Abstract
Uniparental reproduction in diploids, via asexual reproduction or selfing, reduces the independence with which separate loci are transmitted across generations. This is expected to increase the extent to which a neutral marker is affected by selection elsewhere in the genome. Such effects have previously been quantified in coalescent models involving selfing. Here we examine the effects of background selection and balancing selection in diploids capable of both sexual and asexual reproduction (i.e., partial asexuality). We find that the effect of background selection on reducing coalescent time (and effective population size) can be orders of magnitude greater when rates of sex are low than when sex is common. This is because asexuality enhances the effects of background selection through both a recombination effect and a segregation effect. We show that there are several reasons that the strength of background selection differs between systems with partial asexuality and those with comparable levels of uniparental reproduction via selfing. Expectations for reductions in Ne via background selection have been verified using stochastic simulations. In contrast to background selection, balancing selection increases the coalescence time for a linked neutral site. With partial asexuality, the effect of balancing selection is somewhat dependent upon the mode of selection (e.g., heterozygote advantage vs. negative frequency-dependent selection) in a manner that does not apply to selfing. This is because the frequency of heterozygotes, which are required for recombination onto alternative genetic backgrounds, is more dependent on the pattern of selection with partial asexuality than with selfing.
Keywords: coalescence, asexual reproduction, self-fertilization, effective population size, background selection, balancing selection
IN addition to forming the basis for much of modern evolutionary inference, coalescence theory provides a rigorous approach to predicting how various biological phenomena will affect patterns of genomic variation. Although the coalescence of neutral alleles is simple in ideal populations, the process is altered by a myriad of biological realities such as population structure, changes in population size, and non-Poisson variance in reproductive success. Existing theory has tackled these and many other important deviations from the simplest case (Wakeley 2008). In addition to ecological factors such as those above, selection at one site in the genome can profoundly influence the coalescence times of linked neutral sites, thereby affecting patterns of genomic diversity. Although selective sweeps can dramatically reduce coalescence times (Maynard Smith and Haigh 1974; Kaplan et al. 1989), here we focus on two other forms of selection: background and balancing selection.
Background selection refers to selection on deleterious alleles occurring in the genetic background of a focal neutral allele and its resulting consequences for coalescence (reviewed in Charlesworth 2012). Assuming selection is strong relative to drift, deleterious alleles are doomed to extinction and linked neutral sites will share the same fate unless they can escape via recombination (Charlesworth et al. 1993). Looking backward in time, this means that extant copies of a neutral site must trace their ancestry, either avoiding deleterious backgrounds or recombining onto them only in the relatively brief period prior to their selective elimination. Thus, there are fewer potential ancestors for a neutral site than the actual number of individuals present in the preceding generations. In other words, background selection reduces the effective population size, shortening coalescence times. As this logic suggests, the consequences of background selection are much stronger if there is tight linkage between the focal neutral site and the locus experiencing deleterious mutations (Hudson and Kaplan 1995; Nordborg et al. 1996a). Because of heterogeneity across the genome in the local deleterious mutation rate, variation in the strength of background selection is believed to be a major source of the variation in sequence diversity across chromosomes in species such as Drosophila melanogaster (Charlesworth and Campos 2014; Elyashiv et al. 2014) and humans (McVicker et al. 2009).
Balancing selection preserves variation at a selected site, allowing two selected alleles to persist much longer than expected under drift alone. The same logic applies to two copies of a neutral site linked to alternative alleles at the selected site (Hudson and Kaplan 1988; Kaplan et al. 1988; Hey 1991). By increasing coalescence times, balancing selection can increase levels of neutral diversity at sites closely linked to targets of selection as seen for self-incompatibility alleles (Richman et al. 1996; Kamau and Charlesworth 2005) and major histocompatibility loci (Hughes and Nei 1988).
Most coalescent models implicitly assume that organisms are either haploid or diploid but obligately sexual and randomly mating. However, some of the most reproductively prolific diploids reproduce uniparentally much of the time via selfing or asexual reproduction (e.g., duckweeds, monogonant rotifers, ostracods, Daphnia, and many grasses and fungi). Asexuality and selfing involve very different types of inheritance and, consequently, their “direct” effects on coalescence times are in opposite directions; asexuality increases coalescence time whereas selfing reduces it (see below). Yet both forms of uniparental reproduction reduce the independence with which two alleles at separate loci within the same individual trace their ancestry backward in time. As a consequence of this effectively higher degree of linkage, uniparental inheritance is expected to magnify the importance of linked selection on coalescence. Indeed, previous work has demonstrated that the effects of both background and balancing selection on coalescence are heightened by high rates of selfing (Nordborg 1997).
Here we develop coalescent models for organisms capable of both sexual and asexual reproduction. Like selfing, asexuality enhances the consequences of selection elsewhere across the genome, sometimes dramatically so. Partial asexuality and selfing are often lumped together, at least colloquially, because both are systems involving uniparental inheritance with reduced rates of genetic mixing. However, these alternative modes of uniparental reproduction heighten linked selection in somewhat different ways and we highlight the differences, although we do not present any original results with respect to selfing.
Theory and Results
Direct effects
In the absence of any linked selection, the expected coalescence time for two randomly sampled alleles in a diploid sexually outcrossing species is E[T] = 2N (Wakeley 2008). For a species that produces a fraction σ of its offspring sexually and the remainder asexually, the expected coalescence time of two alleles sampled from different individuals is
| (1) |
as shown by Ceplitis (2003) (see also Bengtsson 2003 and Hartfield et al. 2015). It takes, on average, N generations for the two alleles to trace their ancestry into the same individual. In diploids, there is only a 1/2 chance that they will coalesce within that individual. This means that two alleles pass through the same individual, on average, two separate times before coalescing, thus giving rise to the 2N. After entering the same individual the first time (but not coalescing), the two alleles must be separated into different individuals via sex before they can “try” again. The waiting time to be split by sex is 1/σ.
As shown by Nordborg and Donnelly (1997), for a species that produces a fraction o of its offspring via outcrossing and the remainder by selfing, the expected coalescence time of two alleles sampled from different individuals is
| (2) |
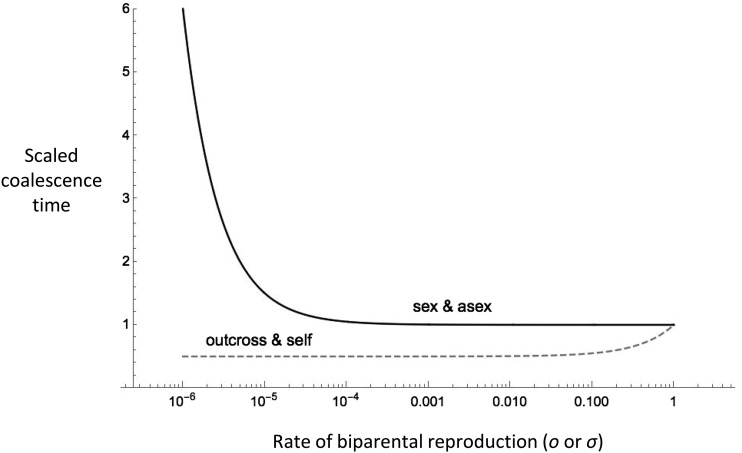
Contrasting (1) and (2), we see the direct effects of selfing and asexuality differ in several ways (Figure 1). Increasing the rate of uniparental reproduction (i.e., reducing o or σ) reduces coalescence time in the case of selfing but has the opposite effect with partial asexuality. With selfing, coalescence time decreases linearly with declining rates of outcrossing. In contrast, the effect of partial asexuality on coalescence time is negligible unless the rate of sex is very small [i.e., the direct effect of partial asexuality can be ignored unless σ ∼ O(1/N)]. Whereas very high selfing (o → 0) reduces the coalescent time by a maximum of 50%, the maximal increase in E[T] with very high asexuality (σ → 0) is unlimited in principle. However, if gene conversion is incorporated in the partial sex model, increases in coalescent time from asexuality are much more limited and can even be reversed (Hartfield et al. 2015).
Figure 1.
The direct effect of uniparental reproduction via asexuality or selfing on the scaled coalescence time (E[T]/2N). Partial asexuality has a negligible direct effect unless the rate of sex (σ) is on the order of the reciprocal of the population size; here N = 105. High selfing reduces Ne to 50% of N.
We refer to the effects described above as the “direct” effects of uniparental reproduction to distinguish them from the “indirect” effects of uniparental reproduction in mediating the consequences of linked selection.
Background Selection
The effects of background selection in sexual outcrossing species were first modeled in classic articles by Hudson and Kaplan (1994, 1995) and by Nordborg et al. (1996a), using different approaches. To examine background selection in partially asexual species, we follow an approach similar to that used by Nordborg (1997), who studied background and balancing selection in partial selfers. The main idea is that two sampled alleles can be found in a variety of different states (e.g., in the same or different individuals, in wild-type or mutant backgrounds). Based on transition probabilities, we can calculate the expected time to leave the current state and the probability with which the process will enter each of the other states, given that it has left its current state. If there are k states (other than coalescence), then we can create a system of k linear equations of the form
| (3) |
where vi,j is the probability of the process transitioning from state i to state j (moving backward in time). The system can be solved for the expected coalescent times from each state.
We make assumptions similar to those of previous models (Hudson and Kaplan 1994, 1995; Nordborg et al. 1996a; Nordborg 1997). We consider a single selected locus that can mutate from the wild-type A1 to the deleterious allele A2 at rate µ. The neutral locus is at a recombination distance r from the selected locus. Two neutral alleles could be in the same individual or two separate individuals. However, for each neutral allele we also need to consider its genetic background, both the selected allele present on the same chromosome and the selected allele on the homolog. We assume that A1/A2 individuals have a fitness of 1 – hs and that A2/A2 individuals have fitness 1 – s but are sufficiently rare that they can be ignored. The frequency of the deleterious allele is q = µ/hs. Because the allele frequency is assumed to be stable, this requires N is sufficiently large so that Nq >> 1. Technically, we assume 1/N ∼ O(ξ3), and hs, q ∼ O(ξ), where 0 < ξ << 1.
For fully sexual outcrossing species, diploid genotypes are transient so only haplotypes need to be considered. This “haplotype perspective” means that there are only three (noncoalesced) states in which two samples can exist: (i) both samples are found on A1 haplotypes, (ii) both samples are found on A2 haplotypes, and (iii) the two samples are on alternative haplotypes. With selfing there is the potential difficulty of needing to consider samples in the same or different individuals, potentially complicating the model with additional states. However, Nordborg (1997) used a separation of timescales argument to avoid this difficulty. With selfing, two samples from within the same individual either coalesce or transition into separate individuals at rates close to instantaneous with respect to coalescent times. Thus, it is possible to accommodate selfing into a system involving just three states (the haplotype perspective) by the appropriate adjustments of the transition probabilities.
In contrast, the separation of timescales assumption is not applicable with partial asexuality as two samples may remain noncoalesced in the same individual for an extended period if rates of sex are low. Consequently, the system becomes considerably more complicated as we must keep an explicitly diploid perspective. Assuming homozygous mutant (A2/A2) genotypes are sufficiently rare that they can be ignored, there are eight (noncoalesced) states that must be considered: (i) both samples in a single A1/ A1 individual; (ii) both samples in a single A1/A2 individual; (iii) both samples in separate A1/A1 individuals; (iv) one sample in an A1/A1 individual and the other on the A1-haplotype of an A1/A2 individual; (v) one sample in an A1/A1 individual and the other on the A2 haplotype of an A1/A2 individual; (vi) both samples on the A1 haplotype of separate A1/A2 individuals; (vii) the samples in separate A1/A2 individuals, with one on an A1 haplotype and the other on an A2 haplotype; and (viii) both samples on the A2 haplotype of separate A1/A2 individuals. These states are depicted schematically in Supporting Information, Figure S1. A complete model considering all the additional possibilities yields the same results (not shown). The transition probabilities (vi,j) among these states, allowing for both sex and asexual reproduction, are given in File S1.
If we assume that sex is common (σ ∼ O(1)) and linkage is tight (r ∼ O(ξ)), then the mean coalescence time is
| (4) |
The term in parentheses represents the reduction in the effective population size due to background selection. This result is equivalent to the classic result (Hudson and Kaplan 1994, 1995; Nordborg et al. 1996a) but with the effective rate of recombination σr replacing r in the previous models, which assumed obligate sex. It is worth noting that partial asexuality (σ < 1) has an indirect effect on coalescence via background selection [represented by the parenthetical term in (4)], even though its direct effect of asexuality [as represented in (1)] is negligible in magnitude and not shown in this approximation. In this sense, the indirect effects of asexuality are much more important than the direct effects [unless rates of sex are very low, i.e., σ ∼ O(1/N)]. The result above is very similar to the result of Roze (2014) for partially asexual haploids. When the rate of sex is high, the shared history of homologs within a diploid is very brief relative to the coalescence time so it is unsurprising that the diploid model behaves like a haploid one.
If we assume that sex is rare but not too rare (σ ∼ O(ξ), Nσ >> 1) and that linkage is loose (r ∼ O(1)), then
| (5) |
where
| (6) |
and . Under these assumptions, sex is still sufficiently common that the direct effect of asexual reproduction on coalescence time (via the effect highlighted in Equation 1) is negligible. However, through the factor Ψ in (5), the reduced frequency of sex enhances the effect of background selection beyond the effect shown in (4), i.e., reducing r to σr. We now turn our attention to biologically interpreting Ψ.
This additional effect of low sex is clearer if we redo the analysis, assuming tight linkage (r ∼ O(ξ)) and even lower sex (σ ∼ O(ξ2)); then we find
| (7) |
In this approximation, we also see the direct effect of sex on increasing the coalescent time, 1/σ [technically, (7) gives the coalescence time for two alleles sampled from separate individuals; see Ceplitis 2003; Bengtsson 2003; Hartfield et al. 2015]. The reduction in Ne due to background selection is captured by the factor (1 – 2q). This result is different from that of Nordborg et al. (1996a) who, based on their result similar to Equation 4, showed that with no recombination the background selection term is (1 – q). The difference arises because the Nordborg et al. (1996a) result assumes no recombination but obligate sex. Our result can be understood using the insight of Charlesworth et al. (1993) that, without genetic mixing, the effective population size is reduced to mutation-free individuals. The frequency of mutation-free haplotypes is 1 – q but the frequency of mutant-free diploid genotypes is 1 – 2q. We can take (7) to mean that, in diploids with little sex, a focal neutral site is affected by the presence of a deleterious allele on its homolog just as much as it would be by one on its own chromosome, thus accounting for the 2q in place of q. With somewhat higher rates of sex, the neutral site is more likely to be freed by segregation from its homolog and thus will be less affected by mutations there.
The extent to which a focal site will be affected by mutations on the homolog with partial asexuality is captured by Ψ. This can be seen most easily when r is small [and σ ∼ O(ξ)] so that
| (8) |
We can interpret PHI (as in [8]) as representing the chance that a neutral locus will escape a deleterious allele on the homolog via sex, relative to the chance it is eliminated by selection because represents the persistence time of a deleterious allele (1/hs) relative to the waiting time for sex (1/σ). From the equation above, it is easy to see that Ψ → 1 as φ → ∞ (i.e., hs << σ << 1) and Ψ → 2 as φ → 0 (i.e., σ << hs << 1). In other words, background selection will depend as much on mutations on the homolog as on the focal chromosome (Ψ → 2) if the neutral site is unlikely to escape a deleterious allele on the homolog via sex before that allele is eliminated by selection.
Returning to the full version of Ψ in Equation 6, we find the somewhat surprising result that the sensitivity of background selection to mutation on the homolog increases with recombination; i.e., ∂Ψ/∂r = 4φ(1 + 2rφ)/(1 + φ)2 > 0. This can be understood as follows. When r = 0, the focal neutral site is guaranteed to escape the effects of a deleterious allele on the homolog via segregation if sex occurs. However, as r increases, the focal site is more likely to recombine onto the deleterious homolog during sex rather than to escape it. In fact, for unlinked loci (r = 1/2), we have Ψ = 2 (regardless of φ), indicating the focal site is just as strongly affected by mutations on the homolog as on its own chromosome. Although the segregation effect captured by Ψ is maximized for unlinked loci, it is important to note that, overall, background selection is stronger with tighter linkage (Figure 2) for the reasons discussed in the original models on this topic (Hudson and Kaplan 1994, 1995; Nordborg et al. 1996a).
Figure 2.
The scaled coalescence time (E[T]/2N or, equivalently, Ne/N) as a function of the recombination distance between the focal neutral locus and the selected locus in the background selection model with partial asexuality. (A) The reduction in coalescence times with low rates of sex is particularly strong at higher rates of recombination; hs = 0.005. (B) The effects of background selection are larger with weak selection when recombination is low. Mutation rate: µ = 10−6.
In sum, asexual reproduction affects background selection in two ways: (i) via a “recombination effect” (reducing the effective rate of recombination from r to σr) and (ii) via a “segregation effect” (captured by Ψ, which mediates how much neutral alleles on one haplotype are affected by mutations on the homolog). As a result of these effects, background selection can be much stronger in facultative asexuals than in obligately sexual species; this is particularly notable at larger recombination distances (Figure 2). In the absence of deleterious mutations, partial asexuality substantially alters coalescence time only when sex is extremely rare; that is, the direct effect of asexuality is strong only when sexual events across the entire population are rare, i.e., Nσ ∼ O(1) or smaller. Through background selection, partially asexuality can affect coalescence even when sex is common at a population level (Nσ >> 1), especially if sex is rare per capita (σ << 1).
Extension to multiple loci
Following earlier models (Hudson and Kaplan 1995; Nordborg et al. 1996a), we can extend the single-locus background selection results to many loci under the assumption of no disequilibria and no epistasis. Let Bi be the background selection factor for a single locus (i.e., the term in parentheses in Equation 5). Across many loci, the total effect of background selection is . Using Equation 5, assuming that selected sites are densely packed so that the sum can be approximated as an integral, we can calculate
| (9) |
where the integration is over all positions x in the genome. R[x] is the recombination rate between the focal locus and the position x and Ψ′ is Ψ as given by (6) but replacing r with R[x]. Here q[x] is the probability density of deleterious alleles at site x, although we assume below that the density is constant across the genome q[x] = q.
Although (9) does not reduce to a simple analytical expression applicable to any genomic architecture, it can be evaluated numerically. Moreover, we can decompose the total background selection coefficient into components arising due to strongly linked, loosely linked, and unlinked genes: B = BTightlyLinked BLooselyLinked BUnlinked. Useful analytical approximations can be obtained for BTightlyLinked and BUnlinked.
Here, we use “tightly linked” to refer to sites that are within a distance mmax, measured in Morgans, of the focal site, where mmax is the maximum distance over which the map distance (m) is approximately linear with recombination rate. In other words, mmax is the maximum value of m where m ≈ r; for typical mapping functions, mmax < 0.05. We consider a region 2mmax wide with the focal site at its center as “the tightly linked region” and assume deleterious mutations fall uniformly across this region. When the rate of sex is high relative to selection (σ >> hs), the background selection coefficient from the tightly linked region is
| (10a) |
where UTightlyLinked is the diploid mutation rate over the tightly linked region. For the case of σ = 1, this result is the same as equation 8 of Hudson and Kaplan (1995). When the rate of sex is low and selection is weak (σ, hs ∼ O(ξ)), a much better approximation is
| (10b) |
where Ψr∼0 is given by (8) (see File S1).
Except in genomes with very small maps, most genes will be unlinked (r = 1/2) to the focal neutral site. Unlinked loci make a small contribution to the total effect of background selection in species where sex is high. With full sex (σ = 1) it has been shown by other means (Charlesworth 2012) that
| (11) |
where UUnlinked is the diploid rate of deleterious mutation over all sites unlinked to the neutral focal site. However, when sex is low (σ << 1), then the background selection coefficient due to unlinked sites based on (5) and (9) is
| (12) |
where UUnlinked is the diploid rate of deleterious mutation over all sites unlinked to the neutral focal site. It is clear from Equations 10–12 that the effects of background selection are much greater when sex is low, especially the contribution from unlinked loci.
To explore background selection further, we assume that the focal chromosome exists in the center of a 1 Morgan chromosome and that the total map length of the genome is L Morgans (where L ≥ 1). Deleterious mutations are uniformly distributed across the genome; i.e., µ = 1/2 U/L, where U is the diploid genome-wide mutation rate. This means that UTightlyLinked = 2Ummax/L and UUnlinked = U(L – 1)/L. The background selection coefficient due to loosely linked (mmax < r < 1/2) sites is
| (13) |
This integral can be expressed in closed form, using Haldane’s mapping function (R[x] = exp[1 – 2x]/2), but the expression is rather complicated and is left for File S1.
Figure 3 illustrates that the genome-wide effects of background selection can be substantial when σ < 1, even in genomes with long maps (such that gene density is low). If sex is low (σ < 1%), effective population size may be reduced by one or more orders of magnitude with relatively mild mutation rates (U = 0.1). In a moderate to large population (N > 104), there would be a negligible direct effect on coalesce time of asexual reproduction with σ = 10% or 1% (Bengtsson 2003; Ceplitis 2003; Hartfield et al. 2015), but the indirect effect through background selection can be large. The effect of background selection is more sensitive to the strength of selection in long maps than in short maps, with stronger selection resulting in larger effects of background selection, provided σ is not too low. With lower sex, a larger proportion of total background selection is attributable to unlinked loci (Figure 3B). This proportion is greater in larger genomes (as more loci will be unlinked) and when selection is strong, as strongly selected loci do not need to be closely linked to exert their effects (Nordborg et al. 1996a).
Figure 3.
Genome-wide background selection in systems with sexual and asexual reproduction. (A) The background selection coefficient B as a function of the rate of sex, considering mutations across the entire genome. Results for genomes with relatively short and long maps (L = 2 Morgans vs. 20 Morgans) are shown. The longer map implies lower gene density as the mutation rate is held constant (U = 0.1); mutations are assumed to be uniformly distributed across the genome. (B) The background selection coefficient for different genomic regions (tightly linked, loosely linked, and unlinked sites) as well as the total. Parameters are L = 10, hs = 0.005, and U = 0.1. Tightly linked loci make the largest contribution to background selection when rates of sex are very high but unlinked loci make the largest contribution when sex is rare.
Simulation results
The extrapolation to multiple loci used in (9) and in previous work (Hudson and Kaplan 1995; Nordborg et al. 1996a; Glémin and Ronfort 2012) relies on the assumption that loci are independent (i.e., no linkage disequilibrium) and that each selected locus is close to its deterministic equilibrium (q = µ/hs). These assumptions become increasingly suspect as the effective rate of recombination declines and as background selection becomes sufficiently strong that Ne (≈ BN) is substantially reduced. When BNhs is predicted to be low (<1), then we expect that analytical approximations will overestimate the strength of background selection. Under these conditions the analytical approximation is expected to overestimate the reduction in Ne due to background selection, presumably because when drift is strong, then polymorphism is lost from selected sites; i.e., q = 0 < µ/sh, making background selection weaker than predicted. Previous simulation studies have observed that analytical approximations tend to overestimate the strength of background selection when selection is weak or recombination is low (Nordborg et al. 1996a; Kaiser and Charlesworth 2009; Kamran-Disfani and Agrawal 2014).
To test the analytical approximations presented here, we extended the simulation of Kamran-Disfani and Agrawal (2014) to consider a population where individuals can reproduce asexually with a probability σ, via Wright–Fisher dynamics. We measured Ne by tracking a linked quantitative trait over time; when an individual reproduced, the value of this trait was perturbed by a value drawn from a normal distribution with mean 0 and variance 1. The steady-state variance of this trait is equal to the effective population size (Lynch and Hill 1986; Keightley and Otto 2006).
For a given net mutation rate U and heterozygote selection coefficient sh, mutations were initially distributed among the population according to mutation–selection balance. Unless stated otherwise, the population then reproduced for 5N generations, to ensure that neutral markers reached a steady state (Keightley and Otto 2006). The population-wide Ne was then measured at 200 equally spaced intervals for a further 5N generations. The simulation was repeated for 192 burn-ins. The mean Ne from each time series was determined, and simulation points are calculated as the means of these values along with 95% confidence intervals. Simulations are written in C++ and are available online (http://github.com/MattHartfield/FacSexBGSSims).
In general there is a good match between simulation results and analytical approximations (Figure 4A). The effects of background selection tend to be overestimated if sex is low (σ ∼ 0.001), where background selection is predicted to be very strong. Because the analytical approximation for B is expected to fail as BNhs becomes small, we examined the correspondence between simulations and the predicted value of B for different population sizes. As expected, the observed value of B is much larger than the analytical approximation [i.e., the strength of background selection is overestimated by (9)] when N is small but the discrepancy declines with larger N (Figure 4, B and C). This implies that the analytical approximations work well provided N is “sufficiently” large. Although imperfect, the approximations offer good insight into the effects of background selection. Nonetheless, it is important to remember that the extremely strong effects of background selection predicted under low rates of sex (e.g., B < 10−5) are unlikely to be realized unless applied to species with a massive census size. Although the most obvious problem is that the analytical approximation overestimates background selection when N is not sufficiently large, we also find instances where the analytical approximation underestimates the strength of background selection when N is large (Figure 4B). This could suggest a weak effect arising from multilocus associations not captured in the extrapolation of the single-locus analysis to genome-wide background selection given by (9). This effect does not appear to be sensitive to h (not shown) and so is unlikely to be a direct result of segregation load.
Figure 4.
Simulation comparisons of background selection. (A) Simulation results (symbols) and analytical approximations (lines) of B = Ne/N as a function of the rate of sex. Parameters are N = 104, U = 0.02, s = 0.02, h = 0.25, and L = 1 (solid symbols and line) or L = 10 (shaded symbols and dashed line). Bars on simulation symbols represent 95% confidence intervals. (B) Background selection effect for σ = 0.01 as a function of the population size. Parameters are U = 0.02, s = 0.02, h = 0.25, and L = 10. The horizontal shaded line is the theoretical expectation given with Equation 9. (C) Same as B but for σ = 0.001.
Comparison with selfing
Another mode of uniparental reproduction, selfing, also enhances the effect of background selection. As shown by Nordborg (1997), the coalescence time with selfing is
| (14) |
Here F is the inbreeding coefficient that can be written as function of the rate of outcrossing F = (1 – o)/(1 + o). The fraction 1/(1+F) is the direct effect of selfing on reducing Ne [shown in (2) as (1 + o)/2] and is not considered further. The term in parentheses in (14) is the background selection coefficient. With pure outcrossing, the background selection coefficient is the same as the parenthetical term from (4) with σ = 1. Relative to this, selfing changes background selection in two ways. First, it increases the average strength of selection against a deleterious mutation from sh to , where s is selection against the mutation in the homozygous state. This also changes the equilibrium frequency of the deleterious allele to . More importantly, selfing reduces the effective rate of recombination from r to . When selfing rates are high (o << 1), the background selection coefficient simplifies to
| (15) |
There are three important differences between this result and the equivalent term for background selection in partial asexuals (Equation 5). First, the segregation effect captured by Ψ effectively doubles the relevant mutation rate for partial asexuals relative to selfers. Second, a bit of outcrossing in a highly selfing system results in effectively twice as much recombination as the equivalent amount of sex in a highly asexual system [i.e., σr in (5) vs. 2or in (15)]. In sex/asex systems, recombination can occur only with sexual reproduction so the effective recombination rate depends directly on σ. In outcross/self systems, recombination can happen during the production of selfed or outcrossed progeny. However, the only parental genotypes in which recombination is relevant are double heterozygotes, which are created at a rate proportional to the rate of outcrossing, o. However, once a double heterozygote is created via outcrossing, a relevant recombination event can occur in any future generation via selfing as long as the genotype remains a double heterozygote. Because it takes, on average, two generations of selfing before heterozygosity at a site is lost, there are two generations in which meaningful recombination can occur following each outcrossing event [so we have 2or in (15) but σr in (5)]. However, Denis Roze (D. Roze, personal communication) has pointed out that (14) is derived from (4), which assumes tight linkage (r << 1). He has found that the “2” is reduced for loosely linked loci, becoming “1” for unlinked loci. That is, the recombinational difference between selfing and partial asexuals will disappear for unlinked loci.
In addition to these segregation and recombination differences, a third difference arises because the effective strength of selection is greater with selfing than with asexual reproduction (s vs. hs). Stronger selection reduces the frequency of deleterious alleles. As there are fewer deleterious backgrounds to avoid in tracing back a focal neutral site’s ancestry, background selection is made weaker. However, stronger selection also means there is less opportunity to recombine away from a deleterious allele before it is removed by selection, enhancing background selection. Overall, stronger selection reduces background selection if the rate of biparental reproduction (and the opportunity for recombinational escape) is high relative to selection.
As a consequence of the three differences between (15) and (5), background selection can be much stronger in partial sexuals than in selfers when the rate of biparental reproduction is low (Figure 5). However, the reverse may be possible if the net effect of stronger selection in selfers is to increase background selection, which can happen when the rate of biparental reproduction is high relative to selection.
Figure 5.
Background selection in systems with different forms of uniparental reproduction (asexuality and selfing). The background selection coefficient B is shown for systems with sexual and asexual reproduction (solid symbols and line) and systems with outcrossing and selfing (shaded symbols and lines). Analytical approximations (lines) are contrasted with simulation results (symbols; error bars are 95% confidence intervals. The analytical approximation for selfing is based on an extension of (14) and is given in Kamran-Disfani and Agrawal (2014). Parameters: U = 0.02, s =0.02, h = 0.25, L = 10 (N = 104 for simulations).
Balancing Selection
With balancing selection, such as heterozygote advantage or negative frequency dependence, two alleles (A1 and A2) can be maintained indefinitely; i.e., balancing selection prevents normal coalescent processes from operating at the selected site and this affects coalescence at linked sites. Assuming the frequencies of the A1 and A2 alleles (p and q, respectively) are stable, Hudson and Kaplan (1988) showed that closely linked neutral sites have an extended coalescence time under the assumption of obligate sexual reproduction. As mentioned in the Background Selection section, the analysis becomes more complicated when the rate of sex is low because we must use an explicitly diploid perspective rather than a classic haploid perspective. Here we cannot assume that the frequency of the A2/A2 genotype is negligible (Figure S2). As a result, there are 13 states in which we could find two alleles. In addition to the 8 listed in the Background Selection section, the additional 5 states are (ix) both samples in a single A2/A2 individual, (x) both samples in separate A2/A2 individuals, (xi) one sample in an A2/A2 individual and the other on the A1 haplotype of an A1/A2 individual, (xii) one sample in an A2/A2 individual and the other on the A2 haplotype of an A1/A2 individual, and (xiii) one sample in an A1/A1 individual and the other sample in an A2/A2 individual.
In the Background Selection section we ignored gene conversion for simplicity and because the persistence time of any given copy of the deleterious allele is short, limiting the opportunity for gene conversion to be relevant. Here we include gene conversion because A1 and A2 are maintained indefinitely by selection so movement between genetic backgrounds via gene conversion may become important over long timescales, especially when the rate of sex is low, so there is little chance for exchange via meiotic recombination. Specifically, we assume that (unbiased) mitotic gene conversion occurs at rate γ. This rate refers to gene conversion events that cover either the selected site or the neutral site but not both. (The model is built with an additional parameter for gene conversion events that cover both sites but such events do not affect the coalescence time). Throughout, we use “recombination” to refer to (meiotic) crossover recombination, in contrast to gene conversion.
In this analysis, we assume that the equilibrium frequencies of A1/A1, A1/A2, and A2/A2 genotypes are P1/1, P1/2, and P2/2. These frequencies are assumed to be stable and to change little in frequency within a generation (i.e., fitness differences at equilibrium are ignored). Without loss of generality, we use P1/1 = p2 + CA/A, P1/2 = 2(pq – CA/A), and P2/2 = q2 + CA/A, where CA/A is the covariance in allelic state. Note CA/A measures the same genetic property as Fpq but we use separate symbols as we use the relationship F = (1 – o)/(1 + o) for species without asexuality because the inbreeding coefficient is determined only by outcrossing rates, to a good approximation, provided selection is not too strong. With obligate sex (and no selfing) CA/A = 0 but when there is partial asexuality, CA/A can be either positive or negative.
Assuming σ and r are O(ξ) and γ and 1/N are O(ξ2), the expected coalescent time of two alleles sampled from a single A1/A1 individual or from two separate A1/A1 individuals is found to be
| (16) |
When both alleles are sampled from the A2 background, the coalescence time is the same as (16) but with p and q reversed. When two alleles are sampled from alternate backgrounds (A1 and A2), the expected coalescence time
| (17) |
For the sake of discussion, we focus on this latter result. In the case of obligate sex (σ = 1, P1/2 = 2pq), (17) simplifies to E[T1/2] = 2N + 1/(r + γ). The coalescent time is extended by the waiting time for the neutral site to move from one background to the other. This longer coalescent time should allow for the accumulation of more neutral variation surrounding sites under balancing selection (Hudson and Kaplan 1988). When the physical distance is small, r may be very small and this has led to the belief that there may be a strong signature of balancing selection when gene conversion is ignored. However, for short physical distances, gene conversion may be much larger than recombination (γ >> r) and may play a more important role in coalescence (Andolfatto and Nordborg 1998; Wiuf and Hein 2000). There are few estimates of mitotic gene conversion (per base pair): ∼10−7 in the yeast Saccharomyces cerevisiae (reviewed by Mandegar and Otto 2007), 10−5–10−6 in the bdelloid rotifer Adineta vaga (Flot et al. 2013), and ∼10−6 in the fly D. melanogaster (N. P. Sharp and A. F. Agrawal, unpublished results). As implied by (17), gene conversion weakens the signature of balancing selection and this is particularly true in partial asexuals (Figure 6). At very short physical distances, mutation between A1 and A2 provides an important alternate route by which a neutral allele can switch sites, but this is not considered here. Our results apply to physical distances where σr + γ/2 >> µ.
Figure 6.
Coalescence times near a site under balancing selection. The expected time to coalescence (scaled to 2N generations) is shown as a function of the distance (in base pairs) from a site under balancing selection via heterozygote advantage (W1/1 = W2/2 = 1 – s and W1/2 = 1). The population is assumed to be fully sexual and outcrossing (Equation 17, σ = 1; thick shaded line), mostly asexual (Equation 17, σ = 0.01; solid line), or mostly selfing (Equation 19b, o = 0.01; thin shaded line). Symbols represent the expected coalescence time based on simulations (see File S3). Error bars are 95% confidence intervals; in some cases error bars are too small to be visible. We assume r = ρd, where d is the distance in base pairs from the selected site. For the partial asexual case, results are shown with and without gene conversion. For gene conversion, the model of Andolfatto and Nordborg (1998) is used: γ = gd/L for d < L and γ = g for d ≥ L, where L is the length of gene conversion tracts. For the fully sexual case, the results with gene conversion are visually indistinguishable from those with γ = 0 for the parameter values used here. The selfing case assumes no gene conversion, but gene conversion is not expected to affect coalescence in selfers (see Discussion); simulations including gene conversion support this conjecture (File S3). The selfing line asymptotes at 1/2 rather than 1 because of the direct effect of selfing on coalescence (Figure 1). These results ignore mutation at the selected site and thus overestimate the coalescence time for extremely tightly linked sites (e.g., d << 100 if µ = 10−9, see text). Parameter values: s = 0.1, N = 5 × 104, ρ = 2 × 10−8 (Ashburner 1989), g = 2 × 10−6 (N. P. Sharp and A. F. Agrawal, unpublished results), and L = 1400 (Preston and Engels 1996).
As expected, we find in (17) that partial asexuality reduces the effective rate of recombination from r to σr. However, there is an additional effect of asexuality that is not explicit in (17); partial asexuality changes the equilibrium frequency of A1/A2 (i.e., P1/2 ≠ 2pq). The precise nature of this change depends on the mechanism of balancing selection. Consider balancing selection due to heterozygote advantage with symmetrical selection (i.e., W1/2 = 1, W1/1 = W2/2 = 1 – s). With obligate sex, at equilibrium p = q = 1/2 and P1/2 = 1/2. With partial asexuality and assuming the rate of sex is low, p = q = 1/2 but P1/2 = 2(pq – CA/A), where
| (18) |
For example, with s = σ = 0.01, P1/2 ≈ 0.71. In this case, the frequency of heterozygotes (and thus the opportunity for recombination) is increased from 0.5 with full sex to 0.7 with partial sex. Even though P1/2 is higher, this does not cause an increase in the coalescence time because of the 99% reduction in meiotic recombination events when σ = 0.01. Equilibrium genotype frequencies under a more general model of balancing selection are provided in File S2.
Nordborg (1997) studied balancing selection in species with selfing. He found (ignoring gene conversion) that the expected coalescence time for sites on alternate backgrounds (A1 and A2) is
| (19a) |
Assuming that the rate of selfing is high (i.e., weak outcrossing, o << 1), this becomes
| (19b) |
Again, the term in parentheses is the direct effect of selfing. The second term is the increase in coalescent time due to linkage to the target of balancing selection. Contrasting this to the comparable term from the partial asexual result in (17), there are two differences. As noted in the background selection case, a little bit of outcrossing is twice as effective at allowing recombination as a little bit of sex (i.e., the effective rates of recombination are 2or vs. σr). Second, in partial asexuals the frequency of heterozygotes cannot be well approximated from the rate of biparental reproduction and allele frequencies alone (as it is with selfing). Rather CA/A, which determines the frequency of heterozygotes through P1/2, is a function of both the rate of sex and the selection coefficients [as exemplified in (18)].
An important point missing from the comparison above is the likelihood of balancing selection. Consider the model of heterozygote advantage with W1/2 = 1, W1/1 = 1 – s, and W2/2 = 1 – αs, where we assume α > 1 so that W1/1 > W2/2. Ignoring drift, polymorphism is always maintained in sex/asex systems. In contrast, with selfing, the maintenance of polymorphism requires that outcrossing not be too low, specifically that o > (α – 1)/(α + 1); otherwise fixation of the more fit homozygote A1/A1 is stable (see File S2 for details). If one homozygote is more fit than another, variation will not be maintained by balancing selection in highly selfing species but will be in species with partial asexuality.
Even in the symmetric case (α = 1) when polymorphism should be maintained in a deterministic model, we observed that the expected coalescent time from simulations was less than that predicted by the analytical approximations (Figure 6). This is likely due to fluctuations in the allele frequency at the selected locus around its predicted equilibrium due to drift (which make heterozygotes rarer, on average). In Figure 6, we show the results for N = 5 × 104 and the simulation results are only slightly less than the predicted values. With N = 104, allele frequency fluctuations are larger and the simulation results are considerably lower than the predicted values (not shown). This change in N has little effect on results for partial asexuality and complete biparental reproduction as allele frequency of the selected locus is more stably maintained close to its equilibrium.
Of course, balancing selection can occur through mechanisms other than heterozygote advantage and in these other cases the contrast between partial asexuality and sex will be somewhat different. For example, consider a simple model of frequency-dependent selection where the fitness of each genotype is negatively related to its frequency as W1/1 = 1 – (p – α)s, W2/2 = 1 + (p – α)s, and W1/2 = 1. Regardless of the reproductive system, at equilibrium p = α and the polymorphism is stable (i.e., in contrast to heterozygote advantage, there is no difference in the likelihood of balancing selection in this model between partial asexuality and selfing systems; see File S2). However, with partial asexuality, there is no excess of heterozygotes (), meaning that coalescence is affected only by the recombination effect (i.e., the reduction from r to σr) and there is no segregation effect. With other forms of negative frequency-dependent selection (File S2), it is possible for there to be a deficit of heterozygotes with partial asexuality (CA/A > 0), decreasing the opportunity for recombination beyond the change from r to σr, thereby further extending coalescence time.
Discussion
We have examined how linked selection, either background or balancing selection, alters coalescence for pairs of alleles in partial asexuals. As expected, the lower level of genetic mixing in partial asexuals results in much stronger effects of linked selection. In diploids, partial asexuality affects coalescence both via segregation and via recombination. The most obvious effect is through a reduction in the effective rate of recombination. In the case of background selection, the segregation effect can be understood as mediating how much a focal site is affected by deleterious alleles occurring on the homolog in addition to those on its own chromosome. If sex is low, it is equally affected by mutations on either chromosome, effectively doubling the mutation rate. In the case of balancing selection, the segregation effect is manifest in the way that partial asexuality affects the equilibrium frequency of heterozygotes at the selected locus, which regulates the opportunity for a neutral site to move between alternative selected haplotypes via recombination.
Ignoring linked selection, the direct effect of partial asexuality on coalescence is negligible unless the rate of sex is extremely small [i.e., on the order of 1/N (Bengtsson 2003; Ceplitis 2003; Hartfield et al. 2015)]. In contrast, there are strong effects of partial asexuality through background selection even when rates of sex are >>1/N. If sex is <10%, large reductions in Ne through background selection are expected if U is ≥0.1; mutation rates of this magnitude are common among plants and animals (reviewed in Baer et al. 2007 and Halligan and Keightley 2009).
When sex is high, only those mutations that are tightly physically linked make a substantial contribution to background selection. Across the genome of a highly sexual species, variation in gene density resulting in variation in the local deleterious mutation rate leads to variation in the strength of background selection, which contributes to variation in neutral diversity (Charlesworth and Campos 2014; Elyashiv et al. 2014). These local differences may be detectable at the within-gene level [i.e., stronger background selection at the center of genes than on the edges (Loewe and Charlesworth 2007; Zeng and Charlesworth 2011)]. When sex is low, deleterious alleles across the whole genome contribute to background selection. In fact, the majority of background selection experienced by a focal site is due to unlinked loci (Figure 3B) but this is simply because unlinked sites are much more numerous. Even when sex is low, background selection is stronger from tightly linked sites than from unlinked ones (Figure 2B) so one might still expect to find differences in neutral diversity associated with gene density. However, the relationship between background selection and r is much weaker when sex is low, so gene density–diversity patterns are likely to be severely muted.
Because background selection can be so strong with uniparental inheritance, it may be more important than demographic history in determining within-population diversity. As has recently been discussed with respect to selfers (Brandvain et al. 2013; Barrett et al. 2014), transitions to uniparental inheritance can be associated with population bottlenecks but it can be difficult to infer whether reduced diversity in an evolutionarily young population is due to a small number of founders or the strong effects of background selection. More sophisticated approaches, beyond looking for a reduction in diversity, are required to disentangle bottlenecks from very strong background selection (e.g., Brandvain et al. 2013).
The analytical approximations not only quantify how much coalescence times are altered by the reproductive system, but also show they are biologically interpretable. Hence these derivations provide insight into how segregation, recombination, and selection interact to affect average coalescence time. However, the approach used here cannot be used to make other, more detailed, predictions about n-sample genealogies. For example, although the major effect of background selection can be interpreted as a reduction in Ne (as done here), it has been shown that background selection also alters the shape of genealogies with relatively long external branches, leading to negative values of Tajima’s D (Charlesworth et al. 1995; Zeng and Charlesworth 2011; Nicolaisen and Desai 2012). Presumably, these effects would be magnified by low rates of sex.
The reduction in the effective rate of recombination with uniparental inheritance should make it much easier to detect balancing selection. Work by Nordborg et al. (1996b) and Nordborg (1997) showed that, in selfers, this is because (i) the physical distance over which the effects of balancing selection are realized is expanded and (ii) there is lower diversity within each selected allelic class because of stronger background selection. These arguments also apply to partial asexuals but there are differences between these alternative modes of uniparental reproduction. First, there is a higher effective rate of recombination with selfing than with partial asexuality for the same rate of uniparental reproduction (2or vs. σr, as discussed in Results), which will make signatures of balancing selection weaker in selfers than in partial asexuality (Figure 6). There are also differences in the frequency of heterozygotes in partial asexuals compared to selfers but this likely plays a more minor role, conditional on the selected polymorphism being maintained. However, with heterozygote advantage, the conditions for a stable polymorphism are much broader in partial asexuals than in selfers.
Another difference between partial asexuals and selfers is the importance of gene conversion. The frequency of heterozygotes puts an upper limit on the rate of exchange between allelic backgrounds via any genetic mechanism. Selfing directly reduces the frequency of heterozygotes, so gene conversion is not expected to be an important process (although it was not formally included in the model of Nordborg 1997). In partial asexuals, heterozygotes can be very common even if sex is rare (especially if there is heterozygote advantage). Through asexual descent, a lineage can persist in a heterozygous state for many generations, providing numerous opportunities for movement of linked sites between backgrounds via gene conversion. As implied by (17), gene conversion becomes the primary force erasing the signature of balancing selection when σr << γ, which must be true if sex is sufficiently rare. Because of this, the signature of balancing selection may be weaker in partial asexuals than in selfers if the rate of biparental reproduction is very small (Figure 6), even though the reverse is expected at less extreme levels of uniparental reproduction because of the difference in effective rates of recombination (2or vs. σr).
For both background and balancing selection, we have contrasted species with partial asexuality to highly selfing species as alternative modes of uniparental reproduction. However, some systems with partial asexuality may also have selfing. This can occur because asexual reproduction can involve very limited dispersal so that neighboring individuals are often clones; thus, mating between clones, which is genetically equivalent to selfing, could be common unless there are explicit mechanisms of inbreeding avoidance (e.g., self-incompatibility). If sex is rare and when it does occur involves selfing (or its genetic equivalent), then the effective rate of recombination will be extremely low, magnifying the effects of linked selection beyond what we have modeled here.
Here we explored how background selection and balancing selection affect coalescence times but we did not attempt to examine selective sweeps. In sexual populations, beneficial alleles spreading to fixation can drastically reduce coalescence times and diversity levels at closely linked sites (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Stephan et al. 1992; Barton 1998; Hermisson and Pennings 2005) and are believed to play an important role in shaping genomic patterns of diversity, at least in some species (Elyashiv et al. 2014; Williamson et al. 2014). As with other forms of linked selection, we expect that the signature of selective sweeps will be more dramatic in systems with high rates of uniparental reproduction because of their reduced effective rates of recombination. For example, in the highly selfing Caenorhabditis elegans, sweeps are believed to have reduced variation across entire chromosomes (Andersen et al. 2012). Theoretical models for haploids have shown how low rates of sex affect the probability of sweeps and alter patterns of coalescence (reviewed by Neher 2013). Sweeps in haploids cause rapid coalescence of multiple lineages, distorting genealogies. The region of the genome influenced by a sweep is governed by the degree of linkage, thus depending on the rate of sex and the recombination distance. As with background and balancing selection, we speculate a recombination effect will not be the whole story in diploid partial asexuals. With low rates of sexes, a segregation effect is likely because, under asexuality, the selected diploid genotypes are inherited intact (rather than their haploid gametes), altering the trajectory of the selected allele from that in the standard sexual (or haploid) model, which will have implications for surrounding diversity. The full consequences of partial asexuality for diploids with respect to selective sweeps remain a challenge for future work.
Although the consequences of sweeps are likely to be larger in systems with uniparental inheritance, sweeps are less likely to occur in such systems. Both theory (Glémin and Ronfort 2012; Kamran-Disfani and Agrawal 2014) and data (Hough et al. 2013; Burgarella et al. 2015) show that reductions in Ne due to background selection render selection less efficient, reducing the probability of fixation of beneficials, in selfers compared to outcrossers. A qualitatively similar effect is expected in partial asexuals.
A number of interesting and important diploid species reproduce predominantly asexually but these have been studied less by evolutionary geneticists than selfing species. Although there are similarities between partial asexuality and selfing, it is misleading to consider these two forms of uniparental inheritance as conceptually interchangeable. As population genomic data become easier to obtain, the hope is that such data will allow us to study the evolutionary consequences of different forms of uniparental reproduction and, ideally, gain insight into the evolutionary forces acting on reproductive mode. Understanding the coalescent properties of systems with uniparental inheritance is key to interpreting population genomic data and much work remains, especially with respect to partial asexuality.
Acknowledgments
We thank Stephen I. Wright for extensive discussion and Denis Roze for sharing his unpublished result on background selection in selfers. This work was supported by the Natural Sciences and Research Council of Canada (A.F.A.) and by a Marie Curie International Outgoing Fellowship, grant MC-IOF-622936 project SEXSEL (to M.H.).
Footnotes
Communicating editor: J. Hermisson
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.181024/-/DC1.
Literature Cited
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R., et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Nordborg M., 1998. The effect of gene conversion on intralocus associations. Genetics 148: 1379–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., 1989 Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Baer C. B., Miyamoto M. M., Denver D. R., 2007. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 8: 619–631. [DOI] [PubMed] [Google Scholar]
- Barrett S. C. H., Arunkumar R., Wright S. I., 2014. The demography and population genomics of evolutionary transitions to self-fertilization in plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133. [Google Scholar]
- Bengtsson B., 2003. Genetic variation in organisms with sexual and asexual reproduction. J. Evol. Biol. 16: 189–199. [DOI] [PubMed] [Google Scholar]
- Brandvain Y., Slotte T., Hazzouri K. M., Wright S. I., Coop G., 2013. Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 9: e1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgarella C., Gayral P., Ballenghien M., Bernard A., David P., et al. , 2015. Molecular evolution of freshwater snails with contrasting mating systems. Mol. Biol. Evol. 32: 2403–2416. [DOI] [PubMed] [Google Scholar]
- Ceplitis A., 2003. Coalescence times and the Meselson effect in asexual eukaryotes. Genet. Res. 82: 183–190. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2012. The effects of deleterious mutations on evolution at linked sites. Genetics 190: 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Campos J. L., 2014. The relations between recombination rate and patterns of molecular variation and evolution in Drosophila. Annu. Rev. Genet. 48: 383–403. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Morgan M. T., 1995. The pattern of neutral molecular variation under the background selection model. Genetics 141: 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyashiv, E., S. Sattath, T. T. Hu, A. Strustovsky, G. McVicker et al., 2014 A genomic map of the effects of linked selection in Drosophila. arXiv: 1408.5461v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flot J.-F., Hespeels B., Li X., Noel B., Arkhipova I., et al. , 2013. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500: 453–457. [DOI] [PubMed] [Google Scholar]
- Glémin S., Ronfort J., 2012. Adaptation and maladaptation in selfing and outcrossing species: new mutations vs. standing variation. Evolution 67: 225–240. [DOI] [PubMed] [Google Scholar]
- Halligan D. L., Keightley P. D., 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40: 151–172. [Google Scholar]
- Hartfield, M., S. I. Wright, and A. F. Agrawal, 2015 Coalescent times and patterns of genetic diversity in species with facultative sex: effects of gene conversion, population structure and heterogeneity. Genetics 297–312. [DOI] [PMC free article] [PubMed]
- Hermisson J., Pennings P., 2005. Soft sweeps molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J., 1991. A multi-dimensional coalescent process applied to multi-allelic selection models and migration models. Theor. Popul. Biol. 39: 30–48. [DOI] [PubMed] [Google Scholar]
- Hough J., Williamson R. J., Wright S. I., 2013. Patterns of selection in plant genomes. Annu. Rev. Ecol. Evol. Syst. 44: 31–49. [Google Scholar]
- Hudson R. R., Kaplan N. L., 1988. The coalescent process in models with selection and recombination. Genetics 120: 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1994. Gene trees with background selection, pp. 140–153 in Non-Neutral Evolution, edited by Golding G. B. Chapman & Hall, New York. [Google Scholar]
- Hudson R. R., Kaplan N. L., 1995. The coalescent process and background selection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 349: 19–23. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Nei M., 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335: 167–170. [DOI] [PubMed] [Google Scholar]
- Kaiser V. B., Charlesworth B., 2009. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 25: 9–12. [DOI] [PubMed] [Google Scholar]
- Kamau E., Charlesworth D., 2005. Balancing selection and low recombination affect diversity near the self-incompatibility loci of the plant Arabidopsis lyrata. Curr. Biol. 15: 1773–1778. [DOI] [PubMed] [Google Scholar]
- Kamran-Disfani A., Agrawal A. F., 2014. Selfing, adaptation and background selection in finite populations. J. Evol. Biol. 27: 1360–1371. [DOI] [PubMed] [Google Scholar]
- Kaplan N. L., Darden T., Hudson R. R., 1988. The coalescent process in models with selection. Genetics 120: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Otto S. P., 2006. Interference among deleterious mutations favours sex and recombination in finite populations. Nature 443: 89–92. [DOI] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., 2007. Background selection in single genes may explain patterns of codon bias. Genetics 175: 1381–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Hill W. G., 1986. Phenotypic evolution by neutral mutation. Evolution 40: 915. [DOI] [PubMed] [Google Scholar]
- Mandegar M. A., Otto S. P., 2007. Mitotic recombination counteracts the benefits of genetic segregation. Proc. Biol. Sci. 274: 1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- McVicker, G., D. Gordon, C. Davis, and P. Green, 2009 Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5: e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R. A., 2013. Genetic draft, selective interference, and population genetics of rapid adaptation. Annu. Rev. Ecol. Evol. Syst. 44: 195–215. [Google Scholar]
- Nicolaisen L. E., Desai M. M., 2012. Distortions in genealogies due to purifying selection. Mol. Biol. Evol. 29: 3589–3600. [DOI] [PubMed] [Google Scholar]
- Nordborg M., 1997. Structured coalescent processes on different time scales. Genetics 146: 1501–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Donnelly P., 1997. The coalescent process with selfing. Genetics 146: 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Charlesworth B., Charlesworth D., 1996a The effect of recombination on background selection. Genet. Res. 67: 159–174. [DOI] [PubMed] [Google Scholar]
- Nordborg M., Charlesworth B., Charlesworth D., 1996b Increased levels of polymorphism surrounding selectively maintained sites in highly selfing species. Proc. Biol. Sci. 263: 1033–1039. [Google Scholar]
- Preston C. R., Engels W. R., 1996. P-element-induced male recombination and gene conversion in Drosophila. Genetics 144: 1611–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman A. D., Uyenoyama M. K., Kohn J. R., 1996. Allelic diversity and gene genealogy at the self-incompatibility locus in the solanaceae. Science 273: 1212–1216. [DOI] [PubMed] [Google Scholar]
- Roze D., 2014. Selection for sex in finite populations. J. Evol. Biol. 27: 1304–1322. [DOI] [PubMed] [Google Scholar]
- Stephan W., Wiehe T. H. E., Lenz M. W., 1992. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theor. Popul. Biol. 41: 237–254. [Google Scholar]
- Wakeley J., 2008. Coalescent Theory: An Introduction. Roberts & Company, Greenwood Village, CO. [Google Scholar]
- Williamson R. J., Josephs E. B., Platts A. E., Hazzouri K. M., Haudry A., et al. , 2014. Evidence for widespread positive and negative selection in coding and conserved noncoding regions of Capsella grandiflora. PLoS Genet. 10: e1004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf C., Hein J., 2000. The coalescent with gene conversion. Genetics 155: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., Charlesworth B., 2011. The joint effects of background selection and genetic recombination on local gene genealogies. Genetics 189: 251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]