Abstract
A major goal for HIV-1 vaccine development is the production of an immunogen to mimic native, functional HIV-1 envelope trimeric spikes (Env) on the virion surface. We lack a reliable description of a native, functional trimer, however, because of inherent instability and heterogeneity in most preparations. We describe here two conformationally homogeneous Envs derived from difficult-to-neutralize primary isolates. All their non-neutralizing epitopes are fully concealed and independent of their proteolytic processing. Most broadly neutralizing antibodies (bnAbs) recognize these native trimers. Truncation of their cytoplasmic tail has little effect on membrane fusion, but it diminishes binding to trimer-specific bnAbs while exposing non-neutralizing epitopes. These results yield a more accurate antigenic picture than hitherto possible of a genuinely untriggered and functional HIV-1 Env; they can guide effective vaccine development.
HIV-1 envelope glycoprotein (Env) fuses viral and cell membranes, allowing entry of the virus into host cells to initiate infection. The Env polypeptide chain is produced as a precursor, gp160, which trimerizes to (gp160)3 and then undergoes cleavage into two noncovalently associated fragments: the receptor-binding fragment gp120 and the fusion fragment gp41 (1). Three copies each of gp120 and gp41 form the mature envelope spike (gp120/gp41)3, which is the major viral surface antigen and therefore a critical target for vaccine development. Gp120 binds to host primary receptor CD4 and then to coreceptor (e.g., CCR5 or CXCR4), triggering large conformational changes and a cascade of refolding events in gp41 that lead to membrane fusion (2, 3) (fig. S1).
The failure of monomeric gp120 as a vaccine candidate in a large efficacy trial (4, 5) led to the notion that an immunogen mimicking the native, functional envelope trimer would be needed to induce effective, broadly neutralizing antibody (bnAb) responses by vaccination. In particular, bnAbs [except those recognizing the membrane-proximal external region (MPER) (6)] were thought to bind only the untriggered, native Env trimer (7). Attempts to produce such an Env preparation have met with only limited success (8, 9). Moreover, we lack an accurate standard for a native, functional trimer because most Env preparations, both soluble and membrane-bound, including those on the surface of infectious virions, show considerable structural instability and heterogeneity, leading to conflicting interpretations. For instance, based on virus-capturing assays, some groups conclude that certain “non-neutralizing” (including strain-specific neutralizing) epitopes are exposed on the native, functional Env trimer, whereas others believe that there are both functional and nonfunctional Envs present on the surface of infectious viral particles (10–13). Furthermore, the uncleaved ectodomain of trimeric (gp160)3, designated gp140, is often considered to mimic the native state of Env. Recombinant gp140 trimers derived from selected strains are stable and homogeneous, with certain desired antigenic properties (14–16), but we cannot know how closely they resemble functional and untriggered Env spikes without a good native-trimer reference. Are these soluble gp140 trimers—all with certain non-neutralizing epitopes (e.g., V3 loop) exposed—really the best surrogate for a native Env trimer. If not, how can we improve them? Recent work on conformational dynamics of the Env spikes on the virion surface suggests that the native trimer transitions among three distinct prefusion conformations (17). If this is true for difficult-to-neutralize clinical isolates, how can the functional trimer limit access to the non-neutralizing epitopes that overlap with the functionally important sites, such as the CD4 binding site and the V3 loop?
We have previously screened many HIV-1 primary isolates and identified two (clade A 92UG037.8 and clade C C97ZA012) that yield stable, homogeneous gp140 trimers (6, 14). The two Envs have about 74% sequence identity. Their divergence, typical for cross-clade comparisons, samples a range of Env diversity. Additional stable, clade-C trimers have since been reported (18), but we have not yet detected a clear “stability signature.” Our previous immunogenicity studies using either gp120 or gp140 immunogens derived from these two isolates failed to show any autologous neutralizing antibody responses, although V3-specific antibodies were present in the sera of immunized animals (19, 20). We surmised that the non-neutralizing V3 epitopes must not be exposed on the native Env trimers of these strains, despite their accessibility on the corresponding soluble gp140 trimers (14). Indeed, V3 accessibility is the one antigenic characteristic of the stable uncleaved gp140s (14) and of the BG505 SOSIP.664 trimer (15, 21, 22) that does not correlate with neutralization. The V3 loop is only slightly exposed in the disulfide-stabilized SOSIP crystals structure (21), but the molecule in solution presumably visits a wider range of conformations when not bound by one or more conformation-specific antibodies, as in crystal and cryogenic electron microscopy structures.
To study the antigenic characteristics of intact, native Env trimers, we generated 293T cell lines stably transfected with either 92UG037.8 or C97ZA012 gp160 (fig. S2). Efficient fusion with TZM.bl cells (23), completely blocked by T20 (1 mM) (fig. S3A), showed that there were fully functional envelope trimers on the cell surfaces. About 50% of gp160s were cleaved into gp120 and gp41, both inside the cells and on the cell surfaces (fig. S3B).
We analyzed antibody binding to cell-surface Env by a fluorescence-activated cell sorting (FACS) assay (see table S1 for antibodies used and references) (24). Antibody 2G12, which recognizes a glycan-dependent epitope on gp120, bound the cell-surface Env equally well with and without CD4, indicating no significant CD4-induced gp120 shedding (Fig. 1 and fig. S4). CD4 binding site (bs) bnAbs all bound the cell-surface Env tightly, consistent with their neutralization potency; soluble CD4 effectively competed with all of them (Fig. 1, figs. S5A and S6, and table S2). A CD4bs antibody, b6, that does not neutralize the two isolates used here, showed no binding at all, suggesting that its epitope is inaccessible on both cleaved (fusion-competent) (gp120/gp41)3 and uncleaved (not fusion-competent) (gp160)3. Binding of non-neutralizing CD4i (CD4-induced) antibodies 17b and 412d was likewise independent of the cleavage but completely dependent on CD4 engagement. Another non-neutralizing CD4i antibody, A32, failed to bind these Envs under any circumstances. The trimer-specific bnAbs recognized only the native, untriggered Env trimer with high affinity, but not the CD4-bound form (Fig. 1, fig. S5A, and table S2). Another two bnAbs, PGT128 and 10–1074, which target a glycan-dependent epitope in the V3 stem, also bound the native Env but not the CD4-liganded form.
Fig. 1. Antigenic characteristics of the 92UG037.8 Env trimer presented on cell surfaces.
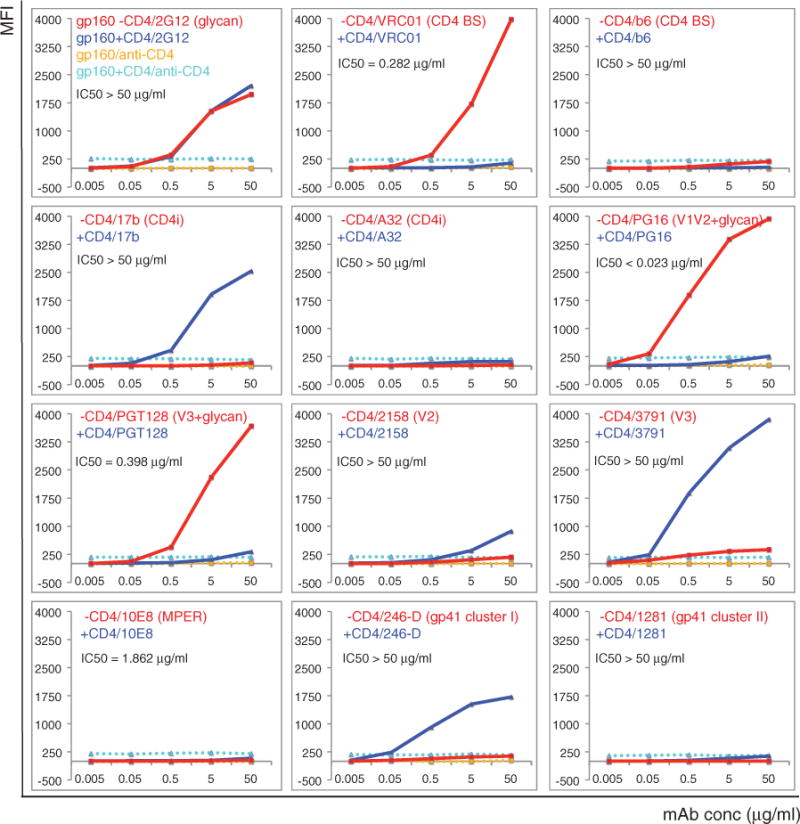
Plots of antibody binding to the Env trimer on the 92UG037.8 gp160 cell surfaces in the absence (red) or presence (blue) of soluble CD4. Fluorescent signal for bound CD4 is shown in the presence of CD4 (cyan) or in the absence of CD4 (orange). Antibodies and their epitopes are indicated. The median inhibitory concentration (IC50) values were determined in a luciferase-based virus neutralization assay using 92UG037.8 gp160 and purified antibodies. Unless specified, all antibodies used are Fab fragments. Original flow cytometry histograms are shown in fig. S4. Extensive control experiments were carried out to ensure the binding specificity, and the experiments were repeated at least twice with almost identical results.
Non-neutralizing epitopes, including V3 loop (3791 and 19b), V2 (2158), gp41 cluster I epitopes (246-D, 240-D, and 7B2), and gp41 cluster II epitopes (1281 and 167-D) were inaccessible on the cell-surface trimer (Fig. 1 and fig. S5A), explaining why these antibodies do not neutralize. Their occlusion was independent of the cleavage between gp120 and gp41, as ~50% of the Env on the cell surfaces remained uncleaved (fig. S3B). Upon CD4 binding, the V3 loop, the cluster I epitope, and (to a lesser extent) the V2 loop all became accessible. Thus, the antibodies recognizing these epitopes might be better categorized as CD4i antibodies. MPER-directed bnAbs did not bind the Env trimer, consistent with our previous work showing that they target a fusion-intermediate conformation of gp41 (6, 25, 26). Isolate 92UG037.8 resists neutralization by two antibodies, PGT151 and 152, with epitopes at the gp120-gp41 interface; these antibodies indeed fail to bind native Env trimers (fig. S5A and table S2). Two other interface-directed bnAb (27) bound only weakly (fig. S5A). These results are reproducible under different conditions (28). In summary, all the Env trimers, cleaved or uncleaved, on the cell surfaces adopted a defined, homogeneous conformation (or small range of conformations) that was recognized by bnAbs only and not by any of the non-neutralizing antibodies tested.
These cell-surface Env trimers have antigenic properties distinct from those of the corresponding gp140 trimers (14, 20), which lack the cytoplasmic tail (CT) and the transmembrane segment (TM). Does the CT influence the antigenicity of the ectodomain? We produced a stable line expressing the 92UG037.8 gp160 with the entire CT deleted (a form designated gp160ΔCT) and replaced by a His tag (fig. S2). This cell line also efficiently fused with TZM.bl cells (fig. S9A). The expression level of gp160ΔCT was much higher than that of intact gp160 (Fig. 2), as reported previously (29). As with the gp160 cell line, both cleaved and uncleaved gp160ΔCT trimers were present on the cell surface (fig. S9B), but with a smaller proportion of cleaved molecules; gp160ΔCT without the His tag showed the same extent of cleavage as did intact gp160 (fig. S10).
Fig. 2. Antigenic properties of the 92UG037.8 gp160-ΔCT.
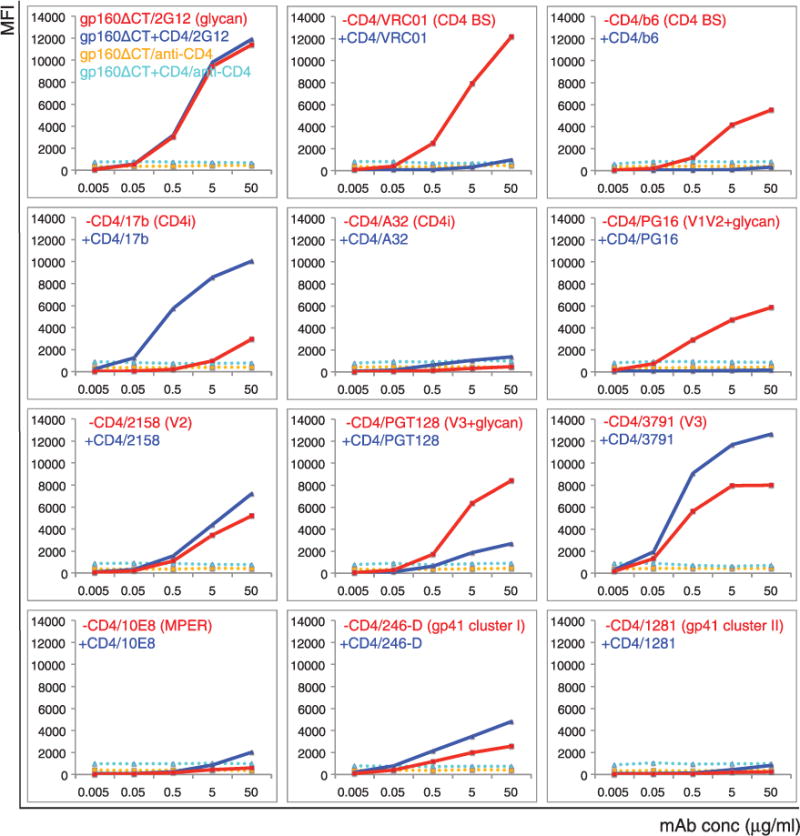
Plots of antibody binding to the Env trimer on the 92UG037.8 gp160-ΔCT cell surfaces in absence (red) or presence (blue) of soluble CD4. Fluorescent signal for bound CD4 is shown in the presence of CD4 (cyan) or in the absence of CD4 (orange). Antibodies and their epitopes are indicated. Unless specified, all antibodies used are Fab fragments. Original flow cytometry histograms are shown in fig. S11. The experiments were repeated at least twice with almost identical results.
Binding of 2G12 to gp160ΔCT was unaffected by CD4, indicating no significant CD4-induced gp120 shedding (Fig. 2 and fig. S11). CD4bs bnAbs bound gp160ΔCT as tightly as they did intact gp160, but the non-neutralizing antibody, b6, also bound detectably to gp160ΔCT (Fig. 2 and figs. S12A and S13), suggesting that the CD4 bs is more exposed when the CT is deleted. The CD4i antibodies, 17b and 412d, also showed weak, but detectable, binding to gp160ΔCT, whereas binding by the trimer-specific bnAbs was significantly reduced when normalized by the Env expression level. The non-neutralizing V3 and V2 epitopes were much more exposed and less dependent on CD4 binding for recognition by antibody. In particular, the V3 antibody 3791 blocked cell-cell fusion mediated by gp160ΔCT but not by gp160 (fig. S14), indicating that removal of the CT can expose the V3 loop on a functional (i.e., fusogenic) trimer. PGT128 binding to gp160ΔCT likewise depended less on CD4, and 10–1074 became totally CD4-independent (Fig. 2 and fig. S12A). In addition, although the MPER epitopes and the cluster II epitopes remained inaccessible, the non-neutralizing cluster I epitopes were exposed even in the absence of CD4. Both His-tagged and nontagged gp160ΔCT constructs had almost identical antigenic profiles despite the different extent of cleavage (figs. S12B and S15), suggesting that the cleavage between gp120 and gp41 does not have a major effect on the trimer antigenicity. Overall, these data indicate that the CT has minimal effect on the membrane fusion function of the Env trimer but that it has an unexpectedly large influence on the antigenic properties of the ectodomain on the other side of the membrane (table S2). Thus, the membrane-fusion capacity of an Env trimer does not depend on strict retention of native antigenic characteristics. The same conclusion can be drawn from comparison of another pair of gp160 and gp160ΔCT, derived from the isolate C97ZA012 (figs. S16A, S16B, S17, and S18).
We constructed four deletion mutants—gp160-CT120, gp160-CT90, gp160-CT60, and gp160-CT30— with gradually decreasing CT lengths (figs. S2 and S10). All these deletion constructs were functional in a cell-cell fusion assay, but shortening the CT led to diminished binding by the trimer-specific bnAbs and increasing exposure of the non-neutralizing epitopes (Fig. 3; fig. S19, A to D; and figs. S20 to S23). Introducing other modifications (30) (fig. S2), previously assumed to be “harmless,” caused even greater changes in antigenicity. These constructs were all nonfunctional (Fig. 3), and we detected much greater exposure of non-neutralizing epitopes, including those of b6, 17b, A32, 2158, 3971, and 246-D. Binding to the trimer-specific bnAbs, PG16 and PGT145, was completely lost (Fig. 3; fig. S19, E to H; and figs. S24 to S27). The MPER and cluster II epitopes remained concealed even in these nonfunctional Envs.
Fig. 3. Effect of the gp41 cytoplasmic tail on antigenic properties of the ectodomain of Env.
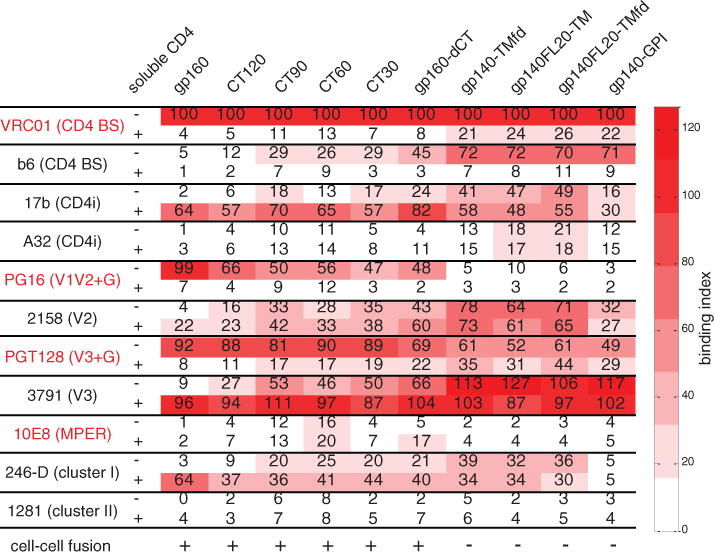
A tabulated summary of antibody binding to various 92UG037.8 Env constructs, including gp160, gp160-CT120, gp160-CT90, gp160-CT60, gp160-CT30, gp160-ΔCT, gp140-TMfd, gp140FL20-TM, gp140FL20-TMfd, and gp140-GPI. Epitopes targeted by the antibodies include CD4bs, CD4 binding site; CD4i, CD4-induced; V1V2+G, the V1V2 loop and glycans; V3+G, the V loop and glycans; MPER, membrane proximal external region; and gp41, cluster I and cluster II. Binding index is normalized by VRC01 binding to each untriggered Env construct in the absence of CD4, and it is defined as the ratio between the maximum MFI of a given antibody binding and the maximum MFI of VRC01 binding to the same Env construct. Cell-cell fusion capacity of each Env construct was monitored by syncytium formation when mixing Env-expressing cells and TZM-bl cells.
Our results demonstrate that a native Env trimer can indeed adopt a defined and homogeneous conformational state without contamination by any irrelevant forms of Env (12). This native, un-triggered conformation seems to be independent of the cleavage between gp120 and gp41. It is particularly surprising that the Env CT has a large effect on the antigenicity of the ectodomain on the other side of the membrane. Various gp160-ΔCT constructs, often with an increased yield, are still widely considered to be faithful substitutes for the full-length gp160, despite published evidence suggesting that the CT may influence epitope exposure (31). Our results demonstrate that truncation of the CT affects the antigenic characteristics of the native Env trimer, even in the absence of the matrix protein, but not its fusogenicity. Thus, a “functional” Env may not have a fully “native” antigenic surface (defined by neutralization), because even a fusion-competent Env trimer can expose non-neutralizing epitopes (Fig. 3). Those apparently “harmless” modifications, such as CT deletion—an approach widely used in various vaccine strategies to enhance Env yield and/or stability—can have a strong effect on trimer structure, antigenicity, and potentially immunogenicity.
Strain-specific neutralizing epitopes, such as the V3 loop and the V1V2 loop, are well protected on the native trimer derived from the hard-to-neutralize strains we have studied, explaining why it is difficult to induce autologous neutralizing antibody responses against such isolates. The antigenic properties of gp160 and gp160-ΔCT may represent, respectively, those of the extreme cases of the difficult-to-neutralize primary isolates that cannot induce autologous neutralizing responses and the easy-to-neutralize, laboratory-adapted strains that induce strong autologous responses, whereas other isolates (of intermediate susceptibility to neutralization) may adopt conformations in between, like those of our partially truncated gp160-CT constructs (Fig. 4). A few mutations within the entire gp160 sequence can convert one form into another, perhaps even within a single patient, just as primary and laboratory-adapted simian HIV or HIV-1 isolates interconvert during in vitro and in vivo passages (32, 33). Interconversion thus allows Envs in a closely related swarm of viruses to sample, at a population level, a much greater dynamic range than previously appreciated (17). In an infected individual, immune pressure might drive any particular isolate to evolve into one that is difficult to neutralize and does not induce autologous neu-tralizing responses (Fig. 4). Such viruses substantially raise the barrier to vaccine development.
Fig. 4. A proposed relationship among all HIV-1 isolates based on their Env properties.
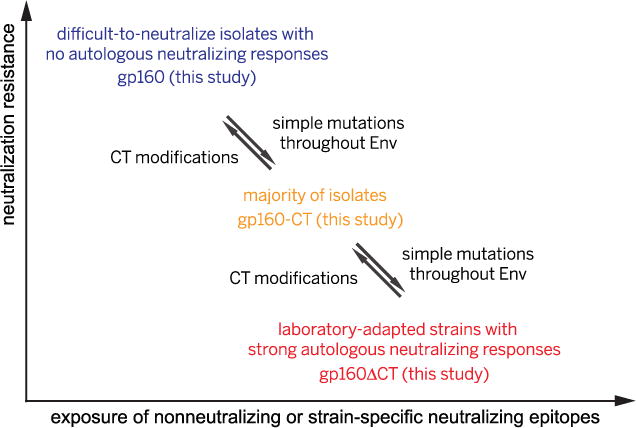
Our gp160 and gp160-ΔCT may represent Envs of the difficult-to-neutralize primary isolates that cannot induce autologous neutralizing antibody responses and the easy-to-neutralize, laboratory-adapted strains that induce strong autologous neutralizing responses, respectively. Other isolates (of intermediate susceptibility to neutralization) may adopt intermediate conformations, like those of our partially truncated gp160-CT constructs. A few mutations can easily convert one form into another, perhaps even within a single patient, and thus an effective vaccine may need to protect against the viruses described in blue.
In summary, the data presented here paint a more accurate antigenic picture than hitherto available of a genuinely native and functional HIV-1 envelope spike from clinically relevant viruses. They provide an excellent reference for studies on the prefusion conformation of HIV-1 Env trimers and serve as guides for assessing how well potential immunogens mimic a native viral spike.
Supplementary Material
Acknowledgments
We thank S. Harrison, H. Ha, and G. Frey for generous advice and assistance; D. Barouch, B. Haynes, and A. Carfi for critical reading of the manuscript; and the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH for reagents. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. This work was supported by NIH grants AI084794 (to B.C. and Dan H. Barouch), GM083680 (to B.C.), AI106488 (to B.C.), Collaboration for AIDS Vaccine Discovery (CAVD) grant OPP1040741 (to Dan H. Barouch from the Bill and Melinda Gates Foundation), and the Center for HIV/AIDS Vaccine Immunology–Immunogen Design AI-100645 (to Barton F. Haynes).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/349/6244/191/suppl/DC1
Materials and Methods
References (34–75)
REFERENCES AND NOTES
- 1.Harrison SC. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan DC, Fass D, Berger JM, Kim PS. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 3.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 4.Flynn NM, et al. J Infect Dis. 2005;191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 5.Pitisuttithum P, et al. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 6.Frey G, et al. Proc Natl Acad Sci USA. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.By “triggered,” we mean any of the sequence of conformations that Env adopts after CD4 binding, on the pathway to fusion; by “native,” those with antigenic properties fully consistent with antibody neutralization.
- 8.Schiffner T, Sattentau QJ, Dorrell L. Retrovirology. 2013;10:72. doi: 10.1186/1742-4690-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCoy LE, Weiss RA. J Exp Med. 2013;210:209–223. doi: 10.1084/jem.20121827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu P, et al. J Virol. 2014;88:5165–5170. doi: 10.1128/JVI.03765-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.York J, et al. J Virol. 2001;75:2741–2752. doi: 10.1128/JVI.75.6.2741-2752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore PL, et al. J Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong T, Crooks ET, Osawa K, Binley JM. J Virol. 2012;86:3574–3587. doi: 10.1128/JVI.06938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacs JM, et al. Proc Natl Acad Sci USA. 2014;111:18542–18547. doi: 10.1073/pnas.1422269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders RW, et al. PLOS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenaga J, et al. PLOS Pathog. 2015;11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munro JB, et al. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bricault CA, et al. J Virol. 2015;89:2507–2519. doi: 10.1128/JVI.03331-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nkolola JP, et al. J Virol. 2010;84:3270–3279. doi: 10.1128/JVI.02252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovacs JM, et al. Proc Natl Acad Sci USA. 2012;109:12111–12116. doi: 10.1073/pnas.1204533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pancera M, et al. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugach P, et al. J Virol. 2015;89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.In the FACS assay, bound primary antibodies were detected by a fluorescently labeled secondary antibody. Stably transfected cell lines for all Env constructs were necessary for obtaining high-quality data with mostly single sharp peaks in histograms; data that met these criteria gave reliable readings of MFI (mean fluorescence intensity) of a relatively homogeneous cell population (fig. S3C).
- 25.Alam SM, et al. Proc Natl Acad Sci USA. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, et al. J Virol. 2014;88:1249–1258. doi: 10.1128/JVI.02664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, et al. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.We obtained the same results regardless of whether we used a Fab or IgG form of the antibody or a 2-domain or 4-domain soluble CD4 (figs. S5A, S6, S5B, and S7). The antigenic properties of gp160 remained largely the same when the entire experiments were performed at 37°C, except that we saw significant CD4-induced gp120 shedding, as indicated by a lower level of 2G12, 17b, and 3791 binding when CD4 was present and by an increase in 246-D binding (figs. S5C and S8).
- 29.LaBranche CC, et al. J Virol. 1995;69:5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The modifications include replacing TM and CT with a glycosylphosphatidylinositol (GPI) anchor (gp140-GPI), replacing CT with a foldon trimerization tag (gp140-TM-fd), inserting a flexible linker between gp120 and gp41 (gp140-FL20-TM), and adding both the foldon tag and the flexible linker (gp140-FL20-TM-fd).
- 31.Edwards TG, et al. J Virol. 2002;76:2683–2691. doi: 10.1128/JVI.76.6.2683-2691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye Y, Si ZH, Moore JP, Sodroski J. J Virol. 2000;74:11955–11962. doi: 10.1128/jvi.74.24.11955-11962.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beaumont T, et al. J Virol. 2001;75:2246–2252. doi: 10.1128/JVI.75.5.2246-2252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


