Abstract
Puccinia psidii sensu lato (s.l.) is the causal agent of eucalyptus and guava rust, but it also attacks a wide range of plant species from the myrtle family, resulting in a significant genetic and physiological variability among populations accessed from different hosts. The uredospores are crucial to P. psidii dissemination in the field. Although they are important for the fungal pathogenesis, their molecular characterization has been poorly studied. In this work, we report the first in-depth proteomic analysis of P. psidii s.l. uredospores from two contrasting populations: guava fruits (PpGuava) and eucalyptus leaves (PpEucalyptus). NanoUPLC-MSE was used to generate peptide spectra that were matched to the UniProt Puccinia genera sequences (UniProt database) resulting in the first proteomic analysis of the phytopathogenic fungus P. psidii. Three hundred and fourty proteins were detected and quantified using Label free proteomics. A significant number of unique proteins were found for each sample, others were significantly more or less abundant, according to the fungal populations. In PpGuava population, many proteins correlated with fungal virulence, such as malate dehydrogenase, proteossomes subunits, enolases and others were increased. On the other hand, PpEucalyptus proteins involved in biogenesis, protein folding and translocation were increased, supporting the physiological variability of the fungal populations according to their protein reservoirs and specific host interaction strategies.
Introduction
Puccinia psidii sensu lato (s.l.) is a biotrophic rust fungi native to South America, where it was first described in guava [1], causing severe infections resulting in losses up to100% of the fruits [2]. Eucalyptus rust is apparently a specialization of P. psidii genotype evolved from rust occurring in Myrtaceae in South America, enabling it to ‘host jump’ and infect Eucalyptus, an introduced Myrtaceae species to Brazil at the end of the 19th century [3, 4]. Likely, some isolates from guava did not infect eucalyptus and vice versa [4] indicating an occurrence in the evolution of a host-specific genotype. Since then, rust has become one of the most important eucalyptus diseases, principally attacking nurseries and young plants, decreasing productivity, and in some cases, causing the death of the highly susceptible plants [5].Guava and other Myrtaceae from South America most likely served as the sources of inoculum during the adaptation of P. psidii to eucalyptus [6].
Recently, the genetic variability in P. psidii s.l. populations infecting different host plants was evaluated and proved that, in general, populations collected from guava had a greater level of diversity than populations from Eucalyptus spp. [6]. The classification of this fungus has been deeply debated. Morphological and molecular studies indicate that Puccinia is polyphyletic group [7, 8, 9], named P. psidii s.l. complex that is able to infect a great range of hosts from the Mytaceaes family. Thus, the disease caused by the fungus currently treated as P. psidii s.l. is best referred to as Myrtle rust [10].
P. psidii s.l. is considered an autoecious species with an incomplete life cycle (the spermogonia stage may be absent).The aecial state is extremely rare [5].The uredia are the most common and principal P. psidii s.l. dissemination structure and are constantly produced during natural and artificial inoculations [4]. Moreover, the uredosporic pustules are the only visual rust symptom that can be used to diagnose infection under field conditions [5, 11]. The uredospore protein profile may play a key role in the establishment of the disease, because they are the first fungal structures that have contact with the host before infection starts, and may carry some key proteins with respect to the host, because the rust uredopores germination is strongly dependent on physical and chemical signals from the host [12].
The physiological variability of P. psidii s.l.uredospores according to the host has been described previously [5, 13, 14]. Coelho et al. [13] identified three groups of rust biotypes, each pathogenic on different host combinations: Psidium guajava only, Eucalyptus grandis and P. guajava or E. grandis and Sizygium jambos. Despite the importance of the uredospore as the primary source of fungal inoculum, little is known about the molecular features of P. psidii s.l. uredospores or how they relate to its many host organisms, for instance guava and eucalyptus.
Global proteomic analysis can provide the framework for an in-depth understanding of the cellular processes and organization of a particular organism. The complexity of the proteome exceeds that of the genome when protein isoforms, alternative splicing variants, and post-translational modifications are taken into account [15]. However, proteomic analysis of filamentous fungi is still lacking in this field [16]. Among the filamentous fungi, the proteomes of the Ascomycota have been studied more intensively than those of Basidiomycota, most likely due to the number of pathogenic fungi that are found in Ascomycota [17]. Gradually, the proteomes of filamentous fungi are being revealed, including the mycelia proteome [18, 19, 20], secretomes [20, 21] and sub-proteomes [22] of a wide range of species, such as Aspergillus spp. [23, 24, 25, 26], Botrytis cinerea [17], Neurospora crassa [22], Penicillium expansum [27], Pleurotus sapidus [28], Sclerotinia sclerotiorum [20], Terebralia palustris [29], P. triticina [30], Blumeria graminis [31], Verticillium albo-atrum [32], and others.
In the present work, we report the first label free proteomic quantification of P. psidii s.l. uredospores from two contrasting fungal populations with known physiological variability that reflects specific molecular interactions directed by the host. The results obtained allowed the identification of pathogenicity or virulence factors of this important pathogen and their correlation with the fungal physiological variation as a possible specific host interaction strategy during the infection process.
Materials and Methods
Pathogen material
P. psidii uredospores were collected from E. grandis leaves (PpEucalyptus) in the experimental area belonging to FuturaGene, Itapetininga-SP, Brazil in accordance with Dr. Esteban R. Gonzalez (FuturaGene Corporation Manager). Uredospores from P. guajava (PpGuava) where isolated from fruits of trees located in the Campus of ESALQ/USP, Piracicaba-SP, Brazil in accordance with the Chief of the Intitution Dr. José Caixeta Filho. Both fields do not involve endangered or protected species. Infected leaves and fruits from E. grandis and P. guajava tree`s were individually collected. We sampled three plants per species, a bulk of leaves or fruits from each plant considered as an independent biological replicate, totalizing three biological replicates per population sample. The uredospores samples from each replicate were divided, and half was immediately frozen at -80°C for proteomic analysis. The remaining sample was dehydrated in silica gel for 48 h at 4°C and frozen at -80°C for light microscopy analysis and for infection assay in E. grandis.
Light microscopy analysis
The morphology of the PpEucalyptus and PpGuava uredospores and the germination viability assays were performed in water-agar medium (8 g/L). The uredospores from each population were suspended in mineral oil (Invitrogen) and 400 μL of each suspension (104 uredospores/mL), spread onto the medium and incubated at 22°C in the dark for 24 h. The uredospores were analyzed using a light microscope (Axiophot II; Zeiss). For morphological analysis, we measured the length and width of 50 uredospores from each population, and the viability of the uredospores was confirmed by the presence of germ tubes after 24 h.
Plant-infection assay
Eighteen highly susceptible E. grandis plantlets (monoprogenie D901) were grown under greenhouse conditions for 120 days and were then transferred to a controlled growth chamber (E15, Conviron),where they were maintained under a 12 h photoperiod (200 μmol m-1 s-1) at 20°C for acclimatization. After seven days, the PpEucalyptus and PpGuava uredospore suspensions containing 104 uredospores mL-1 and 0.05% Tween 20 were sprayed onto six plantlets, two plants per biological sample. To maintain a high level of humidity, the plants were enclosed in transparent plastic bags for the first 48 h, with the first 24 h occurring in complete darkness at 20°C. Thereafter, the plants were returned to the previously described growth conditions. The control plantlets were treated in the same way, but were sprayed only with the 0.05% Tween solution. The evolution of the infection was monitored every two days for two weeks.
Protein extraction and trypsin digestion
Proteins were extracted from uredospores according to the protocol described by Damerval et al. [33] with some modifications. Uredospore samples (1 g) were ground into a powder in liquid nitrogen and homogenized in 30 mL of TCA solution (10% (w/v) trichloroacetic acid and 0.07% (v/v) 2-mercaptoethanol in acetone) per biological replicate. The samples were then incubated for 1 h at -20°C, followed by centrifugation for 20 min at 16,000 g at 2°C. The supernatant was discarded, and the pellet was suspended in 30 mL of 100% cold acetone containing 0.07% (v/v) 2-mercaptoethanol. The pellets were incubated at -20°C, for one h, followed by centrifugation for 20 min at 16,000 g at 2°C. The pellet was again suspended in 30 mL of 100% cold methanol and incubated for one hour at -80°C. After centrifugation at 16,000 g for 20 min at 2°C, the supernatant was discarded, and the resulting pellet was lyophilized.
The dried pellets were solubilized in 800 μL of TCT buffer [7 M urea, 2 M thiourea, 10 mM DTT and 0.4% (v/v) Triton X-100]. Complete protein solubilization was achieved by vigorous shaking using a vortex. Protein extracts were desalinized using Amicon Ultra-0.5 mL 3K-NMWL filter devices (Millipore Corporation).
The total protein concentration was determined using an Agilent Bioanalyzer 2100 (Agilent) and the 230 kDa protein kit. Fifty micrograms of protein was denatured with 25 μL of 0.2% RapiGest (RapiGest SF, Waters), reduced with 2.5 μL of 100 mM dithiothreitol and alkylated with 2.5 μL of 300 mM iodoacetamide. Trypsin digestion was performed with sequencing Grade Modified Trypsin (Promega) at a 1:100 (w/w) enzyme: protein ratio. After digestion, 10 μL of 5% (v/v) trifluoroacetic acid was added to the digested mixture to hydrolyze the RapiGest. The peptide mixture was then desalted using PepClean C 18-columns (Thermo-Fisher Scientific). The final volume of 50 μL was obtained by the addition of 20 mM ammonium formate (pH 10) solution containing 1 pM rabbit phosphorylase (internal standard to data normalization and label free protein quantification- P00489) to the lyophilized, desalted peptide sample.
MSE analysis
Mass spectra of the peptide fragments were acquired by reverse-phase ultraperformance liquid chromatography (2D Technology nanoACQUITY-Waters). First dimension separation was achieved in an XBridge BEH130 C18 5μm 300 μm x 50 mm column. Elution was performed using 5 different binary gradients with 20 mM pH 10 ammonium formate in acetonitrile, at a flow rate of 2 μL/min. Eluted peptides from the first dimension column were trapped in a Symmetry C18 5 μm 180 μm x 20 mm column and diluted, online, with acetonitrile containing 0.1% formic acid. Second dimension separation was performed in a HSS T3 1.8 μm 75 μm x 100 mm column, using a binary gradient from 3 to 85% of acetonitrile with 0.1% formic acid, during 52 min, at a flow rate of 350 μL/min.
Mass spectrometry acquisition was achieved in a Synapt G2 HDMS mass spectrometer equipped with an ion mobility cell and a nanolockspray source in the positive ion and ‘V’ mode. The doubly-charged ion ([M+2H]2+) was used for initial single-point calibration and MS/MS fragment ions of GFP [Glu 1]-Fibrinopeptide B with a [M + 2H]2+ = 785,84206 m/z was used as lock mass, to obtain the final instrument calibration. Data-independent scanning (MSE) experiments were performed by switching between low (3 eV) and elevated collision energies (15–50 eV), applied to the trap ‘T-wave’ cell filled with argon. Scan time of 0.8 s were used for low and high energy scans from m/z 50 to 2000 [34].
Processing Parameters and Database Search
The raw data processing, protein identification and relative quantitative analyses were all performed using the ProteinLynx Global Server (PLGS- v 2.5.1). The processing parameters included: automatic tolerance for precursor and product ions, minimum of 3 fragment ions matched per peptide, minimum of 7 fragment ions matched per protein, minimum of 2 peptides matched per protein, 1 possible trypsin missed cleavage, carbamidomethylation of cysteine as fixed modification and oxidation of methionine as variable modification, and a maximum false positive discovery rate (FDR) of 4% that was determined based on the search of a reversed database, which was generated automatically using PLGS 2.5.1 by reversing the sequence of each entry.
To identify and quantify the proteins, the intensities of the spectra were calculated by the stoichiometric method, according to the internal standard, the sequence of rabbit phosphorylase (Uniprot entry: P00489), by MSE analysis [34] and normalized using the PLGS auto normalization function. The amount and sequence of the matched peptide and protein fmols were obtained, based on the ratio of its three most abundant peptides (“High Top 3” method) determined in each individual experiment [34], considering the data from 3 biological replicates for each sample. All protein hits were identified with a confidence of > 95%. Only those proteins identified in at least two out of 3 replicates performed for each biological sample, were regarded as having undergone a significant change.
Protein identifications were obtained with the embedded ion accounting algorithm of PLGSsoftware searching into the UniProtKB/Swiss (http://www.uniprot.org/ release Version 2015_01) from Puccinia spp proteins (28,122 entries) appended in the internal standard. The final expression data was screened according to the following criteria: statistical significant fold change as well as standard deviation (SD) and their correspondent p value: p≤0.05 and p≥ 0.95 were considered whose abundance was statistically increased in the PpEucalyptus and PpGuava populations, respectively. Data labeled as PpEucalyptus and PpGuava represent proteins that were only identified in that population.
After the PLGS 2.5.1 expression analysis the data was manually inspected, and some parameters obtained using the MassPivot v 101.jar were included: (i) the average amount (fmol) of protein, (ii) average score of proteins and (iii) average amount of matched peptides to each protein.
Functional Analysis
The protein sequences that were matched with ionized peptides were functionally categorized by the Gene Ontology Consortium (GO) [35] (http://www.geneontology.org). The terms were obtained using the Blast2GO software with the default parameters specified by the program [36]. These annotations were previously simplified using the GO Slim feature. The EC numbers (Enzyme Commission) were obtained and the prediction of the subcellular localization of the proteins performed using the Signal P (http://www.cbs.dtu.dk/services/SignalP/) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) programs by the InterProScan tool (http://www.ebi.ac.uk/interpro/).
Results
Light microscopy analysis and plant infection
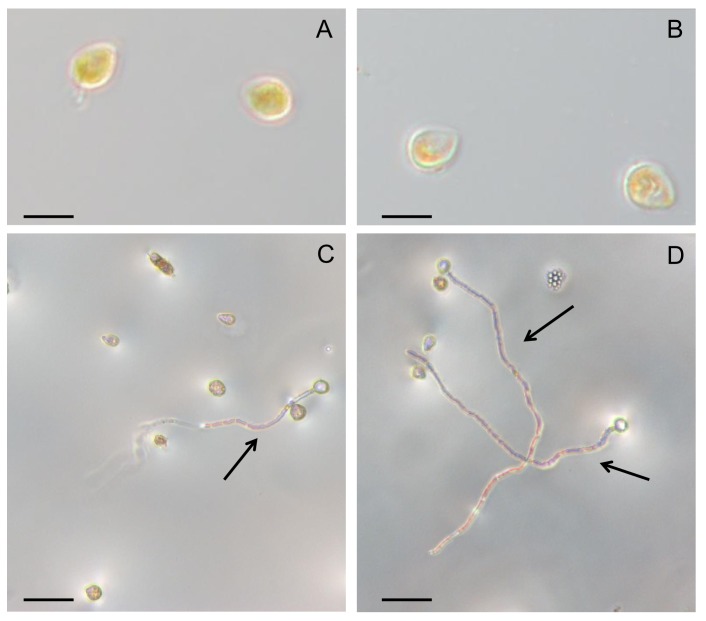
The morphological analysis showed that the size and shape of the uredospores were similar for both the PpEucalyptus and PpGuava populations (Fig 1A and 1B). The average length of the uredospores were 19.5 ± 2.2 μm and 19.9 ± 0.4 μm, respectively, for the PpEucalyptus and PpGuava populations, and their respective average widths were 13.5 ± 1.9 μm and 11.7 ± 0.7 μm, suggesting that there was no relationship between the uredospore size or shape and virulence or physiological variability of the populations. The populations also showed similar uredospore viability under in vitro conditions, with approximately 25% of the spores germinating on water-agar media (Fig 1C and 1D).
Fig 1. Morphological and viability analysis of Puccinia psidii uredospores.
P. psidii uredospores from E. grandis (A) and P. guajava (B) exhibit similar morphology and germination viability, respectively (C and D).The arrows indicate the fungal germ tube in both uredospore populations, 24 hours after inoculation in water-agar medium. Light microscopy images of PpEucalyptus and PpGuava uredospores are shown at 100 X (A and B) and 200 X (C and D) magnification. Scale bar: 20 μm in A and B, 50 μm in C and D.
Two weeks after inoculating E. grandis plantlets, uredospore pustules were visible on the host leaves inoculated with PpEucalyptus (Fig 2A), whereas the plantlets inoculated with PpGuava (Fig 2B) and the control plantlets (Fig 2C) did not exhibit any rust symptoms. The absence of pustules, chlorotic flecks or leaf deformation, demonstrate the existence of physiological variability and pathogenic specificity between the sampled populations.
Fig 2. Eucalyptus infection by guava and eucalyptus rust.

Symptoms induced by inoculation of P. psidii uredospores from PpEucalyptus (A) on E. grandis variety D901. This clone is rust susceptible when grown under field conditions. The white arrows indicate the fungal pustules. The leaves are shown 15 days after inoculation.The PpGuava populations (B) and control (C) did not show typical rust symptoms.
Overview of the P. psidii uredospore proteome
In total, 340 proteins were identified using the UniProt Puccinia genera sequences (UniProt database). Using spectral counting based label free quantification, we observed changes in relative protein abundance and a greater amount of proteins up regulated in PpGuava (S1 Table). Seventy five proteins were found exclusively in PpGuava and 29 in PpEucalyptus. Among the common proteins, 25 and 120 were increased in PpEucalyptus and PpGuava respectively (Fig 3).
Fig 3. Proteins identified from P. psidii uredospore populations.
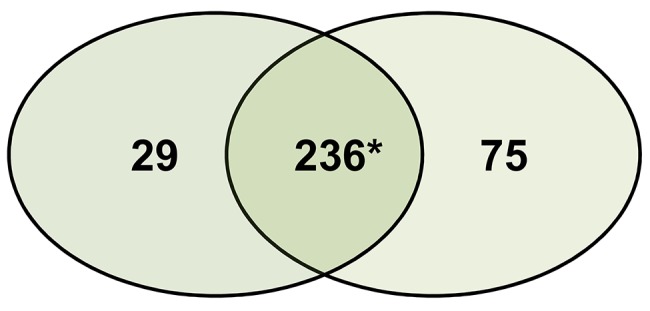
The proteins exclusively found in PpGuava (right) and PpEucalyptus (left) and common to both populations (center). Of the common proteins, 25 and 120 whose abundance were increased in PpEucalyptus and PpGuava, respectively.
Table 1 shows some significant differences between PpGuava and PpEucalyptus protein profiles. Proteins commonly found in fungal proteome studies were identified in both fungal populations, such as ribosomal subunits, elongation factors and tubulins, as well as proteins related to energy generation, such as ATP synthase subunits.
Table 1. Identification of proteins that were differentially represented in the P. psidii uredospore populations from E. grandis (PpEucalyptus) and Psidium guajava (PpGuava).
| Accessiona | Description | Scoreb | Ratio c | Log Ratio d | SDe | p valuef |
|---|---|---|---|---|---|---|
| E3L509 | 1 4 alpha glucan branching enzyme | 480.40 | 1.28 | 0.25 | 0.23 | 0.98 |
| E3L9C1 | 14 3 3 family protein | 12200.67 | 1.42 | 0.35 | 0.12 | 1.00 |
| E3L0L2 | 2 isopropylmalate synthase | 147.74 | 1.20 | 0.18 | 0.10 | 1.00 |
| E3KG66 | 2 methylcitrate dehydratase | 399.00 | 1.42 | 0.35 | 0.25 | 0.99 |
| E3KHM1 | 26S protease regulatory subunit 8 | 2686.01 | 1.12 | 0.11 | 0.14 | 0.95 |
| E3K1H4 | 26S protease subunit rpt4 | 2005.47 | 1.27 | 0.24 | 0.16 | 0.98 |
| E3K877 | 26S proteasome non-ATPase regulatory subunit 14 | 1971.74 | 1.51 | 0.41 | 0.16 | 1.00 |
| E3KKA1 | 26S proteasome regulatory subunit 6A | 2758.75 | 1.75 | 0.56 | 0.13 | 1.00 |
| E3KZT9 | 26S proteasome regulatory subunit 7 | 708.37 | 1.22 | 0.20 | 0.19 | 0.96 |
| E3JZ59 | 3 isopropylmalate dehydratase | 483.41 | 1.25 | 0.22 | 0.16 | 1.00 |
| E3L5E7 | 40S ribosomal protein S1 | 801.36 | PpGuava | PpGuava | PpGuava | PpGuava |
| E3JWG6 | 40S ribosomal protein S11 | 2195.29 | 0.31 | -1.16 | 0.24 | 0.00 |
| E3KBT0 | 40S ribosomal protein S16 | 1362.80 | 1.70 | 0.53 | 0.23 | 1.00 |
| E3K5L6 | 40S ribosomal protein S19 A | 7449.16 | 0.64 | -0.44 | 0.16 | 0.00 |
| E3JVN0 | 40S ribosomal protein S21 | 4844.32 | 1.62 | 0.48 | 0.16 | 1.00 |
| E3L8H2 | 40S ribosomal protein S3 | 675.32 | 1.60 | 0.47 | 0.21 | 1.00 |
| E3KJL1 | 40S ribosomal protein S5 | 506.24 | 0.66 | -0.42 | 0.27 | 0.00 |
| E3K826 | 60S acidic ribosomal protein L10 | 1395.34 | 2.41 | 0.88 | 0.16 | 1.00 |
| E3KKV5 | 60S ribosomal protein L14 | 1806.10 | 0.20 | -1.59 | 0.44 | 0.00 |
| E3KGR6 | 60S ribosomal protein L23 | 3152.83 | PpGuava | PpGuava | PpGuava | PpGuava |
| E3KZR5 | 60S ribosomal protein L9e | 687.84 | 1.37 | 0.32 | 0.29 | 0.98 |
| E3KVF5 | Acetyl CoA carboxylase biotin carboxylase | 465,35 | PpEucalyptus | PpEucalyptus | PpEucalyptus | PpEucalyptus |
| E3L3M4 | Acetyl CoA acetyltransferase | 5467.85 | 1.71 | 0.54 | 0.13 | 1.00 |
| E3KLM7 | Acetyl-CoA acyltransferase | 2220,71 | 1.62 | 0.48 | 0.15 | 1.00 |
| E3KL13 | Acetylglutamate kinase | 256.81 | 1.14 | 0.37 | 0.19 | 0.99 |
| E3K4I1 | Actin | 25906.00 | 1.16 | 0.15 | 0.05 | 1.00 |
| E3JQQ0 | Acyl coenzymeA oxidase | 384.27 | 1.46 | 0.38 | 0.15 | 1.00 |
| E3JUV4 | Acyl-CoA dehydrogenase | 259.56 | 1.80 | 0.59 | 0.22 | 1.00 |
| E3JPY7 | ADP ribosylation factor | 1302.03 | 1.39 | 0.33 | 0.23 | 1.00 |
| E3KWD3 | ADP.ATP carrier protein | 6064.67 | 1.27 | 0.24 | 0.09 | 1.00 |
| E3KIY1 | AGC/AKT protein kinase | 392.41 | 1.36 | 0.31 | 0.24 | 0.99 |
| E3JT53 | Argininosuccinate synthase | 2357.95 | 1.30 | 0.26 | 0.15 | 1.00 |
| E3L109 | ASF1 like histone chaperone | 521.43 | PpEucalyptus | PpEucalyptus | PpEucalyptus | PpEucalyptus |
| E3KQV7 | Aspartate aminotransferase * | 3436.00 | 1.45 | 0.37 | 0.11 | 1.00 |
| E3KQV8 | Aspartate aminotransferase * | 1152.40 | 1.62 | 0.48 | 0.18 | 1.00 |
| E3K898 | Aspartate tRNA ligase | 397.95 | 1.31 | 0.27 | 0.21 | 0.99 |
| E3KZB7 | ATP synthase subunit alpha | 15253.20 | 1.32 | 0.28 | 0.05 | 1.00 |
| E3K357 | ATP synthase subunit beta | 24197.30 | 1.28 | 0.25 | 0.06 | 1.00 |
| E3K775 | Autocrine motility factor receptor | 878.26 | 2.01 | 0.70 | 0.21 | 1.00 |
| E3KLJ3 | Calmodulin | 2518.71 | 1.45 | 0.37 | 0.19 | 0.99 |
| E3JYU7 | Capping protein (Actin filament) muscle Z-line. beta | 484.72 | PpGuava | PpGuava | PpGuava | PpGuava |
| E3KXV8 | Catalase | 1294.27 | 1.22 | 0.20 | 0.12 | 1.00 |
| E3KRP0 | cell division cycle protein 48 | 10137.00 | 1.25 | 0.22 | 0.06 | 1.00 |
| E3L2Y3 | Citrate synthase * | 3648.19 | 1.45 | 0.37 | 0.07 | 1.00 |
| E3KIG5 | Citrate synthase * | 287.31 | PpGuava | PpGuava | PpGuava | PpGuava |
| E3KQN1 | CK1/CK1/CK1-D protein kinase | 356.34 | PpGuava | PpGuava | PpGuava | PpGuava |
| E3K4L0 | Coronin | 335.26 | 1.28 | 0.25 | 0.17 | 1.00 |
| E3JW29 | Cytochrome c | 1794.11 | 1.17 | 0.16 | 0.12 | 1.00 |
| E3L8N2 | Cytochrome c oxidase subunit Va | 939.33 | 1.79 | 0.58 | 0.46 | 0.98 |
| E3KXK0 | Enolase | 13598.00 | 1.36 | 0.31 | 0.08 | 1.00 |
| E3JRG6 | Epsin 3 | 210.54 | PpEucalyptus | PpEucalyptus | PpEucalyptus | PpEucalyptus |
| E3JUT0 | Fructose-bisphosphate aldolase. class II | 2168.36 | 0.68 | -0.38 | 0.19 | 0.00 |
| E3K026 | GDP mannose 4 6 dehydratase | 230.21 | 1.48 | 0.39 | 0.22 | 1.00 |
| E3KEB5 | Glucose 1-dehydrogenase | 363.08 | PpEucalyptus | PpEucalyptus | PpEucalyptus | PpEucalyptus |
| E3K5I2 | Glucose 6 phosphate 1 dehydrogenase | 4756.44 | 1.25 | 0.22 | 0.21 | 0.99 |
| E3L278 | Glutathione reductase (NADPH) | 679.20 | 1.27 | 0.24 | 0.21 | 1.00 |
| E3L363 | Glycerol 3 phosphate dehydrogenase | 1383.98 | 1.43 | 0.36 | 0.12 | 1.00 |
| E3KA23 | Glycyl-tRNA synthetase | 443.03 | 1.16 | 0.15 | 0.14 | 0.99 |
| E3K2A8 | GTP-binding nuclear protein spi1 | 3836.23 | 1.35 | 0.30 | 0.09 | 1.00 |
| E3KXR7 | Guanine nucleotide-binding protein subunit beta-like protein | 9323.70 | 1.36 | 0.31 | 0.11 | 1.00 |
| E3K5H8 | Heat shock 70kDa protein 4 | 1721.81 | 1.75 | 0.56 | 0.09 | 1.00 |
| E3KZR1 | Heat shock protein 60 | 7677.27 | 1.54 | 0.43 | 0.06 | 1.00 |
| E3JVS0 | Heat shock protein 83 | 16505.40 | 1.17 | 0.16 | 0.04 | 1.00 |
| E3KYT3 | Heat shock protein HSS1 * | 15718.00 | 1.16 | 0.15 | 0.05 | 1.00 |
| Q01877 | Heat shock protein HSS1 * | 11951.00 | PpGuava | PpGuava | PpGuava | PpGuava |
| E3K643 | Heat shock protein SSB | 12675.20 | 1.27 | 0.24 | 0.06 | 1.00 |
| E3K1Q1 | Heme-binding peroxidase | 6757.64 | 1.73 | 0.55 | 0.14 | 1.00 |
| E3L1S2 | Histone H2A | 7513.68 | 0.92 | -0.08 | 0.08 | 0.03 |
| E3KEI6 | Histone H2B | 6731.25 | 0.63 | -0.47 | 0.14 | 0.00 |
| E3JQ71 | Histone-binding protein RBBP4 | 790.83 | PpGuava | PpGuava | PpGuava | PpGuava |
| E3KZR0 | Hsp10 like protein | 1463.97 | 1.45 | 0.37 | 0.15 | 1.00 |
| E3JT24 | Hsp70-like protein | 2010.73 | 1.45 | 0.37 | 0.10 | 1.00 |
| E3JX32 | Hydroxymethylglutaryl CoA synthase | 271.48 | 1.42 | 0.35 | 0.20 | 1.00 |
| E3KHG9 | Inorganic pyrophosphatase | 8350.30 | 1.22 | 0.20 | 0.09 | 1.00 |
| E3K023 | Isocitrate lyase | 821.33 | 1.19 | 0.17 | 0.10 | 1.00 |
| E3K387 | Ketol-acid reductoisomerase, mitochondria | 3191.20 | 1.57 | 0.45 | 0.11 | 1.00 |
| E3L7N3 | Long chain fatty acid CoA ligase 1 | 269.38 | PpEucalyptus | PpEucalyptus | PpEucalyptus | PpEucalyptus |
| E3K352 | Malate dehydrogenase * | 2264.81 | 1.82 | 0.60 | 0.17 | 1.00 |
| E3L321 | Malate dehydrogenase * | 4555.28 | 1.82 | 0.60 | 0.10 | 1.00 |
| E3KNM2 | Mannose 1 phosphate guanyltransferase | 2499.70 | 1.39 | 0.33 | 0.13 | 1.00 |
| E3JQQ5 | Minichromosome maintenance protein 3 | 315.62 | 1.43 | 0.36 | 0.16 | 1.00 |
| E3JQ11 | Mitochondrial processing peptidase subunit beta | 597.65 | 1.55 | 0.44 | 0.17 | 1.00 |
| E3KGF0 | Myo-inositol-1-phosphate synthase | 290.58 | 1.43 | 0.36 | 0.27 | 0.99 |
| E3K8H6 | NADH dehydrogenase (Ubiquinone) Fe-S protein 3 | 272.99 | 1.68 | 0.52 | 0.33 | 1.00 |
| E3LB27 | NADH dehydrogenase flavoprotein 2 | 1752.80 | 3.49 | 1.25 | 0.39 | 1.00 |
| E3LBN8 | Peptidyl-prolyl cis-trans isomerase | 416.71 | PpEucalyptus | PpEucalyptus | PpEucalyptus | PpEucalyptus |
| E3JVD3 | Phospho 2 dehydro 3 deoxyheptonate aldolase | 553.04 | PpGuava | PpGuava | PpGuava | PpGuava |
| E3L907 | Phosphoenolpyruvate carboxykinase | 5909.17 | 1.73 | 0.55 | 0.11 | 1.00 |
| E3K5I5 | Phosphoglucomutase | 1874.05 | 1.40 | 0.34 | 0.12 | 1.00 |
| E3KDN8 | Phosphomannomutase | 2918.24 | 1.63 | 0.49 | 0.13 | 1.00 |
| E3KGF8 | Prohibitin 1 | 1398.25 | 1.84 | 0.61 | 0.16 | 1.00 |
| E3K554 | Proteasome subunit alpha type | 2947.92 | 1.43 | 0.36 | 0.20 | 1.00 |
| E3KIE7 | Proteasome subunit beta type | 386.94 | PpEucalyptus | PpEucalyptus | PpEucalyptus | PpEucalyptus |
| E3JXB4 | Protein transporter SEC23 | 218.15 | 1.73 | 0.55 | 0.29 | 1.00 |
| E3KY42 | Pyridoxine biosynthesis protein | 3497.74 | 1.22 | 0.20 | 0.12 | 1.00 |
| E3KTX9 | Pyruvate carboxylase | 1507.30 | 1.12 | 0.11 | 0.07 | 1.00 |
| E3KSI6 | Pyruvate dehydrogenase E1 component subunit alpha | 211.05 | 1.43 | 0.36 | 0.17 | 1.00 |
| E3KF45 | Rab family protein | 413.58 | 1.70 | 0.53 | 0.32 | 1.00 |
| E3KHK5 | Ribose 5 phosphate isomerase | 205.02 | 1.54 | 0.43 | 0.18 | 1.00 |
| E3KNA3 | S25 ribosomal protein | 1314.31 | PpEucalyptus | PpEucalyptus | PpEucalyptus | PpEucalyptus |
| E3KNU8 | S-adenosylmethionine synthase | 1648.47 | 1.67 | 0.51 | 0.17 | 1.00 |
| E3JXY4 | Serine/threonine-protein phosphatase PP1 | 187.58 | 1.63 | 0.49 | 0.41 | 1.00 |
| E3JS02 | Small COPII coat GTPase | 1604.51 | 1.57 | 0.45 | 0.25 | 1.00 |
| E3KYA0 | Small nuclear ribonucleoprotein D3 | 852.35 | PpEucalyptus | PpEucalyptus | PpEucalyptus | PpEucalyptus |
| E3JQK5 | Small nuclear ribonucleoprotein E | 2077.40 | PpGuava | PpGuava | PpGuava | PpGuava |
| E3K8I4 | Stress induced phosphoprotein 1 | 532.99 | 1.36 | 0.31 | 0.26 | 0.99 |
| E3KK64 | Succinate dehydrogenase flavoprotein subunit mitochondrial | 2231.15 | 1.49 | 0.40 | 0.14 | 1.00 |
| E3KMN8 | Thioredoxin reductase | 529.23 | 1.79 | 0.58 | 0.13 | 1.00 |
| E3JSQ1 | Transaldolase | 3002.86 | 1.30 | 0.26 | 0.13 | 1.00 |
| E3JPZ9 | Triosephosphate isomerase | 7959.44 | 1.32 | 0.28 | 0.09 | 1.00 |
| E3KK52 | Tryptophan synthase | 218.58 | 1.27 | 0.24 | 0.13 | 1.00 |
| E3JT05 | Tubulin alpha-1A chain | 5271.32 | 1.16 | 0.15 | 0.10 | 0.99 |
| E3KPL3 | Tubulin beta chain | 18116.00 | 1.45 | 0.37 | 0.04 | 1.00 |
| E3K479 | tubulin binding cofactor A | 689.77 | 1.51 | 0.41 | 0.21 | 1.00 |
| E3KA35 | U6 snRNA-associated Sm-like protein LSm3 | 1905.08 | 1.36 | 0.31 | 0.33 | 0.97 |
| E3KBL0 | Ubiquitin-activating enzyme E1 | 515.66 | 1.26 | 0.23 | 0.14 | 1.00 |
| E3KLK1 | Ubiquitin-conjugating enzyme E2 N | 1681.96 | 1.52 | 0.42 | 0.21 | 1.00 |
| E3K7I4 | UDP-glucose 4-epimerase | 358.18 | 0.77 | -0.26 | 0.22 | 0.03 |
| E3K0X4 | UDP-glucose 6-dehydrogenase | 3766.77 | 1.19 | 0.17 | 0.08 | 1.00 |
| E3L4X0 | UTP-glucose-1-phosphate uridylyltransferase | 2906.12 | 1.32 | 0.28 | 0.07 | 1.00 |
| E3L015 | Vacuolar type proton ATPase catalytic subunit A | 6515.19 | 1.20 | 0.18 | 0.08 | 1,00 |
a Access ID is the access identification of the protein in the Uniprot database (http://www.uniprot.org/ release Version 2014_08).
b The score of protein expression analysis
c and d Ratio and the Log (PpGuava/PpEucalyptus) of quantified proteins respectively
e Standard Deviation (SD) of the Log of PpGuava/PpEucalyptus ratio and fp-value of Log of PpGuava/PpEucalyptus ratio obtained from PLGS 2.5.1.
A p value ≤0.05 identifies proteins less abundant, whereas a p value ≥0.95 identifies proteins more abundant in PpGuava than in PpEucalyptus; p value identifies proteins that were unique to either the PpEucalyptus or PpGuava P. psidii uredospore populations isolated from eucalyptus and guava, respectively.
* Variant proteins.
The functional analysis showed a clear predominance of proteins related to stress response, such as heat shock proteins, catalases that were more abundant in PpGuava than in PpEucalyptus. Proteins related to fungal pathogenesis such as, citrate synthase and enolase, among other, were also unique or increased in PpGuava population. Additionally, fatty acid synthase was unique in the PpEucalyptus population.
Some proteins were found to have one or more variants, with different abundance levels. Our results suggest that there are a variety of protein homologs in the uredospores; for example, there were matches for 4 different malate dehydrogenase proteins and 2 matches for pyruvate carboxylase. Moreover, 167 proteins were described as uncharacterized proteins with different degrees of abundance (S1 Table).
Functional categorization of identified proteins
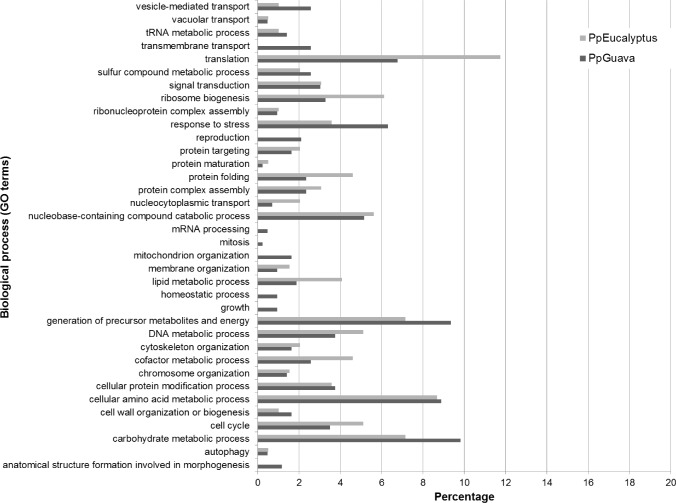
Proteins were categorized according to biological process using the Blast2GO software and major categories were present in both populations, such as cellular amino acid metabolic process, carbohydrate metabolism process, generation of precursor metabolites and energy, cellular protein modification process, signal transduction, vacuolar transport and others (Fig 4). However some GO terms, such as anatomical structure formation involved in morphogenesis, growth, mitosis, reproduction, transmembrane transport, mRNA processing, homeostatic process, mitochondrion organization, were only represented by the unique or more abundant from the PpGuava than PpEucalyptus population.
Fig 4. Gene ontology of biological process terms in the proteomic analysis.
Bar graph represents the ratio of % composition of term in the proteomic data.
This information and more details about the proteins as the Enzyme Code, Signal P and the presence of transmembrane helice domain (TMHMM), as well as proteins scores and peptide sequence are listed in S2 Table.
Discussion
P. psidii s.l. is unusual rust in that it exhibits a wide host range and is known to infect over 33 plant genera and 129 species, primarily in the Myrtaceae [37, 38, 39]. Formerly, P. psidii s.l. is conspecific and has been known as guava or eucalyptus rust [4, 40, 41, 42] due to the importance of these two hosts to the pathogen.
Despite the rust symptoms in the field being more pronounced in guava, cross-infection using PpGuava uredospores did not result in visible rust symptoms on E. grandis. However, light microscopy analysis of the PpEucalyptus and PpGuava uredospores showed the ability of both to produce germ tubes under in vitro conditions and probably having the potential to infect the host. Differences in the pathogenesis of P. psidii population’s were first reported by Maclachlan [43]. Since then, several cross-infection studies involving several hosts and P.psidii, collected from different plants of the Myrtle family, have shown clear differences in the pathogenicity of the organism [4, 13, 14, 44, 45, 46]. Aparecido et al. [14] inoculated five different hosts (Corymbia citriodora, P. guajava, S. jambos, Eugenia sp., and Eugenia uvalha) with uredospores collected from eight different host species.From this experiment, the authors described four distinct pathogenic groups. In our study, the PpGuava did not infected E. grandis, or there were no visible symptoms. It suggests the existence of a host-specific interaction, as a natural physiological variability among the uredospore populations. The differences observed in their protein profiles may be explained by the natural genetic variability within the investigated fungal populations. Our data is corroborated by recent molecular analysis of P. psidii populations that clearly proved that there is a correlation between the genetic variability of the P. psidii populations and the host from which they were collected [6].
Although rust is an economically important disease in eucalyptus and in other myrtle cultures, very little is known about the changes in the proteome of different populations of P. psidii s.l. Moreover, the monitoring of the changes in the uredospores protein profile, according to original host, is a new perspective to better understanding the plant-microbe interactions.
We evaluated two populations of the same fungal species, and observed a large number of proteins whose levels changed between the samples, as well as the presence of unique proteins in both samples. We opted to perform fungal population protein analysis based on the fact that the P. psidii has a biotrophic life style. There is no report of its growth and multiplication on synthetic media [6]. Being unable to grow outside of the host, it is difficult to characterize the proteome from other fungal structure. Therefore, it is laborious to produce spores using susceptible plants for growth and purification of the isolates. During this process, artificial selection may occur potentially producing a more aggressive pathogen depending on the host.
Some studies, which used different methods and equipments, found larger number of proteins than the 340 described for P. psidii. For instance, Bindschedler et al. [47] found 441 proteins from ungerminated spore of Blumeria graminis f. sp. Hordei. Cooper et al. [48] classified approximately 400 proteins from asexual uredospores of Uromyces appendiculatus and Liang et al. [49] observed the presence of 719 proteins from the three stages of sclerotia formation of Sclerotinia sclerotiorum. The lack of a P. psidii database was a strong challenge in the proteomic studies. The protein spectrums were matched with those from Puccinia genus from Uniprot. P. psidii has a non-complete genome sequenced. There are just few proteins sequences from this organism [50]. Many of these proteins are beta tubulins and transcription factors, which points a difficulty for protein identification of P. psidii uredospores. Even though, apart from the difficulties, we could have a good view of the protein roles in the uredospores between two contrasting P. psidii populations.
Using uredospores from populations that have been previously described as different biotypes [13], we identified proteins that are potentially related to the degree of variability and aggressiveness of the P. psidii allowing us to make some inferences regarding fungal diversity. A larger number of unique and/or abundant proteins were found in the PpGuava uredospores; this is most likely to be related to the higher genetic diversity within PpGuava, resulting in a broader range of proteins produced by this population, as well as a larger number of possible isoforms, which may be justified by the probable higher number of alleles in this population. These data are in agreement with a recent study that compared rust populations from eucalyptus and other myrtle species in Brazil and revealed a higher genetic diversity in the guava populations [6].The authors suggested that the pathogen recently migrated from a native South American host. When the fungus moved to other plant species, such as the exotic Eucalyptus spp., a new selection process was introduced by the new host [6]. Graça et al. [39] did not prove that P. psidii jumped from guava to eucalyptus host, but the existence of host-associated biotypes of P. psidii in Brazil, indicates that this diversity must be considered. Morin et al. [51] investigated the host-range of the P. psidii s.l. present in Australia. The authors concluded that the development of the disease in one host species in a tribe did not mean that a related species or genus also developed the disease. Moreover, they demonstrated the inability of the Australian rust accession to develop uredinia on P. guajava compared to the accession from Florida [52], supporting the hypothesis of physiological diversity between these populations.
Interestingly, during the uredospore phase, we identified many proteins related to fungal pathogenicity, such as peptidases, proteases, and proteins that are able to modify host factors [53, 54, 55, 56, 57], that were unique or more abundant only in the PpGuava than in PpEucalyptus population, for instance, proteassomes. Proteasomes are multi-subunit and multicatalytic proteolytic complexes that are found in the cells of most organisms, and these complexes play important roles in protein turnover in both the cytosol and the nucleus. They selectively degrade intracellular proteins in eukaryotic cells, particularly via the ubiquitin-proteasome pathway [58], and are involved in multiple specific functions, such as cell cycle regulation, cell signaling, the selective elimination of abnormal proteins, and the flux of substrates through metabolic pathways [59, 60]. As previously suggested by Kim et al.[61], the wax coated hydrophobic surfaces, which naturally occur on the host surface, may activate the 20S proteasome to mobilize the storage proteins that are present in uredospores. Chaperones are responsible for the initial folding and maintenance of proteins structure compared to the conventional heat shock proteins (HSPs), which are present only under conditions of high temperature or stress. Some studies suggested that these proteins also act to prevent protein aggregation, thus controlling protein turnover, which will increase its longevity and virulence [62]. Hodgetts et al. [63] observed that the up-regulation of HSP90 in Saccharomyces cerevisiae resulted in an increase of virulence in infected mice. We identified many HSP classes, some with peptide signals for secretion, similar to the data reported by Song et al [30]. The authors found high level of HSP in P. triticina haustorium. All identified HSPs were less abundant in the PpEucalyptus than in PpGuava population, most likely because the uredospores from the guava fruit are under more stressfull conditions [47] than those on PpEucalyptus, suggesting a differential response to the host organism. Larger amounts of HSPs were also identified in the Uromyces appendiculatus uredospores, which cause bean rust [48]. More investigation into the expression of HSPs is needed to better understand the roles of these proteins in the infection process during the P. psidii s.l. life cycle.
Tubulin and actin proteins were also increased in abundance in PpGuava. Similarly, Mandelc et al. [32] also observed that these proteins were increased in abundance in the lethal phytotype of V. albo-atrum. Tubulins and actins are the major proteins involved in the formation of the microtubules and microfilaments. GTPases regulates cellular polarization and motility, which affects the cytoskeleton, and are involved in the regulation of the secretion process and endocytosis [64]. The enolases identified in this study were also higher in abundance in the PpGuava population. Enolases participate in other cellular activities and can even act as HSPs; they can bind to cytoskeletal and chromatin structures to modulate transcription, thus playing a crucial role in pathophysiological processes [65, 66]. Enolase proteins increased it abundance in a virulent isolate of Pyrenophora tritici-repentis, the causal agent of tan spot in wheat [67]. Another example is the increased abundance of malate dehydrogenase in PpGuava. This protein is related to fungal virulence, the secretion of this compound acidifies the environment, signaling the expression and secretion of virulence factors and phytotoxins [17, 32].
Another interesting protein class, not previously related to fungal virulence, is prohibitin. This protein, with homologues in animals, fungi, plants, and unicellular eukaryotes was first reported as a negative regulator of cell proliferation. However, prohibitin is expressed as two transcripts with varying lengths of the 3' untranslated region. The longer one is present at higher levels in proliferating cells, suggesting that this longer 3' untranslated region sequence may function as a trans-acting regulatory RNA [68, 69].
Acetyl CoA carboxylase biotin carboxylase is abiotin-dependent enzyme involved in the biosynthesis, as well as, in the metabolism of fatty acids [70, 71]. Fatty acid synthase catalyzes the formation of long-chain fatty acids from acetyl-CoA, malonyl-CoA and NADPH [72]. Both were found as PpEucalyptus unique proteins., maybe in PpGuava these proteins may be present in lower abundance.
Our data also support the model proposed for U. appendiculatus by Cooper et al. [48], which suggests that the uredospore exists in a suspended translational state. Uredospores can produce proteins very quickly without having to assemble de novo translational components. Moreover, the peptide chains dispersed in the ribosome are protected by heat shock proteins and can resist adverse conformational changes caused by desiccation or high temperatures that the uredospores might be subjected to, once released into the environment [48].Corroborating this hypothesis, we identified a large number of ribosomal proteins subunits with different level of abundance, which may be related to the beginning of the infection process by the pathogen.
Additionally, categorization by protein function showed increased abundance of proteins related to translation, ribosome biogenesis and protein folding from the PpEucalyptus sample. Response to stress, generation of precursor metabolites and energy and carbohydrate metabolic process were more abundant in PpGuava. The proteins involved in signal transduction were present in similar proportion in both populations.
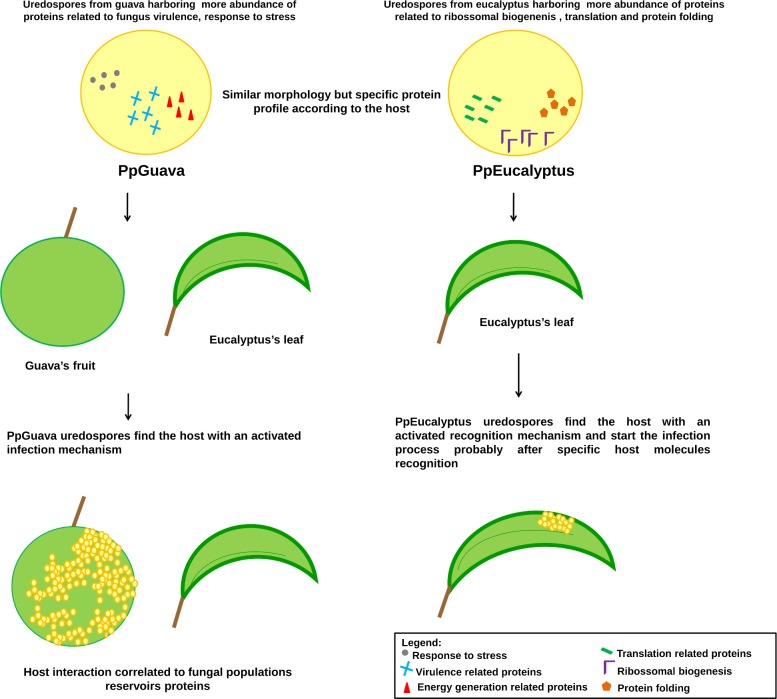
In turn, uredospores populations have specific reservatory of proteins according to the host from supporting the physiological variability of these two populations. The virulent proteins from PpEucalyptus were less abundant when compared to PpGuava. In guava, close to 100% of the fruits can be lost due to severe infections most likely because the uredospores reservatory of proteins of this population; related with virulence and stress response. Likely, PpEucalyptus uredospores need to process of proteins of virulence to start the host infection process, its reservatory is composed by biogenesis, protein folding and translation (Fig 5). It is commonly observed that uredopores from a specific host are less aggressive when infecting a host from other species. The aggressiveness increases after successive cycles of infection in the new host. One hypothesis is the modulation of the expression of some genes according to the host, resulting in different protein reservoirs in each population.
Fig 5. Protein profiles in PpGuava and PpEucalyptus and the correlation with their physiological variability.
While proteins correlated to fungal virulence and stress response had the abundance increased in PpGuava, proteins related to biogenesis, protein folding and translation had the abundance increased in PpEucalyptus.
The proteomic information generated in the present work contributes to a better understanding of the mechanisms underlying the pathogenicity of P. psidii s.l. and its interaction with its host organism. More comprehensive proteomic analyses of the P. psidii-host interaction, combined with other studies, will be important for elucidating the molecular basis of the interactions in this intriguing pathosystem.
Supporting Information
Access ID is the access identification of the protein in Uniprot database (http://www.uniprot.org/ release Version 2015_02). The score of protein expression analysis, ratio and Log (PpGuava/PpEucalyptus) of quantified proteins, standard deviation (SD) of the PpGuava/PpEucalyptus ratio log and their p-values of the Log of the PpGuava/PpEucalyptus ratio obtained from PLGS 2.5.1. A p value ≤ 0.05 identifies proteins whose abundance decrease, whereas a p value ≥ 0.95 identifies proteins whose abundance increased in PpGuava; p value identifies proteins that were unique to either the PpEucalyptus or PpGuava P. psidii uredospore populations isolated from eucalyptus and guava, respectively (XLS).
(XLSX)
Organism source, GO biological process, Enzyme Code, Signal P, Trans-membrane helice domain (TMHMM), as well as protein score, average amount (fmol) of protein, average score of proteins and peptide sequences and average amount of matched peptides to each protein.
(XLSX)
Acknowledgments
We thank Dr. Esteban R. Gonzalez (FuturaGene Corporation) for providing the uredospores from P. psidii populations.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by FAPESP (Grant 2008/50361-1). The authors would also like to thank FAPESP for the fellowships to MCQ (Proc. no. 2010/50445-0), APB (2013/07596-6), FGM (2011/17455-5), IGFB (2012/22227-4) and SGG (2010/15417-6).
References
- 1.Winter G. Repertorium. Rubenhorstii fungi europaei et extraeuropaei.Cent XXXI et XXXII. Hedwigin. 1884; 23, 164–172. [Google Scholar]
- 2.Ferrari JT, Nogueira EMC, Santos AJT. Control of rust (Puccinia psidii) in guava (Psidium guajava). Acta Hort. 1997; 452: 55–57. [Google Scholar]
- 3.Gonçalves S. Preliminary list of plant diseases in Espirito Santo State Rio de Janeiro, Ministério da Agricultura, 1929; 1–12. [Google Scholar]
- 4.Ferreira FA. Ferrugem do Eucalipto. Revista Árvore. 1983; 7: 91–109. [Google Scholar]
- 5.Glen M, Alfenas AC, Zauza EAV, Wingfield MJ, Mohammed C. Puccinia psidii: A threat to the Australian environment and economy–A review. Australas Plant Path. 2007; 36: 1–16. [Google Scholar]
- 6.Quecine MC, Bini AP , Romagnoli EM, Andreote FD, Moon DH, Labate CA. Genetic variability of Puccinia psidii populations revealed by PCR-DGGE and T-RFLP markers. Plant Dis. 2014; 98: 16–23. [DOI] [PubMed] [Google Scholar]
- 7.Simpson JA, Thomas K, Grgurinovic CA. Uredinales species pathogenic on species of Myrtaceae. Australas Plant Pathol. 2006; 35: 549–562. [Google Scholar]
- 8.Maier W, Begerow D, Weis M, Oberwinkler F. Phylogeny of the rust fungi: an approach using the nuclear large subunit ribosomal DNA sequences. Can J Bot. 2003; 81: 12–23. [Google Scholar]
- 9.Walker J. Pacific mycogeography: deficiencies and irregularities in the distribution of plant parasitic fungi. Aust J Bot. 1983; 10: 89–136. [Google Scholar]
- 10.Roux J, Greyling I, Coutinho TA, Verleur M, Wingfield MJ. The myrtle rust pathogen, Puccinia psidii, discovered in Africa. IMA Fungus. 2013; 4: 155–159. 10.5598/imafungus.2013.04.01.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueiredo MB. Life cycle and ecology of Puccinia psidii. O Biol. 2001; 63: 69–71. [Google Scholar]
- 12.Wiethölter N, Horn S, Reisige K, Beike U, Moerschbacher BM. In vitro differentiation of haustorial mother cells of the wheat stem rust fungus, Puccinia graminis f. sp. tritici, triggered by the synergistic action of chemical and physical signals.Fungal Genet Biol. 2003; 38: 320–326. [DOI] [PubMed] [Google Scholar]
- 13.Coelho L, Alfenas AC, Ferreira FA. Variabilidade fisiológica de Puccinia psidii—Ferrugem do Eucalipto. Summa Phytopathol. 2001; 27: 295–300. [Google Scholar]
- 14.Aparecido CC, Figueiredo MB, Furtado EL. Groups of physiological variability in Puccinia psidii populations. Summa Phytopathol. 2003; 29: 234–238. [Google Scholar]
- 15.Gokce E, Franck WL, Oh Y, Dean RA, Muddiman DC. In-Depth analysis of the Magnaporthe oryzae conidial proteome. J. Proteome Res. 2012; 11: 5827−5835. 10.1021/pr300604s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y, Nandakumar MP, Marten MR. Proteomics of filamentous fungi. Trends Biotechnol. 2007; 25: 395–400. [DOI] [PubMed] [Google Scholar]
- 17.Doyle S. Fungal proteomics: from identification to function. FEMS Microbiol Lett. 2011; 321: 1–9. 10.1111/j.1574-6968.2011.02292.x [DOI] [PubMed] [Google Scholar]
- 18.Ebstrup T, Saalbach G, Egsgaard HA. Proteomics study of In vitro germination and appressoria formation in Phytophthora infestans. Proteomics. 2005; 5: 2839–2848. [DOI] [PubMed] [Google Scholar]
- 19.Fernández-Acero FJ, Jorge I, Calvo E, Vallejo I, Carbú M, Camafeita E, et al. Proteomic analysis of phytopathogenic fungus Botrytis cinerea as a potential tool for identifying pathogenicity factors, therapeutic targets and for basic research. Arch Microbiol. 2007; 187: 207–215. [DOI] [PubMed] [Google Scholar]
- 20.Yajima W, Kav NNV. The proteome of the phytopathogenic fungus Sclerotinia sclerotiorum. Proteomics. 2006; 6: 5995–6007. [DOI] [PubMed] [Google Scholar]
- 21.Paper JM, Scott-Craig JS, Adhikari ND, Cuomo CA, Walton JD. Comparative proteomics of extracellular proteins in vitro and in planta from the pathogenic fungus Fusarium graminearum. Proteomics. 2007; 7: 3171–3183. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt S, Prokisch H, Schlunck T, Camp DG, Ahting U, Waizenegger T, et al. Proteome analysis of mitochondrial outer membrane from Neurospora crassa. Proteomics. 2006; 6: 72–80. [DOI] [PubMed] [Google Scholar]
- 23.Kniemeyer O, Lessing F, Scheibner O, Hertweck C, Brakhage AA. Optimisation of a 2-D gel electrophoresis protocol for the human-pathogenic fungus Aspergillus fumigatus. Curr Genet. 2006; 49: 178–189. [DOI] [PubMed] [Google Scholar]
- 24.Melin P, Schnürer J, Wagner EG. Proteome analysis of Aspergillus nidulans reveals proteins associated with the response to the antibiotic concanamycin a, produced by Streptomyces species. Mol. Genet. Genomics. 2002; 267: 695–702. [DOI] [PubMed] [Google Scholar]
- 25.Carberry S, Neville CM, Kavanagh KA, Doyle S. Analysis of major intracellular proteins of Aspergillus fumigatus by MALDI mass spectrometry: identification and characterization of an elongation factor 1b protein with glutathione transferase activity. Biochem Biophys Res Commun. 2006; 341: 1096–1104. [DOI] [PubMed] [Google Scholar]
- 26.Oda K, Kakizono D, Yamada O, Iefuji H, Akita O, Iwashita K. proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 2006; 72: 3448–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin G, Tian S, Chan Z, Li B. Crucial role of antioxidant proteins and hydrolytic enzymes in pathogenicity of Penicillium expansum: analysis based on proteomic approach. Mol. Cell. Proteomics. 2007; 6: 425–438. [DOI] [PubMed] [Google Scholar]
- 28.Zorn H, Peters T, Nimtz M, Berger RG. The secretome of Pleurotus sapidus. Proteomics. 2005; 5: 4832–4838. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu M, Wariishi H. Development of a sample preparation method for fungal proteomics. FEMS Microbiol Lett. 2005; 247: 17–22. [DOI] [PubMed] [Google Scholar]
- 30.Song X, Rampitsch C, Soltani B, Mauthe W, Linning R, Banks T, et al. Proteome analysis of wheat leaf rust fungus, Puccinia triticina, infection structures enriched for haustoria. Proteomics. 2011; 11: 944–963. 10.1002/pmic.201000014 [DOI] [PubMed] [Google Scholar]
- 31.Godfrey D, Zhang Z, Saalbach G, Thordal-Christensen H. A proteomics study of barley powdery mildew haustoria. Proteomics. 2009; 9: 3222–3232. 10.1002/pmic.200800645 [DOI] [PubMed] [Google Scholar]
- 32.Mandelc S, Radisek S, Jamnik P, Javornik B. Comparison of mycelial proteomes of two Verticillium albo-atrum pathotypes from hop. Eur J Plant Pathol. 2009;125: 159–171. [Google Scholar]
- 33.Damerval C, Vienne D, Zivy M Thiellement H. Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling protein. Electrophoresis. 1986; 7: 53–54. [Google Scholar]
- 34.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006; 5: 144–156. [DOI] [PubMed] [Google Scholar]
- 35.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. the geneontology consortium. Nat Genet. 2000; 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005; 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 37.Carnegie AJ, Lidbetter JR. Rapidly expanding host range for Puccinia psidii sensu lato in Australia. Australas Plant Pathol. 2012; 41: 13–29. [Google Scholar]
- 38.Tommerup IC, Alfenas AC, Old KM. Guava rust in Brazil–a threat to Eucalyptus and other Myrtaceae. New Zealand J Forest Sci. 2003; 33: 420–428. [Google Scholar]
- 39.Graça RN, Ross-Davis AL, Klopfenstein NB, Kim MS, Peever TL, Cannon PG, et al. Rust disease of eucalypts, caused by Puccinia psidii, did not originate via host jump from guava in Brazil. Mol Ecol. 2013; 22: 6033–6047. 10.1111/mec.12545 [DOI] [PubMed] [Google Scholar]
- 40.Joffily J. Ferrugem do eucalipto. Bragantia. 1944; 4: 475–487. [Google Scholar]
- 41.Coutinho TA, Wingfield MJ, Alfenas AC, Crous PW. Eucalyptus rust: A disease with the potential for serious international implications. Plant Dis. 1998; 82: 819–825. [DOI] [PubMed] [Google Scholar]
- 42.Kriticos DJ, Morin L, Leriche A, Anderson RC, Caley P. Combining a climatic niche model of an invasive fungus with its host species distributions to identify risks to natural as sets: Puccinia psidii Sensu Lato in Australia. PLoS One. 2013; 8:e64479 10.1371/journal.pone.0064479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maclachlan JD. A Rust of the pimento tree in jamaica, BWI. Phytopathol. 1938; 28: 157–170. [Google Scholar]
- 44.Marlatt RB, Kimbrough JW. Rust (Puccinia psidii) of Allspice (Pimenta dioica) Appears in Florida. P Fl St Hort Soc. 1980; 93: 111. [Google Scholar]
- 45.Castro HA, Krugner TL, Idehira CHF, Cappello MSC, Marchi AB. Inoculação cruzada de eucalyptus, goiaba (Psidium Guajava) e jambeiro (Sizygium Jambos) com Puccinia psidii. Fitopatol Bras. 1983; 8: 491–497. [Google Scholar]
- 46.Castro HA, Bergamim FA, Krugner TL. Padrão de produção de uredosporos em mudas de eucalyptus spp inoculadas artificialmente com Puccinia psidii. Summa Phytopathol. 1984; 10: 155–170. [Google Scholar]
- 47.Bindschedler LV, Burgis TA, Mills DJ, Ho JTC, Cramer R, Spanus PD. In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei. Mol Cell Proteomics. 2009; 8: 2368–2381. 10.1074/mcp.M900188-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooper B, Garrett WM, Campbell KB. Shotgun identification of proteins from uredospores of the bean rust Uromyces appendiculatus. Proteomics. 2006; 6: 2477–2484. [DOI] [PubMed] [Google Scholar]
- 49.Liang Y, Rahman MH, Strelkov SE, Kav NNV. Developmentally induced changes in the sclerotial proteome of Sclerotinia sclerotiorum. Fungal Biol. 2010; 114: 619–627. 10.1016/j.funbio.2010.05.003 [DOI] [PubMed] [Google Scholar]
- 50.Tan MK, Collins D, Chen Z, Englezou A, Wilkins MR. A brief overview of the size and composition of the myrtle rust genome and its taxonomic status. Mycology. 2014; 5: 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morin L, Aveyard R, Lidbetter JR, Wilson PG. Investigating the host-range of the rust fungus Puccinia psidii sensu lato across tribes of the family myrtaceae present in Australia. PLoS One. 2012; 7:e35434 10.1371/journal.pone.0035434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rayachhetry MB, Van TK, Center TD, Elliott ML. Host range of Puccinia psidii, a potential biological control agent of Melaleuca quinquenervia in Florida. Biol Control. 2001; 22: 38–45. [Google Scholar]
- 53.Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. P Natl Acad Sci USA. 2005; 102: 6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia Y. Proteases in pathogenesis and plant defence. Cel. Microbiol. 2004; 6: 905–913. [DOI] [PubMed] [Google Scholar]
- 55.Roden J, Eardley L, Hotson A, Cao Y, Mudgett MB. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol Plant Microbe In. 2004; 17: 633–643. [DOI] [PubMed] [Google Scholar]
- 56.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. Embo J. 2000; 19: 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Debono M, Goedee RS. Antibiotics that inhibit fungal cell wall development. Annu Rev Microbiol. 1994; 48: 471–497. [DOI] [PubMed] [Google Scholar]
- 58.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998; 92: 367–380. [DOI] [PubMed] [Google Scholar]
- 59.Monaco JJ, Nandi D. The genetics of proteasomes and antigen processing. Annu Rev Genet. 1995; 29: 729–754. [DOI] [PubMed] [Google Scholar]
- 60.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996; 65: 801–847. [DOI] [PubMed] [Google Scholar]
- 61.Kim ST, Yu S, Kim SG, Kim HJ, Kang SY, Hwang DH, et al. Proteome analysis of rice blast fungus (Magnaporthe grisea) proteome during appressorium formation. Proteomics. 2004; 4: 3579–3587. [DOI] [PubMed] [Google Scholar]
- 62.Burnie JP, Carter TL, Hodgetts SJ, Matthews RC. Fungal heat-shock proteins in human disease. FEMS Microbiol Rev. 2006; 30: 53–88. [DOI] [PubMed] [Google Scholar]
- 63.Hodgetts S, Matthews R, Morrissey G, Mitsutake K, Piper P, Burnie J. Over-expression of Saccharomyces cerevisiae hsp90 enhances the virulence of this yeast in mice. FEMS Immunol Med Microbiol. 1996; 16: 229–234. [DOI] [PubMed] [Google Scholar]
- 64.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006; 16: 522–529. [DOI] [PubMed] [Google Scholar]
- 65.Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cell Mol Life Sci. 2001; 58: 902–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pancholi V, Chhatwal GS. Housekeeping enzymes as virulence factors for pathogens. Int J Med Microbiol. 2003; 293: 391–401. [DOI] [PubMed] [Google Scholar]
- 67.Cao T, Kim YM, Kav NN, Strelkov SE. A proteomic evaluation of Pyrenophora tritici-repentis, causal agent of tan spot of wheat, reveals major differences between virulent and avirulent isolates. Proteomics. 2009; 9: 1177–1196. 10.1002/pmic.200800475 [DOI] [PubMed] [Google Scholar]
- 68.Van Aken O, Pecenkova T, van de Cotte B, De Rycke R, Eeckhout D, Fromm H, et al. Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 2007; 52: 850–864. [DOI] [PubMed] [Google Scholar]
- 69.Mishra S, Murphy LC, Murphy LJ. The prohibitins: emerging roles in diverse functions. J Cell Mol Med. 2006; 10: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell. Mol. Life Sci. 2005; 62: 1784–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brownsey RW, Zhande R, Boone AN. Isoforms of acetyl-CoA carboxylase: structures, regulatory properties and metabolic functions. Biochem Soc Trans. 1997; 25: 1232–1238. [DOI] [PubMed] [Google Scholar]
- 72.Niwa H, Katayama E, Yanagida M, Morikawa K. Cloning of the fatty acid synthetase beta subunit from fission yeast, coexpression with the alpha subunit, and purification of the intact multifunctional enzyme complex. Protein Expr Purif.1998; 13:403–413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Access ID is the access identification of the protein in Uniprot database (http://www.uniprot.org/ release Version 2015_02). The score of protein expression analysis, ratio and Log (PpGuava/PpEucalyptus) of quantified proteins, standard deviation (SD) of the PpGuava/PpEucalyptus ratio log and their p-values of the Log of the PpGuava/PpEucalyptus ratio obtained from PLGS 2.5.1. A p value ≤ 0.05 identifies proteins whose abundance decrease, whereas a p value ≥ 0.95 identifies proteins whose abundance increased in PpGuava; p value identifies proteins that were unique to either the PpEucalyptus or PpGuava P. psidii uredospore populations isolated from eucalyptus and guava, respectively (XLS).
(XLSX)
Organism source, GO biological process, Enzyme Code, Signal P, Trans-membrane helice domain (TMHMM), as well as protein score, average amount (fmol) of protein, average score of proteins and peptide sequences and average amount of matched peptides to each protein.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.