Abstract
The production of high-affinity antibodies by B cells is essential for pathogen clearance. Antibody affinity for antigen is increased through the affinity maturation in germinal centers (GCs). This is an iterative process in which B cells cycle between proliferation coupled with the acquisition of mutations and antigen-based positive selection, resulting in retention of the highest-affinity B cell clones. The posttranscriptional regulator microRNA-155 (miR-155) is critical for efficient affinity maturation and the maintenance of the GCs; however, the cellular and molecular mechanism by which miR-155 regulates GC responses is not well understood. Here, we utilized a miR-155 reporter mouse strain and showed that miR-155 is coexpressed with the proto-oncogene encoding c-MYC in positively selected B cells. Functionally, miR-155 protected positively selected c-MYC+ B cells from apoptosis, allowing clonal expansion of this population, providing an explanation as to why Mir155 deletion impairs affinity maturation and promotes the premature collapse of GCs. We determined that miR-155 directly inhibits the Jumonji family member JARID2, which enhances B cell apoptosis when overexpressed, and thereby promotes GC B cell survival. Our findings also suggest that there is cooperation between c-MYC and miR-155 during the normal GC response, a cooperation that may explain how c-MYC and miR-155 can collaboratively function as oncogenes.
Introduction
Germinal centers (GCs) form in B cell follicles of secondary lymphoid organs upon extensive proliferation of antigen-activated B cells that respond to T cell help. They are essential for the production of plasma cells that secrete high-affinity antibodies and high-affinity memory B cells. Despite their importance for vaccine- and infection-induced protection (1, 2), there is limited understanding of the molecular program that leads to the selection of high-affinity B cell clones within the GC.
Affinity maturation is the result of somatic hypermutation (SHM) of the B cell receptor (BCR) genes during intensive B cell division in the dark zone (DZ) (3), followed by rounds of affinity-based selection in the light zone (LZ), where B cells are either positively selected or die (4). This selection process is considered to be dependent on the affinity of the newly mutated BCR. Positively selected GC B cells can migrate back to the DZ, where they proliferate and undergo further SHM. This bidirectional interzonal migration cycle was postulated in the cyclic reentry model (5–7), and it is believed to be essential for efficient affinity maturation (4). Ultimately, positively selected B cells differentiate into memory B cells or plasma cells and exit the GC.
At the molecular level, the master regulator of GCs, BCL6, is upregulated in DZ B cells and represses genes involved in cell cycle arrest, the DNA damage response, and plasma cell differentiation (8). This allows SHM to take place, which requires high expression of AID in DZ B cells (9). As DZ B cells migrate toward the LZ, BCL6 expression is downregulated and B cells become dependent on extrinsic signals arising from interactions with antigen, follicular DCs, and T cells. As a result of such signaling events, a fraction of LZ B cells is positively selected. Recent studies have shown that c-MYC is expressed in those positively selected LZ B cells and is a critical regulator in GC maintenance (10, 11).
Among the genes repressed by BCL6 is the microRNA-155 (miR-155) (8), a well-established regulator of activated B cells (8, 12–15). Despite the known role for miR-155 in regulating the GC response, the mechanisms by which it acts are only beginning to be understood. It has been suggested that BCL6, by inhibiting miR-155 in DZ B cells, positively regulates the expression of miR-155 target genes (8). However, it remains to be learned what cellular processes and molecular targets miR-155 regulates while it is expressed in GC B cells.
Here, we uncover a dynamic regulation of miR-155, which is expressed in a small subset of LZ B cells. The miR-155+ subset is enriched in cycling cells and coexpresses c-MYC, demonstrating that miR-155 expression is linked to positively selected B cells. Functionally, we observed that expression of miR-155 protects c-MYC+ LZ B cells from apoptosis and thus plays a critical role in the maintenance of the GC response and in affinity maturation. One of the molecular targets that miR-155 directly inhibits is JARID2, whose overexpression promotes apoptosis of LZ B cells. Overall, our results reveal a mechanism of affinity selection by functionally linking c-MYC and miR-155.
Results
miR-155 deficiency decreases the number of DZ and LZ B cells.
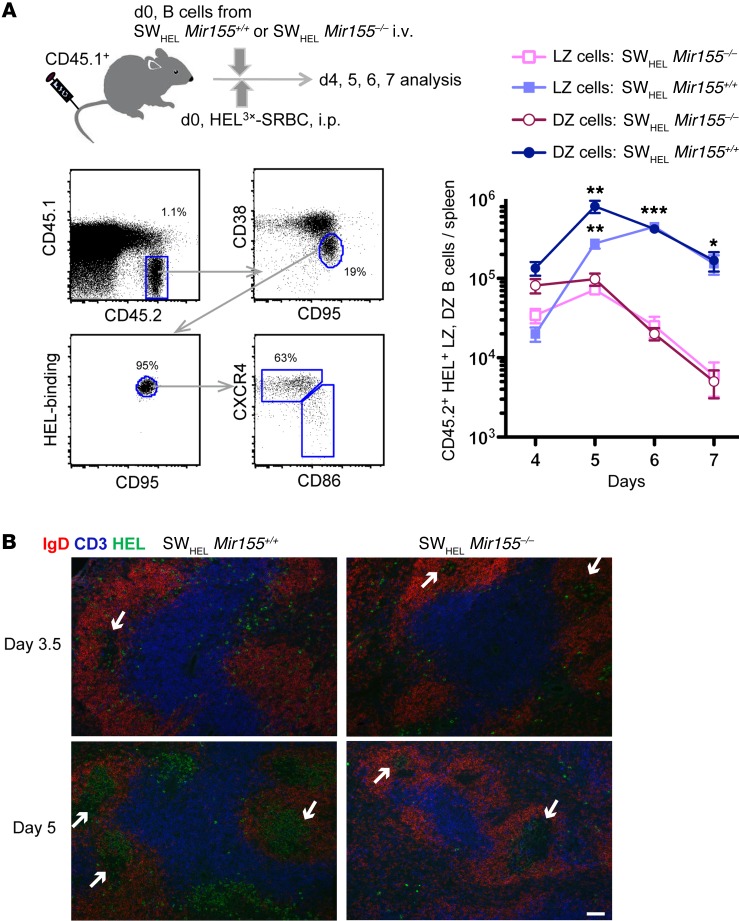
To further understand the defects in GC responses caused by miR-155 deficiency in a B cell–intrinsic manner, we utilized the SWHEL mouse model. SWHEL mice have the heavy and light chains of the HyHEL10 BCR that recognizes hen egg lysozyme (HEL) knocked in to the endogenous Bcr locus. This enables tracking of class-switch recombination and SHM of the transgenic BCR during the GC response (16). SWHEL Mir155+/+ or SWHEL Mir155–/– B cells were adoptively transferred into CD45.1+ congenic recipients, which were immunized with HEL3X, a HEL mutant with a Ka of approximately 1.5 × 106 M–1 affinity for HyHEL10 (17, 18), coupled to sheep red blood cells (SRBC). HEL+ GC B cells were gated as described in Figure 1A (left). We observed the peak of DZ B cells at day 5 and the peak of LZ B cells at day 6 (Figure 1A, right). Although the absence of miR-155 had no effect on the number of GC B cells until day 4, cell numbers were severely reduced: ~6-fold at day 5, ~19-fold at day 6, and ~28-fold at day 7 (Figure 1A, right). Immunohistological analysis confirmed these results and further revealed that B cell migration was not altered by miR-155 deficiency (Figure 1B). These results indicate that miR-155 is required to maintain the GC response.
Figure 1. miR-155 in B cells is required for the maintenance of the GC response.
SWHEL Mir155+/+ or SWHEL Mir155–/– donor B cells were adoptively transferred into CD45.1+ congenic recipient mice that were then injected with HEL3X-SRBC. Spleen cells from these mice were analyzed by flow cytometry at the indicated days. (A)The gating strategy and experimental design are shown (left). The FACS plot analyzing SWHEL Mir155+/+ donor cell at day 4 is shown as an example. The number of donor-derived (CD45.1–CD45.2+) HEL-specific GC B cells was calculated based on the proportion of cells stained as CD38loCD95+ and HEL-binders and further divided as CXCR4hiCD86lo for DZ B cells and CXCR4loCD86hi for LZ B cells (right). Data represent 1 out of 3 independent experiments. The mean ± SEM is shown (SWHEL Mir155+/+; d4, n = 6; d5, n = 4; d6, n=5; and d7, n = 4. SWHEL Mir155–/–; d4, n = 6; d5, n = 4; d6, n = 8; and d7, n = 4). Two-tailed unpaired t test, SWHEL Mir155+/+ LZ vs. SWHEL Mir155–/– LZ, at d5, d6, and d7. *P < 0.05, **P < 0.005, ***P < 0.0005, and the same for SWHEL Mir155+/+ DZ vs. SWHEL Mir155–/– DZ. (B) Splenic sections containing adoptively transferred B cells of the indicated genotype were stained for IgD in red, CD3 in blue, and HEL binding in green at d3.5 or d5. Arrows indicate GCs. Scale bar: 50 μm.
Cycling LZ B cells express high levels of miR-155.
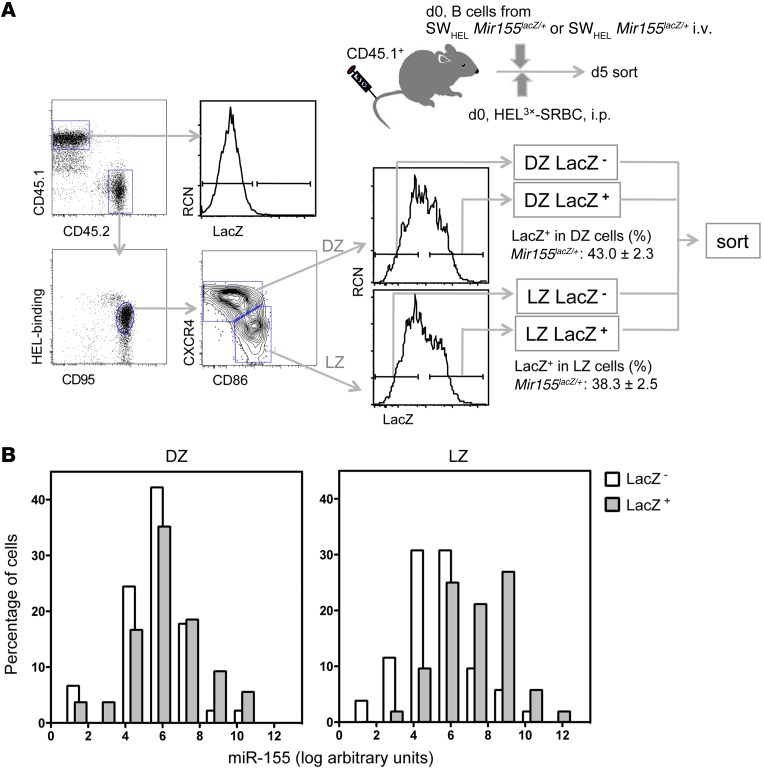
Having established that miR-155 is required for the maintenance of GC B cells in a B cell–intrinsic manner, we next assessed its expression pattern to understand where it exerts its function in the GC response. To identify miR-155 expressing cells, we used the miR-155–LacZ reporter strain, which replaces the Mir155 coding sequence with a LacZ cassette, producing a nonfunctional allele (13). The transcriptional activity of the Mir155 gene is monitored by detecting LacZ activity. If transcription is coupled to processing, LacZ activity may mirror mature miR-155 abundance. We tested whether LacZ expression was associated with miR-155 expression in single cell–sorted GC B cells. We adoptively transferred SWHEL Mir155LacZ/+ B cells into CD45.1+ congenic mice and injected them with HEL3X-SRBC. Five days later, we sorted GC B cells as DZ or LZ further divided by LacZ status, generating 4 subsets: DZ LacZ–, DZ LacZ+, LZ LacZ–, and LZ LacZ+ (Figure 2A). LacZ+ cells tend to express higher levels of miR-155 than LacZ– cells (Figure 2B), although the discrimination is much clear for LZ B cells. This is likely due to the lower level of miR-155 in DZ B cells than LZ B cells. These results indicate that miR-155 is mainly expressed at the highest levels in a fraction of LZ B cells.
Figure 2. A subset of LZ B cells highly expresses miR-155.
SWHEL Mir155LacZ/+ B cells were adoptively transferred into CD45.1+ congenic recipient mice, which were then injected with HEL3X-SRBC and analyzed after 5 days. (A) GC B cell gating strategy to define 4 GC subsets. First, DZ and LZ B cells were identified as explained in Figure 1 and then LacZ activity was visualized with fluorescein di-V-galactoside (FDG). The percentages of LacZ+ cells are shown (mean ± SEM of independent sorting and flow cytometric analyses [n = 16]). (B) The 4 subsets of GC B cells indicated in A were single cell sorted from 4 mice and analyzed for the expression of miR-155 by qPCR using U6 for normalization (DZ LacZ–, n = 45; DZ LacZ+, n = 54; DZ LacZ–, n = 52; DZ LacZ+, n = 52). Histograms show the distribution of cells on the basis of miR-155 expression in log-transformed arbitrary units. Bars are nudged to prevent overlapping. Background level was established by assessing the expression of miR-155 in Mir155–/– GC B cells, which was negligible (not shown). Also, miR-155 in naive B cells was very low (not shown).
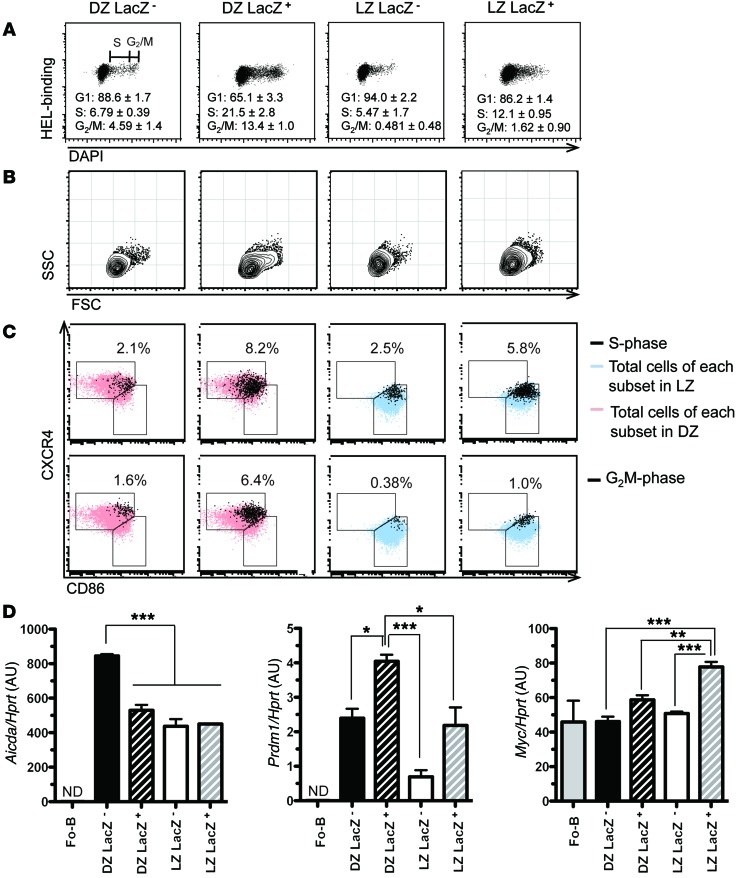
We next assessed the association between cells expressing high levels of miR-155 and proliferation. According to the cyclic reentry GC model, positively selected B cells start cell cycling in the LZ and then either differentiate into memory/plasma cells and exit GC or reenter the DZ and actively proliferate. Thus, the cell cycle status of miR-155+ LZ B cells may allow us to assess the role being played by miR-155. In agreement with published data (4), we found that the DZ is the major site for proliferation (Figure 3A). Hardly any LZ B cell were in G2/M-phase, but LZ LacZ+, which express the greatest amount of miR-155, had a higher proportion of S-phase cells (12.1% ± 0.95%) than LZ LacZ– cells (5.47% ± 1.7%); Figure 3A). In the DZ, LacZ positivity also defined a highly proliferative subset with a consistent increase in size (Figure 3, A and B). Interestingly, S and G2/M cells were clustered at the border of the DZ and LZ regions delineated in the FACS plots on the basis of CXCR4 and CD86 (Figure 3C). Altogether, these results indicate that GC B cells are a more heterogeneous population than originally thought.
Figure 3. miR-155 expression is associated with cell cycle progression of GC B cells.
SWHEL Mir155LacZ/+ B cells were adoptively transferred into CD45.1+ congenic recipient mice, which were then injected with HEL3X-SRBC and analyzed after 5 days. LacZ reporter staining is incompatible with permeabilization and, therefore, cells were sorted first using the strategy explained in Figure 2A. Subsequently, cell cycle was analyzed in the 4 subsets of GC B cells by DAPI staining. The mean of the indicated fraction ± SEM from 3 independent experiments is shown. (A) HEL-binding vs. DAPI. (B) Forward scatter (FSC) vs. side scatter (SSC). (C) S-phase and G2/M-phase B cells for each of the sorted subsets were gated as shown in A. The upper panel shows the CXCR4/CD86 distribution of cells in S-phase cells (black dots) superimposed on DZ cells (pink) or LZ cells (light blue) of the indicated subset. The lower panel is similar to the upper panel, except that G2/M-phase cells (black) are shown. The percentage of black dots over total cell per gate is shown. A representative FACS plot of the indicated subset out of 3 independent experiments is shown (A–C). (D) The indicated subsets were sorted at day 5 after HEL3x-SRBC immunization. Aicda, Prdm1, and Myc transcripts levels were analyzed by qPCR and normalized by Hprt. Naive follicular B cells (Fo-B) were sorted as B220+AA4.1–CD21/35intCD23hi cells from 2 naive mice. The experiment shown is representative of 3 independent sorting experiments. In each experiment, cDNA was prepared from sorted cells after pooling 5–10 mice. The mean ± SEM is shown. *P < 0.05, **P < 0.01, ***P < 0.001 using 1-way ANOVA followed by a Tukey’s multiple comparisons post-test.
To decipher the directionality of the interzonal DZ-LZ cycling, we looked at the proportions of S and G2/M cells in the 4 different subsets. In the LZ, a higher proportion of LacZ+ cells than LacZ– cells were in S-phase, suggesting that the LacZ+ cells are enriched in positively selected cells in transit to the DZ. As the DZ LacZ– cells contained a decreased proportion of S- and G2/M-phases compared with the vigorously proliferating DZ LacZ+ cells (Figure 3A), we concluded that they were completing their rounds of cell division. Collectively, our working model is that cycling cells in the LZ express high levels of miR-155 (LZ LacZ+) and they migrate to the DZ, where they proliferate actively (DZ LacZ+) and start downregulating miR-155 expression. They then start to exit the cell cycle (DZ LacZ–) and, upon completion of the cell cycle, they return to the LZ (LZ LacZ–).
To substantiate this model, we next looked at the expression pattern of some key GC molecules. We measured activation-induced cytidine deaminase (Aicda) transcripts because this gene is known to be highly expressed in DZ B cells (19) in order to enable SHM. Aicda expression was highest in the DZ LacZ– subset (Figure 3D). Cells in this subset are dividing but are already past their most active stage of proliferation (Figure 3A), which may allow them to increase the number of BCR mutations before they return to the LZ. Elevated expression of Prdm1 was observed in the DZ LacZ+ subset (Figure 3D), in agreement with the findings showing that GC B cells differentiate into plasma cells and leave GCs through the DZ (20). We next looked at Myc expression because it is upregulated in proliferative positively selected LZ B cells (10, 11). As expected, Myc expression was low in DZ B cells, and its greatest expression was detected in the LZ LacZ+ subset, which is high in miR-155 (Figure 2B, Figure 3D, and Supplemental Figure 2A; supplemental material available online with this article; doi:10.1172/JCI82914DS1). Although these analyses were carried out at day 5, highest expression of both miR-155 and c-MYC in LZ LacZ+ was also observed at day 4 (Supplemental Figure 2, A and B). Together, these results suggest that c-MYC and miR-155 are coexpressed in positively selected LZ B cells.
miR-155 provides survival signals to positively selected c-MYC+ LZ B cells.
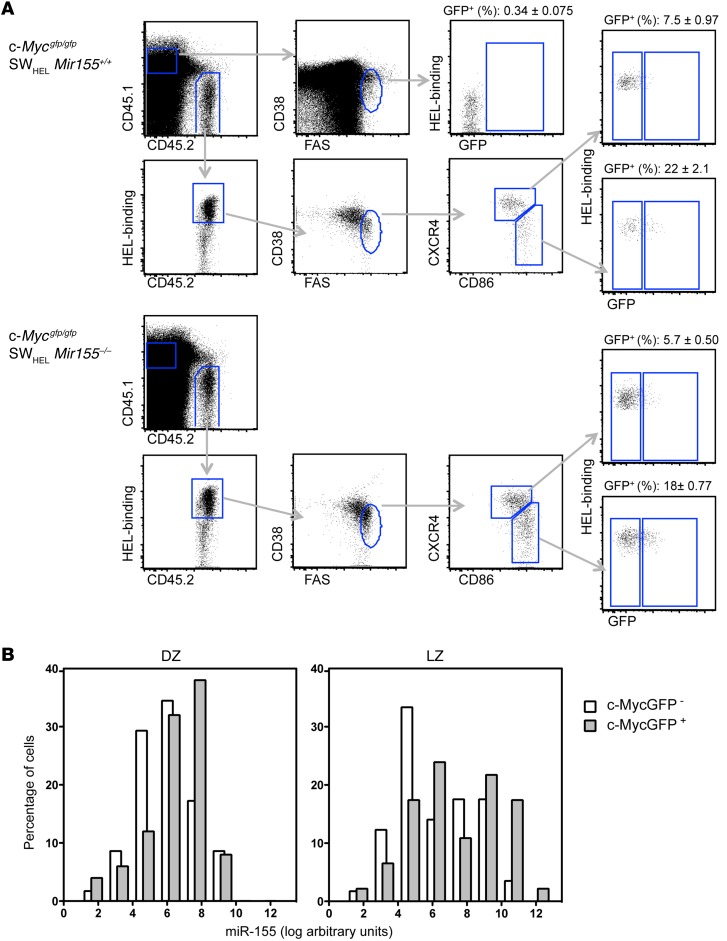
To characterize further the expression pattern of miR-155 and c-MYC in LZ B cells, we made use of Mycgfp/gfp reporter mice (21), which we crossed to SWHEL mice. After adoptive transfer of Mycgfp/gfp SWHEL B cells into CD45.1+ congenic mice followed by HEL3X-SRBC injection, we single cell–sorted c-MYC+ LZ, c-MYC– LZ, c-MYC+ DZ, and c-MYC– DZ B cells from these mice (Figure 4A) and measured the expression of miR-155 by quantitative PCR (qPCR). c-MYC+ cells expressed higher levels of miR-155 than c-MYC– cells, both in the DZ and the LZ, indicating that c-MYC is a strong predictor of miR-155 expression (Figure 4B). In agreement with our previous analysis (Figure 2B), on average, DZ B cells expressed lower levels of miR-155 than LZ B cells. Consequently LZ B cells coexpressed c-MYC and miR-155 at its highest level (Figure 4B). To examine whether miR-155 deficiency affected the expression of c-MYC, we compared the proportions of c-MYC+ GC B cells originating from adoptively transferred miR-155–sufficient and –deficient B cells. As shown in Figure 4A, disruption of miR-155 did not affect the emergence of c-MYC+ LZ B cells. Thus, induction of c-MYC does not require miR-155.
Figure 4. miR-155 is coexpressed with c-MYC in a subset of LZ B cells.
Splenic B cells derived from Mycgfp/gfp SWHEL Mir155+/+ or Mycgfp/gfp SWHEL Mir155–/– mice were adoptively transferred into CD45.1+ congenic recipients, which were injected with HEL3X-SRBC. (A) Four days later, CD45.1–CD45.2+CD38loCD95+ HEL–specific donor-derived splenic GC B cells were gated as DZ or LZ and further divided based on GFP expression. The mean ± SEM from 2 independent sorting experiments is shown (n = 6). (B) Four days after adoptive transfer of Mycgfp/gfp SWHEL Mir155+/+ cells, DZ GFP–, DZ GFP+, LZ GFP–, and LZ GFP+ cells were single cell sorted according to the gates shown in A from 3 mice, and miR-155 expression was measured by qPCR using U6 for normalization (DZ GFP–, n = 58; DZ GFP+, n = 50; LZ GFP–, n = 57; LZ GFP+, n = 46). Histograms show the distribution of cells on the basis of miR-155 expression in log-transformed arbitrary units. Bars are nudged to prevent overlapping. Background level was established by assessing the expression of miR-155 in Mir155–/– GC B cells.
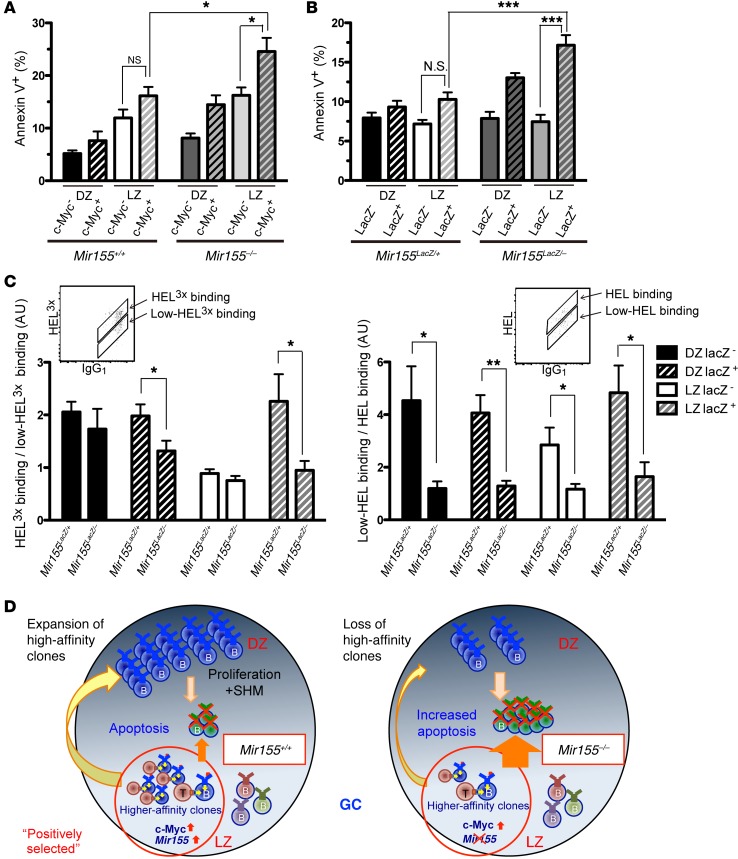
Having established that c-MYC and miR-155 are mainly coexpressed, we then assessed whether the 2 molecules are functionally linked. We split GC B cells on the basis of c-MYC expression in combination with DZ and LZ delineation. Apoptosis was measured in miR-155–sufficient and –deficient subsets by AnnexinV staining, in combination with DAPI. We chose AnnexinV combined with DAPI staining to detect early apoptotic cells, as — unlike the detection of caspase 3 or cleaved poly-ADP ribose polymerase (PARP) — its measurement does not diminish the fluorescent signal for LacZ activity. We observed that c-MYC+ cells are more prone to apoptosis in the absence of miR-155 in both LZ and DZ, whereas c-MYC– Mir155–/– cells were similar to their Mir155+/+ counterparts (Figure 5A). To further validate these results, we measured apoptosis after delineating DZ and LZ B cells on the basis of LacZ expression in the presence or absence of miR-155. Then we generated miR-155–deficient mice in which one allele was replaced by a LacZ cassette (13) and the second allele was a previously described null allele (12), thus producing a Mir155LacZ/– genotype. Using the same adoptive transfer scheme described in previous sections, we observed similar levels of LacZ activity (based on the geometric mean fluorescence intensity [gMFI]) among sorted DZ and LZ GC B cells (Supplemental Figure 1A). In addition, LacZ activity was similar in DZ and LZ GC B cells derived from SWHEL Mir155LacZ/+ or SWHEL Mir155LacZ/– mice (Supplemental Figure 1B), indicating that transcriptional activity of the Mir155 promoter is not dependent on the expression of miR-155. The Mir155LacZ/– strain was then compared with Mir155LacZ/+ and so allowed us to assess how B cells that would have expressed miR-155 (LacZ+) were affected by miR-155 loss. As shown in Figure 5B, the subset of LZ B cells enriched in miR-155 expression in WT cells was the most susceptible to apoptosis in the absence of miR-155, followed by DZ LacZ+ cells. These results suggest that miR-155 provides survival signals mainly to c-MYC+ positively selected B cells, which is consistent with the highest levels of miR-155 expression on those cells. These results did not exclude a role for miR-155 in regulating the cell cycle. However, when we measured cell cycle dynamics in the 4 subsets in miR-155–sufficient or –deficient B cells, we detected no more than a small decrease in S-phase B cells in DZ LacZ+ Mir155–/– relative to their Mir155+/+ counterparts (Supplemental Table 1), suggesting that cell cycle regulation is not a major function of miR-155.
Figure 5. Positively selected LZ B cells gain antiapoptotic signals via miR-155.
(A) Splenic B cells derived from Mycgfp/gfp SWHEL Mir155+/+ or Mycgfp/gfp SWHEL Mir155–/– mice were adoptively transferred into CD45.1+ congenic recipients, which were injected with HEL3X-SRBC. Four days later, spleen cells from these mice were analyzed. The graph shows the percentage of AnnexinV+7-AAD– cells determined by flow cytometry in DZ and LZ B cells further divided on the basis of c-MYC expression. The mean ± SEM from 2 independent experiments is shown (Mycgfp/gfp Mir155+/+, n = 13; Mycgfp/gfp Mir155–/–, n = 13). *P < 0.05 using 1-way ANOVA followed by a Tukey’s multiple comparisons post-test. (B) SWHEL Mir155LacZ/+ or SWHEL Mir155LacZ/– B cells were adoptively transferred into CD45.1+ congenic recipients, which were injected with HEL3X-SRBC. Spleen cells from these mice were analyzed 5 days later. The graph shows the percentage of AnnexinV+7-AAD– cells determined by flow cytometry in DZ and LZ B cells further divided on the basis of LacZ positivity. The mean ± SEM from 2 independent experiments is shown (Mir155LacZ/+, n = 10; Mir155LacZ/–, n = 11). ***P < 0.001 using 1-way ANOVA followed by a Tukey’s multiple comparisons post-test. (C) Using the same HEL3X-SRBC injection as in B, IgG1+ donor–derived GC B cells were analyzed for their binding ability against HEL3X and HEL. Two independent experiments were performed, and the mean ± SEM of a representative experiment is shown (Mir155LacZ/+, n = 7; Mir155LacZ/–, n = 6). *P < 0.05, **P < 0.005 using 2-tailed unpaired t test (C). (D) Summary illustrations showing the role of miR-155 in the GC response. The absence of miR-155 expression in positively selected c-MYC+ LZ B cells increase apoptosis in the population. Consequently, GC cellularity is decreased, and concomitantly, affinity maturation is abrogated.
In the framework of the cyclic reentry model, we would predict that failure to provide survival signals to positively selected clones would impair affinity maturation. We tested affinity maturation in miR-155–deficient B cells according to DZ, LZ, and LacZ activity 8 days after HEL3X-SRBC injection by staining with HEL3X and HEL (18). We chose this time point because, by this stage, affinity maturation has started in this model system (22) and we can still detect miR-155–deficient GC B cells. As affinity maturation progresses, binding to HEL3X becomes brighter concomitant with a dimmer HEL-binding. Therefore, we assessed the ability of the BCR to bind HEL3X and HEL by calculating the ratio of high/low HEL3X–binding B cells (Figure 5C and Supplemental Figure 3) and the ratio of low/high HEL–binding B cells. In the FACS plots shown in Figure 5C, the top diagonal gate indicates SWHEL B cells that carry BCRs with higher binding to HEL3X or HEL, respectively. In the same plots, the bottom gate corresponds to SWHEL B cells that carry BCRs with lower binding to HEL3x or HEL, respectively (18). In both cases, the higher ratio represents higher affinity. Based on the HEL3X staining, we observed decreased affinity maturation due to miR-155 deficiency mainly in the LacZ+ LZ B cells (Figure 5C, left), which is consistent with increased apoptosis in this fraction (Figure 5B) and with previous reports showing impaired affinity maturation without miR-155 (13, 14). The HEL stain showed a similar pattern with higher differences between miR-155–sufficient and –deficient B cells (Figure 5C, right), again in accordance with published data. There are minor differences in the antigen-binding ratios between HEL3x and HEL, which is explained as follows. Immunization with HEL3x frequently leads to loss of HEL binding by the BCRs expressed by SWHEL B cells. As the response progresses, SWHEL B cells acquire higher affinity for HEL3x and thus are brightly stained with HEL3x (top diagonal gate in HEL3x staining), whereas the rest of SWHEL B cells are detected in the bottom diagonal gate (Figure 5C). However, with HEL staining, SWHEL B cells that retain HEL binding are detected in the top diagonal gate, and the remainder of SWHEL B cells carrying high-affinity BCRs that bind to HEL3x or that do not bind to either HEL3x or HEL with high affinity fall into the bottom diagonal gate. Taken together, our results suggest that miR-155 expression is required for the survival of c-MYC+ LZ B cells and that failure of c-MYC+ LZ B cells to receive miR-155–dependent survival signals leads to decreased GC cellularity with concomitant impaired affinity maturation (Figure 5D).
In silico simulations are consistent with the hypothesis that miR-155 regulates the survival of positively selected LZ B cells.
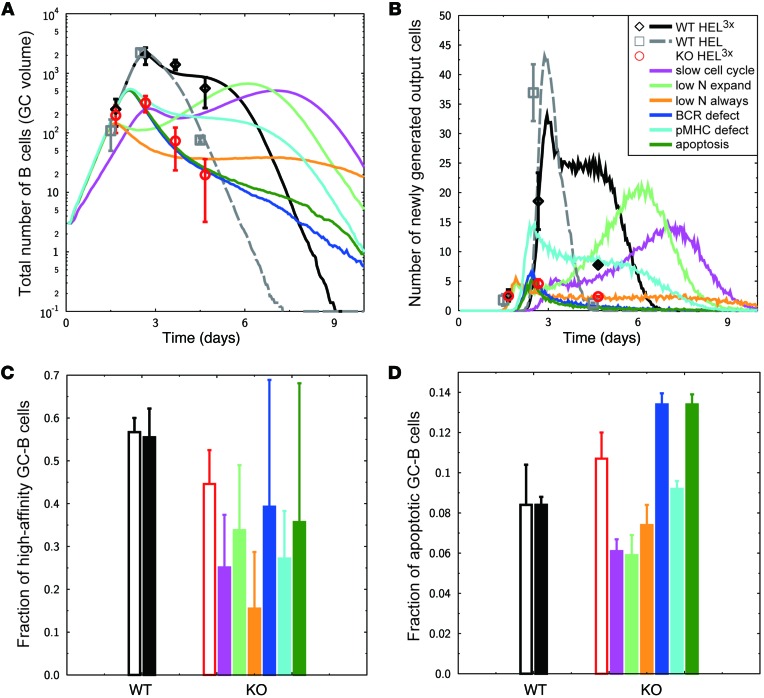
To further substantiate our conclusions, we next asked whether the impaired survival of positively selected GC B cells would be sufficient to explain the complex phenotype of miR-155–deficient GC B cells, which includes a reduced GC size, a shorter duration of the GC reaction, reduced affinity maturation, and increased apoptosis of GC B cells. We extended the previously established mathematical model of the GC reaction (20) by affinity-dependent numbers of divisions (3, 23) and by antibody feedback (24), and we validated the model with WT data for GC population kinetics, plasma cell generation, affinity maturation, and apoptosis using in silico immunization with HEL3X or HEL (Figure 6, A–D, black lines and black-filled bar or gray dashed lines, respectively). Subsequently, different hypotheses for the effect of miR-155 deficiency were tested for their ability to reproduce the in vivo phenotype (Figure 6, A–D, colored lines and bars). When the effect of miR-155 deficiency was assumed to change division parameters, the in silico phenotype was in contradiction with the measured GC population kinetics (Figure 6, A–C, magenta, light green, orange lines and bars). This supports the in vivo result that miR-155 would not be a major regulator of the cell cycle (Supplemental Table 1). Similarly, a defect in peptide-MHC presentation was too weak to induce the observed fast shutdown of the GC reaction and thus unable to explain the phenotype (Figure 6, A–C, cyan lines and bars). However, when defective BCR signaling resulting in reduced antigen uptake (Figure 6, A–D, and Supplemental Figure 4, blue lines and bars) or when increased apoptosis of the subset of positively selected GC B cells were considered (Figure 6, A–D, and Supplemental Figure 4, green lines and bars), we were able to qualitatively reproduce the phenotype observed in the in vivo experiment. This is quantitatively supported by the best agreement between these 2 models and in vivo data (Supplemental Table 2). While the in silico results are consistent with the hypothesis that miR-155 regulates the survival of positively selected GC B cells, the in vivo data did not suffice to exclude or support a deregulation of BCR signaling.
Figure 6. GC phenotype due to miR-155 deficiency in B cells in silico.
(A–D) Simulations of WT GC immunized with HEL3x (black lines and black-filled bars) or HEL (gray dashed lines) were used for validation of the reference simulations with in vivo experimental data (black open diamonds [A and B] and black open bars [C and D] for HEL3x, and gray open squares [A and B] for HEL). In silico miR-155 KO experiments were performed and compared with in vivo experimental results (red open circles [A and B] and red open bar [C and D]) assuming various effects of miR-155 KO: reduced cell cycle speed (magenta lines [A and B] and magenta-filled bar [C and D]), reduced number of divisions during GC expansion (light-green lines [A and B] and light-green–filled bar [C and D]), and in addition, during GC selection phase (orange lines [A and B] and orange-filled bar [C and D]), defect of BCR leading to reduced antigen collection (blue lines [A and B] and blue-filled bar [C and D]), defect in antigen processing or presentation on peptide-MHC (cyan lines [A and B] and cyan-filled bar [C and D]), or increased apoptosis rate in the B cell subset positively selected by follicular T helper cells (green lines [A and B] and green-filled bar [C and D]). The GC phenotype is characterized by the total GC size (A), the number of output cells generated by one GC in the last hour (B), the fraction of high-affinity GC B cells at day 7 after immunization (C), and the fraction of apoptotic GC B cells at day 5 after immunization (D). All lines and bars are the mean of 50 simulations. Standard deviations for A and B and time courses for C and D are provided in Supplemental Figure 4.
The miR-155 target JARID2 regulates the apoptosis of GC B cells.
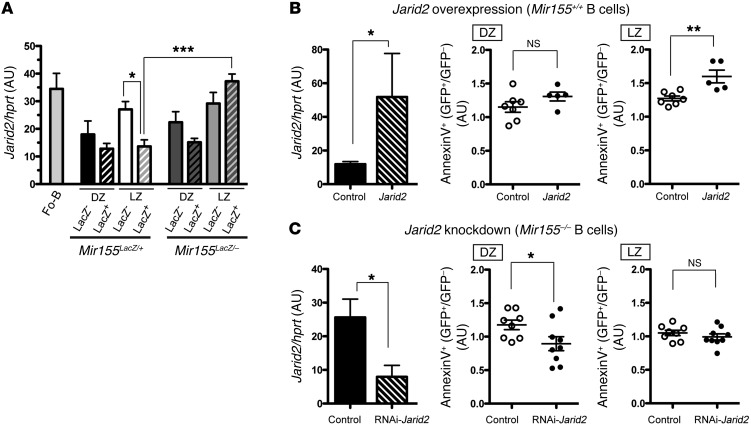
The in silico modeling of the GC response showed that a 60% increase in the apoptosis of positively selected LZ B cells is sufficient to explain the steady decrease in GC numbers and loss of affinity maturation due to miR-155 deficiency. One of the miR-155 target genes that is consistently modulated by miR-155 is Jarid2 (12, 14, 15, 25, 26). JARID2 is a DNA-binding protein that belongs to the Jumonji (jmj) family (27) and has 2 miR-155 binding sites in its 3′-UTR (28, 29). As its downregulation by miR-155 promotes the survival of chicken fibroblasts (28), we examined JARID2 expression in GC B cell subsets. In miR-155–sufficient B cells, Jarid2 mRNA was downregulated in the transition from the LZ LacZ– subset to the LZ LacZ+ subset (Figure 7A). However, miR-155–deficient B cells failed to downregulate Jarid2 mRNA in this transition (Figure 7A). This expression pattern suggests that JARID2 is a target of miR-155 in GC B cells. We next investigated whether JARID2 could be a miR-155 mediator. Mir155+/+-activated B cells were transduced with a JARID2-IRES-GFP–expressing retrovirus (~4-fold overexpression, Figure 7B) or a control virus expressing only GFP. In addition, Mir155–/–-activated B cells were transduced with an RNAi-JARID2-IRES-GFP expression (~3-fold reduced expression, Figure 7C) or GFP control. The retrovirally transduced B cells were adoptively transferred into HEL-SRBC–injected CD45.1+ congenic mice (Supplemental Figure 5), and apoptosis was measured in DZ and LZ B cells (Figure 7B, JARID2 overexpression; Figure 7C, JARID2 knockdown). Samples were normalized relative to the GFP– counterparts to correct for potential biases across cultures, although there were no statistically significant differences in the level of apoptosis of GFP– cells from control- or JARID2-transduced B cells (not shown). Ectopic expression of JARID2 in Mir155+/+ GC B cells significantly increased the percentage of AnnexinV+ cells in the LZ, and there is a trend of increased apoptosis in the DZ B cells, although it is not statistically significant at the 5% level (Figure 7B). In contrast, reducing JARID2 expression in Mir155–/– GC B cells promoted the survival of DZ B cells but had not affected LZ B cell survival (Figure 7C). Taken together, these results suggest that JARID2 expression regulates the apoptosis of GC B cells in a miR-155–dependent manner.
Figure 7. The miR-155 target gene Jarid2 regulates the apoptosis of GC B cells.
(A) SWHEL Mir155LacZ/+ or SWHEL Mir155LacZ/– B cells were adoptively transferred into CD45.1+ congenic recipients that were injected with HEL3X-SRBC. After 5 days, donor cells were sorted as described in Figure 2A. Jarid2 transcripts were measured by qPCR using Hprt for normalization. The mean ± SEM is presented from a representative data of 2 independent experiments (n = 6). *P < 0.05, ***P < 0.001 using 1-way ANOVA followed by a Tukey’s multiple comparisons post-test. (B) SWHEL Mir155+/+ activated B cells were transduced with JARID2/IRES-GFP virus or IRES-GFP control (empty vector). The GFP+ cells were sorted, and gene overexpression efficiency was evaluated by qPCR. The mean ± SEM of experiments is shown (n = 4) (left). Retrovirally transduced SWHEL B cells were adoptively transferred into HEL-SRBC–injected CD45.1+ congenic mice (Supplemental Figure 5). Four days later, apoptosis was measured using AnnexinV and 7-AAD in GC HEL-binders split on the bases of GFP expression and DZ/LZ status as described in Supplemental Figure 6. The ratio of apoptosis was calculated as the relative values of the GFP+ to the GFP– counterparts to correct for potential biases across cultures. Data are pooled from 2 independent experiments and presented as the mean ± SEM. Each symbol represents a mouse. (C) The same method as B was used, except that activated SWHEL Mir155–/– B cells were retrovirally transduced with RNAi-JARID2/IRES-GFP or control vector, and Jarid2 silencing efficiency was evaluated by qPCR. Data are pooled from 2 independent experiments and presented as mean ± SEM. Each symbol represents a mouse. *P < 0.05, **P < 0.005 using two-tailed unpaired t test (B and C).
Discussion
It has been known that miR-155 is required for GC function under physiological conditions (30); however, its precise mode of action remained unknown. Several lines of evidence in both mouse and human have indicated that miR-155 is expressed by a subset of GC B cells (13, 31). However, the exact GC B cell population that expresses miR-155 has been not identified. Here, we show that positively selected B cells express the highest levels of miR-155 within the GC. Previous work has identified BCL6 as a transcriptional repressor of miR-155 that ensures its shut-down in DZ B cells. Interestingly, BCL6 also represses c-MYC expression (10, 11), suggesting that the 2 molecules may be cosuppressed when LZ B cells reenter the DZ. Whether c-MYC and miR-155 induction is coincidental or one molecule precedes the other remains to be tested.
The in silico modeling of the GC supports our in vivo results showing that elevated apoptosis of positively selected GC B cells due to the Mir155 deletion is sufficient to explain the loss of cellularity, reduced antibody secretion, and reduced affinity maturation. The simulation results also suggest that the in vivo phenotype of miR-155–deficient mice may be caused by defective BCR signaling, which leads to decreased antigen uptake. However, while no reports so far show that miR-155 influences antigen uptake in B cells, we cannot exclude this possibility. Of interest, the in silico modeling also suggests that miR-155 may play a role in the initiation of the GC reaction. This is an area that needs further investigation but can also point toward synergy between c-MYC and miR-155, as c-MYC also plays a role in the initiation of the GC response (10, 11).
Our data suggest that JARID2 is one of the miR-155 targets that needs to be silenced in order to control apoptosis. As predicted, JARID2 overexpression in WT B cells increased the apoptosis of LZ B cells and, to a lesser extent, DZ B cells. However, its downregulation in miR-155–deficient B cells only promoted the survival of DZ B cells. Although we achieved 60% JARID2 knockdown in cultured B cells, we cannot assume that JARID2 silencing is equally efficient in vivo. Alternatively, ectopically silenced JARID2 in a subset of LZ B cells that highly express JARID2 in the miR-155–sufficient background (e.g., LZ LacZ–, Figure 7A) may have caused deleterious effects on cell survival. JARID2 regulates cell proliferation and differentiation in several tissues (27, 32, 33), and recent reports have shown that JARID2 is a part of polycomb repressive complex 2 (PRC2) (32, 34, 35). Excessively low expression of JARID2 in these GC B cells may have disturbed PRC2 recruitment to its targets, including survival/apoptosis-related genes (36). In this scenario, the protective survival effect conferred by lowering JARID2 in LZ LacZ+ B cells may have been diluted or became undetectable. Disentangling this issue would require generating mice carrying Jarid2 alleles unresponsive to miR-155. The identity of JARID2 target genes in GC B cells and whether JARID2 function in GC B cells is PRC2 dependent remains unknown. It also remains unknown whether targets of miR-155, in addition to JARID2, are important in the regulation of apoptosis in c-MYC+ GC B cells.
Expression of c-MYC in positively selected GC B cells is essential to maintain the GC response, although its role is uncertain. c-MYC regulates a large quantity of transcription of genes and thus controls cell growth and proliferation; however, this molecule also functions to sensitize cells to apoptosis (37–39). We showed that miR-155 expression in GC B cells mainly protect c-MYC+ positively selected LZ B cells from apoptosis and, to a lesser extent, c-MYC+ DZ B cells. Given these results, we suggest that miR-155 mitigates the proapoptotic effects caused by c-MYC in positively selected GC B cells, and thus, we provide a basis for functional cooperation between c-MYC and miR-155 in the normal course of the GC reaction. Understanding the physiology of the GC response also informs on B cell lymphomagenesis. Although miR-155 host transcript BIC and c-MYC have been shown to cooperate in promoting cellular transformation using the chicken system (40, 41), how these molecules achieve this has been unknown for a long time. The functional interaction between c-MYC and miR-155 we show here may explain the longstanding enigma of the 2 cooperating oncogenes: high expression of miR-155 provides extra survival advantage to transformed c-MYC+ cells. Interestingly, c-MYC acquires the ability to bind to regulatory elements of Jarid2 upon initiation of cellular transformation and concomitantly inhibits its expression (42). Thus, it is possible that one of the points of molecular convergence between miR-155 and c-MYC is JARID2 and that repression of this molecule by both miR-155 and c-MYC may be required for survival of transformed B cells. In fact, this regulation may also be relevant to activated B cell–like diffuse large B cell lymphoma cells (ABC-DLBCL), as ABC-DLBCL cells overexpress both c-MYC (43–45) and miR-155 (46, 47).
ABC-DLBCL resembles post–GC B cells that are blocked in their process of differentiation toward plasma cells (48). Given that positively selected GC B cells are enriched in plasmablast progenitors (4) and express high levels of miR-155 and c-MYC, these cells may represent the normal counterpart of ABC-DLBCL. In contrast, low expression of miR-155 has been reported in another DLBCL subtype, GCB-DLBCL, and Burkitt’s lymphoma, both of which resemble LZ B cells according to the analyses of their gene-expression profiles (19). This is in agreement with our results showing low expression of miR-155 in the DZ and in most LZ GC B cells.
Collectively, we have shown that miR-155 expression is dynamically regulated in GC B cells. Only a small fraction of proliferating positively selected c-MYC+ LZ B cells express the mature form of the miRNA. At the functional level, we observed that miR-155 expression protects c-MYC+ LZ B cells from apoptosis via JARID2, and thus it is critical for the maintenance of both the GC response and affinity maturation. We demonstrate the functional link between c-MYC and miR-155 in GC B cells, which may also be relevant to activated ABC-DLBCL. In the long-term, our results open up the possibility that blocking the functional interaction between miR-155 and c-MYC, by use of small inhibitors, may be a potential route for improved lymphoma therapy.
Methods
Mice.
CD45.1+ congenic mice were bred and maintained in the Babraham Research Campus small animal facility. miR-155–deficient mice, SWHEL mice, and Mycgfp/gfp mice have been described previously (12, 16, 21). SWHEL mice were a gift from R. Brink (Garvan Institute of Medical Research/University of New South Wales), and Mycgfp/gfp mice were a gift from B.P. Sleckman (Washington University School of Medicine, St. Louis, Missouri, USA). miR-155–LacZ reporter mice (13) were purchased from The Jackson Laboratory. All mice were maintained on the C57BL/6 background.
Adoptive transfer.
SRBCs were conjugated to recombinant HEL (Sigma-Aldrich) or HEL3X (a gift from R. Brink [Garvan Institute of Medical Research/University of New South Wales]) with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (Sigma-Aldrich) as previously described (49). CD45.2+ spleen B cells from SWHEL donor mice were purified using mouse B cell Isolation Kit (Miltenyi Biotec). HEL-binding B cells (5 × 104) were i.v. injected into CD45.1+ congenic recipients, followed by injection with 2 × 108 SRBC conjugated to a specific recombinant HEL protein. For analyses, CD45.2+ donor splenocytes were enriched by CD45.1 negative selection using an autoMACS pro separator (Miltenyi Biotec).
Immunohistology.
Splenic cryosections were fixed in acetone and blocked with 30% normal horse serum. HEL-binding cells were detected by incubation with 100 ng/ml HEL followed by polyclonal rabbit anti-HEL serum and goat anti–rabbit IgG-FITC (Southern Biotechnology Associates). T cells were stained with anti–CD3 biotin (clone 500A2, eBioscience) followed by streptavidin Alexa Fluor 550 (Invitrogen), and follicular B cells with anti–IgD 647 (clone 11-26c.2a, BD Biosciences). Slides were analyzed using a Zeiss Axiovert 200M microscope with a ×10 objective and processed using Adobe Photoshop software.
Flow cytometry and cell sorting.
Multicolor flow cytometry for analyses or for sorting was performed on a LSR Fortessa or FACSAria (BD Biosciences), respectively. Single cell suspensions of splenocytes were blocked with anti-CD16/32 mAb (clone 2.4G2, in house), and followed by staining with the following antibodies: anti-B220 (clone RA3-6B2), anti-CD38 (clone 90), anti-CD45.2 (clone 104), anti-CXCR4 (clone 2B11), and AA4.1, from eBioscience; anti-CD86 (clone GL1, BioLegend); and anti-CD95 (clone Jo2) and anti-IgG1 (clone A85-1), from BD Biosciences. HEL-binding B cells were stained as described previously (17). LacZ activity was visualized using the FluoReporter-LacZ-Flow cytometry kit (Invitrogen). For cell cycle analyses, cells of each sorted subset containing more than 5 × 104 cells were fixed and permeabilized using a Cytofix/Cytoperm Kit (BD Biosciences), followed by staining with 10 μg/ml DAPI. Cell cycle was calculated by FlowJo Dean/Jett/Fox algorithms. To quantify protein expression, we used the gMFI to account for the log-normal behavior of flow data. The data were analyzed with FlowJo software (Tree Star).
qPCR.
Relative quantifications of gene expression were determined by Hprt expression. Single cells were directly sorted into 96-well plates containing 5 μl water and 25 ng carrier RNA (QIAGEN) per well. miR-155 and U6 were reverse transcribed with Super Script III (Invitrogen) and Taqman MicroRNA assay set (Applied Biosystems). The primers used for Bcl6 were 5′-CAGCAACATCTACTCGCCCAA-3′ and 5′-ATGGAGCATTCCGAGCAGAAG-3′; the remainder of the primers were as described elsewhere: Prdm1 (50), Aicda (15), Myc (11), and Jarid2 (32).
Retroviral transduction of B cells.
Mouse Jarid2 cDNA (Thermo Scientific) and JARID2-specific RNAi oligonucleotide (32) were subcloned into the pMigRI-IRES-GFP vector. Retroviral vectors were introduced into Plat-E packaging cells, and retroviral supernatants were collected 48 hours after transfection. For B cell activation, SWHEL mice were i.p. injected with 4 mg HEL, and after 6 hours, mice were sacrificed and spleens were collected. B cells were cultured in RPMI supplemented with 10% FBS, 2 mM GlutaMAX (Invitrogen), 50 U/ml penicillin, 50 μg/ml streptomycin, and 50 μM 2-ME in the presence of 1 μg/ml anti-CD40 (clone HM40-3, eBioscience) for 16 hours before spin infection. Twenty-eight to 30 hours after infection, the cells were adoptively transferred into CD45.1+ congenic recipients that had been i.p. injected with 2 × 108 HEL-SRBC 30 hours earlier.
Mathematical model.
For mathematical modeling of the GC reaction, an agent-based model combined with a model for soluble antibodies was employed (20, 24). The model was validated with WT data and then used to simulate different hypothetical effects of miR-155. The first data point of WT mice immunized with HEL or with HEL3X determined the time of GC onset after immunization. The best fit was achieved by GC onset 2 days after immunization with HEL and 2 days and 8 hours after immunization with HEL3X. The parameters used were the result of various test runs and represent the best choice in terms of RSS values (see Supplemental Table 2) for each of the miR-155 impact points. Each simulation was repeated 50 times. The mean time course is shown in Figure 6, and the standard deviation for the best solutions is represented in Supplemental Figure 4.
Statistics.
Statistical analyses were performed by a 2-tailed unpaired Student’s t test in Microsoft Excel or 1-way ANOVA test for multiple group comparisons followed by a Tukey’s multiple comparisons post-test in GraphPad Prism software. A P value less than 0.05 was considered statistically significant.
Study approval.
All animal procedures complied with UK Home Office regulations (London, United Kingdom) and EU Directive 2010/63/EU (European Union, Brussels, Belgium) and were approved by the Babraham Research Campus Animal Welfare, Experimentation and Ethics Committee (Cambridge, United Kingdom).
Author contributions
RN and EV designed research studies. RN, RL, DL, GA, TGP, and EV conducted experiments and acquired and/or analyzed data. MMH performed and interpreted the mathematical simulations. RB provided reagents and training. MT interpreted data and critically revised the manuscript. RN and EV wrote the manuscript.
Supplementary Material
Acknowledgments
The authors acknowledge the staff of the Babraham Institute small animal facility for technical support, Arthur Davis for FACS, and Geoff Butcher and Michelle Linterman for critical reading of the manuscript. R. Nakagawa was supported by a Marie Curie Incoming Fellowship from the European Union’s Seventh Framework Programme for research, technological development, and demonstration. E. Vigorito was supported by the Medical Research Council grants G1001781 and G0700287 and by the Biotechnology and Biological Sciences Research Council. M. Meyer-Hermann was supported by the German Federal Ministry of Education and Research within the Measures for the Establishment of Systems Medicine, project SYSIMIT (BMBF eMed project SYSIMIT, FKZ: 01ZX1308B) and by the Human Frontier Science Program (RGP0033/2015).
Footnotes
Rinako Nakagawa’s present address is: Immunity and Cancer Laboratory, The Francis Crick Institute, London, United Kingdom.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(1):377–388. doi:10.1172/JCI82914.
See the related Commentary beginning on page 1.
References
- 1.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 2.Nieuwenhuis P, Opstelten D. Functional anatomy of germinal centers. Am J Anat. 1984;170(3):421–435. doi: 10.1002/aja.1001700315. [DOI] [PubMed] [Google Scholar]
- 3.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509(7502):637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143(4):592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kepler TB, Perelson AS. Cyclic re-entry of germinal center B cells and the efficiency of affinity maturation. Immunol Today. 1993;14(8):412–415. doi: 10.1016/0167-5699(93)90145-B. [DOI] [PubMed] [Google Scholar]
- 6.Meyer-Hermann M, Deutsch A, Or-Guil M. Recycling probability and dynamical properties of germinal center reactions. J Theor Biol. 2001;210(3):265–285. doi: 10.1006/jtbi.2001.2297. [DOI] [PubMed] [Google Scholar]
- 7.Oprea M, Perelson AS. Somatic mutation leads to efficient affinity maturation when centrocytes recycle back to centroblasts. J Immunol. 1997;158(11):5155–5162. [PubMed] [Google Scholar]
- 8.Basso K, et al. BCL6 positively regulates AID and germinal center gene expression via repression of miR-155. J Exp Med. 2012;209(13):2455–2465. doi: 10.1084/jem.20121387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/S0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 10.Calado DP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012;13(11):1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominguez-Sola D, et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol. 2012;13(11):1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 14.Vigorito E, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27(6):847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu D, et al. The miR-155-PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. J Exp Med. 2014;211(11):2183–2198. doi: 10.1084/jem.20140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phan TG, et al. B cell receptor-independent stimuli trigger immunoglobulin (Ig) class switch recombination and production of IgG autoantibodies by anergic self-reactive B cells. J Exp Med. 2003;197(7):845–860. doi: 10.1084/jem.20022144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan TD, Gatto D, Wood K, Camidge T, Basten A, Brink R. Antigen affinity controls rapid T-dependent antibody production by driving the expansion rather than the differentiation or extrafollicular migration of early plasmablasts. J Immunol. 2009;183(5):3139–3149. doi: 10.4049/jimmunol.0901690. [DOI] [PubMed] [Google Scholar]
- 18.Paus D, Phan TG, Chan TD, Gardam S, Basten A, Brink R. Antigen recognition strength regulates the choice between extrafollicular plasma cell and germinal center B cell differentiation. J Exp Med. 2006;203(4):1081–1091. doi: 10.1084/jem.20060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, Nussenzweig MC. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120(11):2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer-Hermann M, Mohr E, Pelletier N, Zhang Y, Victora GD, Toellner KM. A theory of germinal center B cell selection, division, and exit. Cell Rep. 2012;2(1):162–174. doi: 10.1016/j.celrep.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38(2):342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 22.Phan TG, et al. High affinity germinal center B cells are actively selected into the plasma cell compartment. J Exp Med. 2006;203(11):2419–2424. doi: 10.1084/jem.20061254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer-Hermann M. Overcoming the dichotomy of quantity and quality in antibody responses. J Immunol. 2014;193(11):5414–5419. doi: 10.4049/jimmunol.1401828. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Germinal center B cells govern their own fate via antibody feedback. J Exp Med. 2013;210(3):457–464. doi: 10.1084/jem.20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabani MM, et al. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010;38(13):4466–4475. doi: 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205(3):585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi T, et al. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9(10):1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- 28.Bolisetty MT, Dy G, Tam W, Beemon KL. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J Virol. 2009;83(23):12009–12017. doi: 10.1128/JVI.01182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahlke C, et al. A microRNA encoded by Kaposi sarcoma-associated herpesvirus promotes B-cell expansion in vivo. PLoS One. 2012;7(11): doi: 10.1371/journal.pone.0049435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253(1):146–157. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 31.Tan LP, et al. miRNA profiling of B-cell subsets: specific miRNA profile for germinal center B cells with variation between centroblasts and centrocytes. Lab Invest. 2009;89(6):708–716. doi: 10.1038/labinvest.2009.26. [DOI] [PubMed] [Google Scholar]
- 32.Pasini D, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235(9):2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- 34.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen X, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caganova M, et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J Clin Invest. 2013;123(12):5009–5022. doi: 10.1172/JCI70626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6(10):1915–1922. [PubMed] [Google Scholar]
- 38.Evan GI, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69(1):119–128. doi: 10.1016/0092-8674(92)90123-T. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y, Glynn JM, Guilbert LJ, Cotter TG, Bissonnette RP, Green DR. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science. 1992;257(5067):212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- 40.Clurman BE, Hayward WS. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol Cell Biol. 1989;9(6):2657–2664. doi: 10.1128/MCB.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tam W, Hughes SH, Hayward WS, Besmer P. Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding RNA, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis. J Virol. 2002;76(9):4275–4286. doi: 10.1128/JVI.76.9.4275-4286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabò A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511(7510):488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 44.Hu S, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121(20):4021–4031. doi: 10.1182/blood-2012-10-460063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson NA, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30(28):3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kluiver J, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207(2):243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 48.Shaffer AL. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodnow CC, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334(6184):676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 50.Kaji T, et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J Exp Med. 2012;209(11):2079–2097. doi: 10.1084/jem.20120127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.