Abstract
Protein expression level is one of the strongest predictors of protein sequence evolutionary rate, with high-expression protein sequences evolving at slower rates than low-expression protein sequences largely because of constraints on protein folding and function. Expression evolutionary rates also have been shown to be negatively correlated with expression level across human and mouse orthologs over relatively long divergence times (i.e., ∼100 million years). Long-term evolutionary patterns, however, often cannot be extrapolated to microevolutionary processes (and vice versa), and whether this relationship holds for traits evolving under directional selection within a single species over ecological timescales (i.e., <5000 years) is unknown and not necessarily expected. Expression is a metabolically costly process, and the expression level of a particular protein is predicted to be a tradeoff between the benefit of its function and the costs of its expression. Selection should drive the expression level of all proteins close to values that maximize fitness, particularly for high-expression proteins because of the increased energetic cost of production. Therefore, stabilizing selection may reduce the amount of standing expression variation for high-expression proteins, and in combination with physiological constraints that may place an upper bound on the range of beneficial expression variation, these constraints could severely limit the availability of beneficial expression variants. To determine whether rapid-expression evolution was restricted to low-expression proteins owing to these constraints on highly expressed proteins over ecological timescales, we compared venom protein expression levels across mainland and island populations for three species of pit vipers. We detected significant differentiation in protein expression levels in two of the three species and found that rapid-expression differentiation was restricted to low-expression proteins. Our results suggest that various constraints on high-expression proteins reduce the availability of beneficial expression variants relative to low-expression proteins, enabling low-expression proteins to evolve and potentially lead to more rapid adaptation.
Keywords: protein expression, selective constraints, evolutionary rates, adaptation
THE expression level of a protein is one of the strongest predictors of protein sequence evolutionary rate; sequences of highly expressed proteins evolve more slowly than low-expression proteins (Duret and Mouchiroud 1999; Pal et al. 2001; Gout et al. 2010; Yang et al. 2012; Nabholz et al. 2013; Park et al. 2013). This relationship may be a function of specific selective constraints on sequences to avoid protein misfolding (Drummond et al. 2005; Geiler-Samerotte et al. 2011), protein misinteractions (Yang et al. 2012), a decrease in protein function (Cherry 2010; Gout et al. 2010), and/or messenger RNA (mRNA) misfolding (Park et al. 2013). Analyses of microarray data have shown that expression evolutionary rate is also negatively correlated with expression level across human and mouse orthologs (Liao and Zhang 2006). Although the selective constraints imposed on the sequences of highly expressed proteins are well documented (Zhang and Yang 2015), the mechanistic basis of the negative correlation between expression evolutionary rate and expression level remains unclear (Liao and Zhang 2006). This relationship between expression level and expression evolutionary rate has only been documented when comparing orthologous genes across species with relatively long divergence times [e.g., human and mouse diverged approximately 100 million years ago (Liao and Zhang 2006)]. Long-term evolutionary patterns, however, are often unpredictable/intractable because of stochastic environmental fluctuations and irregular ecological changes (Grant and Grant 2002). Microevolutionary processes, however, often can be predicted simply based on selection and heritability (Grant and Grant 2002) and, therefore, present an opportunity to identify the mechanisms of divergence that often become lost in macroevolutionary patterns [e.g., species differences vs. speciation-generating changes (Coyne and Orr 2004)]. Although this relationship between expression level and expression evolutionary rate has been documented as a long-term evolutionary pattern (Liao and Zhang 2006), whether this relationship holds for traits evolving under directional selection within a single species over ecological timescales (i.e., microevolution, <5000 years) is unknown.
Expression is a metabolically costly process requiring energy for transcription, translation, and mobilization of the translational machinery (Dekel and Alon 2005; Gout et al. 2010). The expression level of a protein is predicted to be a tradeoff between the beneficial effects of its function and the energetic costs of its expression (Cherry 2010; Gout et al. 2010), and selection should drive the expression level of all proteins close to values that maximize fitness (Dekel and Alon 2005; Nabholz et al. 2013). Although most proteins will be expressed near their optimal levels, abundantly expressed proteins should be highly optimized owing to the increased energetic cost of production (Gout et al. 2010; Vishnoi et al. 2010). Stabilizing selection on expression level, therefore, should be stronger for high-expression proteins, and this constraint should reduce the amount of standing expression variation for high-expression proteins relative to low-expression proteins. High-expression proteins also could eventually reach an upper bound on expression because of physiological and biophysical constraints (i.e., only so much of a particular protein can be made per cell or tissue). Because high-expression proteins are closer to the upper bound than low-expression proteins, the range of beneficial expression variation available to these abundant proteins is reduced. We therefore may expect that over short ecological timescales, adaptive divergence in expression level would be limited to low-expression proteins because of reductions in the standing expression variation for high-expression proteins owing to stabilizing selection, and the range of beneficial expression variation available to high-expression proteins because of the upper-bound constraint.
Rapid adaptation is often associated with strong directional selection in novel environments following colonization events or dietary changes (Reznick and Ghalambor 2001). Therefore, most studies of adaptation on ecological timescales involve translocations, trait manipulations, or other perturbations to assess the speed of adaptation (Reznick and Ghalambor 2001; Fraser et al. 2011) and may not accurately reflect natural conditions. Focusing on populations inhabiting young barrier islands, however, could alleviate these concerns because of the colonization of a novel environment, changes in resource availability, and potentially limited gene flow (Doley et al. 2008; Vincent et al. 2009; Kolbe et al. 2012; Spurgin et al. 2014). Comparative studies of sympatric taxa can identify which evolutionary processes produced the observed patterns of differentiation (Gomulkiewicz et al. 2007). Several genera of North American pit vipers inhabit barrier islands of the southeastern United States, making them ideal for studying rapid adaptation.
Snake venoms are comprised of approximately 20–100 toxic peptides and proteins (Calvete et al. 2010; Margres et al. 2014, 2015a) that collectively function in predation and defense. Although most quantitative traits are the products of developmental pathways where changes in expression level may have effects mediated through complex interaction networks, toxin expression variation directly changes the phenotype because relative amounts of venom components determine venom efficacy. Expression is typically measured at the mRNA level (Rokyta et al. 2012; Margres et al. 2013; Rokyta et al. 2013). The proteome, however, is more representative of the actual phenotype (Diz et al. 2012), particularly for venoms (Casewell et al. 2014), and the specialization of the venom gland makes venom genetically tractable (Margres et al. 2014, 2015a). Because venom is a secretion (Gibbs et al. 2009), protein expression can be measured directly by reversed-phase high-performance liquid chromatography (RP-HPLC).
To determine whether rapid-expression evolution was restricted to low-expression proteins because of constraints on highly expressed proteins, we compared protein expression across mainland and island populations for three species of pit vipers native to the southeastern United States: the eastern diamondback rattlesnake (Crotalus adamanteus), the pygmy rattlesnake (Sistrurus miliarius), and the cottonmouth (Agkistrodon piscivorus). Because of the young age of the islands [e.g., <5000 years; see Materials and Methods (Lopez and Rink 2007)] and the selective constraints potentially limiting the evolvability of highly expressed proteins, we predicted that abundant proteins would exhibit less differentiation in expression than low-expression proteins, with low-expression proteins exhibiting patterns of rapid, adaptive differentiation.
Materials and Methods
Sampling
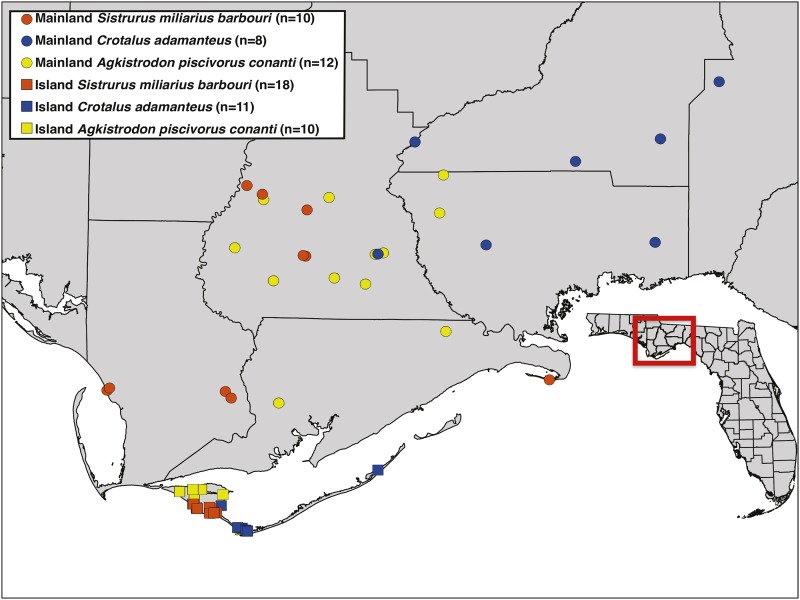
We collected venom and blood samples from eight C. adamanteus, 10 S. miliarius, and 12 A. piscivorus from the Florida mainland and 11 C. adamanteus, 18 S. miliarius, and 10 A. piscivorus from St. Vincent, Little St. George, and St. George islands (Figure 1). These islands are Holocene formations (<5000 years old) located in the Gulf of Mexico 7 km from the mouth of the Apalachicola River delta (Lopez and Rink 2007). All C. adamanteus were used in the analyses of Margres et al. (2015a). We recorded sex, snout-vent length, and total length for each individual. We limited our analyses to adults to avoid the potentially confounding effects of ontogenetic protein expression variation, which has been documented previously (Mackessy 1988; Calvete et al. 2010; Durban et al. 2013; Margres et al. 2015b). Samples were collected under the following permits: Florida Fish and Wildlife Conservation Commission (FWC) LSSC-13-00004 and LSSC-09-0399 and St. Vincent National Wildlife Refuge Permit 41650-2012-08. The sampling procedures were approved by the Florida State University Institutional Animal Care and Use Committee (IACUC) under protocols 0924 and 1333.
Figure 1.
Sampling of island and mainland populations across three genera of pit vipers. We collected venom and blood samples from 19 C. adamanteus, 28 S. miliarius, and 22 A. piscivorus across island and mainland populations. Locations of the study sites within the state of Florida are indicated on the inset map.
Transcriptomic analysis
We followed the exact approach of Rokyta et al. (2013) for transcriptomic assembly and analysis. Briefly, we performed two de novo assemblies using the Extender program with 1000 merged reads and three additional de novo assemblies using NGen. We identified and annotated toxin sequences following BlastX searches against the National Center for Biotechnology Information (NCBI) nonredundant protein database. Only complete protein-coding sequences were retained. All raw reads were deposited in the NCBI Short Read Archive (SRA), and the toxin transcripts were deposited in the NCBI Transcriptome Shotgun Assembly (TSA) database.
Mass spectrometry analysis
Chromatographic separation and tandem mass spectrometry (MS/MS) of the 25 RP-HPLC C. adamanteus peaks have been analyzed previously (Margres et al. 2014, 2015a). Thirty-five RP-HPLC peaks for S. miliarius and 42 RP-HPLC peaks for A. piscivorus were collected as described previously (Margres et al. 2014, 2015a). Briefly, samples were run in triplicate on an externally calibrated Thermo LTQ Orbitrap Velos nLC-ESI-LIT-Orbitrap. MS/MS spectra were extracted by Proteome Discoverer v1.4.0.288, and Sequest v1.4.0.288 (Thermo Fisher Scientific, San Jose, CA) was used to search the species-specific transcriptome databases with signal peptides removed and assuming the digestion enzyme trypsin, allowing one missed digestion site. Scaffold v4.3.2 (Proteome Software, Portland, OR) was used to validate MS/MS-based peptide and protein identifications for both species. Peptide identifications were accepted if they could be established at greater than 95% probability by the Scaffold local false discovery rate (FDR) algorithm, contained at least two identified peptides, and possessed unique peptide evidence. To identify the major toxins within each peak, only proteins with >10% of the total spectral matches within each peak were reported. All data for S. miliarius and A. piscivorus are shown in Supporting Information, Table S5. The raw proteomic data for C. adamanteus have been published previously (Margres et al. 2014, 2015a).
Protein quantification and statistical analyses
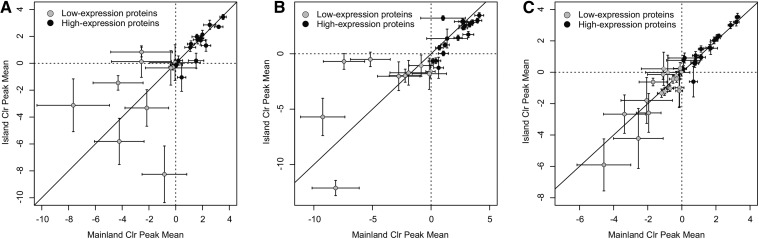
RP-HPLC was performed on a Beckman System Gold HPLC (Beckman Coulter, Fullerton, CA) equipped with Beckman 32 Karat Software v8.0 for peak quantification, as described by Margres et al. (2014, 2015a) for 30 μg of total protein for S. miliarius (Figure S1A), 100 μg of total protein for C. adamanteus (Figure S1B), and 50 μg of total protein for A. piscivorus (Figure S1C). The raw data are contained in Table S6. This approach produces compositional data subject to constant-sum constraints and inherently biased toward negative correlation among components (Aitchison 1986). Therefore, we followed the approach of Margres et al. (2015a) and Wray et al. (2015) and used centered log ratio (clr) and isometric log ratio (ilr) transformations (Egozcue et al. 2003), when appropriate, to transform the data using the robCompositions package (Templ et al. 2011) in R prior to statistical analysis (Filzmoser et al. 2009). We used the multiplicative replacement strategy (Martin-Fernandez et al. 2003) implemented in the R package zCompositions assuming a detection threshold of 0.01% (the smallest measured value) to resolve the issue of zeros. We used the adonis function from the vegan package (Oksanen et al. 2007) in R and Euclidean distances to perform a permutational or nonparametric multivariate analysis of variance [MANOVA (McCardle and Anderson 2001)] (McArdle and Anderson 2001) on the ilr-transformed data to test for significant protein expression variation, as described previously (Margres et al. 2015a). To determine whether the variation detected was restricted to low-abundance proteins and whether highly expressed proteins were conserved, we divided the RP-HPLC peaks for each species into low- and high-abundance data sets prior to conducting the nonparametric MANOVA as described earlier. Here we first calculated the percent mean for each peak and then clr transformed these values. If the mean for an individual peak was less than the geometric mean, it was classified as low expression. If the mean for an individual peak was greater than the geometric mean, it was classified as highly expressed. All statistical analyses of high and low expression were performed following this approach. For Figure 2, however, we first clr transformed the raw percentage data for all samples and then calculated the mean directly from the clr values because this allowed us to estimate the SE shown in the figure. We performed a linear discriminant function analysis using the lda function in R on the ilr-transformed data for each species to assess group membership placement probabilities across populations, as described previously (Margres et al. 2015a).
Figure 2.
Expression differentiation and variation were constrained to low-expression proteins. We plotted the clr mean for each RP-HPLC peak across mainland (x-axis) and island (y-axis) populations for C. adamanteus (A), S. miliarius (B), and A. piscivorus (C). High-expression proteins were highly correlated across populations across all species, and low-expression proteins exhibited a much larger degree of differentiation and variance within populations, particularly for the two species that exhibited significant differentiation (A and B). The larger variance for low-expression proteins relative to high-expression proteins in A. piscivorus (C), despite a lack of population differentiation, was strong evidence supporting our expectation that strong stabilizing selection would reduce the amount of standing expression variation for high-expression proteins. Bars indicate SE, solid line indicates a perfect agreement, dashed lines indicate the origin (i.e., the geometric mean), and proteins less than these values were considered low-expression proteins.
DNA sequencing
S. miliarius and A. piscivorus DNA was extracted from whole-blood samples drawn from the caudal vein using the Omega Bio-Tek E.Z.N.A Tissue DNA Kit according to the manufacturer’s protocol. A fragment of cytochrome b, 841 and 1003 bp, respectively, was amplified in 25-μl PCR runs using the H16064 and L14910 primers and thermal cycling protocol described by Burbrink et al. (2000). An 1018-bp fragment for C. adamanteus (accession numbers KJ730289, KJ730300, KJ730314, KJ730321, KJ730327, KJ730342, KJ730344, KJ730345, KJ730351, KJ730357, KJ730358, KJ730362, KJ730364, KJ730370, KJ730377, KJ730383, KJ730389, KJ730394, and KJ730396) and C. horridus (outgroup; accession number KJ730366) were taken from Margres et al. (2015b) following the same approach. PCR products were purified using the QIAGEN QIAquick PCR Purification Kit, and sequencing was on the Applied Biosystems 3730 Genetic Analyzer.
All individuals from both populations across all species were used in species-specific phylogenetic analyses. For each species, sequences were aligned using the MegAlign module of the DNA STAR Lasergene 11 software suite. Model selection was performed using jModelTest 0.1.1 under default settings (Guindon and Gascuel 2003; Darriba et al. 2012), with the Akaike information criterion used to determine the most appropriate model for each species (Akaike 1974). A maximum-likelihood (ML) analysis was run in PAUP* 4.0b10 (Swofford 1998) using a heuristic search with 100 stepwise random-addition sequence replicates and the tree bisection-reconnection method. To assess support for the ML tree, we also performed a nonparametric bootstrap analysis using 1000 pseudoreplicates with 10 stepwise random-addition sequence replicates. Base frequencies, rate matrix, proportion of invariable sites, and shape were estimated from the data.
Data availability
Peptide reports for all mass spectrometry analyses are in Table S5, and the raw liquid chromatography data are in Table S6. Sanger sequences were submitted to the National Center for Biotechnology Information (NCBI) Trace Archive under accession numbers KP881369–KP881418. Annotated transcriptome sequences were submitted to the GenBank Transcriptome Shotgun Assembly database under accession number GDBJ02000000 for S. miliarius and GDAZ02000000 for A. piscivorus.
Results and Discussion
Rapid differentiation in protein expression following the colonization of an island
We performed a nonparametric MANOVA comparing protein expression levels across island and mainland populations separately for each species (Figure 1) and detected significant expression differentiation in S. miliarius (P < 0.01) and C. adamanteus (P = 0.05) but not A. piscivorus (P = 0.93). This significant differentiation in expression represents rapid (i.e., <5000 years) phenotypic divergence in S. milarius and C. adamanteus. As mentioned earlier, comparative studies of sympatric taxa can help to identify which evolutionary processes produced the observed patterns of differentiation (Gomulkiewicz et al. 2007). The lack of expression differentiation in A. piscivorus highlights the significance of the differentiation in S. miliarius and C. adamanteus and suggests that different evolutionary processes are responsible for these different patterns. The geographic variation in protein expression for C. adamanteus and S. miliarius is consistent with local adaptation as a result of variable selective pressures owing to genotype-by-genotype-by-environment interactions, and the lack of expression variation in A. piscivorus may be a result of diffuse selection owing to its generalist diet (Vincent et al. 2004) or high levels of gene flow (but see later). We next examined whether this differentiation in expression level for C. adamanteus and S. miliarius was biased toward high- or low-expression proteins.
Expression differentiation and variation are constrained to low-expression proteins
To determine whether the variation detected in S. miliarius and C. adamanteus was restricted to low-abundance proteins and highly expressed proteins were conserved, we divided the RP-HPLC peaks for each species into low- and high-abundance data sets based on the clr-transformed mean for each peak and conducted a nonparametric MANOVA as described earlier. If the mean for an individual peak was less than the geometric mean, it was classified as low expression. If the mean for an individual peak was greater than the geometric mean, it was classified as highly expressed. Of the 25 peaks in C. adamanteus and 28 peaks in S. miliarius, 13 and 14 were classified as low-expression proteins, respectively. We detected significant expression variation only in the low-expression data sets for both S. miliarius (Plow < 0.01, Phigh = 0.25) and C. adamanteus (Plow = 0.05, Phigh = 0.29). As expected, neither class exhibited significant variation in A. piscivorus (Plow = 0.92, Phigh = 0.83).
We next looked at the covariance matrix of the clr-transformed data sets to identify the most variable peaks relative to the classification of each protein as high or low expression, as described earlier. Low-expression proteins accounted for 95.4% of the variance in C. adamanteus, 86.2% of the variance in A. piscivorus, and 57.7% of the variance in S. miliarius (Table S1), indicating that differentiation in protein expression and/or standing expression variation, especially in C. adamanteus, was restricted to low-expression loci. The lower proportion of variance accounted for by low-expression proteins in S. miliarius may have been a reflection of the high amount of variation detected in peak 26 (16.1%; Table S1). This peak was classified as highly expressed but had the lowest expression level of any highly expressed protein.
We then compared the clr mean for each RP-HPLC peak across mainland (x-axis) and island (y-axis) populations for all three species (Figure 2). We calculated the coefficient of determination R2 and found a good fit for all three species in high-expression proteins (), indicating that high-expression proteins were conserved across populations in all three species. Consistent with our previous analyses, low-expression proteins exhibited significant differentiation in C. adamanteus (R2 = 0.019) and S. miliarius (R2 = 0.328) but not A. piscivorus (R2 = 0.746).
To determine whether the increased variance in low-expression proteins was biological or a result of technical biases, we conducted six RP-HPLC analyses on a single venom sample from a mainland C. adamanteus pit viper and plotted the clr mean (x-axis) and variance (y-axis) for all RP-HPLC peaks (Figure S2). If the increased variance in low-expression proteins was because of a limitation of our approach to accurately quantify low-abundance peaks, we would expect to see a significant negative correlation (i.e., a substantial reduction in variance as expression increased). We found a lack of correlation between the clr mean and variance among all peaks (R2 = 0.0426, R = −0.2063, P = 0.3224; Figure S2A) and even less so following removal of a single outlying low-abundance peak (R2= 0.0052, R = −0.0720, P = 0.7380; Figure S2B; R2 is the coefficient of determination, and R is Pearson’s correlation coefficient), indicating that the increased variance in low-expression proteins was not an artifact of our method but rather biological and consistent with our expectations for stabilizing selection.
Overall, abundant proteins exhibited significantly less differentiation in expression than low-expression proteins, with the latter exhibiting patterns of rapid differentiation. These results are consistent with previous work that found a negative correlation between expression evolutionary rate and expression level between human and mouse orthologs (Liao and Zhang 2006). Our results, however, demonstrate that this pattern holds over ecological timescales as well as 100 million years of divergence. We predicted that stabilizing selection on expression level should be stronger for high-expression proteins and reduce the amount of standing expression variation relative to low-expression proteins. We found strong evidence supporting this expectation (Figure 2), especially in A. piscivorus (Figure 2C), despite a lack of expression differentiation. Our results suggest that the expression level of highly expressed proteins evolves under considerable constraints, potentially because these proteins are already expressed at or near their physiological maxima, and the expression level of a protein is a strong predictor of both protein sequence and protein expression evolutionary rates. These constraints may limit the range of beneficial expression variation available to high-expression proteins, indicating that rapid, adaptive divergence would be restricted to low-expression proteins over ecological timescales.
The process driving expression differentiation in low-expression proteins
The differentiation in protein expression detected may be a result of genetic drift following a relaxation of evolutionary constraints (Khan et al. 2013), selection following the colonization of a novel environment (Fraser et al. 2011), founder effects (Kolbe et al. 2012), and/or phenotypic plasticity (Hunt et al. 2011). We did not specifically test for plasticity in this study because venom expression differences have been shown repeatedly to be under genetic control and not environmentally induced (Daltry et al. 1996; Gibbs et al. 2011; Holding et al. 2015; Margres et al. 2015b), and the feeding ecology of venomous snakes makes adaptive plasticity unlikely. Because venom is stored for long periods of time (e.g., over winter and between infrequent feeding events) and previous meals may not be robust predictors of future meals, plasticity would be unlikely to provide any adaptive advantage for this trait. Gibbs et al. (2011) fed different groups of S. miliarius different prey items over extended time periods and did not find any significant changes in venom expression; Margres et al. (2015b) documented the ontogenetic shift in venom expression in C. adamanteus in laboratory-raised individuals and found that geographic differences in venom expression held over long periods of time despite the animals being raised under identical conditions in captivity; and Holding et al. (2015) recently showed that prey preference is also under genetic control and not affected by previously fed upon items in S. miliarius. Because these studies failed to identify any plastic changes in venoms in the two species in our study that exhibited significant expression differentiation, we rejected the hypothesis that plasticity played any role in generating the observed variation in venom expression.
Although relaxed purifying selection on protein expression levels is believed to be rare, a recent study by Khan et al. (2013) proposed a theoretical framework for determining whether expression differentiation was consistent with directional selection or the relaxation of evolutionary constraints. The authors stated that a shift in mean expression level associated with high within-lineage variation is indicative of drift following a relaxation of constraints, and a shift in mean expression level associated with low within-lineage variation is indicative of directional selection within that particular population. The nonparametric MANOVA identified mean differences in the expression levels of low-expression proteins between island and mainland C. adamanteus and S. miliarius populations, and we again used the covariance matrix of the clr-transformed data sets to examine within-population variances across island and mainland populations for both species. According to Khan et al. (2013), if the variance for a particular protein or class of proteins is greater within the mainland population, this would suggest directional selection in the island population. To compare the magnitude of the differences in variances across island and mainland populations, we calculated the total variances across all peaks in both populations for all three species. For the two species with significant mean expression differences (C. adamanteus and S. miliarius), the total variances across all peaks were much larger in the mainland populations than in the island populations (313 mainland vs. 228 island in C. adamanteus, 341 mainland vs. 239 island in S. miliarius). We did not find a significant mean expression difference for A. piscivorus, and the total variances across all peaks were nearly identical (204 mainland vs. 192 island). In C. adamanteus, the total variance for low-expression proteins also was greater within the mainland population than within the island population (301 mainland vs. 214 island), and 9 of the 13 low-expression proteins had greater variances within the mainland population than within the island population (although this frequency was not significantly different from what we would expect by chance; P = 0.17; Figure 2). Similarly, in S. miliarius, the total variance for low-expression proteins was greater within the mainland population than within the island population (199 mainland vs. 128 island), and 11 of the 14 low-expression proteins had greater variances within the mainland population than within the island population (P = 0.03; Figure 2). Again, in A. piscivorus, the total variances for low-expression proteins across mainland and island populations were nearly identical (172 mainland vs. 171 island). We next performed the multivariate analogue to a Levene’s test to test for homogeneity of group variances dispersions across island and mainland populations for all peaks, high-expression peaks only, and low-expression peaks only. To determine whether the variances of the island and mainland populations were significantly different, we ran an ANOVA to compare the distances of group members (e.g., an island individual) to the group centroid (e.g., island population centroid) across populations. We failed to identify any significant differences in these comparisons, although the low-expression analysis did approach significance in S. miliarius (P = 0.07), and we typically saw a reduction in the P-value in the low-expression comparisons relative to the high-expression-only analyses (e.g., in C. adamanteus, Phigh = 0.69, Plow = 0.32). Power analyses suggested that these nonsignificant results, however, may have been the result of small sample size; for a one-way ANOVA comparing two groups with a moderate effect size (f = 0.25) and a significance level of 0.05, we would need a shared sample size of approximately 63 individuals to obtain a power (i.e., confidence) of 0.80. Our largest shared sample size was n = 10 (i.e., 10 island individuals and 10 mainland individuals) in S. miliarius and A. piscivorus. The shared sample size in C. adamanteus was n = 8. Even when assuming a large effect size (f = 0.40) and reducing the power to 0.75, we still have less than half the adequate sample size required (n = 23) to detect a significant result.
The significant differentiation in mean protein expression across island and mainland populations, along with the reduction in intra-island expression variation, also could be explained by an alternative selection model where selection would be maintaining diversity on the mainland rather than driving expression differentiation on the island. If we assume that dietary variation on the mainland is much larger than on the island, a reasonable assumption because islands typically exhibit a significant reduction in species diversity (MacArthur and Wilson 1967), selection could maintain the higher variation in venom expression on the mainland because of the higher variation in diet. Therefore, under this hypothesis, the significant mean differences in expression between island and mainland populations would be the result of founder effects, and the larger variance on the mainland would be the result of diversifying selection. This alternative hypothesis, however, would require some sort of biogeographic structure in the mainland population for selection to maintain this expression variation. Our sampling is from a very small region of contiguous and uniform habitat. The only potential biogeographic barriers on the mainland are the Ochlockonee and Apalachicola rivers. Although the latter has been repeatedly documented as a biogeographic barrier to a number of organisms (Baer 1998; Burbrink et al. 2000), the former has never been known, to the best of our knowledge, to impede gene flow in squamates, and most of our sampling was east of the Apalachicola River (i.e., only 4 of the 10 mainland S. miliarius were collected west of the Apalachicola River). Additionally, the island system sits at the mouth of the Apalachicola River, and immigration is equally likely to have occurred from either side of the Apalachicola River. Margres et al. (2015b) recently sequenced a 986-bp fragment of ND5 and a 1018-bp fragment of cytochrome b for C. adamanteus and found two haplotypes in panhandle Florida. Both haplotypes were present in island and mainland populations and were found on either side of both rivers with no obvious frequency differences across the Ochlockonee River. Considering that we failed to detect any population structure within the mainland population in C. adamanteus when sequencing two loci with dense sampling (i.e., 70 individuals in panhandle Florida), we find it reasonable to assume a lack of population structure within the mainland populations for all species and, therefore, that neither river is a barrier that could restrict gene flow to the degree necessary for this alternative model.
Overall, our results are most consistent with the predictions of Khan et al. (2013) for directional selection in the island populations of C. adamanteus and S. miliarius. Lower within-population variance in the island populations, however, is not surprising given the likelihood of a smaller effective population size than on the mainland, and this may reflect founder effects rather than directional selection.
Founder effects, genetic or phenotypic changes in a population as a result of being initially colonized by relatively few individuals, can cause divergence among populations, particularly island populations (Kolbe et al. 2012; Spurgin et al. 2014). Although the variance analysis suggested that directional selection produced the expression differentiation identified in low-expression proteins across populations (Khan et al. 2013), this divergence in phenotype, along with the lack of within-population variance, could be a result of the island being initially colonized by a small number of individuals. To determine whether the identified differentiation in protein expression between island and mainland populations was a result of demographic histories, we sequenced cytochrome b for all sampled individuals. A reduction in genetic diversity within the island populations relative to the mainland populations would indicate founder effects, whereas similar genetic diversity across island and mainland populations would indicate a lack of founder effects owing to multiple colonization events and/or continuous gene flow. We found a complete lack of genetic diversity across C. adamanteus island and mainland populations (0.0% sequence divergence), a single variable site across S. miliarius island and mainland populations (0.1% sequence divergence; Figure S3), and five variable sites across A. piscivorus island and mainland populations (0.3% sequence divergence; Figure S4). We did identify a monophyletic clade of island A. piscivorus, although not all island specimens were in this clade. Determining the demographic histories of these populations was difficult because of the lack of genetic diversity within C. adamanteus and S. miliarius. This absence of genetic variation across island and mainland individuals could indicate ongoing gene flow (i.e., a lack of founder effects) or could be a result of the young age of these island populations (i.e., 5000 years was not enough time for neutral differentiation to occur). Identifying a genetic bottleneck following a founding event has been known to be sensitive to the number of loci examined, and only using a single locus can result in a type II error (Spurgin et al. 2014). Therefore, we currently cannot rule out founder effects, although strong selection can overwhelm founder effects over ecological timescales (Kolbe et al. 2012), and the only species that exhibited any degree of neutral population differentiation (i.e., A. piscivorus) did not significantly differ in mean expression.
Demonstrating that this expression differentiation was a result of selection and not founder effects or another neutral process would require fitness comparisons across populations. In the absence of fitness data, comparing the differentiation of traits under putative selection with that of neutral markers may allow the identification of adaptive variation (Savolainen et al. 2013). The differentiation in protein expression among S. miliarius and C. adamanteus populations that can be accounted for by divergence at neutral markers may reflect neutral processes, but variation that exceeds this neutral divergence may be indicative of directional selection (Whitehead and Crawford 2006; Richter-Boix et al. 2010; Margres et al. 2015b). To determine whether the identified differentiation in protein expression between island and mainland populations was a result of adaptive or neutral processes, we used the sequence data discussed earlier to compare neutral divergence and protein expression differentiation. The lack of neutral differentiation and the significant phenotypic divergence across S. miliarius and C. adamanteus populations suggest that the observed protein expression differentiation was a result of directional selection (Margres et al. 2015b), potentially despite gene flow, although founder effects currently cannot be ruled out.
Expression variation is typically attributed to cis-regulatory mutations (Carroll 2008). cis-regulatory mutations, however, are not the only mechanism for altering the amounts of protein produced (Hastings et al. 2009). Polymorphisms at a much larger genomic scale, such as gene duplications and deletions (Stranger et al. 2007), also can alter the expression level of a particular protein (Nguyen et al. 2006). The correlation between gene copy-number differences and changes in gene expression has been documented previously (Cheng et al. 2005; Freeman et al. 2006; Nair et al. 2008), including in venoms (Margres et al. 2015b), and venom protein families are believed to be the result of gene duplication and positive selection (Casewell et al. 2011) via the birth-and-death model of protein evolution (Fry et al. 2008). The significant expression variation we detected therefore could be the result of variation in copy number, assuming that variation in copy number would affect low-expression (and presumably low-copy) genes more than high-expression, high-copy genes (e.g., the difference between 10 and 12 copies for a particular protein may not be significant, but the difference between 2 and 4 copies may be). Genomic drift (i.e., the random duplication and deletion of genes) has been shown to play an important role in generating copy-number variation (Nozawa et al. 2007, McCarroll et al. 2008; Nei et al. 2008), and it has even been proposed that neutral processes are responsible for maintaining the vast majority of all identified variations in copy number (Nozawa et al. 2007). Genomic drift, however, is stochastic and would only affect our estimates of variance, not the mean. Therefore, regardless of the mechanism (e.g., cis-regulatory mutation, copy-number variation, micoRNA regulation, or translational efficiency), our results suggest that the identified expression differentiation and variation were the result of selection rather than neutral processes, although, again, founder effects cannot be ruled out.
The rate of fixation of expression levels
The significant variation detected in C. adamanteus and S. miliarius demonstrated that, on average, expression levels for low-expression proteins differed between island and mainland populations. To determine whether this protein expression variation was fixed within each population, we used a linear discriminant function analysis to assess group membership placement probabilities (Margres et al. 2015a). If the expression phenotypes have been fixed in the island populations, we would expect placement probabilities near 100% for island S. miliarius and C. adamanteus. This analysis, however, is problematic if the sample size (i.e., number of individuals per species) does not exceed the number of variables (i.e., RP-HPLC peaks). For S. miliarius, the number of variables equaled the sample size (n = 28), while the number of variables (var = 25) exceeded the sample size (n = 19) in C. adamanteus. Therefore, we performed the analysis on the low- and high-abundance data sets independently. Based on our previous analyses, we would expect relatively low placement probabilities for the high-abundance data sets and higher placement probabilities for the low-abundance data sets given that low-expression proteins explained most of the variation in our data.
Analysis of the high-abundance data sets accurately assigned 45.5% of island and 50.0% of mainland C. adamanteus and 77.8% of island and 70.0% of mainland S. miliarius. Analysis of the low-abundance data sets accurately assigned 81.2% of island and 75.0% of mainland C. adamanteus and 77.8% of island and only 60.0% of mainland S. miliarius. The slightly lower placement probability for mainland S. miliarius in the low-expression data set may have been a reflection of the high amount of variation detected in high-abundance peak 26, as discussed earlier. We did see, however, a significant improvement in placement probabilities for C. adamanteus in the low-abundance data set, as expected. These placement probability percentages, although indicative of significant population differentiation in the expression of low-expression proteins, demonstrated that the island expression patterns were not yet fixed in either species. Because the island phenotypes were not fixed, founder effects (discussed earlier) were unlikely to cause the observed differentiation in expression levels because following a severe bottleneck with a relatively short recovery period (<5000 years), we would expect near fixation of the expression phenotype. Therefore, our results suggest that these expression differences were the result of directional selection. Local adaptation is predicted to act as a barrier to migration owing to reduced immigrant fitness, and subsequent genetic drift eventually will result in neutral genetic structure across populations (Spurgin et al. 2014). The age of the islands (<5000 years), however, may be insufficient to allow completion of this process. Therefore, this isolation by adaptation (Spurgin et al. 2014) may be incipient, and the lack of expression-level fixation and neutral differentiation despite, on average, significant variation in low-abundance protein expression simply may reflect the young age of the island populations. This also could indicate ongoing gene flow in these two species, which is predicted to increase the probability of successful establishment and persistence in novel environments (Forsman 2014) as well as potentially promote local adaptation in coevolutionary contexts (North et al. 2010).
Protein Identification
To identify the individual proteins present in each RP-HPLC peak, we used the approach of Margres et al. (2014, 2015a) to correlate specific toxin transcripts with specific venom proteins. We identified 122 and 157 unique putative toxin transcripts in the venom-gland transcriptomes of S. miliarius (GenBank Transcriptome Shotgun Assembly accession number GDBJ02000000) and A. piscivorus (GenBank Transcriptome Shotgun Assembly accession number GDAZ02000000), respectively, and these toxin transcripts were grouped into 63 and 76 clusters on the basis of <1% nucleotide divergence in their coding sequences, as described previously (Rokyta et al. 2012, 2013; Margres et al. 2013, 2015a). The venom-gland transcriptome for C. adamanteus was previously assembled and annotated [NCBI SRA accession number SRA050594, GenBank Transcriptome Shotgun Assembly accession number GBEX01000000 (Rokyta et al. 2012; Margres et al. 2014, 2015a)]; 76 unique putative toxin transcripts that grouped into 44 clusters were identified (Margres et al. 2015a). Following transcriptome assembly and analysis, we identified unique proteomic evidence for 24 of the 63 S. miliarius toxin clusters (Table S2 and Figure S1A) and 30 of the 76 A. piscivorus toxin clusters (Table S3 and Figure S1C). Proteomic analysis of C. adamanteus venom also was described previously (Margres et al. 2014, 2015a). We reanalyzed these data using different parameters (see Materials and Methods) and identified 18 of the 44 C. adamanteus toxin clusters (Table S4 and Figure S1B). Table S5 contains peptide reports for S. miliarius and A. piscivorus.
Our previous analyses demonstrated that high-expression proteins were conserved across populations and that low-expression proteins exhibited significant expression variation. To determine whether particular protein families were over- or underrepresented in high- (i.e., less variable) and low-expression (i.e., more variable) RP-HPLC peaks across all three species (Table S1), we compared protein family presence/absence across expression classes. Cysteine-rich secretory proteins (CRISPs) were only identified in low-expression peaks 16 and 17 in A. piscivorus (Table S3). These peaks were the fourth most and most variable peaks, respectively, suggesting that CRISP expression was not only biased toward low-expression in A. piscivorus but also exhibited the most variation within populations. Myotoxin (peaks 1b and 2) and a single phospholipase A2 protein (peak 10) were only identified in high-expression peaks in C. adamanteus. These proteins were the third and second least variable, respectively, indicating that these proteins were highly expressed with little variation. The C-type lectin protein family was represented by a single protein in peaks 28, 29, and 32 in S. miliarius. All these peaks were highly expressed but possessed very different variances. Peak 29 was the fifth least variable peak, peak 32 was the thirteenth least variable peak, but peak 28 was the second most variable peak. Detecting the same toxic protein in multiple peaks suggests that this protein undergoes post-transcriptional modifications (Casewell et al. 2014; Margres et al. 2015a), and the expression variation (or lack thereof) detected in this protein appears to be post-transcriptional variant specific. Overall, the lack of bias toward a particular expression level for the most diverse protein families (e.g., snake venom metalloproteinases and snake venom serine proteinases) indicated that expression variation was locus specific or sometimes post-transcriptional variant specific rather than gene-family specific.
Conclusion
We compared the rates of expression evolution for high- and low-expression proteins and found that, over ecological timescales, expression levels of abundant proteins were significantly conserved and rapid-expression evolution was restricted to low-expression proteins. Our results are consistent with microarray studies examining human and mouse orthologs (Liao and Zhang 2006) and suggest that stabilizing selection on high-expression proteins reduced the amount of standing expression variation in these abundant proteins. This reduction in standing variation, in combination with the upper-bound constraint, limited the rate at which adaptive expression variation was generated in high-expression proteins. Therefore, the expression level of a protein is a strong predictor of both protein expression and protein sequence evolutionary rate. Gibbs et al. (2009) found that highly expressed proteins exhibited less variation in presence-absence variation than low-expression proteins in Sistrurus rattlesnakes. The authors speculated that highly expressed proteins performed generic killing functions and that low-expression proteins were not only prey specific but also more evolvable, consistent with our findings. The greater evolvability of low-expression proteins relative to high-expression proteins may allow them to respond more rapidly to novel selective pressures, and although the optimal expression of these proteins is relatively low, the fitness effects of a regulatory mutation affecting expression may be high (Gout et al. 2010).
Although our results are consistent with theoretical predictions (Gout et al. 2010) and previous work in mammalian systems (Liao and Zhang 2006; Zhang and Yang 2015), these findings contradict previous work showing that venom loci expressed at all levels contribute to protein expression divergence among adult C. adamanteus (Margres et al. 2015a). This study, however, used range-wide sampling with much older divergence times than the current study [i.e., 1.27 million years vs. <5000 years (Margres et al. 2015b)]. Together these results suggest that although both high- and low-expression venom proteins exhibited significant expression variation over large spatial and temporal scales (Margres et al. 2015b), rapid-expression evolution was confined to low-expression venom proteins. We may see different patterns of expression differentiation in venoms over different timescales because of the selective and physiological constraints acting on high-expression proteins. These constraints may reduce the potential of highly expressed proteins to generate beneficial sequence and expression variation for a given venom protein, suggesting that rapid, adaptive divergence would be restricted to low-expression venom proteins over ecological timescales. Given enough time, however, beneficial expression variation can arise in high-expression venom proteins (Margres et al. 2015a). Our results suggest that in the context of proteins evolving under directional selection, the initial steps in the adaptive process may be restricted to mutations affecting low-expression proteins owing to constraints on highly expressed proteins, with expression differentiation in proteins expressed at higher levels occurring over larger temporal scales. Therefore, this microevolutionary bias in expression evolutionary rate may result in the long-term evolutionary pattern previously documented (Liao and Zhang 2006; Margres et al. 2015a), suggesting that short-term processes can, at least occasionally, be extrapolated to a macroevolutionary level.
Acknowledgments
We thank Pierson Hill, Jacob Loyacano, Joe Pfaller, Mark S. Margres, and Flavio Morrissiey for help in acquiring venom samples. We thank Megan Lamb, Danielle Jones, Jennifer Wanat, and Rebecca Bernard with the Florida Department of Environmental Protection and the Apalachicola National Estuarine Research Reserve and Bradley Smith and Shelley Stiaes with the U.S. Fish and Wildlife Service and the St. Vincent National Wildlife Reserve for access to field sites. Samples were collected under the following permits: Florida Fish and Wildlife Conservation Commission (FWC) LSSC-13-00004 and LSSC-09-0399, Florida Department of Environmental Protection permit number 04101310, and St. Vincent National Wildlife Refuge permit number 41650-2012-08. Sample collection was approved by the Florida State University Institutional Animal Care and Use Committee (IACUC) under protocols 0924 and 1333. This work was supported by the National Science Foundation (DEB-1145987 to D.R.R.), Florida State University (to M.J.M.), and the Gopher Tortoise Council (to M.J.M.).
Footnotes
Communicating editor: J. J. Bull
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180547/-/DC1
Literature Cited
- Aitchison J., 1986. The Statistical Analysis of Compositional Data. Chapman and Hall, London. [Google Scholar]
- Akaike H., 1974. A new look at statistical model identification. IEEE Trans. Automat. Contr. 19: 716–723. [Google Scholar]
- Baer C., 1998. Species-wide population structure in a southeastern US freshwater fish Heterandria formosa: gene flow and biogeography. Evolution 52: 183–193. [DOI] [PubMed] [Google Scholar]
- Burbrink F. T., Lawson R., Slowinski J., 2000. Mitochondrial DNA phylogeography of the polytypic North American rat snake (Elaphe obsoleta): a critique of the subspecies concept. Evolution 54: 2107–2118. [DOI] [PubMed] [Google Scholar]
- Calvete J. J., Sanz L., Cid P., de la Torre P., Flores-Díaz M., et al. , 2010. Snake venomics of the Central American rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J. Proteome Res. 9: 528–544. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Casewell N. R., Wagstaff S. C., Harrison R. A., Renjifo C., Wüster W., 2011. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 28: 2637–2649. [DOI] [PubMed] [Google Scholar]
- Casewell N. R., Wagstaff S. C., Wuster W., Cook D., Bolton F., et al. , 2014. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl. Acad. Sci. USA 111: 9205–9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Ventura M., She X., Khaitovich P., Graves T., et al. , 2005. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437: 88–93. [DOI] [PubMed] [Google Scholar]
- Cherry J., 2010. Expression level, evolutionary rate, and the cost of expression. Genome Biol. Evol. 2: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Daltry J. C., Wüster W., Thorpe R. S., 1996. Diet and snake venom evolution. Nature 379: 537–540. [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G., Doallo R., Posada D., 2012. jModelTest2: more models, new heuristics and parallel computing. Nat. Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel E., Alon U., 2005. Optimality and evolutionary tuning of the expression level of a protein. Nature 436: 588–592. [DOI] [PubMed] [Google Scholar]
- Diz A., Martinez-Fernandez M., Rolan-Alvarez E., 2012. Proteomics in evolutionary ecology: linking the genotype with the phenotype. Mol. Ecol. 21: 1060–1080. [DOI] [PubMed] [Google Scholar]
- Doley R., Tram N., Reza M., Kini R., 2008. Unusual accelerated rate of deletions and insertions in toxin genes in the venom glands of the pygmy copperhead (Austrelaps labialis) from Kangaroo Island. BMC Evol. Biol. 8: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D., Bloom J., Adami C., Wilke C., Arnold F., 2005. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. USA 102: 14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban J., Perez A., Sanz L., Gomez A., Bonilla F., et al. , 2013. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genomics 14: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D., 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egozcue J. J., Pawlowsky-Glahn V., Mateu-Figueras G., Barceló-Vidal C., 2003. Isometric logratio transformations for compositional data analysis. Math. Geol. 35: 279–300. [Google Scholar]
- Filzmoser P., Hron K., Reimann C., 2009. Principal component analysis of compositional data with outliers. Environmetrics 20: 621–632. [Google Scholar]
- Forsman A., 2014. Effects of genotypic and phenotypic variation on establishment are important for conservation, invasion, and infection biology. Proc. Natl. Acad. Sci. USA 111: 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser D., Weir L., Bernatchez L., Hansen M., Taylor E., 2011. Extent and scale of local adaptation in salmonid fishes: review and meta-analysis. Heredity 106: 404–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J., Perry G., Feuk L., Redon R., McCarroll S., et al. , 2006. Copy number variation: new insights in genome diversity. Genome Res. 16: 949–961. [DOI] [PubMed] [Google Scholar]
- Fry B. G., Scheib H., van der Weerd L., Young B., McNaughtan J., et al. , 2008. Evolution of an arsenal. Mol. Cell. Prot. 7: 215–246. [DOI] [PubMed] [Google Scholar]
- Geiler-Samerotte K., Dio M., Budnik B., Wang S., Hartl D., et al. , 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc. Natl. Acad. Sci. USA 108: 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs H. L., Sanz L., Calvete J. J., 2009. Snake population venomics: proteomics-based analyses of individual variation reveals significant gene regulation effects on venom protein expression in Sistrurus rattlesnakes. J. Mol. Evol. 68: 113–125. [DOI] [PubMed] [Google Scholar]
- Gibbs H. L., Sanz L., Chiucchi J. E., Farrell T. M., Calvete J. J., 2011. Proteomic analysis of ontogenetic and diet-related changes in venom composition of juvenile and adult dusky pigmy rattlesnakes (Sistrurus miliarius barbouri). J. Proteomics 74: 2169–2179. [DOI] [PubMed] [Google Scholar]
- Gomulkiewicz R., Drown D., Dybdahl M., Godsoe W., Nuismer S., et al. , 2007. Dos and don’ts of testing the geographic mosaic theory of coevolution. Heredity 98: 249–258. [DOI] [PubMed] [Google Scholar]
- Gout J.-F., Kahn D., Duret L., Paramecium Post-Genomics Consortium , 2010. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 6: e1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P., Grant B., 2002. Unpredictable evolution in a 30-year study of Darwin’s finches. Science 296: 707–711. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hastings P., Lupski J., Rosenberg S., Ira G., 2009. Mechanisms of change in gene copy number. Nat. Rev. Genet. 10: 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding M., Kern E., Denton R., Gibbs H., 2015. Fixed prey cue preferences among dusky pigmy rattlesnakes (Sistrurus miliarius barbouri) raised on different long-term diets. Evol. Ecol. •••: 1–7. [Google Scholar]
- Hunt B., Ometto L., Wurm Y., Shoemaker D., Soojin V. Y., et al. , 2011. Relaxed selection is a precursor to the evolution of phenotypic plasticity. Proc. Natl. Acad. Sci. USA 108: 15936–15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z., Ford M., Cusanovich D., Mitrano A., Pritchard J., et al. , 2013. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 342: 1100–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe J., Leal M., Schoener T., Spiller D., Losos J., 2012. Founder effects persist despite adaptive differentiation: a field experiment with lizards. Science 335: 1086–1089. [DOI] [PubMed] [Google Scholar]
- Liao B., Zhang J., 2006. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during Mammalian evolution. Mol. Biol. Evol. 23: 1119–1128. [DOI] [PubMed] [Google Scholar]
- Lopez G., Rink W., 2007. 2006 Characteristics of the burial environment related to quartz SAR-OSL dating at St. Vincent Island, NW Florida, USA. Quat. Geochronol. 2: 65–70. [Google Scholar]
- MacArthur R., Wilson E., 1967. The Theory of Island Biogeography, Vol. 1 Princeton University Press, Princeton, NJ. [Google Scholar]
- Mackessy S. P., 1988. Venom ontogeny in the Pacific rattlesnakes Crotalus viridis helleri and C. v. oreganus. Copeia 1988: 92–101. [Google Scholar]
- Margres M. J., Aronow K., Loyacano J., Rokyta D. R., 2013. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics 14: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres M. J., McGivern J. J., Wray K. P., Seavy M., Calvin K., et al. , 2014. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). J. Proteomics 96: 145–158. [DOI] [PubMed] [Google Scholar]
- Margres M. J., McGivern J. J., Seavy M., Wray K. P., Facente J., et al. , 2015a Contrasting modes and tempos of venom expression evolution in two snake species. Genetics 199: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres M. J., Wray K. P., Seavy M., McGivern J. J., Sanader D., et al. , 2015b Phenotypic integration in the feeding system of the eastern diamondback rattlesnake (Crotalus adamanteus). Mol. Ecol. 24: 3405–3420. [DOI] [PubMed] [Google Scholar]
- Martin-Fernandez J., Barcelo-Vidal C., Pawlowsky-Glahn V., 2003. Dealing with zeros and missing values in compositional data sets using nonparametric imputation. Math. Geol. 35: 253–278. [Google Scholar]
- McArdle B., Anderson M., 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82: 290–297. [Google Scholar]
- McCarroll S., Kuruvilla F., Korn J., Cawley S., Nemesh J., et al. , 2008. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 40: 1166–1174. [DOI] [PubMed] [Google Scholar]
- Nabholz B., Ellegren H., Wolf J., 2013. High levels of gene expression explain the strong evolutionary constraint of mitochondrial protein-coding genes. Mol. Biol. Evol. 30: 272–284. [DOI] [PubMed] [Google Scholar]
- Nair S., Miller B., Barends M., Jaidee A., Patel J., et al. , 2008. Adaptive copy number evolution in malaria parasites. PLoS Genet. 4: e1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Niimura Y., Nozawa M., 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9: 951–963. [DOI] [PubMed] [Google Scholar]
- Nguyen D., Webber C., Ponting C., 2006. Bias of selection on human copy-number variants. PLoS Genet. 2: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North A., Pennanen J., Ovaskainen O., Laine A., 2010. Local adaptation in a changing world: the roles of gene-flow, mutation, and sexual reproduction. Evolution 65: 79–89. [DOI] [PubMed] [Google Scholar]
- Nozawa M., Kawahara Y., Nei M., 2007. Genomic drift and copy number variation of sensory receptor genes in humans. Proc. Natl. Acad. Sci. USA 104: 20421–20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J., R. Kindt, P. Legendre, B. O‘Hara, M. H. H. Stevens et al., 2007 vegan: Community Ecology Package. R package version 1.17-0; available at: http://cran.r-project.org/web/packages/vegan/.
- Pal C., Papp B., Hurst L., 2001. Highly expressed genes in yeast evolve slowly. Genetics 158: 927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Chen X., Yang J., Zhang J., 2013. Differential requirements for mRNA folding partially explain why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. USA 110: E678–E686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick D., Ghalambor C., 2001. The population ecology of contemporary adaptations: what empirical studies reveal about the conditions that promote adaptive evolution. Genetica 112–113: 183–198. [PubMed] [Google Scholar]
- Richter-Boix A., Teplitsky C., Rogell B., Laurila A., 2010. Local selection modifies phenotypic divergence among Rana temporaria populations in the presence of gene flow. Mol. Ecol. 19: 716–731. [DOI] [PubMed] [Google Scholar]
- Rokyta D. R., Lemmon A. R., Margres M. J., Aronow K., 2012. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genomics 13: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D. R., Wray K. P., Margres M. J., 2013. The genesis of an exceptionally deadly venom in the timber rattlesnake (Crotalus horridus) revealed through comparative venom–gland transcriptomics. BMC Genomics 14: 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen O., Lascoux M., Merila J., 2013. Ecological genomics of local adaptation. Nat. Rev. Genet. 14: 807–820. [DOI] [PubMed] [Google Scholar]
- Spurgin L., Illera J., Jorgensen T., Dawson D., Richardson D., 2014. Genetic and phenotypic divergence in an island bird: isolation by distance, by colonization or by adaptation? Mol. Ecol. 23: 1028–1039. [DOI] [PubMed] [Google Scholar]
- Stranger B., Forrest M., Dunning M., Ingle C., Beazley C., et al. , 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L., 1998. Phylogenetic Analysis Using Parsimony* (PAUP*), Version 4.0. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Templ M., Hron K., Filzmoser P., 2011. robCompositions: An R-Package for Robust Statistical Analysis of Compositional Data, pp. 341–355 in Compositional Data Analysis: Theory and Applications. Wiley, Chichester, UK. [Google Scholar]
- Vincent S., Herrel A., Irschick D., 2004. Sexual dimorphism in head shape and diet in the cottonmouth snake (Agkistrodon piscivorus). J. Zool. 264: 53–59. [Google Scholar]
- Vincent S., Brandley M., Kuriyama T., Mori A., Herrel A., Hasegawa M., 2009. Insular gigantism and dwarfism in a snake, adaptive response or spandrel to selection on gape size? Nature Preced. hdl:10101/npre.2009.3360.1.
- Vishnoi A., Kryazhimskiy S., Bazykin G., Hannenhalli S., Plotkin J., 2010. Young proteins experience for variable selection pressures than old proteins. Genome Res. 20: 1574–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A., Crawford D., 2006. Neutral and adaptive variation in gene expression. Proc. Natl. Acad. Sci. USA 103: 5425–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray K. P., Margres M. J., Seavy M., Rokyta D. R., 2015. Early significant ontogenetic changes in snake venoms. Toxicon 96: 74–81. [DOI] [PubMed] [Google Scholar]
- Yang J., Liao B., Zhuang S., Zhang J., 2012. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc. Natl. Acad. Sci. USA 109: E831–E840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yang J., 2015. Determinants of the rate of protein sequence evolution. Nat. Rev. Genet. 16: 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Peptide reports for all mass spectrometry analyses are in Table S5, and the raw liquid chromatography data are in Table S6. Sanger sequences were submitted to the National Center for Biotechnology Information (NCBI) Trace Archive under accession numbers KP881369–KP881418. Annotated transcriptome sequences were submitted to the GenBank Transcriptome Shotgun Assembly database under accession number GDBJ02000000 for S. miliarius and GDAZ02000000 for A. piscivorus.