Abstract
Conventional high-grade osteosarcoma is the most common primary bone cancer with relatively high incidence in young people. Recurrent and metastatic tumors are difficult to treat. We performed a kinase inhibitor screen in two osteosarcoma cell lines, which identified MEK1/2 inhibitors. These inhibitors were further validated in a panel of six osteosarcoma cell lines. Western blot analysis was performed to assess ERK activity and efficacy of MEK inhibition. A 3D culture system was used to validate results from 2D monolayer cultures. Gene expression analysis was performed to identify differentially expressed gene signatures in sensitive and resistant cell lines. Activation of the AKT signaling network was explored using Western blot and pharmacological inhibition. In the screen, Trametinib, AZD8330 and TAK-733 decreased cell viability by more than 50%. Validation in six osteosarcoma cell lines identified three cell lines as resistant and three as sensitive to the inhibitors. Western blot analysis of ERK activity revealed that sensitive lines had high constitutive ERK activity. Treatment with the three MEK inhibitors in a 3D culture system validated efficacy in inhibition of osteosarcoma viability. MEK1/2 inhibition represents a candidate treatment strategy for osteosarcomas displaying high MEK activity as determined by ERK phosphorylation status.
Keywords: osteosarcoma, MEK, pharmacological inhibition, 3D culture, ERK phosphorylation
INTRODUCTION
Osteosarcoma is the most common primary malignant bone tumor occurring predominantly in children and adolescents, as well as in people older than 40 years of age. It is thought to arise from mesenchymal stem cells that are capable of producing osteoid [1, 2]. At the moment of diagnosis, 10-20% of the patients present with metastasis. About 30-40% of the patients with localized osteosarcoma will present with relapse mainly as lung metastasis. Patients with recurrence have very poor prognosis with 23-33% 5-year overall survival [3]. Therefore, new effective therapies are urgently needed to improve the prognosis of osteosarcoma patients.
Screening a kinase inhibitor library of pre-clinical or clinically approved drugs provides the possibility of identifying novel candidate treatments for osteosarcoma that can be translated to the clinic.
Receptor tyrosine kinases (RTKs) are frequently hyperactive through mutation or overexpression in cancer causing aberrant activation of downstream signaling cascades, including Ras/Raf/MEK/ERK [4]. This cascade is known to be involved in cell survival, proliferation, and differentiation by regulating the activation of transcription factors such as c-Myc, c-Fos, Ets, and Elk-1 [5, 6]. In osteosarcoma, RTKs such as EGFR [7], KIT [8], FGFR1 [9], and IGFR-1 [10] were found to be amplified or upregulated. Furthermore, ERK pathway activation was reported in 67% of osteosarcomas analyzed [11]. Genetic screens also identified PI3K/AKT signaling as a driver of osteosarcoma [12, 13] and inhibition of this pathway is a potential treatment for osteosarcoma [14]. Inhibition of overexpressed or mutant RTKs is a candidate for cancer treatment but often leads to compensatory activation of another RTK driving cancer cell survival and proliferation [15]. Inhibition of common downstream effector kinases could be an effective way to circumvent such resistance. In this study, we performed a kinase inhibitor screen to identify candidate targets for human osteosarcoma, and identified MEK inhibitors as possible therapeutic targets in cells with constitutive ERK activation.
RESULTS
Kinase inhibitor screen for inhibition of osteosarcoma cell viability identifies MEK inhibitors
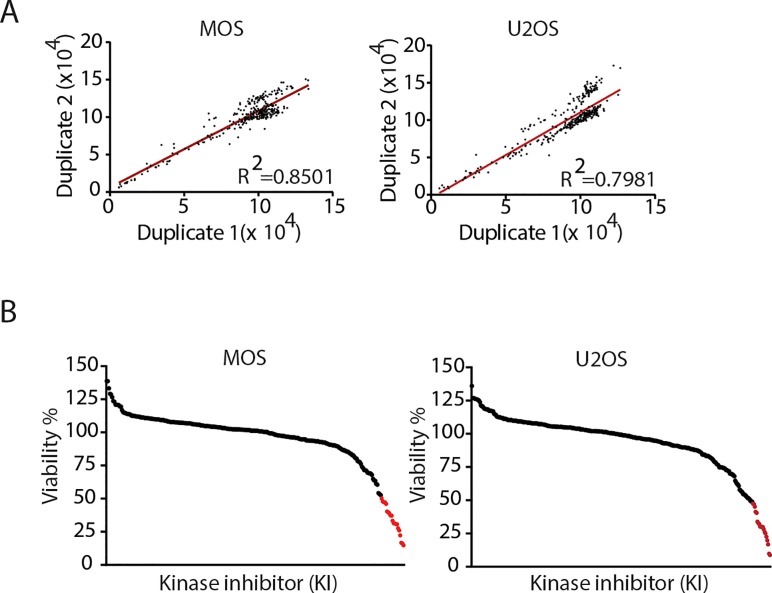
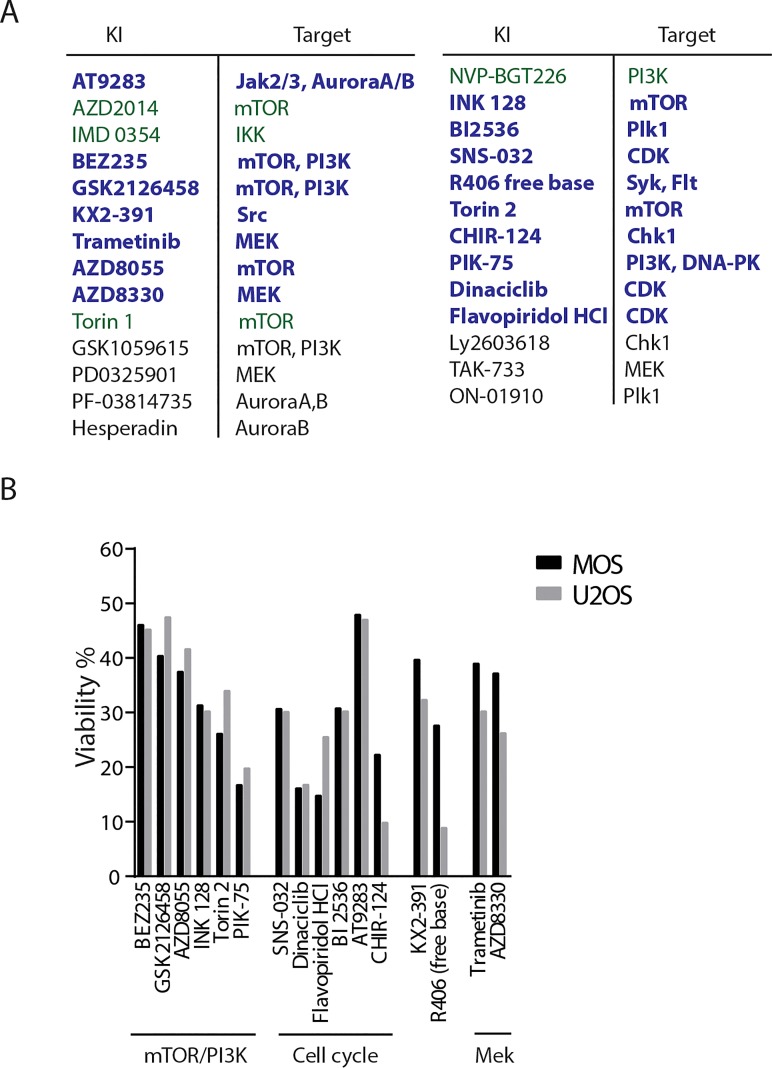
A library composed of 273 kinase inhibitors was used to screen for inhibitors that, as a single agent, decreased viability of osteosarcoma cells. MOS and U2OS were exposed to a concentration of 1μM for 72 hours and viability was determined by measuring ATP production. Each screen was performed in duplicate with a goodness of fit (R2) of 0.8501 for the screen in MOS and 0.7981 in U2OS (Figure 1A). All values were normalized to DMSO condition, and the candidates that exhibited less than 50% viability were considered a hit (Figure 1B). Under this criterium, we identified 16 inhibitors in common for MOS and U2OS of which, six targeted the PI3K/mTOR pathway (BEZ235, GSK2126458, AZD8055, Torin 2, INK-128, PIK-75), six targeted the cell cycle (AT9283, BI2536, SNS-032, CHIR-124, dinaciclib and flavopiridol HCl), one targted Src (KX-391), one targeted Syk and Flt (R406 free base), and two were MEK1/2 inhibitors (Figure 2A, B). The PI3K/mTOR pathway has been implicated in osteosarcoma cell survival and proliferation in vivo [16]. Dinaciclib and flavoripirol were previously reported to induce apoptosis in osteosarcoma cells [17, 18]. Plk1 inhibition has been shown to cause cell death in osteosarcoma cells and its expression correlates with overall survival in osteosarcoma patients [19-21]. Here we focused on three MEK1/2 inhibitors: Trametinib and AZD8330, which were common in MOS and U2OS, and TAK-733, which was a hit in U2OS (in MOS treatment with TAK-733 showed 71% remaining viability).
Figure 1. Kinase inhibitor screen in two human osteosarcoma cell lines.
A) The screen was performed in MOS and U2OS cell lines in duplicate. The graphs represent the goodness of fit of the screens. B) All results were normalized to DMSO and hits are defined by <50% viability (red).
Figure 2. Selection of hits in two human osteosarcoma cell lines.
A) List of hits common to both cell lines (bold blue), only in MOS (green) and only in U2OS cells (black). B) Bar graphs representing the hits common to both cell lines, their viability score relative to DMSO, and known biological activity.
MEK1/2 inhibition leads to apoptosis in cells with constitutive ERK activation
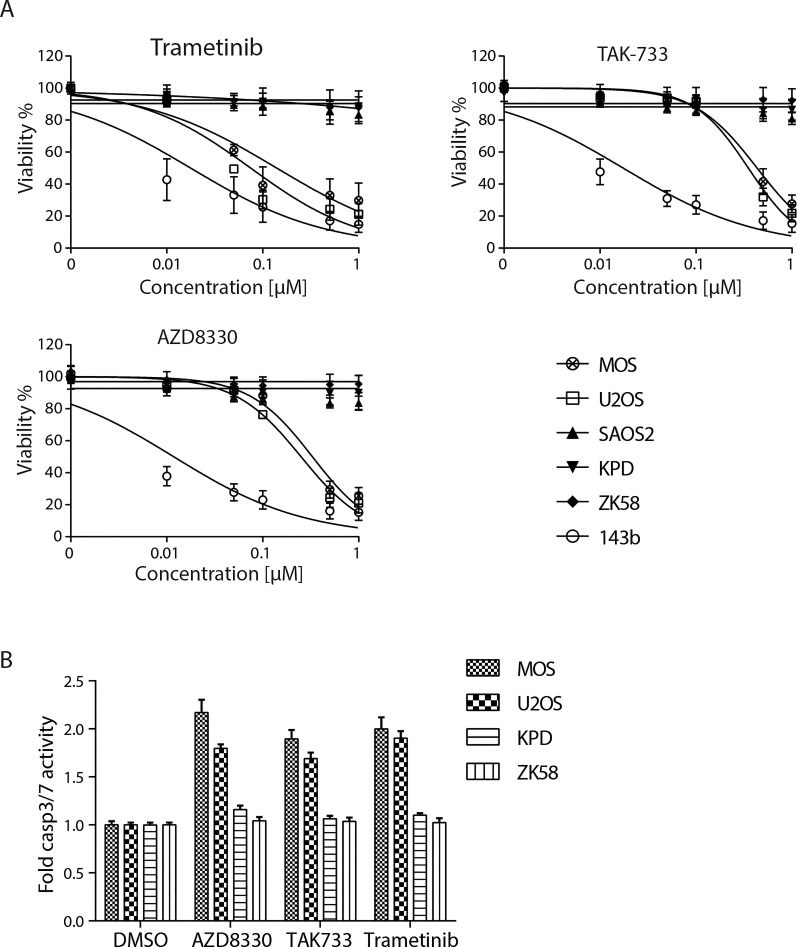
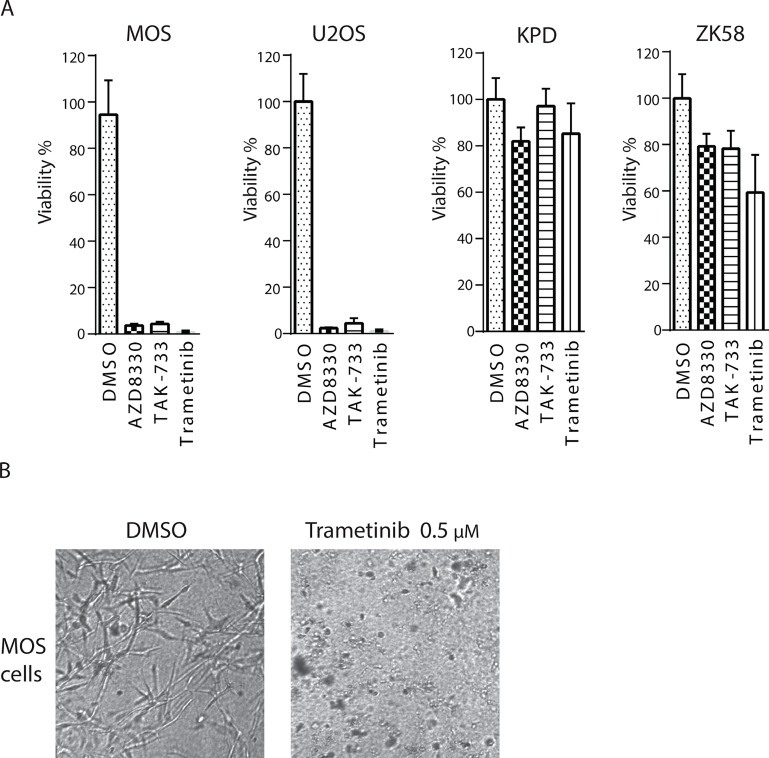
The activity of these three inhibitors was tested using concentration ranges in six osteosarcoma cell lines: MOS, U2OS, KPD, ZK58, 143b and Saos-2 (Figure 3A). All three inhibitors decreased viability of MOS and U2OS and strongly affected 143b. By contrast, viability of KPD, ZK58 and Saos-2 was not affected by any of the three inhibitors. A capase3/7 activity assay confirmed that exposure to 0.5μM of each of the drugs induced apoptosis in MOS and U2OS, but not in KPD and ZK58 cells (Figure 3B).
Figure 3. Validation of three MEK inhibitors in 6 osteosarcoma cell lines.
A) Dose response curves for Trametinib, AZD8330 and TAK-733 in 6 osteosarcoma cell lines as indicated. Cells were exposed for 72 hours. Each graph represents mean±s.e.m. of three replicates. B) Caspase 3/7 activity in presence of indicated inhibitors relative to DMSO in 4 osteosarcoma cell lines. The graph is a representative experiment of 3 independent experiments, each performed in triplicate. Mean±s.d. is shown.
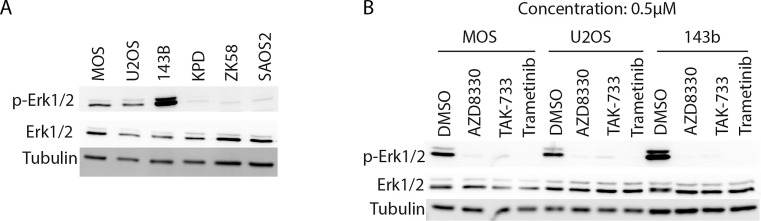
Next, we asked if the observed differences in the response to MEK inhibition was related to the status of MEK activity, as measured by phosphorylation of the MEK target, ERK. Indeed, 143b, which was the most sensitive cell line, is Ki-ras+ transformed [22] and showed the most prominent ERK phosphorylation, followed by the other two sensitive cell lines, MOS and U2OS (Figure 4A). The resistant cell lines KPD, ZK58 and Saos-2 showed no constitutive ERK activation. Exposing MOS, U2OS and 143b to a concentration of 0.5μM of Trametinib, AZD8330 or TAK-733 for 6 hours, led to loss of ERK phosphorylation indicating effective MEK inhibition (Figure 4B).
Figure 4. Western blot analysis of ERK phosphorylation in 6 osteosarcoma cell lines and effect of MEK inhibition.
A) Western blot analysis of total ERK and phospho-ERK in 6 osteosarcoma cell lines. B) Western blot analysis of total ERK and phospho-ERK in MOS, U2OS and 143b osteosarcoma cell lines after 6 hours treatment with DMSO or 0.5μM of the indicated MEK inhibitors.
Validation of MEK inhibition in a 3D cell culture system
We made use of 3D cultures of identified sensitive and resistant cell lines to further validate the effect of Trametinib, AZD8330 and TAK-733. MOS, U2OS, KPD, and ZK58 were suspended in a collagen-matrigel mixture, and exposed 24 hours later to 0.5μM of each inhibitor for a period of 72 hours. As observed in 2D cultures, MOS and U2OS cells died in the presence of each of the three inhibitors whereas KPD and ZK58 were not affected (Figure 5A, 5B).
Figure 5. Validation of sensitivity to MEK inhibitors in a 3D culture system.
A) MOS, U2OS, KPD and ZK58 cells were re-suspended in a collagen-matrigel mix and 3D cultures were subsequently exposed to 0.5μM of the MEK inhibitors for 72 hours. Graphs are a representative experiment of two replicates, each performed in quadruplicate. Mean±s.d is shown. B) Representative images of 3D MOS cultures in absence or presence of Trametinib.
Potential mechanisms of resistance in cell lines not sensitive to MEK inhibition
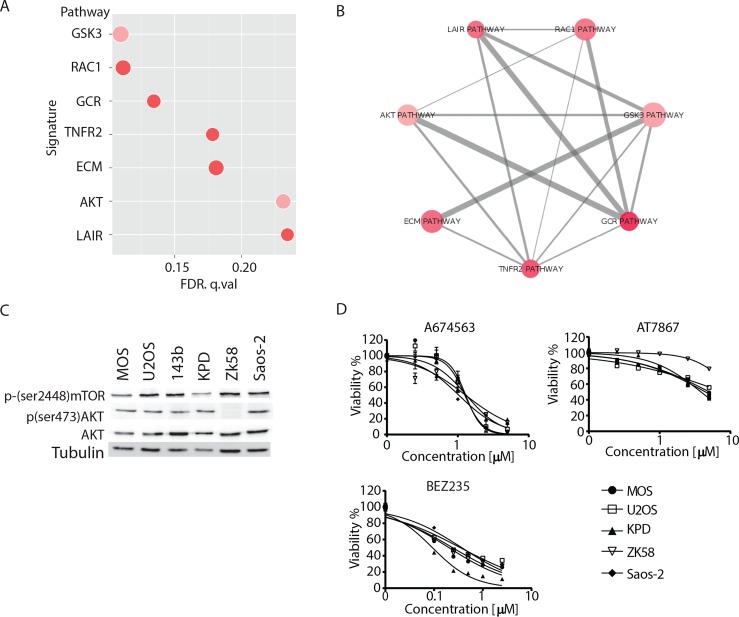
Our data indicated that MEK1/2 inhibition could be used to treat osteosarcomas that present with constitutive ERK activation but not in cases where MEK activity is low. Ras/Raf mutations are strong predictors for sensitivity to MEK inhibition [23, 24] explaining sensitivity of 143b. We searched for mutations in exons or splice sites in the genes MEK1, MEK2, A-Raf, B-Raf, C-Raf, EGFR, FGFR, IGFR1, K-Ras, H-Ras and N-Ras in all cell lines used, employing a previously published method [25] but could not identify mutations that may explain high constitutive ERK phosphorylation in MOS or U2OS (data not shown). Next, we performed a pathway analysis on gene expression differences in sensitive (MOS, U2OS and 143b) versus resistant (KPD, ZK58 and Saos-2) cell lines [26]. This analysis revealed 7 signatures with enrichment of differentially expressed genes (Figure 6A). One of the signatures was the AKT pathway, which had positive fold change for 15/22 genes upregulated in the resistant cell lines (Figure 6C). However, Western blot analysis of phospho-AKT(Ser473) showed active AKT in all cell lines except ZK58 (Figure 6B). Similarly, mTOR, a downstream target of AKT signaling, was not differentially activated between sensitive and resistant cell lines (Figure 6C). In agreement, all cell lines responded similarly to inhibition of AKT signaling using A674563 (inhibits AKT1 selectively) or AT7867 (inhibits AKT1/2/3) and were highly sensitive to a dual PI3K/mTOR inhibitor, BEZ235 (Figure 6D). These data indicate that other differentially activated signaling pathways, rather than the predicted difference in AKT activity underlie differential sensitivity of the osteosarcoma cell lines to MEK inhibition.
Figure 6. Analysis of AKT pathway and its pharmacological inhibition.
A) Plot representing the 7 signatures that were significantly enriched (FDR< 0.25) in the cell lines resistant to MEK inhibition based on gene expression data. Pink/red represents the enrichment score (red>pink), and size represents the gene set size of the signature. B) Schematic representation of similarity between the 7 signatures. Pink/red represents the enrichment score (red>pink), and the line width represents the number of genes shared between signatures. C) Western blot analysis of total AKT, phosho(Ser473)-AKT and phospho-(Ser2448)-mTOR in the indicated osteosarcoma cell lines. D) Dose response curves for the indicated AKT-mTOR inhibitors in the indicated osteosarcoma cell lines. Mean±s.d for experiment performed in triplicate is shown.
DISCUSSION
To identify new candidate avenues for therapeutic intervention for osteosarcoma we performed a kinase inhibitor screen in two human osteosarcoma cell lines. Our screen confirms previously reported findings (e.g. PI3K-AKT-mTOR inhibition), thereby validating our screen. It also identifies new drugs in the context of osteosarcoma that are in the clinic for other malignancies and hence may be candidates for repurposing.
PI3K-Akt-mTOR pathway is a network that controls many cellular processes such as cell proliferation, survival, metabolism and genomic integrity [27]. It has been shown that osteosarcoma strongly depends on this pathway for cell survival and proliferation and pathway inhibition triggers cell death [13, 28]. The expression of mTOR is correlated with event-free survival and cancer progression in osteosarcoma [29]. Our screen confirms mTOR signaling as a potential target to treat osteosarcoma.
The main characteristic of tumor cells is uncontrolled cell proliferation and cell cycle regulators are key players in cancer growth. Our screen identifies several inhibitors targeting this hallmark of cancer, including inhibitors of cyclin-dependent kinases and spindle checkpoints. Cyclin-dependent kinases 2,4 and 6 are altered in 80-90% of tumors[30]. In osteosarcoma, the Rb/p16/CDK4 axis is often deregulated with mutations or deletions in these genes [31, 32]. Aurora and polo-like kinases are critical regulators of the mitotic spindle and have been implicated in various cancers [33]. Several studies have shown that inhibition of Aurora kinases leads to cell death in osteosarcoma [34, 35]. The Aurora kinase A inhibitor Alisertib (not present in our library) is undergoing testing in a phase II clinical trial of refractory solid tumors (NCT01154816). Inhibition of polo like kinase (Plk) 1 causes growth inhibition in various cancers [5, 36]. In osteosarcoma, Plk1 show higher expression in tumor samples compared to normal tissue, and its inhibition with NMS-P397 (not present in our library) leads to growth arrest and apoptosis [22].
The Ras-Raf-MEK-ERK mitogen activated protein kinase cascade is known to be involved in cell proliferation, apoptosis, differentiation and development. It integrates signals from cell surface receptors to activate ERK, which in turn enters the nucleus and activates transcription factors such as c-Myc, c-Fos, Ets, and Elk-1 [37]. This pathway is often deregulated in tumors due to mutations or overexpression of upstream signaling components. B-Raf and Ras are frequently mutated in melanoma, colorectal cancer, ovarian cancer, lung cancer and pancreatic cancer among others [38, 39]. In osteosarcoma, ERK pathway activity was reported to occur in 67% of the cases analyzed, and mutations in B-Raf were only found in 13% of the cohort [11]. We identify three MEK inhibitors in the osteosarcoma cell viability screen: Trametinib is a selective allosteric inhibitor of MEK1/2 designed to treat tumors with overactive MEK-ERK pathway, which is found in tumors with B-Raf mutations [40]. It was approved for melanoma, and it has also been tested in patients with pancreatic cancer, colorectal cancer and other solid tumors with B-Raf mutations [41]. AZD8330 and TAK-733 are two selective allosteric MEK1/2 inhibitors [42, 43]. TAK-733 has shown good antitumor activity in melanoma cells [44] as well as in human lung cancer [45].
Our findings imply that MEK1/2 inhibition is a candidate approach to treat osteosarcomas harboring high ERK activity. Strikingly, while ERK phosphorylation status predicts sensitivity to MEK inhibition, mutation analysis of upstream components of this pathway does not identify candidate predictive mutations. Hence, ERK phosphorylation in tumor tissue as identified by immunohistochemistry may be a more accurate biomarker predicting sensitivity to MEK1/2 inhibitors than genomic analyses. We have not identified an alternative pathway selectively driving viability/growth of cell lines that are resistant to MEK inhibition. An enriched set of genes in the lines points to differential activation of the AKT pathway but based on AKT and mTOR phosphorylation status this pathway is active in all lines and, in agreement, all cell lines are similarly sensitive to AKT-mTOR inhibition. Interestingly, this indicates that three independent cell lines showing strong activity of MEK as well as AKT depend on the activity of both pathways. I.e., inhibition of either pathway is sufficient to cause loss of viability rather than these pathways compensating for each other.
To our knowledge, we are the first to describe the efficacy of MEK inhibition in osteosarcoma cells with high ERK phosphorylation. Recently, a Phase I clinical trial (NCT02124772) started enrolling patients with solid tumors, including osteosarcoma, to study the efficacy of trametinib in combination with dabrafenib. In this setting, such association between ERK phosphorylation status and response to trametinib may be investigated.
MATERIALS AND METHODS
Reagents and antibodies
The kinase inhibitor library (L1200), Trametinib, AZD8330 and TAK-733 inhibitors were purchased from SelleckChem (Huissen, Netherlands). The ERK (9102), phospho(44/42)-ERK (137F5), phospho(Ser2448)-mTOR (D9C2), phospho(Ser473)-AKT (#9271) and AKT (#9272) antibodies were from Cell Signaling (Bioké, Leiden, Netherlands). The antibody against tubulin (T-9026) was from Sigma Aldrich (Zwijndrecht, The Netherlands).
Cell culture
Human osteosarcoma cell lines MOS, U2OS, 143B, ZK58, KPD and Saos-2 were previously described[46, 47]. Cells were grown in RPMI1640 medium supplemented with 10% fetal bovine serum and 25 U/mL penicillin and 25 μg/mL of penicillin-streptomycin. All cells were cultured in a humidified incubator at 37°C with 5% CO2.
Immunoblotting
Cells were lysed with SDS protein buffer (125mM Tris/HCl pH 6.8, 20% glycerol, 4% SDS and 0.2% bromophenol blue). Proteins were resolved by SDS-PAGE and transferred to polyvinylidine difluoride membrane. Membranes were blocked in 5% BSA-TBST (TRIS-0.05% Tween20), followed by overnight incubation with primary antibodies and 45 minutes incubation with HRP-conjugated secondary antibodies. Chemiluminescence was detected with a Typhoon 9400 imager (GE Healthcare).
Cell viability and caspase3/7 activity
Cells were processed using the ATPlite 1Step kit (Perkin Elmer) according to the manufacturer's instructions, followed by luminescence measurement on a plate reader. Caspase 3/7 activity was assessed with Caspase-Glo® 3/7 from Promega (Leiden, The Netherlands) according to manufacturer's protocol, and luminescence measurement on a plate reader.
3D culture assay
MOS and U2OS cells were cultured in 384-well plates (Greiner μclear) in a hydrogel containing Matrigel (Beckton Dickinson) and collagen I, supporting invasive growth of both cell lines. Cells in culture were trypsinized and directly added to the cooled gel solution. Using a robotic liquid handler (CyBio Selma 96/60), 14.5μL of gel-cell suspension was transferred to each well of a 384-well plate (2000 cells/well). After polymerization for 30 minutes at 37°C in an atmosphere of 5% CO2, growth medium was added on top of the gel. After 24 hours, the cells were exposed to the compounds in quadruplicate for a period of 72 hours. For measuring cell viability in 3D, ATPlite was used as indicated by the manufacturer and luminescence was measured using a FluoroStar plate reader. Percentage viability was thereafter calculated by normalization of all conditions to DMSO. Results are presented as means ± SD. Images of 3D cultures were taken with a BD Pathway 855 (BD Biosciences).
Pathway analysis
We used a previously published dataset of mRNA expression of 19 osteosarcoma cell lines [Namlos et al PLoSOne 7, e48086, 2012] and performed a LIMMA analysis [48] of sensitive (MOS, U2OS and 143b) versus resistant (KPD, ZK58 and Saos-2) cell lines [26]. We then ran a pre-ranked gene set enrichment analysis [49] using MSigDb v5.0 BioCarta (http://www.biocarta.com) signatures on the Benjamini and Hochberg False Discovery Rate corrected p-values obtained from LIMMA. Statistically significant signatures were defined as signatures with FDR<0.25.
Statistical analysis
Dose response curve fitting and statistical analyses were performed with GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA).
Footnotes
CONFLICTS OF INTEREST
L.S. Price is founder and co-owner of OcellO B.V. a contract research company that offers screening services using 3D tissues. This role has had no bearing on the content of the manuscript.
REFERENCES
- 1.Anninga JK, Gelderblom H, Fiocco M, Kroep JR, Taminiau AH, Hogendoorn PC, Egeler RM. Chemotherapeutic adjuvant treatment for osteosarcoma: where do we stand? European journal of cancer. 2011;47(16):2431–2445. doi: 10.1016/j.ejca.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg AE, Cleton-Jansen A-M, Pinieux Gd, Deyrup AT, Hauben E, Squire J. WHO Classification of Tumours of Soft Tissue and Bone. Lyon: IARC; 2013. Conventional osteosarcoma; pp. 282–288. [Google Scholar]
- 3.Buddingh EP, Anninga JK, Versteegh MI, Taminiau AH, Egeler RM, van Rijswijk CS, Hogendoorn PC, Lankester AC, Gelderblom H. Prognostic factors in pulmonary metastasized high-grade osteosarcoma. Pediatric blood & cancer. 2010;54(2):216–221. doi: 10.1002/pbc.22293. [DOI] [PubMed] [Google Scholar]
- 4.Schlessinger J. Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harb Perspect Biol. 2014;6(3) doi: 10.1101/cshperspect.a008912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reagan-Shaw S, Ahmad N. Silencing of polo-like kinase (Plk) 1 via siRNA causes induction of apoptosis and impairment of mitosis machinery in human prostate cancer cells: implications for the treatment of prostate cancer. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19(6):611–613. doi: 10.1096/fj.04-2910fje. [DOI] [PubMed] [Google Scholar]
- 6.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14(19):2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen YH, Koeppen H, Garcia R, Chiriboga L, Tarlow BD, Peters BA, Eigenbrot C, Yee H, Steiner G, Greco MA. Epidermal growth factor receptor in osteosarcoma: expression and mutational analysis. Hum Pathol. 2007;38(8):1184–1191. doi: 10.1016/j.humpath.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Entz-Werle N, Gaub MP, Lavaux T, Marcellin L, Metzger N, Marec-Berard P, Schmitt C, Brugiere L, Kalifa C, Tabone MD, Pacquement H, Gentet P, Lutz P, Oudet P, Babin A. KIT gene in pediatric osteosarcomas: could it be a new therapeutic target? Int J Cancer. 2007;120(11):2510–2516. doi: 10.1002/ijc.22593. [DOI] [PubMed] [Google Scholar]
- 9.Fernanda Amary M, Ye H, Berisha F, Khatri B, Forbes G, Lehovsky K, Frezza AM, Behjati S, Tarpey P, Pillay N, Campbell PJ, Tirabosco R, Presneau N, Strauss SJ, Flanagan AM. Fibroblastic growth factor receptor 1 amplification in osteosarcoma is associated with poor response to neo-adjuvant chemotherapy. Cancer Med. 2014;3(4):980–987. doi: 10.1002/cam4.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacEwen EG, Pastor J, Kutzke J, Tsan R, Kurzman ID, Thamm DH, Wilson M, Radinsky R. IGF-1 receptor contributes to the malignant phenotype in human and canine osteosarcoma. J Cell Biochem. 2004;92(1):77–91. doi: 10.1002/jcb.20046. [DOI] [PubMed] [Google Scholar]
- 11.Pignochino Y, Grignani G, Cavalloni G, Motta M, Tapparo M, Bruno S, Bottos A, Gammaitoni L, Migliardi G, Camussi G, Alberghini M, Torchio B, Ferrari S, Bussolino F, Fagioli F, Picci P, et al. Sorafenib blocks tumour growth, angiogenesis and metastatic potential in preclinical models of osteosarcoma through a mechanism potentially involving the inhibition of ERK1/2, MCL-1 and ezrin pathways. Molecular cancer. 2009;8:118. doi: 10.1186/1476-4598-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriarity BS, Otto GM, Rahrmann EP, Rathe SK, Wolf NK, Weg MT, Manlove LA, LaRue RS, Temiz NA, Molyneux SD, Choi K, Holly KJ, Sarver AL, Scott MC, Forster CL, Modiano JF, et al. A Sleeping Beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet. 2015;47(6):615–624. doi: 10.1038/ng.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry JA, Kiezun A, Tonzi P, Van Allen EM, Carter SL, Baca SC, Cowley GS, Bhatt AS, Rheinbay E, Pedamallu CS, Helman E, Taylor-Weiner A, McKenna A, DeLuca DS, Lawrence MS, Ambrogio L, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111(51):E5564–5573. doi: 10.1073/pnas.1419260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuijjer ML, van den Akker BE, Hilhorst R, Mommersteeg M, Buddingh EP, Serra M, Burger H, Hogendoorn PC, Cleton-Jansen AM. Kinome and mRNA expression profiling of high-grade osteosarcoma cell lines implies Akt signaling as possible target for therapy. BMC Med Genomics. 2014;7:4. doi: 10.1186/1755-8794-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu AM, Huang PH. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010;70(10):3857–3860. doi: 10.1158/0008-5472.CAN-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gobin B, Battaglia S, Lanel R, Chesneau J, Amiaud J, Redini F, Ory B, Heymann D. NVP-BEZ235, a dual PI3K/mTOR inhibitor, inhibits osteosarcoma cell proliferation and tumor development in vivo with an improved survival rate. Cancer letters. 2014;344(2):291–298. doi: 10.1016/j.canlet.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Tanaka K, Li X, Okada T, Nakamura T, Takasaki M, Yamamoto S, Oda Y, Tsuneyoshi M, Iwamoto Y. Cyclin-dependent kinase inhibitor, flavopiridol, induces apoptosis and inhibits tumor growth in drug-resistant osteosarcoma and Ewing's family tumor cells. Int J Cancer. 2007;121(6):1212–1218. doi: 10.1002/ijc.22820. [DOI] [PubMed] [Google Scholar]
- 18.Fu W, Ma L, Chu B, Wang X, Bui MM, Gemmer J, Altiok S, Pledger WJ. The cyclin-dependent kinase inhibitor SCH 727965 (dinacliclib) induces the apoptosis of osteosarcoma cells. Mol Cancer Ther. 2011;10(6):1018–1027. doi: 10.1158/1535-7163.MCT-11-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi U, Honda K, Satow R, Kobayashi E, Nakayama R, Ichikawa H, Shoji A, Shitashige M, Masuda M, Kawai A, Chuman H, Iwamoto Y, Hirohashi S, Yamada T. Functional genome screen for therapeutic targets of osteosarcoma. Cancer science. 2009;100(12):2268–2274. doi: 10.1111/j.1349-7006.2009.01310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan Z, Ji D, Weinstein EJ, Liu X, Susa M, Choy E, Yang C, Mankin H, Hornicek FJ. Lentiviral shRNA screen of human kinases identifies PLK1 as a potential therapeutic target for osteosarcoma. Cancer letters. 2010;293(2):220–229. doi: 10.1016/j.canlet.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Morales AG, Brassesco MS, Pezuk JA, Oliveira JC, Montaldi AP, Sakamoto-Hojo ET, Scrideli CA, Tone LG. BI 2536-mediated PLK1 inhibition suppresses HOS and MG-63 osteosarcoma cell line growth and clonogenicity. Anti-cancer drugs. 2011;22(10):995–1001. doi: 10.1097/CAD.0b013e32834a16d4. [DOI] [PubMed] [Google Scholar]
- 22.Sero V, Tavanti E, Vella S, Hattinger CM, Fanelli M, Michelacci F, Versteeg R, Valsasina B, Gudeman B, Picci P, Serra M. Targeting polo-like kinase 1 by NMS-P937 in osteosarcoma cell lines inhibits tumor cell growth and partially overcomes drug resistance. Invest New Drugs. 2014;32(6):1167–1180. doi: 10.1007/s10637-014-0158-6. [DOI] [PubMed] [Google Scholar]
- 23.Jing J, Greshock J, Holbrook JD, Gilmartin A, Zhang X, McNeil E, Conway T, Moy C, Laquerre S, Bachman K, Wooster R, Degenhardt Y. Comprehensive predictive biomarker analysis for MEK inhibitor GSK1120212. Mol Cancer Ther. 2012;11(3):720–729. doi: 10.1158/1535-7163.MCT-11-0505. [DOI] [PubMed] [Google Scholar]
- 24.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, Golub TR, Sebolt-Leopold J, Sellers WR, Rosen N. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439(7074):358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Eijk R, Licht J, Schrumpf M, Talebian Yazdi M, Ruano D, Forte GI, Nederlof PM, Veselic M, Rabe KF, Annema JT, Smit V, Morreau H, van Wezel T. Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle aspirates from non-small-cell lung cancer using allele-specific qPCR. PLoS One. 2011;6(3):e17791. doi: 10.1371/journal.pone.0017791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuijjer ML, Rydbeck H, Kresse SH, Buddingh EP, Lid AB, Roelofs H, Burger H, Myklebost O, Hogendoorn PC, Meza-Zepeda LA, Cleton-Jansen AM. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosomes Cancer. 2012;51(7):696–706. doi: 10.1002/gcc.21956. [DOI] [PubMed] [Google Scholar]
- 27.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nature reviews Drug discovery. 2014;13(2):140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupte A, Baker EK, Wan SS, Stewart E, Loh A, Shelat AA, Gould CM, Chalk AM, Taylor S, Lackovic K, Karlstrom A, Mutsaers AJ, Desai J, Madhamshettiwar PB, Zannettino AC, Burns C, et al. Systematic Screening Identifies Dual PI3K and mTOR Inhibition as a Conserved Therapeutic Vulnerability in Osteosarcoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(14):3216–29. doi: 10.1158/1078-0432.CCR-14-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q, Deng Z, Zhu Y, Long H, Zhang S, Zhao J. mTOR/p70S6K signal transduction pathway contributes to osteosarcoma progression and patients' prognosis. Medical oncology. 2010;27(4):1239–1245. doi: 10.1007/s12032-009-9365-y. [DOI] [PubMed] [Google Scholar]
- 30.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nature reviews Cancer. 2001;1(3):222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 31.Mohseny AB, Tieken C, van der Velden PA, Szuhai K, de Andrea C, Hogendoorn PC, Cleton-Jansen AM. Small deletions but not methylation underlie CDKN2A/p16 loss of expression in conventional osteosarcoma. Genes Chromosomes Cancer. 2010;49(12):1095–1103. doi: 10.1002/gcc.20817. [DOI] [PubMed] [Google Scholar]
- 32.Wei G, Lonardo F, Ueda T, Kim T, Huvos AG, Healey JH, Ladanyi M. CDK4 gene amplification in osteosarcoma: reciprocal relationship with INK4A gene alterations and mapping of 12q13 amplicons. Int J Cancer. 1999;80(2):199–204. doi: 10.1002/(sici)1097-0215(19990118)80:2<199::aid-ijc7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Molecular cancer research : MCR. 2007;5(1):1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 34.Tavanti E, Sero V, Vella S, Fanelli M, Michelacci F, Landuzzi L, Magagnoli G, Versteeg R, Picci P, Hattinger CM, Serra M. Preclinical validation of Aurora kinases-targeting drugs in osteosarcoma. British journal of cancer. 2013;109(10):2607–2618. doi: 10.1038/bjc.2013.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Z, Jiang J, Yang H, Ge Z, Wang Q, Zhang L, Wu C, Wang J. Silencing of Aurora kinase A by RNA interference inhibits tumor growth in human osteosarcoma cells by inducing apoptosis and G2/M cell cycle arrest. Oncology reports. 2014;31(3):1249–1254. doi: 10.3892/or.2014.2986. [DOI] [PubMed] [Google Scholar]
- 36.Bu Y, Yang Z, Li Q, Song F. Silencing of polo-like kinase (Plk) 1 via siRNA causes inhibition of growth and induction of apoptosis in human esophageal cancer cells. Oncology. 2008;74(3-4):198–206. doi: 10.1159/000151367. [DOI] [PubMed] [Google Scholar]
- 37.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell research. 2002;12(1):9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 38.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26(22):3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 39.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, Stivala F, Libra M, Basecke J, Evangelisti C, Martelli AM, Franklin RA. Roles of the Raf/MEK/ ERK pathway in cell growth, malignant transformation and drug resistance. Biochimica et biophysica acta. 2007;1773(8):1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe H, Kikuchi S, Hayakawa K, Iida T, Nagahashi N, Maeda K, Sakamoto J, Matsumoto N, Miura T, Matsumura K, Seki N, Inaba T, Kawasaki H, Yamaguchi T, Kakefuda R, Nanayama T, et al. Discovery of a Highly Potent and Selective MEK Inhibitor: GSK1101. ACS Med Chem Lett. 2011;2(4):320–324. doi: 10.1021/ml200004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright CJ PL. Trametinib: first global approval. Drugs. 2013;73(11):1245–1254. doi: 10.1007/s40265-013-0096-1. [DOI] [PubMed] [Google Scholar]
- 42.Dong Q, Dougan DR, Gong X, Halkowycz P, Jin B, Kanouni T, O'Connell SM, Scorah N, Shi L, Wallace MB, Zhou F. Discovery of TAK-733, a potent and selective MEK allosteric site inhibitor for the treatment of cancer. Bioorganic & medicinal chemistry letters. 2011;21(5):1315–1319. doi: 10.1016/j.bmcl.2011.01.071. [DOI] [PubMed] [Google Scholar]
- 43.Cohen RB, Aamdal S, Nyakas M, Cavallin M, Green D, Learoyd M, Smith I, Kurzrock R. A phase I dose-finding, safety and tolerability study of AZD8330 in patients with advanced malignancies. European journal of cancer. 2013;49(7):1521–1529. doi: 10.1016/j.ejca.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 44.von Euw E, Atefi M, Attar N, Chu C, Zachariah S, Burgess BL, Mok S, Ng C, Wong DJ, Chmielowski B, Lichter DI, Koya RC, McCannel TA, Izmailova E, Ribas A. Antitumor effects of the investigational selective MEK inhibitor TAK733 against cutaneous and uveal melanoma cell lines. Mol Cancer. 2012;11:22. doi: 10.1186/1476-4598-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishino S, Miyake H, Vincent P, Mori I. Evaluation of the therapeutic efficacy of a MEK inhibitor (TAK-733) using F-fluorodeoxyglucose-positron emission tomography in the human lung xenograft model A549. Ann Nucl Med. 2015 doi: 10.1007/s12149-015-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohseny AB, Machado I, Cai Y, Schaefer KL, Serra M, Hogendoorn PC, Llombart-Bosch A, Cleton-Jansen AM. Functional characterization of osteosarcoma cell lines provides representative models to study the human disease. Lab Invest. 2011;91(8):1195–1205. doi: 10.1038/labinvest.2011.72. [DOI] [PubMed] [Google Scholar]
- 47.Ottaviano L, Schaefer KL, Gajewski M, Huckenbeck W, Baldus S, Rogel U, Mackintosh C, de Alava E, Myklebost O, Kresse SH, Meza-Zepeda LA, Serra M, Cleton-Jansen AM, Hogendoorn PC, Buerger H, Aigner T, et al. Molecular characterization of commonly used cell lines for bone tumor research: a trans-European EuroBoNet effort. Genes Chromosomes Cancer. 2010;49(1):40–51. doi: 10.1002/gcc.20717. [DOI] [PubMed] [Google Scholar]
- 48.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 49.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]