Abstract
To grow, insects must periodically shed their exoskeletons. This process, called ecdysis, is initiated by the endocrine release of Ecdysis Trigger Hormone (ETH) and has been extensively studied as a model for understanding the hormonal control of behavior. Understanding how ETH regulates ecdysis behavior, however, has been impeded by limited knowledge of the hormone’s neuronal targets. An alternatively spliced gene encoding a G-protein-coupled receptor (ETHR) that is activated by ETH has been identified, and several lines of evidence support a role in ecdysis for its A-isoform. The function of a second ETHR isoform (ETHRB) remains unknown. Here we use the recently introduced “Trojan exon” technique to simultaneously mutate the ETHR gene and gain genetic access to the neurons that express its two isoforms. We show that ETHRA and ETHRB are expressed in largely distinct subsets of neurons and that ETHRA- but not ETHRB-expressing neurons are required for ecdysis at all developmental stages. However, both genetic and neuronal manipulations indicate an essential role for ETHRB at pupal and adult, but not larval, ecdysis. We also identify several functionally important subsets of ETHR-expressing neurons including one that coexpresses the peptide Leucokinin and regulates fluid balance to facilitate ecdysis at the pupal stage. The general strategy presented here of using a receptor gene as an entry point for genetic and neuronal manipulations should be useful in establishing patterns of functional connectivity in other hormonally regulated networks.
Keywords: behavior, ecdysis, hormones, neural circuit, transgene targeting
HORMONES are major determinants of behavior, playing essential roles in mating, feeding, stress response, and other activities related to survival and reproduction. Identifying the neural circuits through which hormones act, however, has been complicated by the fact that many hormones directly enter the central nervous system and exert their effects at broadly dispersed sites. Establishing the “connectome” of hormonal action thus clearly requires tools different from those used to study synaptic connectivity in neural circuits. Pfaff and his colleagues pioneered the strategy of using sites of hormone binding as a guide to mapping behavioral circuits: By determining the principal sites of estrogen binding in female rat brains they elucidated the network underlying the rodent lordosis response (Pfaff and Keiner 1973; Pfaff et al. 1994). Receptor mapping has similarly provided key insights into the networks underlying other behaviors in both vertebrates and invertebrates, such as feeding (Wu et al. 2003; Scott et al. 2009), sleep and circadian rhythms (Marcus et al. 2001; Im and Taghert 2010), offspring care (Insel 1990), and pair bonding (Young et al. 1997). In general, however, the labor-intensive nature of receptor mapping and the complexity of most hormonally governed neural networks has made them difficult to fully unravel.
A comparatively tractable neuroendocrine network that has been extensively characterized in insects governs the shedding of the exoskeleton at the time of molting, a process called ecdysis (Zitnan and Adams 2012; White and Ewer 2014). This process is initiated by two related peptides (ETH1 and ETH2) encoded by the Ecdysis Triggering Hormone (ETH) gene. The ETH peptides activate a stereotyped sequence of motor programs that varies with species and developmental stage. The execution of the motor programs is accompanied by the successive release of several factors, including eclosion hormone (EH), crustacean cardioactive peptide (CCAP), and bursicon. In Drosophila, neurons that secrete these factors have been shown by calcium imaging to become sequentially active after exposure to ETH peptides—most potently by ETH1—suggesting that they directly coordinate motor programming. In addition, these neurons have been shown by in situ hybridization to express the A isoform of an identified ETH receptor (ETHRA; Kim et al. 2006a,b).
The ETHR gene encodes an alternatively spliced G-protein coupled receptor first identified in the Drosophila genome in 2001 (Hewes and Taghert 2001). Splicing of the gene into two receptor isoforms appears to be highly conserved in insects (Roller et al. 2010), and in Drosophila the ETHRA and ETHRB splice isoforms are known to differ both in their affinities for the ETH peptides (Iversen et al. 2002; Y. Park et al. 2003) and their expression patterns within the nervous system at the pupal stage as determined by in situ hybridization (Kim et al. 2006b). The in vivo functions of ETHR and its two splice variants have not been characterized in detail in any insect. In the red flour beetle, Tribolium, global RNAi knockdown of ETHRA, but not ETHRB, has been shown to produce ecdysis deficits (Arakane et al. 2008). More recently in Drosophila, targeted ETHRA overexpression and tandem knockdown of both splice isoforms have been used to demonstrate that ETHR expression levels influence the timing of execution of ecdysis motor programs (Kim et al. 2015). Manipulation of ETHR expression in neurons that express both CCAP and bursicon alters the onset of the ecdysis motor program, while duration of the pre-ecdysis motor program is influenced by ETHR expression levels in a second group of neurons that express the peptide leucokinin (Lk).
To more comprehensively investigate the function of both the ETHR gene and the neurons that express it in Drosophila, we here use the recently introduced Trojan exon method of genetic targeting (Diao et al. 2015). Our results demonstrate that neurons that express the ETHRA isoform are required for ecdysis at all developmental stages and that functionally distinct subsets regulate not only behavior, but also fluid balance at the time of the pupal molt. In contrast, we find that ETHRB and the neurons that express it are dispensable for larval ecdysis, but are required at both pupal and adult ecdysis. Our results demonstrate that Trojan exon-mediated receptor mapping is a powerful approach to establishing connectivity and function within a hormonally governed circuit.
Materials and Methods
Reagents
All restriction enzymes used were from New England BioLabs Inc. All gene synthesis was performed by Epoch Life Science, Inc. (Sugar Land, TX), all primer synthesis by Integrated DNA Technologies, Inc. (Coralville, Iowa), and sequencing by Macrogen USA (Rockville, MD). qRT–PCR was performed using the Platinum Quantitative RT–PCR ThermoScript One-Step System (Invitrogen, Carlsbad, CA) and all qRT–PCR reactions were run in a DNA Engine Opticon 2 thermocyler (BioRad, Hercules, CA). Synthetic ETH conformed to the sequence of the Drosophila ETH1 peptide and was synthesized by Bachem Americas (Torrance, CA). Tyramine and all other chemicals were from Sigma-Aldrich (St. Louis, MO).
Molecular Biology
Generation of expression constructs:
The constructs used to make transgenic lines with expression specifically in ETHRA- or ETHRB-expressing neurons were designed for insertion into the MI00949 MiMIC site and were modified from synthesized starting constructs that consisted of the region of the ETHR genomic locus from the MI00949 insertion point to the 3′ end of the exon 4b, but included a BamHI site placed immediately before the stop codon of either exon 4a and/or exon 4b of the ETHR gene. In addition, an Hsp70 polyadenylation signal was incorporated into each of the starting constructs at a position just 3′ to the endogenous polyadenylation signal of exon 4a and/or exon 4b to ensure robust transcription termination of the expressed effector. All starting constructs were inserted into the SalI site of pBS-KS-attB1-2 and then modified by inserting effector transgenes into the BamHI sites in exon 4a or exon 4b. The effector transgenes were amplified by PCR from appropriate Trojan exons (Diao et al. 2015) and included fusions of the T2A coding sequence to the sequences encoding: Gal4, Gal80, p65AD, or QF2. (The latter fragment was inserted into a BglII site placed in exon 4b rather than BamHI to allow the BamHI site placed in exon 4a to be used to insert a T2A–Gal4-encoding fragment.)
The UAS–ETHRA rescue construct was made by amplifying the ETHRA sequence by RT–PCR from Drosophila central nervous system (CNS) RNA and cloning it first into the pGEM-Teasy vector and then into pUAST (Drosophila Genomics Resource Center, Indiana University, Bloomington, IN). The UAS–ETHRB rescue construct was synthesized from the GenBank coding sequence (accession no. NM_206533.2) and cloned into EcoRI and Asc I sites of a pH-Stinger vector (Drosophila Genomics Resource Center, Indiana University, Bloomington, IN), which was modified to include an attB site (inserted between the AvrII and XbaI restriction sites) and the 5XUAS–hsp70 minimal promoter sequence of pUAST (inserted between the SphI and EcoRI sites).
qRT–PCR:
RNA was extracted using TRIzol (Life Technologies, Grand Island, NY) from first-instar larvae with the genotypes w; +; ETHRMI00949–Gal4/Df(3R)ED10838 or w;+; ETHRMI06847–Gal4/ Df(3R)ED10838, and relative quantification of gene expression by real-time RT–PCR was used to evaluate the knockout of ETHR. The transcriptional levels of ETHRA and ETHRB were normalized to those of the housekeeping gene β-actin gene Act57B, as done previously by Bahadorani et al. (2010), in both experimental and controls (genotype: w1118; +; +). The expression levels of ETHRA and ETHRB in experimental animals were then calculated as percentage changes relative to those in controls using the method (Livak and Schmittgen 2001). The primers used for qRT–PCR were ETHR exon3 sense, GTACTTGGCCACGAGATG; ETHR exon 4a antisense, TACTCGGCCACCCACAGAAT; and exon 4b antisense, TAGGTGGATATGGCGATGAT; Act57B sense, GCTCTGGTCGTTGACAAT; Act57B antisense, GGTCTCAAACATGATCTG;
Fly lines
Flies were typically raised on standard cornmeal–molasses food at 25° and 65% humidity. For phenotypic analysis, animals were grown on apple juice/agar plates supplemented with yeast paste.
ETHR-specific effector lines were generated by inserting T2A–Gal4, T2A–Gal4DBD, or T2A–3xGal80 Trojan exons into the MI00949 or MI06847 MiMIC sites by plasmid injection as described previously (Diao et al. 2015). To generate Gal4, Gal80, and p65AD effector lines with expression specifically in ETHRA- or ETHRB-expressing cells, we used ΦC31 to insert synthetic constructs (shown in Figure 1A and described below) into the MI00949 MiMIC site. Transgenic fly lines were made by embryonic germline injections and convertants were isolated using the screening procedure described previously (Diao et al. 2015). Insertions into the MI00949 site were homozygous lethal except for ETHRMI00949–Gal4DBD, in which case ∼5% of animals survived to adulthood, perhaps due to inefficient splicing of the Trojan exon. The UAS–ETHRA rescue line was made by P-element transgenesis using standard techniques and the UAS–ETHRB rescue line was made by ΦC31-mediated cassette exchange at the attP40 site on chromosome using the UAS–ETHRB rescue construct described above.
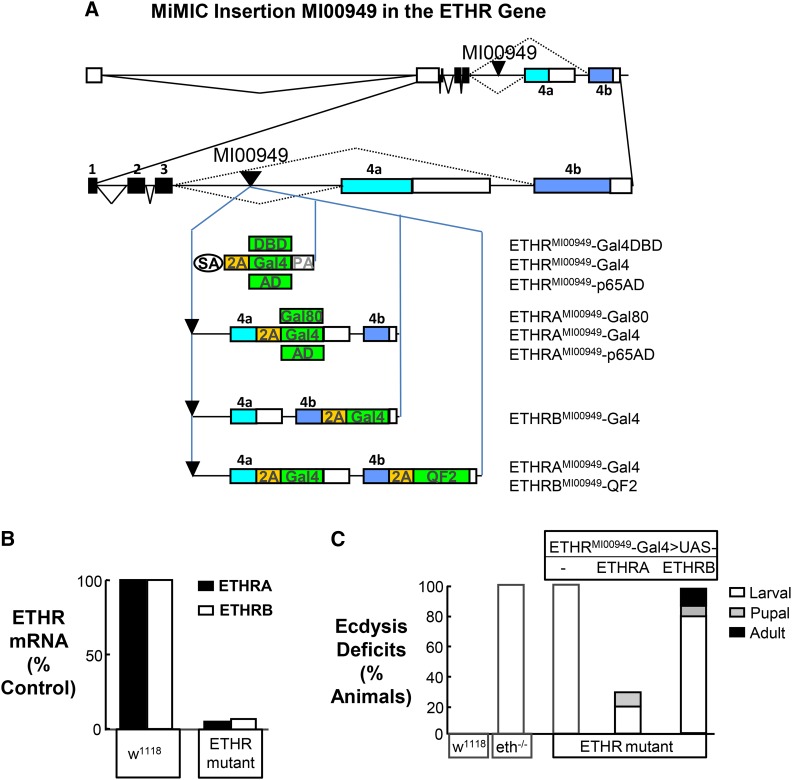
Figure 1.
The ETHR is essential for larval ecdysis. (A) Schematics of the ETHR gene locus and the constructs inserted into the MI00949 MiMIC site to generate the indicated effector lines. Top: entire ETHR locus showing all introns (lines) and exons (solid boxes, coding; open boxes, noncoding). Bottom: inset of the coding region, including alternatively spliced exons 4a (light blue) and 4b (dark blue), followed by the individual constructs integrated into the MI00949 site using ΦC31 integrase. Effector coding sequences are indicated by green boxes, and for similar constructs differing only in the effector sequence, the green box indicating the effector alone is shown. (B) Relative mRNA levels of ETHRA and ETHRB in control animals and in mutant animals bearing the ETHRMI00949–Gal4 insertion over a deficiency allele [i.e., Df(3R)ED10838)] that spans the ETHR locus. The actin transcript is used as reference. The relative levels of ETHRA and ETHRB transcripts in w1118 control flies was set to be 100%. (C) Bar graph showing the percentage of animals of each genotype that died exhibiting ecdysis deficits at the larval (open), pupal (shaded), and/or adult (solid) stage. ETHR mutation was accomplished as in B, and with UAS–ETHRA and UAS–ETHRB driven by ETHRMI00949–Gal4. The ETH null mutant used was eth25a. Larval ecdysis deficits represent the cumulative lethality resulting from deficits at both larval molts. N ≥ 100 in all cases.
The eth25a mutant was the kind gift of Michael Adams (Park et al. 2002) and the following lines were described in J. H. Park et al. (2003): w;CCAP–Gal4;+, w;+; Sb1/TM3,Ser,act–GFP, w; Sp/CyO,act–GFP; Dr/ TM3,Ser,act–GFP, w;+;UAS–rpr,UAS–lacZ/TM3,Ser,Act–GFP; w;+; UAS–LacZ. w;CCAP–Gal80;+ was described in Luan et al. (2006a); w;CCAP–Gal4DBD;+ was described in Luan et al. (2006b), and the Trojan exon lines ChaTMI04508–Gal4DBD, VGlutMI04979–Gal4DBD, amonMI00899-p65AD, and Gad1MI09277–p65AD were described in Diao et al. (2015) All other lines were obtained from the Bloomington Stock Center at Indiana University, including those containing the MI00949 and MI06847 MiMIC insertions in the ETHR gene (Bloomington Stocks 32718 and 41110, respectively), which were generated by the Drosophila Gene Disruption Project, http://flypush.imgen.bcm.tmc.edu/pscreen/mimic.html (Nagarkar-Jaiswal et al. 2015). Lk-Gal4 was Bloomington Stock 51993.
Genetic manipulations
ETHR and ETHRB loss-of-function:
The interruption of the ETHR gene by the T2A–Gal4 Trojan exon placed in the MI00949 locus produces a homozygous lethal, loss-of-function allele of ETHR. To analyze the loss-of-function phenotypes of this allele and to ensure that they did not derive from second-site mutations in the chromosome bearing the ETHRMI00949–Gal4 insertion, we analyzed animals in which this chromosome was paired with a chromosome bearing the Df(3R)ED10838 deficiency (Bloomington Stock no. 9485), which deletes ETHR and 18 neighboring genes. To accomplish this, w;+; ETHRMI00949–Gal4/TM3, Ser, Act–GFP females were crossed with w/>;+; Df(3R)ED10838/TM3, Ser, Act–GFP males (or w1118/>;+;+ for controls) and embryos were collected every 4 hr on apple juice agar plates supplemented with yeast paste. Newly hatched larvae lacking GFP fluorescence were transferred to new apple juice/yeast plates and examined daily for dead larvae, which were scored for ecdysis deficits. Loss-of-function mutants with a ETHRMI06847–Gal4/ Df(3R)ED10838 genotype were similarly generated and analyzed.
Genetic rescue experiments were conducted in parallel with loss-of-function experiments using the same crossing scheme except that animals with the Df(3R)ED10838 deficiency also carried a UAS–ETHRA or UAS–ETHRB rescue transgene. (Because the chromosome bearing the UAS–ETHRA transgene is homozygous lethal, this chromosome was placed over a CyO, act–GFP chromosome.) Larval ecdysis deficits were similarly analyzed in eth25a mutants (kind gift of Michael Adams) bearing null mutations in the ETH gene. Homozygotes from the stable line eth25a/TM3, Ser, Act–GFP were transferred to apple juice agar plates and observed for the larval ecdysis deficits.
Neuronal suppression and ablation:
All neuronal suppression experiments, with one exception, were conducted using two copies of the UAS–Kir2.1 transgene using a line containing inserts on both the second and third chromosomes (i.e., yw; UAS–Kir2.1; UAS–Kir2.1) to ensure the maximal possible expression. This is because transgene expression driven by the ETHR driver lines is developmentally dynamic and therefore episodic. The exception involved the cross of the ChATMI04508–Gal4DBD and ETHRMI00949-p65AD hemidrivers, which used a single copy of UAS–Kir2.1 because both Kir2.1 transgenes could not be genetically recombined with the hemidrivers. The temperature-sensitive Gal4 inhibitor Gal80ts expressed under the control of the ubiquitous tubulin promoter (i.e., tub–Gal80ts) was used in experiments requiring temporally restricted expression of Kir2.1 for conditional suppression (Mcguire et al. 2003). Temperature shifts from 18° to 31° were performed as indicated in the figure legends. Cell-type-specific ablation was carried out as described previously by J. H. Park et al. (2003). Briefly, expression of the cell death gene reaper was targeted to neurons to be ablated together with β-galactosidase (encoded by LacZ), which was used to monitor the success of ablation by immunohistochemistry.
Scoring of ecdysis deficits
In all experiments in which the developmental phenotypes were assessed except those involving manipulation of the CCAP- and Lk-expressing neurons, crosses were conducted on apple juice agar plates supplemented with yeast paste and eggs were collected every 4 hr. Newly hatched larvae were transferred to fresh apple juice plates and scored daily under a stereomicroscope for mortality and ecdysis phenotypes. (In experiments in which CCAP- or Lk-expressing neurons were manipulated animals were grown up and observed in standard food vials.) Larvae exhibiting the “buttoned up” phenotype described by Park et al. (2002), which is characterized by the presence of double mouth hooks, double vertical plates, thick tracheal trunks, and/or partially shed cuticle were scored as having larval ecdysis deficits. Similarly, pupae that died without fully everting their heads and/or appendages, or retained an air bubble in the abdomen indicating failure to initiate pre-ecdysis behavior, were scored as having pupal ecdysis deficits. Although animals with disrupted function of ETHRB or the neurons that express it often everted their heads and appendages before dying, they were also scored as having pupal ecdysis deficits based on their abnormal behavioral and physiological phenotypes as described in the text. Pharate adults that failed to eclose were scored as having adult ecdysis deficits.
Assay of body fluid volume and tyramine feeding
Extractable body fluid volume was measured in batches of 40–45 Lk–Gal4 > 2XUAS–Kir2.1 pupae (or control animals lacking the Lk–Gal4 driver) at 8 and 48 hr APF. The collected animals were weighed and then crushed on a Freeze ‘N Squeeze gel extraction spin column (BioRad, Hercules, CA) using a pellet pestle and centrifuged at 3000 RPM in an Eppendorf microcentrifuge. The volume of fluid collected after centrifugation was measured and the ratio of this volume to the starting weight calculated for each genotype. In those cases where animals were treated with the diuretic agent, tyramine, the compound was administered either by raising first-instar larvae on food containing 10 mg/ml tyramine or by transferring third-instar feeding larvae onto food containing 20mg/ml tyramine.
Microscopy
Immunohistochemistry:
Nervous system wholemounts were excised from either freshly ecdysed second instar larvae or from stage 4 pupae with an air bubble (Bainbridge and Bownes 1981) and prepared for immunolabeling essentially as described previously, using normal donkey or goat serum in the blocking solution (Luan et al. 2006a). Rabbit anti-CCAP (Jena Biosciences, Jena, Germany) was used at 1:1000 dilution, rabbit anti-Burs antibody (Luan et al. 2006a) was used at 1:5000 dilution as previously described, and similarly, anti-Lk, anti-Crz, anti-DMS, anti-DH31, and anti-NPF antibodies were used as described previously (Park et al. 2008). Rabbit or guinea pig secondary antibodies were conjugated to either Alexafluor 488 or Alexafluor 555 (Invitrogen, Carlsbad, CA), or Cy3 (Jackson ImmunoResearch, Inc., West Grove, PA). The pattern of ETHR expression was visualized using either UAS–6XEGFP or by UAS–EYFP. In the latter cases, the immunofluorescence signal was enhanced using a mouse monoclonal anti-GFP antibody (Life Technologies, Grand Island, NY) used at 1:500 dilution. Visualizing Kir2.1 expression was similarly accomplished by amplifying the intrinsic fluorescence of its N-terminal EGFP tag (Baines et al. 2001) using the anti-GFP monoclonal. ETH-immunoreactivity was visualized using a rabbit anti-Drosophila ETH1 antibody (kind gift of Michael Adams) used at 1:2000. In the reaper-mediated cell ablation experiments, mouse anti-β-galactosidase (Promega, Madison, WI) was used at 1:2000 dilution to verify cell killing. Confocal imaging was performed using either an Olympus FV500 or a Nikon C2 confocal microscope using a 20× objective. Unless otherwise noted, the images presented are volume-rendered images of a Z-stack collected through the entire preparation. To quantify the coincidence of ETHRAMI00949–Gal4 expression and anti-CCAP immunostaining, 8 CNS preparations were stained and imaged and individual image planes of a Z-stack were scored for presence or absence of ETHRAMI00949–Gal4-mediated expression of UAS–EYFP. The labeling was assigned a numerical score of 0 (absent), weak (1), moderate (2), or strong (3), with weak labeling typically representing signal that was not detectable in the volume-rendered images. Average scores for each neuron were then calculated and represented as indicated in Figure 4B. Note that paired neurons in hemisegments of the abdominal neuromeres were not individually distinguished in this analysis.
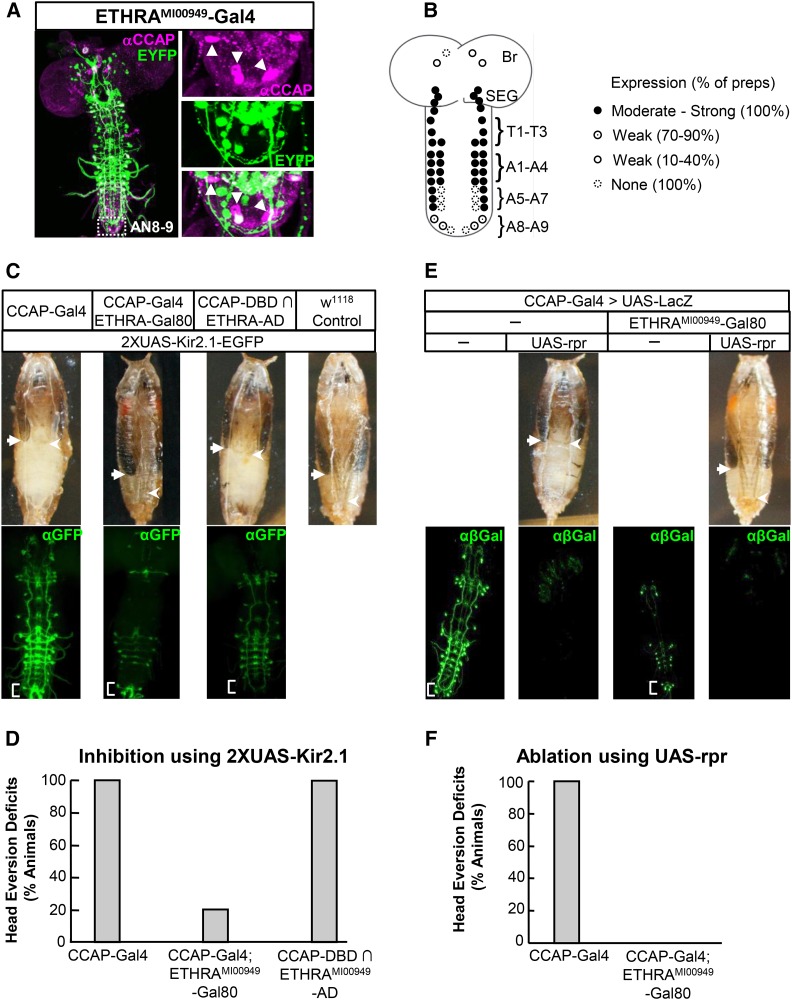
Figure 4.
ETHRA-expressing neurons that coexpress CCAP are required for head eversion at pupal ecdysis. (A) Volume-rendered confocal image of a CNS whole mount from an ETHRAMI009049–Gal4 animal (left) expressing UAS–EYFP (green) double labeled with anti-CCAP antibody (magenta). Right: close-ups of anti-CCAP (top), ETHRA (middle), or overlapping (bottom) labeling of the inset region containing the AN8-9 CCAP-expressing neurons. Three of the five CCAP-expressing neurons in this preparation express little to no ETHRA (arrowheads). (B) Schematic summarizing expression of the ETHRAMI00949–Gal4 line in CCAP-expressing neurons. Levels of expression were analyzed as described in text for eight preparations. (C) Images showing the pupal phenotypes of animals with the indicated genotypes (top), all of which include two copies of the UAS–Kir2.1 transgene. Ecdysis deficits, including failure of head eversion and unextended legs and wings, are seen in pupae in which all CCAP-expressing neurons are suppressed (CCAP–Gal4) or in which the subset that also expresses ETHRA (CCAP–DBD ∩ ETHRA–AD) is suppressed. Maximum extension of the uneverted wings (arrows) and legs (arrowheads) are indicated. The corresponding neuronal patterns of Kir2.1 expression (enhanced by anti-GFP immunostaining) from representative animals of each genotype except the control (which does not express the EGFP-tagged Kir2.1 construct) are shown in volume-rendered confocal micrographs of CNS whole mounts (bottom). (D) Bar graphs showing the percentage of animals of the indicated genotypes dying with head eversion deficits. N ≥ 100 for each genotype. (E) Images showing the pupal phenotypes of two representative animals with the indicated genotypes (top), all of which express a UAS–LacZ transgene under the control of CCAP–Gal4. Ecdysis deficits are seen only in pupae in which all CCAP-expressing neurons are ablated by coexpression of the cell death gene reaper (UAS–rpr), but not if Gal80 is expressed in the ETHRA pattern (ETHRAMI00949–Gal80). Arrows and arrowheads as in C. The LacZ expression in CCAP-expressing neurons was visualized by anti-β-galactosidase immunostaining in animals of each genotype, as shown in the representative volume-rendered confocal micrographs of CNS whole mounts (bottom). (F) Bar graph showing the percentage of animals with the indicated genotypes dying with head eversion deficits.
GCaMP Imaging:
Ca2+ imaging was carried out essentially as described in Kim et al. (2006b). Cryptocephalic pupae expressing GCaMP6s (Chen et al. 2013) under the control of ETHRMI00949–Gal4 were dissected under cold phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 2 mM KH2PO4, pH 7,3) ∼4 hr prior to pupal ecdysis as determined by appearance of the abdominal gas bubble. Central nervous systems excised during dissection were mounted either dorsal or ventral side up and minimally embedded in 200 µl of 1.5% low-melting-temperature agarose solution and left to harden for 45 min in a 10–15° humidified chamber. CNS preparations were then covered with Schneider´s insect medium and GCaMP6s fluorescence in the ventral nerve cord (VNC) was imaged every 3 sec for 60 min using an Olympus DSU Spinning Disc microscope with the disc removed and a 40× W NA 0.80 immersion lens. Each 3-sec image was a composite representing the maximum projection of five images (acquired in five steps of 7 µm at 200 ms per image with 2 sec required for processing and storage). By this means it was possible to record from most cells in the ventral or dorsal half of the VNC being imaged. Preparations were first imaged for 5 min to measure baseline activity, prior to addition of 600 nM synthetic ETH1. Resulting recordings were analyzed using Cell^R Olympus Imaging Software (v. 2.6). Image series were collapsed using Fiji (Schindelin et al. 2012) and additional analysis was carried out using ImageJ (Schneider et al. 2012).
Videorecording of pupal ecdysis behviors:
Cryptocephalic pupae were placed in a humidified chamber approximately 0.5 hr before pupal ecdysis and videorecorded from the ventral side. As originally described by Robertson (1936), animals approaching pupal ecdysis exhibit a prominent abdominal bubble and vigorous movement of the gut, in which the yellow body has appeared and around which the larval fat body begins to break up. To record pupal ecdysis behavior, puparia were coated with a mixture of halocarbon oil and water (∼2:1) applied with a brush to increase their transparency. Puparia were then placed ventral side down on a cover glass, which was attached to a glass slide with double-stick tape to form a small chamber. Behavior was recorded for ∼2 hr at 20× magnification using a Sony NEX VG20 camcorder mounted on an Olympus SZX12 stereomicroscope. Videorecords were then imported into iMovie (Apple Inc., Cupertino, CA) software for analysis. Behaviors corresponding to pre-ecdysis, ecdysis, and post-ecdysis were identified as described by Kim et al. (2006b).
Data availability
All constructs and fly lines described here for manipulating the ETHR gene and the cells that express it are available upon request.
Results
The gene encoding the two ETHR isoforms is essential for ecdysis
To characterize ETHR function, we took advantage of a MiMIC transposon (MI00949) located in the intron preceding the alternatively spliced exons of the ETHR gene, 4a and 4b (Figure 1A; Venken et al. 2011). We have previously shown that replacing such MiMIC insertions with a “Trojan exon” encoding an in-frame T2A-Gal4 construct conveniently provides genetic access to cells expressing the gene in which the MiMIC transposon is inserted by driving expression of the Gal4 transcription factor in the same pattern as the endogenous gene (Diao et al. 2015).
Replacement of MI00949 with the T2A–Gal4 module is thus expected to produce an ETHRMI00949–Gal4 allele together with a nonfunctional ETHR translation product containing only the first four of the seven transmembrane domains found in the mature receptor (i.e., those encoded by exons 1–3). Obtaining such a product requires that splicing of the Trojan exon is efficient. To determine the efficiency of Trojan exon splicing, we performed real-time PCR on RNA isolated from first-instar larvae bearing the ETHRMI00949–Gal4 heterozygous with a chromosomal deficiency that deletes all of the ETHR locus (Df (3R) ED10838). We find that mature transcripts encoding both isoforms of ETHR are present at levels <5% of those found in w1118 control animals (Figure 1B). The ETHRMI00949–Gal4 allele thus substantially knocks out ETHR gene function.
To determine the effects of ETHR gene knockout we analyzed the phenotypes of animals bearing the ETHRMI00949–Gal4 allele heterozygous with the Df (3R) ED10838 deficiency. We found that 99% of these animals die as larvae with double mouth hooks and double vertical plates, phenotypes that are diagnostic for ecdysis failure and similar to those seen in animals lacking a functional ETH gene, as has been described previously (Figure 1C; Park et al. 2002). We took advantage of the fact that the ETHRMI00949–Gal4 line should express Gal4 throughout the expression pattern of the ETHR gene to drive expression of UAS–ETHRA and UAS–ETHRB constructs in the mutant background. We found that the highly penetrant larval ecdysis deficits observed in the mutants were substantially rescued by selectively restoring expression of UAS–ETHRA and that 65% of these animals successfully completed all molts and developed to the adult stage (Figure 1C). In contrast, selectively expressing UAS–ETHRB failed to rescue the larval ecdysis deficit, with only 20% of animals surviving the larval stage. Approximately half (n = 12/22) of the survivors subsequently died at pupal or adult ecdysis.
The phenotypes that we observe from mutating the ETHR locus are consistent with a lesion in the ETH signaling pathway and strongly argue that the encoded ETHR proteins are the sole mediators of ETH peptide actions in ecdysis. Our rescue experiments further suggest that the ETHRA, but not the ETHRB, isoform is essential for larval ecdysis. In addition, the substantial success of rescue at all developmental stages observed with UAS–ETHRA indicates that this isoform can compensate for the loss of ETHRB and that the ETHRMI00949–Gal4 line drives expression of the Gal4 in all cells essential for ecdysis.
Neurons that express the ETHR gene are essential for ecdysis
To further analyze the fidelity of the ETHRMI00949–Gal4 line we examined its expression in the pupal CNS, where ETHR expression has been previously characterized by in situ hybridization (Kim et al. 2006b). Consistent with previous studies, expression of a UAS–6XGFP reporter is driven in neurons in many parts of the CNS (Figure 2A), and specifically in neurons known to express ETHRA mRNA, including those that synthesize EH, Lk, and bursicon (Figure 2B). The pattern of expression observed with fluorescent reporters is considerably broader than that of ETHR message as detected by in situ hybridization—likely due to the much higher sensitivity of the Gal4-amplified reporter expression—and indicates that a large number of central neurons respond to the ETH peptides, ETH1 and ETH2.
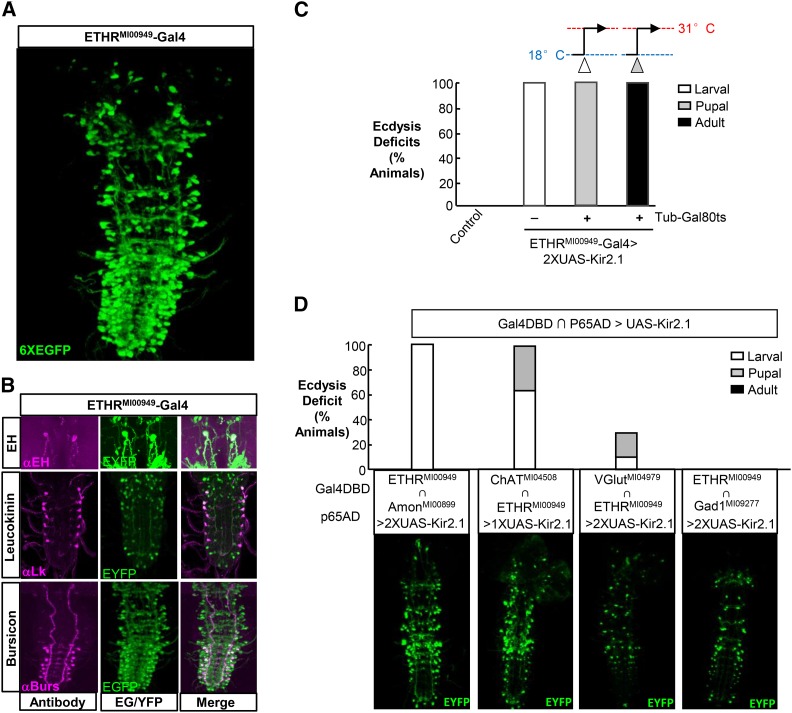
Figure 2.
ETHR-expressing neurons are required for ecdysis throughout development. (A) Volume-rendered confocal image of the expression pattern of the ETHRMI00949–Gal4 line revealed by a UAS–6XEGFP reporter. (B) Neurons that express EH, Lk, and bursicon are included in the expression pattern of ETHRMI00949–Gal4 as revealed by immunostaining specific for each factor. UAS–2xEYFP was used as a reporter for double labeling with anti-EH (top) and anti-Lk (middle), while UAS–6xEGFP was used for anti-Burs double labeling (bottom). (C) Bar graph showing the results of suppressing ETHR-expressing neurons at different developmental stages using 2X UAS–Kir2.1. Complete larval lethality due to ecdysis failure was observed in animals raised at 25°. To examine the effects of suppression on pupal or adult ecdysis, tub–Gal80ts was used to prevent Kir2.1 expression through the larval or pupal stages, with animals shifted from 18° to 31° at the feeding L3 stage (open arrowhead) or 7d APF (shaded arrowhead). N ≥ 50 for each genotype. (D) Effects of suppressing subsets of neurons within the ETHRMI00949–Gal4 pattern that signal via: neuropeptides, acetylcholine, glutamate, or GABA. Top: bar graph showing the percentage of animals dying of ecdysis deficits at the indicated stage in animals expressing UAS–Kir2.1 under the control of an ETHR hemidriver in conjunction with the hemidriver expressing in the pattern of the prohormone convertase, Amontillado (Amon), choline acetyltransferase (ChAT), the vesicular glutamate transporter (VGlut), or glutamic acid decarboxylase (Gad1). Four parallel control crosses in which flies lacked the ETHR hemidriver showed no deficits. Bottom: volume-rendered confocal images showing the expression patterns for the hemidriver pair shown above each image visualized with UAS–EYFP. N = 4–6 for each genotype.
To directly examine the responsiveness to ETH peptides of neurons within the ETHRMI00949–Gal4 pattern, we used this driver to selectively express the genetically encoded calcium indicator UAS–GCaMP6s and monitored changes in intracellular Ca2+ in these neurons in excised pupal nervous systems after application of ETH1. This manipulation has been previously shown to induce long-lasting changes in activity in known ensembles of ETHRA-expressing neurons that coexpress particular peptides, including EH and CCAP (Kim et al. 2006b). We find similar changes in activity in a broad subset of neurons within the ETHRMI00949–Gal4 expression pattern (Supporting Information, Figure S1, File S1, and File S2). These ETH1-induced changes consist of either large Ca2+ spikes (Figure S1B, neurons 97 and 35) or sustained periods of elevated Ca2+ (Figure S1B, neuron 142). Weaker oscillations in Ca2+ levels were also observed in many neurons and may also represent responses to ETH1 (Figure S1B, neuron 21), although future work in which it is possible to identify individual neurons and monitor their activity across preparations will be required to assess the reproducibility of responses to the hormone. However, control preparations to which no ETH1 had been added had similar percentages of neurons with spontaneous Ca2+ oscillations (24% vs. 21% seen in a representative experimental sample, Figure S1C) and contained relatively few neurons that experienced substantial changes in Ca2+ levels (File S3). The results presented here are thus consistent with the conclusion that many, if not most, neurons in the ETHRMI00949–Gal4 expression pattern respond to ETH1.
To directly assess whether the neurons within the ETHRMI00949–Gal4 expression pattern are required for ecdysis, we used this driver to express the inward rectifier K+ channel, Kir2.1, a potent inhibitor of neuronal excitability (Paradis et al. 2001), and examined the effects of this manipulation on viability. We find that suppression of neuronal activity in the ETHRMI00949–Gal4 pattern, similar to the abrogation of ETHR gene function, results in larval lethality with ecdysis defects in 99% of animals (Figure 2C). To determine whether ETHRMI00949–Gal4 expressing neurons are likewise required for ecdysis at later developmental stages, we used a temperature-sensitive allele of the Gal4 inhibitor, Gal80, under the control of the tubulin promoter (i.e., tub–Gal80ts) to permit animals to develop through the larval and/or pupal stages before turning on expression of UAS–Kir2.1(McGuire et al. 2003). Suppression of the ETHR-expressing neurons only during the pupal and adult molts caused lethality with ecdysis deficits in nearly 100% of animals (Figure 2C).
Neuropeptidergic neurons that express the ETHR gene are essential for ecdysis
Taken together, the above data demonstrate that the ETHRMI00949–Gal4 line faithfully targets neurons that express the ETHR and support the conclusion that Trojan exons inserted into the MI00949 site efficiently capture the expression pattern of the ETHR gene. We took advantage of this specificity of expression to also make Trojan exon lines that express the Split Gal4 components, Gal4DBD and p65AD (Luan et al. 2006b; Pfeiffer et al. 2010), in ETHR-expressing neurons (Figure 1A) and used these lines in conjunction with complementary hemidrivers to identify functionally important subpopulations of the ETHR-expressing neurons (Figure 2D). To do so, we used UAS–Kir2.1 to selectively suppress ETHR-expressing subpopulations that use neuropeptides, acetylcholine, glutamate, or GABA as signaling molecules. This involved pairing either ETHRMI00949–Gal4DBD or ETHRMI00949–p65AD with the following previously described Trojan exon hemidrivers: AmonMI00899–p65AD, ChATMI04508–Gal4DBD, VGlutMI04979–Gal4DBD, and Gad1MI09277–p65AD (Amontillado, Amon; Diao et al. 2015). Consistent with the known role of neuropeptidergic neurons in generating the ecdysis sequence, we find substantial larval ecdysis deficits in animals in which Kir2.1 is expressed in neurons that express both ETHR and Amon, a prohormone convertase required for peptide processing (Rayburn et al. 2003). Profound deficits were similarly seen after silencing the cholinergic subset of ETHR-expressing neurons, which appears to overlap considerably with the Amontillado-expressing, peptidergic component (Figure 2D, bottom). Suppression of the subset of ETHR-expressing neurons that coexpress the vesicular glutamate transporter, VGlut, results in more modest ecdysis deficits, which nevertheless suggests some role for glutamatergic neurotransmission within the ecdysis network. Interestingly, although there is strong evidence that activity within the ecdysis circuit is regulated by inhibitory inputs (Baker et al. 1999; Zitnan and Adams 2000; Fuse and Truman 2002), we found no indication that ETHR-expressing neurons themselves use the inhibitory transmitter GABA as an essential signaling molecule. However, the existence of neurons that express both ETHR and Gad1 (Figure 2D, bottom) suggests that GABAergic inhibition may nevertheless shape the output of the ecdysis network in ways that do not result in lethality. It is also possible that for the GABAergic subset, as for the other subsets, suppression was not complete. However, for all hemidriver combinations except the one targeting the cholinergic subset of ETHR-expressing neurons, we used two copies of the UAS-Kir2.1 transgene.
The expression patterns of ETHRA and ETHRB are largely distinct
Previous observations indicating that the ETHR splice variants are expressed in different subsets of neurons at the pupal stage (Kim et al. 2006b) suggests that the two ETHR isoforms may also mediate distinct functions. To directly examine the expression patterns and functions of ETHRA and ETHRB, we selectively targeted them using a modified version of the Trojan exon technique. We introduced constructs into the MI00949 site that duplicated the ETHR gene sequence 3′ to this site except for the fusion of T2A–Gal4 coding sequences to exons 4a or 4b, just prior to the stop codons for ETHRA or ETHB, respectively (Figure 1A). Insertion of these constructs is expected to maintain the normal pattern of alternative splicing of the modified exons and result in the production of full-length ETHRA and ETHRB translation products fused to T2A peptides accompanied by the coexpression of Gal4. To facilitate comparison of the ETHRA and ETHRB expression patterns we similarly used a modified Trojan exon bearing a T2A–Gal4 fusion to the coding sequence of ETHRA and a T2A–QF2 fusion to the coding sequence of ETHRB to allow independent expression of different fluorescent protein reporters in ETHRA- and ETHRB-expressing neurons (Figure 1A).
The expression patterns of the ETHRAMI00949–Gal4 and ETHRBMI00949–Gal4 lines, as assessed using a UAS–EYFP reporter, reveal that both ETHRA and ETHRB are broadly expressed in the pupal CNS (Figure 3A). Consistent with what has been noted previously, the ETHRAMI00949–Gal4 expression pattern contains many peptidergic neurons, including previously identified subsets known to express the peptides FMRFamide, Lk, and CCAP as well as EH (data not shown) (Kim et al. 2006b, 2015). Double labeling with a palette of antipeptide antibodies revealed additional small subsets of peptidergic neurons in the central brain within the ETHRAMI00949–Gal4 expression pattern (Figure S2), including neurons expressing the factors corazonin (Crz), myosuppressin (MS), diuretic hormone 31 (DH31), and neuropeptide F (NPF). ETHRB expression, as revealed by use of the dual-ETHR line, ETHRAMI00949–Gal4/ETHRBMI00949–QF2, is largely distinct from that of ETHRA (Figure 3A, right), with overlapping expression observed in only four bilateral pairs of neurons, which are located in the central brain, the subesophageal ganglion (SEG), the third thoracic ganglion (T3) of the ventral nerve cord, and in one of the abdominal neuromeres (AN). Based on their position and morphology, the neurons in the central brain appeared to correspond to the EH-expressing Vm neurons, which we confirmed by anti-EH immunostaining (data not shown). The other three pairs express the neuropeptide CCAP, but unlike the Vm neurons, they are only inconsistently observed in the expression patterns of the ETHRBMI00949–Gal4 line and may not robustly express ETHRB.
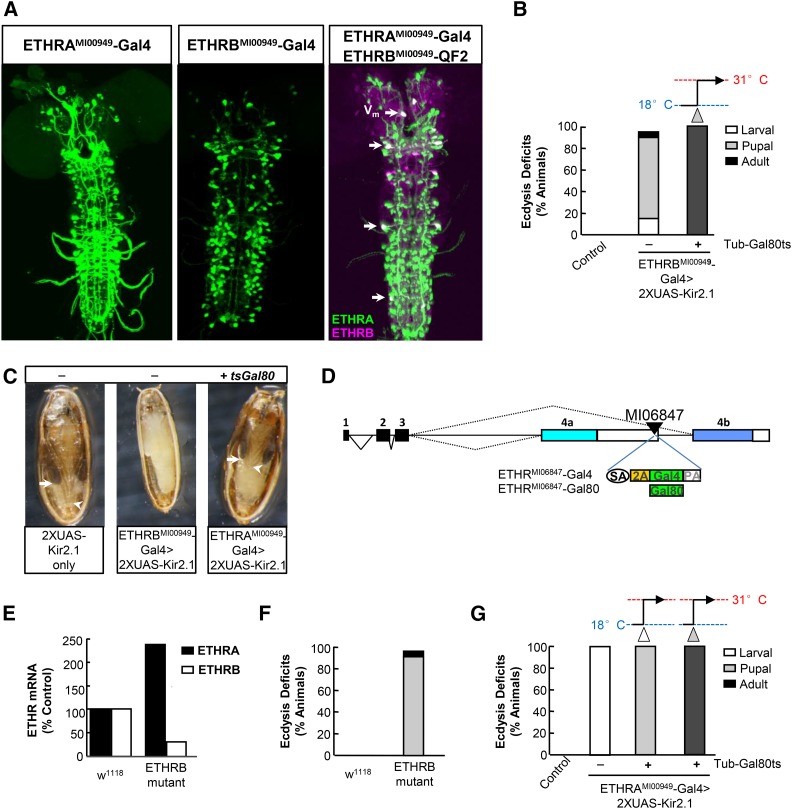
Figure 3.
ETHRA- and ETHRB-expressing neurons play different developmental roles in ecdysis. (A) Volume-rendered confocal images of CNS whole mounts showing the expression patterns of drivers selective for ETHRA- (left) and ETHRB-expressing (middle) neurons or both (right). Drivers were made as described in Figure 1A. Arrows, neurons that express both ETHR isoforms. Vm, EH-expressing ventromedial neurons. (B) Bar graph showing the percentage of animals dying with ecdysis deficits at the indicated developmental stage when neurons within the ETHRBMI00949–Gal4 expression pattern are suppressed using 2X UAS–Kir2.1. Animals raised at 25° died at the pupal stage and to examine the effects of suppression at the adult stage, tub–Gal80ts was used and animals were shifted from 18° to 31° at d7 APF (gray arrow). N ≥ 50 for each genotype. (C) Photomicrographs of control pupae (2X UAS–Kir2.1 only, left) and pupae that have died due to suppression of the ETHRB-expressing (middle) or ETHRA-expressing (right) neurons. Suppression of ETHRA-expressing neurons at the pupal stage was accomplished using tub–gal80ts as described in the legend of Figure 2C. Maximum extension of the uneverted wings (arrows) and legs (arrowheads) are indicated. (D) Schematic of the coding regions of the ETHR gene locus similar to that shown in Figure 1A, indicating the site of the MI06847 MiMIC insertion and the two constructs integrated into that site using ΦC31 integrase. As in A, the two constructs share the same design and differ only in the effector (green box). (E) Relative mRNA levels of ETHRA and ETHRB in control animals and in mutant animals bearing the ETHRMI06847–Gal4 insertion over a deficiency allele [Df(3R)ED10838] that spans the ETHR locus. The transcript level of actin is used as reference. The relative levels of ETHRA and ETHRB transcripts in control flies was set to be 100%. (F) Bar graph showing the effects of mutating ETHRB by placing the ETHRBMI06847–Gal4 insertion over the Df(3R)ED10838 deficiency allele. Percentage of animals dying with pupal (shaded) and adult (solid) ecdysis deficits are shown. N ≥ 100 for each genotype. (G) Bar graph showing the percentage of animals dying with ecdysis deficits at the indicated developmental stage when neurons within the ETHRAMI00949–Gal4 expression pattern are suppressed using 2X UAS–Kir2.1. Animals raised at 25° died at the larval stage and to examine the effects of suppression at the pupal and adult stages, tub–Gal80ts was used and animals were shifted from 18° to 31° as feeding L3 larvae (open arrow) or at d7 APF (shaded arrow), respectively. N ≥ 50 for each genotype.
Neurons that express ETHRB are essential for pupal and adult, but not larval, ecdysis
While several classes of ETHRA-expressing neurons have been implicated in mediating ecdysis, the role of ETHRB-expressing neurons is, in general, unknown. To examine the function of these neurons, we used the ETHRBMI00949–Gal4 line to drive expression of UAS–Kir2.1 and found that suppression of ETHRB-expressing neurons leads to substantial developmental lethality, with most animals dying as pupae (Figure 3B). The cause of death was difficult to assign, but these animals exhibited numerous abnormalities at pupal ecdysis. A common marker used to stage animals for pupal ecdysis—i.e., the bubble that normally forms in the abdomen and disappears early in the ecdysis sequence (Robertson 1936; Bainbridge and Bownes 1981)—was not always present. In addition, behavioral expression was disrupted leaving animals pushed toward the anterior end of the puparium at the end of pupal ecdysis where they remained shrunken and white until death (Figure 3C, middle). This was in contrast to control animals, which pull back to the posterior end of the puparium and eventually darken (Figure 3C, left). A small fraction of animals in which the ETHRB-expressing neurons were suppressed developed to the adult stage, but then failed to eclose. Use of Gal80ts to restrict neuronal suppression to just prior to adult emergence confirmed that ETHRB-expressing neurons are required for adult ecdysis (Figure 3B).
In contrast to its effects on both pupal and adult ecdysis, suppressing the ETHRB-expressing neurons caused only limited lethality at the larval stage (Figure 3B). However, the levels of transgene expression driven by the ETHRBMI00949–Gal4 line are weak compared to those driven by the ETHRMI00949– and ETHRAMI00949–Gal4 lines (data not shown), raising the possibility that the levels of suppression achievable with this line might be insufficient to completely block larval ecdysis. We therefore interfered with ETHRB function by an alternative approach, taking advantage of a MiMIC transposon (MI06847) fortuitously inserted into the 3′ UTR of exon 4a (Figure 3D), which we reasoned should prevent normal splicing of the ETHRB isoform after insertion of a T2A–Gal4 Trojan exon.
We confirmed that the ETHRMI06847–Gal4 insertion substantially reduces expression of ETHRB by real-time PCR, although the insertion also elevates levels of ETHRA expression (Figure 3E), perhaps due to aberrant splicing induced by the interruption of exon 4a. Most animals bearing the ETHRMI06847–Gal4 allele over the Df(3R)ED10838 deficiency survive the larval stage, but 95% die as pupae with deficits similar to those observed in ETHRBMI00949–Gal4 > UAS–Kir2.1 animals (Figure 3F). Neuronal suppression in the ETHRMI06847–Gal4 pattern resulted in more severe pupal ecdysis deficits than those seen in ETHRBMI00949–Gal4 > UAS–Kir2.1 animals (data not shown), and several lines of evidence indicate that ETHRMI06847–Gal4 pattern includes ETHRA-expressing neurons (see results with ETHRMI06847–Gal80 below). Unlike the ETHRBMI00949–Gal4 line, the ETHRMI06847–Gal4 line thus appears to only approximate the native distribution of the ETHRB isoform at the larval and pupal stages, but taken together the results obtained with the two lines argue that ETHRB and the neurons that express it are essential for pupal and adult, but not larval, ecdysis.
Neurons that express ETHRA are essential for ecdysis at all developmental stages
In contrast to the ETHRB-expressing neurons, neurons that express ETHRA are required for larval ecdysis: all ETHRAMI00949–Gal4 > UAS–Kir2.1 animals die at the larval stage with ecdysis deficits (Figure 3G). In an effort to isolate essential subsets of neurons required for larval ecdysis, we blocked Gal4 activity in various subsets of ETHRA-expressing neurons using Gal80-expressing constructs, including one made by insertion of a 3XGal80 Trojan exon into the MI06847 site (Figure 3D). Use of this construct, ETHRMI06847–Gal80, together with one that expresses Gal80 in all CCAP-expressing neurons (CCAP–Gal80), substantially restricted the larval ETHRAMI00949–Gal4 expression pattern to ∼20 bilateral pairs of neurons in the CNS (Figure S3A). Suppression within this restricted pattern still blocked larval ecdysis (data not shown). Using antipeptide antibodies, we were able to positively identify both NPF and Lk-immunopositive neurons among this group (Figure S3, B and C), but UAS–Kir2.1-mediated suppression using Gal4 lines that drive expression in cells that make these two neuropeptides failed to inhibit larval ecdysis. However, the line specific for leucokinin-expressing neurons (Lk–Gal4) did yield pronounced pupal ecdysis deficits (see below). We conclude that CCAP-, Lk-, and NPF-expressing neurons are dispensable for larval ecdysis, but have yet to identify those neurons within the ETHRAMI00949–Gal4 expression pattern that are required for this process.
In addition to larval ecdysis, ETHRA-expressing neurons are also required for pupal ecdysis: restricting suppression of this group to the pupal stage using Gal80ts resulted in 100% lethality with failure of head eversion (Figure 3G). Despite this deficit, and in contrast to animals in which the ETHRB-expressing neurons are suppressed, these animals continue to develop, acquiring characteristics of pharate adult animals such as darkened wings and pigmented eyes (Figure 3C, right). These animals did not eclose, however, and selective suppression of ETHRA-expressing neurons at the pharate adult stage blocked eclosion, or adult ecdysis, demonstrating that ETHRA-expressing neurons are required for ecdysis at all developmental stages (Figure 3G).
ETHRA-expressing neurons that express CCAP are required for head eversion
The block of head eversion observed upon suppression of ETHRA-expressing neurons is consistent with the known requirement for CCAP-expressing neurons in pupal ecdysis (J. H. Park et al. 2003; Kim et al. 2006b; Luan et al. 2006a). Interestingly, however, the ETHRAMI00949–Gal4 expression pattern did not appear to consistently include all CCAP-expressing neurons. In particular, late-differentiating CCAP-expressing neurons in the posterior segments of the ventral nerve cord (AN5-9) were often missing. These neurons have previously been shown to be required for pupal ecdysis, with the subset in segments AN8-9 specifically implicated in mediating head eversion (Veverytsa and Allan 2012). However, the latter neurons are not among those reported to express ETHRA based on in situ hybridization studies (Kim et al. 2006b), and we likewise find that they are mostly absent from the expression patterns of our ETHR Gal4 lines when visualized by a UAS–EYFP reporter alone. Immunostaining the fluorescent reporter to determine the full extent of labeling reveals weak and inconsistent expression in two-thirds of neurons (Figure 4, A and B). The feeble and sporadic ETHRA expression in these neurons was surprising, given the essential functional role ascribed to them, and we therefore sought to confirm the importance of these neurons to pupal ecdysis using intersectional methods.
To selectively manipulate neurons that express CCAP, but not ETHRA, we used CCAP–Gal4 together with an ETHRAMI00949–Gal80 line, created using a 3XGal80 Trojan exon (Figure 1A). Although the ETHRAMI00949–Gal80 does not completely suppress CCAP–Gal4 activity in ETHRA-expressing neurons it substantially reduces it while retaining high UAS–Kir2.1 expression in the AN8-9 CCAP-expressing neurons (Figure 4C, bracket). Despite this, Kir2.1 expression does not block head eversion, indicating that CCAP-expressing neurons outside of the ETHRA pattern are dispensable for that process (Figure 4, C and D). Indeed, animals subjected to this manipulation typically complete development and eclose as adults. To exclude the possibility that head eversion deficits were not observed due to insufficient Kir2.1-mediated suppression of electrical activity, we also ablated the same set of neurons using the cell death gene reaper (UAS–rpr) and likewise observed no overt impairment of head eversion (Figure 4, E and F).
In addition, we selectively suppressed the complementary set of CCAP- and ETHRA-expressing neurons. To do so, we created an ETHRAMI00949-p65AD hemidriver using the modified Trojan exon method (Figure 1A) and paired it with a CCAP–Gal4DBD hemidriver to drive UAS–Kir2.1. We found that suppression of neurons that express both CCAP and ETHRA results in animals with severe head eversion deficits that fail to complete pupal ecdysis (Figure 4, C and D). Together, the results from these intersectional experiments demonstrate that only those CCAP-expressing neurons that are direct targets of ETH are essential for pupal ecdysis, a population that does not consistently include the AN8-9 CCAP-expressing neurons.
ETHRA-expressing neurons that co-express Leucokinin are required for pupal ecdysis
As noted above, neurons that express the Lk neuropeptide are also direct targets of ETH and express the ETHRA isoform. Although Lk is known primarily for promoting diuresis in insects (Halberg et al. 2015), this peptide has also been proposed to play a role in ecdysis-related behaviors in the hawkmoth, Manduca sexta (Kim et al. 2006a), and more recently, Lk-expressing neurons have been directly implicated in regulating the timing of pupal pre-ecdysis in Drosophila (Kim et al. 2015). To confirm that Lk-expressing neurons in Drosophila are activated by ETH, we used Lk–Gal4 to drive expression of UAS–GCaMP6s and examined responses to ETH in CNS preparations excised just prior to pupal ecdysis. We consistently observe rapid—albeit comparatively weak—induction of activity in the Lk-expressing neurons under these conditions (Figure 5A and File S4). The average latency from the time of ETH application is short (7.8 ± 0.3 min, SE; n = 167), and the average duration of response is 18.5 ± 0.9 min (SE, n = 167), with activity in the Lk-expressing neurons of AN7 typically being the most sustained (Figure 5B).
Figure 5.
Impairment of pupal ecdysis with suppression of leucokinin-expressing neurons. (A) Ca2+ levels in Lk-expressing neurons of the abdominal ganglion (indicated by segment) as measured by GCaMP6s at the time of ETH addition (t = 0, left) and 5 min later (right). Relative Ca2+ levels increase with color spectrum from blue to red to white. (B) Time traces of GCaMP6s activity seen in responding Lk neurons from excised CNS preparations treated with ETH1. (C) Bar graph showing the percentage of animals with the indicated genotypes that exhibited failure of wing and leg extension at pupal ecdysis. Most animals in which the Lk-expressing neurons were suppressed with two copies of the UAS–Kir2.1 transgenes using Lk–Gal4, showed such deficits, but not when they were administered tyramine (TY) throughout development. (D) Photomicrographs of pharate adults in which two copies of the UAS–Kir2.1 transgene were either not expressed (control, left) or expressed under control of Lk–Gal4 to inhibit Lk-expressing neurons (right). The heads of the bloated Lk–Gal4 > Kir2.1 animals are squeezed forward to the anterior edge of the puparium and have unextended wings and legs compared with the control animal. White arrows and arrowheads, extension of wings and legs, respectively; black arrowhead, accumulated fluid in the abdomen; gray arrowhead, position of the eyes. (E) Bar graphs showing the accumulation of extracellular fluid in the puparium of control animals and animals in which the Lk-expressing neurons have been silenced. Fluid levels are already increased in Lk–Gal4 > 2X Kir2.1 animals at 8 hr (white bars) after puparium formation (APF) and remain elevated at 48 hr (black bars) APF.
To determine the role of Lk-expressing neurons in pupal ecdysis, we next examined the effects of neuronal suppression using UAS–Kir2.1. We found that 80% of animals in which the Lk-expressing neurons were suppressed died at the pupal stage. Although none of these animals had head eversion deficits, 90% failed to fully extend their legs and wings, two processes that immediately follow head eversion during pupal ecdysis (Figure 5, C and D). An interesting feature of the pupae in which the Lk-expressing neurons were suppressed was that they seemed bloated, with their heads filling the anterior end of the puparium, which is normally occupied by air (Figure 5D; right). We assayed the accumulation of extracellular fluid by directly measuring the extractable volume of liquid normalized to body weight. We find that fluid accumulation is elevated at 8 hr after puparium formation and continues to increase through the period of pupal ecdysis, which occurs ∼12 hr after puparium formation (Figure 5E).
Careful observation revealed several other abnormalities in animals in which Lk-expressing neurons were suppressed. The bubble that appears in the abdomen several hours prior to pupal ecdysis is smaller in these animals and simply floats to the anterior surface instead of moving laterally and then posteriorly. To determine whether this abnormality is associated with deficits in pre-ecdysis behavior, we videorecorded animals during the period of pupal ecdysis and observed that all motor patterns previously described appear to be executed (Kim et al. 2006b). However, the movements are greatly attenuated—perhaps due to swelling of the body within the fluid-filled puparia (File S5). These observations suggest that the ecdysis deficits we observed might be secondary to deficits in regulating fluid balance.
A primary target of Lk peptide in the regulation of fluid balance is the Malpighian tubules, which secrete fluid into the hindgut (Terhzaz et al. 1999). The neurotransmitter tyramine has been shown to act on the Malpighian tubules by a similar mechanism to promote diuresis (Cabrero et al. 2013), and we therefore sought to reverse the effects of Lk-neuron suppression by feeding larvae with tyramine, either chronically or starting at the third-instar larval stage. We found that both treatments effectively prevented fluid accumulation in the pupae and, in addition, eliminated the previously observed pupal ecdysis deficits (Figure 5C). These results support the conclusion that the observed pupal ecdysis deficits are a secondary consequence of dysregulation of Lk’s primary role in regulating fluid balance.
Discussion
To characterize the neural circuit that governs ecdysis in Drosophila, we have exploited the Trojan exon technique to map and manipulate the ETH signaling pathway and have analyzed the effects of genetic and neuronal loss-of-function at the level of the ETHR gene, its splice variants, and the cells that express them. We find that genetic disruption of ETHR expression phenocopies ETH loss-of-function, indicating that the ETHR gene encodes the sole receptor for ETH peptides in mediating ecdysis. Consistent with this, we find that suppression of ETHR-expressing neurons blocks ecdysis at all developmental stages. Both the genetic and neuronal loss-of-function data further reveal distinct developmental requirements for the two ETHR isoforms and the neurons that express them, with ETHRB-, but not ETHRA-expressing neurons dispensable for larval ecdysis. Finally, we demonstrate that ETHR-expressing neurons regulate both behavioral and physiological processes at pupal ecdysis.
ETHRA and ETHRB are differentially required during development
Previous observations have shown that ETH initiates ecdysis at all stages in Drosophila development (Park et al. 1999, 2002; Clark et al. 2004; Kim et al. 2006b), but apart from work done on a small subset of neurons known to express ETHRA, the broader ecdysis circuit targeted by ETH and the functional roles of its receptors have remained largely uncharacterized. The data presented here confirm the importance of ETHRA in ecdysis, as previously demonstrated in Tribolium using RNAi knockdown (Arakane et al. 2008). In addition, they demonstrate, for the first time in any insect species, an essential function for ETHRB in that selective knockdown of ETHRB expression substantially blocks pupal ecdysis. The fact that larval ecdysis is largely unimpaired by this manipulation—together with the observation that restoration of ETHRA, but not ETHRB, expression compensates for loss of ETHR function at larval ecdysis—strongly argues that ETHRA and ETHRB have distinct functional roles and that their contribution to ecdysis is differentially dependent on developmental stage. These conclusions are consistent with the results of neuronal suppression using ETHRA- and ETHRB-specific Gal4 drivers: ETHRA-expressing neurons are required for ecdysis throughout development, whereas ETHRB-expressing neurons are required only after the larval stage. In addition, the differing phenotypes of pupal lethality seen with suppression of ETHRB- vs. ETHRA-expressing neurons argues that the two receptor isoforms mediate different processes.
Our results also confirm and extend the conclusions drawn previously from in situ hybridization studies that ETHRA and ETHRB are expressed in distinct populations of neurons (Kim et al. 2006b). That this is also the case in the hawkmoth, Manduca (Kim et al. 2006a) suggests that this feature, like the generation of ETHRA and ETHRB splice isoforms itself, is highly conserved. Our finding that ETHRA and ETHRB serve distinct functions that are mediated by different populations of neurons thus seems likely to represent an evolutionarily ancient characteristic of insect ecdysis circuits.
Interestingly, an exception to the mutually exclusive expression of ETHRA and ETHRB occurs in the Vm neurons, which secrete EH and occupy a unique role in the ecdysis circuit. EH acts in a well-characterized positive feedback loop between the Vm neurons and the ETH-secreting Inka cells (Ewer et al. 1997; Kingan et al. 1997; Clark et al. 2004). ETH and EH are initially released at low levels, but each reinforces the other’s secretion to cause the later, massive release of both hormones at high levels, an event that is thought to drive the progression of the ecdysis sequence. It is possible that the ETHRB isoform, which exhibits higher sensitivity to ETH than ETHRA (Iversen et al. 2002; Y. Park et al. 2003), mediates the initial Vm response to ETH causing low-level EH release, while expression of the ETHRA isoform participates in the late, high-level response. Further work will, however, be required to test this hypothesis.
ETHRA-expressing neurons regulate multiple processes at pupal ecdysis
While the function of ETHRB-expressing neurons has been enigmatic, several groups of peptidergic neurons known to express ETHRA have been previously implicated in governing ecdysis. The results presented here refine and expand our knowledge of two such cell groups, those that express CCAP or Lk.
CCAP-expressing neurons as a group have been shown to be required for head eversion, a defining event of pupal ecdysis (J. H. Park et al. 2003). The results presented here provide direct evidence that the subset of CCAP-expressing neurons that coexpresses ETHRA is the one required for head eversion. A previous study of a late-differentiating CCAP-expressing neurons concluded that cells in abdominal neuromeres AN8-9 are required for head eversion (Veverytsa and Allan 2012). However, these neurons are not observed to express ETHRA mRNA (Kim et al. 2006b), and we were surprised to likewise find them inconsistently represented in the expression patterns of our ETHRMI00949– and ETHRAMI00949–Gal4 lines. Our finding that both electrical suppression and ablation of these neurons leaves head eversion unimpaired forces us to conclude that CCAP-expressing neurons in AN8-9 are not responsible for this process. This conclusion is consistent with a recent report that activation of CCAP-expressing neurons that also express bursicon (and therefore do not include the AN8-9 neurons) is sufficient to induce head eversion (Kim et al. 2015). It is possible that the correlational nature of earlier results (Veverytsa and Allan 2012), which were based on the effects of stochastic ablation of subsets of all CCAP-expressing neurons, may have inadvertently suffered from sampling errors that biased the interpretation to the opposite conclusion. In general, our demonstration that only those CCAP-expressing neurons that coexpress ETHRA block ecdysis underscores the ability of the receptor-based mapping approach described here to correctly identify critical nodes in hormonally controlled behavioral circuits.
A second critical node revealed by our analysis is the subset of ETHRA-expressing neurons that express the diuretic factor leucokinin. Based on neuronal suppression experiments using two copies of the UAS–Kir2.1 transgene, we demonstrate that Lk-secreting neurons maintain fluid balance to support behavioral execution. We find that ecdysis deficits induced by neuronal suppression are reversed together with fluid imbalance by feeding of tyramine suggesting that these phenotypes share a common cause in the dysregulation of fluid secretion at the level of the Malpighian tubules (Terhzaz et al. 1999). A report that appeared during revision of this manuscript indicates that Lk-expressing neurons also specifically regulate the timing of pre-ecydsis behavior (Kim et al. 2015), which we did not assay. It will be interesting to determine whether these deficits are independent of fluid imbalance and persist in tyramine-fed animals. Curiously, neither fluid imbalance nor the overt deficits in appendage extension that we observe are described by Kim et al. (2015) as a consequence ablating the Lk-expressing neurons. It is possible that this apparent inconsistency results from the use of different Lk–Gal4 drivers, or from the differential efficacy of neuronal suppression by UAS–Kir2.1 vs. cell killing by UAS–rpr, but in either case, the two studies support the conclusion that Lk-expressing neurons regulate physiological and/or behavioral processes important for pupal ecdysis.
Multiple functional subsets of ETHR-expressing neurons
Consistent with what has been previously found (Kim et al. 2006a,b; Zitnan and Adams 2012), our analysis suggests that peptidergic neurons, in general, are well represented within the ETHR expression pattern and are essential to the ecdysis circuit. We also provide evidence that subsets of ETHR-expressing neurons that use the neurotransmitters acetylcholine and glutamate are functionally important. Suppression of the cholinergic subset potently blocks ecdysis at both the larval and pupal stages and may well include many neurons that also express peptides. The glutamatergic subset, however, is likely to be distinct from the cholinergic group based on the previously reported nonoverlapping expression of the cholinergic marker, ChaT, and the glutamatergic marker, VGlut (Diao et al. 2015). Interestingly, electrical suppression of a GABAergic subset of ETHR-expressing neurons does not result in overt ecdysis failure. It thus seems likely that inhibitory inputs previously shown to regulate the execution of different phases of the ecdysis sequence in Manduca and thought to also function in Drosophila (Baker et al. 1999; Zitnan and Adams 2000; Fuse and Truman 2002) do not derive from ETHR-expressing neurons, are not GABAergic, or are not strictly essential for ecdysis. We favor the last possibility and note that we have focused here only on gross ecdysis deficits. More subtle defects that affect behavioral coordination, execution, or timing and do not result in lethality will require closer analysis. Our preliminary results, however, suggest that many of the ETHR-expressing neurons identified in this study can be expected to play specific roles in ecdysis at some developmental stage.
The efficacy of Trojan exon-mediated receptor mapping
The Trojan exon methodology used here to identify, manipulate, and parse the patterns of ETHR expression represents a systematic and versatile strategy for mapping functional connectivity within hormone-mediated neural circuits. In the case of the ecdysis circuit, this strategy has not only facilitated analysis of the neural substrates of behavior and physiology, but has revealed unanticipated developmental differences in the importance of the two ETHR isoforms. In the fly as in other insects, the motor patterns that mediate ecdysis vary considerably across developmental stages to accommodate differences in body plan and environmental context. However, the changes that occur in the ecdysis circuit over development remain largely unknown. The tools developed here should provide the basis for a thorough-going investigation of this, and other, important issues.
Acknowledgments
We thank Michael Adams for the eth25a mutant and anti-ETH antibody, Haojiang Luan, Amicia Elliott, and Rodrigo Mancilla for technical assistance, and Haojiang Luan and Amicia Elliott for critical reading of the manuscript. This work was supported by the Intramural Research Program of the National Institute of Mental Health (B.H.W.) and by grants from FONDECYT (no. 1141278) and Centro Interdisciplinario de Neurociencia de Valparaíso (P09-022-F), which is supported by the Millennium Scientific Initiative of the Ministerio de Economía, Fomento y Turismo (J.E.), and by National Institutes of Health (NIH) grants from the National Institute of Neurological Disease and Stroke (R01NS021749) and the National Institute of Mental Health (R01MH067122) (P.T.). In addition, we thank the Bloomington Stock Center and the Drosophila Gene Disruption Project which generated the MiMIC lines used here. The authors declare no conflicts of interest.
Footnotes
Communicating editor: H. J. Bellen
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.182121/-/DC1.
Sequence data from this article have been deposited with the GenBank Data Libraries under accession no. NM_206533.2.
Literature Cited
- Arakane Y., Li B., Muthukrishnan S., Beeman R. W., Kramer K. J., et al. , 2008. Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mech. Dev. 125: 984–995. [DOI] [PubMed] [Google Scholar]
- Bahadorani S., Bahadorani P., Marcon E., Walker D. W., Hilliker A. J., 2010. A Drosophila model of Menkes disease reveals a role for DmATP7 in copper absorption and neurodevelopment. Dis. Model. Mech. 3: 84–91. [DOI] [PubMed] [Google Scholar]
- Bainbridge S. P., Bownes M., 1981. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66: 57–80. [PubMed] [Google Scholar]
- Baines R. A., Uhler J. P., Thompson A., Sweeney S. T., Bate M., 2001. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci. 21: 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J. D., McNabb S. L., Truman J. W., 1999. The hormonal coordination of behavior and physiology at adult ecdysis in Drosophila melanogaster. J. Exp. Biol. 202: 3037–3048. [DOI] [PubMed] [Google Scholar]
- Cabrero P., Richmond L., Nitabach M., Davies S. A., Dow J. A. T., 2013. A biogenic amine and a neuropeptide act identically: tyramine signals through calcium in Drosophila tubule stellate cells. Proceedings of the Royal Society B-Biological Sciences, Vol. 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. W., Wardill T. J., Sun Y., Pulver S. R., Renninger S. L., et al. , 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. C., Del Campo M., Ewer J., 2004. Neuroendocrine control of larval ecdysis behavior in Drosophila: complex regulation by partially redundant neuropeptides. J. Neurosci. 24: 4283–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F., Ironfield H., Luan H., Diao F., Shropshire W. C., et al. , 2015. Plug-and-play genetic access to Drosophila cell types using exchangeable exon cassettes. Cell Rep. 10: 1410–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer J., Gammie S. C., Truman J. W., 1997. Control of insect ecdysis by a positive-feedback endocrine system: roles of eclosion hormone and ecdysis triggering hormone. J. Exp. Biol. 200: 869–881. [DOI] [PubMed] [Google Scholar]
- Fuse M., Truman J. W., 2002. Modulation of ecdysis in the moth Manduca sexta: the roles of the suboesophageal and thoracic ganglia. J. Exp. Biol. 205: 1047–1058. [DOI] [PubMed] [Google Scholar]
- Halberg K. A., Terhzaz S., Cabrero P., Davies S. A., Dow J. A. T., 2015. Tracing the evolutionary origins of insect renal function. Nat. Commun. DOI: 10.1038/ncomms7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewes R. S., Taghert P. H., 2001. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 11: 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im S. H., Taghert P. H., 2010. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J. Comp. Neurol. 518: 1925–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel T. R., 1990. Regional changes in brain oxytocin receptors postpartum: time-course and relationship to maternal-behavior. J. Neuroendocrinol. 2: 539–545. [DOI] [PubMed] [Google Scholar]
- Iversen A., Cazzamali G., Williamson M., Hauser F., Grimmelikhuijzen C. J. P., 2002. Molecular identification of the first insect ecdysis triggering hormone receptors. Biochem. Biophys. Res. Commun. 299: 924–931. [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Han M.-R., Lee G., Lee S. S., Kim Y.-J., et al. , 2015. Rescheduling behavioral subunits of a fixed action pattern by genetic manipulation of peptidergic signaling. PLoS Genet. 11: e1005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Zitnan D., Cho K. H., Schooley D. A., Mizoguchi A., et al. , 2006a Central peptidergic ensembles associated with organization of an innate behavior. Proc. Natl. Acad. Sci. USA 103: 14211–14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Zitnan D., Galizia C. G., Cho K. H., Adams M. E., 2006b A command chemical triggers an innate behavior by sequential activation of multiple peptidergic ensembles. Curr. Biol. 16: 1395–1407. [DOI] [PubMed] [Google Scholar]
- Kingan T. G., Gray W., Zitnan D., Adams M. E., 1997. Regulation of ecdysis-triggering hormone release by eclosion hormone. J. Exp. Biol. 200: 3245–3256. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(− ΔΔC) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Luan H., Lemon W. C., Peabody N. C., Pohl J. B., Zelensky P. K., et al. , 2006a Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J. Neurosci. 26: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H., Peabody N. C., Vinson C. R., White B. H., 2006b Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus J. N., Aschkenasi C. J., Lee C. E., Chemelli R. M., Saper C. B., et al. , 2001. Differential expression of orexin receptors 1 and 2 in the rat brain. J. Comp. Neurol. 435: 6–25. [DOI] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L., 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302: 1765–1768. [DOI] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal S., Lee P. T., Campbell M. E., Chen K. C., Anguiano-Zarate S., et al. , 2015. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife DOI: 10.7554/eLife.08469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S., Sweeney S. T., Davis G. W., 2001. Homeostatic control of presynaptic release is triggered by postsynaptic membrane depolarization. Neuron 30: 737–749. [DOI] [PubMed] [Google Scholar]
- Park D., Veenstra J. A., Park J. H., Taghert P. H., 2008. Mapping peptidergic cells in Drosophila: Where DIMM fits in. PLoS One 3: e1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Schroeder A. J., Helfrich-Forster C., Jackson F. R., Ewer J., 2003. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development 130: 2645–2656. [DOI] [PubMed] [Google Scholar]
- Park Y., Zitnan D., Gill S. S., Adams M. E., 1999. Molecular cloning and biological activity of ecdysis-triggering hormones in Drosophila melanogaster. FEBS Lett. 463: 133–138. [DOI] [PubMed] [Google Scholar]
- Park Y., Filippov V., Gill S. S., Adams M. E., 2002. Deletion of the ecdysis-triggering hormone gene leads to lethal ecdysis deficiency. Development 129: 493–503. [DOI] [PubMed] [Google Scholar]
- Park Y., Kim Y. J., Dupriez V., Adams M. E., 2003. Two subtypes of ecdysis-triggering hormone receptor in Drosophila melanogaster. J. Biol. Chem. 278: 17710–17715. [DOI] [PubMed] [Google Scholar]
- Pfaff D., Keiner M., 1973. Atlas of estradiol-concentrating cells in central nervous system of female rat. J. Comp. Neurol. 151: 121–157. [DOI] [PubMed] [Google Scholar]
- Pfaff D. W., Schwartz-Giblin S., McCarthy M. M., Kow L.-M., 1994. Cellular and Molecular Mechanisms of Female Reproductive Behaviors. Raven Press, New York. [Google Scholar]
- Pfeiffer B. D., Ngo T. T., Hibbard K. L., Murphy C., Jenett A., et al. , 2010. Refinement of tools for targeted gene expression in Drosophila. Genetics 186: 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayburn L. Y. M., Gooding H. C., Choksi S. P., Maloney D., Kidd A. R., et al. , 2003. amontillado, the Drosophila homolog of the prohormone processing protease PC2, is required during embryogenesis and early larval development. Genetics 163: 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C. W., 1936. The metamorphosis of Drosophila melanogaster, including an accurately timed account of the principal morphological changes. J. Morphol. 59: 351–399. [Google Scholar]
- Roller L., Zitnanova I., Dai L., Simo L., Park Y., et al. , 2010. Ecdysis triggering hormone signaling in arthropods. Peptides 31: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. M., Lachey J. L., Sternson S. M., Lee C. E., Elias C. F., et al. , 2009. Leptin targets in the mouse brain. J. Comp. Neurol. 514: 518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhzaz S., O’Connell F. C., Pollock V. P., Kean L., Davies S. A., et al. , 1999. Isolation and characterization of a leucokinin-like peptide of Drosophila melanogaster. J. Exp. Biol. 202: 3667–3676. [DOI] [PubMed] [Google Scholar]
- Venken K. J., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veverytsa L., Allan D. W., 2012. Temporally tuned neuronal differentiation supports the functional remodeling of a neuronal network in Drosophila. Proc. Natl. Acad. Sci. USA 109: E748–E756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, B. H., and J. Ewer, 2014 Neural and hormonal control of postecdysial behaviors in insects. Annu. Rev. Entomol. 59: 363–381. [DOI] [PMC free article] [PubMed]
- Wu Q., Wen T. Q., Lee G., Park J. H., Cai H. N., et al. , 2003. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron 39: 147–161. [DOI] [PubMed] [Google Scholar]
- Young L. J., Winslow J. T., Nilsen R., Insel T. R., 1997. Species differences in V(1)a receptor gene expression in monogamous and nonmonogamous voles: behavioral consequences. Behav. Neurosci. 111: 599–605. [DOI] [PubMed] [Google Scholar]
- Zitnan D., Adams M. E., 2000. Excitatory and inhibitory roles of central ganglia in initiation of the insect ecdysis behavioural sequence. J. Exp. Biol. 203: 1329–1340. [DOI] [PubMed] [Google Scholar]
- Zitnan D., Adams M. E., 2012. Neuroendocrine regulation of ecdysis, pp. 253–309 in Insect Endocrinology, edited by Gilbert L. I. Elsevier, New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All constructs and fly lines described here for manipulating the ETHR gene and the cells that express it are available upon request.