Abstract
Many diploid organisms undergo facultative sexual reproduction. However, little is currently known concerning the distribution of neutral genetic variation among facultative sexual organisms except in very simple cases. Understanding this distribution is important when making inferences about rates of sexual reproduction, effective population size, and demographic history. Here we extend coalescent theory in diploids with facultative sex to consider gene conversion, selfing, population subdivision, and temporal and spatial heterogeneity in rates of sex. In addition to analytical results for two-sample coalescent times, we outline a coalescent algorithm that accommodates the complexities arising from partial sex; this algorithm can be used to generate multisample coalescent distributions. A key result is that when sex is rare, gene conversion becomes a significant force in reducing diversity within individuals. This can reduce genomic signatures of infrequent sex (i.e., elevated within-individual allelic sequence divergence) or entirely reverse the predicted patterns. These models offer improved methods for assessing null patterns of molecular variation in facultative sexual organisms.
Keywords: facultative sex, migration, gene conversion, genealogies, heterogeneity in sex
FACULTATIVE sexual species, which have both parthenogenetic and sexual stages in their life cycles, are widespread in nature. They have been a focus for empirical studies of the role of sex in evolution (Goddard et al. 2005; D’Souza and Michiels 2008; King et al. 2009; Morran et al. 2009; Becks and Agrawal 2010; Hojsgaard and Hörandl 2015). The biology of facultative sexual organisms is also a research field with broad applications because many organisms of evolutionary, medical, and agricultural importance undergo both sexual and asexual reproduction (Chang et al. 2013; Ankarklev et al. 2014; Hand and Koltunow 2014; Yoshida et al. 2014). However, our understanding of the structuring of genetic diversity in such organisms remains limited.
For effective population genetic analysis of facultative sexual individuals to be made, there needs to be a theoretical basis for predicting how neutral diversity is affected under various demographic and reproductive scenarios. This is so that the two are not confounded, especially because they are strongly intertwined [reviewed by Halkett et al. (2005) and Arnaud-Haond et al. (2007)]. A classic prediction for organisms with very low rates of sex is that owing to the resulting lack of segregation, diploid asexual organisms tend to accumulate extensive diversity within individuals, because the two alleles from a single individual remain isolated by descent in the absence of sex. This phenomenon has been labeled the Meselson effect because it was used by Meselson and colleagues as evidence for a lack of sex in bdelloid rotifers (Mark Welch and Meselson 2000; Butlin 2002) [but see Mark Welch et al. (2008) for an alternative explanation for their original findings].
One theoretical approach to quantify this effect has been to derive how traditional diversity measures, such as the effective population size Ne and F-statistics, are affected when sex is rare (Balloux et al. 2003; Yonezawa et al. 2004; de Meeûs and Balloux 2005). Such studies are based on pairwise sample comparisons, with heightened within-individual diversity not becoming apparent unless sex is extremely rare (with frequency less than 1/N, for N the total population size). Combining these summary statistics with information on clonal or genotypic diversity can improve inference when rates of sex are not too low (Balloux et al. 2003; Halkett et al. 2005; Arnaud-Haond et al. 2007).
An alternative approach is to analyze polymorphisms using coalescent theory to recreate the possible genealogies of samples (Kingman 1982; Wakeley 2009). Basic calculations were carried out by Brookfield (1992) and Burt et al. (1996) to determine how partial sex affects diversity in DNA fingerprinting analyses and isolates of a human pathogen, respectively. More complete analyses then were performed by Bengtsson (2003) and Ceplitis (2003) to determine expected coalescent times for pairs of samples taken either from the same or different individuals. These analyses confirmed that the rate of sex has to be less than the reciprocal of the population size for mean coalescent times to be substantially altered compared with the classic case of obligate sex.
With the advent of high-throughput genome sequencing technologies, there are growing opportunities to characterize both the demographic and reproductive histories of partially sexual populations. However, formal theory does not yet exist for genealogies of facultative sexual organisms that take into account demographic factors, gene conversion, and temporal and spatial heterogeneity in the rate of sexual reproduction. The potential importance of gene conversion is especially pertinent because there is growing recognition that this process may be an important homogenizing force in diploid asexual populations (Crease and Lynch 1991; Schön et al. 1998; Normark 1999; Schön and Martens 2003; Schaefer et al. 2006; Flot et al. 2013). In addition, previous theoretical studies have not accounted for cases where the rates of sex change in either space or time. It is well known that several organisms adjust their rate of sex in a stressful or dense environment. This mechanism is well supported both theoretically (Redfield 1988; Hadany and Otto 2007) and empirically (Grishkan et al. 2003; Snell et al. 2006; Levin and King 2013), but the overall consequences for patterns of genetic diversity are not clear. Previously published theory also suffers from ambiguities or typographical errors (explained later) that make interpretation of existing results difficult. In addition, no algorithm yet exists for how to recreate genealogies for more than two sequences with a range of sexual reproduction rates, which is essential for simulating the distribution of neutral variation under complex scenarios.
Here we outline theory to rectify this issue. We rederive equations of the coalescent time for pairs of samples taken from either different or the same individuals using a structured coalescent framework (Nordborg 1997), clarifying discrepancies that appear between Bengtsson (2003) and Ceplitis (2003). We further show how additional phenomena, including self-fertilization, gene conversion, population subdivision, and heterogeneity in rates of sex, can be included in the coalescent with partial sex. These results can be used to derive estimators for mutation, migration, and sex rates based on mean pairwise diversity. These initial analyses then will inform on how to create a simulation program in which there exists an arbitrary rate of sex that can be used to efficiently create genealogies for more than two chromosomes.
Materials and Methods
The basic model
We begin by considering a population with an arbitrary rate of sex but is otherwise ideal. Bengtsson (2003) and Ceplitis (2003) derived mean coalescent times for two alleles (two-sample mean coalescent times) from this basic model. However, we reexamine this model to clarify discrepancies between the earlier studies, as well as to lay the groundwork for further extensions.
Model outline
We consider a diploid Wright-Fisher population of size N individuals so that there are 2N gene copies in total. Then σ is the rate of sex; that is, with probability σ, an offspring is produced sexually, otherwise, it is produced asexually. For a specific individual, one can imagine that each of its gene copies inhabits a separate genomic deme within the individual, pertaining to the left- and right-hand allele copy respectively (Ceplitis 2003). We choose two of these gene copies and track their genealogy back in time until they reach their common ancestor. These allele pairs can be sampled from either different individuals or the same individual. With obligate sex, this distinction is irrelevant owing to the random segregation and union of gametes. However, if sex is infrequent, then pairs of alleles can remain “packaged” within an individual lineage as a left and right haplotype for long periods of time. This structuring and the low level of exchange between haplotypes when sex is infrequent create a fundamental difference from the standard coalescent.
Two sampled alleles can be found in one of three states: (1) one allele in each of two separate individuals, (2) two separate alleles within the same individual, and (3) coalesced. Ceplitis (2003) argued that it is necessary to divide the first state into two separate states: both alleles in the same deme (either the left or right demes of separate individuals), or one allele in the left deme and one in the right deme. However, it can be shown that this partitioning is unnecessary as long as the appropriate transition probabilities are applied, which average over these possible states. We calculate the mean coalescent time by first determining the transition probabilities among the three states mentioned earlier. We assume that N is large, so terms of order 1/N2 and higher are discarded.
The transition matrix for the process is
| (1) |
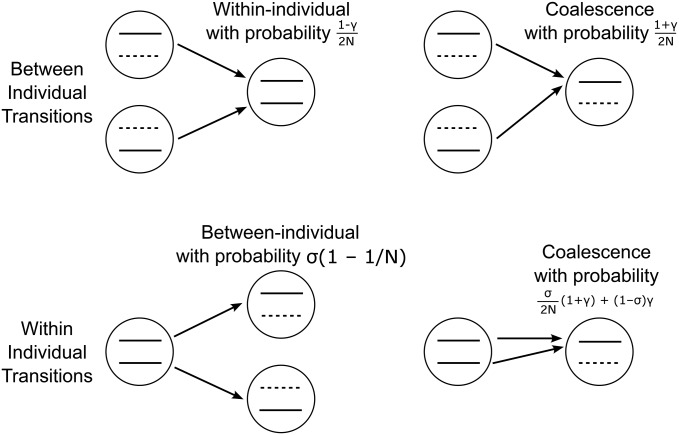
Further details of all transitions and analysis are outlined in Supporting Information, File S2A; a schematic of the basic transitions is shown in Figure 1.
Figure 1.
A schematic of the transition probabilities between states for the coalescent with infrequent sex (with rate σ) and gene conversion (rate γ). Only those states where the configurations have changed are shown. Focal samples are shown as solid black lines, whereas dashed black lines represent placeholder homologous alleles that are not sampled. Transitions from the within-individual state to the between-individual state require sex, whereas the reverse transition can occur via sexual or asexual reproduction.
To find the expected coalescent times, we follow the method of Slatkin (1991). Note that the 2 × 2 submatrix G, representing the top-left corner of T (Equation 1) is a matrix of noncoalescence; that is, it denotes the probability that two samples do not coalesce in a single generation. The probability that two alleles from either state have not coalesced t generations in the past is given by , where (here the T denotes vector transposition).
Conversely, the probability that two alleles have coalesced by time t is . For now, we will focus on the mean coalescent time; Slatkin (1991) showed that it can be calculated by
| (2) |
where is the vector of mean between- and within-individual mean coalescent times, and I is the identity matrix. The noncoalescence matrix G is crucial for determining the expected coalescent times. It can be decomposed into the relative contributions of sexual and asexual reproduction as follows:
| (3) |
This decomposition makes it easier to see how other complexities (e.g., selfing and gene conversion) can be included in this framework.
To make subsequent calculations more convenient, we shift to working in continuous time by rescaling time by the population size, i.e., τ = 2Nt. A formal way to rescale in continuous time is to write ; i.e., A captures transitions over “fast” timescales [which occur with probability ], and B captures transitions over “slow” timescales [which occur with probability ]. Möhle (1998) proved that if the matrix is written in this manner, the continuous time analogue of the transition matrix T, which we denote Q, can be written in the form
| (4) |
where
and
If we partition T (Equation 1) while assuming that , then we can show that the continuous-time transition matrix tends to the obligate sex case (File S1A). If , then A is equal to the identity matrix; thus , where
| (5) |
Here , which is equal to twice the expected number of sexually reproduced offspring produced per generation. This matrix shows that with rare sex, two samples from the same individual (the state represented by the second row of matrix B) cannot coalesce. Instead, the samples have to be placed within different individuals before coalescence can occur, which requires at least one bout of sex.
Where possible, subsequent analyses will be performed using the discrete transition matrix T, and the resulting expressions are derived assuming that N is large and are expressed in units of 2N generations; i.e., using the coalescent timescale () and compound parameters [e.g., ]. Nevertheless, we will sometimes use unscaled parameters when investigating rates of sex where .
Mean coalescent times:
Using Equations 2 and 3, we obtain the expected coalescent times for two samples taken from different individuals and from the same individual in units of 2N generations
| (6) |
Results are similar in the unscaled case, so they generally hold even if . Ceplitis (2003, Equation 2) derived equivalent equations; Bengtsson (2003, Equations 1 and 2) presented results in terms of the eigenvalues of G, although there appears to be typographical errors in the way these eigenvalues are presented. File S1A shows how equivalent results can be derived using the eigenvalues of G and how they compare with what is presented in Bengtsson (2003).
Visual inspection of Equation 6 shows some key properties of the within- and between-individual mean coalescent times. They verify that the rate of sex will significantly affect the expected coalescent time only if it is very rare, at least , as found in previous studies. Second, the within-individual expected coalescent time is greater than that between individuals, formalizing the Meselson effect (Mark Welch and Meselson 2000; Butlin 2002). Two samples from within a single individual are, by definition, members of different left/right sides. Sex is thererfore required to put them in the same side before coalescence can occur (on average, two sex events are needed). Only 50% of two-allele samples from different individuals will be on different left/right sides, so the increase in mean coalescent time resulting from low rates of sex between individual samples is only half as large.
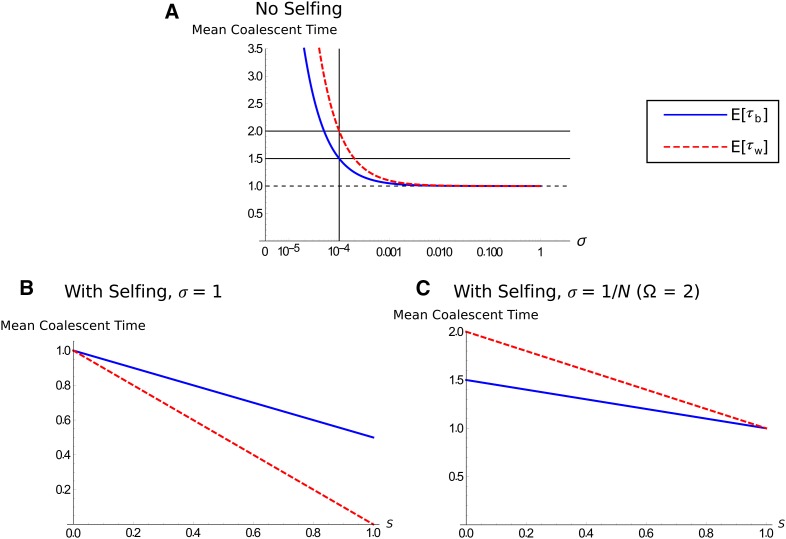
Figure 2A demonstrates how the mean coalescent times increase rapidly as σ becomes significantly less than . If we set (i.e., ) in Equation 6, then and . That is, the between-individual mean coalescent times are 1.5 times higher than in Kingman (obligate sexual) coalescent, and the within-individual mean coalescent times are twofold higher. We can also use analytical methods to determine the variance and probability distribution of the coalescent process; this analysis will be left until gene conversion is added so that its effect with rare sex can be quantified.
Figure 2.
Expected coalescent times with different reproductive systems. The population size for all plots is N = 10,000. (A) Expected mean coalescent time according to Equation 6 in coalescent time units as a function of sex σ. The blue line represents between-individual mean coalescent time , and the red line is within-individual mean coalescent time . The vertical line represents (), and the horizontal solid lines show the predicted mean coalescent times for this value (; on the coalescent timescale), whereas the horizontal dashed line shows . (B and C) Expected coalescent time if selfing is also included, according to Equation 9. Line colors are the same as in A, with (B) σ = 1 or (C) σ = 1/N ().
Multisequence coalescent simulations
The two-sample results are useful for clarifying how neutral drift should operate in facultative sexual populations. However, when analyzing empirical data, one would normally obtain many samples from a population and then estimate neutral diversity based on those samples. For obligate sexual organisms, coalescent simulations have proved to be essential for modeling expected neutral diversity levels caused by complex demographic scenarios, such as population bottlenecks or expansions (Hudson 2002). Equivalent analysis on sequence data from facultative sexual species is not possible using existing coalescent simulation software. This is so because gene samples remain paired within individual lineages if sex is rare, a type of genetic isolation not considered in existing simulation packages. Therefore, it is necessary to create a new coalescent algorithm to account for partial rates of sex. Our goal is to develop an algorithm that would remain valid under all rates of sex, including high rates [as opposed to purely under the low sex limit ].
The simulation procedure considers arbitrary rates of sex, as well as gene conversion and migration via an island model (which will be treated analytically in later sections). The main challenge is to consider how samples change state over time (through coalescence, migration, splitting, or forming pairs within individuals) while allowing an arbitrary rate of sex. Most coalescent simulation algorithms assume that each possible event is rare so that when an event does arise, only one event occurs. However, if there are several paired samples and the rate of sex is not very low (, for NT the total population size), many paired samples can split up into different individuals in a single generation; furthermore, an additional action (e.g., coalescence) is then possible in that same generation. The simulation accounts for this issue by determining the probability that a specific number of paired samples are split by sex in a generation and then designating what possible actions subsequently occur, given these splits.
We formalize this algorithm as follows: let there be xj paired samples and yj single samples (i.e., where only one of the two alleles is ancestral to the sample) in deme j, for ; we can also denote the configuration for deme j as . Let and ; then the total number of ancestral allele copies over all demes is , with the configuration for all demes denoted . If the rate of sex in deme j is σj (to account for spatial heterogeneity in rates of sex), then the probability that kj of xj samples are split by sex, with each part of the paired sample assigned to different adults back in time, is drawn from a binomial distribution
This event changes the configuration for all demes by producing 2kj new single samples in that deme: . The kj splits are chosen independently in each deme, without replacement, from the existing paired samples. This process creates a vector of paired samples that split in each deme.
Once this intermediate step is complete, there can be 10 other outcomes, which are outlined in Table 1. All additional outcomes (events 2–9 in Table 1) involve events that are of order and are thus assumed to be rare (including the gene conversion rate γ and the migration rate m), so only up to one additional event can arise. Hence, when an event occurs, it involves at least one paired sample splitting by sex, followed by an additional event. Alternatively, no sex occurs, in which case only events 5–10 in Table 1 can happen.
Table 1. Possible events in facultative sexual coalescent simulation.
| kj samples split by sex in deme j: | |||
|---|---|---|---|
| Event | Description | Configuration change in a deme | Probability |
| 1 | New single samples remain single | 1–all other probabilities | |
| 2 | One of the paired samples is re-created | ||
| 3 | A new single sample coalesces with an existing single sample | ||
| 4 | A new single sample coalesces with an existing paired sample | ||
| 5 | Two existing single samples form a new paired sample | ||
| 6 | Two existing single samples coalesce | ||
| 7 | Two remaining paired samples doubly coalesce asexually | ||
| 8 | One remaining paired sample coalesces with a single sample | ||
| 9 | Paired sample coalesces via gene conversion | ||
| 10 | A sample migrates to another deme | ||
| (before migration) | |||
The listed probabilities for events 1–4 are conditional on sex having occurred and are otherwise zero.
Building on this logic, we unite the preceding actions into a coalescent algorithm as follows:
In a single generation, an event occurs if either at least one paired sample is split by sex or no sex occurs, but one of events 5–10 in Table 1 happens regardless. The probability of no paired samples splitting via sex in any of the demes is . If no sex occurs, we need to consider the possibility that one of events 5–10 in Table 1 has happened. Let pE0 be the sum of the probabilities of events 5–10 (given that kT = 0) across all demes represented by the sample.
The total probability of any event occurring in a generation is . Standard probability theory (Wakeley 2009) tells us that the time until an event occurs is geometrically distributed with mean psum. Hence, psum is used to draw the time until the next event arises, which is then scaled by 2NT generations so that it is on the coalescent timescale.
-
Once the time to the next event is drawn, it involves sex with probability
(7) If sex does occur, the number of sexual events is drawn independently from each deme from a binomial distribution, discarding cases where no paired samples split by sex across all the demes represented by the sample.
Given the vector of paired samples affected by sex k, one then calculates the probability of subsequent events occurring in each deme using the probabilities outlined in Table 1. Let pE,j be the probability that event E occurs in deme j (if E = 1, where split samples remain split, then this event occurs over all demes by definition). The probability that event E occurs in any deme is . Therefore, the probability that event E is chosen is , and the probability that this event occurs in deme j is . These probabilities are used to form multinomial distributions, from which the next event and deme of occurrence then can be drawn.
The configurations of all demes are then changed based on the outcome. If it is a migration event, an individual is chosen from the deme of event occurrence, and its migration target is chosen at random from the d − 1 other demes.
The process is repeated until all samples coalesce into a single common ancestor. Mutations are then added to each branch of the genealogy assuming a Poisson distribution with mean , where , and τi is the length of branch i.
We have implemented this algorithm into a simulation program for R (R Development Core Team 2014), as shown in File S3, or it can be downloaded from http://github.com/MattHartfield/FacSexCoalescent. The simulation performs the preceding process, and for each run it outputs the coalescent times and a genealogy in Newick tree format (Felsenstein 2004). It also drops mutations along the genealogy according to the infinite-sites model, which can be used to determine how accurate summary statistics are at inferring the rate of sex and demography. We have rigorously tested this simulation to ensure that it replicates two-sample behavior. Outputs are similar to those presented by ms (Hudson 2002) to make it easier to compare simulation outputs between the two programs, if needed. Note that ms uses a timescale of 4N units, but our program uses a coalescent timescale of 2N.
In addition, we wrote a variant of the program that does not require population size to be defined as an input and operates solely on scaled coalescent time. This program is faster than the full program but requires that all parameters, including the rates of sex, are assumed to be on the same order as 1/N [i.e., ]. In this case, instead of first resolving the number of sexual reproductions in a generation, a single bout of sex is defined as one of the possible outcomes were an action to arise. To consider any rate of sex, all simulation results presented here were produced using the full simulation, which can take values of σ between 0 and 1.
Results
Simulation results of summary statistics
Using the full simulation, we investigated how traditional population genetics parameters are affected under low rates of sex to determine whether there is a risk that they show spurious signs of selection or demography. We ran our coalescent algorithm while varying the rate of sex and analyzed the mutational outputs to calculate summary statistics. One thousand trees were run for each point for all simulation results throughout. Confidence intervals were calculated using a normal-distribution approximation; similar intervals were produced if 1000 bootstrap samples were used.
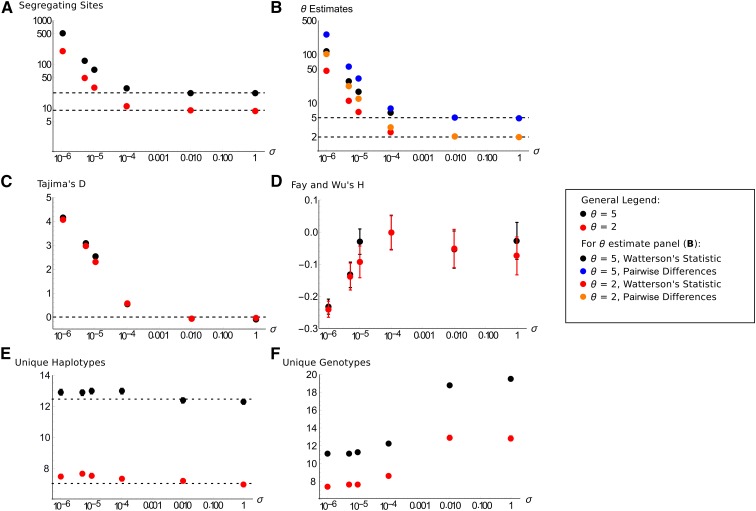
Results are shown in Figure 3 for N = 10,000, θ = 5 with 25 paired samples, although results are nearly identical if 50 single samples were simulated instead (File S2A). We need to specify a fixed population size because we cannot explicitly simulate high rates of sex [i.e., ] in diploids using only the scaled parameter (2Nσ). As the rate of sex decreases below 1/N, the number of polymorphic sites shoots up owing to the increased overall coalescent times, especially within individuals, creating greater diversity (Figure 3A). This leads to an increase in traditional estimates of θ that are much higher than the true mutation rate (Figure 3B). With rare sex, Tajima’s D also increases to high values (Figure 3C), while Fay and Wu’s H drops despite a small increase for (Figure 3D). Both of these behaviors are caused by an increase in the number of intermediate-frequency variants, especially within individuals, as sex becomes rare. Finally, the number of unique haplotypes increases (Figure 3E) and the number of unique genotypes decreases (Figure 3F) as σ drops. Both plots reflect how the lack of sex creates new haplotypes within individuals but homogenizes individual genotypes, in line with theory previously reported by Balloux et al. (2003). These results make clear that if one does not account for rare rates of sex, spurious signatures of selection or demography can arise that can confound analyses. Deeper branches also can lead to an overestimation of the mutation rate or effective population size.
Figure 3.
How classic genomic summary statistics are affected by infrequent sex in a nonsubdivided population. All graphs are as a function of σ; parameters are N = 10,000, θ = 5 (black points by default) or θ = 2 (red points by default), based on 25 paired samples, averaged over 1000 coalescent simulations. Error bars are 95% confidence intervals; if they cannot be seen, they lie within the points. (A) Mean number of segregating sites in a sample; the horizontal bar shows the expectation for σ = 1 (obligate sex). (B) Estimates of θ using Watterson’s θ (black or red points for θ = 5 or 2) or the mean pairwise differences between pairs of samples (blue or orange points for θ = 5 or 2). (C) Tajima’s D. The horizontal line at zero is the unbiased expectation for obligate sex. (D) Fay and Wu’s normalized H statistic. (E) The mean number of unique haplotypes present: the obligate sex expectation (dashed line) is calculated using Equation 3.85 of Ewens (2004). (F) The mean number of unique genotypes present.
Sex with selfing
We can extend the preceding analysis to consider a case where an individual that reproduces sexually can self-fertilize with probability S and outcross with probability 1 − S. The coalescent for obligate sexuality with selfing has been well investigated (Milligan 1996; Nordborg and Donnelly 1997; Nordborg 1997, 2000; Nordborg and Krone 2002); the key result is that the genealogy is equivalent to a Kingman coalescent, but with the population size scaled by the selfing rate so that the expected coalescent time is reduced. We can implement similar methods to determine how inbreeding and asexuality interact to affect coalescent times.
The transition matrix T with selfing and asexuality is written as (see File S1A for further information)
| (8) |
This equation demonstrates that self-fertilization only affects within-individual configurations. If a bout of selfing arises, then the samples coalesce with probability ½; otherwise, they will remain paired within the same individual. Using the matrix-inversion method of Slatkin (1991) and rescaling to the coalescent timescale (), the mean coalescent times are obtained as
| (9) |
This equation makes explicit how selfing reduces the mean coalescent times. Ceplitis (2003, Equation 5) also derived equivalent equations in the limit of low rates of sex. However, there appears to be a typographical error in his term for coalescence for two samples taken from different individuals and different chromosome arms. Similarly, Bengtsson (2003) derived accurate terms for the eigenvalues of G in the limit of weak sex, but plots of the effect of selfing appear to be inaccurate (compare his Figure 2 with our Figure 2C). These differences are discussed further in File S1A.
Selfing and asexual reproduction have opposing effects on the mean coalescent times and hence differently affect and . With selfing, when rates of sex are high, but the reverse is true when rates of sex are low (Figure 2, B and C). With obligate sexual reproduction, complete selfing reduces the between-individual mean coalescent time by half, while the within-individual mean coalescent time is instantaneous (Figure 2B). For , both mean coalescent times equal 1 with complete selfing (Figure 2C).
Note that because the main focus of this study is on the effects of sexual relative to asexual reproduction on diversity patterns, we did not implement self-fertilization in the facultative sexual coalescent simulation.
Gene conversion
Mitotic gene conversion can strongly affect the genetic architecture of organisms and can be especially important in facultative sexual organisms, as shown recently in the genome sequence of bdelloid rotifers (Flot et al. 2013). If a gene conversion event arises in which one allele sample replaces another during reproduction (irrespective of reproduction type), only one of the two samples is effectively passed onto its offspring.
We assume that unbiased gene conversion occurs at rate γ. The transition matrix becomes
| (10) |
We can solve this matrix as usual; we further set S = 0 to simplify the analysis (solutions with selfing are provided in File S1A). Furthermore, we make the substitutions and and rescale to the coalescent timescale [so that ]. The mean coalescent times then can be written as
| (11) |
where . Assuming that φ is not too small, each result in Equation 11 is dominated by the first term on the right-hand side. If sex is rare relative to gene conversion (), the leading terms go to 1 and 0, respectively, for and . For , we must then consider the second term, which goes to as the first term goes to 0. This shows that low sex with comparatively high gene conversion behaves similarly to selfing in that is half the standard value, and is much smaller than . The dependence on the ratio φ means that a reduction in expected coalescent time with gene conversion can occur at much larger rates of sex relative to population size () than the increase in mean coalescent time that we discussed earlier, although the absolute rate of sex must be low ().
The reason that gene conversion is important when sex is low can be understood by considering coalescence from two samples taken from separate individuals. In the absence of gene conversion, two samples are put into the same individual, on average, two separate times (requiring at least one bout of sex) before coalescing (e.g., the first time the two samples descended from different homologous chromosomes, but the second time they descended from the same chromosome). When the rate of sex is low, each time the two samples are put into the same individual, they persist together in that genotype for many asexual generations. If gene conversion is high relative to the rate of sex, the samples are likely to coalesce via gene conversion before the genotype is broken apart by sex. Thus, two samples need only be put into the same individual once before coalescing, so the coalescent time is half what it would be otherwise. If two samples start in the same individual, then the time to coalescence is simply the waiting time until gene conversion . This waiting time can be much lower than that for two alleles in separate individuals to enter the same ancestral individual (equal to half a time unit on the coalescent scale) if .
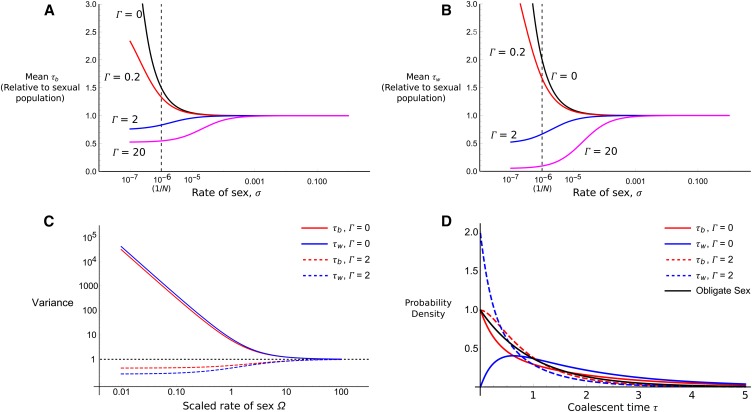
Note that when gene conversion is rare relative to sex (), Equation 11 collapses to the results assuming no gene conversion (Equation 6). The effect of gene conversion on mean coalescent time is shown in Figure 4 (A and B), demonstrating how mean coalescent time is greatly reduced when gene conversion is strong compared to sex.
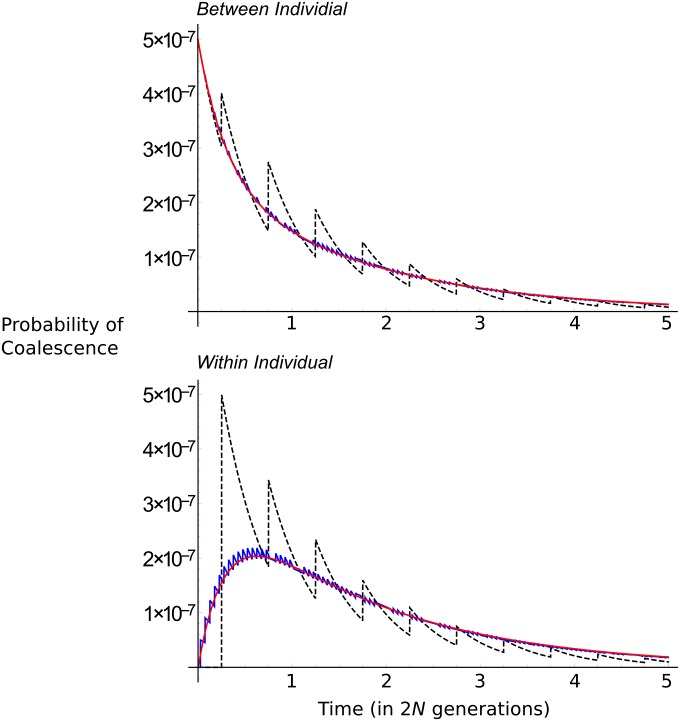
Figure 4.
(A and B) Expected coalescent time as a function of the rate of sexual reproduction. The population is of size N = 106 for two samples taken from separate individuals (A) or the same individual (B). This value of N was used so that 1/N is on the same order as gene conversion rates obtained from empirical data (see text for details). Gene conversion rates of (black), 0.2 (red), 2 (blue), and 20 (magenta) are shown. (C) The variance of the coalescent process as a function of the scaled rate of sex (), scaled to that for the fully sexual coalescent. Red lines are for τb; blue for τw; solid lines for ; dashed line for . Horizontal dashed line is the variation with complete sex. (D) Probability density of the partial sex coalescent (assuming that for red and blue lines and σ = 1 for the black line, i.e., obligate sex) as a function of coalescent time units. Line colors are as in C, with the addition of the obligate sexual distribution (black solid line).
Gene conversion rates are generally weak, with Flot et al. (2013) estimating gene conversion rates of 10−5 to 10−6 per site in bdelloid rotifers. However, these rates are feasibly the same order (or higher) as for most species, which is the same rate at which infrequent sex starts affecting expected coalescent times. Thus, the strong impact of gene conversion when sex is rare can explain why little within-individual divergence has been reported in facultative and obligate sexual genomes despite very low (or zero) rates of sex (Crease and Lynch 1991; Schön et al. 1998; Normark 1999; Schön and Martens 2003; Flot et al. 2013).
As with selfing, gene conversion reduces the mean coalescent times by increasing the probability that two samples within an individual will coalesce. However, there are important distinctions between the two processes and their interaction with asexual reproduction. Selfing occurs as a reproductive alternative to asexual reproduction, and the effects of selfing and low sex on mean coalescent time are additive, as shown in Equation 9. In contrast, gene conversion interacts with asexual reproduction rather than being an alternative to it. Gene conversion is only a significant force when asexual reproduction is sufficiently common that two samples within an individual are kept together for many generations; this is why φ, the ratio of sex to gene conversion, is important. Nonetheless, both selfing and gene conversion reduce coalescence times and increase homozygosity. Distinguishing between them may be possible through examination of multilocus models because the two processes affect linkage disequilibrium differently.
Variance and probability distribution with and without gene conversion:
Asexuality and gene conversion alter not only the mean coalescent time but also other aspects of the probability distribution. Previously, Bengtsson (2003) derived the distribution for the case with partial sex and selfing; here we present a derivation to extend these results to consider gene conversion. We achieve this by following the method of Herbots (1997) and set up a series of linear equations to find the Laplace transform, and therefore the probability distribution, of the coalescent process. The method is outlined in File S2A.
If we denote the random variable of the between- and within-individual expected coalescent times as τb and τw, respectively, the variances of these times are
| (12) |
Furthermore, the probability density functions of each time and can be written as a mixture of two exponential distributions
| (13) |
where are the rate parameters of the exponential functions (where 1 and 2 refer to the plus and minus variants of the second term), and the A and B terms are scaling constants, i.e.,
| (14) |
Figure 4C plots the variance (Equation 12) with and without gene conversion. With no gene conversion, the variance greatly increases as the rate of sex decreases, reflecting the greatly increased and sporadic coalescent times that result with infrequent sex. However, with gene conversion present at rate , the variance is lower than for the obligate sex coalescent. Figure 4D plots the corresponding probability distributions. Without gene conversion, the probability density distributions for τb and τw are qualitatively similar to those for two-allele samples from either different demes or the same deme in a subdivided population model (Herbots 1997). has a mode at zero, whereas has a nonzero mode. This result is indicative of how paired samples have to be split by sex before they are able to coalesce, just as samples from different demes must migrate into the same deme before coalescence can occur. Hence, coalescence at early times is not possible. If gene conversion is present, the distribution of τb is weakly affected by the distribution of τw and becomes strongly skewed with a mode at zero. Both the variance and probability distributions with gene conversion provide further evidence that low rates of gene conversion greatly reduce the coalescent times.
Effect of gene conversion on summary statistics:
In multisample simulations, we found that relatively low rates of gene conversion () generally lead to the opposite behavior of the summary statistics shown in Figure 3 owing to increased homozygosity. That is, most estimators decrease with lower sex, except for Fay and Wu’s H. The number of unique haplotypes also decreases for low rates of sex, as in cases without gene conversion (File S2B). Therefore, the simultaneous reduction in both haplotype and sequence diversity with decreased rates of sex, in contrast to observing allelic sequence divergence, can be used to determine the presence of gene conversion (or a similar effect such as selfing) in facultative sexual organisms.
Sequence-dependent gene conversion
The preceding section assumes that gene conversion is possible, but if enough differences accumulate between alleles, then gene conversion may no longer operate. Here we consider the time needed for two sequences to achieve sufficiently high divergence to halt gene conversion. Using a different approach, we reach the same finding as Walsh (1987). He considered duplicated gene sequences diverging at a certain rate, with possible gene conversion homogenizing the two sequences. If each gene acquires kc mutations, further conversion is prevented, and each sample begins to act independently.
Assume that gene conversion occurs at rate Γ and that is the scaled mutation rate for a gene that can lead to sequence divergence. If kc such mutations occur, then no further gene conversion is possible. We further assume that sex is sufficiently rare (i.e., ) that it does not affect this process. Conditional on an event occurring, the probability that it is a conversion is , and the probability that it is a mutation is (note that the factor of 2 exists because mutation can occur on either of the two gene copies). For complete divergence to occur, kc mutations must arise before gene conversion homogenizes both sequences. Hence, following from a conversion event, the probability pc that sufficient divergence occurs before another gene conversion event is . Thus, pc is equivalent to Equation 6 of Walsh (1987), which was obtained by a different approach.
Given that pc > 0, the critical level of divergence will occur eventually, but we can ask how long we expect this process to take. Let n denote the number of conversion events that happen before divergence eventually occurs. Thus, n follows a geometric distribution with expectation or (Walsh 1987, Equation 7). For example, if and kc = 10, then there are expected to be ∼6 × 107 gene conversion events before sufficient divergence is reached. Given that both and kc are likely to be greater than 10, is likely to be even larger than in this example. Hence, situations where divergence stops gene conversion are highly unlikely to occur.
Coalescent times for an island model
Here we show how a simple demographic model, the island model, also can be implemented into the facultative sexual analysis. The population consists of d demes with Nd individuals in each, so the total population size is . In each generation, an individual can migrate to another deme with probability ; this assumption ensures that no more than one sample migrates per generation. We also can write this migration rate as so that processes can be considered in continuous time. To keep solutions tractable, we do not consider selfing in this analysis.
In this model, two alleles can be found in three noncoalesced states. Pairs of samples can be taken either from different demes, from different individuals within a deme, or from the same individual. File S2A shows how the transition matrix G is formed and solved to obtain mean coalescent times for different demes td, within demes tb, and within individuals tw. Conveniently, in the continuous timescale [], and are exactly as in Equation 11 with , and is the same as but with an additional term to denote isolation by distance (Slatkin 1991) (see also File S1B). Hence, the effects of population structure, as well as the joint effect of gene conversion and low sex, are simply additive. As in a nonsubdivided population, sex has to be on the order of the inverse of the total population size to have a significant impact on the expected coalescent time. If gene conversion is present with rate , then mean coalescent times are greatly reduced. Hence, even a small rate of gene conversion reduces within-individual diversity that otherwise would be caused by low rates of sex. Further information is available in File S1B.
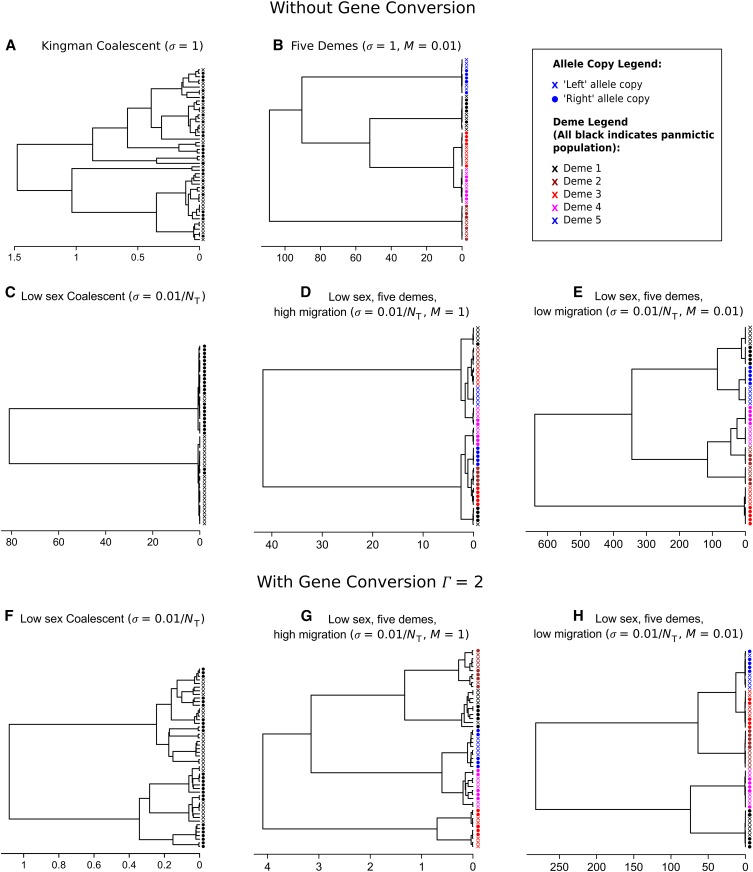
For multisample genealogies without gene conversion, several distinct types of tree topologies can occur depending on the relative rates of sex and migration. A standard genealogy with obligate sex and high migration is shown in Figure 5A. It is well known (Wakeley 2000; Wakeley and Aliacar 2001) that with obligate sex and limited migration, samples separate themselves into subclades principally by their geographic location (Figure 5B), meaning that most of the variation is among demes; the same result occurs with facultative sex, provided that sex is not too rare. However, if sex is low, then there tends to be two deep lineages, present within all demes, representing the long expected coalescent times between left/right haplotypes, assuming that gene conversion is rare relative to sex (Figure 5C). That is, most of the variation is within individuals created by excess heterozygosity. When migration and sex are both low but migration is high relative to sex [i.e., ], then principally two major subclades arise based on the chromosome arm of the sample. Within these major clades, five further clades are apparent owing to the geographic location of the samples (Figure 5D). A large fraction of the genetic variance is within individuals, but there is also a considerable amount of variation among populations. Finally, if migration rates are of the same order or lower than the rate of sex, the opposite pattern is seen: there are five clades owing to geographic separation, with pairs of subclades then forming as a result of the lack of genetic segregation (Figure 5E). The frequency spectra for each of these five examples are shown in File S2C (a–e). With low sex, sequences present in half the allelic samples are very common, representing ongoing divergence at the two different allele copies. Furthermore, once isolation by distance is additionally prevalent, polymorphisms tend to arise in multiples of five (because d = 5 in our example), reflecting segregation both by deme and by allelic copy.
Figure 5.
Examples of typical genealogies produced by facultative sex coalescent simulations. For all simulations, , and 25 paired samples were simulated. Scale bars indicate time in past in units of 2NT generations. Subdivision by either allelic copy or deme is shown in the legend. (A) Traditional (Kingman) coalescent in a nonsubdivided population with obligate sex. (B) Obligate sexual population in an island model, with d = 5 and . (C and F) Nonsubdivided population with low sex . (D and G) Low-sex population () in an island model with a relatively high migration rate (d = 5, ). (E and H) Low-sex population () in an island model with a relatively low migration rate (d = 5, ). No gene conversion is present for A–E, but it is present at a rate in F–H.
With even small rates of gene conversion [], spatial structure remains, but little genetic partitioning is evident, leading to greater mixing of the two chromosome arms (Figure 5, F–H). Such trees give a misleading appearance of either a higher overall rate of sex or spatially heterogeneous rates of sex. File S2C (f and h) shows the relevant site-frequency spectra: low-frequency variants are much more likely to be produced as in the obligate sex case, highlighting how even weak gene conversion creates spurious genomic signatures of frequent sex.
Diversity measurements and parameter estimation:
In the absence of gene conversion and with low rates of sex, it is possible to use the migration model results with Γ = 0 to derive diversity-based estimators for the mutation, migration, and sex rates. File S2A outlines how to derive the following scaled estimators for the mutation rate , sex rate , and migration rate (we use different notation to differentiate Me from the parameter introduced earlier):
| (15) |
where is the average between-deme pairwise diversity, is the average within-deme diversity, and is the average within-individual diversity. We also investigate what sampling strategies should be used to most accurately estimate these parameters. Briefly, paired within-individual samples should be used as much as possible, and if the means of and are taken from multiple chromosomes, the ratio of means should be taken instead of the mean of ratios. However, these estimators assume that within-individual coalescent times are the longest owing to low rates of sex. If gene conversion occurs on the same order or greater than the rate of sex, within-individual diversity would be removed, so these estimators perform badly. Hence, such estimators would work only in the case where , which may not apply to many partially asexual species that have large population sizes.
Heterogeneity in rates of sex
Results so far have assumed that the rate of sex remains constant over genealogic time. However, this assumption may not be realistic for a broad range of taxa. Facultative sexual organisms are known to adjust their rate of sexual reproduction depending on certain biotic and abiotic conditions that can vary in time and space. We describe here how to extend the preceding derivations to account for either temporal or spatial heterogeneity. Unless otherwise stated, we do not consider gene conversion in these derivations. For brevity, we only outline key results, with further explanation available in File S2A.
Temporally heterogeneous sexual reproduction:
Consider a case in which there is a single population, but the rates of sex vary over time. At time t in the past, the rate of sex is σt, and the noncoalescent matrix for that generation Gt is given by Equation 10, replacing σ with σt. The distributions of coalescent times (in generations) for within- and between-individual samples are given by
| (16) |
where
| (17) |
are the probabilities that two samples that are between individuals or within individuals at time t − 1 coalesce at time t.
Numerical evaluation of various scenarios involving temporal heterogeneity indicates that the distribution of coalescent times can be well approximated using the arithmetic average rate of sex and otherwise ignoring temporal heterogeneity. This works well provided that the fluctuations in rates of sex are fast relative to the average coalescent time so that lineages can be expected to experience “average” rates of sex over their coalescent history. When very long periods (∼N generations) of low sex are punctuated by very brief periods of high rates of sex, use of the average rate of sex can be misleading. For example, we consider a species that reproduces exclusively asexually most of the time, but once every g generations a fraction of offspring is produced sexually, giving an average rate of sex of . As illustrated in Figure 6, when = 1/10 and g = N/10 (i.e., ), the distributions closely follow those expected in a population with a constant rate of sex of σ = 1/N. However, the distributions are notably different when = 1 and g = N (i.e., ). During the periodic episodes of sex, within-individual samples are converted to between-individual samples, allowing for their subsequent coalescence; this creates periodic spikes in the distribution of coalescent times.
Figure 6.
Distribution of coalescent times with temporal heterogeneity in rate of sex, assuming no self-fertilization or gene conversion. The rate of sex is assumed to be zero most of the time, but once every g generations, a fraction of offspring is produced sexually, giving an average rate of sex of . Here (N = 106) for all cases, but this is achieved via very high heterogeneity ( = 1 and g = N; black dashed line), moderate heterogeneity ( = 0.1 and g = 0.1N; blue line), or no heterogeneity ( = 1/N and g = 1; red line). The first sexual episode occurs g/2 generations in the past. The upper and lower panels show the distributions of coalescent times for between- and within-individual samples, respectively.
Spatially heterogeneous sexual reproduction:
We next consider the case in which there is an island model, but rates of sex occur at a lower rate σL in some demes and at a higher rate σH in others (and these rates stay constant within each deme over time). There are dL low-sex demes and dH high-sex demes, with . Although we have assumed only two rates of sex, the same approach can be extended to consider an arbitrary number of rates. Given the variation in sex rates considered, we will work with unscaled parameters of σ.
When calculating mean pairwise coalescent times, there are now seven states to consider. For between-deme comparisons, two samples can be taken from either two different low-sex demes, two different high-sex demes, or one from a low-sex deme and the other from a high-sex deme. In addition, for within-deme samples (either from the same or separate individuals), the two samples can be from either a high-sex deme or a low-sex deme.
One can form the 7 × 7 G matrix, but in this case, deriving each term is complicated, and the overall solutions are cumbersome. Hence, the full derivation is saved for File S1D. Simpler equations can be obtained if it is assumed that migration and . These simpler terms are described in File S2A. In particular, elegant terms can be obtained for the difference in mean coalescent times between high- and low-sex demes
| (18) |
These equations illustrate two points regarding the differences in expected coalescent times between samples from low-sex demes compared with those from high-sex demes. The differences increase as migration rates decrease, and they are also independent of the rates of sex. This is so because we have assumed that . Consequently, alleles from different left/right haplotypes cannot or are extremely unlikely to coalesce within the low-sex demes. Such alleles must first move to high-sex demes before coalescence can occur. The difference shown in Equation 18 represents the waiting time for alleles in low-sex demes to migrate to high-sex demes before coalescence can occur. Equation 18 reflects the true differences as long as σL is low enough compared to the migration rate (File S1D).
We can ask how average coalescent times correspond to those expected in a metapopulation without spatial heterogeneity in sex rates but with the same average rate of sex [i.e., using ]. File S2D highlights how using the weighted mean in the homogeneous result underestimates true mean coalescent times. This is so because using an average rate of sex does not account for how sequences taken from low-sex demes contribute disproportionally to the overall coalescent time.
File S2A shows how we apply diversity-based estimators for the rates of sex to simulation data from a spatially heterogeneous case in the absence of gene conversion. If migration is not too high (), then application of these estimators to individual demes can give a decent estimation of the rate of sex within them. Estimates are less accurate if migration is higher (), instead predicting a rate of sex intermediate between the two true values.
Discussion
We have outlined here a comprehensive theoretical treatment to consider coalescent processes with arbitrary rates of sex. After reapplying structured coalescent theory to previous models (Bengtsson 2003; Ceplitis 2003), we show how to calculate expected coalescent times for pairs of samples taken either between or within individuals (Equation 6). We show how this structured coalescent can be further extended to take other cases into account, such as self-fertilization (Equation 9), gene conversion (Equation 11), and migration according to an island model. From the migration case, we further derived statistical estimators for the rates of mutation, migration, and sex (Equation 17).
Our multisample simulation results highlight that with very low rates of sexual reproduction, strong departures from the standard neutral model expectations for summary statistics such as Tajima’s D and Fay and Wu’s H are expected, reflecting the deep genealogical divergence between alleles within individuals. Thus, species with very low rates of sex can exhibit diversity patterns resembling long-term balancing selection and/or population subdivision. This highlights the importance of analysis of within vs. between individual patterns of diversity to help untangle the relative role of demographic history and selection from partial sex on the shape of genealogies.
To more accurately account for demographic effects, we found two solutions. First, newly derived estimators for the mutation, sex, and migration rates based on pairwise diversity (Equation 17) can be accurate if good sampling practice is followed (File S2A). However, these simple estimators can produce misleading results under a wide variety of scenarios that cannot be accounted for because of insufficient degrees of freedom in the equations. These include the presence of gene conversion rates of similar magnitude to sex and spatial or temporal heterogeneity in sex. We therefore suspect that the best manner to account for complex demography would be to use coalescent simulations in an inference framework, such as approximate Bayesian computation (ABC) (Sunnåker et al. (2013)), to predict how summary statistics change under different regimes. Our new simulation package could be used in an ABC-type inference while varying the rate of sex to simultaneously infer sex and demography.
One major result is that once sex becomes on the same order as gene conversion, the latter becomes a powerful force in reducing within-individual diversity and therefore removing genetic signatures of rare sex. Beyond the challenges this presents for estimating rates of sexual reproduction, this result also has important evolutionary consequences because the absence of permanent heterozygosity can influence not only the structuring of neutral diversity but also the exposure of both deleterious and advantageous recessive mutations to selection. Although elevated heterozygosity has been observed in some partial and obligate asexual organisms (Tucker et al. 2013; Hollister et al. 2015), there is increasing evidence that a major source of high within-individual diversity may be hybrid origins of asexual lineages rather than ongoing accumulation of diversity owing to new mutations (Tucker et al. 2013). Under biologically realistic rates of gene conversion, this initially high heterozygosity may be rapidly eroded by gene conversion, decreasing neutral variation within and between individuals (Tucker et al. 2013). Furthermore, these potentially high rates of gene conversion make it unlikely that two allelic copies develop sufficient divergence before conversion resets their independent evolution (see also Walsh 1987).
These analyses provide a more thorough theoretical basis for which the genealogies of samples from facultative sexual organisms can be reconstructed to provide the null distribution of neutral diversity for such populations. We have outlined methodology so that it is clear how the results presented here can be extended to cover even more complex scenarios; nevertheless, there are several apparent routes for extending this work. Although we have shown how to implement demography via an island model, it would be desirable for future models to consider a broader array of demographic scenarios. Real-world populations go through a bewildering array of changes, including extinction, recolonization, population expansion, and bottlenecking (Whitlock and McCauley 1999; Veeramah and Hammer 2014). Advanced coalescent models for obligate sexual organisms have been developed to determine how complex demographic effects affect genealogies (Rosenberg and Nordborg 2002; Hudson 2002); it will certainly be worthwhile to implement similar demographic scenarios into facultative sex coalescent simulations given the known impact of spatial effects on clonal species distribution (Arnaud-Haond et al. 2007).
The other major extension that will prove important is to implement recombination. In this case, the outputs will no longer be a genealogy, but rather a recombination graph (Griffiths 1981, 1991), although it would still be possible to create genealogies for unrecombined genomic segments. Recombination graphs can provide greater power for detecting demographic effects with fewer samples, although it can be more computationally demanding to run relevant analyses (Schiffels and Durbin 2014). Adding recombination will prove important for facultative sexual coalescents because while sex has to be very rare [] to affect the nonrecombining genealogies presented here, differences in recombination graphs might become apparent with higher rates of sex. This is so because it is expected that recombination correlates linearly with sex rate. A similar result was derived for partially selfing organisms by Nordborg (2000). Genomic sequences of facultative sexual organisms can reveal evidence of rare recombination events, providing proof of cryptic sex [examples were shown by Grimsley et al. (2010) for the marine algae Ostreococcus spp. and Signorovitch et al. (2005) for Placozoa]. In addition, ancestral graphs would be able to separate out areas of the genome affected by gene conversion, leading to much more refined measurements of how this force affects genetic architecture. In particular, gene conversion events in the absence of sex will lead to much higher rates of gene conversion relative to crossing over. This will have a strong effect on short- vs. long-range linkage disequilibrium along chromosomes, which may enable accurate estimators of the rate of sex even in the presence of significant rates of gene conversion. Recombination graphs of facultative sexual organisms thus can greatly increase the power by which the effects of infrequent sex and demography can be disentangled.
Acknowledgments
We thank two anonymous reviewers and John Wakeley for comments on the manuscript. M.H. is supported by a Marie Curie International Outgoing Fellowship (grant MC-IOF-622936, project SEXSEL). This work also was supported by Discovery Grants (to A.F.A. and S.I.W.) from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Communicating editor: J. Wakeley
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.178004/-/DC1
Literature Cited
- Ankarklev J., Brancucci N. M. B., Goldowitz I., Mantel P.-Y., Marti M., 2014. Sex: how malaria parasites get turned on. Curr. Biol. 24: R368–R370. [DOI] [PubMed] [Google Scholar]
- Arnaud-Haond S., Duarte C. M., Alberto F., Serrão E. A., 2007. Standardizing methods to address clonality in population studies. Mol. Ecol. 16: 5115–5139. [DOI] [PubMed] [Google Scholar]
- Balloux F., Lehmann L., de Meeûs T., 2003. The population genetics of clonal and partially clonal diploids. Genetics 164: 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becks L., Agrawal A. F., 2010. Higher rates of sex evolve in spatially heterogeneous environments. Nature 468: 89–92. [DOI] [PubMed] [Google Scholar]
- Bengtsson B. O., 2003. Genetic variation in organisms with sexual and asexual reproduction. J. Evol. Biol. 16: 189–199. [DOI] [PubMed] [Google Scholar]
- Brookfield J. F. Y., 1992. DNA fingerprinting in clonal organisms. Mol. Ecol. 1: 21–26. [Google Scholar]
- Burt A., Carter D. A., Koenig G. L., White T. J., Taylor J. W., 1996. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc. Natl. Acad. Sci. USA 93: 770–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin R., 2002. The costs and benefits of sex: new insights from old asexual lineages. Nat. Rev. Genet. 3: 311–317. [DOI] [PubMed] [Google Scholar]
- Ceplitis A., 2003. Coalescence times and the Meselson effect in asexual eukaryotes. Genet. Res. 82: 183–190. [DOI] [PubMed] [Google Scholar]
- Chang H.-H., Moss E. L., Park D. J., Ndiaye D., Mboup S., et al. , 2013. Malaria life cycle intensifies both natural selection and random genetic drift. Proc. Natl. Acad. Sci. USA 110: 20129–20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crease T. J., Lynch M., 1991. Ribosomal DNA variation in Daphnia pulex. Mol. Biol. Evol. 8: 620–640. [Google Scholar]
- de Meeûs T., Balloux F., 2005. F-statistics of clonal diploids structured in numerous demes. Mol. Ecol. 14: 2695–2702. [DOI] [PubMed] [Google Scholar]
- D’Souza T. G., Michiels N. K., 2008. Correlations between sex rate estimates and fitness across predominantly parthenogenetic flatworm populations. J. Evol. Biol. 21: 276–286. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics: 1. Theoretical Introduction (Interdisciplinary Applied Mathematics Series, Vol. 27), Ed. 2 Springer, New York. [Google Scholar]
- Felsenstein J., 2004. Inferring Phylogenies, Vol. 2 Sinauer Associates, Sunderland, MA. [Google Scholar]
- Flot J.-F., Hespeels B., Li X., Noel B., Arkhipova I., et al. , 2013. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500: 453–457. [DOI] [PubMed] [Google Scholar]
- Goddard M. R., Godfray H. C. J., Burt A., 2005. Sex increases the efficacy of natural selection in experimental yeast populations. Nature 434: 636–640. [DOI] [PubMed] [Google Scholar]
- Griffiths R. C., 1981. Neutral two-locus multiple allele models with recombination. Theor. Popul. Biol. 19: 169–186. [Google Scholar]
- Griffiths R. C., 1991. The two-locus ancestral graph. Lect. Notes Monogr. Ser. 18: 100–117. [Google Scholar]
- Grimsley N., Pequin B., Bachy C., Moreau H., Piganeau G., 2010. Cryptic sex in the smallest eukaryotic marine green algae. Mol. Biol. Evol. 27: 47–54. [DOI] [PubMed] [Google Scholar]
- Grishkan I., Korol A. B., Nevo E., Wasser S. P., 2003. Ecological stress and sex evolution in soil microfungi. Proc. Biol. Sci. 270: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadany L., Otto S. P., 2007. The evolution of condition-dependent sex in the face of high costs. Genetics 176: 1713–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkett F., Simon J.-C., Balloux F., 2005. Tackling the population genetics of clonal and partially clonal organisms. Trends Ecol. Evol. 20: 194–201. [DOI] [PubMed] [Google Scholar]
- Hand M. L., Koltunow A. M. G., 2014. The genetic control of apomixis: asexual seed formation. Genetics 197: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbots H. M., 1997. The structured coalescent, pp. 231–255 in Progress in Population Genetics and Human Evolution (IMA Volumes in Mathematics and Its Applications, Vol. 87), edited by Donnelly P., Tavaré S. Springer, New York. [Google Scholar]
- Hojsgaard D., Hörandl E., 2015. A little bit of sex matters for genome evolution in asexual plants. Front. Plant Sci. 6: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister J. D., Greiner S., Wang W., Wang J., Zhang Y., et al. , 2015. Recurrent loss of sex is associated with accumulation of deleterious mutations in Oenothera. Mol. Biol. Evol. 32: 896–905. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- King K. C., Delph L. F., Jokela J., Lively C. M., 2009. The geographic mosaic of sex and the Red Queen. Curr. Biol. 19: 1438–1441. [DOI] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982. On the genealogy of large populations. J. Appl. Probab. 19: 27–43. [Google Scholar]
- Levin T. C., King N., 2013. Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta. Curr. Biol. 23: 2176–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch D. B., Meselson M., 2000. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science 288: 1211–1215. [DOI] [PubMed] [Google Scholar]
- Mark Welch D. B., Mark Welch J. L., Meselson M., 2008. Evidence for degenerate tetraploidy in bdelloid rotifers. Proc. Natl. Acad. Sci. USA 105: 5145–5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan B. G., 1996. Estimating long-term mating systems using DNA sequences. Genetics 142: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhle M., 1998. A Convergence theorem for Markov chains arising in population genetics and the coalescent with selfing. Adv. Appl. Probab. 30: 493–512. [Google Scholar]
- Morran L. T., Parmenter M. D., Phillips P. C., 2009. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature 462: 350–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., 1997. Structured coalescent processes on different time scales. Genetics 146: 1501–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., 2000. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Donnelly P., 1997. The coalescent process with selfing. Genetics 146: 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Krone S. M., 2002. Separation of time scales and convergence to the coalescent in structured populations, pp. 194–232 in Modern Developments in Theoretical Population Genetics, edited by Slatkin M., Veuille M. Oxford University Press, Oxford, UK. [Google Scholar]
- Normark B. B., 1999. Evolution in a putatively ancient asexual aphid lineage: recombination and rapid karyotype change. Evolution 53: 1458–1469. [DOI] [PubMed] [Google Scholar]
- R Development Core Team , 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Redfield R. J., 1988. Evolution of bacterial transformation: Is sex with dead cells ever better than no sex at all? Genetics 119: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Nordborg M., 2002. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat. Rev. Genet. 3: 380–390. [DOI] [PubMed] [Google Scholar]
- Schaefer I., Domes K., Heethoff M., Schneider K., Schön I., et al. , 2006. No evidence for the “Meselson effect” in parthenogenetic oribatid mites (Oribatida, Acari). J. Evol. Biol. 19: 184–193. [DOI] [PubMed] [Google Scholar]
- Schiffels S., Durbin R., 2014. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön I., Butlin R. K., Griffiths H. I., Martens K., 1998. Slow molecular evolution in an ancient asexual ostracod. Proc. Biol. Sci. 265: 235–242. [Google Scholar]
- Schön I., Martens K., 2003. No slave to sex. Proc. Biol. Sci. 270: 827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorovitch A. Y., Dellaporta S. L., Buss L. W., 2005. Molecular signatures for sex in the Placozoa. Proc. Natl. Acad. Sci. USA 102: 15518–15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., 1991. Inbreeding coefficients and coalescence times. Genet. Res. 58: 167–175. [DOI] [PubMed] [Google Scholar]
- Snell T. W., Kubanek J., Carter W., Payne A. B., Kim J., et al. , 2006. A protein signal triggers sexual reproduction in Brachionus plicatilis (rotifera). Mar. Biol. 149: 763–773. [Google Scholar]
- Sunnåker M., Busetto A. G., Numminen E., Corander J., Foll M., et al. , 2013. Approximate Bayesian computation. PLOS Comput. Biol. 9: e1002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A. E., Ackerman M. S., Eads B. D., Xu S., Lynch M., 2013. Population-genomic insights into the evolutionary origin and fate of obligately asexual Daphnia pulex. Proc. Natl. Acad. Sci. USA 110: 15740–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramah K. R., Hammer M. F., 2014. The impact of whole-genome sequencing on the reconstruction of human population history. Nat. Rev. Genet. 15: 149–162. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 2000. The effects of subdivision on the genetic divergence of populations and species. Evolution 54: 1092–1101. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 2009. Coalescent Theory: An Introduction, Vol. 1 Roberts & Company Publishers, Greenwood Village, CO. [Google Scholar]
- Wakeley J., Aliacar N., 2001. Gene genealogies in a metapopulation. Genetics 159: 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J. B., 1987. Sequence-dependent gene conversion: can duplicated genes diverge fast enough to escape conversion? Genetics 117: 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M. C., McCauley D. E., 1999. Indirect measures of gene flow and migration: . Heredity 82: 117–125. [DOI] [PubMed] [Google Scholar]
- Yonezawa K., Ishii T., Nagamine T., 2004. The effective size of mixed sexually and asexually reproducing populations. Genetics 166: 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Burbano H. A., Krause J., Thines M., Weigel D., et al. , 2014. Mining herbaria for plant pathogen genomes: back to the future. PLoS Pathog. 10: e1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]