Abstract
Background Roots are essential organs for higher plants. They provide the plant with nutrients and water, anchor the plant in the soil, and can serve as energy storage organs. One remarkable feature of roots is that they are able to adjust their growth to changing environments. This adjustment is possible through mechanisms that modulate a diverse set of root traits such as growth rate, diameter, growth direction and lateral root formation. The basis of these traits and their modulation are at the cellular level, where a multitude of genes and gene networks precisely regulate development in time and space and tune it to environmental conditions.
Scope This review first describes the root system and then presents fundamental work that has shed light on the basic regulatory principles of root growth and development. It then considers emerging complexities and how they have been addressed using systems-biology approaches, and then describes and argues for a systems-genetics approach. For reasons of simplicity and conciseness, this review is mostly limited to work from the model plant Arabidopsis thaliana, in which much of the research in root growth regulation at the molecular level has been conducted.
Conclusions While forward genetic approaches have identified key regulators and genetic pathways, systems-biology approaches have been successful in shedding light on complex biological processes, for instance molecular mechanisms involving the quantitative interaction of several molecular components, or the interaction of large numbers of genes. However, there are significant limitations in many of these methods for capturing dynamic processes, as well as relating these processes to genotypic and phenotypic variation. The emerging field of systems genetics promises to overcome some of these limitations by linking genotypes to complex phenotypic and molecular data using approaches from different fields, such as genetics, genomics, systems biology and phenomics.
Keywords: Root, root development, root growth, genetics, root patterning, systems biology, modelling, systems genetics, networks, Arabidopsis thaliana
INTRODUCTION
Mostly hidden from view, below the ground, roots constitute an essential organ for higher plants. They provide the plant with nutrients and water, anchor the plant in the soil, and can serve as energy storage organs. The ability of roots to acquire minerals and water from the soil determines, to a large extent, the ability of a plant to grow (Zobel, 1986). The root system is frequently exposed to a multitude of environmental constraints (such as drought, extreme temperature, lack of essential nutrients, exposure to toxic minerals, and soil compaction) and it is one remarkable feature of roots that they are able to adjust their growth to such changing environments (Malamy, 2005). This adjustment is realized by the tuning of a diverse set of root traits such as growth rate, diameter, growth direction and lateral root (LR) formation. These traits shape the root system architecture (RSA), which is the spatial configuration of roots in the soil. The RSA shows a significant degree of plasticity in response to the heterogeneous distribution of soil resources and variations of soil conditions (Lynch, 1995a). Understanding which regulatory processes and underlying genetic components regulate root growth, and thereby RSA, is not only a fascinating question of basic biology but also key to breed and engineer better performing plants. However, it has become clear that root growth regulation is a highly complicated process and is controlled at many different levels by complex actions of gene networks in both time and space. In this review we will describe fundamental work that has shed light on the basic principles in root growth and development, will proceed with the emerging complexities and how they have initially been addressed, and finally describe and argue for a systems-genetics approach. Throughout this review, due to reasons of simplicity and extent, we will mostly limit ourselves to the work in the model plant arabidopsis (Arabidopsis thaliana), for which much of the research in root growth regulation at the molecular level has been conducted.
THE ROOT SYSTEM
The distribution and spatial configuration of the roots in the soil – the RSA – is a fundamentally important physiological parameter for plants. It is determined by the shapes, sizes and three-dimensional distribution of roots, as well as by the branching arrangement of the primary and higher order roots. Other important factors are the root hair density and length; increases in length and/or number of root hairs can dramatically increase the root–soil interface. Essentially, RSA determines the volume of soil that is explored by the roots (given by the depth and breadth of the extent of the root system) and the root surface area that interfaces with the soil. The volume of soil that is explored determines the zone from which mobile nutrients can be acquired, and the root surface area determines the zone from which immobile nutrients can be acquired (Bray, 1954). RSA varies between species and also displays significant natural variation within a species (Hochholdinger and Tuberosa, 2009; Giehl et al., 2012; Gruber et al., 2013; Rosas et al., 2013).
In dicots such as arabidopsis, the root system consists of a single primary root (PR) of embryonic origin, which often remains active throughout the plant’s life cycle and can develop several orders of LRs (Osmont et al., 2007), and post-embryonically derived junction roots formed at the collet, the junction between the hypocotyl and root (Falasca and Altamura, 2003). In monocots such as maize (Zea mays) and rice (Oryza sativa), the root system contains embryonic primary and seminal roots (SRs), and post-embryonic shoot-borne roots and LRs (Hochholdinger and Tuberosa, 2009). The PRs are the first root to emerge in both dicots and monocots, and are derived from embryonically formed meristematic tissue at the root tip called the root apical meristem (RAM). The RAM consists of a basal stem cell pool around an organizing centre called the quiescent centre (QC; Dolan et al., 1993). Root hairs are formed in the differentiation zone of the root from specialized epidermal cells called trichoblasts, and vastly increase the root surface area and contribute to the acquisition of immobile nutrients (Bates and Lynch, 1996). LRs are the most important root class for the RSA. In arabidopsis and other dicots, LRs are derived from pericycle cells adjacent to the xylem tissue (xylem-pole pericycle) (Dubrovsky et al., 2000). In monocots such as maize and rice, they are initiated in the phloem-pole pericycle, and cells derived from the pericycle and endodermis contribute to the LRs (De Smet et al., 2006).
THE GENETIC BASIS OF ROOT GROWTH AND DEVELOPMENT
The root system and RSA are the results of continuous root growth and development. Our understanding of root growth regulation and development, and their consequence, RSA, is most advanced in the model plant arabidopsis. The available genome sequence (Arabidopsis Genome Initiative, 2000) together with the short generation time made it perfectly suited for genetic approaches. Moreover, the ease of genetic and molecular manipulation and the steadily growing number of genetic, biochemical and computational materials in the arabidopsis research community have resulted in making arabidopsis the most thoroughly investigated plant species. Root research in this model has profited from the root’s optical transparency, its small diameter and simplicity of organization, the rigid order of cell divisions and unusually invariant cellular lineages, and, finally, the ability to grow it easily in large numbers on sterile agar plates. Consequently, the arabidopsis root has been used to address not only questions related to root physiology but also basic questions in developmental and cell biology. In this section, we review genetic and molecular processes underlying root development, and then highlight how these mechanisms impact root growth. We note that in this section, we mainly restrict ourselves to highlight seminal work identifying key genes and mechanisms with classical approaches and will review more recent work that used systems-biology approaches for the same purpose in a later section.
Cell fate specification in the root
The function of the root is highly dependent on its cellular architecture, with precisely defined cell types radially arranged in cell files around the central axis of the root. A key question for root biology was therefore how this precise patterning is regulated. Landmark studies conducted clonal analyses to reveal the origin of the cell files, identifying initial cells that give rise to specific cell lineages (Dolan et al., 1994; Scheres et al., 1994). While the regularity of cell divisions (Dolan et al., 1993; Baum and Rost, 1996) and the rigid cellular organization were suggestive of lineage-based determinants strictly controlling cell fate (van den Berg et al., 1995; Kidner et al., 2000), laser ablation studies showed that positional cues are critical for cell fate determination (van den Berg et al., 1995). In particular, the ablation of specific cells enabled neighbouring cells from another cell file to occupy the freed space and adopt the developmental fate of the removed cells, as revealed by the expression of tissue-specific marker genes (van den Berg et al., 1995). These studies highlighted one of many examples of the developmental plasticity of root cells.
Ground tissue.
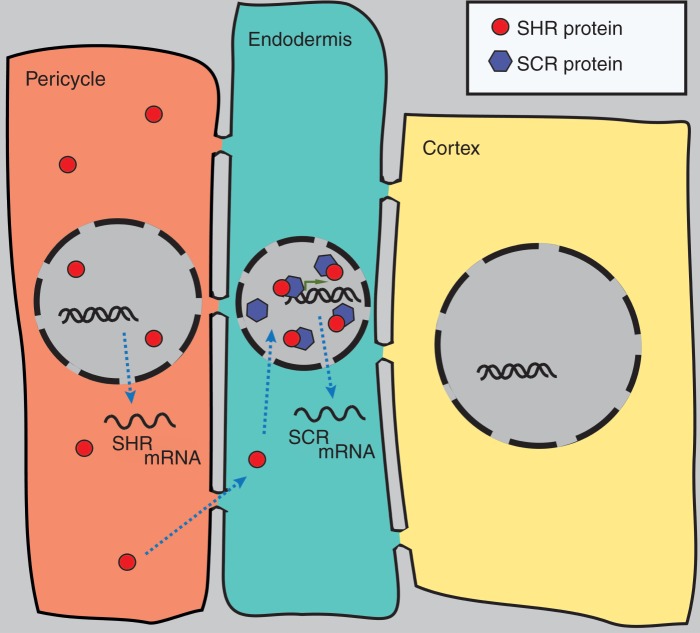
Cell fate specification is largely governed by the activity of key transcription factors. For instance, roots of the GRAS family transcription factor mutants shortroot (shr) and scarecrow (scr) contain only one ground tissue layer that usually consists of the endodermis and cortex cell layers (Benfey et al., 1993; Scheres et al., 1995). While the shortroot ground tissue layer is entirely lacking endodermal differentiation markers (Benfey et al., 1993), the ground tissue in scarecrow has features of both cortex and endodermis (Scheres et al., 1995; Di Laurenzio et al., 1996). Subsequent studies indicated that SHR as well as SCR are both necessary for the periclinal cell division of the daughter cell of the cortex endodermis initial (CEI) that gives rise to both cell types of ground tissue initial daughter cells (Scheres et al., 1995). However, only SHR is required for the specification of endodermal cell fate (Benfey et al., 1993; Helariutta et al., 2000; Nakajima et al., 2001). Intriguingly, SHR mRNA is not expressed in the ground tissue but only in the stele (pericycle and vascular tissues). This non-cell-autonomous function of SHR is due to the movement of the SHR protein from the stele to the ground tissue (Nakajima et al., 2001). There, SHR directly induces the expression of SCR (Levesque et al., 2006). In turn, SCR sequesters SHR to the endodermal cell nucleus, thereby preventing SHR movement (Fig. 1) and further upregulating SCR expression, thus giving rise to a positive feedback loop that ensures the appropriate timing and location of the cell division (Cui et al., 2007). Further studies, using tools of systems biology, identified additional key genes involved in the SHR/SCR regulatory circuit and eventually led to a mathematical model of this patterning process (see later). Apart from these efforts, other ground tissue patterning genes have been identified. For instance, SCHIZORHIZA (SCZ), a gene that belongs to the heat shock factor family of transcription factors, is required for the establishment of ground tissue stem cells in the embryonic root (Pernas et al., 2010). The scz mutant develops supernumerary layers of ground tissue and root hairs originating from the sub-epidermal layer (Mylona et al., 2002), implying a role for SCZ in suppressing both extranumerary periclinal cell divisions and epidermal cell fate in CEI daughter cells (Mylona et al., 2002).
Fig. 1.
Model for SHR/SCR activity in ground tissue patterning. SHR mRNA and protein are expressed in the pericycle. SHR protein moves to the endodermis, where it increases SCR expression and is in turn sequestered by SCR to the nucleus. This constitutes a positive feedback loop, reinforcing the inhibition of further cell–cell movement of SHR.
Epidermis.
Cell fate specification of cells within one tissue has been studied intensely in the epidermis – a root tissue consisting of two cell types, hair-forming trichoblasts and hairless atrichoblasts. The hair cell files are located adjacent to the anticlinal cell wall between two cortical cell files (Fig. 2). Each hairless cell file is located above a single cortical cell file, thus not spanning anticlinal cell boundaries. The epidermal cell pattern is already established during embryogenesis (Costa and Dolan, 2003). Both cell types are distinguishable as trichoblasts and atrichoblasts (Fig. 3A) long before the emergence of root hairs (Dolan et al., 1994; Galway et al., 1994; Berger et al., 1998a). Laser ablation experiments showed that cell fate determination of hair-bearing and hairless cells is dependent on position-dependent cell–cell communication, rather than lineage-related determinants (Berger et al., 1998b). Surgical experiments in radish (Bünning, 1951), another member of the Brassicaceae family, pointed to the hair cell fate being the default fate of epidermal cells.
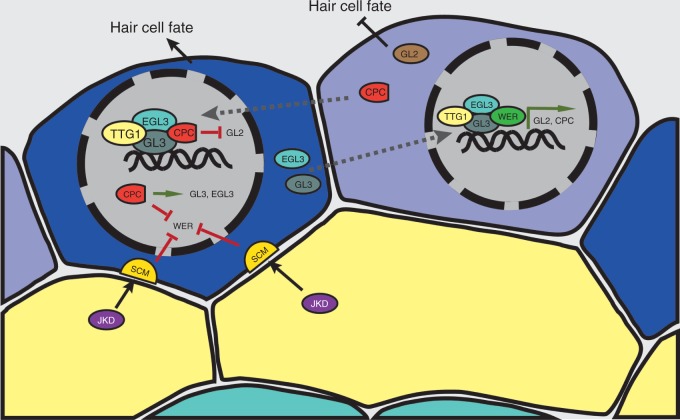
Fig. 2.
Model for epidermis cell fate specification. The TTG1/GL3/EGL3/WER complex directly activates expression of GL2 and CPC in atrichoblast cells (in light blue). CPC moves circumferentially to neighbouring cells, there outcompetes WER in binding to TTG1/GL3/EGL3 and results in the downregulation of GL2 and promotion of hair cell identity (trichoblast in dark blue). JKD and SCM provide positional cues from the underlying cortex cell layer by repressing WER and tipping the balance in favour of trichoblast cell fate.
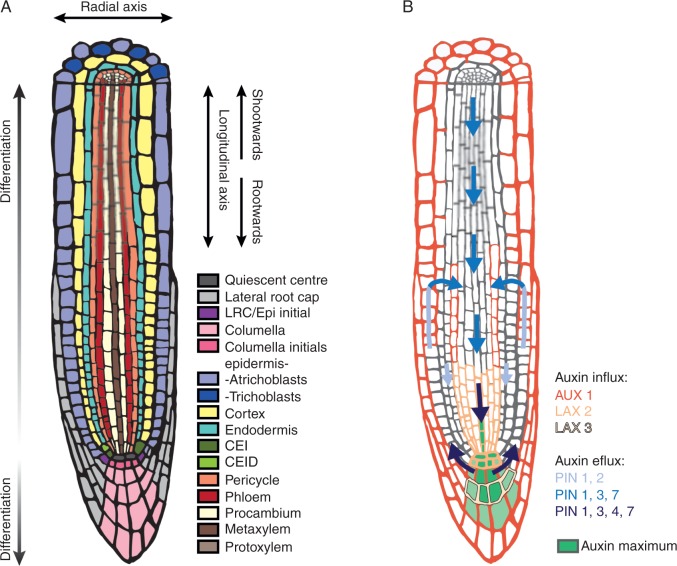
Fig. 3.
Root apical–basal patterning. (A) Schematic of the root apical patterning. (B) A simplified auxin reflux loop in the root apex. The direction of the auxin flow is mediated by PIN efflux carriers.
Genetic screens provided insight into the specification of hair vs. hairless cells, and numerous mutants with impaired root epidermal cell fate determination have been discovered. These include genes required for hairless cell fate such as WEREWOLF (WER), TRANSPARENT TESTA GLABRA (TTG1) and GLABRA2 (GL2) (Galway et al., 1994; Rerie et al., 1994; Masucci et al., 1996; Lee and Schiefelbein, 1999) and the redundantly acting GLABRA3 (GL3) and ENHANCER OF GLABRA 3 (EGL3) (Bernhardt et al., 2003). Single loss-of-function mutants of the last two genes show only slight increases in root hair production, while gl3 egl3 double mutants form ectopic hair cells (Bernhardt et al., 2003). In contrast to hairless cell fate-specifying genes, CAPRICE (CPC) promotes hair cell fate specification, cpc having irregularly distributed and dramatically reduced numbers of root hairs (Wada et al., 1997).
In atrichoblast cells, the hairless cell fate-promoting genes (WER, GL3, EGL3 and TTG1) act upstream of GL2 and CPC (Hung et al., 1998; Lee and Schiefelbein, 1999; Bernhardt et al., 2003). Both GL3 and EGL3 directly interact with WER (Bernhardt et al., 2003) and TTG1 in yeast two-hybrid (Y2H) assays (Payne et al., 2000; Esch et al., 2003; Zhang et al., 2003). This evidence of protein–protein interaction suggested an activator complex consisting of WER, GL3, EGL3 and TTG1 (Zhang et al., 2003). This complex binds to the promoter regions of GL2 and CPC, and directly activates their transcription (Koshino-Kimura et al., 2005). While GL2 specifies hairless cell fate (Masucci et al., 1996) and therefore acts cell autonomously, CPC is required for hair cell fate determination (Wada et al., 1997) and acts non-autonomously (Wada et al., 2002) by moving circumferentially to neighbouring cells (Wada et al., 2002; Kurata et al., 2005). Since CPC interacts with both GL3 and EGL3 in yeast cells (Bernhardt et al., 2003), a competition model between the R2R3-type MYB transcription factor WER and the R3-type MYB transcription factor CPC was suggested (Lee and Schiefelbein, 1999) and later on experimentally confirmed (Bernhardt et al., 2005; Tominaga et al., 2007). While TTG1/GL3/EGL3 interact in hairless cells with WER, interaction with the mobile CPC protein that accumulates in hair cells generates an inactive complex (Bernhardt et al., 2005). This leads to the downregulation of GL2 and results in hair cell fate. Moreover accumulation of CPC leads to downregulation of WER and upregulation of GL3 and EGL3 (Bernhardt et al., 2005), respectively. In turn, GL3 and EGL3 move from root hair cells to neighbouring hairless cells and facilitate the assembly of the active transcription complex TTG1/GL3/EGL3/WER which leads to GL2 upregulation and reinforces the hairless cell fate (Bernhardt et al., 2005). Overall, this complex regulation constitutes a bilateral inhibition feedback loop that leads to a high stability of epidermal patterning (Savage et al., 2008). Two genes are key factors for providing the initial positional information for the epidermal cell patterning from the underlying cortex cell layer. These genes encode a receptor-like kinase SCRAMBLED (SCM; Kwak and Schiefelbein, 2008; Schiefelbein et al., 2009) and a zinc finger protein JACKDAW (JKD; Hassan et al., 2010). SCM is epistatic to JKD, and both single mutants show ectopic expression of WER, GL2 and CPC proteins (Hassan et al., 2010). Furthermore, systems-biology approaches have identified further components for epidermal patterning (see later).
Stele.
Of all tissues in the root, the stele, a composite tissue that consists of several cell types and contains the vascular bundles, displays the highest degree of complexity during patterning and differentiation. This enables drastic terminal differentiation events that lead to highly specialized vasculature cells, such as dead vessel elements or sieve elements lacking a nucleus. Many important molecular pathways and regulators that are involved in stele patterning and differentiation of the vasculature have been identified. These involve a variety of factors and pathways such as hormones, transcription factors and microRNAs (miRNAs; a very comprehensive summary of the current state in the field is given in Lucas et al., 2013). Here we highlight several key patterning events in the stele.
Different cell types in the stele are generated through a set of asymmetric cell divisions. wooden leg 1/cytokinin response 1 (wol/cre1) mutants contain a stele that has a reduced number of cell files (Scheres et al., 1995) since these plants are impaired in executing a number of periclinal asymmetric cell divisions. This results in the specification of all vascular cells in wol/cre1 mutants as protoxylem cells (Mähönen et al., 2000). The reduced number of vascular cylinder cells can be phenocopied by decreasing cytokinin levels specifically in the procambium using a tissue-specifically expressed version of the CYTOKININ OXIDASE2-encoded enzyme (Mähönen et al., 2006). Consistent with that, a suppressor screen for the wol/cre1 phenotype identified ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6), a negative regulator of cytokinin signalling, supporting the key role of cytokinin signalling in phloem and metaxylem vascular tissue morphogenesis (Mähönen et al., 2006). Interestingly, non-cell-autonomous movement of small RNAs also conveys positional information and contributes to the patterning of the stele. In particular, the SHR/SCR module induces the expression of two miRNA genes MIR165a and MIR166b in the endodermis. MicroRNAs 165 and 166 subsequently move to the stele. There, they downregulate the expression of the class III HD-ZIP transcription factor PHABULOSA. This activity is necessary for xylem (Carlsbecker et al., 2010), pericycle and ground tissue patterning (Miyashima et al., 2011). phb mutants, expressing miRNA-resistant PHB transcripts with insertion in the miR165/166 target site, contain a stele with fewer cell files and rare extranumerary cortex cell files (Miyashima et al., 2011), demonstrating the critical role of miRNAs in cell fate determination. Moreover, the centripetal movement of miRNAs 165 and 166 results in differential distribution of their target PHB and subsequent dosage-dependent specification of the xylem cell types. Low levels of PHB determine protoxylem, and higher levels of PHB determine metaxylem cell fate (Carlsbecker et al., 2010). Another critical gene for controlling the number of cell files in the vascular bundle is LONESOME HIGHWAY. It does not appear to be necessary for any particular cell type determination, but for the bilateral symmetry of the root vasculature. lhw roots are formed with only one xylem and phloem pole (Ohashi-Ito and Bergmann, 2007). LHW forms hetordimers with another basic helix–loop–helix (bHLH) protein, TARGET OF MONOPTEROS5 (TMO5) (De Rybel et al., 2013), and both are critical for the periclinal cell divisions necessary for the establishment of correct patterning of the vascular bundle. Light was shed on phloem cell fate specification by characterization of the altered phloem development (apl) mutant. apl seedlings exhibit finite root growth as well as arrested shoot development. Ultimately this mutation is lethal for seedlings. Detailed analysis of the defects in apl mutants revealed impaired asymmetric cell divisions. Phloem cell differentiation is also disrupted in apl mutants, with cells in the position of the phloem adopting xylem features. This implies a role for APL in promoting phloem cell identity determination as well as repressing xylem specification at phloem poles in the root vasculature (Bonke et al., 2003).
Root cap.
The root cap constitutes a protective layer of cells in front of the RAM. A remarkable feature of this tissue is that root cap cells, during their differentiation, are continuously removed from the root tip. The removal of root cap cells is a combination of shedding of living cells at the root tip and programmed cell death (PCD) in the shootward part of the LR cap (Fendrych et al., 2014). Cells of the root cap originate from the root cap initial cells. Key genes for root cap cell fate determination are the NAC domain transcription factor genes FEZ and SOMBRERO (SMB; Willemsen et al., 2008). These genes control the timing and orientation of divisions of the root cap initials. FEZ is expressed in the epidermis/LR cap initial cell and columella initial cell prior to its periclinal cell division. FEZ activates expression of SMB in the daughter cells of the root cap initials. SMB in turn mediates the repression of FEZ, thereby closing a feedback loop and thus suppressing further periclinal divisions in the root cap daughter cells (Willemsen et al., 2008). Together with two other NAC transcription factor genes, BEARSKIN1 (BRN1) and BEARSKIN2 (BRN2), SMB controls the maturation of the root cap (Bennett et al., 2010). Lateral root cap cells are not cleared off in the smb mutant line due to the critical role of SMB in PCD of these cells (Fendrych et al., 2014), while in the brn1 brn2 double mutant columella cells do not detach and continue to divide. In the smb brn1 brn2 triple mutant, all root cap cells fail to mature (Bennett et al., 2010).
Cellular proliferation and differentiation in the root
Root cells develop along the root’s longitudinal axis (Fig. 3A). Cell proliferation takes place in the root meristem, a zone that is situated in the root tip directly shootward of the root cap. The columella is the only tissue in which cells differentiate only rootwards (in the direction of root growth). All other tissues are formed by cells that differentiate shootwards (in the direction opposite to root growth). In the course of their differentiation, root cells rapidly expand longitudinally, thereby pushing the root tip along the growth direction of the root. Clonal analyses traced the origin of all root cells back to root initials (Dolan et al., 1994; Scheres et al., 1994) that are organized around the QC (Fig. 3A). Laser ablation studies showed that in order to maintain stem cell-like status, the root initials need to be in contact with the QC cells (van den Berg et al., 1997). QC cell identity is linked to the expression of the WUSCHEL-RELATED HOMEOBOX 5 (WOX5) transcription factor gene (Sarkar et al., 2007). The expression of WOX5 is restricted to the QC cells by the presence of the WOX5 repressor, ACR4, in the neighbouring cells. ACR4 is a receptor-like kinase gene that is expressed in the neighbouring columella initials and columella cells (De Smet et al., 2008). WOX5 repression by ACR4 is dependent on the signal peptide CLE40 (Fiers et al., 2005) that is expressed in differentiated columella cells and represents the signal perceived by ACR4 leading to a repression of WOX5 and stem cell fate (Stahl and Simon, 2009). Therefore, QC identity and homeostasis are governed by the WOX5–ACR4–CLE40 pathway. Several additional gene modules have also been shown to be involved in QC maintenance. SCR (Di Laurenzio et al., 1996), originally described as a regulator of radial patterning of the ground tissue (see above), is also necessary to maintain QC cells (Sabatini et al., 2003), and the cell cycle regulator RETINOBLASTOMA-RELATED PROTEIN 1 (RBR1) acts downstream of SCR in this process (Wildwater et al., 2005). Besides these, PLETHORAs (PLTs), AP-2 type transcription factors, are also crucial for QC maintenance (Aida et al., 2004). Expression of PLT genes is regulated by the auxin maxima in the root meristem (Aida et al., 2004; Petersson et al., 2009). In turn, PLT genes regulate the expression of PINFORMED (PIN) auxin efflux genes (Blilou et al., 2005), thereby contributing to the maintenance of the auxin maxima. Apart from the WOX5–ACR4–CLE40, the SHR/SCR/RBR (Cruz-Ramírez et al., 2012) and the auxin–PLT pathways, other less well-characterized pathways including TYROSYLPROTEIN SULFOTRANSFERASE and ROOT MERISTEM GROWTH FACTORS are also required to maintain a functional QC (Matsuzaki et al., 2010; Zhou et al., 2010).
Balancing the proliferation and differentiation rate in the root is key in determining root growth as root elongation is largely determined by the number of cell divisions of stem cell progenitors and their subsequent cellular expansion (Beemster and Baskin, 1998). Key for regulating the proliferation of cells in the RAM are general cell cycle genes including members of CYCLIN-DEPENDENT KINASES and CYCLINS (Inagaki and Umeda, 2011). Multiple pathways impact cell cycle regulation including reactive oxygen species (ROS) (Tsukagoshi, 2012), DNA damage (Culligan et al., 2004; Cools et al., 2011; Spadafora et al., 2011) and plant hormones (Takatsuka and Umeda, 2014). Not only cell proliferation, but also cell elongation can be regulated by cell cycle regulators through regulation of polyploidy via endoreduplication, a DNA replication process which repeats G1 and S phases without G2 and M phases and is not accompanied by cell division (Kondorosi et al., 2000). Cells with a higher level of ploidy grow to a larger size, thereby leading to increased root elongation (Hayashi et al., 2013). The transition to the endoreduplication cycle is suppressed by auxin (Ishida et al., 2010). Various other pathways impact cell elongation, such as cell wall formation by cellulose synthase activity (Chen et al., 2010) and re-orientation of cellulose fibres (Anderson et al., 2010).
The most prominent role in setting the rate of proliferation and differentiation in the root is that of the cross-talk between two major plant hormones, auxin and cytokinin. Examination of mutants of auxin transporters showed that the correct localization and intensity of the auxin response maximum is necessary for regulating proliferation and differentiation (Sabatini et al., 1999). The localization of this auxin response maximum is, to a large part, determined by the auxin reflux loop in the root apex (Fig. 3B). This reflux loop is maintained through membrane-localized auxin transport proteins: AUXIN RESISTANT1 (AUX1), LIKE-AUX2 (LAX2), LIKE-AUX3 (LAX3) (Bennett et al., 1996; Swarup et al., 2001; Péret et al., 2012b; Band et al., 2014), P-GLYCOPROTEIN ABC transporter family members (Geisler and Murphy, 2006) and the PIN proteins (Blilou et al., 2005). In particular, PIN proteins are critical for polar auxin transport. While single pin mutants exhibit subtle phenotypes in the primary root (Gälweiler et al., 1998; Müller et al., 1998; Friml et al., 2002a, b, 2003; Blilou et al., 2005), most double mutants generated for PIN1, PIN2, PIN3, PIN4 and PIN7 show additive effects on the orientation of cell division, root meristem size and root length (Blilou et al., 2005). However, pin1pin2 double mutants, and all triple and quadruple mutants containing pin2, display epistatic behaviour (Blilou et al., 2005). Overall, these data underscore the importance of PINs and polar auxin transport in patterning and setting the rate of proliferation and differentiation in the root. Consequently, the exogenous application of auxin leads to promotion of cell division and an increase in meristem size (Blilou et al., 2005; Dello Ioio et al., 2007); however, cell elongation is reduced, which altogether results in shorter roots (Rahman et al., 2007). In contrast to auxin, the exogenous application of cytokinin causes a decrease in the root meristem size, while the cell division rate remains unchanged (Dello Ioio et al., 2007). Accordingly, cytokinin biosynthesis mutants have longer meristems (i.e. an increased number of cells from the QC to the first elongated cell), which is phenocopied by the constitutive expression of the cytokinin-inactivating enzyme AtCKX1 in the vasculature tissue at the transition zone. This indicates that cytokinins promote differentiation in the root meristem (Dello Ioio et al., 2007) while auxin promotes cell division. These opposing functions are crucial to determine the balance of proliferation and differentiation and therefore the location of the transition zone. At the molecular level, auxin and cytokinin interact by regulating in opposing ways the abundance of SHY2/IAA3 protein, a member of the auxin-induced Aux/IAA family (Dello Ioio et al., 2008). Cytokinin signalling mediates transcription of SHY2 through the AHK3/ARR1 signalling pathway. In turn, SHY2 downregulates expression of the auxin transport genes PIN1, PIN3 and PIN7. On the other hand, auxin promotes degradation of SHY2 via the SCFTIR1 pathway, which derepresses transcription of the cytokinin biosynthesis gene IPT5 (Dello Ioio et al., 2008). Overall, this balance between auxin and cytokinin signalling sets the balance between cell division and cell differentiation at the root transition zone, thus regulating meristem size and consequently root growth rate. Besides these very well characterized interactions, many regulatory steps exist for cytokinin as well as auxin pathways. These are present at almost every level, including their biosynthesis, transport, signalling and metabolism, each of which has a major impact on root development. Therefore, to generate accurate models of hormonal action and interaction at the molecular level, such as for auxin and cytokinin in the root tip, knowledge of the cellular and sub-cellular distribution of signalling components and hormones is needed. Initial steps in these directions have been made. For instance, high-resolution maps of the intracellular distribution of auxin (Petersson et al., 2009) and cytokinin (Antoniadi et al., 2015) in the root apex have been generated. In the case of auxin, these data supported the relatively recent concept that auxin is not exclusively shoot derived but that local auxin biosynthesis substantially contributes to auxin homeostasis in the root tip (Ljung et al., 2001; Bhalerao et al., 2002; Petersson et al., 2009).
While much has been learned about the pathways governing stem cell homeostasis, and the balance of proliferation and differentiation in the past years (see also later), many questions remain, such as at which growth stage, in which cellular environment and in which environmental context different pathways actually contribute, which stimuli impinge on which pathways, and how specifically these pathways interact with each other.
Lateral root formation
In the seed, only the PR is present as the radicle in the plant embryo (Grunewald et al., 2007). Higher order roots are the result of post-embryonic LR formation events that represent de novo organogenesis (Dubrovsky et al., 2006). LR formation is a key process that significantly contributes to shape the RSA (Lynch, 1995b). In arabidopsis and most dicots, LRs are formed from pericycle cells, which are adjacent to the xylem pole (Casimiro et al., 2003; Péret et al., 2009). LRs are usually spaced along the PR in a regular left–right alternating pattern (De Smet et al., 2007). LR formation can be partitioned into several stages (Malamy and Benfey, 1997) and involves four key events: priming, initiation, primordium formation and emergence. Priming and specification of future LR primordium sites are correlated with increased auxin signalling in the basal meristem (De Smet et al., 2007; Moreno-Risueno et al., 2010). These primed LR sites are xylem pole pericycle cells. These are different from other pericycle cells and show downregulation of Kip-Related Protein genes (KRP1 and KRP2), which are inhibitors of the G1 to S transition (Himanen et al., 2002). Auxin accumulation in these ‘founder cells’ precedes LR initiation. Subsequent maintenance of these auxin maxima is necessary for proper LR organogenesis (Dubrovsky et al., 2008). The auxin maximum is regulated by auxin reflux between the endodermis and the pericycle, mediated by the auxin efflux carrier PIN3 (Marhavy et al., 2013). The resulting auxin gradient is necessary for the activation of a key module in LR priming, the SOLITARY ROOT/ARF7/ARF19 genes (Fukaki et al., 2002; Vanneste et. al., 2005). This module activates transcription of LBD16/ASL18 and LBD29/ASL16, whose activities are all required for LR formation (Okushima et al., 2007). After priming, an asymmetric cell division takes place, marking LR initiation. Here, the receptor-like kinase ACR4 was identified as a critical factor for ensuring the correct specification of the LR primordium (De Smet et al., 2008). A further auxin module, BODENLOS/IAA12-MONOPTEROS/ARF5 is another key module during this stage of LR formation (De Smet et al., 2010). All these events occur in the pericycle, an internal layer of the root. Consequently, the newly formed LR primordium has to break through the ground tissue (endodermis, cortex and epidermis layers). This emergence process is highly regulated and, in particular, LR primordium-derived auxin is crucial for reprogramming adjacent cells. The co-ordinated action of the auxin influx carriers AUX1 and LAX3 plays a key role in this (De Smet et al., 2007; Swarup et al., 2008). Specifically, the auxin-induced expression of LAX3 in the cortical and epidermal cells that directly border the new primordium leads to the induction of cell-wall-remodelling enzymes that facilitate proper loosening and separation of the overlaying layers during the emergence of the newly formed LR (Swarup et al., 2008). During this process, direct communication of the pericycle and the endodermis, involving SHY2-mediated auxin signalling, is crucial for LR initiation (Vermeer et al., 2014).
While much of the LR formation is driven by auxin, other hormones impact LR formation, mainly by modulating auxin-dependent processes. Similar to the RAM, cytokinins act antagonistically to auxin during LR initiation by modulating auxin transport and preventing the formation of auxin maxima (Laplaze et al., 2007). Consequently, repressed cytokinin responses were observed in the founder cells, while enhanced cytokinin responses took place in the pericycle cells between two existing primordia (Bielach et al., 2012). Additionally, younger LR primordia are more sensitive to perturbations in the cytokinin pathway compared with those in later developmental stages (Bielach et al., 2012). Cytokinin biosynthetic genes play an important role in suppressing the initiation of new LRs in the neighbouring cells. Mutations of these genes caused a reduction of cytokinin in the cells adjacent to the LR primordium, which led to an abnormal positioning of LRs in an ACR4-independent fashion (Chang et al., 2015).
Abscisic acid (ABA) is another hormone that modulates LR formation through auxin signalling. ABA inhibits LR formation immediately after LR emergence in a reversible manner (De Smet et al., 2003). The ABI3 (ABA INSENSITIVE3) member of the ABA signalling pathway is induced by auxin in the LR primordium, and loss-of-function abi3 plants show reduced response to auxin and auxin transport inhibitors (Brady et al., 2003). Another key transcription factor linking auxin and ABA is MYB77. This transcription factor interacts with ARF7 (Shin et al., 2007) and PYL8, an ABA receptor (Zhao et al., 2014).
Lateral root formation, and therefore root architecture, is strongly impacted by nutrient availability (Lopez-Bucio et al., 2003). For example, LR emergence of arabidopsis grown on high nitrate concentrations is systematically inhibited (Walch-Liu et al., 2006). In contrast, a local increase of nitrate concentration has a stimulatory effect on LR growth, which is abolished in the auxin-resistant mutant axr4 (Zhang et al., 1999). Several other molecular links between nitrate and auxin pathways in regulating LR development were identified (Guo et al., 2002; Gifford et al., 2008; Vidal et al., 2010; Rosas et al., 2013). Strikingly, the nitrate transporter NRT1.1 also facilitates auxin transport in the LR primordium, thus controlling root branching (Krouk et al., 2010). When nitrate is absent, NRT1.1 blocks auxin accumulation and growth of the LR by supporting basipetal auxin transport. At higher nitrate concentrations (e.g. 1 mm), the action of NRT1.1 is blocked, leading to accumulation of auxin in the LR primordium and permitting its growth (Krouk et al., 2010). Nitrogen interferes with other hormonal pathways as well. For instance, nitrate application induces the expression of genes involved in cytokinin biosynthesis (Sakakibara et al., 2006), which are important for proper LR patterning (Chang et al., 2015). Nitrate and ABA pathways are connected by ABI4 and ABI5, as the two ABA-insensitive mutants lines abi4 and abi5 display a lesser degree of reduction of LR formation by high nitrate concentration (Signora et al., 2001). Phosphorus is another example of how nutrients affect hormonal signalling in order to modulate LR formation, and thereby RSA. In phosphate-deficient conditions, expression of the auxin receptor gene TRANSPORT INHIBITOR RESPONSE1 (TIR1) is stimulated, which in turn promotes the activation of downstream ARF transcription factors that are involved in the control of LR formation (Pérez-Torres et al., 2008). Finally, iron-dependent LR growth promotion is dependent on rootward auxin transport. In particular, the auxin influx carrier AUX1 modulates this response, since iron-stimulated LR elongation is lost in aux1 mutants (Giehl et al., 2012)
Control of root growth direction
The direction in which a root grows is a key parameter for soil exploration and the response to environmental cues. The directional characteristics of growth are termed tropisms and, depending on whether plants grow towards a signal or away from it, the tropism is defined as positive or negative, respectively. There are multiple tropisms including gravitropism (gravity), phototropism (light), hydrotropism (water) and thigmotropism (touch) (Esmon et al., 2005).
Gravitropism is highly regulated by auxin transport mechanisms (Baldwin et al., 2013). Polar localization of PIN proteins creates an asymmetrical auxin distribution and allows gravitropic bending by elongation of only the side of the root that is furthest from the direction of the vector of gravity (Wisniewska et al., 2006). Other genes that have been shown to be important regulators for the gravity response include the DnaJ-like protein gene ALTERED RESPONSE TO GRAVITY (Sedbrook et al., 1999), an E3 ligase family gene WAVY GROWTH 3 (Sakai et al., 2012) and PHOSPHOLIPASE C (Andreeva et al., 2010). In comparison with gravitropism, molecular mechanisms of other root tropisms are not well understood. Root phototropism has been known for a long time. While usually not directly exposed to light, roots are frequently exposed to light through ambient diffusion or soil upheaval (Galen et al., 2007). Moreover, photons can be efficiently conducted through the vasculature (Sun et al., 2003). The occurrence of phototropism is species dependent, and it has been reported that almost 43 % of examined plant species showed a negative root phototropism (Kutschera and Briggs, 2012). A blue light receptor gene NONPHOTOTROPIC HYPOCOTYL 1/PHOTOTROPIN 1 (Liscum and Briggs, 1995) and red light receptor genes PHYTOCHROME A and B (Kiss et al., 2003) are involved in negative and positive root phototropism, respectively. Examples of molecular mechanisms and genes involved in hydrotropism are, for instance, the cytokinin-dependent involvement of ALTERED HYDROTROPIC RESPONSE 1 (Saucedo et al., 2012), the ABA-dependent involvement of NO HYDROTROPIC RESPONSE 1 (Ponce et al., 2008) and the land-plant-specific gene MIZU-KUSSEI 1 (MIZ1) (Kobayashi et al., 2007). Overall, it is thought that these components modulate how the root cap senses and responds to water availability (Cassab et al., 2013). MILDEW RESISTANCE LOCUS O4 (Chen et al., 2009) and ENDOBINDING 1 (Gleeson et al., 2012) have been identified as thigmotropism regulators that when mutated, displayed intense responses to touching surfaces.
Since multiple stimuli occur simultaneously in nature, it is expected that multiple tropism mechanisms impact each other. While the signalling mechanisms of different tropisms are independent [for instance the absence of hydrotropism in miz-1 mutants did not have any effect on gravitropism (Kobayashi et al., 2007)], there is significant cross-talk. For instance, gravitropism and hydrotropism affect each other (Takahashi et al., 2009), and, while hydrotropism of arabidopsis is dominant to gravitropism (Takahashi et al., 2002), that of pea roots is overcome by gravitropism (Takahashi et al., 1992). Overall, this shows that multiple signals coming from largely independent signalling pathways are integrated, but which tropism will dominate depends on the genetic constitution of the plant.
APPROACHING COMPLEXITY IN ROOT GROWTH CONTROL USING SYSTEMS BIOLOGY
With the discovery of an ever-increasing number of genes that are involved in regulating root growth and their complex interactions, it became clear that more holistic approaches were needed to comprehend the regulation of root growth. An area of biology that has emerged to approach such challenges is systems biology. Here, neither single genes nor proteins are at the focus of the research; instead, the focus is on the systems that are defined by interactions of biologically relevant entities such as genes, proteins, cells, tissues, organs or organisms. The study of root growth in arabidopsis has long been at the forefront of plant systems biology (Hill et al., 2013). We will highlight multiple areas in which it has significantly expanded our comprehension of complex biological processes that are important in the context of root growth and development.
Understanding networks at the level of cell types
One of the first milestones in the plant systems-biology field was the generation of a cellular atlas of gene expression by combining fluorescence-activated cell sorting (FACS) of green fluorescent protein (GFP)-labelled cell populations and microarray analysis (Birnbaum et al., 2003). This allowed, for the first time in plants, an insight into cell type expression patterns of the >20 000 genes that were present on the ATH1 microarray platform. In the subsequent years, this expression atlas was refined by including more cell types and distinct developmental zones of the root (Brady et al., 2007), as well as by applying stress to the roots before FACS (Dinneny et al., 2008; Long et al., 2010; Iyer-Pascuzzi et al., 2011). Together, these large-scale approaches enabled the discovery of cell-type-specific dominant expression patterns of large numbers of genes, as well as the discovery of stress response centres and the impact of stress on cell fate decisions (Dinneny et al., 2008; Long et al., 2010; Iyer-Pascuzzi et al., 2011). Importantly, these approaches and tools, due to their easy accessibility (Brady et al., 2007; Winter et al., 2007), constituted highly useful resources for a whole field, enabling rapid insights into gene expression patterns. Overall, they have paved the way for popularizing in silico gene expression analysis and have also provided a valuable resource for identifying novel molecular mechanisms and regulatory models. For instance, screening for transcription factors that were specifically expressed in the transition zone, where cell proliferation ceases and cell differentiation starts, resulted in the discovery of UPB1 and a novel mechanism regulating the balance of ROS between the zones of cell proliferation and the zone of cell elongation where differentiation begins (Tsukagoshi et al., 2010). Indeed, the transcriptional regulation of ROS by UPB1 controls the transition from proliferation to differentiation in the root (Tsukagoshi et al., 2010). A novel mechanism for the periodic patterning of the root was also discovered based on the Brady transcriptome data. In particular, transcriptome data from two individual roots were subjected to methods capable of identifying periodic responses. The results of this analysis, together with long-term imaging, revealed that the position of LRs and root bending are periodic responses, which appear to be regulated by a mechanism resembling an endogenous clock (Moreno-Risueno et al., 2010).
Motivated by such successes, cell-type-specific atlases have been created for other molecules such as small RNAs (Breakfield et al., 2012), proteins (Petricka et al., 2012) and metabolites (Moussaieff et al., 2013). However, while profiling of mRNA is relatively straightforward, the other atlases faced technical difficulties, such as the large number of cells required for metabolite measurements or the incomplete sampling of the proteome by GeLC-MS/MS (in-gel tryptic digestion followed by liquid chromatography-tandem mass spectrometry).
The root gene expression atlas has also been the starting point for generating gene network models that try to infer relationships of gene regulation. For instance, transcription factors and miRNAs that were preferentially expressed in the stele were subjected to systematic yeast one-hybrid (Y1H) and Y2H assays to assess the binding of transcription factors to transcription factor and miRNA-promoters and the interaction between transcription factors at the protein level (Brady et al., 2011). These interaction data, in conjunction with expression data, were then used to generate a directed network model. The accuracy of this model was verified by testing predicted interactions using chromatin immunoprecipitation (ChIP) for transcription factors and predicted direct targets, as well as expression and phenotypic analysis of mutant lines of targets. Interestingly, 65 % of the transcription factor knockout lines tested showed molecular phenotypes, but only 16 % showed morphological phenotypes (Brady et al., 2011). Overall, these results highlighted the robustness of biological regulation and the difficulty in studying complex gene regulation at the systems level using only single-gene knockout lines. Along similar lines, a network model of secondary cell wall synthesis was recently generated (Taylor-Teeples et al., 2014). For this, genes were selected using the root expression atlas as well as annotations; molecular interactions of these genes were then mapped by Y1H, and a network constructed from all of the data. This network model of secondary cell wall synthesis provided specific insights into the gene regulation underlying cell wall synthesis, as well as its modulatory capacity under stress conditions (Taylor-Teeples et al., 2014).
Overall, these recent studies show the power of cell-type-resolved experimental data sets to uncover complex regulatory relationships between large sets of genes, yielding models with predictive power.
Understanding specific regulatory networks
While cell-type-specific gene expression maps allow for the identification of genes and the networks and pathways that they act in, similar inferences can be made from transcriptome data sets that are centred around specific genes or regulatory processes. For instance, a cell sorting approach combined with a transcriptome analysis of the LR regulator S-phase kinase-associated protein 2 (SKP2B), followed by the identification and analysis of genes that were enriched in SKP2B-expressing cells, revealed an important role for redox-related genes and ROS-dependent mechanisms in LR development (Manzano et al., 2014). Transcriptome analyses identified GATA23 as a key factor defining LR founder cell identity (De Rybel et al., 2010), and the receptor-like kinase ACR4 was identified as a key regulator for the formative cell divisions of LR organogenesis (De Smet et al., 2008). A transcriptome approach analysing FACS-sorted root hair epidermal and non-root hair epidermal cells from multiple mutant lines displaying either more or fewer root hairs than the wild type, in conjunction with a modelling approach, generated a comprehensive network model of 208 core epidermal genes (Bruex et al., 2012).
Studies analysing time courses of transcriptome data acquired after induction of biological or developmental processes have great power to resolve complex biological processes at the molecular level. For instance, chemical induction of LR formation revealed specific transcriptional stages that precede cell divisions in the process of LR formation (Himanen et al., 2004). A similar study identified 60 distinctly responding genes clusters, and underscored cell wall remodelling as an essential feature of LR development (Lewis et al., 2013). A high-resolution time course experiment after root bending revealed that a large number of genes showed an oscillatory pattern during LR formation. Further analysis of these data suggested that the circadian clock was rephased during LR formation and that this process is necessary for gating auxin signalling during LR development to facilitate organ emergence (Voß et al., 2015). The same time course data were reanalysed with a custom time-delay correlation algorithm (TDCor) to generate a comprehensive gene regulatory network (GRN) in order to identify genes and their interactions controlling LR primordium initiation and patterning. Notably, this GRN model revealed that cell fate decisions during early LR patterning are dependent on the mutual inhibition between the ARF7 and ARF5 regulator modules (Lavenus et al., 2015). Another study in the context of cell fate decisions in the RAM used dense time course transcriptome data coupled with genome-wide detection of direct targets for SHR to uncover the temporal dynamics of regulation of the formative cell division of the CEI, including the discovery of a specific cell cycle gene (CYCD6;1) of crucial importance for this particular cell division (Sozzani et al., 2010).
Such data can be used as an excellent starting point for capturing complex regulatory processes in space and time via mathematical modelling approaches. For instance, based on the regulation of CYCD6;1 by SCR/SHR and a potential link to the RBR protein, an iterative approach combining genetics, cell biology and mathematical modelling was able to identify the specific molecular interactions that determine the precise spatio-temporal pattern of the CEI formative cell division (Cruz-Ramirez et al., 2012). Key to this success was a procedure in which predictions of alternative mathematical models of interactions of multiple key components were compared with experimental results (Cruz-Ramirez et al., 2012). Another highly successful approach that combined genetic analysis and mathematical modelling was the identification of the genetic network and molecular mechanism that generates distinct hormonal response zones in the vasculature; the results sufficiently explained vascular tissue growth and patterning (De Rybel et al., 2014).
Dissecting the complex networks of plant hormonal signalling
Plant growth and development are co-ordinated and controlled by highly complex hormonal signalling pathways that act locally as well as systemically. The intrinsic complexities of these pathways are difficult to comprehend by reductionist approaches. Modelling approaches have turned out to be particularly useful in approaching this complexity. A landmark study in modelling hormonal signalling in the root was a model of auxin distribution that incorporated diffusion and PIN-facilitated auxin transport across cells using a simplified root layout. It was able to recreate experimentally observed auxin distribution patterns and morphogenesis over a broad range of time scales (Grieneisen et al., 2007). Further refinement was achieved by incorporating real-world root cell geometries and experimentally determined localization of auxin transporters into the model. Experimentally tested using the DII-VENUS auxin sensor and computational image analysis, this model led to the insight that auxin efflux carriers alone cannot generate the experimentally observed auxin distribution at the root tip. Rather, non-polar auxin influx transporters control auxin abundance, and the polar PIN efflux transporters control the direction of auxin transport in these tissues (Band et al., 2014). An auxin signalling reporter and mathematical modelling were used to quantify auxin redistribution after a gravitropic stimulus. Interestingly, auxin was redistributed very rapidly and returned to the normal distribution after the root tip had changed its angle to 40 ° to the horizontal. This led to the postulation of a tipping point mechanism that reverses the auxin flow after bending of the root has reached an angle threshold (Band et al., 2012). Most recently, an iterative mathematical modelling approach that incorporated diverse parameters such as growth, gene expression, protein turnover and movement, auxin levels and response, and experimental testing of the model’s predictions, led to insights into the question of how auxin can concurrently mediate rapid responses such as gravitropism, and yet regulate stable developmental zonation (Mähönen et al., 2014).
The complex interplay of developmental regulation by auxin and RSA was illustrated by a model which combined cell shape and auxin transport, revealing that the local curvature of the root has an intricate impact in mediating changes in auxin transport and therefore local concentration (Laskowski et al., 2008). A mathematical modelling approach that took into account three-dimensional cell and tissue geometries, as well as expression of auxin transporters and their transport activity, revealed the necessity for a particular activation sequence of the LAX3 auxin influx carrier and PIN3 auxin efflux carrier for LR emergence (Péret et al., 2013). Using a mathematical model describing the biophysical properties of the root, in particular the LR primordium, as well as the effects of water flows mediated by the PIP2;1 aquaporin, important predictions of LR emergence in pip2;1 mutants and overexpressors were confirmed, showing the importance of the spatial and temporal aquaporin-dependent transport of water through root tissue (Péret et al., 2012a).
While these studies had been focused largely on auxin distribution in the root, another study focused on the impact of cell identity on auxin-dependent gene expression responses. Here, transcriptome analysis in four distinct tissues of the arabidopsis root demonstrated that tissue identity and developmental stage made important contributions to auxin responses, with auxin-dependent genes showing down- or upregulation depending on the tissue context (Bargmann et al., 2013). Modelling even more specific aspects of molecular auxin signalling components, such as the Aux/IAA system, predicted that the ratios between protein and mRNA turnover rates are key determinants of the systems properties (Middleton et al., 2010).
While auxin signalling has received the most attention, other hormonal signalling pathways have also been investigated. Two examples include insights into the distinct time scales and feedback loops of different components involved in gibberellin signalling (Middleton et al., 2012), and the importance of receptor complex oligomerization states of the ABA receptor for modulating hormonal responses (Dupeux et al., 2011). Moreover, the interplay between different hormone pathways was also studied. Modelling of a regulatory circuit centred around POLARIS (PLS), which mediates cross-talk between auxin, ethylene and cytokinin in arabidopsis, revealed that this cross-talk is mainly shaped by PLS controlling the relative contribution of auxin transport and biosynthesis (Liu et al., 2010). Overall, systems-biology approaches have increased our comprehension of complex regulatory systems far beyond the level of what could have been achieved using traditional forward genetic approaches.
OUTLOOK: MOVING TOWARDS ROOT SYSTEMS GENETICS
Systems-biology approaches are highly successful in shedding light on complex biological processes. However, there are significant limitations for many of these methods for capturing dynamic processes. For instance, while time courses of transcriptome data are becoming more common, there are still limits with regard to how many time points can be acquired, especially with cell type resolution (partly due the cost of these approaches and partly due to technical reasons). Moreover, transcriptome approaches measure only transcriptional responses, and important regulatory levels such as protein abundance or protein localization changes cannot be captured using these approaches. With many other data types with less spatial resolution, such as most protein–DNA binding profiles (ChiP-chip/ChIPseq), or data acquired in yeast such as protein–protein (Y2H), and promoter–transcription factor interactions (Y1H), it is often not clear how to interpret these data in the context of a multicellular organism with processes occurring over time in different cell and tissue contexts.
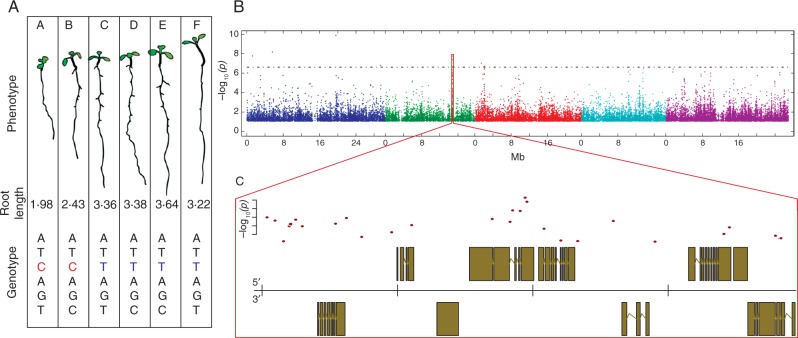
In contrast, phenotypic analysis of mutant lines or natural accessions can provide very high spatial and temporal resolution that can be acquired in various conditions, but it cannot be immediately related to complex molecular processes. However, recent advances have made it possible to link complex molecular data with complex phenotypic data. In particular, large-scale phenotyping approaches in combination with genome-wide association mapping [genome-wide association studies (GWAS)] allow for the mapping of complex and highly resolved phenotypes to the whole genome, and to identify the loci that determine these phenotypes (Fig. 4). If combined with systems-biology-type data, GWAS provide the possibility to study how genetic information is translated by molecular, cellular and physiological networks to shape complex phenotypes (Nadeau and Dudley, 2011). Such efforts have been defined as the subject of the emerging field of systems genetics (Ayroles et al., 2009; Mackay et al., 2009; Nadeau and Dudley, 2011; Civelek and Lusis, 2014). Systems genetics is a synthesis of multiple fields, including genetics, genomics, systems biology and phenomics (Markowetz et al., 2015). Much like systems biology, it aims to study the relationships of multiple components and how interactions of these components give rise to biological function. However, systems genetics specifically aims to address the question of how genetic variation leads to phenotypic (trait) variation. This promises to approach similarly complex questions to those approached using systems biology, but firmly anchors these approaches in the genotype to phenotype problem, thus adding a genetic and phenotypic/physiological axis. Likewise, it extends traditional genetics since it adds functional insights into intermediate layers between genotype and phenotype. Moreover, due to the ability to measure many phenotypes of the same genotypes in many conditions, such approaches are not restricted to one trait but are ideally extended to multiple traits, their dependence on each other, and their interaction with the environment. For instance, a systems-genetics approach can unravel which molecular consequences of a particular genotype, under certain environmental conditions, lead to changes at the cellular level and how these cellular changes relate to specific organismal phenotypes. This would lead to not only the prediction of molecular components that would alter these phenotypes when targeted by breeding, specific drugs or treatments, but also to mathematical models predicting phenotypes from genotypes. While systems genetics is still in its infancy, there has been recent progress in this direction in the root. For instance, a recent study systematically assessed cellular root traits in 201 arabidopsis accessions. This led to the identification of strong correlations between different cellular traits such as meristem size and mature cell size, indicating a tight genetic control of proliferation and differentiation in the root. Using expression data for the genes in the associated genomic region, it was possible to identify the causal gene and its alleles for the two highly correlated root cellular traits (Meijón et al., 2013). Importantly, these cellular changes translated into changes in root length. Overall, this demonstrated that by combining GWAS and expression information, it was possible to identify an unknown regulator and its alleles that regulate two highly correlated cellular traits, thereby regulating root growth, an organ-level phenotype. Another study approached the relationship between traits, growth conditions and genotypes using GWAS, genome-wide expression analysis and phenomics. This study not only uncovered the fact that most root traits, such as the length of primary and LRs or LR density, are independently controlled in a genotype- and environment-dependent manner, but also mapped a large number of potentially causal genes underlying this remarkable plasticity (Gifford et al., 2013). Due to the increasing number of highly efficient phenotyping pipelines for roots, the growing popularity of GWAS for root traits and the excellent functional genomics resources for roots, much more progress in this area can be expected.
Fig. 4.
Schematic of an exemplary GWAS. (A) Phenotypes of various genotyped strains (upper part) are quantified for a trait (in this example root length; mid-part). GWAS is conducted by using the quantified phenotypes and their genotypes to find significant associations of single nucleotide polymorphisms (SNPs) and the phenotype. Exemplary sequences that highly associate with the example root length values are shown in the lower part. (B) Example of a Manhattan plot that depicts the genome-wide associations {x-axis, SNP position along the genome; y-axis, significance of association [–log10(P-value)]; associations coloured by chromosome}. (C) Genomic region surrounding a significant GWA peak. Top: −log10(P-values) of associations of the SNPs. Bottom: gene models in genomic regions. The x-axis represents the position on the chromosome.
ACKNOWLEDGEMENTS
We apologize that many important contributions to the field could not be cited because of space constraints. We thank Klaus Brackmann and Elke Barbez for critically reading the manuscript, and Matt Watson for editing the manuscript. Work in the Busch laboratory is supported by funds from the Austrian Academy of Science through the Gregor Mendel Institute and grants by the Austrian Science Foundation (FWF) and the Vienna Science and Technology Fund (WWTF).
LITERATURE CITED
- Aida M, Beis D, Heidstra R, et al. 2004. The PLETHORA genes mediate patterning of the arabidopsis root stem cell niche. Cell 119: 119–120. [DOI] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C. 2010. Real-time imaging of cellulose reorientation during cell wall expansion in arabidopsis roots. Plant Physiology 152: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva Z, Barton D, Armour WJ, et al. 2010. Inhibition of phospholipase C disrupts cytoskeletal organization and gravitropic growth in arabidopsis roots. Planta 232: 1263–1279. [DOI] [PubMed] [Google Scholar]
- Antoniadi I, Plačková L, Simonovik B, et al. 2015. Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. The Plant Cell 27: 1955–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, et al. 2009. Systems genetics of complex traits in Drosophila melanogaster. Nature Genetics 41: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin KL, Strohm AK, Masson PH. 2013. Gravity sensing and signal transduction in vascular plant primary roots. American Journal of Botany 100: 126–142. [DOI] [PubMed] [Google Scholar]
- Band LR, Wells DM, Larrieu A, et al. 2012. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proceedings of the National Academy of Sciences, USA 109: 4668–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Fozard JA, et al. 2014. Systems analysis of auxin transport in the Arabidopsis root apex. The Plant Cell 26: 862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann B, Vanneste S, Krouk G, et al. 2013. A map of cell type-specific auxin responses. Molecular Systems Biology 9: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. 1996. Stimulation of root hair elongation in Arabidopsis thailiana by low phosphorus availability. Plant, Cell and Environment 19: 529–538. [Google Scholar]
- Baum SF, Rost TL. 1996. Root apical organization in Arabidopsis thaliana 1. Root cap and protoderm. Protoplasma 192: 178–188. [Google Scholar]
- Beemster GT, Baskin TI. 1998. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiology 116: 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. 1993. Root development in arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, et al. 1996. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950. [DOI] [PubMed] [Google Scholar]
- Bennett T, van den Toorn A, Sanchez-Perez GF, et al. 2010. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. The Plant Cell 22: 640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. 1995. Cell fate in the arabidopsis root meristem determined by directional signalling. Nature 378: 62–65. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. 1997. Short-range control of cell differentiation in the arabidopsis root meristem. Nature 390: 287–289. [DOI] [PubMed] [Google Scholar]
- Berger F, Haseloff J, Schiefelbein J, Dolan L. 1998a. Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Current Biology 8: 421–430. [DOI] [PubMed] [Google Scholar]
- Berger F, Hung CY, Dolan L, Schiefelbein J. 1998b. Control of cell division in the root epidermis of Arabidopsis thaliana. Developmental Biology 194: 235–245. [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. 2003. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the arabidopsis root. Development 130: 6431–6439. [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J. 2005. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the arabidopsis root epidermis. Development 132: 291–298. [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G. 2002. Shoot-derived auxin is essential for early lateral root emergence in arabidopsis seedlings. The Plant Journal 29: 325–332. [DOI] [PubMed] [Google Scholar]
- Bielach A, Podlesakova K, Marhavy P, et al. 2012. Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. The Plant Cell 24: 3967–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, et al. 2003. A gene expression map of the Arabidopsis root. Science 302: 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, et al. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y. 2003. APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186. [DOI] [PubMed] [Google Scholar]
- Bünning E. 1951. Über die differenzierungsvorgänge in der Cruciferenwurzel. Planta 39: 126–153. [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P. 2003. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. The Plant Journal 34: 67–75. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee J, et al. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Sci4ence 318: 801–806. [DOI] [PubMed] [Google Scholar]
- Brady SM, Zhang L, Megraw M, et al. 2011. A stele-enriched gene regulatory network in the Arabidopsis root. Molecular Systems Biology 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray RH. 1954. A nutrient mobility concept of soil plant relationships. Soil Science 104: 9–22. [Google Scholar]
- Breakfield NW, Corcoran DL, Petricka JJ, et al. 2012. High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Research 22: 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, et al. 2012. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genetics 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee J, Roberts CJ, et al. 2010. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, et al. 2003. Dissecting Arabidopsis lateral root development. Trends in Plant Science 8: 165–171. [DOI] [PubMed] [Google Scholar]
- Cassab GI, Eapen D, Campos ME. 2013. Root hydrotropism: an update. American Journal of Botany 100: 14–24. [DOI] [PubMed] [Google Scholar]
- Chang L, Ramireddy E, Schmülling T. 2015. Cytokinin as a positional cue regulating lateral root spacing in Arabidopsis. Journal of Experimental Botany 66: 4759–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR. 2010. Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proceedings of the National Academy of Sciences, USA 107: 17188–17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Noir S, Kwaaitaal M, et al. 2009. Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in arabidopsis root thigmomorphogenesis. The Plant Cell 21: 1972–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. 2014. Systems genetics approaches to understand complex traits. Nature Reviews Genetics 15: 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T, Iantcheva A, Weimer AK, et al. 2011. The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. The Plant Cell 23: 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S, Dolan L. 2003. Epidermal patterning genes are active during embryogenesis in Arabidopsis. Development 130: 2893–2901. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I. 2012. A bistable circuit involving SCARECROW–RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, et al. 2007. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425. [DOI] [PubMed] [Google Scholar]
- Culligan K, Tissier A, Britt A. 2004. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. The Plant Cell 16: 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, et al. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, et al. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, et al. 2010. A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Current Biology 20: 1697–1706. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Möller B, Yoshida S, et al. 2013. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Developmental Cell 24: 426–437. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Adibi M, Breda AS, et al. 2014. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345: 1255215. [DOI] [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang HM. 2003. An abscisic acid-sensitive checkpoint in lateral root development of arabidopsis. The Plant Journal 33: 543–555. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inzé D, Beeckman T. 2006. Lateral root initiation or the birth of a new meristem. Plant Molecular Biology 60: 871–887. [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of arabidopsis. Development 134: 681–690. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, et al. 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597. [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, et al. 2010. Bimodular auxin response controls organogenesis in arabidopsis. Proceedings of the National Academy of Sciences, USA 107: 2705–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, et al. 1996. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, et al. 2008. Cell identity mediates the response of arabidopsis roots to abiotic stress. Science 320: 942–945. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, et al. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, et al. 1994. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474. [Google Scholar]
- Dubrovsky JG, Doerner PW, Colón-Carmona A, Rost TL. 2000. Pericycle cell proliferation and lateral root initiation in arabidopsis. Plant Physiology 124: 1648–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Gambetta GA, Hernandez-Barrera A, Shishkova S, Gonzalez I. 2006. Lateral root initiation in arabidopsis: developmental window, spatial patterning, density and predictability. Annals of Botany 97: 903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, et al. 2008. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proceedings of the National Academy of Sciences, USA 105: 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Santiago J, Betz K, et al. 2011. A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO Journal 30: 4171–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch JJ, Chen M, Sanders M, et al. 2003. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130: 5885–5894. [DOI] [PubMed] [Google Scholar]
- Esmon CA, Pedmale UV, Liscum E. 2005. Plant tropisms: providing the power of movement to a sessile organism. International Journal of Developmental Biology 49: 665–674. [DOI] [PubMed] [Google Scholar]
- Falasca G, Altamura MM. 2003. Histological analysis of adventitious rooting in Arabidopsis thaliana (L.) Heynh seedlings. Plant Biosystems 137: 265–273. [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, et al. 2014. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in arabidopsis. Current Biology 24: 931–940. [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, et al. 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. The Plant Cell 17: 2542–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, et al. 2002a. AtPIN4 mediates sink-driven auxin gradients and root patterning in arabidopsis. Cell 108: 661–673. [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. 2002b. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in arabidopsis. Nature 415: 806–809. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, et al. 2003. Efflux-dependent auxin gradients establish the apical–basal axis of arabidopsis. Nature 426: 147–153. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. 2002, Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. The Plant Journal 29: 153–168. [DOI] [PubMed] [Google Scholar]
- Galen C, Rabenold JJ, Liscum E. 2007. Light-sensing in roots. Plant Signaling and Behavior 2: 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. 1994. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Developmental Biology 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, et al. 1998. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230. [DOI] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. 2006. The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Letters 580: 1094–1102. [DOI] [PubMed] [Google Scholar]
- Giehl RFH, Lima JE, von Wirén N. 2012. Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1-mediated auxin distribution. The Plant Cell 24: 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. 2008. Cell-specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences, USA 105: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Banta JA, Katari MS, et al. 2013. Plasticity regulators modulate specific root traits in discrete nitrogen environments. PLoS Genetics 9: e1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson L, Squires S, Bisgrove SR. 2012. The microtubule associated protein END BINDING 1 represses root responses to mechanical cues. Plant Science 187: 1–9. [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B. 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. [DOI] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N. 2013. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiology 163: 161–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Parizot B, Inzé D, Gheysen G, Beeckman T. 2007. Developmental biology of roots: one common pathway for all angiosperms? International Journal of Plant Developmental Biology 1: 212–225. [Google Scholar]
- Guo FQ, Wang RC, Crawford NM. 2002. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots. Journal of Experimental Botany 53: 835–844. [DOI] [PubMed] [Google Scholar]
- Hassan H, Scheres B., Blilou I. 2010. JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 137: 1523–1529 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Hasegawa J, Matsunaga S. 2013. The boundary of the meristematic and elongation zones in roots: endoreduplication precedes rapid cell expansion. Scientific Reports 3: 2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, et al. 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567. [DOI] [PubMed] [Google Scholar]
- Hill K, Porco S, Lobet G, et al. 2013. Root systems biology: integrative modeling across scales, from gene regulatory networks to the rhizosphere. Plant Physiology 163: 1487–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T. 2002. Auxin-mediated cell cycle activation during early lateral root initiation. The Plant Cell 14: 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, et al. 2004. Transcript profiling of early lateral root initiation. Proceedings of the National Academy of Sciences, USA 101: 5146–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Tuberosa R. 2009. Genetic and genomic dissection of maize root development and architecture. Current Opinion in Plant Biology 12: 172–177. [DOI] [PubMed] [Google Scholar]
- Hung CY, Lin Y, Zhang M, Pollock S, Marks MD, Schiefelbein J. 1998. A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiology 117: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S, Umeda M. 2011. Cell-cycle control and plant development. International Review of Cell and Molecular Biology 291: 227–261 [DOI] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K. 2010. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71. [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jackson T, Cui H, et al. 2011. Cell identity regulators link development and stress responses in the Arabidopsis root. Developmental Cell 21: 770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C, Sundaresan V, Roberts K, Dolan L. 2000. Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211: 191–199. [DOI] [PubMed] [Google Scholar]
- Kiss JZ, Mullen JL, Correll MJ, Hangarter RP. 2003. Phytochromes A and B mediate red-light-induced positive phototropism in roots. Plant Physiology 131: 1411–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Takahashi A, Kakimoto Y, et al. 2007. A gene essential for hydrotropism in roots. Proceedings of the National Academy of Sciences, USA 104: 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E, Roudier F, Gendreau E. 2000. Plant cell-size control: growing by ploidy? Current Opinion in Plant Biology 3: 488–492. [DOI] [PubMed] [Google Scholar]
- Koshino-Kimura Y, Wada T, Tachibana T, Tsugeki R, Ishiguro S, Okada K. 2005. Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant and Cell Physiology 46: 817–826 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, et al. 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell 18: 927–937. [DOI] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, et al. 2005. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Briggs WR. 2012. Root phototropism: from dogma to the mechanism of blue light perception. Planta 235: 443–452. [DOI] [PubMed] [Google Scholar]
- Kwak SH, Schiefelbein J. 2008. Regulated accumulation of the SCRAMBLED receptor and position-dependent cell type patterning in Arabidopsis. Current Biology 18: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, et al. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. The Plant Cell 19: 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, et al. 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biology 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Guyomarc’h S, et al. 2015. Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. The Plant Cell 27: 1368–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. 1999. WEREWOLF, a MYB-related protein in arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483. [DOI] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, et al. 2006. Whole-genome analysis of the short-root developmental pathway in Arabidopsis. PLoS Biology 4: 739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK. 2013. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. The Plant Cell 25: 3329–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Briggs WR. 1995. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. The Plant Cell 7: 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Mehdi S, Topping J, Tarkowski P, Lindsey K. 2010. Modelling and experimental analysis of hormonal crosstalk in arabidopsis. Molecular Systems Biology 6: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G. 2001. Sites and homeostatic control of auxin biosynthesis in arabidopsis during vegetative growth. The Plant Journal 28: 465–474. [DOI] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. 2010. The bHLH transcription factor POPEYE regulates response to iron deficiency in arabidopsis roots. The Plant Cell 22: 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bucio J, Cruz-Ramirez A, Herrera-Estrella L. 2003. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology 6: 280–287. [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Groover A, Lichtenberger R, et al. 2013. The plant vascular system: evolution, development and functions. Journal of Integrative Plant Biology 55: 294–388. [DOI] [PubMed] [Google Scholar]
- Lynch JP. 1995a. Rhizoeconomics: the roots of shoot growth limitations. HortScience 42: 1107–1109. [Google Scholar]
- Lynch J. 1995b. Root architecture and plant productivity. Plant Physiology 109: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. 2009. The genetics of quantitative traits: challenges and prospects. Nature Reviews Genetics 10: 565–577 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. 2000. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes and Development 14: 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, et al. 2006. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98. [DOI] [PubMed] [Google Scholar]
- Mähönen AP, ten Tusscher K, Siligato R, et al. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44. [DOI] [PubMed] [Google Scholar]
- Malamy JE. 2005. Intrinsic and environmental response pathways that regulate root system architecture. Plant, Cell and Environment 28: 67–77. [DOI] [PubMed] [Google Scholar]
- Manzano C, Pallero-Baena M, Casimiro I, et al. 2014. The emerging role of reactive oxygen species signaling during lateral root development. Plant Physiology 165: 1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavy P, Vanstraelen M, De Rybel B, et al. 2013. Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO Journal 32: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowetz F, Boutros M, Pons C, et al. 2015. Systems genetics. Linking genotypes and phenotypes. Cambridge: Cambridge University Press. [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, et al. 1996. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in arabidopsis. Science 329: 1065–1067. [DOI] [PubMed] [Google Scholar]
- Meijón M, Satbhai SB, Tsuchimatsu T, Busch W. 2013. Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nature Genetics 46: 77–81. [DOI] [PubMed] [Google Scholar]
- Middleton AM, King JR, Bennett MJ, Owen MR. 2010. Mathematical modelling of the Aux/IAA negative feedback loop. Bulletin of Mathematical Biology 72: 1383–1407. [DOI] [PubMed] [Google Scholar]
- Middleton AM, Úbeda-Tomás S, Grif J, et al. 2012. Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proceedings of the National Academy of Sciences, USA 109: 7571–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Koi S, Hashimoto T, Nakajima K. 2011. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138: 2303–2313. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A, Rogachev I, Brodsky L, et al. 2013. High-resolution metabolic mapping of cell types in plant roots. Proceedings of the National Academy of Sciences, USA 110: E1232–E1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, et al. 1998. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO Journal 17: 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P, Linstead P, Martienssen R, Dolan L. 2002. SCHIZORIZA controls an asymmetric cell division and restricts epidermal identity in the Arabidopsis root. Development 129: 4327–4334. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Dudley AM. 2011. Systems genetics. Science 331: 1015–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. 2001. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. 2007. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 134: 2959–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. The Plant Cell 19: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. 2007. Hidden branches: developments in root system architecture. Annual Review of Plant Biology 58: 93–113. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. 2000. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, et al. 2009. Arabidopsis lateral root development: an emerging story. Trends in Plant Science 14: 399–408. [DOI] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, et al. 2012a. Auxin regulates aquaporin function to facilitate lateral root emergence. Nature Cell Biology 14: 991–998. [DOI] [PubMed] [Google Scholar]
- Péret B, Swarup K, Ferguson A, et al. 2012b. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. The Plant Cell 24: 2874–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Middleton AM, French AP, et al. 2013. Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Molecular Systems Biology 9: 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, et al. 2008. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. The Plant Cell 20: 3258–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas M, Ryan E, Dolan L. 2010. SCHIZORIZA controls tissue system complexity in plants. Current Biology 20: 818–823. [DOI] [PubMed] [Google Scholar]
- Petersson S V, Johansson AI, Kowalczyk M, et al. 2009. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. The Plant Cell 21: 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Schauer MA, Megraw M, et al. 2012. The protein expression landscape of the Arabidopsis root. Proceedings of the National Academy of Sciences, USA 109: 6811–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce G, Rasgado FA, Cassab GI. 2008. Roles of amyloplasts and water deficit in root tropisms. Plant, Cell and Environment 31: 205–217. [DOI] [PubMed] [Google Scholar]
- Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor EB, Baskin TI. 2007. Auxin, actin and growth of the Arabidopsis thaliana primary root. The Plant Journal 50: 514–528. [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. 1994. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes and Development 8: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Rosas U, Cibrian-Jaramillo A, Banta JA, et al. 2013. Integration of responses within and across Aarabidopsis natural accessions uncovers loci controlling root systems architecture. Proceedings of the National Academy of Sciences, USA 110: 15133–15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes and Development 17: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Mochizuki S, Haga K, et al. 2012. The WAVY GROWTH 3 E3 ligase family controls the gravitropic response in Arabidopsis roots. The Plant Journal 70: 303–314. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N. 2006. Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends in Plant Science 11: 440–448. [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, et al. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814. [DOI] [PubMed] [Google Scholar]
- Saucedo M, Ponce G, Campos ME, et al. 2012. An altered hydrotropic response (ahr1) mutant of Arabidopsis recovers root hydrotropism with cytokinin. Journal of Experimental Botany 63: 3587–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage NS, Walker T, Wieckowski Y, Schiefelbein J, Dolan L, Monk NAM. 2008. A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biology 6: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, et al. 1994. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120: 2475–2487. [Google Scholar]
- Scheres B, Dilaurenzio L, Willemsen V, et al. 1995. Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62. [Google Scholar]
- Schiefelbein J, Kwak SH, Wieckowski Y, Barron C, Bruex A. 2009. The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. Journal of Experimental Botany 60: 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Chen R, Masson PH. 1999. ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proceedings of the National Academy of Sciences, USA 96: 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H. 2001. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. The Plant Journal 28: 655–662. [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, et al. 2007. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. The Plant Cell 19: 2440–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, et al. 2010. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora ND, Doonan JH, Herbert RJ, et al. 2011. Arabidopsis T-DNA insertional lines for CDC25 are hypersensitive to hydroxyurea but not to zeocin or salt stress. Annals of Botany 107: 1183–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Simon R. 2009. Is the arabidopsis root niche protected by sequestration of the CLE40 signal by its putative receptor ACR4? Plant Signaling and Behavior 4: 634–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Yoda K, Suzuki M, Suzuki H. 2003. Vascular tissue in the stem and roots of woody plants can conduct light. Journal of Experimental Botany 54: 1627–1635. [DOI] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, et al. 2001. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the arabidopsis root apex. Genes and Development 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benkova E, Swarup R, et al. 2008. The auxin influx carrier LAX3 promotes lateral root emergence. Nature Cell Biology 10: 946–954. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Brown CS, Dreschel TW, Scott TK. 1992. Hydrotropism in pea roots in a porous-tube water delivery system. HortScience 27: 430–432. [PubMed] [Google Scholar]
- Takahashi N, Goto N, Okada K, Takahashi H. 2002. Hydrotropism in abscisic acid, wavy, and gravitropic mutants of Arabidopsis thaliana. Planta 216: 203–211. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Miyazawa Y, Fujii N. 2009. Hormonal interactions during root tropic growth: hydrotropism versus gravitropism. Plant Molecular Biology 69: 489–502. [DOI] [PubMed] [Google Scholar]
- Takatsuka H, Umeda M. 2014. Hormonal control of cell division and elongation along differentiation trajectories in roots. Journal of Experimental Botany 65: 2633–2643. [DOI] [PubMed] [Google Scholar]
- Taylor-Teeples M, Lin L, de Lucas M, et al. 2014. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R, Iwata M, Okada K, Wada T. 2007. Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. The Plant Cell 19: 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. 2010. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H. 2012. Defective root growth triggered by oxidative stress is controlled through the expression of cell cycle-related genes. Plant Science 197: 30–39. [DOI] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster G T, et al. 2005. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. The Plant Cell 17: 3035–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JEM, von Wangenheim D, Barberon M, et al. 2014. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, et al. 2010. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107: 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voß U, Wilson M H, Kenobi K, et al. 2015. The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nature Communications 6: 7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. 1997. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R. 2002. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419. [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan YB, Remans T, Forde BG. 2006. Nitrogen regulation of root branching. Annals of Botany 97: 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, et al. 2005. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349. [DOI] [PubMed] [Google Scholar]
- Willemsen V, Bauch M, Bennett T, et al. 2008. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Developmental Cell 15: 913–922. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, et al. 2006. Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Jennings A, Barlow PW, Forde BG. 1999. Dual pathways for regulation of root branching by nitrate. Proceedings of the National Academy of Sciences, USA 96: 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. 2003. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Xing L, Wang XG, et al. 2014. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Science Signaling 7: ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Wei L, Xu J, et al. 2010. Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. The Plant Cell 22: 3692–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel RW. 1986. Rhizogenetics (root genetics) of vegetable crops. HortScience 21: 956–959. [Google Scholar]