Abstract
Purpose
Adenocarcinoma, the most common non-small cell lung cancer is a leading cause of death worldwide, with a low overall survival (OS) despite increasing attempts to achieve an early diagnosis and accomplish surgical and multimodality treatment strategies. Cellular retinol binding protein-1 (CRBP-1) regulates retinol bioavailability and cell differentiation, but its role in lung cancerogenesis remains uncertain.
Experimental design
CRBP-1 expression, clinical outcome and other prognostic factors were investigated in 167 lung adenocarcinoma patients. CRBP-1 expression was evaluated by immunohistochemistry of tissue microarray sections, gene copy number analysis and tumor methylation specific PCR. Effects of CRBP-1 expression on proliferation/apoptosis gene array, protein and transcripts were investigated in transfected A549 lung adenocarcinoma cells.
Results
CRBP-1High expression was observed in 62.3% of adenocarcinomas and correlated with increased tumor grade and reduced OS as an independent prognostic factor. CRBP-1 gene copy gain also associated with tumor CRBP-1High status and dedifferentiation. CRBP-1-transfected (CRBP-1+) A549 grew more than CRBP-1− A549 cells. At >1μM concentrations, all trans-retinoic acid and retinol reduced viability more in CRBP-1+ than in CRBP-1− A549 cells. CRBP-1+ A549 cells showed up-regulated RARα/ RXRα and proliferative and transcriptional genes including pAkt, pEGFR, pErk1/2, creb1 and c-jun, whereas RARβ and p53 were strongly down-regulated; pAkt/pErk/ pEGFR inhibitors counteracted proliferative advantage and increased RARα/RXRα, c-jun and CD44 expression in CRBP-1+ A549 cells.
Conclusion
CRBP-1High expression in lung adenocarcinoma correlated with increased tumor grade and reduced OS, likely through increased Akt/Erk/EGFR-mediated cell proliferation and differentiation. CRBP-1High expression can be considered an additional marker of poor prognosis in lung adenocarcinoma patients.
Keywords: lung cancer, CRBP-1, Akt, Erk, EGFR, prognostic marker, survival
INTRODUCTION
Non-small-cell lung cancer (NSCLC) is a leading cause of cancer death worldwide [1]. Lung cancer is divided in two major categories according to histological features and response to conventional therapies. More than 85% of lung malignant tumors are NSCLC [2] and adenocarcinoma frequence largely prevails among lung NSCLCs hystotypes [3]. Despite the recent advances in diagnostic and therapeutic procedures, including the development of computed tomography-based screening programs for early detection of lung cancer in higher risk populations, overall survival (OS) in lung adenocarcinoma patients remains poor with a heterogeneous and as yet suboptimal response rates to both surgical and multimodality chemo-radiation therapeutic approaches [3]. Research effort has been focused on identifying new biomarkers and those molecular pathways influencing critically NSCLC progression. The discovery of activating EGFR mutations and the subsequent development of tyrosine kinase inhibitors led to a revolution in the treatment of NSCLC patients [4]. Recently, the screening of other prognostic genetic factors or biomarkers influencing DNA repair mechanisms and inflammatory response have been suggested to predict recurrence or metastasis of NSCLC [5]. The discovery of additional markers of NSCLC heterogeneity remains a goal to suggest new therapeutic perspectives and clinical trials to improve individual therapeutic response. Vitamin A (retinol) and its metabolites are essential for many biological processes and influence epithelial cell differentiation and proliferation [6]. Biological activity of retinol is normally mediated by specific receptors. Retinol mediates pleiotropic and transcriptional effects of retinoids through the binding to nuclear receptors, namely the retinoic acid receptors (RARα, β, and γ) and retinoid X receptors (RXRα, β, and γ) [7]. Recently, increased interest has been focused on the role of cellular retinol and retinoic acid binding proteins (CRBPs and CRABPs) in carcinogenesis. Physiologically, CRBPs and CRABPs regulate intracellular retinoid trafficking and retinoid-induced cell activities. CRBP-1 is a 15 kDa cytosolic binding protein crucial for the uptake and subsequent esterification of retinol, so regulating of its bioavailability and transcriptional activities [6] [8]. CRBP-1 is indispensable for embryonic development, growth, vision and survival of vertebrates and in lung transient CRBP-1 expression is reported during pre-natal alveolus formation [9]. A potential role of CRBP-1-driven aberrant intracellular retinoid signaling in non-lung cancer carcinogenesis has been highlighted [10-13]. Natural and synthetic retinoids are effective for the treatment of skin proliferative disorders and also represent chemopreventive agents [14,15]. Therapeutic employment of retinoids in NSCLC patients gave controversial results and a link to smoking habits suggested [16]. Here, we aimed to define if CRBP-1 expression can influence tumor progression and OS in lung adenocarcinoma patients. CRBP-1 expression and its relationship with survival and other prognostic factors was investigated in a series of 167 adenocarcinoma patients. The effects of CRBP-1 transfection on proliferation, transcription, dedifferentiation and sensitivity to retinoids in A549 adenocarcinoma cells were also reported and discussed.
RESULTS
CRBP-1 expression and clinicopathological characteristics
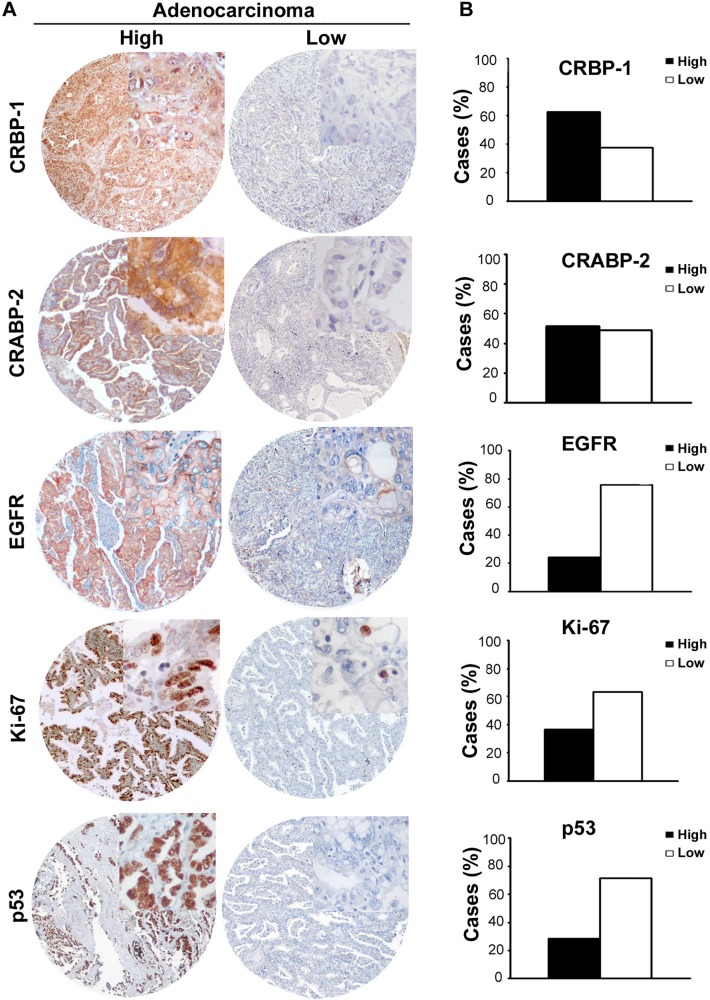
Representative immunostainings of CRBP-1 expression and other lung adenocarcinoma biomarkers are reported in figure 1A. As shown in figure 1B, CRBP-1High expression was detected in 62.3%. EGFR and Ki-67 expression were comparable to the literature [17,18]. CRABP-2High expression was detected in 51.85% of tumors. Immunohistochemistry also documented that CRBP-1 expression was absent in non-neoplastic lung parenchyma and present in bronchial epithelium (Supplementary Figure S1A-F). CRBP-1 expression varied according to the hystopathological subtype (Supplementary Figure S1G). In particular, acinar, micropapillary, solid and mucinous subtypes resulted more frequently CRBP-1High, whereas CRBP-1Low expression prevailed among lepidic and papillary subtypes. Patients' clinicopathological characteristics and their relationship with CRBP-1 expression are reported in table 1. Smokers status (current or former smokers) showed a significant association with tumor CRBP-1High expression (p<0.001). As concerning the tumor grade, CRBP-1High status associated to the loss of differentiation of lung adenocarcinomas (p<0.001).
Figure 1. Immunostaining and semiquantitative evaluation of CRBP-1 and other biomarkers in TMA sections of lung adenocarcinomas.
A, representative images of “High” and “Low” CRBP-1, CRABP-2, EGFR, Ki-67 and p53 expression in lung adenocarcinoma. B, bar graphs showing the percentages of tumors with “High” and “Low” expression. Diaminobenzidine is used as chromogen. Criteria for the definition of “High” and “Low” tumors are reported in the Patients and Methods section. Original magnification, 40X; the inset highlighting the staining at higher magnification, 400X.
Table 1. Clinicopathologic characteristics by CRBP-1 expression and factors influencing survival in lung adenocarcinoma patients.
| Total Number | CRBP-1 expression | |||
|---|---|---|---|---|
| Low (%) | High (%) | |||
| Variable * | 167 (%) | 63 (37.7) | 104 (62.3) | P |
| Sex | ||||
| male | 128 (76.6) | 48 | 80 | 0.91 |
| female | 39 (23.4) | 15 | 24 | |
| Age | ||||
| < 67 years | 85 (50.9) | 32 | 53 | 0.98 |
| > 67 years | 82 (49.1) | 31 | 51 | |
| Smoking status | ||||
| current or former smokers | 138 (82.6) | 40 | 98 | 0.001 |
| no-smokers | 29 (17.4) | 23 | 6 | |
| Histologic grade | ||||
| G1 | 18 (10.8) | 13 | 5 | 0.001 |
| G2+G3 | 149 (89.2) | 50 | 99 | |
| pT | ||||
| T1+T2 | 134 (80.2) | 49 | 85 | 0.37 |
| T3+T4 | 33 (19.8) | 14 | 19 | |
| Disease stage | ||||
| I | 112 (65.6) | 43 | 69 | 0.37 |
| II+III+IV | 55 (34.4) | 20 | 35 | |
| Variable ** | P | HRa | 95% Clb |
|---|---|---|---|
| CRBP-1 | 0.02 | 0.43 | 0.21-0.89 |
| EGFR | 0.001 | 3.18 | 0.67-6.05 |
| CRABP-2 | 0.13 | 1.61 | 0.86-3.00 |
| Ki-67 | 0.3 | 0.72 | 0.38-1.33 |
| RARβ | 0.09 | 0.58 | 0.31-1.08 |
| RARα | 0.68 | 0.87 | 0.45-1.69 |
| Smokers/CRBP-1High | 0.07 | 0.50 | 0.64-1.06 |
| Coexpression EGFR/CRBP-1 | 0.24 | 0.56 | 0.21-1.46 |
χ2 test was used for relationships;
Multivariate Cox Regression Model;
Hazard Ratio;
Confidence Interval
CRBP-1High expression in lung adenocarcinoma associates with reduced overall survival
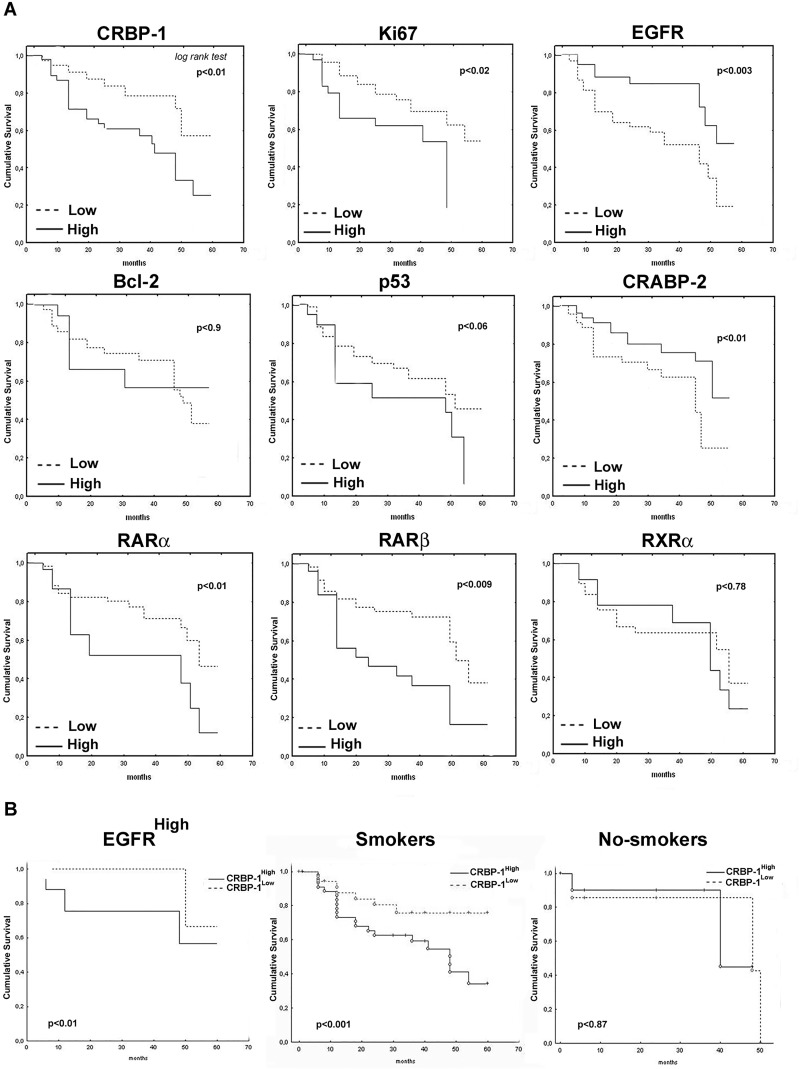
As reported in figure 2A, CRBP-1High expression in lung adenocarcinoma associated with reduced patients' OS (p<0.01). High RARα, RARβ and Ki-67 (p<0.01; p<0.009 and p<0.02, respectively) and low CRABP-2 and EGFR expression (p<0.01 and p<0.003, respectively) also correlated with lower OS, similarly to that reported in the literature [19]. As reported in figure 2B, tumor subgroup with EGFRHigh and CRBP-1High coexpression associated with reduced OS (p<0.01). CRBP-1High expression strongly also associated with reduced OS in smoker (p<0.001) but not in no-smoker patients. Multivariate analysis (Table 1) documented CRBP-1High and EGFRHigh expression as independent prognostic factors in lung adenocarcinoma patients (HR=0.43 and 3.18; p<0.02 and p<0.001, respectively).
Figure 2. Overall survival and CRBP-1 and other biomarker expression in lung adenocarcinoma patients.
A, CRBP-1 and other biomarker expression and overall survival in lung adenocarcinoma patients (n=167). B, survival rate of patients with tumor CRBP-1High and EGFRHigh coexpression and smoking habits in lung adenocarcinoma patients. Significance is calculated by Log rank test.
Relationship between CRBP-1 and other lung adenocarcinoma markers
CRBP-1High correlated positively with EGFRHigh expression (rho=0.38, p<0.01), and negatively with p53 expression (rho=−0.30, p<0.01). CRABP-2High inversely correlated with bcl-2High expression (rho=−0.25, p<0.05) and patients' age (rho=−0.28, p<0.05). An inverse correlation between EGFR and bcl2 expression (rho=−0.31, p<0.01) also existed. As reported in supplementary figure S1H, we compared keratins and nox4 expression according to CRBP-1 expression. In general, expression of keratins 1 and 5/6 was focal, whereas keratin 14 was not expressed. The percentages of tumor keratin 1 and 5/6 in CRBP-1High was increased and almost double compared with CRBP-1Low tumors. Instead the percentage of nox4 expression was similar. Screening for TK domain (exons 18-21; Supplementary figure S2A) documented that 19.8% of lung adenocarcinomas were EGFR-mutated, mostly in 19 and 21 exons, according to the literature [20,21]. We did not find mutations in 18 and 20 exons in our patients' cohort. A tendential correlation between EGFR-mutated status and CRBP-1High expression was observed, although the association was not statistically significant (rho=0.21, p=0.15).
CRBP-1 gene copy number and methylation in lung adenocarcinoma
As reported in supplementary figure S2B, 45.3% of lung adenocarcinomas showed 3-6, 42.85% two and 11.85% less than two copies of CRBP-1 gene. Increased CRBP-1 gene copy number associated with tumor CRBP-1High status and dedifferentiation (rho=0.31; p<0.05). Since CRBP-1 expression was absent in a subset of tumors, we analyzed the methylation of the promoter region flanking the CRBP-1 gene (Supplementary figure S2C, D). Promoter methylation was present in 21.4% of adenocarcinomas and a correlation between CRBP-1Low status and methylation gene expression was also documented (rho=−0.36, p<0.01).
CRBP-1 transfection increased proliferation and retinoid sensitivity of A549 adenocarcinoma cells
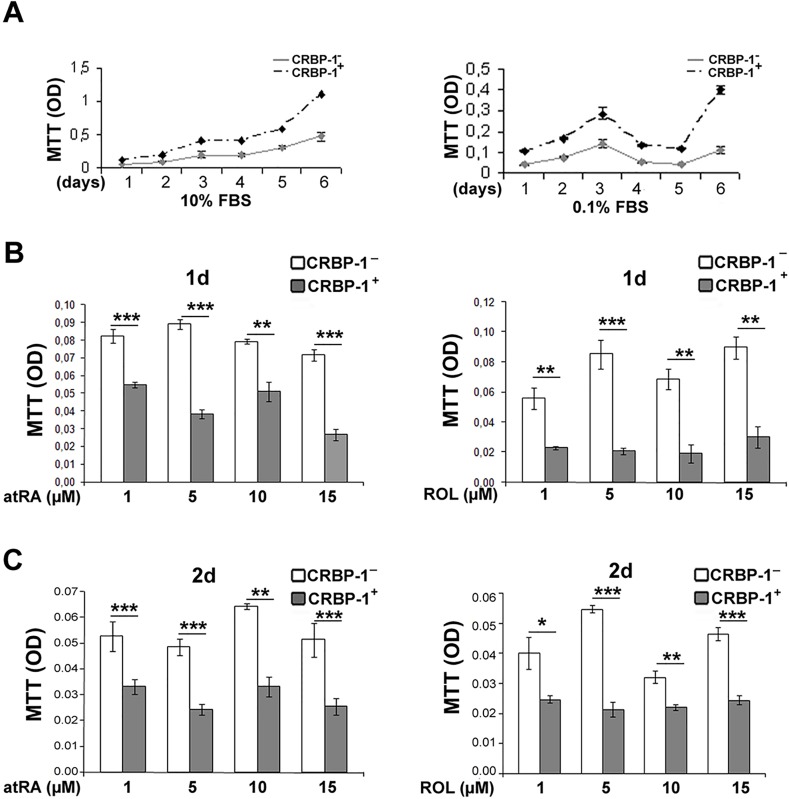
A mammalian CRBP-1-expressing vector was used to generate stable transfectant A549 lung adenocarcinoma cell lines. Wild A549 cells did not display appreciable CRBP-1 mRNA and protein levels (Supplementary figure S3A, B). After 6 days (figure 3A), MTT assay showed that CRBP-1+ grew more than CRBP-1− A549 cells. Cell count gave similar results (not shown). At concentrations >1μM (figure 3B, C), atRA and ROL reduced viability more in CRBP-1+ than in CRBP-1− A549 cells. Finally, after 2 weeks of culture with 10% FBS, the ability of CRBP-1+ A549 cells to form colonies (66.0% ± 3.04%) slightly increased compared to CRBP-1− cells (58.13% ± 2.92%), although the difference was not significant. Treatments with atRA and ROL reduced clonogenicity in both CRBP-1+ and CRBP-1− A549 compared to control cultures (data not shown).
Figure 3. Viability and retinoid-related survival of CRBP-1-transfected A549 adenocarcinoma cells.
A, Serum-dependent of CRBP-1+ A549 cells growth is increased compared to CRBP-1− cells. B-C, MTT assay shows reduced viability of CRBP-1+ compared to CRBP-1− A549 cells after 1 and 2 days of atRA and ROL treatment in the presence of 0.1% FBS. Values are expressed as means ± SEM of three different experiments; *p < 0.05, **p < 0.005, ***p <<0.0005.
CRBP-1 transfection influences differentiation and RAR/RXR signaling of A549 adenocarcinoma cells
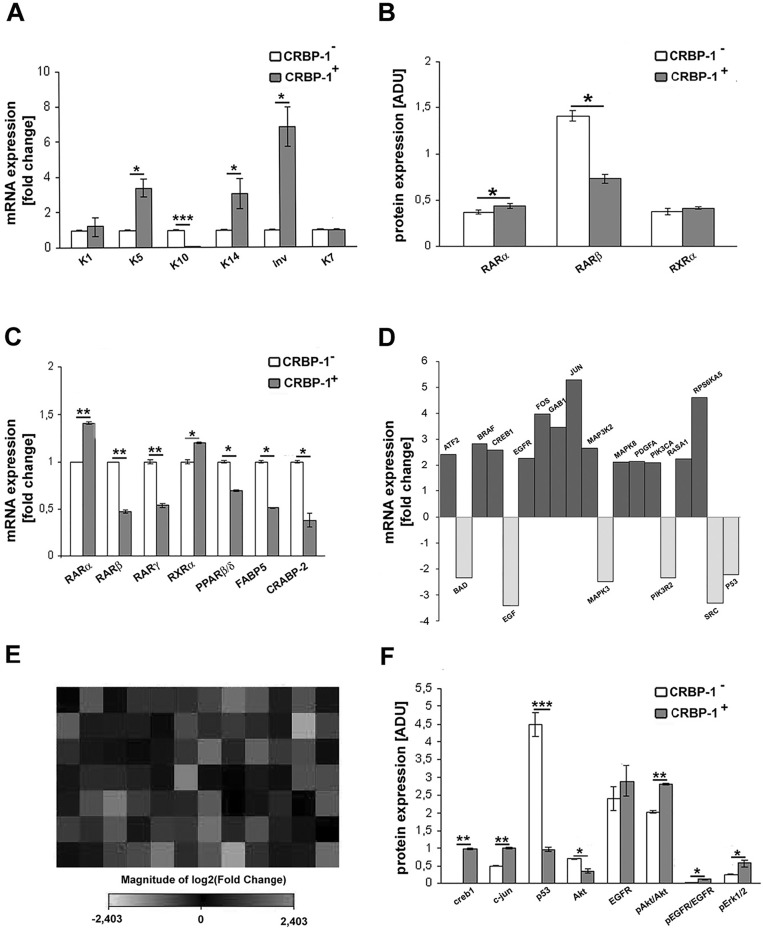
In order to further investigate the effects on phenotype of CRBP-1 expression we performed proliferative, epithelial and epithelial to mesenchymal markers expression in A549 cells. Real-time PCR (figure 4A) showed the up-regulation of epithelial proliferative markers as keratin 1, 5 and 14 and involucrin and the down-regulation of keratin 10 transcripts in CRBP-1+ compared to CRBP-1− A549 cells; keratin 7 level was unchanged. Keratin 5 and 14 up-regulation was confirmed by blots (data not shown). CRBP-1+ A549 cells also showed the strong up-regulation of nox4 and CD44 expression compared to CRBP-1− A549 cells (figure 5A). The expression of other epithelial-to-mesenchymal transition markers such as vimentin, nestin, smad4, nanog, sox2, snail, TGFβ and MMP9 did not change (Supplementary figure S3C). We also evaluated the influence of CRBP-1 expression on retinoid signalling by blots and real time PCR. As reported in figure 4B and C, RARα and RARβ expression were up-regulated and down-regulated, respectively at both protein and transcriptional level in CRBP-1+ compared to CRBP-1− A549 cells; moreover, PPARβ/δ, FABP5 and CRABP-2 transcripts resulted down-regulated.
Figure 4. CRBP-1 transfection influences transcriptional pathways and differentiation of A549 adenocarcinoma cells.
A, bar graph after real-time PCR showing keratin (K) 1, 5, 14 and involucrin up-regulation, K10 down-regulation and unmodified K7 transcription level in CRBP-1+ compared to CRBP-1− A549 cells. B, densitometric analysis of RARα, RARβ and RXRα protein expression by blot analysis. C, bar graph of RARs, RXRα, PPARβ/δ, FABP5 and CRABP-2 transcripts. D, bar graph and E, heat map of RT2 profilerTM PCR assay of EGF/PDGF signaling-specific genes in CRBP-1+ A549 cells. Up-regulated and down-regulated genes are in dark grey and light grey, respectively. F, densitometric analysis of creb1, c-jun, p53, pAkt/Akt, pEGFR/EGFR and pErk1/2 protein expression by blots. Columns are means ± SEM of three different experiments. *p < 0.05, **p < 0.005 and ***p <0,001. Abbreviations: ADU, arbitrary densitometric units.
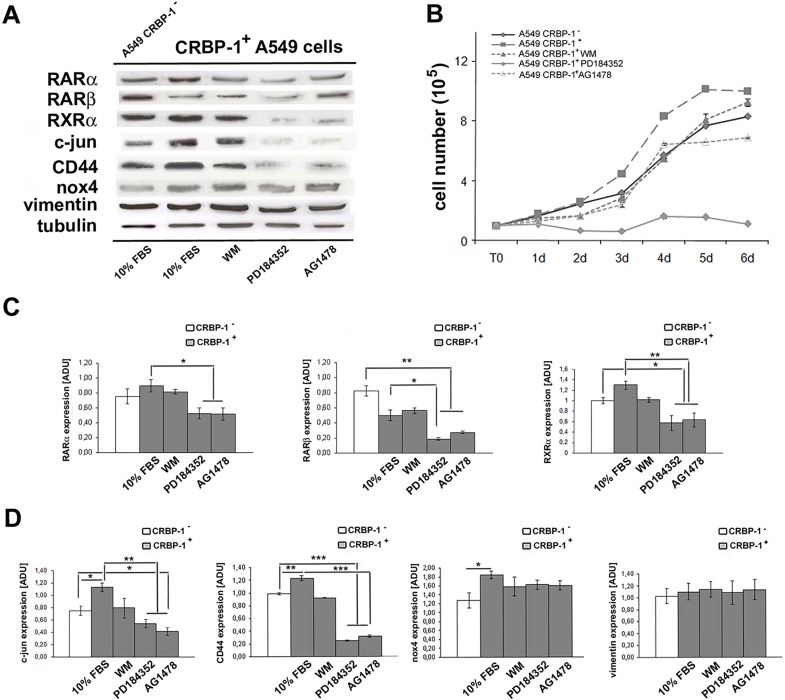
Figure 5. Effects of inhibition of AKT, MAPKK and EGFR activity in CRBP-1+ A549 cells.
A, Representative blots of RARs, RXRα, c-jun, CD44, nox4 and vimentin protein expression in CRBP-1+ A549 treated with inhibitors for 48 hours compared with CRBP-1− A549 cells. B, cell growth graph of CRBP-1− and CRBP-1+ A549 cells maintained with 10% of FBS and treated with wortmannin (WM), PD184352 and AG1478, as Akt, MAPKK and EGFR inhibitors up to 6 days. Densitometric analysis (C) RARs, RXRα, (D) c-jun, CD44, nox4 and vimentin protein expression. Membranes were reblotted with anti-tubulin to ensure equal loading. Results are mean values ± SEM of the three different experiments. *p < 0.05, **p < 0.005 and ***p <0,001. Abbreviations: ADU, arbitrary densitometric units.
CRBP-1 transfection associated with up-regulation growth and transcriptional gene levels in A549 adenocarcinoma cells
As reported in figure 4D and E, PCR array documented a series of up-regulated genes influencing cell growth and survival (BRAF, EGFR, GAB1, MAP3K2, MAPK8, PDGFA, PIK3CA, RASA1) and transcriptional activators (ATF2, CREB1, FOS, JUN, RPS6KA5) in CRBP-1+ compared to CRBP-1− A549 cells. Instead, BAD, EGF, MAPK3, PIK3R2, SRC and P53 gene levels resulted down-regulated. Blot analysis (figure 4F) also documented the up-regulation of transcriptional factors creb1 and c-jun and of activity of survival and proliferative pathways, including pAkt, pEGFR and pErk1/2 in CRBP-1+ compared to CRBP-1− A549 cells; instead p53 was down-regulated.
Akt/Erk/EGFR inhibitors influence proliferative and epithelial-to-mesenchymal transition marker expression in CRBP-1+ A549 adenocarcinoma cells
In order to better identify those pathways regulated by CRBP-1 expression, we performed inhibition experiments. As reported in figure 5B, PD184352 MAPK inhibitor drastically reduced proliferation of CRBP-1+ A549 cells. Similar effects on proliferation were observed after wortmannin and AG1478 treatment. Moreover, as reported in figure 5C and D, PD184352 and AG1478 inhibitors strongly reduced RARβ, c-jun and CD44 expression in CRBP-1+ A549 cells. Also wortmannin-induced inhibition of pAKT reduced RARβ, RXRα, c-jun and CD44 expression to levels similar to CRBP-1− A549 cells. All inhibitors did not change nox4 and vimentin protein expression.
DISCUSSION
Our study results have shown that in a cohort of lung adenocarcinoma patients undergoing radical surgical treatment, tumor CRBP-1High expression was associated with an unfavorable OS. In normal cells, CRBP-1 regulates intracellular retinol trafficking and bioconversion, so facilitating its biological functions [6]. In particular, retinol contributes to epithelial cell proliferation and differentiation. CRBP-1High expression in lung adenocarcinomas was parallel to the increase of CRBP-1 gene copy number and also associated with increased tumor grade and keratin 1 and 5/6 expression, suggesting that CRBP-1High expression reflects a more aggressive and dedifferentiated phenotype of adenocarcinoma cells. This was confirmed from the increased proliferation and dedifferentiation markers expression in CRBP-1- transfected A549 cells. Current paradigms retain that lung cancer arises from pluripotential stem cells capable of differentiation into one or several histological cell types, with the activation of genes recapitulating embryonic lung development. Lung adenocarcinoma has been associated with de novo gene expression of developmental terminal sac and alveolar stages, with a prevalence of genes influencing differentiation and signal transduction [22]. Our data are in line with the link between aberrant CRBPs expression and carcinogenesis described in non-lung districts, including laryngeal and hepatic cancer [10,12,13]. The same was reported in high-grade gliomas, where CRBP-1High expression also associated with poor prognosis [23]. Aberrant CRBP-1 expression also occurred in non-epithelial malignant tumors, such as leiomyosarcomas [24]. Aberrant CRBP-1 expression was not univocal in non-lung epithelial malignancies. Loss of CRBP-1 expression has been reported in human dedifferentiated breast, endometrial and ovarian cancers [8,10,11,25]. It is likely that the prevalence of CRBP-1High phenotype in lung adenocarcinoma facilitates intracellular retinoid level accumulation and trafficking supporting tumor cell proliferation and dedifferentiation in response to oncogenetic stimuli [6].
Over-expression of oncogenes or inactivation of tumor suppressor genes have been identified in a significant number of NSCLC patients [26]. Besides, clinical prognostic factors, such as stage, sex, and performance status, tumor molecular markers have been recognized to influence OS in NSCLC patients [27]. Our results strongly suggest that CRBP-1High expression can be considered as an additional phenotypic marker of lung adenocarcinomas with a more aggressive clinical course. We also documented that RARαHigh, RARβHigh and CRABP-2Low expression associates with reduced OS. We also documented that EGFRHigh positively correlated with CRBP-1High expression, strongly supporting an interaction between CRBP-1-mediated retinoid and EGFR pathways [28]. This finding is apparently in contrast with the overall reduced survival in patients with EGFRLow expression. The literature contains conflicting data on the relationship between EGFR expression and survival in lung cancer. Variability and discrepancy of results may be due to heterogeneity of study population related to EGFR status at time of primary diagnosis, EGFR mutational status and/or chemotherapy [17]. CRBP-1-mediated increased transport of retinol to intracellular related enzymatic milieu is likely to amplify RAR-mediated transcriptional signals [29]. In epithelial cells, atRA and RAR-selective ligands specifically down-regulated EGFR-dependent activities and atRA regulated cell growth and differentiation [28]. Nevertheless, retinoids are known to control genes that do not contain classical RARE motifs, and to activate directly intracellular signaling molecules or transcription factors, such as Erk signaling components [30].
Our present data help to better understand the complex role and the contrasting opinions concerning the efficacy of retinoid-related in chemotherapeutic regimens in lung cancer patients. Retinoids are successfully used in preneoplastic or neoplastic skin diseases, head and neck cancer, neuroblastoma and cutaneous T-cell lymphoma. The use of retinoids for the treatment and prevention of lung cancer gave controversial results [31]. Preliminary evidence including preclinical and observational studies reported promising results, but these effects were not fully translated in human interventional settings. Encouraging results derived from efficacy of oral administration of high-dose vitamin A in reducing the number of primary tumors related to tobacco consumption and improving the disease-free interval in patients resected for stage I lung cancer [32]. Conversely, other trials found a significantly increased risk of lung cancer in current and former smokers. In particular, retinoids reduced tumor occurrence and mortality in non-smokers and were beneficial in former smokers, but increased the risk of lung cancer in smokers [16]. Our data demonstrating that OS was particularly reduced in smoker patients with CRBP-1High tumor expression is highly suggestive. Although other studies are needed to clarify mechanisms responsible of higher incidence of lung cancers in smokers who received retinoids, it is likely that CRBP-1High expression favors retinoid-induced proliferation of tumors cells. As matter of fact, CRBP-1 transfection increased proliferation, up-regulated RAR-α and down-regulated RARβ expression in A549 adenocarcinoma cells.
Gene array and blot documented that CRBP-1 transfection A549 cells induced the increase of transcription and activity of proliferative, transcriptional and dedifferentiative genes, including pEGFR/pAkt/pErk pathways, RARα, c-jun and CD44. CRBP-1 transfection effects were partially reverted by specific inhibitors of EGFR/Akt/Erk pathways. C-jun up-regulation has been reported to influence carcinogenesis and tumour progression in lung adenocarcinoma cells [33]. Our hypothesis is that CRBP-1 alone or homodimerized or heterodimerized with RARα or RARα/RXRα could interact and activate Akt and down-regulate related transcriptional, proliferative and dedifferentiative genes. This hypothesis is consistent with the previously reported finding that RARα over-expression increases activity and co-localizes with Akt at cell membrane level likely by interacting with PI3k [34]. Our in vitro results showed increased keratin 5, 14 and involucrin expression in CRBP-1+ A549 cells. Most of poorly differentiated adenocarcinomas express focally keratins 5, 6, 14 and 17 [35]. Coexpression of keratin 14, a basal cell marker of squamous and glandular epithelia, keratin 5 and involucrin are reported to represent a stem cell or progenitor cell phenotype in cancer cells [36]. Other experiences are needed to better clarify through with pathway CRBP-1 favors epithelial to mesenchymal transition in adenocarcinoma cells. In this light, we also described the increased expression of CD44 and nox4 in CRBP-1+ A549 cells. Increased CD44 expression was described to be associated with a poor outcome in lung adenocarcinoma patients and tumor progression [37].
In conclusion, in the present study we documented that CRBP-1High expression in lung adenocarcinomas associates with a poor survival and increased tumor grade, likely influencing the activity of Akt/EGFR gene pathways. Further studies are needed to verify the possibility of CRBP-1-related therapeutic intervention aimed to reduce NSCLC progression for a more personalized chemotherapeutic regimens.
PATIENTS AND METHODS
For the study purpose, 167 NSCLC patients who underwent surgical resection with histologic diagnosis of adenocarcinoma either at the Policlinic of Tor Vergata University of Rome and at the Santa Maria della Misericordia Hospital of Perugia, Italy, between 2003 and 2009 were included. Patients' written informed consent was obtained. The study was approved by the Local Ethics Committee. Tumor classification was in accordance with WHO criteria and the most diffuse immunohistochemical panel [38,39]. Tumor subtyping, grading and staging were in accordance with the international tumor-node-metastasis system (TNM) [39,40]. Criteria of exclusion were pre-operative radiation and/or chemotherapy and inadequate amount of tumor tissue for correct routinary processing and diagnosis (at least two tissue cores).
Tissue microarray construction
For tissue microarray (TMA) construction, tissue samples from diagnostic biopsies and operative procedures were obtained from representative paraffin blocks maintaining patients' anonymity. All tumor slides were reviewed by light microscopy examination of Haematoxylin&Eosin (H&E)-stained sections. The most representative tumor areas were carefully selected and TMA constructed using positive and negative controls [8]. Serial 4 μm-thick sections were stained with H&E or employed for immunohistochemistry.
Immunohistochemical study
For immunohistochemistry, sections were incubated with mouse monoclonal anti-human Ki-67 (clone 30-9), bcl2 (clone 124), p53 (clone DO-07), EGFR (clone 3C6), keratin 5/6 (clone D5/16B4) and keratin 14 (clone SP53) antibodies using an automatic immunostaining device (Ventana-Roche Diagnostics Milan, Italy) [41]. Serial sections were also incubated for 1 h with rabbit polyclonal anti-CRBP-1 (1:200; clone FL-135, Santa Cruz Biotechnology, Heidelberg, Germany), anti-CRABP-2 (1:300; Bethyl Laboratories, Montgomery, USA), anti-RARα (1:500; clone sc-551, Santa Cruz Biotechnology), anti-RARβ (cytoplasmic isoform β4; 1:100; clone ab53161 Abcam, Cambridge, UK) and anti-RXRα antibodies (1:500; clone sc-553, Santa Cruz Biotechnology), anti-keratin 1 (1:750; clone ab24643 Abcam), anti-Nox4 (1:500; H-300, Santa Cruz Biotechnology). Diaminobenzidine was used as final chromogen. Slides were also stained with a mouse monoclonal anti-CRBP-1 antibody (1:10, gifted from Dr ML Bochaton, University of Geneva, Switzerland), that gave similar results (not shown).
CRBP-1 gene copy number variation and methylation
CRBP-1 gene copy number was analyzed by using real-time PCR and TaqMan Genotyping Master Mix (n. 4317355), with RNase control reagents (n. 4316844) and CRBP-1 (Hs00443703, Applied Biosystem, Foster City, CA, USA) as probe. PCR amplification was performed in an ABI PRISM 7500 (Applied Biosystem) according to manufacturer's instructions. Data analysis was performed using the manufacturer's integrated web-based software package. Genomic DNA, methylation specific PCR and copy number assay were performed as reported [8]. Briefly, DNA was isolated by using a FFPE tissue kit (Qiagen, Hilden, Germany) and PCR carried out using AmpliTaq Gold DNA polymerase (Applied Biosystem) and specific primers, as reported [8].
EGFR mutational status
EGFR mutational status was analized by pyrosequencing [42]. The TK domain of the EGFR coding sequence involving exons 18-21 was amplified by using the EGFR TKI Response PQ (Diatech Pharmacogenetics, Jesi AN, Italy) and then sequenced using PyroMark Q24 instrument (Qiagen).
Cell transfection
Human A549 adenocarcinoma cells (Sigma-Aldrich, St. Louis, USA) maintained in RPMI 1640 (Lonza Bio Pharma AG, Switzerland) were transfected ([43]) by using a vector pTargeT Mammalian expression system carrying the whole sequence of CRBP-1 gene (NM_002899) and the gene for the resistance to G418 (CRBP-1+, Promega, Italy), or the G418-resistance gene alone (CRBP-1-). After 20 days, stable transfected clones were collected and tested by PCR and western blot. The correct plasmid sequence was confirmed by Sanger sequencing. Experimental procedures were repeated by using two different transfected clones, which gave similar results (not shown).
Cell growth and viability
For proliferation studies, overnight serum-starved cells were treated with different concentrations of atRA and ROL (Sigma-Aldrich) in 0.1% FBS up to 6 days. For cell viability, 3-(4,5 dimethylthiazol-2-yl)-2,5diphenyl-tetrazolimbromide assay (MTT; Sigma-Aldrich) was carried out in triplicate ([44]. In some instances, CRBP-1+ cells were pre-treated with selective EGFR (AG1478; Sigma-Aldrich; 10 μM), phosphatidylinositol 3-kinase/ AKT (Wortmannin; Sigma-Aldrich; 10 μM) and the mitogen-activated protein kinase kinase (MAPKK, PD184352; Sigma-Aldrich; 2μM) inhibitors.
Clonogenic assay
For the clonogenic assay ([45], cells were seeded and treated with different concentrations of atRa and ROL. Colonies arising from survival cells were fixed and stained with 1% methylene blue (Sigma-Aldrich) in 0.1% methanol and their percentages as plating efficiency (PE) calculated.
Western blot analysis
After isolation, content determination and electrophoresis, proteins were elettroblotted [46] and incubated with a polyclonal rabbit anti-CRBP-1, anti-Creb1, anti-CD44, anti-c-Jun, anti-Nox4, anti-p53, anti-RXRα, anti-RARα (Santa Cruz Biotechnology), anti-RARβ, (Abcam), anti-phosphorylated v-akt murine Jesi AN, Italy) and then sequenced using PyroMark Q24 thymoma viral oncogene homolog (pAkt Ser473), anti-AKT (pan), anti-phosphorylated extracellular-signal-regulated kinases (pErk1/2), anti-phosphorylated epidermal growth factor receptor (anti-EGFR Thr669), anti-EGFR antibody (Cell Signaling Technology, Danvers, MA, USA), anti-keratin 5 (clone H-40, Santa Cruz Biotechnology), mouse anti-vimentin (clone J144, Abcam), anti-keratin 14 (LL001, Santa Cruz Biotechnology) and anti-total tubulin antibody (Sigma-Aldrich). Revelation and densitometric blot analysis were performed in three independent experiments and Akt and EGFR activity expressed as phospho/total protein ratio [47].
Gene expression analysis
Total RNA was extracted [48], reverse-transcribed and a commercially available RT profiler PCR array of 86 genes human EGF/PDGF signaling (PAHS-040Z, Qiagen) and Real time PCR performed according to the manufacturer' instruction and primers listed in supplementary table S1; Δ2-microglobulin, Δ-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as housekeeping genes. Data analysis was performed by using the integrated web-based software package using ∆∆Ct fold-change calculation (Qiagen) in triplicate experiments.
Semiquantitative and statistical analysis
CRBP-1, CRABP-2, nox4 and keratin expression was estimated at 400X magnification by two of the Authors by using the following semiquantitative grading system: absent (0), weakly positive <50% (0.5), moderately positive <50% or weakly positive >50% (1), strongly positive <50% or moderately positive >50% (2), strongly positive >50% (3), as reported [25]. Inter-observer reproducibility was >95%. Tumors with 0-0.5 CRBP-1 and CRABP-2 score were arbitrarily grouped as “Low” and those with 1-3 score as “High”. For p53, Ki-67, bcl-2 and RAR/RXR, tumors were arbitrarily considered as “Low” when expression was less than 20%. Continuous moderate or marked membranous staining in >50% of cells was required for the definition of EGFRHigh expression [49]. For each case, the ratio of the score and the number of analyzed fields was calculated. Results were analyzed by means of Student's t test. Univariate analysis of relationship among CRBP-1 and clinicopathological variables was performed by using χ2 test, whereas correlations between CRBP-1 and the other biomarkers by Spearman's rank correlation test. Survival curves were analyzed by Kaplan-Meier method and significant differences between subgroups were calculated by the log-rank test. Independent prognostic factors were identified by multivariate analysis using Cox proportional hazards model. Only factors showing prognostic significance in univariate analysis were adopted in multivariate analysis. Differences were considered statistically significant for value of p<0.05. SPSS 16 software program (Spss inc. Chicago, IL, USA) was used for statistical analysis.
SUPPLEMENTARY MATERIAL FIGURES AND TABLE
Acknowledgments
The authors thank S. Cappelli, L. Santangelo, R. Bernabei and C. Semproni for their technical assistance, and Dr. F. Tacconi for the collection of clinical data.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Beasley MB, Brambilla E, Travis WD. The 2004 World Health Organization classification of lung tumors. Semin Roentgenol. 2005;40:90–97. doi: 10.1053/j.ro.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, Powell CA, Beer D, Riely G, Garg K, Austin JH, Rusch VW, Hirsch FR, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc. 2011;8:381–385. doi: 10.1513/pats.201107-042ST. [DOI] [PubMed] [Google Scholar]
- 3.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102:464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Napoli JL. Biosynthesis and metabolism of retinoic acid: roles of CRBP and CRABP in retinoic acid: roles of CRBP and CRABP in retinoic acid homeostasis. J Nutr. 1993;123:362–366. doi: 10.1093/jn/123.suppl_2.362. [DOI] [PubMed] [Google Scholar]
- 7.Zhuang Y, Faria TN, Chambon P, Gudas LJ. Identification and characterization of retinoic acid receptor beta2 target genes in F9 teratocarcinoma cells. Mol Cancer Res. 2003;1:619–630. [PubMed] [Google Scholar]
- 8.Doldo E, Costanza G, Ferlosio A, Passeri D, Bernardini S, Scioli MG, Mazzaglia D, Agostinelli S, Del Bufalo D, Czernobilsky B, Orlandi A. CRBP-1 expression in ovarian cancer: a potential therapeutic target. Anticancer Res. 2014;34:3303–3312. [PubMed] [Google Scholar]
- 9.Dirami G, Massaro GD, Clerch LB, Ryan US, Reczek PR, Massaro D. Lung retinol storing cells synthesize and secrete retinoic acid, an inducer of alveolus formation. Am J Physiol Lung Cell Mol Physiol. 2004;286:L249–256. doi: 10.1152/ajplung.00140.2003. [DOI] [PubMed] [Google Scholar]
- 10.Kuppumbatti YS, Bleiweiss IJ, Mandeli JP, Waxman S, Mira Y, Lopez R. Cellular retinol-binding protein expression and breast cancer. J Natl Cancer Inst. 2000;92:475–480. doi: 10.1093/jnci/92.6.475. [DOI] [PubMed] [Google Scholar]
- 11.Orlandi A, Ferlosio A, Ciucci A, Francesconi A, Lifschitz-Mercer B, Gabbiani G, Spagnoli LG, Czernobilsky B. Cellular retinol binding protein-1 expression in endometrial hyperplasia and carcinoma: diagnostic and possible therapeutic implications. Mod Pathol. 2006;19:797–803. doi: 10.1038/modpathol.3800586. [DOI] [PubMed] [Google Scholar]
- 12.Peralta R, Baudis M, Vazquez G, Juarez S, Ortiz R, Decanini H, Hernandez D, Gallegos F, Valdivia A, Piña P, Salcedo M. Increased expression of cellular retinol-binding protein 1 in laryngeal squamous cell carcinoma. J Cancer Res Clin Oncol. 2010;136:931–938. doi: 10.1007/s00432-009-0735-9. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt-Graff A, Ertelt V, Allgaier HP, Koelble K, Olschewski M, Nitschke R, Bochaton-Piallat ML, Gabbiani G, Blum HE. Cellular retinol-binding protein-1 in hepatocellular carcinoma correlates with beta-catenin, Ki-67 index, and patient survival. Hepatology. 2003;38:470–480. doi: 10.1053/jhep.2003.50321. [DOI] [PubMed] [Google Scholar]
- 14.Gatti S, Carrozzo AM, Orlandi A, Nini G. Treatment of inflammatory linear verrucous epidermal naevus with calcipotriol. Br J Dermatol. 1995;132:837–839. doi: 10.1111/j.1365-2133.1995.tb00744.x. [DOI] [PubMed] [Google Scholar]
- 15.Orlandi A, Bianchi L, Costanzo A, Campione E, Spagnoli LG, Chimenti S. Evidence of increased apoptosis and reduced proliferation in basal cell carcinomas treated with tazarotene. J Invest Dermatol. 2004;122:1037–1041. doi: 10.1111/j.0022-202X.2004.22414.x. [DOI] [PubMed] [Google Scholar]
- 16.Omenn GS, Goodman GE, Thornquist MD, Balmes J, Cullen MR, Glass A, Keogh JP, Meyskens FL, Valanis B, Williams JH, Barnhart S, Hammar S. Effects of a combination of beta carotene and vitamin A on lung cancer and cardiovascular disease. N Engl J Med. 1996;334:1150–1155. doi: 10.1056/NEJM199605023341802. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch FR, Varella-Garcia M, Bunn PA, Di Maria MV, Veve R, Bremmes RM, Barón AE, Zeng C, Franklin WA. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 18.Oka S, Uramoto H, Shimokawa H, Iwanami T, Tanaka F. The expression of Ki-67, but not proliferating cell nuclear antigen, predicts poor disease free survival in patients with adenocarcinoma of the lung. Anticancer Res. 2011;31:4277–4282. [PubMed] [Google Scholar]
- 19.Van Dyke AL, Cote ML, Prysak GM, Claeys GB, Wenzlaff AS, Murphy VC, Lonardo F, Schwartz AG. COX-2/EGFR expression and survival among women with adenocarcinoma of the lung. Carcinogenesis. 2008;29:1781–1787. doi: 10.1093/carcin/bgn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegelin MD, Borczuk AC. Epidermal growth factor receptor mutations in lung adenocarcinoma. Lab Invest. 2014;94:129–137. doi: 10.1038/labinvest.2013.147. [DOI] [PubMed] [Google Scholar]
- 21.Reck M, Popat S, Reinmuth N, De Ruysscher D, Kerr KM, Peters S. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii27–39. doi: 10.1093/annonc/mdu199. [DOI] [PubMed] [Google Scholar]
- 22.Borczuk A, Gorenstein L, Walter K, Assaad AA, Wang L, Powell CA. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol. 2003;163:1949–1960. doi: 10.1016/S0002-9440(10)63553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campos B, Centner FS, Bermejo JL, Ali R, Dorsch K, Wan F, Felsberg J, Ahmadi R, Grabe N, Reifenberger G, Unterberg A. Aberrant expression of retinoic acid signaling molecules influences patient survival in astrocytic gliomas. Am J Pathol. 2011;178:1953–1964. doi: 10.1016/j.ajpath.2011.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlandi A, Francesconi A, Clement S, Ropraz P, Spagnoli LG, Gabbiani G. High levels of cellular retinol binding protein-1 expression in leiomyosarcoma: possible implications for diagnostic evaluation. Virchows Arch. 2002;441:31–40. doi: 10.1007/s00428-001-0576-7. [DOI] [PubMed] [Google Scholar]
- 25.Orlandi A, Ferlosio A, Ciucci A, Sesti F, Lifschitz-Mercer B, Gabbiani G, Spagnoli LG, Czernobilsky B. Cellular retinol-binding protein-1 expression in endometrial tromal cells: physiopathological and diagnostic implications. Histopathology. 2004;45:511–517. doi: 10.1111/j.1365-2559.2004.01963.x. [DOI] [PubMed] [Google Scholar]
- 26.Schiller JH, Adak S, Feins RH, Keller SM, Fry WA, Livingston RB, Hammond ME, Wolf B, Sabatini L, Jett J, Kohman L, Johnson DH. Lack of prognostic significance of p53 and K-ras mutations in primary resected non-small-cell lung cancer on E4592: a Laboratory Ancillary Study on an Eastern Cooperative Oncology Group Prospective Randomized Trial of Postoperative Adjuvant Therapy. J Clin Oncol. 2001;19:448–457. doi: 10.1200/JCO.2001.19.2.448. [DOI] [PubMed] [Google Scholar]
- 27.Solis LM, Behrens C, Raso MG, Lin HY, Kadara H, Yuan P, Galindo H, Tang X, Lee JJ, Kalhor N, Wistuba II, Moran CA. Histologic patterns and molecular characteristics of lung adenocarcinoma associated with clinical outcome. Cancer. 2012;118:2889–2899. doi: 10.1002/cncr.26584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sah JF, Eckert RL, Chandraratna RA, Rorke EA. Retinoids suppress epidermal growth factor-associated cell proliferation by inhibiting epidermal growth factor receptor-dependent ERK1/2 activation. J Biol Chem. 2002;277:9728–9735. doi: 10.1074/jbc.M110897200. [DOI] [PubMed] [Google Scholar]
- 29.Lin B, Chen GQ, Xiao D, Kolluri SK, Cao X, Su H, Zhang XK. Orphan receptor COUP-TF is required for induction of retinoic acid receptor beta, growth inhibition, and apoptosis by retinoic acid in cancer cells. Mol Cell Biol. 2000;20:957–970. doi: 10.1128/mcb.20.3.957-970.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canon E, Cosgaya JM, Scsucova S, Aranda A. Rapid effects of retinoic acid on CREB and ERK phosphorylation in neuronal cells. Mol Biol Cell. 2004;15:5583–5592. doi: 10.1091/mbc.E04-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritz H, Kennedy D, Fergusson D, Fernandes R, Doucette S, Cooley K, Seely A, Sagar S, Wong R, Seely D. Vitamin A and retinoid derivatives for lung cancer: a systematic review and meta analysis. PLoS One. 2011;6:e21107. doi: 10.1371/journal.pone.0021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pastorino U, Infante M, Maioli M, Chiesa G, Buyse M, Firket P, Rosmentz N, Clerici M, Soresi E, Valente M. Adjuvant treatment of stage I lung cancer with high-dose vitamin A. J Clin Oncol. 1993;11:1216–1222. doi: 10.1200/JCO.1993.11.7.1216. [DOI] [PubMed] [Google Scholar]
- 33.Eckhoff K, Flurschutz R, Trillsch F, Mahner S, Janicke F, Milde-Langosch K. The prognostic significance of Jun transcription factors in ovarian cancer. J Cancer Res Clin Oncol. 2013;139:1673–1680. doi: 10.1007/s00432-013-1489-y. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Regalado A, Vargas M, Garcia-Carranca A, Arechaga-Ocampo E, Gonzalez-De la Rosa CH. Activation of Akt pathway by transcription-independent mechanisms of retinoic acid promotes survival and invasion in lung cancer cells. Mol Cancer. 2013;12:44. doi: 10.1186/1476-4598-12-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karantza V. Keratins in health and cancer: more than mere epithelial cell markers. Oncogene. 2011;30:127–138. doi: 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knosel T, Emde V, Schluns K, Schlag PM, Dietel M, Petersen I. Cytokeratin profiles identify diagnostic signatures in colorectal cancer using multiplex analysis of tissue microarrays. Cell Oncol. 2006;28:167–175. doi: 10.1155/2006/354295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y, Wu H, Zhang M, Ding L, Meng F, Fan X. Expression of the epithelial-mesenchymal transition-related proteins and their clinical significance in lung adenocarcinoma. Diagn Pathol. 2013;8:89. doi: 10.1186/1746-1596-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Travis WD, Rekhtman N, Riley GJ, Geisinger KR, Asamura H, Brambilla E, Garg K, Hirsch FR, Noguchi M, Powell CA, Rusch VW, Scagliotti G, Yatabe Y. Pathologic diagnosis of advanced lung cancer based on small biopsies and cytology: a paradigm shift. J Thorac Oncol. 2010;5:411–414. doi: 10.1097/JTO.0b013e3181d57f6e. [DOI] [PubMed] [Google Scholar]
- 39.Yoshizawa A, Motoi N, Riely GJ, Sima CS, Gerald WL, Kris MG, Park BJ, Rusch VW, Travis WD. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011;24:653–664. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- 40.Rami-Porta R, Ball D, Crowley J, Giroux DJ, Jett J, Travis WD, Tsuboi M, Vallières E. proposals for the revision of the T descriptors in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol. 2007;2:593–602. doi: 10.1097/JTO.0b013e31807a2f81. [DOI] [PubMed] [Google Scholar]
- 41.Ferlosio A, Arcuri G, Doldo E, Scioli MG, De Falco S, Spagnoli LG, Orlandi A. Age-related increase of stem marker expression influences vascular smooth muscle cell properties. Atherosclerosis. 2012;224:51–57. doi: 10.1016/j.atherosclerosis.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 42.Normanno N, Pinto C, Taddei G, Gambacorta M, Castiglione F, Barberis M, Clemente C, Marchetti A. Results of the First Italian External Quality Assurance Scheme for somatic EGFR mutation testing in non-small-cell lung cancer. J Thorac Oncol. 2013;8:773–778. doi: 10.1097/JTO.0b013e31828c2b08. [DOI] [PubMed] [Google Scholar]
- 43.Campagnolo L, Costanza G, Francesconi A, Arcuri G, Moscatelli I, Orlandi A. Sortilin expression is essential for pro-nerve growth factor-induced apoptosis of rat vascular smooth muscle cells. PLoS One. 2014;9:e84969. doi: 10.1371/journal.pone.0084969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cervelli V, Scioli MG, Gentile P, Doldo E, Bonanno E, Spagnoli LG, Orlandi A. Platelet-rich plasma greatly potentiates insulin-induced adipogenic differentiation of human adipose-derived stem cells through a serine/ threonine kinase Akt-dependent mechanism and promotes clinical fat graft maintenance. Stem Cells Transl Med. 2012;1:206–220. doi: 10.5966/sctm.2011-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 46.Matera MG, Calzetta L, Passeri D, Facciolo F, Rendina EA, Page C, Cazzola M, Orlandi A. Epithelium integrity is crucial for the relaxant activity of brain natriuretic peptide in human isolated bronchi. Br J Pharmacol. 2011;163:1740–1754. doi: 10.1111/j.1476-5381.2011.01339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stasi MA, Scioli MG, Arcuri G, Mattera GG, Lombardo K, Marcellini M, Riccioni T, De Falco S, Pisano C, Spagnoli LG, Borsini F, Orlandi A. Propionyl-L-carnitine improves postischemic blood flow recovery and ateriogenetic revascularization and reduces endothelial NADPH-oxidase 4-mediated superoxide production. Arterioscler Thromb Vasc Biol. 2010;30:426–435. doi: 10.1161/ATVBAHA.109.201533. [DOI] [PubMed] [Google Scholar]
- 48.Orlandi A, Ciucci A, Ferlosio A, Pellegrino A, Chiariello L, Spagnoli LG. Increased expression and activity of matrix metalloproteinases characterize embolic cardiac myxomas. Am J Pathol. 2005;166:1619–1628. doi: 10.1016/S0002-9440(10)62472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitamura A, Hosoda W, Sasaki E, Mitsudomi T, Yatabe Y. Immunohistochemical detection of EGFR mutation using mutation-specific antibodies in lung cancer. Clin Cancer Res. 2010;16:3349–3355. doi: 10.1158/1078-0432.CCR-10-0129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.