Abstract
DNA double-strand breaks (DSBs) represent one of the most threatening lesions to the integrity of genomes. In yeast Saccharomyces cerevisiae, NuA4, a histone acetylation complex, is recruited to DSBs, wherein it acetylates histones H2A and H4, presumably relaxing the chromatin and allowing access to repair proteins. Two subunits of NuA4, Yng2 and Eaf3, can interact in vitro with methylated H3K4 and H3K36 via their plant homeodomain (PHD) and chromodomain. However, the roles of the two domains and how they interact in a combinatorial fashion are still poorly characterized. In this study, we generated mutations in the PHD and chromodomain that disrupt their interaction with methylated H3K4 and H3K36. We demonstrate that the combined mutations in both the PHD and chromodomain impair the NuA4 recruitment, reduce H4K12 acetylation at the DSB site, and confer sensitivity to bleomycin that induces DSBs. In addition, the double mutant cells are defective in DSB repair as judged by Southern blot and exhibit prolonged activation of phospho-S129 of H2A. Cells harboring the H3K4R, H3K4R, K36R, or set1Δ set2Δ mutant that disrupts H3K4 and H3K36 methylation also show very similar phenotypes to the PHD and chromodomain double mutant. Our results suggest that multivalent interactions between the PHD, chromodomain, and methylated H3K4 and H3K36 act in a combinatorial manner to recruit NuA4 and regulate the NuA4 activity at the DSB site.
Keywords: Saccharomyces cerevisiae, DSB repair, NuA4, histone modification, multivalent interaction
IN eukaryotic cells, DNA repair occurs in the context of chromatin (Morrison and Shen 2005; van Attikum and Gasser 2009). The chromatin is composed of nucleosomes that are formed by an octamer of histones containing two copies each of H2A, H2B, H3, and H4, wrapped in 146 bp of DNA (Rando 2012). These histones are evolutionarily conserved and are subjected to post-translational modifications such as acetylation, methylation, phosphorylation, and ubiquitination (Jenuwein and Allis 2001). These post-translational modifications of histones alter chromatin structure, thus facilitating the binding between domains and histones that regulate the accessibility of DNA during DNA replication, transcription, and repair. To date, a large number of domains that can associate with acetylated, methylated, and phosphorylated histones have been characterized. For example, chromodomain associates mainly with methylated K36 and K4 of histone H3 (Joshi and Struhl 2005; Xu et al. 2008). On the other hand, the plant homeodomain (PHD) associates mainly with methylated H3K4, although its association with methylated H3K36 has also been reported (Gozani et al. 2003; Li et al. 2006; Shi et al. 2007). Although many histone-binding domains have been characterized, how these domains act in a combinatorial fashion within chromatin in biological processes has not been fully elucidated. Recently, it has been reported that multivalent interactions between domains and histones may not only enhance protein binding specificity, but also play an important role in allosterically regulating activities of the complex (Jenuwein and Allis 2001; Rando 2012).
DNA double-strand breaks (DSBs) represent one of the most threatening lesions to the integrity of genomes. Histone modification complexes such as NuA4 and chromatin remodeling complexes such as RSC, INO80, and SWR1 have been shown to be recruited to the DSB site (Morrison et al. 2004; Chai et al. 2005; Papamichos-Chronakis et al. 2006; Bao and Shen 2007; Shim et al. 2007; Morrison and Shen 2009; Faucher and Wellinger 2010). Their function is to change the structure of chromatin, presumably relaxing it to allow the access of repair proteins. Our knowledge of chromatin modification at the DSB sites is primarily derived from a single DSB introduced at the specific site of a genome such as an HO site in the yeast genome or an I-SceI site in the human genome (Haber 2002; Sugawara and Haber 2006). Specific protein complexes and histone modifications at the DSB can be tested by conducting the chromatin immunoprecipitation (ChIP) experiment. Among all chromatin modifications linked to the DSB damage response, it is clear that phosphorylation of serine 129 of histone H2A (H2A-S129p) occurs within a few minutes after the break (Downs et al. 2004; van Attikum and Gasser 2009). The Η2Α-S129p can spread over >10 kb on each side of the break, thereby making it easily detectable by immunofluorescence or ChIP. H2A-S129p is commonly used as a marker for DSBs (Downs et al. 2004). In addition, a recent study showed that H3K4me3 is enriched at the DSB site, and Set1 is recruited at the DSB site to participate in nonhomologous end joining (NHEJ) (Faucher and Wellinger 2010). H3K4me3 is also found at meiotic DSB sites, indicating the significance of H3K4 methylation at DSB sites (Borde et al. 2009). According to the findings of a recent study, H3K36 methylation is enriched at the DSB site and is critical for DSB checkpoint activation and pathway choice (Jha and Strahl 2014). In addition, H3K36me2 is also found to be induced at DSBs in the human sarcoma cell line; this can improve the association of repair components, such as NBS1 and Ku70, thereby enhancing NHEJ (Fnu et al. 2011).
The NuA4 complex, an essential histone acetyltransferase (HAT), is responsible for the acetylation of histone H2A, H4, and H2AZ (Doyon and Cote 2004). Chromatin immunoprecipitation experiments demonstrate that NuA4 is recruited to the DSB site, and NuA4 acetylates K5, K8, K12, and K16 of histone H4 (Bird et al. 2002). It has been suggested that the recruitment of NuA4 causes the chromatin to be in an “open” state, thereby allowing the repair proteins to access the damaged DNA. NuA4 is not only involved in DSB repair, but it also regulates transcription. NuA4 specifically acetylates nucleosomes at the promoters of actively transcribed genes (Ginsburg et al. 2009). The NuA4 complex consists of 13 subunits of proteins; it has a combined molecular weight of 1 MDa. Esa1 is the catalytic subunit of the NuA4 (Allard et al. 1999; Doyon and Cote 2004). Mutations in several NuA4 subunits such as esa1, epl1, and yng2 are highly sensitive to DNA damaging agents. Interestingly, several subunits of NuA4 contain domains that can recognize and bind to specifically modified histones. Among these are the chromo, plant homeodomain (PHD), actin-related, and YEATS domains. Eaf3 contains a chromodomain, which interacts with methylated H3K4 and H3K36 in vitro (Selleck et al. 2005; Xu et al. 2008). Esa1 also contains the chromodomain, but this domain contains the N-terminal extension that occupies the H3K36me binding site (Huang and Tan 2013). Therefore, the Esa1 chromodomain is thus unlikely to bind H3K36me, but it may bind to nucleosomal DNA and play an important role in nucleosomal HAT activity. Yng2 contains the PHD domain, which interacts with H3K4me3 and methylated H3K36 (Choy and Kron 2002; Boudreault et al. 2003; Shi et al. 2007). Arp4 contains the actin-related domain that interacts with H2A-S129p (Downs et al. 2004). Yaf9 contains the YEATS domain (Schulze et al. 2009, 2010; Wang et al. 2009). A recent study has shown that the Yaf9 YEATS domain can interact with acetylated H3, such as H3K9ac, H3K14ac, and H3K27ac (Li et al. 2014). Although these domain–histone interactions have been identified, it remains unclear how the actual in vivo involvement of each domain acts in the functioning of NuA4 and how these domains function in a combinatorial fashion.
In this study, we dissected the contribution of the PHD and the chromodomain in the functioning of NuA4. We generated mutations in the PHD domain of Yng2 and the chromodomain of Eaf3. With these PHD and chromodomain mutants, we focused on the DSB repair function of NuA4. We demonstrate that mutations in either the PHD or chromodomain do not have any significant effects on the sensitivity to DNA-damaging treatments. However, combined mutations in both the PHD and chromodomain (eaf3-W84A,W88A yng2-phdΔ) cause defects in DSB repair and display the slow growth phenotype. Using ChIP experiments based on the galactose-inducible HO endonuclease-mediated DSB at the MAT HO locus, we reveal that the combined mutations in both the PHD and chromodomain impair the NuA4 recruitment, reduce the NuA4 acetylation activity, and display prolonged H2A-S129p at the HO cleavage site. Cells harboring the H3K4R mutant that cause the H3K4 site to be unable to be methylated, display very similar phenotypes to the PHD chromodomain double mutant. In addition, by in silico analysis of previously published genome-wide datasets, we reveal that the binding occupancy of Esa1, a catalytic subunit of NuA4, is highly correlated with the occupancy of H3K4 methylation. Given that cells harboring set1Δ set2Δ and H3K4R, K36R mutants are all defective in bleomycin treatment, we conclude that the combined interactions between the PHD, chromodomain, and specifically methylated H3K4 and H3K36 play an important role in the recruitment and activation of the NuA4 complex at DSBs, thereby facilitating DSB repair.
Materials and Methods
Yeast strains
All strains used in this study were derived from W303 (MATa,ade2-1, trp1-1, can1-100, leu2-3,112, his3-11,15) and are listed in Table 1. The EAF3 gene was disrupted in the W303 strain by a PCR-based gene deletion strategy (Longtine et al. 1998) using two fusion primers, 5′ GAG GCC TCG TCA CTG GAT TTA CCC TAT TGA AGA ACG TAT ACG GAT CCC CGG GTT AAT TAA 3′ and 5′ CTA AAT ACT AGA AAT AAT CCC AAG CTA GAA TAT AAA CGT CGA ATT CGA GCT CGT TTA AAC 3′, that amplify the KanMX6 cassette. The resulting PCR product was transformed into the W303 strain to generate the eaf3 deletion (eaf3Δ) strain.
Table 1. Yeast strains used in this study.
| Strains | Genotype and carried plasmids |
|---|---|
| yHJL198 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, eaf3::KAN |
| yHJL223 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, yng2::NAT |
| yHJL253 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, eaf3::KAN, yng2::NAT |
| yHJL284 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, pRS415, pRS416 |
| yHJL285 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 eaf3::KAN pRS415-eaf3W84AW88A, pRS416 |
| yHJL286 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yng2::NAT pRS415, pRS416-yng2Δphd |
| yHJL287 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 eaf3::KAN, yng2::NAT, pRS415-eaf3W84AW88A, pRS416-yng2Δphd |
| yHJL292 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, pRS415-HHF1-HHT1 |
| yHJL294 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, pRS415-HHF1-H3K4R |
| yHJL296 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, pRS415-HHF1-H3K36R |
| yHJL298 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, pRS415-HHF1-H3K4R,K36R |
| yHJL310 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15,yng2::NAT, pRS415- YNG2 |
| yHJL312 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, yng2::NAT, pRS415- yng2D239A |
| yHJL314 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, yng2::NAT, pRS415- yng2W247A |
| yHJL316 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, yng2::NAT, pRS415- yng2Y224A |
| yHJL318 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15,yng2::NAT, pRS415- yng2Δphd |
| yHJL326 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, yng2::NAT pRS416-HHF1-HHT1, pRS415-YNG2-myc-his6 pSE271-GAL-HO endonuclease |
| yHJL328 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, yng2::NAT pRS414-HHF1-H3K4R, pRS415- YNG2-myc-his6 YCP50-GAL-HO endonuclease |
| yHJL336 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 eaf3::KAN yng2::NAT pRS414-HO, pRS415-eaf3W84AW88A, pRS416-yng2Δphd |
| yHJL345 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, pRS415-EAF3, eaf3::KAN |
| yHJL346 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, pRS415-eaf3Y81A, eaf3::KAN |
| yHJL347 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, pRS415-eaf3W88A, eaf3::KAN |
| yHJL348 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, pRS415-eaf3W84AW88A, eaf3::KAN |
| yHJL385 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15,SET1-myc::kanMX6, YCP50-GAL-HO endonuclease |
| yHJL389 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15, SET2-TAP::HIS3MX6,YCP50-GAL-HO endonuclease |
| yHJL401 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15,yng2::NAT, ESA1-TAP::HIS3MX6 pRS415- YNG2 pSE271-GAL-HO endonuclease |
| yHJL403 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 eaf3::KAN, yng2::NAT, ESA1-TAP::HIS3MX6 pRS415-eaf3W84AW88A, pRS416-yng2Δphd pSE271-GAL-HO endonuclease |
| yHJL405 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, ESA1-TAP::HIS3MX6 pRS415-HHF1-H3K4R pSE271-GAL-HO endonuclease |
| yHJL416 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 set1::KANMX6 |
| yHJL429 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 set2::KANMX6 |
| yHJL430 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 set1::KANMX6 set2::KANMX6 |
| yHJL433 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 set1::KANMX6 |
| yHJL436 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 set1::KANMX6 pRS313-FLAG-SET1 |
| yHJL438 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 set2::KANMX6 pRS415-SET2 |
| yHJL440 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 set1::KANMX6 set2::KANMX6 pRS313-FLAG-SET1 pRS415-SET2 |
| yHJL442 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, pRS415-HHF1-H3K4R |
| yHJL444 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, pRS415-HHF1-H3K36R |
| yHJL446 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hht1hhf1::KAN, hht2hhf2::KAN, pRS415-HHF1-H3K4R,K36R |
Similarly, the YNG2 gene was disrupted using pFA6a-NAT-NT2 as the template, and two primers, 5′ GGA TAT GCA AGT TTA TAT TGG ACA ACA TAA CCA ATA GAA GCG GAT CCC CGG GTT AAT TAA 3′ and 5′ GTG TAA ATG AGG TCA TTC AGT CTC AAA AAG GTA TTT TTG TGA TGA ATT CGA GCT CGA TTA 3′. The resulting PCR product was transformed into diploid W303 yeast. Then it was sporulated to four spores to obtain the yng2 deletion (yng2Δ) strain.
The eaf3yng2 double deletion strain was generated by mating the eaf3Δ and yng2Δ strains, followed by sporulation to obtain the eaf3yng2 double deletion mutant. The set1 and set2 null strains were generated by the PCR-based gene deletion strategy (Longtine et al. 1998). The set1Δ strain was generated using two primers, 5′ TAT TTG TTG AAT CTT TAT AAG AGG TCT CTG CGT TTA GAG ACG GAT CCC CGG GTT AAT TAA 3′ and 5′ TGT TAA ATC AGG AAG CTC CAA ACA AAT CAA TGT ATC ATC GGA ATT CGA GCT CGT TTA AAC 3′, that amplify the KanMX6 cassette. Similarly, the set2Δ strain was generated using two primers, 5′ TCA AAC CTT TCT CCT TTC CTG GTT GTT GTT TTA CGT GAT CCG GAT CCC CGG GTT AAT TAA 3′ and 5′ GAA AAC GTG AAA CAA GCC CCA AAT ATG CAT GTC TGG TTA AGA ATT CGA GCT CGT TTA AAC 3′, that amplify the KanMX6 cassette. The resulting PCR products were transformed into W303a and W303α strains to generate the set1Δ and set2Δ strains, respectively. The set1Δ set2Δ double deletion strain was generated by mating the set1Δ and set2Δ strains, followed by sporulation to obtain the set1Δ set2Δ double deletion mutant.
The Δhht1hhf1Δhht2hhf2 DY20D strain was a kind gift from Mary Ann Osley and Cheng-Fu Kao (Academia Sinica, Taipei, Taiwan) (Nakanishi et al. 2009). The PCR products containing the hht1hhf1::KAN and hht2hhf2::KAN were transformed into W303a and W303α, respectively. After mating, a plasmid containing the wild-type HHT1HHF1 was transformed into the diploid W303 strain and then sporulated to obtain the hht1hhf1hht2hhf2 deletion strain.
The SET1-myc strain was generated by a PCR-based gene integration strategy using two fusion primers, 5′ TTG TTT ATG TGG AGC ACC TAA TTG TAA AGG TTT CTT GAA CCG GAT CCC CGG GTT AAT TAA 3′ and 5′ TGT TAA ATC AGG AAG CTC CAA ACA AAT CAA TGT ATC ATC GGA ATT CGA GCT CGT TTA AAC 3′ to amplify the pFA6a-myc-KanMX6 plasmid. The resulting PCR product was transformed into the W303a strain to generate the SET1-myc strain. The SET2-TAP strain was generated by a PCR-based gene integration strategy using two primers, 5′ CGG TCT CCC AGT CCC AAA GAC TAG 3′ and 5′ GTC GAG AAG ATG TGT CAC ATT ACC 3′, and the previously generated yeast TAP-fusion ORF [catalog (cat.) no. YSC1178-202231847, Thermo Scientific] as a template (Gavin et al. 2002; Ghaemmaghami et al. 2003). The ESA1-TAP strain was generated by a PCR-based gene integration strategy using two primers, GCT CCA TGA CTT CGA TGA CCA CTA and GGG TAC CGG ATG CAG AAT TGA AGA, and the previously generated yeast TAP-fusion ORF (cat. no. YSC1178-202233516, Thermo Scientific) as a template (Gavin et al. 2002; Ghaemmaghami et al. 2003). The resulting PCR products were transformed into the W303a strain to generate the SET2-TAP and ESA1-TAP strains, respectively. These constructs were verified by both PCR and Western blotting with a specific anti-CBP antibody (Santa Cruz).
For the ChIP experiments, plasmids containing the HO endonuclease (pSE271-GAL-HO or YCP50-GAL-HO; kind gifts from James Haber) were transformed into W303a, H3K4R, eaf3-W84A,W88A yng2-phdΔ, SET1-myc, and SET2-TAP strains.
A complete list of yeast strains is shown in Table 1.
Plasmid construction
EAF3 containing 1 kb upstream and 130 bp downstream of the open reading frame (ORF) was amplified by high proofreading enzyme (Phusion, NEB) and cloned between the SmaI and XbaI sites of vector pRS415. The eaf3-Y81A, eaf3-W88A, and eaf3-W84A,W88A mutants were generated by using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) with two primers 5′ GGG AAG TGT TTT TTC ATA CAT GCG CAA GGC TGG AAG TCG AG 3′ and 5′ CTC GAC TTC CAG CCT TGG GCA TGT ATG AAA AAA CAC TTC CC 3′, 5′ AAG GCT GGA AGT CGA GTG CGG ATG AGT GGG TTG GAT A 3′ and 5′ TAT CCA ACC CAC TCA TCC GCA CTC GAC TTC CAG CCT T 3′, and 5′ TCA TAC ATT ACC AAG GCG CGA AGT CGA GTG CGG ATG AGT GGG TTG GAT A 3′ and 5′ TAT CCA ACC CAC TCA TCC GCA CTC GAC TTC GCG CCT TGG TAA TGT ATG A 3′, respectively. YNG2 containing 200 bp upstream and overall ORF was amplified by high proofreading enzyme (Phusion, NEB) using two primers, 5′ AAA GCG GCC GCG AAT AAG CTA GCA AAT GCT AGA TTA ATT TAG ATT AAG TTT AAC C 3′ and 5′ AAA CTC GAG CGT TAC GTT TTC TTT TCA GTT TGT TTT TTT CCA TCT CAA TTT TAC 3′. Subsequently, it was cloned between the NotI and XhoI sites of vector pcDNA3.1. The resulting recombinant gene YNG2-myc was cut with NotI and PmeI and then cloned between the NotI and SmaI sites of pRS415. The yng2-Y224A, yng2-W247A, and yng2-D239A mutants were generated by using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) using two primers: 5′ GAA GAG GAC AAA ACT TTA GCC TGC TTC TGT CAA AGA GTT TCG 3′ and 5′ CGA AAC TCT TTG ACA GAA GCA GGC TAA AGT TTT GTC CTC TTC 3′, 5′ GTG ATG GAC CCA ACT GTA AAT ATG AAG CGT TTC ATT ATG ATT GTG 3′ and 5′ CAC AAT CAT AAT GAA ACG CTT CAT ATT TAC AGT TGG GTC CAT CAC 3′, and 5′ GTT TGG AGA AAT GGT TGC ATG TGC TGG ACC CAA CTG TAA ATA TG 3′ and 5′ CAT ATT TAC AGT TGG GTC CAG CAC ATG CAA CCA TTT CTC CAA AC 3′, respectively. To generate the yng2-phdΔ−myc mutant, two artifical AflII sites upstream and downstream of the PHD domain were created using QuikChange II XL Site-Directed Mutagenesis Kit. After digestion with AflII and religation, the yng2-phdΔ mutant was generated. All of these constructs were verified by DNA sequencing.
The full length of YNG2 or mutant yng2-phdΔ was cloned into SmaI/SalI sites of pGex6P-1 vector. The full length of wild-type EAF3 or mutant eaf3-W84A,W88A was cloned into EcoRI/XhoI sites of pGex6p-1 vector.
The pRS313-FLAG-SET1 plasmid was a kind gift from Cheng-Fu Kao (Academia Sinica, Taipei, Taiwan). To construct pRS415-SET2, the SET2 gene containing 600 bp upstream and overall ORF was amplified by high proofreading enzyme (Phusion, NEB) using two primers, 5′ AAA GAG CTC GGG AAA ACG TTC AAG AAT CTC TCT 3′ and 5′ AAA CCC GGG ACA GAA AAC GTG AAA CAA GCC 3′. Subsequently, it was cloned between the SacII and XmaI sites of vector pRS415. These clones were verified by DNA sequencing.
The H3K4R, H3K36R, and H3K4R, K36R mutants were generated by using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) with two primers, 5′ GCA AAC AAT GGC CAG AAC AAG GCA AAC AGC AAG A 3′ and 5′ TCT TGC TGT TTG CCT TGT TCT GGC CAT TGT TTG C 3′, and 5′ CGC CCC ATC TAC CGG TGG TGT TAG GAA GCC TCA C 3′ and 5′ GTG AGG CTT CCT AAC ACC ACC GGT AGA TGG GGC G 3′, respectively.
Survival assay
Spot assay:
Yeast cells were cultured in 3 ml of liquid broth for 16 hr and then diluted to OD600 = 0.8, which is about 8 × 106 cells/ml. Ten-fold serial dilutions were made and spotted on agar plates containing the following reagents: 0.006%, 0.01%, or 0.03% of methyl methanesulfonate (MMS); 1 μg/ml or 3 μg/ml of phleomycin; 2 mU/ml, 5 mU/ml, or 10 mU/ml of bleomycin. A total of 3 μg/ml of phleomycin can cause a phenotype that is very similar to that treated with 2 mU/ml of bleomycin. Since bleomycin is a chemotherapy drug widely used to treat cancer, we used bleomycin in most of our study. Plates are also subjected to UV irradiation with the following intensities, 10 J/m2, 20 J/m2, or 30 J/m2.
Cell growth measurement:
Yeast strains, W303, eaf3-W84A,W88A, yng2-phdΔ, and eaf3-W84A,W88A yng2-phdΔ were grown at 30° for 24 hr. The concentration of these cells was determined by OD600 at time intervals of 3, 6, 9, 12, and 24 hr.
qRT-PCR analysis
Total RNA from wild-type or mutant cells was isolated using RNeasy mini kit (Qiagen) and was subjected to reverse transcription using GoScript reverse transcription system (Promega). The resulting cDNA samples were analyzed using the real-time PCR analysis (ABI StepOne Plus Real-Time PCR Systems). The real-time PCR was performed in a 40-μl reaction with 0.8 μl of 10 μM primers, 0.8 μl of 50 mM MgCl2, 20 μl iQSYBR green supermix (BIO-RAD), and 1 μl cDNA. The primer sequences are as follows: RPS11B: 5′ ATG AAA ATGTAACCACGGAGAAA T 3′ and 5′ AAAATTGTTCCGTATGATGGAGAT 3′; RPL2B: 5′ TTT TGT ATC ATG ACA ACA GGG TTC 3′ and 5′ GTA ACC ATG ACG TTC AGC ATA GTC 3′; and RPL19B: 5′ TTG GTA GCA ACA TCC ATA GAA GAA 3′ and 5′ GTA AAG AGC ATA TTC GCA AAG GTT 3′. The real-time PCR was started at 95° for 3 min, which was then followed by 40 cycles at 95° for 15 sec and 55° for 45 sec.
The expression of actin (ACT1) was used as an internal control. The expression level of each gene in wild-type or mutant cells was normalized by the level of ACT1 in each cell.
Purification of GST-Yng2, GST-Eaf3, GST-yng2-phdΔ, and GST-eaf3-W84A, W88A, and in vitro peptide binding assay
The GST fusion proteins were purified according to the manufacturer’s instructions using Glutathione Sepharose 4B (GE Healthcare). The biotin-conjugated histone H3 (residues 1–21, cat. no. 12-403), H3K4me2 (cat. no. 12-460), and H3K4me3 (cat. no. 12-564) were purchased from Millipore. H3 (residues 21–44), H3K36me2, and H3K36me3 peptides were kindly gifted by Mau-Sun Chang (National Taiwan University, Taipei, Taiwan). A total of 1 μg of biotin-conjugated histone peptides and 1 μg of GST fusion proteins were incubated in peptide binding buffer (50 mM Tris-HCl 7.5, 300 mM NaCl, 0.1% NP-40, 1 mM PMSF, and protease inhibitors) for 2 hr at 4° with rotation, followed by the addition of streptavidin beads (Amersham) and incubated for 1 hr at 4° with rotation. After extensive washing with peptide binding buffer, bound GST fusion proteins were analyzed by SDS–PAGE and Western blotting using anti-GST antibody.
Chromatin immunoprecipitation
To carry out ChIP, 1.2 liters of culture was grown with 1% succinate acid and 2% glycerol at 30° until OD600 reached ∼0.5–1, which is ∼5 × 106–107 cells/ml). A total of 200 ml of cell culture was then taken out before adding galactose at 0-min time point. Then, 2% galactose was added to the culture to induce the expression of HO endonuclease. After 1 hr, 2% glucose was added to repress the expression of HO endonuclease. As shown in Figure 4A, cell culture samples were taken out at each time point and subjected to ChIP experiments. Cells were resuspended in crosslinking buffer (10 mM HEPES pH = 7.5, 3 mM MgCl2) and crosslinked with 1% formaldehyde at room temperature for 30 min. To generate spheroplasts, fixed cells were treated with spheroplast buffer (18.2% sorbitol, 1% glucose, 0.2% yeast nitrogen base, 0.2% casamino acids, 0.6% HEPES, 0.6% Tris, and 1 mM DDT) and digested with lyticase. These spheroplasts were treated with PIPES/sorb buffer (20 mM PIPES, 1 mM MgCl2, and 1.2 M sorbitol) and then washed with PBS/0.5 mM PMSF buffer, HEPES/Triton X-100 buffer (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES pH = 6.5, 0.5 mM PMSF, 1 μg/ml pepstatin, and 1 μg/ml leupeptin) and HEPES/NaCl buffer (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES pH = 6.5, 0.5 mM PMSF, 1 μg/ml pepstatin, and 1 μg/ml leupeptin). To fragment the DNA to ∼500 bp, the resulting spheroplasts were resuspended in 1 ml SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH = 8.1, PMSF, leupeptin, and pepstatin) and subjected to sonication (BioruptorTM UCD-200) on ice. After centifugation, the supernatant was transferred into immunoprecipitation (IP) dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris pH = 8.1, 167 mM NaCl, 0.5 mM PMSF, 1 μg/ml pepstatin, and 1 μg/ml leupeptin) to generate the chromatin solution. The immunoprecipitation was performed using 2 μl of anti-H2A-S129p (Abcam, ab15083), H3K36me2 (Millipore, 07-274), H3K4me2 (Millipore, 07-030), H3K4me3 (Abcam, ab8580), c-myc (Abcam, ab9106), or H4K12ac (Millipore, 07-595) antibodies or with IgG-sepharose (GE Healthcare). The DNA samples were analyzed using real-time PCR analysis (ABI StepOnePlus Real-Time PCR Systems) using the following pairs of primers targeted the HO-cleavage site (5′ TCC CCA TCG TCT TGC TCT 3′ and 5′ GCA TGG GCA GTT TAC CTT TA 3′), 2 kb away from the HO-cleavage site (5′ GCC TCT ATG TCC CCA TCT TGT CTC 3′ and 5′ GTG TTC CCG ATT CAG TTT GAC G 3′) and 10 kb away from the HO-cleavage site (5′ TGG ATC ATG GAC AAG GTC CTA C 3′ and 5′ GGC GAA AAC AAT GGC ACT CT 3′) as indicated in Figure 4A.
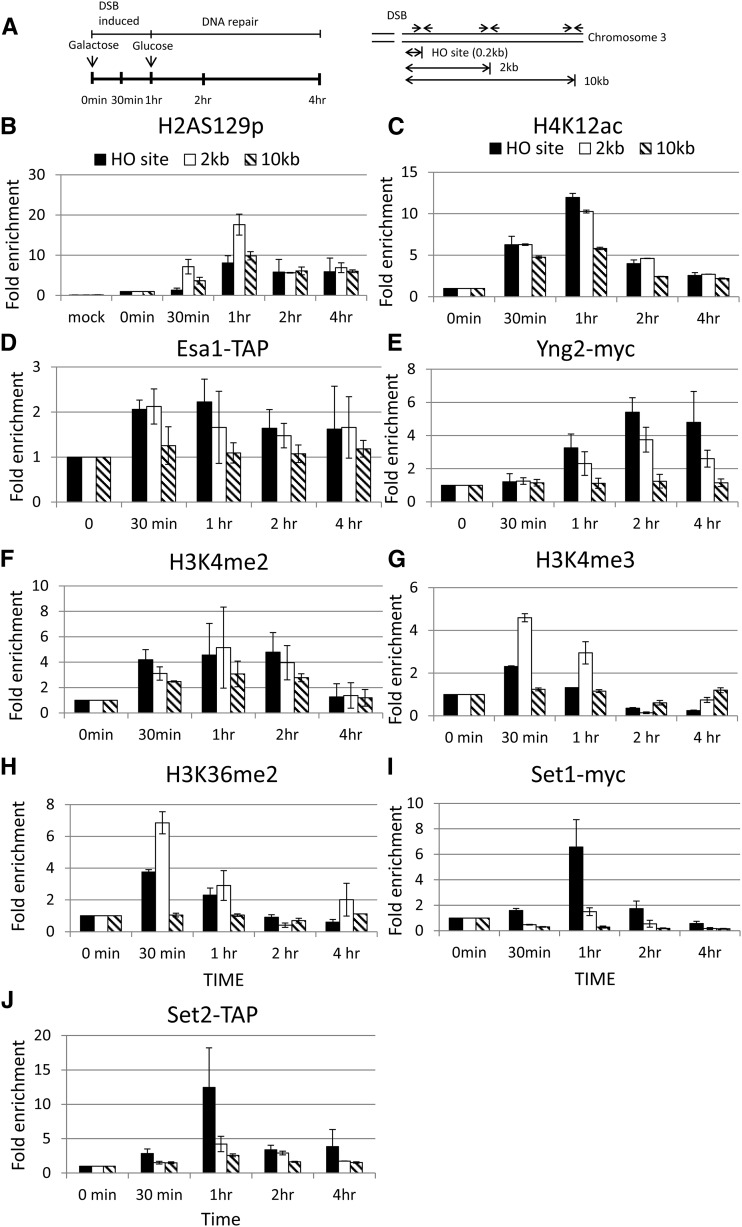
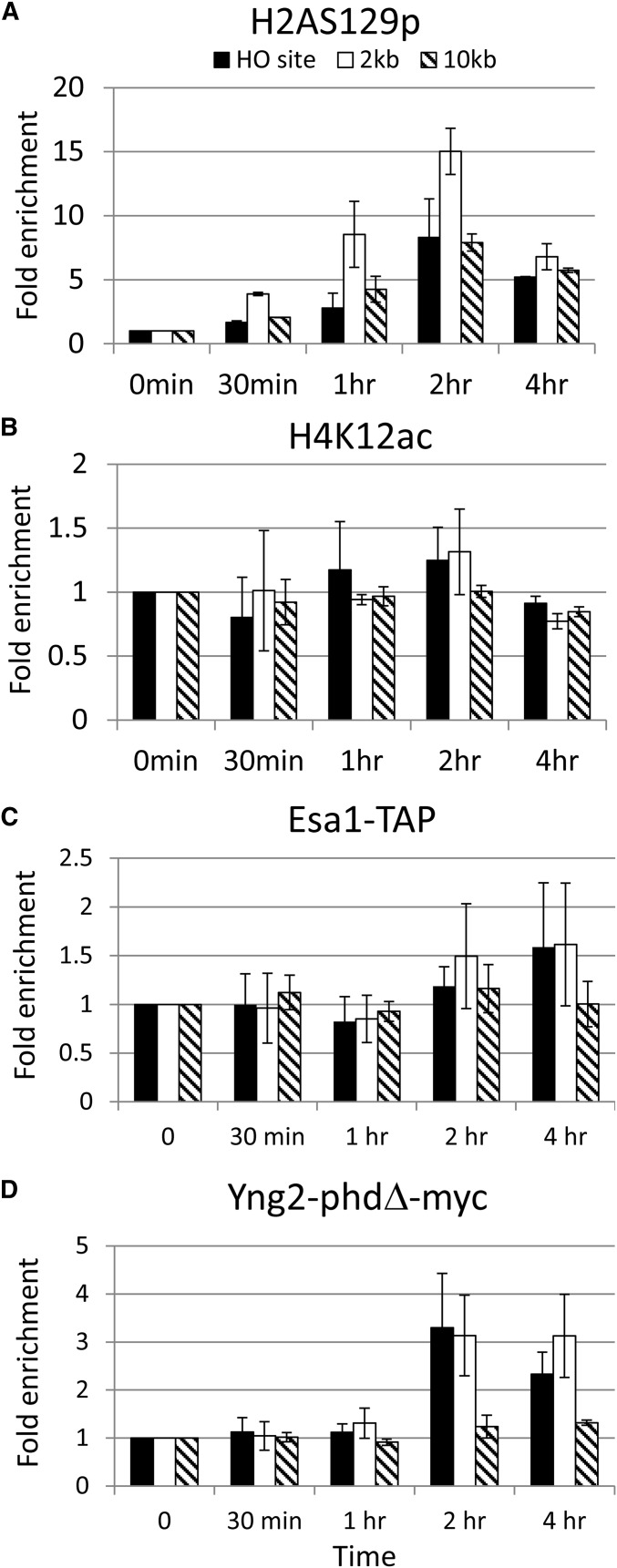
Figure 4.
Histones are specifically modified at the HO DSB site. (A) Schematic of the ChIP experiment. Cells were cultured in SC −Ura −Leu −Trp + 2% glycerol and 1% succinate. The 2% galactose was then added to induce the expression of HO endonuclease at 0-min time point, thus generating a single DSB at the MAT HO locus of chromosome III. One hour later, 2% glucose was added to repress the expression of HO endonuclease, and thus the DSB were repaired. Samples were collected at the indicated time points for the ChIP assay. Specific primers located at HO, 2 kb, and 10 kb from the HO cutting site are indicated as arrows. The ChIP experiments were performed with specific anti-H2A-S129p (B), H4K12ac (C), IgG sepharose beads (to pull down Esa1) (D), myc antibody (to pull down Yng2-myc) (E), H3K4me2 (F), H3K4me3 (G), H3K36me2 (H), myc (to pull down Set1) (I) antibodies, and IgG sepharose beads (to pull down Set2) (J) as indicated.
The real-time PCR was performed in a 40-μl reaction with 0.8 μl each of 10 μM left and right primers (targeting the HO site, and 2 kb and 10 kb away from the HO site), 0.8 μl 50 mM MgCl2, 20 μl iQTM SYBR Green Supermix (BIO-RAD iQTM SYBR Green Supermix kit), and 5 μl ChIP DNA. The PCR cycling was started at 95° for 3 min, followed by 40 cycles at 95° for 15 sec and 55° for 45 sec. The ChIP data were quantified by the following formula: (IP t/input t)/(IP 0/input 0), where IP t and IP 0 are the immunoprecipitated DNA products at time t and time 0, respectively, and input t and input 0 are the input DNA at time t and time 0, respectively. The ChIP data were generated from at least three independent experiments. A similar trend was observed when these experiments were replicated.
The correlation analysis of Esa1 and H3K4 methylation
To obtain the correlation between Esa1 occupancy and H3K4 methylation, we analyzed previously published datasets, which contained the occupancy score of several transcription-related genes, including one of NuA4 subunit, Esa1, and the enrichment ratio of H3K4 methylation in a genome-wide scale (Kirmizis et al. 2007; Guillemette et al. 2011; Venters et al. 2011). By referring to the work of Venters et al. (2011), we fetched those probes that detected the occupancy level of Esa1 exceeding 1.5-fold or 2-fold threshold. Then, we evenly expanded the probe region from the original 70-bp to 1000-bp width. These expanded region sets traversing the whole genome were used to determine whether specific H3K4 methylation patterns (H3K4me3 and H3K4me2) have 1.5-fold or 2-fold enrichment thresholds above the background in that region. The datasets of H3K4 methylation were obtained from the previously published work of Kirmizis et al. (2007) and Guillemette et al. (2011).
Southern blot analysis
Purified genomic DNA was digested with StyI, and the fragments were separated on a 1.2% agarose gel, followed by Southern blot hybridization with a digoxigenin (DIG)-labeled probe specific to the MAT sequence. The DIG-labeled MAT probe was made by PCR using primers, 5′ ATT CTT AGC ATC ATT CTT TGT TC 3′ and 5′ GCC CTT TGA AGA TAT TAC TCT GGG 3′ and DIG-labeling mix (Roche). The DIG-labeled probe targeting the SET2 gene was made by PCR using primers 5′ GGC AGC TTT TCA AAG GGG CTT TCG 3′ and 5′ AAT ATG GCT ATC GGA GCG TAC TGC 3′. Probe-target hybrids were detected with an anti-DIG-alkaline phosphatase antibody (Roche) and a CSPD alkaline phosphatase substrate (Roche) for sensitive chemiluminescent visualization of DIG-labeled probes. The intensities of the bands on the Southern blots corresponding to the probed DNA fragments were analyzed with the GeneTools program (Syngene). The percentage of HO-cut fragments was determined by the amount of HO-cut fragment relative to the SET2 loading control fragment, normalized to the value obtained from an uninduced (0-hr time point) sample. The statistical significance between wild type and mutant was determined by Student’s t-test with P-value <0.05.
Data availability
Strains are available upon request. Figure S1 contains supplemental data in support of Figure 3. Figure S2 contains supplemental data in support of Figure 7.
Figure 3.
H3K4 methylation and H3K36 methylation are important for DSB repair. (A) The H3K4R and H3K36R mutants are sensitive to DNA damage treatment. The hht1hhf1 hht2hhf2 null mutant carrying plasmids containing wild-type (H3) and mutant histone H3 (H3K4R, H3K36R, H3K4R,K36R) were spotted on agar plates and treated with DNA damaging agents as indicated. (B) The set1, set2, and set1 set2 null mutants can be complemented with plasmids carrying wild-type SET1 or SET2 plasmids. (C) Strains carrying the wild-type SET1 SET2, set1, set2, or set1 set2 null mutations were spotted on agar plates and treated with DNA damaging agents as indicated. (D) Strains carrying the wild-type H3, H3K4R, H3K4A, or set1Δ mutations were spotted on agar plates and treated with DNA damaging agents as indicated.
Figure 7.
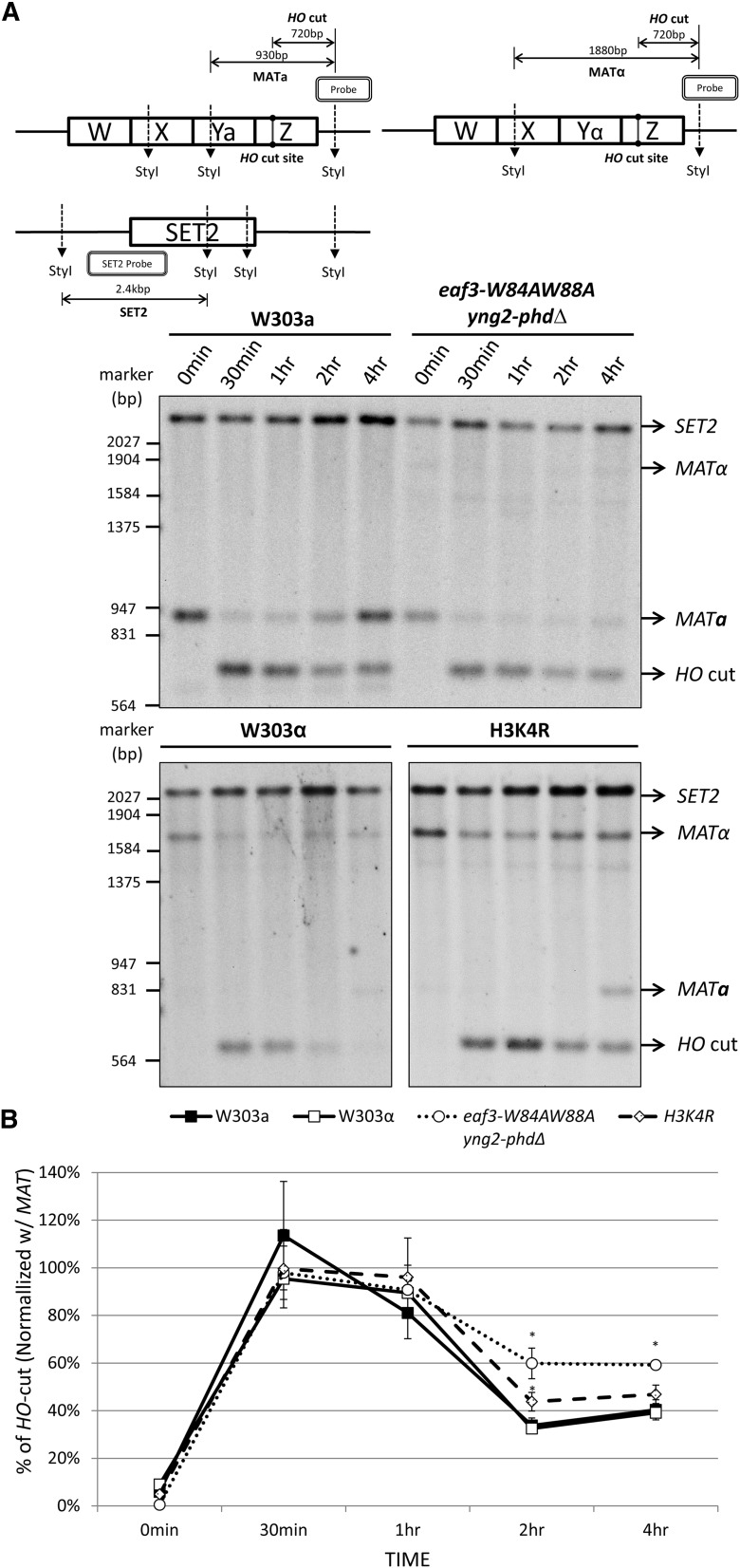
The eaf3-W84A,W88A yng2-phdΔ double mutant and H3K4R mutant cells are defective in DSB repair. (A, top) Diagram of the MAT locus and SET1 locus analyzed is shown. The HO cut site and StyI restriction sites are indicated. (Bottom) Wild-type W303a, W303α, eaf3-W84A,W88A yng2-phdΔ double mutant, and H3K4R mutant cells were induced to generate a HO DSB site, following a protocol similar to the ChIP experiment shown in Figure 4. Genomic DNA was extracted at the time points shown, digested with StyI, and separated on 1.2% agarose gel. Southern blot was hybridized with a DIG-labeled MAT-specific probe and a DIG-labeled SET2-specific probe as a loading control. The size of MATα is larger than that of MATa because MATα lacks a StyI site in the Yα sequence. (B) The percentage of HO cut was determined by the amount of HO cut fragments relative to the SET2 loading control fragments, normalized to the value obtained from an uninduced (0 hr) sample. The asterisk (*) indicates that the difference between wild type and mutants was statistically significant with P-value <0.02. (Student’s t-test, eaf3-W84A,W88A yng2-phdΔ double mutant: P-values 0.001 and 0.002 at 2-hr and 4-hr time points, respectively; and H3K4R: P-value 0.012 at 2-hr time point).
Results
Combined mutations in both the chromodomain and PHD domain result in slow growth phenotype and sensitivity to bleomycin
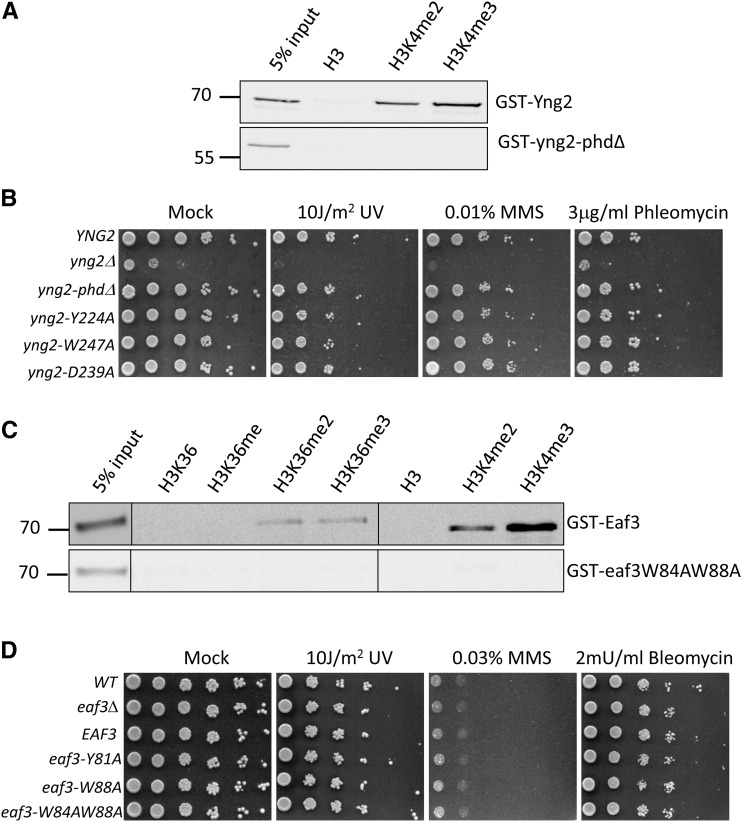
NuA4 contains the PHD and chromodomain that can interact with specifically modified histones (Choy and Kron 2002; Boudreault et al. 2003; Joshi and Struhl 2005; Shi et al. 2007; Xu et al. 2008). Although previous studies have shown that there is high affinity in vitro when the PHD and chromodomain interact with methylated H3K4 (e.g., H3K4me2 and H3K4me3) and methylated H3K36 (e.g., H3K36me2). However, these studies have not been able to determine the contribution of each domain to NuA4 activity, both individually and in combination. To dissect the function of the PHD and chromodomain of NuA4, we generated mutations in the PHD and chromodomain to disrupt their interactions with methylated H3K4 and H3K36. Thereafter, we analyzed the NuA4 activity in DNA repair. Yng2 is the only subunit containing the PHD domain. Previous studies have shown that the PHD domain of Yng2 contained three conserved residues, Y224, D239, and W247, that are important for the interaction with H3K4me3 (Loewith et al. 2000; Shi et al. 2007). We substituted these three residues with alanine and produced yng2-Y224A, yng2-D239A, and yng2-W247A in plasmids. In addition, we generated the yng2-phdΔ, in which the PHD domain is deleted from Yng2. We confirmed that Yng2 protein can interact with H3K4me2 and H3K4me3 peptides (Figure 1A). On the other hand, the yng2-phdΔ protein failed to interact with methylated H3K4 (Figure 1A). As shown in Figure 1B, the yng2 null cells show severe growth defects. The transformation of yng2 null cells with plasmid carrying either the wild-type YNG2 or the yng2 mutants (yng2-Y224A, yng2-D239A, yng2-W247A, and yng2-phdΔ) could fully restore the growth defects caused by the yng2 null mutant. In addition, these yng2 mutants have no significant effects on the sensitivity of DNA damaging agents, such as ultraviolet (UV) light, methyl methanesulfonate (MMS), bleomycin, or phleomycin (Figure 1B). UV induces thymine dimer or six to four crosslinks between bases, while bleomycin or phleomycin induces DSBs. Since bleomycin is a chemotherapeutic drug widely used to treat cancer, we obtained bleomycin from National Cheng Kung University Hospital and used it in the rest of our study. MMS can also induce DSBs, but these DSBs are generated by the stalled replication forks. We conclude that mutations in the PHD domain of yng2 have no effect on NuA4 activity in DNA repair. This finding agrees with that reported in previous studies (Nourani et al. 2001; Selleck et al. 2005).
Figure 1.
Mutations either in the PHD or in the chromodomain show no phenotype to bleomycin-induced DSBs. (A) Peptide binding assay. GST-Yng2 prefers to bind H3K4me2 and H3K4me3. Biotin-conjugated histone peptides are represented in the top of the blot. The pull-down of GST fusion proteins was detected by anti-GST antibody in Western blotting. Input is 5% of GST fusion proteins in the reaction. (B) Mutations in the PHD domain show no sensitivity to DNA damaging agents. Plasmids carrying the wild type (YNG2) or mutants in the PHD domain of YNG2 (yng2-phdΔ, yng2-Y224A, yng2-D239A, yng2-W247A) were transformed into the yng2 null strain (yng2Δ), as indicated. (C) GST-Eaf3 prefers to bind H3K4me2 and H3K4me3, as well as H3K36me2 and H3K36me3. (D) Mutations in the chromodomain of Eaf3 are not sensitive to DNA damaging agents. Plasmids carrying the wild type (EAF3) or mutants in the chromodomain of EAF3 (eaf3-Y81A, eaf3-W88A, eaf3-W84A,W88A) were transformed into the eaf3 null strain (eaf3Δ), as indicated. Cells were subjected to DNA damaging treatment as indicated.
Next, we determined the contribution of chromodomain to NuA4 activity. There are two subunits of NuA4, Eaf3, and Esa1, containing the chromodomain. Since the Esa1 chromodomain is unable to interact with methylated H3K36, Eaf3 is the only subunit that interacts with methylated H3K36 and H3K4 (Selleck et al. 2005; Xu et al. 2008; Huang and Tan 2013). The chromodomain of Eaf3 contains the three residues, Tyr81, Trp84, and Trp88, that are critical for its interaction with H3K36me2 (Xu et al. 2008). Therefore, we generated three mutants, eaf3-Y81A, eaf3-W88A, and eaf3-W84A,W88A, in which the three residues were substituted for alanine to disrupt their interactions with H3K36me2. We confirmed that Eaf3 protein can interact with H3K36me2 and H3K36me3, as well as H3K4me2 and H3K4me3 peptides (Figure 1C). On the other hand, the eaf3-W84A,W88A mutant protein failed to interact with methylated H3K36, as well as methylated H3K4 (Figure 1C). The eaf3 null strains were transformed with plasmids carrying the three mutants. Compared to the wild-type EAF3 cells, deletion of EAF3 had no significant effect on cell growth and DNA damage sensitivity (Figure 1D). The three chromodomain mutants had no effects on the sensitivity of DNA damaging agents, indicating that chromodomain is not required for the NuA4 activity in DNA repair. We used the yng2-phdΔ and eaf3-W84A,W88A for the rest of our study.
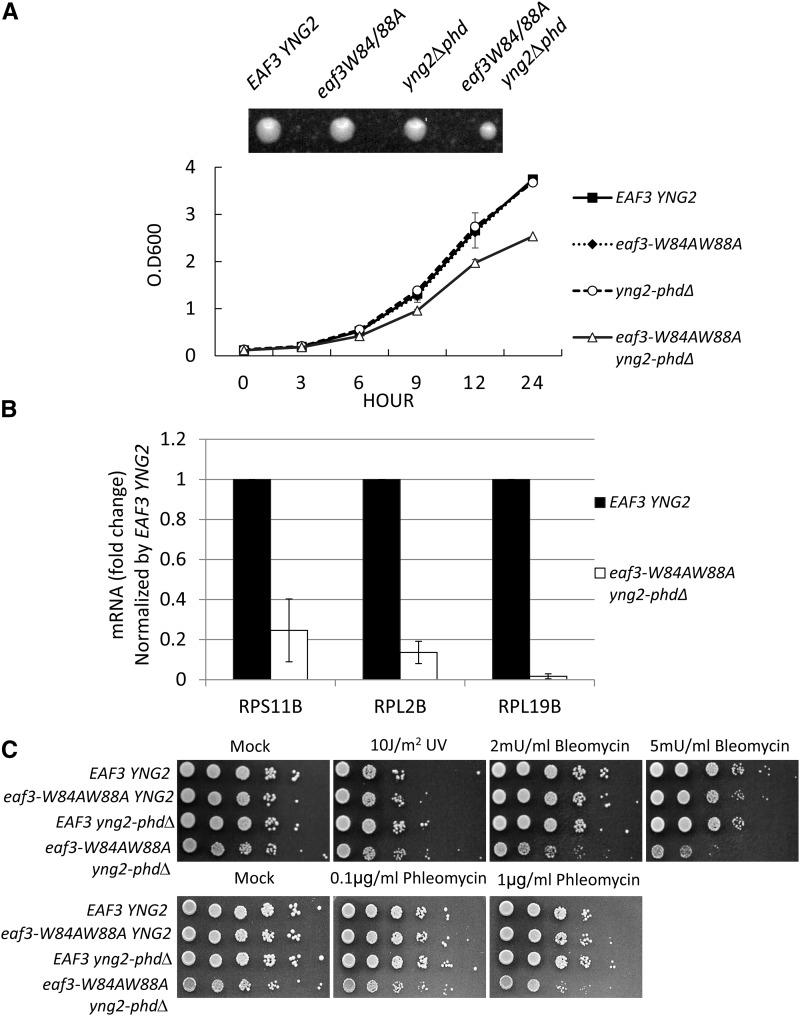
Since a single mutation in either PHD or chromodomain appears to have no impact on the phenotype with regard to growth or defects in DSB repair, it is possible that they work in a combinatorial fashion to facilitate the NuA4 activity. To determine this possibility, we generated a mutant by combining the two mutations, eaf3-W84A,W88A and yng2-phd∆. Here, the chromodomain of Eaf3 and PHD domain of Yng2 were disrupted. The strains containing both chromodomain and PHD mutations (eaf3-W84A,W88A yng2-phd∆) showed a slow growth of phenotype, compared to the wild-type, eaf3-W84A,W88A, or yng2-phd ∆ single mutant cells (Figure 2A). The eaf3-W84A,W88A yng2-phd∆ double mutant had a much smaller colony size than the wild-type or single mutant cells (Figure 2A). Since NuA4 is also required for transcription of many other genes, including ribosomal genes, we also determined the transcription level of ribosomal genes, such as RPS11B, RPL2B, and RPL19B. As shown in Figure 2B, the transcription level of these genes was significantly reduced, when compared with wild-type cells.
Figure 2.
Combined mutations in both the PHD and chromodomain show growth defects and are sensitive to bleomycin-induced DSBs. (A, top) A single cell of each strain was grown at 30° for 2 days. (Bottom) The growth curve of each strain. Strains carrying the wild-type EAF3 YNG2, eaf3-W84A,W88A, yng2-phdΔ, or eaf3-W84A,W88A yng2-phdΔ are indicated. (B) The expression level of ribosomal genes, RPS11B, RPL2B, and RPL19B, was determined by qRT-PCR and normalized by the level in wild-type cells. ACT1 was used as internal control. The expression level of ribosomal genes was normalized by the level of ACT1 in each cell. (C) Combined mutations containing eaf3-W84A,W88A and yng2-phdΔ are sensitive to bleomycin or phleomycin. Strains carrying the wild-type EAF3 YNG2, eaf3-W84A,W88A, yng2-phdΔ, or eaf3-W84A,W88A yng2-phdΔ are indicated.
We further tested the eaf3-W84A,W88A yng2-phd∆ mutant strain with regard to different DNA damage agents, such as UV or bleomycin. The eaf3-W84A,W88A yng2-phd∆ mutant was sensitive to bleomycin or phleomycin treatment, but it was not sensitive to UV (Figure 2C). Since bleomycin or phleomycin treatment can induce DSBs, our results indicate that the PHD domain and chromodomain work in a combinatorial fashion to regulate the NuA4 activity at DSBs, thereby facilitating DSB repair.
H3K4 and H3K36 methylation are important for resistance to bleomycin, a chemotherapeutic drug that induces DSBs
Given that both the PHD and chromodomain of NuA4 could interact with methylated histone H3K4 and H3K36, we tested whether mutations in H3K4 or H3K36 that abrogate H3K4 and H3K36 methylation can result in a phenotype similar to that seen with the eaf3-W84A,W88A yng2-phd∆ mutant. Therefore, to mimic the unmethylated forms of histone H3K4 and H3K36, we generated H3K4R and H3K36R mutants in which the lysine 4 and lysine 36, respectively, of histone H3 were mutated to arginine. As shown in Figure 3A, both H3K4R and H3K36R mutants were sensitive to bleomycin treatment. In addition, the H3K4R, K36R double mutant was more sensitive to bleomycin than single H3K4R or H3K36R mutant alone.
Since SET1 and SET2 are histone methyltransferases that methylate histone H3K4 and H3K36, respectively, we also generated set1, set2, and set1set2 null mutants and tested their DNA damage sensitivity. The set1, set2, and set1set2 null mutants can be fully complemented with plasmids carrying wild-type SET1 and SET2, respectively, suggesting these mutants are indeed set1, set2, and set1set2 null mutantions (Figure 3B). Deletion of either SET1 or SET2 sensitized the cells to bleomycin, which was in good agreement with the H3K4R and H3K36R results (Figure 3C). In addition, the set1 null cells and cells harboring H3K4R or H3K4A mutants were all sensitive to bleomycin (Figure 3D). Interestingly, deletion of both SET1 and SET2 further sensitized the cells to bleomycin (Figure 3C). We conclude that both H3K4 methylation and H3K36 methylation play important roles in DSB repair.
The eaf3-W84A,W88A yng2-phd∆ double mutant fails to induce H4 acetylation at the DSB site
To determine whether the defective phenotype shown in the eaf3-W84A,W88A yng2-phd∆ double mutant was due to an inability to recruit NuA4 at DSB sites, we performed a chromatin immunoprecipitation (ChIP) experiment in the galactose-induced HO system (Figure 4A). In this experiment, galactose was added to induce the expression of HO endonuclease, generating a single cleavage at the MAT locus of chromosome III. After 1 hr, glucose was added to repress the expression of HO endonuclease, thereby allowing the repair of DSB. Specific primers located at 0.17 kb (HO site), 2 kb, or 10 kb away from the HO-cleavage site were used to test the enrichment of these modified histones (Figure 4A). Since previous publications have shown that H2A-S129p and H4K12ac are enriched at the HO DSB site, we therefore used antibodies specific to H2A-S129p and H4K12ac to verify the specificity of this experiment (Downs et al. 2004; Shroff et al. 2004; Faucher and Wellinger 2010; Jha and Strahl 2014). In addition, mock-treated reactions, in which no antibody was added to the ChIP reaction, were also included as a negative control. The mock-treated reactions showed high threshold cycle values (Ct) >28, indicating that a trace amount of DNA was being pulled down. Thus, these reactions represented the background value of ChIP experiments (data not shown). In contrast, Ct values were significantly decreased with specific anti-H2A-S129p and H4K12ac antibodies after HO cleavage, indicating that associated DNA was specifically pulled down. These Ct values were converted to relative concentrations using a standard curve method. To represent the percentage of DNA being pulled down, the resulting IP values were normalized by the concentration of input. These data were further normalized to the value at the time point 0.
As shown in Figure 4B, the trace amount of DNA was pulled down in the mock-treated reactions, indicating a background level of enrichment at the HO site, at 2 kb, and at 10 kb away from the HO cleavage site. In contrast, the levels of H2A-S129p and H4K12ac were significantly enriched at the HO cleavage site after 30 min of HO endonuclease induction and started to decline after 1 hr (Figure 4, B and C). Our data were consistent with previous publications (Downs et al. 2004; Shroff et al. 2004; Faucher and Wellinger 2010; Jha and Strahl 2014). Therefore, our results not only confirm the previous results but also validate our ChIP experiment. To determine whether NuA4 is recruited at the HO cleavage site, we monitored the enrichment of Esa1 and Yng2 at the HO DSB site. We generated the strain harboring the TAP-tagged Esa1 (Esa1-TAP) and myc-tagged Yng2 (Yng2-myc) and performed the ChIP experiment with IgG sepharose and specific anti-myc antibody, respectively. We confirmed that these NuA4 subunits, Esa1 and Yng2, were also recruited to the HO DSB site (Figure 4, D and E). In addition, H3K4me2, H3K4me3, H3K36me2, and their methyltransferases, Set1 and Set2, were also enriched at the HO DSB site (Figure 4, F–J).
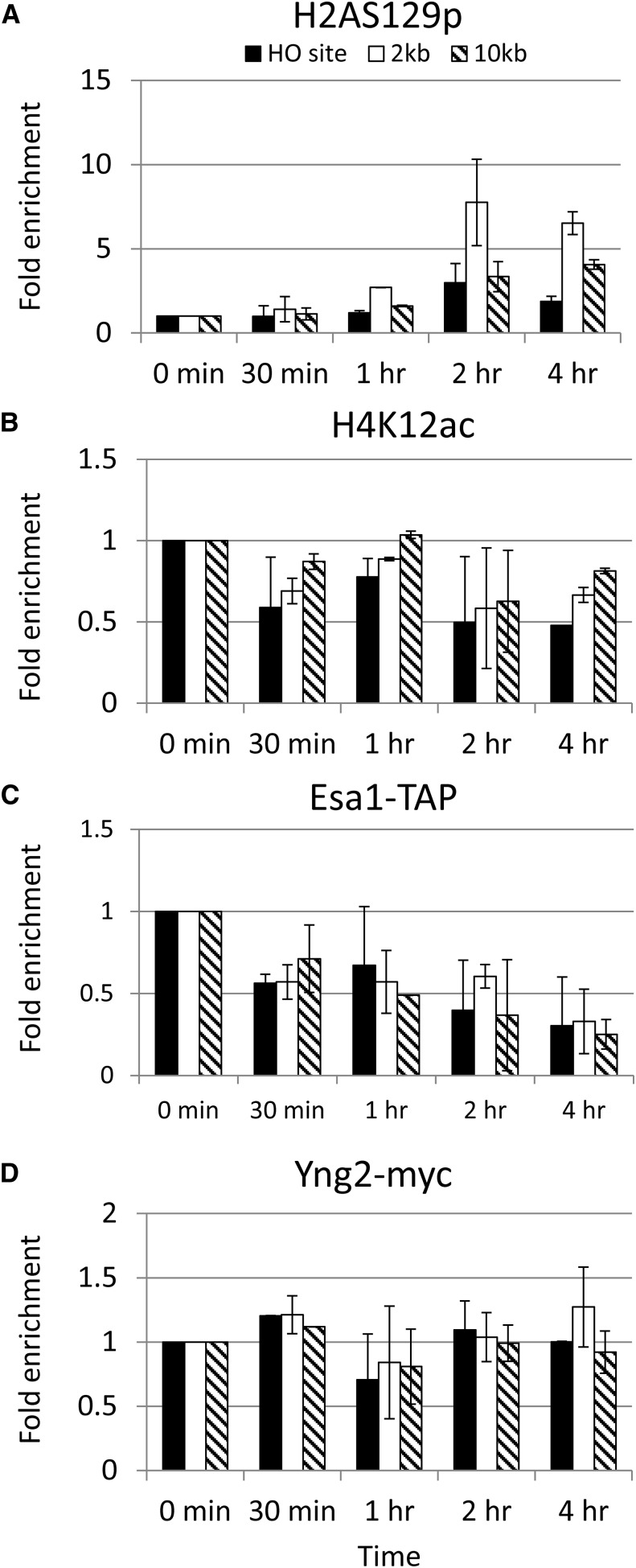
Interestingly, the enrichment of H2A-S129p was delayed in the eaf3-W84A,W88A yng2-phd∆ double mutant cells, reaching its peak at 2 hr and staying high for 4 hr (Figure 5A). In addition, the enrichment of H4K12ac was significantly reduced at the HO DSB site (Figure 5B). To determine whether NuA4 is recruited at the HO cleavage site, we monitored the enrichment of Esa1 (Esa1-TAP) and yng2-phdΔ (yng2-phdΔ-myc) at the HO DSB site via the ChIP experiments with IgG sepharose and specific anti-myc antibodies, respectively. As shown in Figure 5, C and D, the enrichment of Esa1 and yng2-phdΔ was significantly reduced and delayed. Therefore, we have provided the in vivo evidence that the combined interaction between the PHD, chromodomain, and methylated H3K4 and H3K36 played an important role in strengthening the NuA4 recruitment, as well as in the activation of NuA4 activity.
Figure 5.
Histone modifications are altered at the HO DSB site in the eaf3-W84A,W88A yng2-phdΔ mutant. The ChIP experiments were performed with anti-H2A-S129p (A), H4K12ac (B), IgG sepharose beads (to pull down Esa1) (C), and myc (to pull down yng2-phdΔ-myc) (D) antibodies in the eaf3-W84A,W88A yng2-phdΔ mutant as indicated.
The H3K4R abolishes the acetylation of H4 at the HO DSB site
The ChIP experiments were used to further determine whether the H3K4R mutant abolishes the H4 acetylation at the DSB site. Interestingly, the enrichment of H2A-S129p lasted longer, reaching its peak 2 hr after the induction of DSB, similar to that observed with the eaf3-W84A,W88A yng2-phd∆ double mutant cells (Figure 6A). The level of H4K12ac was not enriched after the induction of HO cleavage (Figure 6B).
Figure 6.
The H3K4R mutant fails to induce H4K12ac at the DSB site. The ChIP experiments were performed with anti-H2A-S129p (A), H4K12ac (B), IgG sepharose beads (to pull down Esa1) (C), and myc (to pull down Yng2-myc) (D) antibodies in the H3K4R mutant as indicated.
To determine whether NuA4 complex is recruited to the HO-cleavage site, we monitored the recruitment of Esa1-TAP and Yng2-myc in the ChIP experiment. Both Esa1-TAP and Yng2-myc proteins were unable to recruit to the HO cleavage site (Figure 6, C and D). These results suggest that H3K4 methylation is important for the recruitment of NuA4 complex to the HO DSB site. Consistent with our results, Ginsburg et al. (2014) also recently showed that H3K4 and H3K36 methylation are required for robust NuA4 interactions with nucleosomes.
The eaf3-W84A,W88A yng2-phd∆ double mutant cells are defective in DSB repair
To determine whether the eaf3-W84A,W88A yng2-phd∆ double mutant cells are defective in DSB repair, we monitored the DSB repair by Southern blot analysis. As shown in Figure 7 and Supporting Information, Figure S2, the MATa locus was efficiently cut by the HO endonuclease after induction and subsequently, it was repaired as judged by a reduction in the amount of HO cut fragments and the reappearance of MATa product in wild-type cells. The HO cut fragments of 33% and 40% remained at 2-hr and 4-hr time points, respectively, in wild-type W303a cells (Figure 7B). By contrast, the eaf3-W84A,W88A yng2-phd∆ double mutant was defective in DSB repair, as 59% and 59% of HO cut fragments remained at 2-hr and 4-hr time points, respectively. The differences between wild type and the eaf3-W84A,W88A yng2-phd∆ mutant are significant at 2-hr and 4-hr time points, with P-values of 0.001 and 0.002, respectively. The wild-type W303α shows a similar trend as W303a cells, with 32 and 39% of HO cut fragments at 2-hr and 4-hr time points, respectively. H3K4R mutant shows 44 and 47% of HO-cut at 2-hr and 4-hr time points, with P-values of 0.012 and 0.1, respectively. Although the difference at the 4-hr time point was not significant, we repeated the experiment three times and observed a similar trend each time. Given that H3K4R, K36R and the set1set2 null mutants were even more sensitive to bleomycin than the H3K4R mutant, we also tested the DSB recovery in the H3K4R, K36R and set1set2 null mutants. As shown in Figure S2C, both H3K4R, K36R and set1set2 null mutants were defective in DSB repair, as 57 and 52%, and 54 and 56% of HO-cut remained in 2-hr and 4-hr time points in these two strains, respectively. The differences between wild type and these mutants are statistically significant (H3K4R, K36R: P-values of 0.004 and 0.03 at 2-hr and 4-hr time points, respectively; set1 set2: P-values of 0.007 and 0.01 at 2-hr and 4-hr time points, respectively). Therefore, we conclude that the delayed H2A-S129p in the eaf3-W84A,W88A yng2-phd∆ double mutant and H3K4R mutant cells was due to the defective DSB repair.
Genome-wide overlap of H3K4 methylation and Esa1 binding sites
To further explore the correlation between H3K4 methylation and NuA4 activity, we utilized the previously published datasets and analyzed genome-wide Esa1 binding sites related to H3K4 methylation (Kirmizis et al. 2007; Guillemette et al. 2011; Venters et al. 2011). When the binding occupancy of Esa1 was set to 1.5-fold above the background, the percentages of genes exhibiting H3K4me2 and H3K4me3 enrichment at the same region were 97 and 62%, respectively. When the binding occupancy of Esa1 was set to twofold, the percentages of genes exhibiting H3K4me2 and H3K4me3 enrichment increased to 97.6 and 64%, respectively. Therefore, the binding occupancy of Esa1 is highly correlated with the enrichment of H3K4 methylation, supporting the notion that H3K4 methylation plays an important role in activating the NuA4 complex.
Discussion
Our studies of the NuA4 complex have revealed the key function of the PHD and the chromodomain, which act in a combinatorial way to recruit and regulate the NuA4 complexes at the DSB site (Jenuwein and Allis 2001; Rando 2012). This conclusion is supported by several lines of evidence. First, mutations in either the PHD or chromodomain result in no change of phenotypes in terms of growth or DNA repair, whereas combined mutations in both the PHD and chromodomain show slow growth and sensitivity to bleomycin or phleomycin treatment (Figures 1 and 2). Second, the histone mutants, H3K4R and H3K36R, which mimic the unmethylated form of H3K4 and H3K36, also cause defects in DSB repair (Figure 3A). Third, cells harboring set1Δ or set2Δ mutations are sensitive to bleomycin, with high doses of bleomycin (10 mU/ml) showing more significant phenotypes (Figure 3C). In addition, the set1Δ set2Δ double mutant displays more sensitivity to bleomycin than mutants with either set1Δ or set2Δ alone (Figure 3C). Fourth, H3K4me2, H3K4me3, and H3K36me2 are induced at the HO DSB site, providing binding sites for the PHD and chromodomain of NuA4 (Figure 4 F–H). Fifth, the PHD/chromodomain double mutant that abolishes their interaction with methylated H3K4 and H3K36 is able to recruit Esa1 and yng2-phdΔ to the HO DSB site, albeit less abundantly and in a delayed manner, according to the ChIP analysis (Figure 5, C and D). However, the abundance of H4K12ac is dramatically reduced at the HO DSB site. This suggests that the combined interactions of the PHD and chromodomain with methylated H3K4 and H3K36 not only strengthen the affinity of NuA4 for binding with nucleosomes, but also regulate NuA4 acetylation activity. The PHD and chromodomain double mutant is defective in DSB repair as judged by Southern blot (Figure 7). Consistent with our results, Gingsberg et al. 2014 has also shown that both H3K4 and H3K36 methylation are important for NuA4 recruitment to nucleosomes. This multivalent interaction system provides specificity that robustly targets enzyme complexes to specific loci to regulate biological processes. These multivalent interactions are also observed in the assembly of silent chromatin and in several chromatin remodeling complexes (Liaw and Lustig 2006; Li et al. 2007; Eustermann et al. 2011; Ali et al. 2012; Qiu et al. 2012; Rando 2012; Rothbart et al. 2013).
Specific histone modifications are induced at the DSB sites
Previous studies have shown that H2A-S129p, H4K5ac, H4K8ac, H4K12ac, H4K16ac, H3K4me3, H3K36me2, and H3K36me3 are specifically induced at DSB sites (Downs et al. 2004; Shroff et al. 2004; Faucher and Wellinger 2010; Jha and Strahl 2014). In this study, we further found that H3K4me2 is also induced at the DSB site. By in silico analysis, we found that H3K4me2 is highly correlated with the occupancy of Esa1, reaching 97% correlation. These findings support the notion that H3K4me2 regulates NuA4 activity. In addition, given that H3K4R, H3K36R, set1Δ, and set2Δ mutants are sensitive to bleomycin, and that Set1 and Set2 are recruited to the DSB site, it suggests that the induction of both H3K4 and H3K36 methylation at the DSB site is specific. The methylated H3K4 and H3K36 provide the binding sites for NuA4 through its PHD and chromodomain. These combined domain–histone interactions not only recruit NuA4 robustly, but also activate NuA4 activity at the DSB sites. In support of our findings, several recent publications have also shown that H3K4 and H3K36 methylation play important roles in recruiting and regulating NuA4 activity in DSB repair (Faucher and Wellinger 2010; Ginsburg et al. 2014; Jha and Strahl 2014).
This mechanism can also explain what happens during gene transcription. H3K4 and H3K36 methylation occur at the promoter region, as well as the coding region of actively transcribed genes (Reid et al. 2000; Robert et al. 2004; Liu et al. 2005; Pokholok et al. 2005; Ginsburg et al. 2009; Rando and Winston 2012). Our in silico analysis showed a high correlation between H3K4 methylation and Esa1 occupancy, suggesting that H3K4 methylation can regulate NuA4 activity during transcription. A recent study has also demonstrated that H3K4 and H3K36 methylation is required for NuA4 recruitment to nucleosomes (Ginsburg et al. 2014). In further support of this notion, the eaf3-W84A,W88A yng2-phd∆ mutant that is unable to interact with methylated H3K4 and H3K36 shows decreased transcription of ribosomal genes. Since ribosome synthesis utilizes ∼90% of the energy in the cell, the defective transcription of ribosomal genes could explain the slow growth phenotype in the eaf3-W84A,W88A yng2-phd∆ mutant (Reid et al. 2000; Zaman et al. 2008).
Multivalent domain–histone interactions strengthen the binding affinity and regulate NuA4 activity at the DSB site
The NuA4 complex contains the actin-related, PHD, chromo, and YEATS domains that interact with specifically modified histones (Doyon and Cote 2004). It appears that interactions between the actin-related domain and H2A-S129p, the PHD, and methylated H3K4, and the chromodomain and methylated H3K36 are the most important interactions in regulating NuA4 activity at the HO cleavage site. We found that combined mutations in the PHD and chromodomain of the NuA4 complex are sensitive to bleomycin-induced DSBs. The H2A-S129p activation is delayed and prolonged at the HO cleavage site in the eaf3-W84A,W88A yng2-phd∆ double mutant cells. This phenotype could be due to the altered chromatin structure of DSBs, thus impairing DSB repair. In support of this notion, H4K12ac is dramatically reduced at the HO cleavage site, thus altering end resections or histone evictions.
Given that yng2-phdΔ was still able to recruit to the DSB site, this implies that a single interaction between H2A-S129p and the actin-related domain is the major force in recruiting the NuA4 complex to the DSB site. In addition, previous publications have shown that the NuA4 complex interacts with nucleosomes through the N terminus of Epl1 and Epl1 bridges Yng2 and Esa1 together (Nourani et al. 2001; Boudreault et al. 2003; Selleck et al. 2005). EM analysis also shows that the N terminus of Epl1 and N terminus of Yng2 (PHD domain is not included) contact nucleosomes, whereas Esa1 has no direct contact with nucleosomes (Chittuluru et al. 2011). Therefore, Yng2-phdΔ could also be recruited to the DSB site through Epl1 or through direct contact with nucleosomes. However, to gain a robust recruitment of the NuA4 complex, it appears that it also requires the interaction between the PHD and chromodomain with methylated H3K4 and H3K36. This point of view is also supported by the observation that NuA4 binds methylated H3K4 and H3K36 peptides greater than unmodified H3K4 and H3K36 (Ginsburg et al. 2014). In addition, the binding of NuA4 to nucleosomes isolated from the set1Δset2Δ strain is greatly reduced, compared with nucleosomes isolated from wild type or strains lacking only set1 or set2 (Ginsburg et al. 2014). However, multivalent interactions between the PHD and methylated H3K4, and between the chromodomain and methylated H3K36, can strengthen the recruitment process. Furthermore, these interactions play an important role in activating NuA4 complex. Interestingly, the H3K4R mutant shows a very similar phenotype to the eaf3-W84A,W88A yng2-phd∆ double mutant. However, Yng2 is unable to recruit to the HO-cleavage site in the H3K4R mutant. This indicates that H3K4R can significantly alter the chromatin structure, thus prohibiting the access of the NuA4 complex to the HO-cleavage site. Further studies will be performed to investigate this phenomenon.
Although mutations in NuA4 can also affect transcription, especially the transcription of ribosomal proteins and heat shock proteins (Galarneau et al. 2000; Reid et al. 2000), we think that the defective DSB repair phenotype observed in the PHD and chromodomain double mutant are specific, not indirect, consequences of a change in the transcription of DNA repair or checkpoint genes. First, previous publications have shown that cells harboring esa1 or yng2 mutants do not show significant transcription variation in genes involved in DNA damage checkpoints or repair (Choy et al. 2001; Choy and Kron 2002). Second, cells harboring H4K5, K8, K12, and K16 mutants, a mutated version of H4 that cannot be acetylated, show that none of the genes involved in the DNA DSB repair pathway have substantially decreased expression (Bird et al. 2002). Third, our ChIP analysis has specifically shown that H2A-S129p, H3K4, and H3K36 methylation was induced at the DSB site, and that Esa1 and yng2-phdΔ were recruited to the DSB site, albeit at reduced levels and with some delay (Figure 5, C and D). Therefore, we conclude that multivalent interactions between domains and specifically modified histones play important roles in the assembly and activation of the NuA4 complex at the DSB site, thus facilitating DSB repair.
In conclusion, we propose a model in which DSBs can induce H2A-S129p, which then recruits NuA4 to the DSB site. However, this single interaction is not sufficient to induce a significant amount of histone H4 acetylation adjacent to the DSB site, as it cannot fully activate NuA4 complex at the DSB site. This implies that the PHD and chromodomain of NuA4 are required to strengthen the interaction and fully activate the NuA4 acetylation activity through the combined interactions between the two domains and methylated histone H3K4 and H3K36. These events allow the acetylation of H4 around the DSBs, leading to open chromatin and allowing access to repair proteins that efficiently repair the DSBs.
Acknowledgments
We thank James Haber, Mary Ann Osley, Chuang-Rung Chang, Cheng-Fu Kao, Jun-Yi Leu, Fang-Jen Lee, Shu-Chun Teng (Taiwan Yeast Resources Center), and Huang-Mo Sung for providing strains and technical support; Wu-Chou Su and Ming-Daw Tsai for their generous support; and Shu-Chun Teng and Paul Steed for critical reading of the manuscript. This work was supported by the Taiwan Ministry of Science and Technology (NSC100-2311-B-006-001 and NSC100-2320-B-006-016 to H.L. and NSC-102-2628-B-006-013-MY3 to W.-P.S.); National Cheng Kung University Hospital grant (10408001); and the Taiwan Ministry of Education and the Aim for the Top University Project to the National Cheng Kung University (D100-38B10 to H.L.). Funding for the open access fee was provided by the Ministry of Science and Technology.
Footnotes
Communicating editor: A. Hinnebusch
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.184432/-/DC1.
Literature Cited
- Ali M., Yan K., Lalonde M. E., Degerny C., Rothbart S. B., et al. , 2012. Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J. Mol. Biol. 424: 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard S., Utley R. T., Savard J., Clarke A., Grant P., et al. , 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18: 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Shen X., 2007. Chromatin remodeling in DNA double-strand break repair. Curr. Opin. Genet. Dev. 17: 126–131. [DOI] [PubMed] [Google Scholar]
- Bird A. W., Yu D. Y., Pray-Grant M. G., Qiu Q., Harmon K. E., et al. , 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415. [DOI] [PubMed] [Google Scholar]
- Borde V., Robine N., Lin W., Bonfils S., Geli V., et al. , 2009. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreault A. A., Cronier D., Selleck W., Lacoste N., Utley R. T., et al. , 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17: 1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai B., Huang J., Cairns B. R., Laurent B. C., 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19: 1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittuluru J. R., Chaban Y., Monnet-Saksouk J., Carrozza M. J., Sapountzi V., et al. , 2011. Structure and nucleosome interaction of the yeast NuA4 and Piccolo-NuA4 histone acetyltransferase complexes. Nat. Struct. Mol. Biol. 18: 1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy J. S., Kron S. J., 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 22: 8215–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy J. S., Tobe B. T., Huh J. H., Kron S. J., 2001. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 276: 43653–43662. [DOI] [PubMed] [Google Scholar]
- Downs J. A., Allard S., Jobin-Robitaille O., Javaheri A., Auger A., et al. , 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16: 979–990. [DOI] [PubMed] [Google Scholar]
- Doyon Y., Cote J., 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14: 147–154. [DOI] [PubMed] [Google Scholar]
- Eustermann S., Yang J. C., Law M. J., Amos R., Chapman L. M., et al. , 2011. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 18: 777–782. [DOI] [PubMed] [Google Scholar]
- Faucher D., Wellinger R. J., 2010. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 6: e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fnu S., Williamson E. A., De Haro L. P., Brenneman M., Wray J., et al. , 2011. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 108: 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau L., Nourani A., Boudreault A. A., Zhang Y., Heliot L., et al. , 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5: 927–937. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., et al. , 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., et al. , 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- Ginsburg D. S., Govind C. K., Hinnebusch A. G., 2009. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol. Cell. Biol. 29: 6473–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg D. S., Anlembom T. E., Wang J., Patel S. R., Li B., et al. , 2014. NuA4 links methylation of histone H3 lysines 4 and 36 to acetylation of histones H4 and H3. J. Biol. Chem. 289: 32656–32670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O., Karuman P., Jones D. R., Ivanov D., Cha J., et al. , 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114: 99–111. [DOI] [PubMed] [Google Scholar]
- Guillemette B., Drogaris P., Lin H. H., Armstrong H., Hiragami-Hamada K., et al. , 2011. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 7: e1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., 2002. Uses and abuses of HO endonuclease. Methods Enzymol. 350: 141–164. [DOI] [PubMed] [Google Scholar]
- Huang J., Tan S., 2013. Piccolo NuA4-catalyzed acetylation of nucleosomal histones: critical roles of an Esa1 Tudor/chromo barrel loop and an Epl1 enhancer of polycomb A (EPcA) basic region. Mol. Cell. Biol. 33: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D., 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jha D. K., Strahl B. D., 2014. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat. Commun. 5: 3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A. A., Struhl K., 2005. Eaf3 chromodomain interaction with methylated H3–K36 links histone deacetylation to Pol II elongation. Mol. Cell 20: 971–978. [DOI] [PubMed] [Google Scholar]
- Kirmizis A., Santos-Rosa H., Penkett C. J., Singer M. A., Vermeulen M., et al. , 2007. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449: 928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Gogol M., Carey M., Lee D., Seidel C., et al. , 2007. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316: 1050–1054. [DOI] [PubMed] [Google Scholar]
- Li, H., S. Ilin., W. Wang, E. M. Duncan, J. Wysocka et al., 2006 Molecular basis for site-specific readout of H3 K4 trimethylation by the BPTF PHD finger. Nature 442: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wen H., Xi Y., Tanaka K., Wang H., et al. , 2014. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 159: 558–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw H., Lustig A. J., 2006. Sir3 C-terminal domain involvement in the initiation and spreading of heterochromatin. Mol. Cell. Biol. 26: 7616–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. L., Kaplan T., Kim M., Buratowski S., Schreiber S. L., et al. , 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Meijer M., Lees-Miller S. P., Riabowol K., Young D., 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20: 3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al, 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Morrison A. J., Shen X., 2005. DNA repair in the context of chromatin. Cell Cycle 4: 568–571. [PubMed] [Google Scholar]
- Morrison A. J., Shen X., 2009. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 10: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A. J., Highland J., Krogan N. J., Arbel-Eden A., Greenblatt J. F., et al. , 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775. [DOI] [PubMed] [Google Scholar]
- Nakanishi S., Lee J. S., Gardner K. E., Gardner J. M., Takahashi Y. H., et al. , 2009. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 186: 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani A., Doyon Y., Utley R. T., Allard S., Lane W. S., et al. , 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21: 7629–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Krebs J. E., Peterson C. L., 2006. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20: 2437–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., et al. , 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Liu L., Zhao C., Han C., Li F., et al. , 2012. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 26: 1376–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando O. J., 2012. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr. Opin. Genet. Dev. 22: 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando O. J., Winston F., 2012. Chromatin and transcription in yeast. Genetics 190: 351–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J. L., Iyer V. R., Brown P. O., Struhl K., 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6: 1297–1307. [DOI] [PubMed] [Google Scholar]
- Robert F., Pokholok D. K., Hannett N. M., Rinaldi N. J., Chandy M., et al. , 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart S. B., Dickson B. M., Ong M. S., Krajewski K., Houliston S., et al. , 2013. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 27: 1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J. M., Wang A. Y., Kobor M. S., 2009. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem. Cell Biol. 87: 65–75. [DOI] [PubMed] [Google Scholar]
- Schulze J. M., Wang A. Y., Kobor M. S., 2010. Reading chromatin: insights from yeast into YEATS domain structure and function. Epigenetics 5: 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck W., Fortin I., Sermwittayawong D., Cote J., Tan S., 2005. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 25: 5535–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Kachirskaia I., Walter K. L., Kuo J. H., Lake A., et al. , 2007. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282: 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E. Y., Hong S. J., Oum J. H., Yanez Y., Zhang Y., et al. , 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 27: 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R., Arbel-Eden A., Pilch D., Ira G., Bonner W. M., et al. , 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14: 1703–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E., 2006. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 408: 416–429. [DOI] [PubMed] [Google Scholar]
- van Attikum H., Gasser S. M., 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19: 207–217. [DOI] [PubMed] [Google Scholar]
- Venters B. J., Wachi S., Mavrich T. N., Andersen B. E., Jena P., et al. , 2011. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell 41: 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. Y., Schulze J. M., Skordalakes E., Gin J. W., Berger J. M., et al. , 2009. Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation. Proc. Natl. Acad. Sci. USA 106: 21573–21578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Cui G., Botuyan M. V., Mer G., 2008. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure 16: 1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Zhao X., Broach J. R., 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42: 27–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. Figure S1 contains supplemental data in support of Figure 3. Figure S2 contains supplemental data in support of Figure 7.
Figure 3.
H3K4 methylation and H3K36 methylation are important for DSB repair. (A) The H3K4R and H3K36R mutants are sensitive to DNA damage treatment. The hht1hhf1 hht2hhf2 null mutant carrying plasmids containing wild-type (H3) and mutant histone H3 (H3K4R, H3K36R, H3K4R,K36R) were spotted on agar plates and treated with DNA damaging agents as indicated. (B) The set1, set2, and set1 set2 null mutants can be complemented with plasmids carrying wild-type SET1 or SET2 plasmids. (C) Strains carrying the wild-type SET1 SET2, set1, set2, or set1 set2 null mutations were spotted on agar plates and treated with DNA damaging agents as indicated. (D) Strains carrying the wild-type H3, H3K4R, H3K4A, or set1Δ mutations were spotted on agar plates and treated with DNA damaging agents as indicated.
Figure 7.
The eaf3-W84A,W88A yng2-phdΔ double mutant and H3K4R mutant cells are defective in DSB repair. (A, top) Diagram of the MAT locus and SET1 locus analyzed is shown. The HO cut site and StyI restriction sites are indicated. (Bottom) Wild-type W303a, W303α, eaf3-W84A,W88A yng2-phdΔ double mutant, and H3K4R mutant cells were induced to generate a HO DSB site, following a protocol similar to the ChIP experiment shown in Figure 4. Genomic DNA was extracted at the time points shown, digested with StyI, and separated on 1.2% agarose gel. Southern blot was hybridized with a DIG-labeled MAT-specific probe and a DIG-labeled SET2-specific probe as a loading control. The size of MATα is larger than that of MATa because MATα lacks a StyI site in the Yα sequence. (B) The percentage of HO cut was determined by the amount of HO cut fragments relative to the SET2 loading control fragments, normalized to the value obtained from an uninduced (0 hr) sample. The asterisk (*) indicates that the difference between wild type and mutants was statistically significant with P-value <0.02. (Student’s t-test, eaf3-W84A,W88A yng2-phdΔ double mutant: P-values 0.001 and 0.002 at 2-hr and 4-hr time points, respectively; and H3K4R: P-value 0.012 at 2-hr time point).