elegans gastrulation. These genes include the transcription factor sptf-3 and members of the WAVE complex of actin regulators. These findings suggest that some NTD gene homologs may have broadly shared roles in diverse animal systems in internalizing surface tissues. We speculate that such roles might reflect ancient and conserved roles retained in C. elegans and vertebrates from common ancestors. Alternatively, such roles could reflect convergent uses of similar genes in similar morphogenetic processes. Our approach supports the value of screening in C. elegans to identify genes with shared roles in morphogenesis in diverse animal systems.
sptf-3, an Sp factor, is a novel gastrulation gene in C. elegans
Sp proteins are a family of transcription factors with three conserved C2H2 zinc finger binding domains preceded by a buttonhead (BTD) box, a distinguishing feature of Sp proteins (Suske et al. 2005). There are nine Sp-family members in humans and mice, grouped into three clades from phylogenetic sequence analysis (Schaeper et al. 2010). Four Sp-related factors have been identified in C. elegans (trp-1, sptf-1, sptf-2, and sptf-3); however, only two genes, sptf-2 and sptf-3, contain the three canonical C2H2 zinc finger repeats (Ulm et al. 2011). Phylogenetic analyses of DNA binding domain sequences suggest that Sp factors in C. elegans are associated with the Sp1–Sp4 clade (Ulm et al. 2011), while Sp8 is found within the Sp6–Sp9 clade (Schaeper et al. 2010). In C. elegans, the only Sp factor identified thus far with roles in embryonic development is sptf-3 (Sun et al. 2007; Ulm et al. 2011; Hirose and Horvitz 2013; and our data).
Disruption of sptf-3 in C. elegans has been linked to a range of developmental defects in embryonic morphogenesis, vulva specification, and oocyte production (Ulm et al. 2011). In addition, sptf-3 regulates distinct apoptotic programs of specific cells in the developing worm, controlling both caspase-dependent and -independent pathways (Hirose and Horvitz 2013). In the mouse, Sp8 appears to negatively regulate apoptosis because disruption of Sp8 results in increased apoptosis in neural progenitor and neural crest cells (Zembrzycki et al. 2007; Kasberg et al. 2013).
Our data support an essential role for sptf-3 in embryonic morphogenesis because we have identified a role for sptf-3 in cell internalization during the first stages of gastrulation. When we examined the endoderm fate markers end-1 and end-3 in the sptf-3(RNAi) background, we observed a decrease in their expression, suggesting that, among several distinct roles, sptf-3 plays a role in regulating cell fate decisions in the E cell lineage. Results from published ChIP-seq experiments with SPTF-3 did not identify end-1 or end-3 as direct targets of SPTF-3. However, the ChIP-seq results did identify skn-1 as a target of SPTF-3 (Hirose and Horvitz 2013). skn-1 is a known upstream activator of the end-3 and end-1 genes (reviewed in Maduro and Rothman 2002). Thus, these data suggest that SPTF-3 is part of the network of transcription factors regulating endoderm fate specification in C. elegans.
Disruption of the vertebrate Sp8 results in exencephaly (NTDs in the cranial region) and spina bifida (NTDs in the posterior region) in the mouse and limb truncation defects in the mouse, chick, zebrafish, and X. tropicalis (Bell et al. 2003; Treichel et al. 2003, Kawakami et al. 2004, Chung et al. 2014). Sp8 is part of a transcriptional program that patterns neural progenitors into distinct domains (Griesel et al. 2006; O’Leary et al. 2007; Sahara et al. 2007; Zembrzycki et al. 2007; Kasberg et al. 2013); however, a transcriptional network involving Sp8 during neural tube closure has not yet been identified. It will be interesting to determine whether there are shared targets of sptf-3 and Sp8. In support of this, disruption of Sp8 in zebrafish embryos has been shown to result in decreased expression of gata2, a GATA transcription factor, in the neuroepithelium (Penberthy et al. 2004). The role of gata2 during neural tube closure is not clear because mutations in mice result in early embryonic lethality; however, evidence suggests that gata2 may affect early neuronal differentiation (Nardelli et al. 1999; Craven et al. 2004). Identification of conserved targets during cell internalization in C. elegans gastrulation and neural tube closure may help to identify conserved transcriptional networks that contribute to morphogenesis.
The WAVE complex has early roles in endoderm internalization
The gex mutants (gex-1/wve-1, gex-2, and gex-3) and abi-1 were characterized initially by a failure of endoderm tissue to remain internalized. The term gex refers to gut on the exterior arising from failed hypodermal (epidermal) migration, resulting in a large ventral cleft and exposed endoderm cells at later stages of morphogenesis (Soto et al. 2002; Withee et al. 2004; Patel et al. 2008). Here we provide evidence that WAVE complex mutants have earlier defects in endoderm internalization, which occurs at the 26- to 28-cell stage. We show that knockdown of all known WAVE components resulted in similar endoderm internalization defects, exhibiting late-internalization or failed-internalization phenotypes. The failed-internalization phenotype is a less frequently observed phenotype among gastrulation-defective mutants (Sawyer et al. 2011) and could be influenced by subtle cell positioning defects, as described previously for other C. elegans cells in WAVE complex mutants (Soto et al. 2002; Patel et al. 2008).
Arp2/3, an actin nucleator that can function downstream of the WAVE complex, has essential roles in C. elegans gastrulation (Severson et al. 2002; Roh-Johnson and Goldstein 2009). The small GTPase Rac1 is a known upstream regulator of the WAVE complex, and mutations in ced-10/Rac1 result in ventral enclosure defects similar to the WAVE complex mutants previously characterized (Soto et al. 2002; Patel et al. 2008). ced-10/Rac1 has been implicated in C. elegans gastrulation previously (Roh-Johnson et al. 2012), although whether it functions with the WAVE complex and Arp2/3 during gastrulation is not known.
Studies in C. elegans have suggested that the Arp2/3 complex and members of the WAVE complex have important roles in cell adhesion via the maturation and maintenance of intestinal epithelial junctions (Bernadskaya et al. 2011). Thus, the WAVE complex may have roles in establishing apical junctions in Ea and Ep cells. Apical junctions may contribute to a “clutch” mechanism in which contractions of the apical actomyosin cytoskeleton gradually become coupled to membranes, driving shrinkage of the apical domains of cells (Roh-Johnson et al. 2012). Failure of the E cells to internalize when WAVE complex members are disrupted could feasibly represent a defect in the proposed clutch mechanism. Disruption of the Arp2/3 complex results in a reduction of apical membrane shrinkage, similar to phenotypes observed in gex-3(RNAi) embryos (Roh-Johnson and Goldstein 2009). It is possible that these defects arise because apical junctions are not established correctly or are not attached properly to the actin cortex. In support of WAVE complex members affecting cell adhesion in the E cells, we show that cell compaction is reduced in gex-3 mutant cell isolates.
The WAVE complex has important but poorly understood roles during neural tube closure
Nckap1, Abi1, and Rac1 have been implicated in neural tube closure in mouse as disruption of these genes results in NTDs (Rakeman and Anderson 2006; Yokota et al. 2007; Migeotte et al. 2011; Dubielecka et al. 2011). Mutations in Wave1 and Wave2 do not result in NTDs, and mutations in Wave3 have not been reported (Soderling et al. 2003; Yamazaki et al. 2003; Yan et al. 2003). Although the mechanisms regulated by these WAVE complex members during neural tube closure are not clear, defects in apical actin filaments in the neuroepithelium (Yokota et al. 2007) and lack of lamellipodia protrusions from primary neuroepithelium have been observed (Rakeman and Anderson 2006; Yokota et al. 2007). Nckap1 and Rac1 also have earlier roles during mouse gastrulation: disruption of Nckap1 and Rac1 affect the migration of mesodermal cells away from the primitive streak. This defect is linked to the cardia bifida phenotypes observed in Nckap1 and Rac1 mouse mutants and may also be a contributing factor to the open neural tube in these embryos (Rakeman and Anderson 2006; Migeotte et al. 2011).
Here we present evidence that Nckap1 is required for neural tube closure in Xenopus. When nckap1 function was depleted, neural tube closure was significantly delayed compared to control embryos, and often the neural tube failed to close. The tissue integrity of Nckap1-depleted embryos appeared compromised because lesions or tears in the neural plate tissue formed, and cells dissociated from the neuroepithelium. These phenotypes are similar to those described in cysteine-rich motor neuron protein (Crim1)–depleted Xenopus embryos. Like Nckap1, disruption of Crim1 resulted in cells that dissociated from the neural tube. Crim1 was shown to affect the junctional localization of E- and C-cadherin in the neural plate and the ability of β-catenin to stably associate with junctional complexes (Ponferrada et al. 2012). Although future studies will be needed to determine whether Nckap1 affects the localization or expression of functional adhesion proteins in the neuroepithelium, it seems likely that Nckap1 affects cell adhesion either directly or indirectly. We hope that the use of C. elegans can contribute to identifying candidate cellular mechanisms that might be regulated by nckap1 and other NTD genes during neural tube closure.
Acknowledgments
We thank members of the Goldstein and Conlon laboratories and Gregg Phares for critical reading of and comments on the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40-OD010440). This work was supported by National Institutes of Health grants R01-GM083071 to B.G. and T32-CA009156 and a Howard Hughes postdoctoral fellowship from the Helen Hay Whitney Foundation to D.J.D., National Science Foundation Graduate Research Fellowships to S.C.T. and J.K.H., National Institutes of Health grant R21-HD073044 to F.L.C., American Heart Association Postdoctoral Fellowship 13POST16950044 to P.T., and National Institutes of Health grant F32-GM090447 to J.S.B.
Footnotes
Communicating editor: O. Hobert
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.183137/-/DC1
Literature Cited
- Anders S., Pyl P. T., Huber W., 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. C., Gill J. S., Cinalli R. M., Nance J., 2008. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science 320: 1771–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu P., 1974. Biochemical genetics of Caenorhabditis elegans. Mol. Gen. Genet. 135: 39–44. [Google Scholar]
- Bastiani C., Mendel J., 2006. Heterotrimeric G proteins in C. elegans. WormBook 2006: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. M., Schreiner C. M., Waclaw R. R., Campbell K., Potter S. S., et al. , 2003. Sp8 is crucial for limb outgrowth and neuropore closure. Proc. Natl. Acad. Sci. USA 100: 12195–12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadskaya Y. Y., Patel F. B., Hsu H. T., Soto M. C., 2011. Arp2/3 promotes junction formation and maintenance in the Caenorhabditis elegans intestine by regulating membrane association of apical proteins. Mol. Biol. Cell 22: 2886–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck M. E., Boyle T., Bao Z., Murray J., Mericle B., et al. , 2011. Specific roles for the GATA transcription factors end-1 and end-3 during C. elegans E-lineage development. Dev. Biol. 358: 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S. J., Martin J. S., Polanowska J., Hill D. E., Gartner A., et al. , 2004. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr. Biol. 14: 33–39. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broitman-Maduro G., Lin K. T., Hung W. W., Maduro M. F., 2006. Specification of the C. elegans MS blastomere by the T-box factor TBX-35. Development 133: 3097–3106. [DOI] [PubMed] [Google Scholar]
- Chung H. A., Medina-Ruiz S., Harland R. M., 2014. Sp8 regulates inner ear development. Proc. Natl. Acad. Sci. USA 111: 6329–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven S. E., Lim K. C., Ye W., Engel J. D., de Sauvage F., et al. , 2004. Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development 131: 1165–1173. [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D., Goldstein B., 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200: 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubielecka P. M., Ladwein K. I., Xiong X., Migeotte I., Chorzalska A., et al. , 2011. Essential role for Abi1 in embryonic survival and WAVE2 complex integrity. Proc. Natl. Acad. Sci. USA 108: 7022–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S., Rohatgi R., Podtelejnikov A. V., Mann M., Kirschner M. W., 2002. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418: 790–793. [DOI] [PubMed] [Google Scholar]
- Edgar L. G., Goldstein B., 2012. Culture and manipulation of embryonic cells. Methods Cell Biol. 107: 151–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar L. G., McGhee J. D., 1988. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 53: 589–599. [DOI] [PubMed] [Google Scholar]
- Grana T. M., Cox E. A., Lynch A. M., Hardin J., 2010. SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Dev. Biol. 344: 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R. S., Abitua P. B., Wlodarczyk B. J., Szabo-Rogers H. L., Blanchard O., et al. , 2009. The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nat. Cell Biol. 11: 1225–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesel G., Treichel D., Collombat P., Krull J., Zembrzycki A., et al. , 2006. Sp8 controls the anteroposterior patterning at the midbrain-hindbrain border. Development 133: 1779–1787. [DOI] [PubMed] [Google Scholar]
- Haigo S. L., Hildebrand J. D., Harland R. M., Wallingford J. B., 2003. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 13: 2125–2137. [DOI] [PubMed] [Google Scholar]
- Harland R. M., 1991. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36: 685–695. [DOI] [PubMed] [Google Scholar]
- Harrell J. R., Goldstein B., 2011. Internalization of multiple cells during C. elegans gastrulation depends on common cytoskeletal mechanisms but different cell polarity and cell fate regulators. Dev. Biol. 350: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. J., 2009. Insights into prevention of human neural tube defects by folic acid arising from consideration of mouse mutants. Birth Defects Res. A Clin. Mol. Teratol. 85: 331–339. [DOI] [PubMed] [Google Scholar]
- Harris M. J., Juriloff D. M., 2007. Mouse mutants with neural tube closure defects and their role in understanding human neural tube defects. Birth Defects Res. A Clin. Mol. Teratol. 79: 187–210. [DOI] [PubMed] [Google Scholar]
- Harris M. J., Juriloff D. M., 2010. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res. A Clin. Mol. Teratol. 88: 653–669. [DOI] [PubMed] [Google Scholar]
- Hashimshony T., Feder M., Levin M., Hall B. K., Yanai I., 2015. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature 519: 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J. D., 2005. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J. Cell Sci. 118: 5191–5203. [DOI] [PubMed] [Google Scholar]
- Hildebrand J. D., Soriano P., 1999. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 99: 485–497. [DOI] [PubMed] [Google Scholar]
- Hirose T., Horvitz H. R., 2013. An Sp1 transcription factor coordinates caspase-dependent and -independent apoptotic pathways. Nature 500: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., 2005. Introduction to genetics and genomics, in WormBook, ed. C. elegans Research Community , http://www.wormbook.org. doi/10.1895/wormbook.1.17.1.
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J., 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasberg A. D., Brunskill E. W., Steven Potter S., 2013. SP8 regulates signaling centers during craniofacial development. Dev. Biol. 381: 312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Esteban C. R., Matsui T., Rodriguez-Leon J., Kato S., et al. , 2004. Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Development 131: 4763–4774. [DOI] [PubMed] [Google Scholar]
- Kieserman E. K., Lee C., Gray R. S., Park T. J., Wallingford J. B., 2010 High-magnification in vivo imaging of Xenopus embryos for cell and developmental biology. Cold Spring Harb. Protoc. 2010: pdb prot5427. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer J. S., Bazzicalupo P., Wood W. B., 1980. Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell 19: 569–577. [DOI] [PubMed] [Google Scholar]
- Lee J. D., Silva-Gagliardi N. F., Tepass U., McGlade C. J., Anderson K. V., 2007. The FERM protein Epb4.1l5 is required for organization of the neural plate and for the epithelial-mesenchymal transition at the primitive streak of the mouse embryo. Development 134: 2007–2016. [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Goldstein B., 2003. Mechanisms of cell positioning during C. elegans gastrulation. Development 130: 307–320. [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Marston D. J., Walston T., Hardin J., Halberstadt A., et al. , 2006. Wnt/Frizzled signaling controls C. elegans gastrulation by activating actomyosin contractility. Curr. Biol. 16: 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Komiya Y., Mezzacappa C., Khadka D. K., Runnels L., et al. , 2011. MIM regulates vertebrate neural tube closure. Development 138: 2035–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L. M., Insall R. H., 1998. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8: 1347–1356. [DOI] [PubMed] [Google Scholar]
- Machesky L. M., Mullins R. D., Higgs H. N., Kaiser D. A., Blanchoin L., et al. , 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA 96: 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M. F., Rothman J. H., 2002. Making worm guts: the gene regulatory network of the Caenorhabditis elegans endoderm. Dev. Biol. 246: 68–85. [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Hill R. J., Heid P. J., Newman-Smith E. D., Zhu J., et al. , 2005. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev. Biol. 284: 509–522. [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Broitman-Maduro G., Mengarelli I., Rothman J. H., 2007. Maternal deployment of the embryonic SKN-1→MED-1,2 cell specification pathway in C. elegans. Dev. Biol. 301: 590–601. [DOI] [PubMed] [Google Scholar]
- Migeotte I., Grego-Bessa J., Anderson K. V., 2011. Rac1 mediates morphogenetic responses to intercellular signals in the gastrulating mouse embryo. Development 138: 3011–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H., Suetsugu S., Takenawa T., 1998. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17: 6932–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Soriano V., Belacortu Y., Paricio N., 2012. Planar cell polarity signaling in collective cell movements during morphogenesis and disease. Curr. Genomics 13: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J., Priess J. R., 2002. Cell polarity and gastrulation in C. elegans. Development 129: 387–397. [DOI] [PubMed] [Google Scholar]
- Nance J., Lee J. Y., Goldstein B., 2005. Gastrulation in C. elegans. WormBook 2005: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J., Munro E. M., Priess J. R., 2003. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development 130: 5339–5350. [DOI] [PubMed] [Google Scholar]
- Nardelli J., Thiesson D., Fujiwara Y., Tsai F. Y., Orkin S. H., 1999. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev. Biol. 210: 305–321. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D., Faber J., 1967. Normal Table of Xenopus laevis. North Holland, Amsterdam. [Google Scholar]
- Nishimura T., Takeichi M., 2008. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development 135: 1493–1502. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Honda H., Takeichi M., 2012. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 149: 1084–1097. [DOI] [PubMed] [Google Scholar]
- O’Leary D. D., Chou S. J., Sahara S., 2007. Area patterning of the mammalian cortex. Neuron 56: 252–269. [DOI] [PubMed] [Google Scholar]
- Ono S., Benian G. M., 1998. Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J. Biol. Chem. 273: 3778–3783. [DOI] [PubMed] [Google Scholar]
- Ossipova O., Kim K., Lake B. B., Itoh K., Ioannou A., et al. , 2014. Role of Rab11 in planar cell polarity and apical constriction during vertebrate neural tube closure. Nat. Commun. 5: 3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owraghi M., Broitman-Maduro G., Luu T., Roberson H., Maduro M. F., 2010. Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev. Biol. 340: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel F. B., Bernadskaya Y. Y., Chen E., Jobanputra A., Pooladi Z., et al. , 2008. The WAVE/SCAR complex promotes polarized cell movements and actin enrichment in epithelia during C. elegans embryogenesis. Dev. Biol. 324: 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penberthy W. T., Zhao C., Zhang Y., Jessen J. R., Yang Z., et al. , 2004. Pur alpha and Sp8 as opposing regulators of neural gata2 expression. Dev. Biol. 275: 225–234. [DOI] [PubMed] [Google Scholar]
- Ponferrada V. G., Fan J., Vallance J. E., Hu S., Mamedova A., et al. , 2012. CRIM1 complexes with ss-catenin and cadherins, stabilizes cell-cell junctions and is critical for neural morphogenesis. PLoS One 7: e32635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakeman A. S., Anderson K. V., 2006. Axis specification and morphogenesis in the mouse embryo require Nap1, a regulator of WAVE-mediated actin branching. Development 133: 3075–3083. [DOI] [PubMed] [Google Scholar]
- Rauzi M., Lecuit T., 2009. Closing in on mechanisms of tissue morphogenesis. Cell 137: 1183–1185. [DOI] [PubMed] [Google Scholar]
- Roh-Johnson M., Goldstein B., 2009. In vivo roles for Arp2/3 in cortical actin organization during C. elegans gastrulation. J. Cell Sci. 122: 3983–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh-Johnson M., Shemer G., Higgins C. D., McClellan J. H., Werts A. D., et al. , 2012. Triggering a cell shape change by exploiting preexisting actomyosin contractions. Science 335: 1232–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara S., Kawakami Y., Izpisua Belmonte J. C., O’Leary D. D., 2007. Sp8 exhibits reciprocal induction with Fgf8 but has an opposing effect on anterior-posterior cortical area patterning. Neural Dev. 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J. M., Harrell J. R., Shemer G., Sullivan-Brown J., Roh-Johnson M., et al. , 2010. Apical constriction: a cell shape change that can drive morphogenesis. Dev. Biol. 341: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J. M., Glass S., Li T., Shemer G., White N. D., et al. , 2011. Overcoming redundancy: an RNAi enhancer screen for morphogenesis genes in Caenorhabditis elegans. Genetics 188: 549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeper N. D., Prpic N. M., Wimmer E. A., 2010. A clustered set of three Sp-family genes is ancestral in the Metazoa: evidence from sequence analysis, protein domain structure, developmental expression patterns and chromosomal location. BMC Evol. Biol. 10: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenwolf G. C., Smith J. L., 1990. Mechanisms of neurulation: traditional viewpoint and recent advances. Development 109: 243–270. [DOI] [PubMed] [Google Scholar]
- Severson A. F., Baillie D. L., Bowerman B., 2002. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr. Biol. 12: 2066–2075. [DOI] [PubMed] [Google Scholar]
- Shelton C.A., Bowerman B., 1996. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development 122: 2043–2050. [DOI] [PubMed] [Google Scholar]
- Soderling S. H., Langeberg L. K., Soderling J. A., Davee S. M., Simerly R., et al. , 2003. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl. Acad. Sci. USA 100: 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka S., Amin N. M., Gibbs D., Christine K. S., Charpentier M. S., et al. , 2014. Congenital heart disease protein 5 associates with CASZ1 to maintain myocardial tissue integrity. Development 141: 3040–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnica-Krezel L., Eaton S., 2003. Embryo morphogenesis: getting down to cells and molecules. Development 130: 4229–4233. [DOI] [PubMed] [Google Scholar]
- Soto M. C., Qadota H., Kasuya K., Inoue M., Tsuboi D., et al. , 2002. The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 16: 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Sun H., Nelms B. L., Sleiman S. F., Chamberlin H. M., Hanna-Rose W., 2007. Modulation of Caenorhabditis elegans transcription factor activity by HIM-8 and the related Zinc-Finger ZIM proteins. Genetics 177: 1221–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G., Bruford E., Philipsen S., 2005. Mammalian SP/KLF transcription factors: bring in the family. Genomics 85: 551–556. [DOI] [PubMed] [Google Scholar]
- Takenawa T., Suetsugu S., 2007. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8: 37–48. [DOI] [PubMed] [Google Scholar]
- Tandon P., Showell C., Christine K., Conlon F. L., 2012. Morpholino injection in Xenopus. Methods Mol. Biol. 843: 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon P., Miteva Y. V., Kuchenbrod L. M., Cristea I. M., Conlon F. L., 2013. Tcf21 regulates the specification and maturation of proepicardial cells. Development 140: 2409–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Treichel D., Schock F., Jackle H., Gruss P., Mansouri A., 2003. mBtd is required to maintain signaling during murine limb development. Genes Dev. 17: 2630–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm E. A., Sleiman S. F., Chamberlin H. M., 2011. Developmental functions for the Caenorhabditis elegans Sp protein SPTF-3. Mech. Dev. 128: 428–441. [DOI] [PubMed] [Google Scholar]
- Wallingford J. B., Harland R. M., 2002. Neural tube closure requires Dishevelled-dependent convergent extension of the midline. Development 129: 5815–5825. [DOI] [PubMed] [Google Scholar]
- Wallingford J. B., Niswander L. A., Shaw G. M., Finnell R. H., 2013. The continuing challenge of understanding, preventing, and treating neural tube defects. Science 339: 1222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehman A. M., Poggioli C., Schweinsberg P., Grant B. D., Nance J., 2011. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol. 21: 1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werts A., Roh-Johnson M., Goldstein B., 2011. Dynamic localization of C. elegans TPR-GoLoco proteins mediates mitotic spindle orientation by extrinsic signaling. Development 138: 4411–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withee J., Galligan B., Hawkins N., Garriga G., 2004. Caenorhabditis elegans WASP and Ena/VASP proteins play compensatory roles in morphogenesis and neuronal cell migration. Genetics 167: 1165–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D., Suetsugu S., Miki H., Kataoka Y., Nishikawa S., et al. , 2003. WAVE2 is required for directed cell migration and cardiovascular development. Nature 424: 452–456. [DOI] [PubMed] [Google Scholar]
- Yan C., Martinez-Quiles N., Eden S., Shibata T., Takeshima F., et al. , 2003. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 22: 3602–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Lin X., 2009. Shaping morphogen gradients by proteoglycans. Cold Spring Harb. Perspect. Biol. 1: a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y., Ring C., Cheung R., Pevny L., Anton E. S., 2007. Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron 54: 429–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Horvitz H. R., 1992. The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development 116: 309–320. [DOI] [PubMed] [Google Scholar]
- Zembrzycki A., Griesel G., Stoykova A., Mansouri A., 2007. Genetic interplay between the transcription factors Sp8 and Emx2 in the patterning of the forebrain. Neural Dev. 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Hill R. J., Heid P. J., Fukuyama M., Sugimoto A., et al. , 1997. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11: 2883–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Henzel W. J., Liu X., Lutschg A., Wang X., 1997. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90: 405–413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. Table S1 contains information about the genes we identified as C. elegans homologs to mammalian neural tube defect genes and includes accession numbers.