Abstract
RNA splicing is a major contributor to total transcriptome complexity; however, the functional role and regulation of splicing in heart failure remain poorly understood. Here, we used a total transcriptome profiling and bioinformatic analysis approach and identified a muscle-specific isoform of an RNA splicing regulator, RBFox1 (also known as A2BP1), as a prominent regulator of alternative RNA splicing during heart failure. Evaluation of developing murine and zebrafish hearts revealed that RBFox1 is induced during postnatal cardiac maturation. However, we found that RBFox1 is markedly diminished in failing human and mouse hearts. In a mouse model, RBFox1 deficiency in the heart promoted pressure overload–induced heart failure. We determined that RBFox1 is a potent regulator of RNA splicing and is required for a conserved splicing process of transcription factor MEF2 family members that yields different MEF2 isoforms with differential effects on cardiac hypertrophic gene expression. Finally, induction of RBFox1 expression in murine pressure overload models substantially attenuated cardiac hypertrophy and pathological manifestations. Together, this study identifies regulation of RNA splicing by RBFox1 as an important player in transcriptome reprogramming during heart failure that influence pathogenesis of the disease.
Introduction
RNA splicing is a ubiquitous posttranscriptional process for all multiexon genes in eukaryotes. Alternative mRNA splicing from a single gene yields multiple mature transcripts, which contributes to the total complexity of the human transcriptome and is important for refining cellular identity and function (1, 2). This posttranscriptional regulatory process is essential to cellular specificity, with an estimated approximately 100,000 intermediate to high abundant alternative splice events identified in major human tissues (3). Alternative RNA splicing is regulated by cis-regulatory enhancers and silencers located within pre-mRNAs interacting with trans-acting splicing factors. Misregulated alternative RNA splicing events have a significant role in several human diseases (4–8).
It is well established that alternative mRNA splicing affects a broad spectrum of cardiac genes during normal development as well as pathological manifestation of heart diseases (9–13). For example, the central fructose-metabolizing enzyme in heart is ketohexokinase, which is shown to be regulated during pathological cardiac hypertrophy through the splicing factor SF3B1 (14). Several splicing factors have also been implicated in the regulation of cardiac function. Inactivation of the heterogeneous nuclear ribonucleoprotein U (hnRNP U) in mouse hearts causes lethal dilated cardiomyopathy (15). In addition, mutations of the splicing regulator RBM20 are associated with dilated human cardiomyopathy, and the reduced expression of RBM20 has been suggested to affect several cardiac genes shown to be involved in DCM, including titin (16–19). However, the underlying molecular mechanisms and the specific contribution of RNA splicing to the pathogenesis of stress-induced heart failure (HF) still remain to be fully explored.
In this report, we analyzed the global RNA splicing events in pressure overload–induced failing mouse hearts and identified a muscle-specific isoform of RBFox1 (also known as A2BP1) as a key trans-acting RNA splicing regulator in cardiomyocytes during hypertrophy and HF. RBFox1 has enriched expression in brain and striated muscle cells. RBFox1 expression in brain is associated with autism (20). In addition, RBFox family members, most frequently RBFox1 and RBFox2, appear to be important for heart and skeletal muscle development and function (21, 22). However, the role of RBFox1 in postnatal cardiac function under normal physiological and pathological conditions of induced HF as well as the downstream targets mediated by RBFox1 in the heart remain to be explored. In this study, we found that RBFox1 expression was significantly diminished in both mouse and human failing hearts. RBFox1 expression was essential for normal heart function in the developing zebrafish, and loss of RBFox1 expression significantly aggravated pressure overload–induced cardiac hypertrophy and failure in mice. At a mechanistic level, we found that RBFox1 had a global effect on cardiac mRNA splicing in cardiomyocytes and directly regulated an isoform switch from α1 to α2 in splice variants of the transcription factor family MEF2. RBFox1-mediated Mef2 splicing contributed to cardiomyocyte hypertrophy and pathological gene induction. Most remarkably, cardiac-specific reexpression of RBFox1 could significantly attenuate pressure overload–induced pathological hypertrophy and HF in mice. Therefore, our study reveals for what we believe to be the first time that RBFox1-dependent RNA splicing, in particular an isoform switch of Mef2 gene splice variants, is a regulatory circuit in cardiac transcriptional reprogramming, with a significant effect on the pathogenesis of HF.
Results
RBFox1 is a key splicing regulator in cardiomyocytes, mediating a large number of splicing events during cardiac stress.
In a previous study, we profiled transcriptome-wide alternative RNA splicing events associated with pressure overload–induced HF in mice (23). In an attempt to further analyze the RNA sequencing (RNA-seq) result, a random subset of 32 alternative splicing events (out of 28 genes) identified to be markedly changed in the failing mouse heart was tested by qRT-PCR in neonatal, normal adult, and pressure overload–induced mouse failing hearts (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI84015DS1). All of the splicing events showed reciprocal expression changes during postnatal cardiac maturation versus HF (Supplemental Figure 1). This observation reveals a “fetal-like” alternative RNA splicing pattern in the failing heart parallel to a similar pattern of expression for pathological genes.
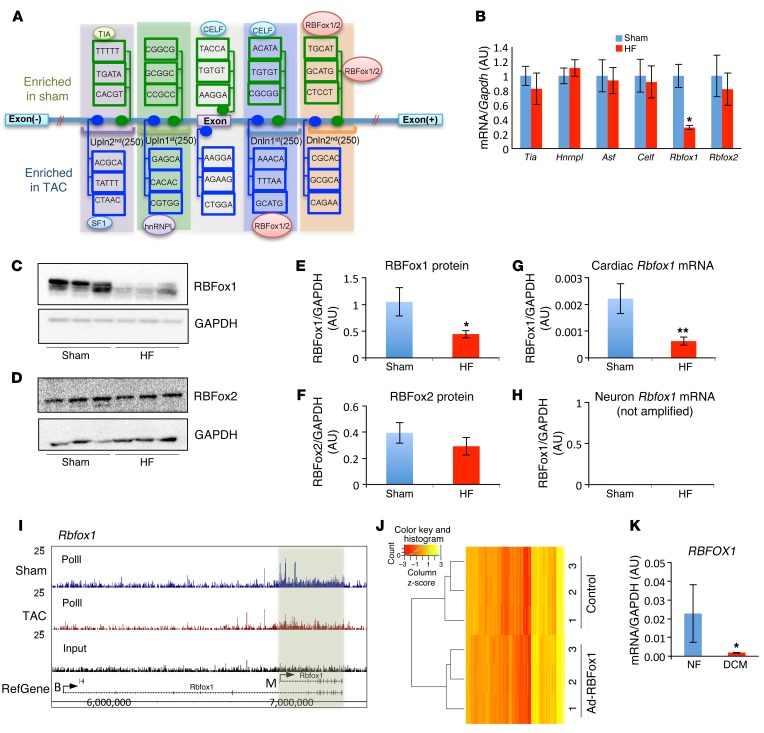
To investigate the underlying molecular basis of such coordinated RNA splicing regulation, we performed a de novo motif discovery analysis for all affected exons in the proximal regions (500 bp of upstream and downstream introns and exons) for the known trans-acting RNA splicing regulators (24). A number of significantly enriched binding motifs, including CELF, TIA, ASF, hnRNPL, and RBFox1/2, were identified (Figure 1A and Supplemental Table 2). The mRNA expression levels for Celf, Tia, Asf, and Hnrnpl were not significantly changed in failing mouse hearts compared with those in normal controls (Figure 1B); neither were expression levels for 22 other known RNA splicing regulators, including those previously implicated in cardiac RNA splicing and the pathogenesis of cardiomyopathy, such as RBM20 (18, 19, 25), SC35 (13), and MBNL2 (ref. 26 and Supplemental Figure 2). Consistent with a recent report (27), RBFox2 expression was only modestly reduced at the protein level in mouse hypertrophic hearts but not significantly affected at the mRNA level (Figure 1, B, D, and F). In contrast, the cardiac isoform of RBFox1 expression showed a significant reduction at both mRNA and protein levels (Figure 1, B, C, E, G, and H). Reduced RNA polymerase II occupancy at the Rbfox1 (but not Rbfox2) locus supports a transcriptional mechanism of RBFox1 repression in the diseased heart (ref. 28 and Figure 1I). In contrast to its diminished expression in the diseased heart, RBFox1 was significantly increased during cardiac development in both zebrafish and mice (Supplemental Figure 3). Therefore, RBFox1 is a potential RNA splicing regulator, with a dynamic expression pattern parallel, with “fetal-like” RNA splicing changes in HF.
Figure 1. RBFox1 is a key splicing regulator repressed in failing hearts.
(A) Schematic of de novo motif discovery from alternative splicing events. A total of five regions — exon; upstream first 250 bp (Upln 1st); upstream second 250 bp (Upln 2nd); downstream first 250 bp (Dnln 1st); and downstream second 250 bp (Dnln 2nd) — were analyzed. The enriched and conserved motif were indicated with their binding protein. (B) Relative mRNA levels of 6 enriched splicing regulators in sham hearts and hearts after TAC (HF) (n = 3 from each group). Data in both C and D were normalized to GAPDH. Western Blot analysis of RBFox1 and RBFox2 expression levels in normal (Sham) and TAC-induced failing hearts (HF) (n = 3 each sample). (E and F) Quantification of protein expression levels of (E) RBFox1 and (F) RBFox2 in sham-operated hearts compared with those in TAC-induced failing hearts based on Western blot shown in C and D. (G and H) Real-time PCR was performed in TAC- and sham-operated mouse hearts using primers (see Supplemental Table 3) designed specifically targeting either (G) cardiac or (H) neuron Rbfox1 splicing variants (n = 3 each sample). (I) RNA polymerase II occupation on the mouse Rbfox1 gene in sham-operated hearts compared with that in hearts 4 days after TAC. A fragment density of 25 is shown throughout. (J) Heatmap depicting sample-scaled expression of 132 exons significantly changed in the RBFox1-expressing NRVMs identified by RNA-seq and RASL-seq. The blue line in the key is a histogram of the values plotted in the heatmap. (K) Quantification of RBFOX1 mRNA expression in nonfailing (NF) and dilated cardiomyopathy (DCM) human heart samples (n = 4 from each group). *P < 0.05, **P < 0.01, Student’s t test (B, E, G, and K).
In order to identify potential downstream RNA splicing events regulated by RBFox1 in the heart, we performed RNA-seq analysis in neonatal rat ventricular myocytes (NRVMs) at basal conditions (with low endogenous RBFox1 expression) and upon adenovirus-mediated RBFox1 expression. A global shift of RNA splicing profile was detected in RBFox1-expressing cardiomyocytes (Figure 1J and Supplemental Table 4). Most relevant to clinical HF, marked loss of RBFOX1 mRNA expression was also observed in human dilated cardiomyopathy hearts (Figure 1K).
RBFox1 directly regulates MEF2 transcription factor mutually exclusive alternative splicing.
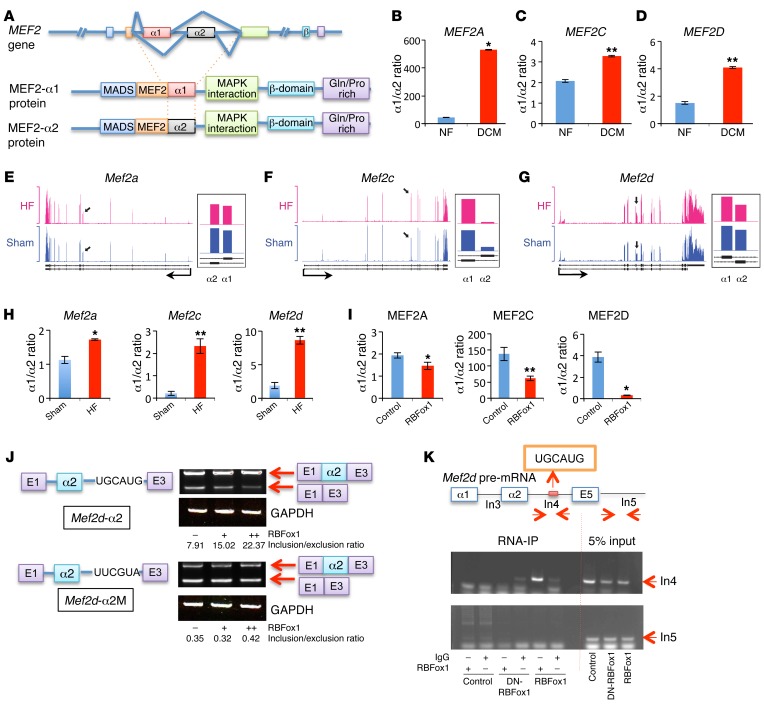
Among the alternatively spliced events identified in both the RBFox1-expressing NRVMs and the failing mouse hearts (23), we found and validated an alternative splicing event for all members of the MEF2 family (MEF2A, MEF2C, and MEF2D) of transcription factors, involving a mutually exclusive utilization of exon α1 versus exon α2. The Mef2 α exons encode a domain 3′ to the MADS/MEF2 motif (refs. 29, 30, and Figure 2A), and the α1- versus the α2-containing isoform of MEF2D was reported to confer differential transcriptional activity in skeletal muscle (31). In failing mouse and human hearts, MEF2A, MEF2C, and MEF2D α1/α2 ratios were increased in association with RBFox1 repression (Figure 2, B–H), while ectopic expression of RBFox1 in NRVMs significantly reduced the α1/α2 ratios for all Mef2 genes (Figure 2I). In contrast, exon β, another alternatively spliced was not consistently affected in the failing mouse heart (Supplemental Figure 4). By motif analysis, we found a consensus RBFox-binding site located near the α1 or α2 exon for the Mef2 genes across multiple vertebrate species (Supplemental Figure 4 and ref. 32). By testing a minigene reporter (33) containing the mouse Mef2d α2 exon/intron sequences in both noncardiomyocytes and NVRMs, RBFox1 expression was shown to be sufficient to promote its splicing inclusion in an RBFox1-binding motif–dependent manner (Figure 2J and Supplemental Figure 5A). A similar effect was also observed for a reporter construct containing the mouse Mef2a α2 exon/intron sequences (Supplemental Figure 5B). Furthermore, cross-linking followed by RBFox1 immunoprecipitation (cross-link RNA immunoprecipitation) and RT-PCR analysis in C2C12 myocytes detected specific enrichment of RBFox1 binding to the endogenous RBFox1-binding motifs near the Mef2a, Mef2c, and Mef2d α2 exons (Figure 2K and Supplemental Figure 5, C and D). All of this evidence suggests that the MEF2 α2 exons are direct downstream targets of RBFox1-mediated splicing regulation in myocytes.
Figure 2. RBFox1 specifically regulates MEF2 α exon inclusion change in failing hearts.
(A) Schematic of the MEF2 gene splicing variants at α exon. (B–D) Quantification of the MEF2 α1/α2 ratio in nonfailing (NF) and dilated cardiomyopathy (DCM) human heart samples (n = 4 from each group). (E–G) Exon-specific RNA reads of the Mef2 genes in failing heart and sham samples based on an RNA-seq data set (43). Black arrows represent the locations of mutually exclusive exons (MXEs) in each gene, and the detail expression profiles (presented as reads per kilobase per million mapped read values) of MXEs at higher magnification are shown to the right. (H) qRT-PCR quantification of Mef2 (Mef2a, Mef2c, and Mef2d) α1 versus α2 transcript ratios from sham hearts (Control) and hearts after TAC (HF) (n = 3 from each group). (I) MEF2 α1/α2 transcript ratio in the control and RBFox1-overexpressing NRVMs (n = 3 from each group). (J) Schematic of the Mef2d α2 exon minigene reporter constructs containing wild-type (Mef2d-α2) or mutated (Mef2d-α2M) RBFox1-binding motif as indicated. Different minigene reporter constructs were transfected alone or in combination with an RBFox1-expressing vector in HEK293 cells. Exon inclusion level was measured by densitometry analysis of RT-PCR products separated by electrophoresis on a 4% agarose gel and indicated as inclusion/exclusion ratio. (K) Cross-link RNA immunoprecipitation assay of Mef2d pre-mRNA sequence. Myoblasts were infected with dominant-negative RBFox1 (DN-RBFox1) and RBFox1 and compared with the mock infected cells. RBFox1 binding to different regions of Mef2d pre-mRNA was detected by semiquantitative RT-PCR as indicated. *P < 0.05, **P < 0.01, Student’s t test (B–D, H, and I).
Genetic inactivation of Rbfox1 promotes cardiac hypertrophy and HF.
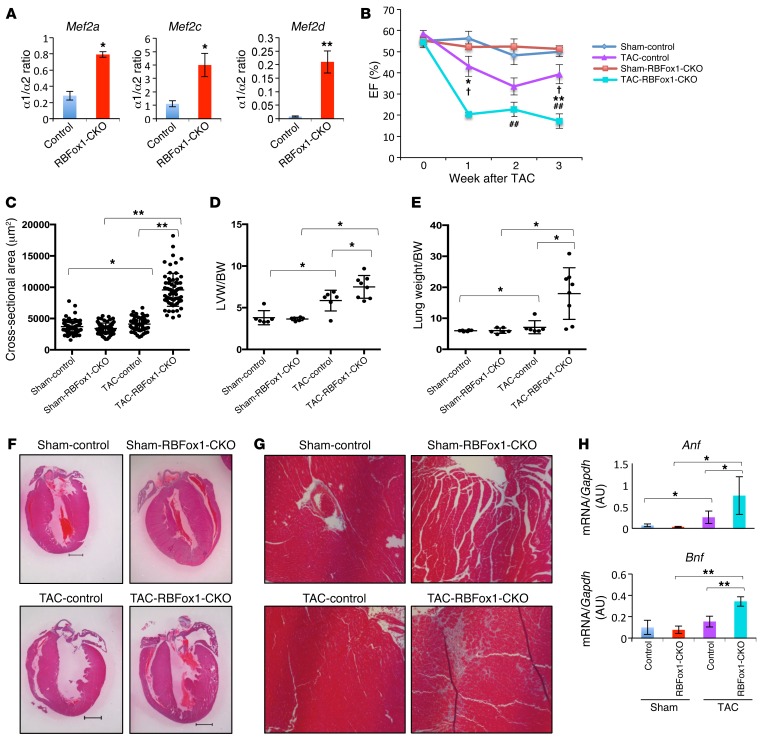
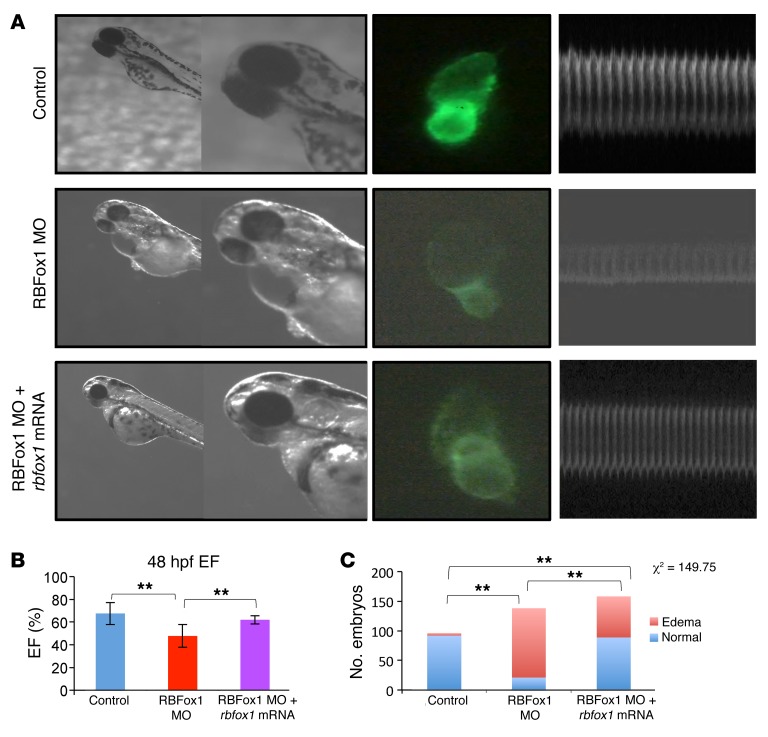
To determine the functional relevance of RBFox1 inactivation in the pathogenesis of cardiac hypertrophy and HF, we generated conditional knockout mice (RBFox1-CKO mice) by cross-breeding Nkx2.5-Cre knockin mice (34) with Rbfox1fl/fl mice (35). Cardiac Rbfox1 expression was markedly diminished in RBFox1-CKO hearts compared with that in control genotypes, without affecting the expression of Rbfox2 (Supplemental Figures 6 and 7). In RBFox1-CKO hearts, the exon α1/α2 ratios were significantly increased for the mouse Mef2a, Mef2c, and Mef2d genes (Figure 3A), phenocopying what was observed in the failing hearts (Figure 2H). In contrast, the inclusion of the Mef2 β exon was not affected (Supplemental Figure 8). RBFox1-CKO mice showed normal cardiac function at 2 months of age but manifested a modest but statistically significant decrease in cardiac function at 6 months of age compared with that of the control genotypes (Supplemental Figure 9A). However, in response to pressure overload induced by transaortic constriction (TAC) at 2 month of age, RBFox1-CKO mice developed HF much earlier and with significantly more severe functional deterioration, ventricular hypertrophy, fibrotic remodeling, and pathologic marker gene expression (Figure 3, B–H; Supplemental Figure 9, B and C; and Supplemental Figure 10). Similar cardiac defects were observed in zebrafish by morpholino inactivation of RBFox1 (Figure 4). These data suggest that loss of RBFox1 expression, as observed in the failing heart, has a direct effect on Mef2 alternative splicing in the intact heart and promotes pressure overload–induced HF in vivo.
Figure 3. Genetic inactivation of Rbfox1 exacerbates cardiac hypertrophy and HF in mice.
(A) Mef2 α1/α2 transcript ratios in 3-month-old wild-type (Control) and RBFox1-CKO mouse hearts at basal state (n = 3 from each group). *P < 0.05, **P < 0.01. (B) Ejection fraction values of sham- and TAC-operated control and RBFox1-CKO littermates measured by echocardiography (Sham-control, n = 6; Sham-RBFox1-CKO, n = 6; TAC-control, n = 6; TAC-RBFox1-CKO, n = 8). *P < 0.05, **P < 0.01, TAC-control vs. TAC-RBFox1-CKO; †P < 0.05, Sham-RBFox1-CKO vs. TAC-RBFox1-CKO; ##P < 0.05, Sham-control vs. TAC-control. (C) Cross-sectional myofiber area in LVs of sham- and TAC-operated control and RBFox1-CKO hearts 3 weeks after TAC. The values were averaged from 60 slides prepared from 3 hearts in each group. *P < 0.05, **P < 0.01. (D) LV weight (LVW) and body weight (BW) ratios from sham- and TAC-operated control and RBFox1-CKO mice 3 weeks after TAC (control, n = 10; RBFox1-CKO, n = 9). *P < 0.05. (E) Lung weight and body weight ratios among the sham, control, and RBFox1-CKO mice 3 weeks after TAC (control, n = 10; RBFox1-CKO, n = 9). *P < 0.05. (F) Representative cross-sectional images of hematoxylin & eosin staining of LV tissues from control and RBFox1-CKO mice following sham surgery or 3 weeks after TAC. Original magnification, ×2. (G) Representative Masson’s trichrome staining of sham- and TAC-treated control and RBFox1-CKO mouse heart sections. Original magnification, ×20. Data are representative of at least 3 independent experiments. (H) Anf and Bnp expression levels in the sham- and TAC-treated hearts from control and RBFox1-CKO mice as indicated (n = 4 each group). *P < 0.05, **P < 0.01. Significant differences between groups were determined by Student’s t test (A) or multiway ANOVA (B–E and H).
Figure 4. Genetic inactivation of rbfox1 exacerbates cardiac hypertrophy and HF in zebrafish.
(A) Zebrafish were injected with control, RBFox1 morpholino (MO) alone, or in combination with zebrafish rbfox1l mRNA. Embryos were analyzed at 48 hours after fertilization (hpf) (original magnification, ×1 [first column]; ×11 [second column]), and hearts were visualized under fluorescent microscope (original magnification, ×11) and recorded to facilitate cardiac function analysis (zebrafish M-mode echocardiography) based on software as described elsewhere (9). (B) Quantification of zebrafish cardiac ejection fraction in the 3 experimental groups (n = 30 per sample). (C) Summary of injection and the number of pericardial edema observed. **P < 0.01, multiway ANOVA (B) or Fisher’s exact test (C).
RBFox1 mediates cardiomyocyte hypertrophy response.
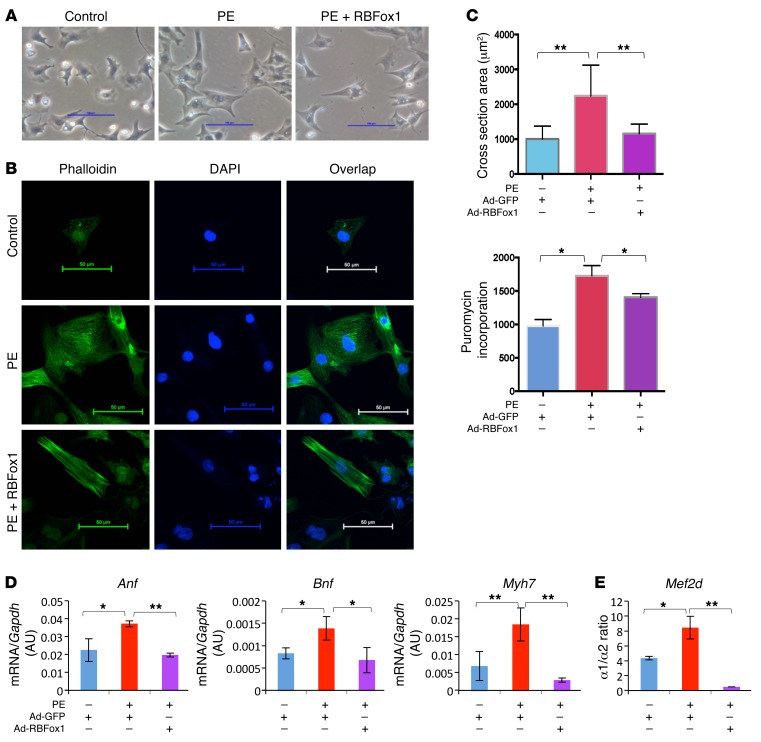
Our data indicate that loss of cardiac RBFox1 expression significantly contributes to pathological hypertrophy and HF. To demonstrate the direct impact of RBFox1 expression in cardiomyocytes, we ectopically expressed RBFox1 in NRVMs and observed attenuated cardiomyocyte hypertrophy in response to phenylephrine (PE) treatment (Figure 5, A–C) and hypertrophic gene expression (Figure 5D) in an RNA splicing–dependent manner (Supplemental Figure 11). Consistently, PE induced α2 to α1 switches for the Mef2d and Mef2c genes, which were also significantly blunted by RBFox1 expression (Figure 5E and Supplemental Figure 12).
Figure 5. RBFox1 plays important role in cardiomyocyte hypertrophy.
(A) Representative bright-field images of NRVMs 48 hours after treatment with PE and infection with GFP (Control) or RBFox1-expressing adenovirus (RBFox1). Original magnification, ×20. (B) Phalloidin staining of NRVMs treated with PE alone or in combination with RBFox1-expressing adenovirus. Green, phalloidin; blue, Hoechst. Original magnification, ×40. Data are representative of at least 3 independent experiments. (C) Cell surface area of NRVMs from the same experimental groups as in A. Cells were stained with wheat germ agglutinin. 100 cells were measured from total of 3 independent experimental samples. Protein synthesis rates as quantified by puromycin incorporation in NRVMs from the same experimental samples as in A following 30 minutes of puromycin labeling (n = 3 each sample). (D) Anf, Bnp, and Myh7 expression in NRVMs following 48 hours of PE treatment in combination with GFP or RBFox1 adenovirus infection (n = 3 each group). (E) Relative Mef2d α1/α2 transcript ratios from the same sample group as in C. *P < 0.05, **P < 0.01, Student’s t test (C–E).
RBFox1/Mef2 gene regulatory circuit in HF.
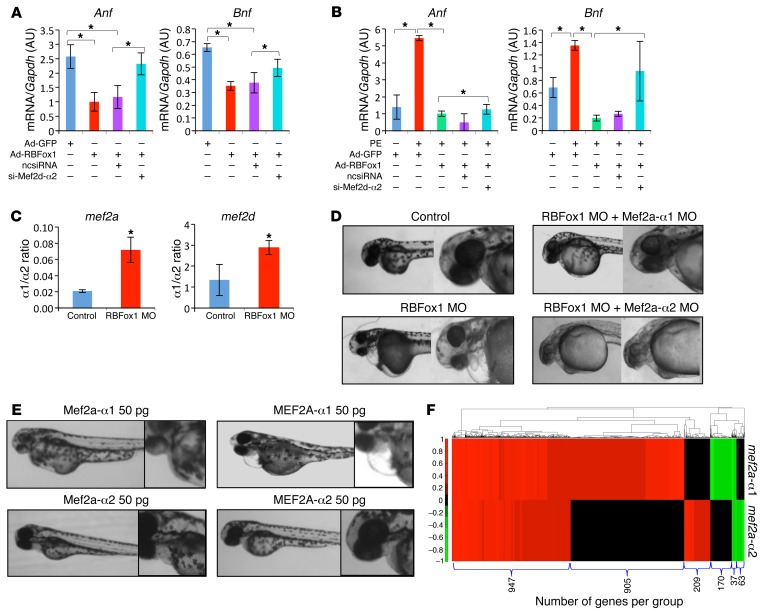
In order to establish the role of the α1 to α2 switch of Mef2 in RBFox1-mediated regulation of cardiac hypertrophy, we first tested the contribution of Mef2 splicing variants in cardiomyocyte hypertrophy in vitro. By using a MEF2D α2–specific siRNA (Supplemental Figure 13), we found that MEF2D α2 variant expression was necessary for RBFox1-mediated attenuation of hypertrophic gene expression in the NRVMs under basal conditions (Figure 6A) as well as PE-treated conditions (Figure 6B). A similar effect was observed for the MEF2C α2 variant (Supplemental Figure 14). Therefore, RBFox1-mediated MEF2C and MEF2D α1 to α2 isoform switch is important for its antihypertrophic effect. In good agreement with an early study in zebrafish (36), inactivation of RBFox1 led to a change of the mef2a and mef2d α1/α2 ratio and HF (Figure 6C). Remarkably, the HF phenotype caused by RBFox1 knockdown was markedly rescued by simultaneous knockdown of the Mef2a α1 variant but not the Mef2a α2 variant (Figure 6D and Supplemental Figure 15). Conversely, overexpression of zebrafish (Mef2a) or mouse (MEF2a) Mef2a α1 but not the α2 splicing variant was sufficient to cause a similar HF phenotype in the developing zebrafish (Figure 6E and Supplemental Figure 16). Finally, RNA-seq analysis from zebrafish expressing α1 or α2 Mef2a variants revealed global differences in gene expression (Figure 6F). All these results suggest that RBFox1 is a potent regulator of cardiac hypertrophy and its downstream splicing variants of the Mef2 genes lead to different effects on the cardiac transcriptome and cardiac pathology.
Figure 6. RBFox1/MEF2 regulatory circuit in cardiomyocyte hypertrophy regulation.
(A) Anf and Bnp expression in NRVMs following GFP or RBFox1 expression, with or without coexpressing a scrambled siRNA (ncsiRNA) or an siRNA targeting Mef2d α2 isoform (si-Mef2d-α2) as indicated (n = 3 each sample). (B) Anf and Bnp expression in NRVMs with or without PE treatment or RBFox1 expression or coexpressing a scrambled siRNA or an siRNA targeted to Mef2d α2 isoform (n = 3 each sample). (C) mef2a and mef2d α1/α2 ratio in RBFox1 morpholino–injected hearts compared with control zebrafish embryo hearts (n = 3 each sample). (D) Zebrafish phenotype upon RBFox1 and Mef2a inactivation. Zebrafish embryos were injected with morpholino targeting RBFox1 alone or in combination with Mef2a α1 or α2 isoform–specific morpholino. Embryo phenotype was analyzed at 48 hours after fertilization. Original magnification, ×1 (first column); ×11 (second column). (E) Zebrafish phenotype upon expression of zebrafish Mef2a α1 or zebrafish Mef2a α2, mouse MEF2A α1, and mouse MEF2A α2 at indicated dose imaged at 48 hours after fertilization. Original magnification, ×1 (first column); ×11 (second column). (F) Gene expression profile in the zebrafish embryos 24 hours following expression of individual mef2a α1 or α2 isoforms analyzed by RNA-seq. The heatmap was generated using significantly upregulated (red) and downregulated (green) genes. *P < 0.05, Student’s t test (A–C).
Restoring RBFox1 expression prevented pathological hypertrophy.
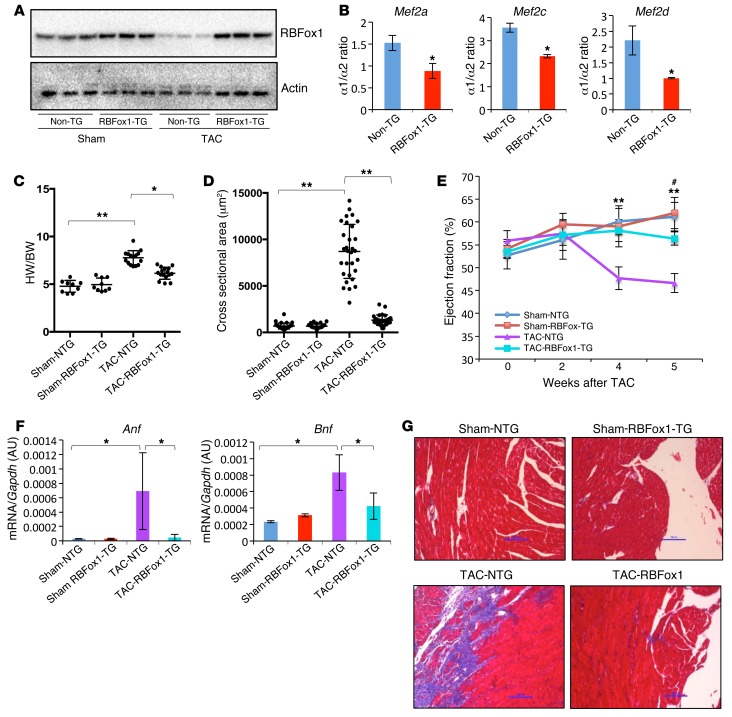
In vitro study suggests that RBFox1 expression can protect cardiomyocytes against pathological hypertrophy. To validate this observation in vivo, we generated a transgenic mouse line with cardiac-specific and inducible expression of RBFox1 (ref. 37, Figure 7A, and Supplemental Figures 17 and 18). Consistent with our earlier in vitro observations, doxycycline-induced RBFox1 expression in the heart also resulted in a substantial and specific enhancement of the α2 exon inclusion for all the Mef2 genes in heart, without affecting the relative inclusion of the β exon (Figure 7B and Supplemental Figure 8). Remarkably, RBFox1 expression significantly attenuated pressure overload–induced cardiac hypertrophy (Figure 7, C and D), contractile dysfunction (Figure 7E), pathological gene expression (Figure 7F), and fibrotic remodeling based on histology and molecular markers (Figure 7G and Supplemental Figure 19). These data support the notion that restoring RBFox1 expression can ameliorate broad features of pathological hypertrophy and HF in the intact hearts.
Figure 7. Restoring RBFox1 prevented pathological hypertrophy in pressure-overloaded mouse hearts.
(A) Western blot analysis of RBFox1 protein in wild-type or single-transgenic hearts (Non-TG) compared with double-transgenic hearts (RBFox1-TG) 6 weeks following sham or TAC operation. (B) Quantification of Mef2 α1/α2 transcript ratio in non-TG and RBFox1-TG hearts after 2 weeks of doxycycline induction (n = 3 each sample). *P < 0.05. (C) Heart weight (HW) and body weight ratios in sham-operated mice and non-TG (NTG) and RBFox1-TG mice 6 weeks after TAC (Sham-NTG, n = 9; Sham-RBFox1-TG, n = 9; TAC-NTG, n = 15; TAC-RBFox1-TG, n = 16). *P < 0.05, **P < 0.01. (D) Cross-sectional area of cardiomyocytes in sham-operated mice and non-TG or RBFox1-TG mice after TAC. Average values were derived from 100 myocytes of from each group. **P < 0.05. (E) Ejection fraction of sham- and TAC-operated non-TG and RBFox1-TG mice measured by echocardiography up to 5 weeks after TAC (Sham-NTG, n = 9; Sham-RBFox1-TG, n = 9; TAC-NTG, n = 15; TAC-RBFox1-TG, n = 16). **P < 0.05, TAC-NTG vs. TAC-RBFox1-TG. #P < 0.05, Sham-NTG vs. TAC-NTG. (F) Anf and Bnp mRNA expression levels in non-TG and RBFox1-TG mice following sham surgery or 5 weeks after TAC (n = 3 each group). *P < 0.05. (G) Representative images of Masson trichrome–stained ventricular sections from the sham- and TAC-operated non-TG and RBFox1-TG mice as indicated. Original magnification, ×20. Data are representative of at least 3 independent experiments. Significant differences between groups were determined by Student’s t test (B) or multiway ANOVA (C–F).
RBFox1 contributed to global RNA splicing reprogramming during HF.
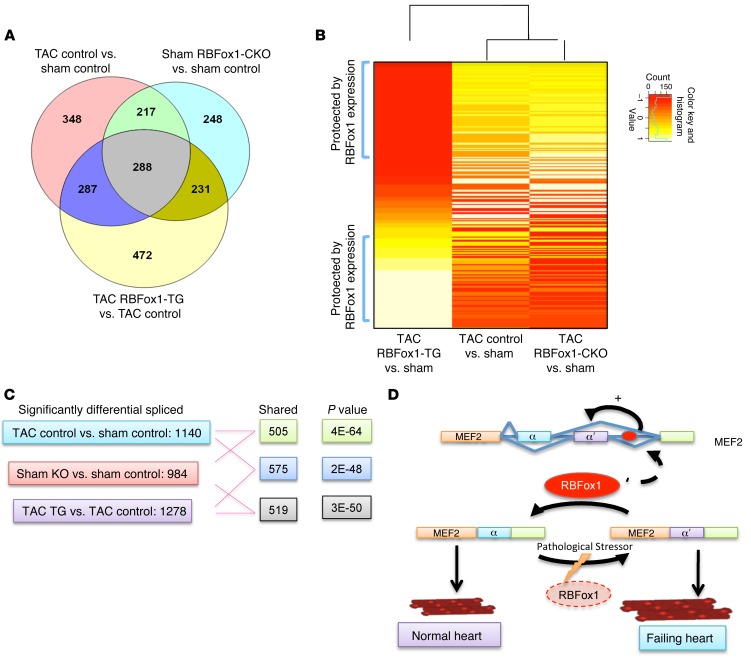
Finally, in order to determine the specific contribution of RBFox1 to global RNA splicing reprogramming during HF, we performed a genome-wide RNA splicing profiling study by RNA annealing, selection, and ligation sequencing (RASL-seq) analysis (38) in cardiac tissues from control, RBFox1-CKO, and RBFox1 transgenic (RBFox1-TG) mice following sham surgery or pressure overload (Supplemental Figure 20). We detected a total of 1,140 significantly changed exon splicing events in the TAC-induced failing mouse hearts compared with that in the controls. Remarkably, 505 of them were also affected by cardiac-specific Rbfox1 knockout (Figure 8, A and C), suggesting that RBFox1 inactivation may contribute up to nearly 44.3% of the exon splicing changes induced by TAC (P < 4E–64). Similarly, 575 of these exons were also affected in RBFox1-TG hearts, again suggesting that RBFox1 reexpression affects 50.4% of TAC-induced exon splicing events (P < 3E–50). Most importantly, clustering analysis from significantly changed splicing events shared among all 3 genotypes showed that RNA splicing profiles from RBFox1-CKO and RBFox1-TG hearts were diagonally located at the opposite direction from those of wild-type hearts following TAC (Figure 8B). While tissues from RBFox1-CKO animals expanded and augmented the RNA splicing changes in wild-type failing hearts, RBFox1-TG tissue showed a reduced or reversed effect on the majority of them. All of these data support the notion that loss of RBFox1-mediated RNA splicing has a major contribution to the global RNA splicing defects observed in the pathologically stressed hearts. Restoring RBFox1 activity can ameliorate a significant degree of global RNA splicing reprogramming, reinforcing its protective effect against hypertrophy and dysfunction.
Figure 8. RBFox1 contributes to global RNA splicing reprogramming during HF.
(A) Venn diagram showing differentially spliced exons in hearts identified by RASL-seq (WT sham, n = 5; WT TAC, n = 7; RBFox1-TG TAC, n = 3; RBFox1-CKO sham, n = 3) (RASL-seq was also performed for RBFox1-TG sham and RBFox1-CKO TAC [data not shown]; RBFox1-TG sham, n = 4; RBFox1-CKO TAC, n = 3). (B) Heatmap depicting the 339 differentially spliced exons between TAC- and sham-operated mice identified in all 3 genotypes (WT, RBFox-TG, and RBFox-CKO). Data are row-scaled, and the blue bar in the key is a histogram of the splicing values plotted in the heatmap. (C) Shared splicing events across different samples together with P values. (D) Graphic abstract.
Discussion
A posttranscriptional gene regulatory circuit we believe is novel in HF.
Although RNA splicing is a well-recognized contributor to transcriptome complexity and alternative RNA splicing variants are commonly observed among many cardiac genes during development and diseases, our knowledge regarding their underlying regulatory mechanisms and the functional impact is still very limited. Alternative RNA splicing is regulated by cis-regulatory enhancers and silencers located within pre-mRNAs interacting with trans-acting splicing factors (1, 2, 39–41). Mutations in several splicing factors, including SC35 and RBM20, have been implicated in dilated cardiomyopathy in mice, rats, and humans (18, 42, 43). Other splicing factors are also shown to play important roles in cardiac development and function, including CUGBP1, ETR-like factors, CELF/Bruno-like RNA-binding proteins, and muscle-blind-like (MBNL) proteins (27, 44–46). However, RNA splicing factors directly involved in stress-induced cardiac hypertrophy and pathological remodeling have not been systematically characterized. In this report, using both unbiased bioinformatics analysis and experimental validations, we have uncovered a muscle-specific form of RNA splicing regulator RBFox1, which has a major contribution to stress-induced cardiac RNA splicing reprogramming. Furthermore, we demonstrate that a mutually exclusive RNA splicing event for the α exon utilization of the transcription factor MEF2 family members is a direct downstream target of RBFox1 and has a critical role in the development of cardiac hypertrophy and HF. While genetic inactivation of Rbfox1 promotes pathological remodeling in stressed mouse hearts and HF in developing zebrafish embryos, RBFox1 expression exerts a marked protective effect on a broad spectrum of cardiac pathological remodeling in pressure-overloaded hearts, including cardiomyocyte hypertrophy and collagen gene expression. In summary, our study uncovered a previously uncharacterized RBFox1/MEF2 regulatory circuit in HF.
RBFox1/MEF2 in transcriptome programming in the heart.
It is known that alternative RNA splicing affects many genes in the heart, including genes encoding structural proteins, such as cardiac tropononin T and titin, or signaling molecules, such as Ca2+/calmodulin-dependent protein kinase (11–13). The mutually exclusive splicing of all the Mef2 genes at their α exon affects a region adjacent in the 3′ direction to the MEF2- and DNA-binding MADS box domains. A recent study reported that the α1 isoform of MEF2D is ubiquitously expressed, whereas the α2 isoform is muscle specific. Furthermore, this study found that the two isoforms have similar DNA-binding profiles across the genome in skeletal muscle but different transcriptional activity due to differential phosphorylation by PKA and binding partners acting as corepressors or coactivators (47). Our data from cardiomyocytes (Figure 6) argue for qualitatively different functions for the MEF2 splice variants containing the α1 exon or the α2 exon. Although MEF2 has a well-established role as a transcriptional activator for cardiogenic and hypertrophic genes during development and disease, our results indicate that the MEF2 α2 isoform contributes to RBFox1-mediated hypertrophic gene repression rather than activation (Figure 6). The underlying molecular basis for these different functions remains to be established but could involve different cofactors and/or targeting to different genes. The same RBFox1-binding motif is present near the targeted Mef2 α2 exon across multiple vertebrate species, including zebrafish, mice, rats, and humans (Supplemental Figure 6), suggesting that the RBFox1/MEF2 regulatory circuit is a highly conserved mechanism in cardiac transcriptome regulation. RBFox2 is reported to regulate Mef2d α exon splicing in skeletal muscle myogenesis (47, 48). A more recent study suggests that loss of RBFox2 expression is also associated with pressure overload–induced hypertrophy and HF (27). Our in vivo analysis indicates that RBFox2 expression is higher in the neonatal heart but markedly reduced in the mature adult mouse heart, in contrast with that after RBFox1 induction in the postnatal hearts. Therefore, RBFox1 and RBFox2 may have both overlapping and unique functions during cardiac maturation and pathogenesis, with RBFox1 being a predominant player in the adult heart.
The RBFox1/MEF2 circuit in HF regulation and therapy.
Our study shows that a single RNA splicing regulator can exert a profound effect on a broad spectrum of pathological features in the diseased heart. Based on both total transcriptome profiling using RNA-seq and RNA splicing profiling by RASL-seq, RBFox1-mediated RNA splicing has a major contribution to the global RNA splicing changes in the diseased heart (Figure 7). This remarkable contribution underscores the significant impact of RBFox1 inactivation or reexpression on the pathogenesis of HF. Rbfox1 deletion is associated with a complex form of congenital cardiomyopathy. Most relevantly, the correlated loss of RBFox1 expression and the splicing changes of the MEF2 α exon are observed in the human DCM hearts (Figures 1 and 2). Therefore, RBFox1-mediated RNA splicing can be a potential new target of intervention against pathological remodeling in the heart and restoring RBFox1 abundance and function can be explored as a novel therapeutic approach for HF.
Methods
Further details are described in the Supplemental Methods.
RBFox1-TG and cardiac-specific knockout mice.
Animals in this study were handled in accordance with the Guide for the Care and Use of Laboratory Animals (8th ed. The National Academies Press. 2011.). The detailed description of the generation of RBFox1-TG and cardiac-specific knockout mice is provided in the Supplemental Methods.
Tissue from human nonfailing and failing hearts.
The failing heart samples (n = 4) were obtained from the left ventricular (LV) anterior wall during heart transplantation or implantation of an LV assist device (49). The nonfailing heart samples (n = 4) were obtained from the LV free wall and procured from the National Disease Research Interchange and the University of Pennsylvania. Nonfailing heart donors had no history of macroscopic or laboratory signs of cardiac disease.
Motif analysis.
Motif analysis was carried out for the exonic regions and 4 intronic regions flanking differentially spliced exons obtained in our previous study (50). The entire exonic region was included. For intronic regions, up to 250 bases from the corresponding exon–intron boundary were included. For introns shorter than 500 bases, only half of the intronic regions were used. Previously described methods (51) were used for motif analysis and are briefly described below.
Motif conservation.
We analyzed sequence conservation of pentamers in the mouse intronic regions to identify potential splicing regulatory elements. The mouse introns were aligned to 7 other mammalian genomes that have at least 5 sequence coverage in the UCSC 28-way multigenome alignment (51). For each pentamer in each region, a conservation rate (CR) was calculated as the fraction of aligned and conserved occurrences among total occurrences. The significance of the CR of each pentamer was evaluated by comparing it with that of 10 other pentamers with similar expected CRs, calculated using the first-order Markov model. This procedure essentially controls for possible sequence bias in the data set. A P value was calculated by using the binomial distribution.
Motif enrichment.
In this analysis, to account for background sequence biases, the introns corresponding to each region were binned according to their GC frequency into 10 groups. Expected pentamer frequency was calculated for each pentamer by using the first-order Markov model in introns of each GC group, respectively. Pentamer enrichment was then evaluated by comparing the occurrence frequency of each pentamer to the overall expected frequency, calculated by summing up the expected counts of all GC groups. A P value was calculated by using the binomial distribution.
Pressure overload model of HF in mice.
LV tissues were collected from male C57BL/6 mice 8 weeks after the TAC procedure (which induced HF) and 1 day after birth (neonatal), respectively, and their corresponding sham controls as described previously (23). Doppler velocity measurements of right and left carotid arteries were obtained from TAC-treated mice to confirm the consistency of the surgery procedure. The HF status of the TAC-treated animals was established based on a significant increase in heart weight and a significant reduction in ejection fractions measured by echocardiography. All experimental procedure and echocardiogram analyses were performed blinded to the mouse genotype.
RNA polymerase II occupancy analysis.
RNA polymerase II occupancy data (28) was visualized at the RBFox1 locus for sham and TAC conditions using the Integrated Genome Browser (52).
Gene expression analysis.
Gene expression analysis via RNA-seq, RASL-seq, qRT-PCR, in-situ hybridization, and luciferase reporter gene assays is described in detail in the Supplemental Methods. RNA-seq and RASL-seq data are available in the NCBI’s BioProject database under accession numbers PRJNA295071 and PRJNA294802, respectively.
Statistics.
Data are expressed as mean ± SD. For comparison between two groups, differences were analyzed by 2-tailed Student’s t test. For comparison of multiple groups, differences were analyzed by 1-way ANOVA. P values ≤ 0.05 were considered as significant.
Study approval.
Human cardiac tissue collection was approved by the UCLA Institutional Review Board (11-001053 and 12-000207). Written consent from participants or their guardians was obtained.
Author contributions
CG conceived the project and designed and performed most of the molecular and genetic studies, data analysis, and interpretation. YW managed funding and participated in experimental design, data analysis, and manuscript preparation. SR performed TAC surgeries and cardiac function studies. JHL, CDR, DJC, MA, TMV, and XX supported bioinformatics analysis and manuscript preparations. JQ, YZ, and XDF contributed RASL-seq analysis. JNC provided support and models for zebrafish analysis. AN provided Nkx2.5-Cre mice as well as support for the phenotypic analysis.
Supplementary Material
Acknowledgments
The authors wish to thank Haiying Pu for her technical assistance and Douglas L. Black (UCLA) for reagents and discussion. This work was supported in part by grants from NIH (HL070079, HL103205, HL108186, and HL110667 to Y. Wang; HL096980 to J. Chen; HL115238 to T.M. Vondriska; R01HG006264 and U01HG007013 to X. Xiao), UCLA Clinical and Translational Science Institute Cardiovascular Pilot Team Research Grant (UL1TR000124 to Y. Wang, X. Xiao, J. Chen, and T.M. Vondriska), an American Heart Association Established Investigator Award (to Y. Wang), an Eli & Edythe Broad Center Predoctoral Fellowship in Stem Cell Science (to C. Gao), the Ruth L. Kirschstein National Research Service Award at UCLA (T32 HL-69766 to D.J. Chapski), and UCLA Cardiovascular Research Laboratories.
Footnotes
Jae-Hyung Lee’s present address is: Department of Life and Nano-pharmaceutical Sciences, Department of Maxillofacial Biomedical Engineering, School of Dentistry, Kyung Hee University, Seoul, South Korea.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(1):195–206. doi:10.1172/JCI84015.
References
- 1.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10(11):741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hallegger M, Llorian M, Smith CW. Alternative splicing: global insights. FEBS J. 2010;277(4):856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 5.Raghavachari N, et al. A systematic comparison and evaluation of high density exon arrays and RNA-seq technology used to unravel the peripheral blood transcriptome of sickle cell disease. BMC Med Genomics. 2012;5(1):28–48. doi: 10.1186/1755-8794-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda A, Valogne Y, Bou Nader M, Brechot C, Faivre J. An intron-retaining splice variant of human cyclin A2, expressed in adult differentiated tissues, induces a G1/S cell cycle arrest in vitro. PLoS One. 2012;7(6): doi: 10.1371/journal.pone.0039249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gang H, et al. A novel hypoxia-inducible spliced variant of mitochondrial death gene Bnip3 promotes survival of ventricular myocytes. Circ Res. 2011;108(9):1084–1092. doi: 10.1161/CIRCRESAHA.110.238709. [DOI] [PubMed] [Google Scholar]
- 8.Yae T, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3: doi: 10.1038/ncomms1892. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, et al. RBM24 is a major regulator of muscle-specific alternative splicing. Dev Cell. 2014;31(1):87–99. doi: 10.1016/j.devcel.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 10.Poon KL, et al. RNA-binding protein RBM24 is required for sarcomere assembly and heart contractility. Cardiovasc Res. 2012;94(3):418–427. doi: 10.1093/cvr/cvs095. [DOI] [PubMed] [Google Scholar]
- 11.Ramchatesingh J, Zahler AM, Neugebauer KM, Roth MB, Cooper TA. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15(9):4898–4907. doi: 10.1128/MCB.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120(1):59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 13.Ding JH, et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004;23(4):885–896. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirtschink P, et al. HIF-driven SF3B1 induces KHK-C to enforce fructolysis and heart disease. Nature. 2015;522(7557):444–449. doi: 10.1038/nature14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye J, et al. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc Natl Acad Sci U S A. 2015;112(23):E3020–E3029. doi: 10.1073/pnas.1508461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maatz H, et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124(8):3419–3430. doi: 10.1172/JCI74523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo W, Pleitner JM, Saupe KW, Greaser ML. Pathophysiological defects and transcriptional profiling in the RBM20–/– rat model. PLoS One. 2013;8(12): doi: 10.1371/journal.pone.0084281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo W, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18(5):766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Methawasin M, et al. Experimentally increasing titin compliance in a novel mouse model attenuates the Frank-Starling mechanism but has a beneficial effect on diastole. Circulation. 2014;129(19):1924–1936. doi: 10.1161/CIRCULATIONAHA.113.005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griswold AJ, et al. Targeted massively parallel sequencing of autism spectrum disorder-associated genes in a case control cohort reveals rare loss-of-function risk variants. Mol Autism. 2015;6(1): doi: 10.1186/s13229-015-0034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedrotti S, et al. The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function. Hum Mol Genet. 2015;24(8):2360–2374. doi: 10.1093/hmg/ddv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frese KS, et al. RNA splicing regulated by RBFOX1 is essential for cardiac function in zebrafish. J Cell Sci. 2015;128(16):3030–3040. doi: 10.1242/jcs.166850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JH, et al. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res. 2011;109(12):1332–1341. doi: 10.1161/CIRCRESAHA.111.249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao X, Wang Z, Jang M, Burge CB. Coevolutionary networks of splicing cis-regulatory elements. Proc Natl Acad Sci U S A. 2007;104(47):18583–18588. doi: 10.1073/pnas.0707349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Refaat MM, et al. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. 2012;9(3):390–396. doi: 10.1016/j.hrthm.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13(12):2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei C, et al. Repression of the central splicing regulator RBFox2 is functionally linked to pressure overload-induced heart failure. Cell Rep. doi: 10.1016/j.celrep.2015.02.013. [published online ahead of print March 3, 2015]. doi: 10.1016/j.celrep.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayed D, He M, Yang Z, Lin L, Abdellatif M. Transcriptional regulation patterns revealed by high resolution chromatin immunoprecipitation during cardiac hypertrophy. J Biol Chem. 2013;288(4):2546–2558. doi: 10.1074/jbc.M112.429449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276(5317):1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6(2):233–244. doi: 10.1016/S1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 31.Sebastian S, et al. Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev. 2013;27(11):1247–1259. doi: 10.1101/gad.215400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin Y, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22(4):905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boutz PL, et al. and Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21(13):1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harmon AW, Nakano A. Nkx2-5 lineage tracing visualizes the distribution of second heart field-derived aortic smooth muscle. Genesis. 2013;51(12):862–869. doi: 10.1002/dvg.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gehman LT, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43(7):706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallagher TL, et al. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol. 2011;359(2):251–261. doi: 10.1016/j.ydbio.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James J, et al. Forced expression of alpha-myosin heavy chain in the rabbit ventricle results in cardioprotection under cardiomyopathic conditions. Circulation. 2005;111(18):2339–2346. doi: 10.1161/01.CIR.0000164233.09448.B1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Qiu J, Fu XD. RASL-seq for massively parallel and quantitative analysis of gene expression. Curr Protoc Mol Biol. 2012;Chapter 4:Unit 4.13.1–9. doi: 10.1002/0471142727.mb0413s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 2010;38(9):2825–2838. doi: 10.1093/nar/gkq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321(5891):956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 41.Bland CS, et al. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 2010;38(21):7651–7664. doi: 10.1093/nar/gkq614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Guo W, Dewey CN, Greaser ML. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 2013;41(4):2659–2672. doi: 10.1093/nar/gks1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linke WA, Bucker S. King of hearts: a splicing factor rules cardiac proteins. Nat Med. 2012;18(5):660–661. doi: 10.1038/nm.2762. [DOI] [PubMed] [Google Scholar]
- 44.Dasgupta T, Ladd AN. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip Rev RNA. 2012;3(1):104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koshelev M, Sarma S, Price RE, Wehrens XH, Cooper TA. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum Mol Genet. 2010;19(6):1066–1075. doi: 10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24(7):653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh RK, et al. Rbfox2-coordinated alternative splicing of Mef2d and Rock2 controls myoblast fusion during myogenesis. Mol Cell. 2014;55(4):592–603. doi: 10.1016/j.molcel.2014.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Runfola V, Sebastian S, Dilworth FJ, Gabellini D. Rbfox proteins regulate tissue-specific alternative splicing of Mef2D required for muscle differentiation. J Cell Sci. 2015;128(4):631–637. doi: 10.1242/jcs.161059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahuja P, et al. Divergent mitochondrial biogenesis responses in human cardiomyopathy. Circulation. 2013;127(19):1957–1967. doi: 10.1161/CIRCULATIONAHA.112.001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JH, et al. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res. 2011;109(12):1332–1341. doi: 10.1161/CIRCRESAHA.111.249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalsotra A, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105(51):20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicol JW, Helt GA, Blanchard SG, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25(20):2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.