Abstract
In the past two decades, aptamers have emerged as a novel class of molecular recognition probes comprising uniquely-folded short RNA or single-stranded DNA oligonucleotides that bind to their cognate targets with high specificity and affinity. Aptamers, often referred to as “chemical antibodies”, possess several highly desirable features for clinical use. They can be chemically synthesized and are easily conjugated to a wide range of reporters for different applications, and are able to rapidly penetrate tissues. These advantages significantly enhance their clinical applicability, and render them excellent alternatives to antibody-based probes in cancer diagnostics and therapeutics. Aptamer probes based on fluorescence, colorimetry, magnetism, electrochemistry, and in conjunction with nanomaterials (e.g., nanoparticles, quantum dots, single-walled carbon nanotubes, and magnetic nanoparticles) have provided novel ultrasensitive cancer diagnostic strategies and assays. Furthermore, promising aptamer targeted-multimodal tumor imaging probes have been recently developed in conjunction with fluorescence, positron emission tomography (PET), single-photon emission computed tomography (SPECT), and magnetic resonance imaging (MRI). The capabilities of the aptamer-based platforms described herein underscore the great potential they hold for the future of cancer detection. In this review, we highlight the most prominent recent developments in this rapidly advancing field.
1. Introduction
Cancer is the second leading cause of death worldwide.1 Therefore, the implementation of highly sensitive and specific imaging modalities for timely cancer diagnosis and progression monitoring are of great clinical significance. To this end, molecular recognition probes such as monoclonal antibodies have significantly improved the performance of routine cancer diagnostics. A plethora of antibody-based methods, such as flow cytometry, cancer biomarker assays, immunohistochemical (IHC) staining, in vivo imaging, and many others have been widely used.2, 3 The suitability of protein antibodies for in vivo cancer applications, however, is severely hampered by several factors, such as their high immunogenicity, thermal instability, laborious and limited methods for chemical modification, and high production cost.4 In this respect, aptamers are considered excellent alternatives to supplement or replace antibody-based methodologies.
In the past two decades, aptamers have emerged as a novel class of oligonucleotide-based molecular recognition probes,5, 6 comprising multifunctional short RNA or single-stranded DNA oligonucleotides (usually 20–80 bases in length, molecular weight ~ 6–30 kDa) with unique three-dimensional structures that can recognize and bind to their targets with high specificity and affinity. Aptamer sequences are developed through an in vitro selection process known as SELEX (systematic evolution of ligands by exponential enrichment), which entails a series of repetitive selection and amplification steps after exposure to the target cell type or ligand.4, 7
Aptamers offer many competitive advantages over protein antibodies that significantly enhance their clinical applicability and suitability. Their most critical attribute, to recognize and bind to their cognate targets with high affinity and specificity, is achieved through the round-by-round SELEX process applied for their development. This target-specific binding takes place through a structural recognition process similar to the one mediating antibody-antigen reactions, and thus aptamers are often referred to as “chemical antibodies”. Their dissociation constants (Kd) are typically in the pico- to nanomolar range, which is comparable to that of antibodies.8–10 Notably, aptamers discriminate between closely related proteins or other molecules,11–14 a highly desirable feature rendering high specificity to aptamer-based diagnostics or therapeutics. Importantly, aptamers recognize a wide range of potential biological targets including ions, drugs, peptides, nucleic acids, proteins, viruses, live cells and tissues,15–22 and have the ability to change conformation upon binding to their targets.23 Furthermore, as small-sized oligonucleotides, aptamers are virtually nonimmunogenic and nontoxic in contrast to protein antibodies.24, 25 Due to their low molecular weights, aptamers penetrate tissue barriers and are rapidly internalized in tumor cells resulting in improved tumor-to-blood and tumor-to-normal tissue ratios, and thus high therapeutic indices or analytical sensitivities.26, 27 Aptamers also exhibit fast renal filtration and short circulating half-lives,28 which are beneficial for in vivo diagnostics as side effects due to fast blood clearance are avoided.29
For in vivo therapeutic purposes, oligonucleotide aptamers are synthesized by simple chemical procedures followed by a series of chemical or structural modifications to improve their bioavailability.4, 7 Aptamers are chemically and thermally stable and are rapidly and reproducibly synthesized at low cost as compared to antibodies. For example, we calculated that the cost per assay with the fluorophore-labeled CD4 aptamer, which was used as a flow cytometric probe in multicolored cell-phenotyping, was about 0.002 dollar per assay, whereas the cost per assay of CD4 antibody was 2 dollars.30 Importantly, aptamers are also easily modified by incorporation of different functional moieties, and aptamer-based analytical reagents can be easily stored or regenerated and reused.31
Many advances have been made in various biomedical fields since the onset of aptamer technology two decades ago. As chemical antibodies, aptamers provide an extremely advantageous alternative or supplement for protein antibodies in disease diagnostics and therapeutics.7 Herein, we highlight recent significant advances on aptamers as promising molecular recognition probes for targeting tumor biomarkers, cancer cell detection, tumor tissue IHC staining, and in vivo tumor imaging. Their applications in theranostics, combining concurrent real-time diagnostic and therapeutic competencies, will not be discussed as they have been presented in detail in one of our recent reviews.32
2. Cancer cell detection and biomarker analysis
Several valuable diagnostic biomarkers, such as circulating tumor cells (CTCs) and soluble tumor-related proteins may be present in the patient’s bloodstream even in the early stages of cancer. Accurate and sensitive detection of these biomarkers is of great significance for timely disease diagnosis and prognosis, evaluation of therapeutic effectiveness, and monitoring cancer recurrence and metastasis. However, it is a technical challenge to detect these biomarkers with high sensitivity due to their ultra-low concentration in the blood.33 In this respect, we and others recently developed an arsenal of detection methods using aptamers as sensitive recognition probes in combination with techniques based on fluorescence, colorimetry, magnetism, electrochemistry, and others. Several significant advances are outlined in this review.
2.1. Circulating tumor cell (CTC) detection
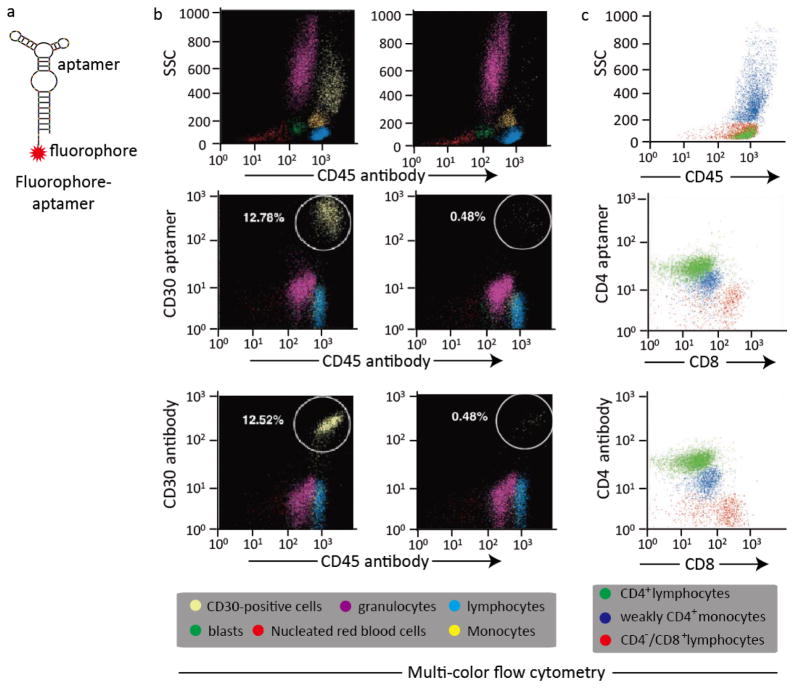
Fluorescent reporters provide one of the most widespread assaying strategies for CTC detection. Due to their high target-specific affinity, fluorophore-labeled aptamers can be used as simple, sensitive and versatile imaging probes not only for CTC detection but for other imaging purposes as well (Fig. 1a). Our recent study was the first to provide compelling evidence that aptamers can be used as alternatives or supplements to antibodies for multi-color flow cytometric analysis of cancer cells.34 In this study, we validated aptamer effectiveness in CTC detection by using the fluorophore Cy5-labeled CD30 aptamer probe to target CD30-positive lymphoma cells by multi-color flow cytometry. Firstly, CD30-positive anaplastic large cell lymphoma (ALCL) cells were mixed with fresh nucleated cells from the bone marrows of healthy donors to mimic a heterogeneous cell specimen, and the proportion of ALCL cells was adjusted to 0.5% and 13%. Then, the Cy5-labeled CD30 aptamer (FITC-labeled CD30 antibody as standard control) and AmCyan-labeled CD45 antibody were simultaneously incubated with the as-prepared cells. After incubation, the stained cells were subjected to flow cytometric analysis by using an LSRII flow cytometry (BD Bioscience, San Jose, CA, USA) with three-color channels. The individual cellular populations in the cell mixture including nucleated red blood cells, blasts, lymphocytes, granulocytes, monocytes and CD30-positive lymphoma cells were separated and gated based on the side scatter (SSC) and CD45 panels. Notably, as depicted in Fig. 1b, the CD30 aptamers could bind and detect target lymphoma cells with nearly identical staining patterns and numbers as the CD30 antibodies.34 In another study, we also investigated the applicability of the CD4 aptamers versus the CD4 antibodies for multicolored cell phenotyping in patients’ pleural fluids.30 In this study, cells from patients’ pleural fluids were collected and incubated simultaneously with different fluorophore-labeled CD4 aptamers (CD4 antibodies as standard control), CD8 and CD45 antibodies, and were then followed by flow cytometric analysis. The distribution of cell populations including CD4+ lymphocytes, weakly CD4+ monocytes and CD4−/CD8+ lymphocytes was gated according to the side scatter (SSC) and CD45 panels. As expected, the CD4 aptamers showed similar staining patterns and numbers for the as-prepared cells as the CD4 antibodies. It was concluded from our studies that aptamers can be used as sensitive and specific probes in a similar fashion to clinically validated antibodies, for flow cytometric analysis (Fig. 1c).30
Fig. 1. Aptamer-based fluorescent probe for multi-color flow cytometric analysis.
(a) Design of a simple, sensitive and versatile fluorophore-aptamer probe for cancer cell detection or in vivo tumor imaging; (b) Multi-color flow cytometric analysis of lymphoma cells by aptamer probes and antibodies. CD30-positive lymphoma cells were mixed with fresh normal marrow cells and stained with AmCyan-labeled CD45 antibodies, and Cy5-labeled CD30 aptamers simultaneously, or FITC-labeled CD30 antibodies as standard control. The individual cellular populations in the cell mixture including nucleated red blood cells, blasts, lymphocytes, granulocytes, monocytes and CD30-positive lymphoma cells, were separated and gated according to the side scatter (SSC) and CD45 panels. Multi-color flow cytometric analysis revealed that both aptamer probes and antibodies detected the same population of CD30-positive lymphoma cells with identical specificities and sensitivities; (c) Cells from patients’ pleural fluids were collected and incubated with different fluorophore-labeled CD4 aptamers (CD4 antibodies as standard control), CD8 and CD45 antibody simultaneously. Multi-color flow cytometric analysis revealed that the CD4 aptamers also showed similar staining patterns and numbers for the as-prepared cells as the CD4 antibodies.
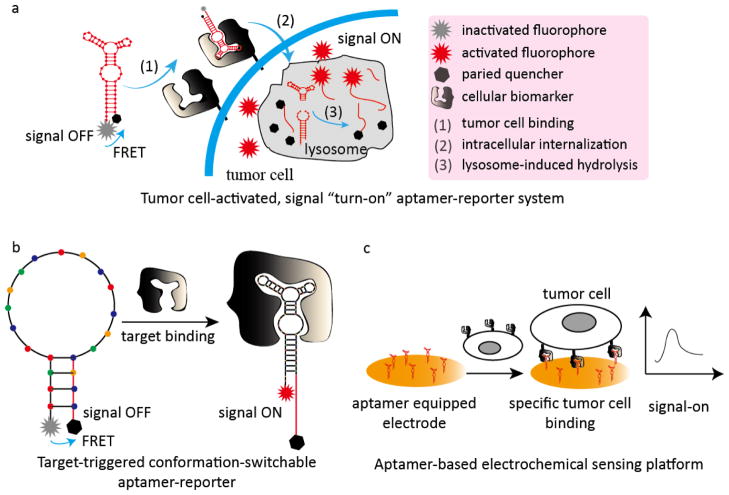
In another recent study, our group developed a cancer cell-activated fluorescent aptamer-reporter system for sensitive detection of CTCs.35 By developing this aptamer probe, we addressed an important issue also encountered with antibody fluorescent imaging probes: the reported ‘always-on’ signal aptamer probes are limited by the high non-specific background noise when the unbound probes are not entirely removed from the assays. Notably, our new signal ‘turn-on’ aptamer probes exhibit high sensitivity but undetectable background noise. These new ‘turn-on’ probes are based on several unique attributes of oligonucleotide aptamers, specifically their switchable-conformation combined with the principle of fluorescent resonance energy transfer (FRET) between the fluorophore and the quencher molecules. In the fluorescent aptamer probe we developed, the 5′- and 3′-termini of the biomarker-specific aptamers were conjugated with the fluorophore reporter and paired quencher molecule, respectively. In the absence of cells of interest, the intact aptamer-reporter remained optically silent (signal OFF-state) due to proximity of the paired quencher to the fluorophore. The aptamer-reporter specifically targeted the CTCs when it was added to whole blood of the patient, and was subsequently internalized into the cell lysosomes. The degradation of the aptamer sequences within the nuclease-rich lysosomes resulted in separation of the fluorophore from the paired quencher molecule, resulting in a bright fluorescent signal (signal ON-state) within minutes with no background noise (Fig. 2a). By using this cancer cell-activated aptamer-reporter system, we demonstrated a rapid one-step and high-throughput assay for CTC detection in whole blood samples, which is an ideal model for point-of-care testing (POCT).35
Fig. 2. Representative aptamer-based analytical models for CTC detection.
(a) Tumor-cell activated, signal ‘turn-on’ aptamer reporter for one-step high-throughput detection of CTCs in patient whole blood samples; (b) Target-triggered conformation-switchable aptamer reporter for CTC detection; (c) A simple aptamer-based electrochemical sensing platform for CTC detection.
In another study, Zhao et al. developed target-triggered analytical or therapeutic entities with superb sensitivity and specificity based on the ability of aptamers to switch conformation upon ligand binding (Fig. 2b).36 In this platform, three aptamers specific for MUC1 (mucin 1), HER2 (human epidermal growth factor receptor 2), and estrogen receptor were used. A fluorophore and a paired quencher molecule were conjugated at the 5′- and 3′-termini of the aptamer sequences, respectively. In the absence of target biomarkers, the paired quencher molecules quenched the fluorescence. The interaction of the aptamer with biomarkers of interest, however, changed the sequence conformation, which induced the release of the fluorophore from the paired quencher molecule, and thus the emission of bright fluorescent signals exclusively in the presence of ligands. The ability of this activatable aptamer-reporter to simultaneously detect three different cellular biomarkers thus confers higher selectivity and sensitivity in cancer cell detection. Remarkably, in the breast cancer cell (MCF-7 cell) study model, the limit of detection (LOD) was as low as 10 cells mL−1.36
Several simple aptamer-based colorimetric assays, which are very promising due to their simplicity for POCT, have also been recently developed for CTC detection,37–39 but their sensitivity is unsatisfactory due to their high LODs (hundreds to thousands of cells). Enhancement of the colorimetric sensitivity was achieved by developing a dual-aptamer nanoparticle (NP)-mediated signal amplification visual strategy and the corresponding assay.40 In this study, MCF-7 breast cancer cells were first separated and concentrated by VEGF (vascular endothelial growth factor) aptamer labeled magnetic beads; then, MUC1 aptamer labeled bimetallic Pt-Au NPs, which catalyze the colorimetric reaction between TMB (3′, 3′, 5′, 5′-tetramethylbenzidine) and H2O2, were incubated with concentrated MCF-7 cells, and sensitive colorimetric signals were generated depending on the number of target cancer cells. Notably, this dual-aptamer based colorimetric platform has a very low LOD (10 cells mL−1 by naked eye), and a wide detection range of 10 – 105 cells mL−1.40
The high sensitivity provided by the previous signal amplification strategy prompted the development of another novel ultrasensitive NP-aptamer-based colorimetric assay with LOD as low as 5 cells in 100 μL binding buffer.41 In this platform, sgc8c aptamers specific for protein tyrosine kinase 7 (PTK7), a biomarker on the surface of leukemia CCRF-CEM cells, were conjugated to iodide-responsive Cu-Au NPs. Upon specific binding of the sgc8c aptamers to the PTK7-positive CCRF-CEM cells, sensitive colorimetric signals were induced by changes in the concentration of the sgc8c aptamer-modified Cu-Au NPs, which affect the colorimetric response of the iodide-catalyzed H2O2-TMB reaction.41
Electrochemistry is another highly sensitive, simple, and stable method that has been widely applied in conjunction with aptamers to detect CTCs. The emerging aptamer-based electrochemical assays have shown great promise in vitro. For example, an ultrasensitive electrochemical assay was developed using a specific aptamer (namely TS11a) for hepatocellular carcinoma cells as a capturing probe, and the gold surface as an electrochemical sensor (Fig. 2c).42 In this study, the amino-labeled TS11a aptamers were conjugated with carboxylic groups on the surface of gold disk electrodes through the EDC/NHS reaction. When the specific hepatocellular carcinoma cells (cell line HepG2 in this study) were captured by immobilized TS11a aptamers, the interfacial electron-transfer resistance between the redox probe and the gold electrode surface was increased dramatically due to blocking direct access between the redox probes and the electrode surface; the changes of resistance were strictly related to the number of captured cells. Thus, the recording of changes in resistance is a sensitive measure of captured cancer cells. As shown in the model, this assay exhibited an ultra-sensitive LOD (2 cells mL−1) and a wide linear detection range (102–106 cells mL−1).42 In another example, an electrochemical assay with superb analytical sensitivity for colon CTCs (LOD of 40 cells mL−1, and a linear range of 1.25 × 102 to 1.25 × 106 cells mL−1) was developed using a MUC1 aptamer as a capture probe and the carbon nanosphere as an electrochemical sensor.43
In addition to the aforementioned analytical strategies, many other aptamer-based signal-readout techniques based on chemiluminescence,44 electrochemiluminescence,45, 46 and photoelectrochemistry47 have been developed and exhibited excellent sensitivity, specificity, and stability for CTC detection. Apart from their detection, capture and isolation of CTC is critical as they are involved in metastasis and recurrence. In this vein, effective CTC capture and isolation can contribute significantly to the advancement of cancer research and treatment. To achieve this aim, aptamer-based capturing and isolation platforms have also been investigated, and have shown optimal efficiency in obtaining CTCs with high purity and viability.48–52 A summary of aptamer-based analytical methods for CTC detection is listed in Table 1.
Table 1.
Aptamer-based analytical platforms for CTC detection
| Aptamer target | Cancer type | Signal-readout technique | Analytical performance | Ref. | |
|---|---|---|---|---|---|
| LOD | Linear range | ||||
| CD30 | ALCL and Hodgkin’s lymphoma | Fluorescent reporter/flow cytometry | No detection | No detection | 34 |
| CD4 | Patient pleural fluid specimen | Fluorescent reporter/flow cytometry | No detection | No detection | 30 |
| CD30 | ALCL and Hodgkin’s lymphoma | Cancer cell activated (signal turn-on) fluorescent reporter | 1 cell in 104 non-specific cells | No detection | 35 |
| Combined detection of MUC1, HER2 and estrogen receptor | Breast cancer | Target-activated fluorescent (signal turn-on) reporter | 10 cells/mL | 10–106 cells/mL | 36 |
| Combined detection of VEGF and MUC1 | Breast cancer | Signal amplification based colorimetry | 10 cells/mL | 10–105 cells/mL | 40 |
| PTK7 | Leukemia | Signal amplification based colorimetry | 50 cells/mL | 50–5000 cells/mL | 41 |
| HepG2 cells | Hepatocellular carcinoma | Electrochemistry | 2 cells/mL | 102–106 cells/mL | 42 |
| MUC1 | Colon cancer | Electrochemistry | 40 cells/mL | 1.25 × 102–1.25 × 106 cells/mL | 43 |
2.2 Detection of tumor-related soluble biomarkers
In addition to CTC detection, the recognition of soluble tumor-related biomarkers such as carcinoembryonic antigen (CEA), epidermal growth factor receptor (EGFR), HER2, prostate specific antigen (PSA), VEGF, and many others, facilitates timely cancer diagnosis and effective therapeutic response monitoring. Similar methods are applied for the construction of aptamer-based analytical scaffolds for CTC and tumor-related soluble biomarker detection. Representative platforms of the tumor-related biomarkers reported in the last 2 years are listed in Table 2.
Table 2.
Aptamer-based analytical platforms for soluble tumor-related protein detection
| Aptamer target | Signal-readout technique | Analytical performance | Ref. | |
|---|---|---|---|---|
| LOD | Liner range | |||
| CEA | Signal amplification based colorimetry | 2 pM | 5 pM–0.5 nM | 53 |
| Chemiluminescence | 4.8 pg/mL | 0.0654–6.54 ng/mL | 54 | |
| Surface-enhanced fluorescence | 3 pg/mL | 0.01–1 ng/mL | 55 | |
| EGFR | Electrochemistry | 50 pg/mL | 1–40 ng/mL | 56 |
| HER2 | Activatable fluorescence | 38 nM | 50–250 nM | 57 |
| MUC1 | Signal amplification-based electrochemistry | 4 pM | 4 pM–1 μM | 58 |
| Electrochemistry | 1 pM | 1–100 nM | 59 | |
| Electrochemistry | 2.2 nM | 8.8–353.3 nM | 60 | |
| PSA | Surface acoustic wave analysis | 10 ng/mL | 10–1000 ng/mL | 61 |
| Electrochemistry | 0.25 ng/mL | 0.25–200 ng/mL | 62 | |
| Chemiluminescence | 1.0 ng/mL | 1.9–125 ng/mL | 63 | |
| Surface-enhanced Raman scattering | 4.8 aM | 0.01–5 fM | 64 | |
| VEGF | Surface-enhanced Raman scattering | 22.6 aM | 0.01–1.0 fM | 65 |
3. Immunohistochemical staining of tumor tissues
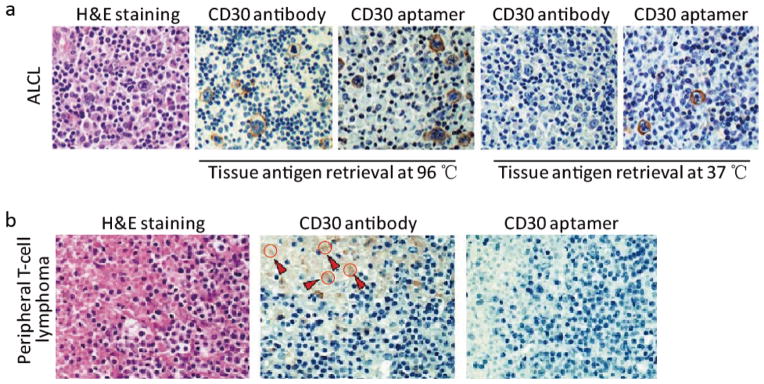
Histopathological evaluation is still the “gold standard” for cancer diagnosis. In addition to traditional staining of tumor tissues with haematoxylin and eosin (H&E staining), IHC study of cellular biomarkers can improve the diagnostic sensitivity, especially for poorly or undifferentiated tumors that cannot be identified by cell morphology alone. Heretofore, antibodies have been the only clinically validated and commercially available probes for IHC staining. In recent years, however, aptamers have been evaluated for their applicability in IHC staining as alternatives or supplements for antibodies. Our group was the first to validate the potential of aptamers for IHC staining on formalin-fixed paraffin embedded (FFPE) patient tissues.66 Utilizing RNA-based CD30 aptamers as recognition probes and CD30 antibodies as standard control, we evaluated the advantages of aptamers versus antibodies for IHC staining of lymphoma tissues. As shown in Fig. 3a, the aptamer achieved IHC staining at simpler reaction conditions than the antibody, such as lower temperature antigen retrieval (37 versus 96 °C for antibody), and shorter probing reaction times (20 versus 90 min for antibody). Importantly, the aptamer probe demonstrated nearly identical but slightly different IHC staining profiles in FFPE lymphoma tissues as compared to the standard antibody. The CD30 antibody staining was concentrated at the cell membrane and Golgi zone; however, the CD30 aptamer stained the cell membrane and the cytoplasm of lymphoma cells. As discussed, this may be due to the fact that (i) small-sized aptamers have better penetration than antibodies, and can access specific targets within the fixed tissues more efficiently, and/or (ii) aptamers and antibodies recognize different epitopes. Notably, the aptamer probe exhibited considerably less background staining in tumor necrotic areas with respect to the standard control antibody. As showed in Fig. 3b, the CD30 antibody showed non-specific staining (brown color) in tumor necrotic areas (depicted in red arrows and circles) whereas the CD30 aptamer did not.66
Fig. 3. Aptamer probes for IHC staining of FFPE tumor tissues.
(a) Tissue sections of CD30-positive anaplastic large cell lymphoma (ALCL) were immunostained with aptamer probes, or antibodies as standard control. After antigen retrieval at 37 °C and probing for 20 min, tissue sections were probed with aptamers and lymphoma cells were specifically immunostained. In contrast, antibody immunostaining of lymphoma cells required higher antigen retrieval temperature (97 °C) for a long probing time (90 min); (b) Aptamer probes showed non-specific staining (brown color) in the tumor necrotic area compared to the antibody stain (depicted with red arrows and circles).
As mentioned, an increasing number of aptamers have been developed as probes for IHC staining.67–69 Dr. Tan’s group developed a fluorophore-labeled, DNA-based EpCAM (epithelial cell adhesion molecule) aptamer to stain frozen and paraffin-embedded sections of colorectal cancer tissues. The study demonstrated that the EpCAM aptamers specifically recognized and stained nests of colorectal cancer tissue with undetectable background noise or cross-reaction with non-specific tissue. Moreover, the reaction time of the EpCAM aptamers was shorter compared to the EpCAM antibodies.70
Apart from cancer cell detection by IHC staining, monitoring the sentinel lymph node is also critical, as the disseminating cancer cells usually move first to this node during metastasis.71 To this end, Dr. Yang’s group developed a panel of aptamers, which specifically targeted lymph node tissue of colon cancer metastatic sites. Furthermore, the fluorescent-labeled aptamer with the best binding affinity for target cells exhibited high detection rates in specifically imaging metastatic tumors and lymph node tissues with metastasis. These results constitute an important stride in the early detection of cancer metastasis.72
In another study, Gupta et al. minimized nonspecific binding of the negatively charged aptamers to positively-charged cellular components such as histones by the addition of an anionic competitor. This development produced a class of novel aptamers, namely the ‘slow off-rate modified aptamers’ (SOMAmer®, SomaLogic Inc., Boulder, CO, USA) with higher binding affinities and slow dissociation rates.73 The presence of an anionic competitor during the selection process provided rapid and specific binding of the fluorophore-labeled HER2 SOMAmer to HER2 in human breast carcinoma cells, and the slow off-rate of the aptamer from the HER2 protein contributed to its selectivity.73
4. In vivo tumor imaging
Currently, the main non-invasive in vivo imaging technologies available for cancer pre- and post-treatment assessment are optical imaging, computed tomography (CT), radionuclide imaging including positron emission tomography (PET) and single-photon emission computed tomography (SPECT), ultrasound (US) imaging, and magnetic resonance imaging (MRI).74 The value of these imaging technologies in cancer diagnosis notwithstanding, they are not cancer- or tumor-type specific.
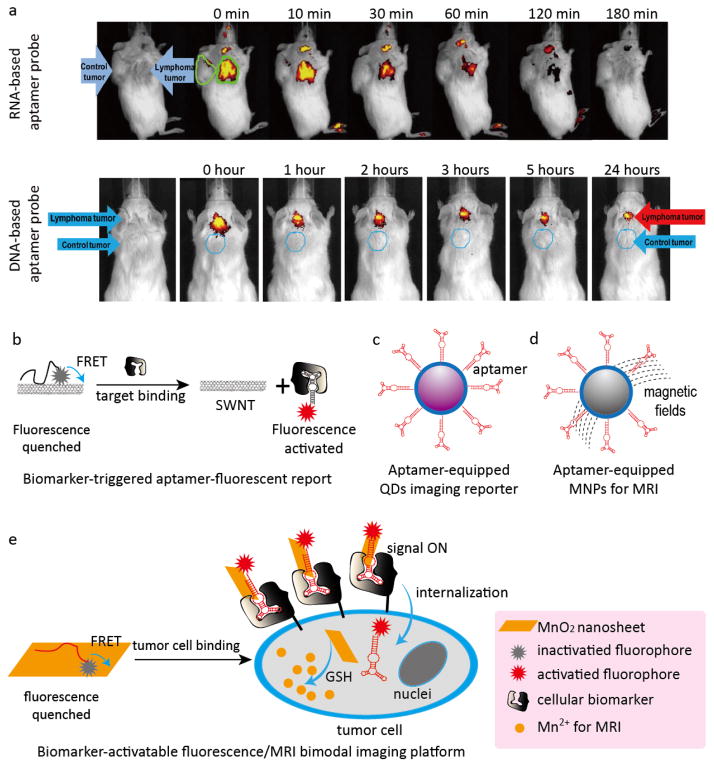
Based on the high resolution and spatial visualization of fluorescent imaging, we developed RNA- and DNA-based aptamer-IRD800CW reporters for in vivo imaging of CD30-positive lymphoma (Fig. 1a). After being systemically administered into a xenograft mouse, the aptamer probes rapidly targeted and accumulated in CD30-positive tumors (<10 min), owing to their small size and simple structure, but they did not react with CD30-negative tumors in the same mouse. The fluorescent signals from the target tumor sites were 4–8 times higher than those in control tumors in the same mouse. The imaging signal was stable for up to 60 min and 24 h for the RNA- and DNA-based aptamer probes, respectively, which may also make them suitable for targeted cancer therapy (Fig. 4a).29 Similar studies have been reported for specific in vivo tumor imaging with the pancreatic ductal adenocarcinoma aptamer-Cy5 reporter,75 the A549 lung adenocarcinoma aptamer-Cy5 reporter,76 and the dual-functional nucleolin aptamer (AS1411) conjugated with the blood-brain barrier targeting peptide aptamer-Cy3 reporter.77 The aforementioned aptamer-fluorescent reporters targeted tumor sites specifically they were retained therein, and showed strong fluorescent signals without significant non-specific accumulation in normal tissues.
Fig. 4. Representative aptamer-based platforms for in vivo tumor imaging.
(a) Studies of RNA- and DNA-based aptamer reporters for in vivo imaging. Both the CD30-positive lymphoma and CD30-negative control tumors were developed in each mouse. RNA-based (upper panel) or DNA-based (lower panel) aptamer probes were systemically administered through the tail veins and whole body imaging was carried out. Aptamer probes specifically highlighted lymphoma tumors but did not react with the control tumor present in the same mouse. (b) Target-triggered fluorescence imaging platform based on the principle of FRET for in vivo imaging; (c) Aptamer-targeted QDs for optical imaging; (d) Aptamer-targeted MNPs for MRI; (e) Dual-activatable, aptamer-based fluorescence/MRI bimodal imaging platform.
As mentioned in the CTC section, this type of aptamer-fluorescent reporter model is considered an ‘always-on’ imaging probe. The construction of specific target-activated, signal ‘turn-on’ imaging systems, similar to those discussed in the CTC section, is a viable method for in vivo imaging. For example, another noteworthy cancer cell-activated fluorescent imaging platform, based on the FRET principle (Fig. 4b) has been developed. In this system, single-walled carbon nanotubes (SWNT) were used as aptamer carriers and fluorescence quenchers whereas Cy5 fluorophore-labeled sgc8c activatable aptamers were used as recognition probes and fluorescent reporters for in vitro cancer cell detection and in vivo cancer imaging.78 Firstly, Cy5 fluorophore-labeled sgc8c aptamers were conjugated through pi-pi stacking interactions with SWNT, which quenched the fluorescent signals. The conjugated Cy5-sgc8c aptamer/SWNTs were injected into tumor-bearing mice, and remained optically silent until reaching the tumor. When they encountered the tumor sites, however, the Cy5-sgc8c aptamers bound to specific cancer cells and separated from SWNT, thus resulting in dramatically enhanced fluorescence signals. This versatile activatable aptamer-fluorescence probing platform based on aptamer/carbon nanotube ensembles significantly improved the fluorescence signal-to-noise ratio in the tumor-bearing mice.78 In another notable approach, a target-triggered fluorescent imaging system was developed based on conformation-switchable sgc8c aptamers labeled with a fluorophore and a paired quencher. The activatable aptamer probe resulted in significantly enhanced image contrast, shortened diagnosis time, and high specificity in differentiating the target cells.79
Although nanomaterials such as quantum dots (QDs) display superb photostability for cancer cell detection as compared to the traditional fluorophores, which are limited by photobleaching,80 the inherent cytotoxicity of QDs hindered their widespread applicability. In order to improve the biocompatibility and reduce the cytotoxicity of QD-based imaging agents, Zhang et al. developed an MUC1 aptamer-functionalized CdTe:Zn2+ doped QD imaging probe for specific in vivo applications (Fig. 4c).81 The in vivo evaluation of the MUC1 aptamer-QDs in tumor-bearing mice revealed enhanced biorecognition and fluorescence sensitivity capabilities, photostability, improved biocompatibility, and reduced cytotoxicity.81
Magnetic nanomaterials based on MRI provide another successful imaging platform. The lack of water-solubility and the suboptimal biocompatibility of magnetic nanoparticles (MNPs) for in vivo imaging were overcome by a similar strategy applied for the construction of aptamer-QD imaging probes: the MNPs were coated with a surfactant (e.g., carboxyl polysorbate 80), and functionalized on the surface with specific aptamers to improve water-solubility, biorecognition, and biocompatibility. Based on this strategy, several aptamer-targeted MNP imaging agents, such as VEGF receptor 2 aptamer-magnetic nanocrystal probes for in vivo imaging of glioblastoma,82 EpCAM aptamer-magnetic nanocrystal probes for in vivo imaging of gastric carcinomas,83 and integrin αvβ3 aptamer-MNPs probes for in vivo imaging of epidermoid carcinoma were recently developed (Fig. 4d).84 All the previous MRI probes showed reduced cytotoxicity, improved biocompatibility even at high concentrations, and importantly, high sensitivity and selectivity in tumor imaging.
Radionuclide imaging including PET and SPECT have excellent applicability for in vivo tumor imaging due to their enhanced sensitivity. To further improve their specific tumor-targeting ability, radionuclide imaging reagents based on highly specific aptamers have been widely developed and evaluated for their clinical applicability. Notable examples include the HER2 aptamer labeled with 99mTc for SPECT imaging of ovarian carcinoma,85 the EGRF variant III aptamer labeled with 188Re for SPECT imaging of glioblastoma,86 and the tenascin-C aptamer labeled with 18F or 64Cu for PET imaging of tenascin-C positive tumors.87 In these studies, the radionuclide-labeled aptamer probes showed great tumor targeting ability and rapid accumulation rates at the tumor sites. In addition, these probes demonstrated fast clearance from the blood resulting in enhanced tumor-to-blood and tumor-to-background ratios, which buttressed their use as highly sensitive and selective imaging contrast agents. Their advantages notwithstanding, the reports of non-specific accumulation of radionuclide-labeled aptamer probes in the liver85, 88 underscored the necessity to further optimize this type of imaging probes.
Aptamer targeted-multimodal imaging probes based on several imaging techniques provide a route to circumvent the specific disadvantages of each one, e.g., poor tissue penetration for fluorescence, low spatial resolution for PET, and suboptimal sensitivity for MRI.74, 89 Recently, Dr. Tan’s group developed a dual-activatable fluorescence/MRI bimodal platform for sensitive and selective cancer imaging (Fig. 4e). In their study, Cy5-labeled sgc8 aptamers were synthesized and used as fluorescent reporters and targeting ligands, and redoxable MnO2 nanosheets were used as aptamer nanocarriers, fluorescence quenchers, and intracellular GSH-activated MRI contrast agents. Subsequently, the Cy5-labeled sgc8 aptamers were conjugated with redoxable MnO2 nanosheets through pi-pi stacking interactions. The resulting fluorescent/MRI probes were inactive in the absence of specific cancer cells. However, when sgc8 aptamers bound to specific cancer cells, the fluorophore Cy5 was activated due to separation from the quencher, and the endocytosed MnO2 nanosheets were reduced by intercellular GSH to generate Mn2+ for MRI detection. Although the imaging capability of this probe was only evaluated for an in vitro cancer cell model, this novel dual activatable fluorescence/MRI bimodal platform showed great potential for highly sensitive and selective in vivo imaging.90 More recently, an AS1411 aptamer-targeted multimodal in vivo imaging probe, capable of simultaneous fluorescence imaging, radionuclide imaging, and MRI, was also developed.91 In this study, the AS1411 aptamer was used as a targeting ligand, and three different imaging agents, including cobalt-ferrite NPs for MRI, fluorescent rhodamine for fluorescence imaging, and the radionuclide 67Ga-citrate for PET imaging were combined for multimodal imaging. The resulting probe specifically targeted tumor sites with high sensitivity but non-specific accumulation in the liver was also reported.91 A summary of aptamer-based analytical platforms for in vivo imaging applications is listed in Table 3.
Table 3.
Aptamer-based analytical platforms for in vivo imaging applications
| Aptamer target | Cancer type | Signal readout technique | Applications | Ref. |
|---|---|---|---|---|
| CD30 | ALCL and Hodgkin’s lymphoma | Fluorescent reporter (signal always-on) | Fluorescent imaging | 29 |
| PL45 cells | Pancreatic ductal adenocarcinoma | Fluorescent reporter (signal always-on) | Fluorescent imaging | 75 |
| A549 cells | Lung cancer | Fluorescent reporter (signal always-on) | Fluorescent imaging | 76 |
| PTK7 | Leukemia | Cancer cell activated fluorescent reporter (signal turn-on) | Fluorescent imaging | 78 |
| PTK7 | Leukemia | Cancer cell activated fluorescent reporter (signal turn-on) | Fluorescent imaging | 79 |
| MUC1 | Lung cancer | Optical nanomaterials QDs | Fluorescent imaging | 81 |
| VEGF receptor 2 | Glioblastoma | Magnetic nanomaterials | MRI | 82 |
| EpCAM | Gastric carcinoma | Magnetic nanomaterials | MRI | 83 |
| Integrin αvβ3 | Epidermoid carcinoma | Magnetic nanomaterials | MRI | 84 |
| HER2 | Ovarian carcinoma | Radionuclide 99mTc | SPECT imaging | 85 |
| EGRF variant III | Glioblastoma | Radionuclide 188Re | SPECT imaging | 86 |
| Tenascin-C | Tenascin-C positive cancer | Radionuclide 18F or 64Cu | PET imaging | 87 |
| PTK7 | Leukemia | Target-activated fluorescent reporter and MRI contrast probe | Multimodal fluorescent imaging and MRI | 90 |
| Nucleolin | Glioma | Magnetic nanoparticles, fluorescent reporters, and the radionuclide 67Ga-citrate | Multimodal MRI, fluorescent and PET imaging | 91 |
5. Current challenges and future directions
Aptamers have emerged as a class of molecules that demonstrate significant potential as versatile biorecognition probes in early cancer diagnosis. Due to their competitive advantages of high target-specific affinity, rapid tissue penetration, nonimmunogenicity, biostability, and ease of modification and production, aptamers have showed superb clinical applicability and suitability over protein antibodies.
Although aptamers have great translational potential, since 2004, only one aptamer-based drug, namely Macugen (pegaptanib sodium, Pfizer/Eyetech), which is a RNA aptamer-based VEGF inhibitor for treatment of age-related macular degeneration, was approved by FDA (US Food and Drug Administration).13 Thus, to accelerate the development of aptamer technology for clinical applications and successful commercialization, the following challenges need to be thoroughly considered:
An obvious impedance for the clinical applications of aptamers is their high nuclease-sensitivity and fast renal filtration due to their small size and oligonucleotide characteristics, which result in short half-life and suboptimal bioavailability. However, this disadvantage can be overcome through different chemical or structural modifications, such as substitutions on the aptamer’s backbone or side chains, incorporation of unnatural nucleotides, addition of caps at the aptamer ends, conjugation of large-sized bioavailable nanomaterials, and many more.4, 7 Actually, many aptamers developed and modified by the aforementioned strategies have significantly improved bioavailability and are thus suitable for clinical applications.4, 92
Another hindrance in the clinical applications of aptamers is that the SELEX process to develop them is time-consuming and suffers from a low success rate. A typical SELEX process consists of 8–20 rounds of repetitive steps with the entire process taking weeks to months, including preparation of the oligonucleotide pool, incubation, partitioning, amplification and finally sequencing. These multiple selection steps have a degree of uncertainty, especially to generate aptamers against “difficult” targets. However, in recent years, many improvements to shorten the selection period and improve the success rate have been achieved such as magnetic beads, affinity chromatography, capillary electrophoresis, microfluidic technology, SOMAmer®, and many others.7, 93–95 These improved techniques can usually shorten the selection period to 4–8 rounds and improve the total success rate by more than 80%.
Although aptamer technology itself has great potential for clinical applications, the parameters of aptamer biodistribution and pharmacokinetics are affected when aptamers are conjugated to functional moieties such as imaging molecules, radionuclides, NPs or drugs. For example, radionuclide-labeled aptamer probes exhibited non-specific accumulation in the liver,85, 88 indicating that the appropriate functional moieties for aptamer conjugation should also be optimized and carefully evaluated.
As of October 2015, there were more than 5000 published articles in the PubMed database including the term “aptamer”. However, according to our statistics, it seems that the researcher interests are mainly focused on how to use aptamers to replace and not to supplement the clinically validated antibodies. Also, many aptamer-based analytical or therapeutic methods are mainly focused on chemical designs or modifications, which to some extent are not in line with the actual clinical necessities. Despite their limitations, antibodies have been clinically validated and commercially available for many years. Focusing in research areas where antibodies are disadvantageous is a viable pathway for aptamers to achieve timely and successful industrialization. Importantly, to achieve the most desired goals, aptamer research and development should be tailored to the actual clinical applications.
In summary, the studies outlined in this review highlight the immense potential of aptamers coupled to nanomaterials such as QDs, SWNTs, and MNPs in constructing aptamer-based biosensors with enhanced recognition abilities. Aptamers used in conjunction with fluorescence, colorimetry, magnetism, electrochemistry or in vivo imaging modalities such as PET and SPECT have provided novel cancer diagnostic strategies with superior performance for early, sensitive, and specific detection of cancer biomarkers. Further development and adaption of the currently available aptamer-based multifunctional tools and their underlying concepts, coupled with the rising interest in this field, will undoubtedly lead to the next generation of aptamer-based recognition probes for cancer detection.
Supplementary Material
Acknowledgments
The authors thank Dr. Helen Chifotides for editorial assistance. This study was partially supported by NIH grants R01CA151955, R33CA173382 and CPRIT grant (RP140315) to YZ.
References
- 1.Siegel R, Naishadham D, Jemal A. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Soori G, Dobleman TJ, Xiao GG. Expert Rev Mol Diagn. 2014;14:97–106. doi: 10.1586/14737159.2014.866039. [DOI] [PubMed] [Google Scholar]
- 3.Souriau C, Hudson PJ. Expert Opin Biol Ther. 2003;3:305–318. doi: 10.1517/14712598.3.2.305. [DOI] [PubMed] [Google Scholar]
- 4.Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Mol Ther Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Sun H, Zu Y. Molecules. 2015;20:11959–11980. doi: 10.3390/molecules200711959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Nat Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 9.Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR, Kim N, Saccomano NA, Wilcox SK, Zichi D, Sanders GM. PLoS One. 2011;6:e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parekh P, Kamble S, Zhao N, Zeng Z, Portier BP, Zu Y. Biomaterials. 2013;34:8909–8917. doi: 10.1016/j.biomaterials.2013.07.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong X, Lv Y, Chen T, Zhang X, Wang K, Tan W. Annu Rev Anal Chem (Palo Alto Calif) 2014;7:405–426. doi: 10.1146/annurev-anchem-071213-015944. [DOI] [PubMed] [Google Scholar]
- 12.Cho EJ, Lee JW, Ellington AD. Annu Rev Anal Chem (Palo Alto Calif) 2009;2:241–264. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 13.Ng EW, Adamis AP. Ann N Y Acad Sci. 2006;1082:151–171. doi: 10.1196/annals.1348.062. [DOI] [PubMed] [Google Scholar]
- 14.Jenison RD, Gill SC, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 15.Ciesiolka J, Gorski J, Yarus M. RNA. 1995;1:538–550. [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Goldstein IJ, Mei HY, Engelke DR. Proc Natl Acad Sci U S A. 1998;95:5462–5467. doi: 10.1073/pnas.95.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoltenburg R, Nikolaus N, Strehlitz B. J Anal Methods Chem. 2012;2012:415697. doi: 10.1155/2012/415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Duan JH, Song YM, Ma J, Wang FD, Lu X, Yang XD. J Transl Med. 2012;10:148. doi: 10.1186/1479-5876-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci U S A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Xu H, Ding H, Huang Y, Cao X, Yang G, Li J, Xie Z, Meng Y, Li X, Zhao Q, Shen B, Shao N. J Pathol. 2009;218:327–336. doi: 10.1002/path.2543. [DOI] [PubMed] [Google Scholar]
- 21.Srisawat C, Engelke DR. Nucleic Acids Res. 2010;38:8306–8315. doi: 10.1093/nar/gkq696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marangoni K, Neves AF, Rocha RM, Faria PR, Alves PT, Souza AG, Fujimura PT, Santos FA, Araujo TG, Ward LS, Goulart LR. Sci Rep. 2015;5:12090. doi: 10.1038/srep12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermann T, Patel DJ. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 24.G Eyetech Study. Retina. 2002;22:143–152. [Google Scholar]
- 25.Ireson CR, Kelland LR. Mol Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 26.Martinez O, Bellard E, Golzio M, Mechiche-Alami S, Rols MP, Teissie J, Ecochard V, Paquereau L. Nucleic Acid Ther. 2014;24:217–225. doi: 10.1089/nat.2013.0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melancon MP, Zhou M, Zhang R, Xiong C, Allen P, Wen X, Huang Q, Wallace M, Myers JN, Stafford RJ, Liang D, Ellington AD, Li C. ACS Nano. 2014;8:4530–4538. doi: 10.1021/nn406632u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne SE, Matsumura I, Ellington AD. Curr Opin Chem Biol. 1997;1:5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 29.Zeng Z, Parekh P, Li Z, Shi ZZ, Tung CH, Zu Y. Theranostics. 2014;4:945–952. doi: 10.7150/thno.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, Zhao N, Zeng Z, Chang CC, Zu Y. Am J Clin Pathol. 2010;134:586–593. doi: 10.1309/AJCP55KQYWSGZRKC. [DOI] [PubMed] [Google Scholar]
- 31.Potyrailo RA, Murray AJ, Nagraj N, Pris AD, Ashe JM, Todorovic M. Angew Chem Int Ed Engl. 2015;54:2174–2178. doi: 10.1002/anie.201411094. [DOI] [PubMed] [Google Scholar]
- 32.Sun H, Zu Y. Small. 2015;11:2352–2364. doi: 10.1002/smll.201403073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong B, Zu Y. Theranostics. 2013;3:377–394. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P, Zhao N, Zeng Z, Feng Y, Tung CH, Chang CC, Zu Y. Lab Invest. 2009;89:1423–1432. doi: 10.1038/labinvest.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Z, Tung CH, Zu Y. Mol Ther Nucleic Acids. 2014;3:e184. doi: 10.1038/mtna.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, Wu P, Zhang H, Cai C. Biosens Bioelectron. 2015;68:763–770. doi: 10.1016/j.bios.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Medley CD, Smith JE, Tang Z, Wu Y, Bamrungsap S, Tan W. Anal Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- 38.Liu G, Mao X, Phillips JA, Xu H, Tan W, Zeng L. Anal Chem. 2009;81:10013–10018. doi: 10.1021/ac901889s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi H, Li D, Xu F, He X, Wang K, Ye X, Tang J, He C. Analyst. 2014;139:4181–4184. doi: 10.1039/c4an00561a. [DOI] [PubMed] [Google Scholar]
- 40.Wang K, Fan D, Liu Y, Wang E. Biosens Bioelectron. 2015;73:1–6. doi: 10.1016/j.bios.2015.05.044. [DOI] [PubMed] [Google Scholar]
- 41.Ye X, Shi H, He X, Wang K, He D, Yan L, Xu F, Lei Y, Tang J, Yu Y. Anal Chem. 2015;87:7141–7147. doi: 10.1021/acs.analchem.5b00943. [DOI] [PubMed] [Google Scholar]
- 42.Kashefi-Kheyrabadi L, Mehrgardi MA, Wiechec E, Turner AP, Tiwari A. Anal Chem. 2014;86:4956–4960. doi: 10.1021/ac500375p. [DOI] [PubMed] [Google Scholar]
- 43.Cao H, Ye D, Zhao Q, Luo J, Zhang S, Kong J. Analyst. 2014;139:4917–4923. doi: 10.1039/c4an00844h. [DOI] [PubMed] [Google Scholar]
- 44.Bi S, Ji B, Zhang Z, Zhang S. Chem Commun (Camb) 2013;49:3452–3454. doi: 10.1039/c3cc39296a. [DOI] [PubMed] [Google Scholar]
- 45.Zhang M, Liu H, Chen L, Yan M, Ge L, Ge S, Yu J. Biosens Bioelectron. 2013;49:79–85. doi: 10.1016/j.bios.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, He Y, Zhang Y, Liu M, Liu Y, Li J. Nanoscale. 2014;6:11196–11203. doi: 10.1039/c4nr03053b. [DOI] [PubMed] [Google Scholar]
- 47.Liu F, Zhang Y, Yu J, Wang S, Ge S, Song X. Biosens Bioelectron. 2014;51:413–420. doi: 10.1016/j.bios.2013.07.066. [DOI] [PubMed] [Google Scholar]
- 48.Viraka Nellore BP, Kanchanapally R, Pramanik A, Sinha SS, Chavva SR, Hamme A, 2nd, Ray PC. Bioconjug Chem. 2015;26:235–242. doi: 10.1021/bc500503e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng F, Cheng Y, Wang J, Lu J, Zhang B, Zhao Y, Gu Z. Adv Mater. 2014;26:7333–7338. doi: 10.1002/adma.201403530. [DOI] [PubMed] [Google Scholar]
- 50.Chen Q, Wu J, Zhang Y, Lin Z, Lin JM. Lab Chip. 2012;12:5180–5185. doi: 10.1039/c2lc40858a. [DOI] [PubMed] [Google Scholar]
- 51.Sheng W, Chen T, Tan W, Fan ZH. ACS Nano. 2013;7:7067–7076. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GS, Phillips JA, Dorfman DM, Karnik R, Karp JM. Proc Natl Acad Sci U S A. 2012;109:19626–19631. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang K, Zhai S, Zhang Z, Fu X, Shao J, Lin Z, Qiu B, Chen GN. Analyst. 2014;139:4330–4334. doi: 10.1039/c4an00417e. [DOI] [PubMed] [Google Scholar]
- 54.Zhou ZM, Feng Z, Zhou J, Fang BY, Qi XX, Ma ZY, Liu B, Zhao YD, Hu XB. Biosens Bioelectron. 2015;64:493–498. doi: 10.1016/j.bios.2014.09.050. [DOI] [PubMed] [Google Scholar]
- 55.Yang X, Zhuo Y, Zhu S, Luo Y, Feng Y, Xu Y. Biosens Bioelectron. 2015;64:345–351. doi: 10.1016/j.bios.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 56.Ilkhani H, Sarparast M, Noori A, Zahra Bathaie S, Mousavi MF. Biosens Bioelectron. 2015;74:491–497. doi: 10.1016/j.bios.2015.06.063. [DOI] [PubMed] [Google Scholar]
- 57.Niazi JH, Verma SK, Niazi S, Qureshi A. Analyst. 2015;140:243–249. doi: 10.1039/c4an01665c. [DOI] [PubMed] [Google Scholar]
- 58.Wen W, Hu R, Bao T, Zhang X, Wang S. Biosens Bioelectron. 2015;71:13–17. doi: 10.1016/j.bios.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Zhang Q, Qian C, Hao N, Xu L, Yao C. Biosens Bioelectron. 2015;64:485–492. doi: 10.1016/j.bios.2014.09.052. [DOI] [PubMed] [Google Scholar]
- 60.Hu R, Wen W, Wang Q, Xiong H, Zhang X, Gu H, Wang S. Biosens Bioelectron. 2014;53:384–389. doi: 10.1016/j.bios.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 61.Zhang F, Li S, Cao K, Wang P, Su Y, Zhu X, Wan Y. Sensors (Basel) 2015;15:13839–13850. doi: 10.3390/s150613839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Z, Kasprzyk-Hordern B, Goggins S, Frost CG, Estrela P. Analyst. 2015;140:2628–2633. doi: 10.1039/c4an02277g. [DOI] [PubMed] [Google Scholar]
- 63.Cha T, Cho S, Kim YT, Lee JH. Biosens Bioelectron. 2014;62:31–37. doi: 10.1016/j.bios.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 64.Ma W, Yin H, Xu L, Wu X, Kuang H, Wang L, Xu C. Chem Commun (Camb) 2014;50:9737–9740. doi: 10.1039/c4cc03734k. [DOI] [PubMed] [Google Scholar]
- 65.Zhao S, Ma W, Xu L, Wu X, Kuang H, Wang L, Xu C. Biosens Bioelectron. 2015;68:593–597. doi: 10.1016/j.bios.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 66.Zeng Z, Zhang P, Zhao N, Sheehan AM, Tung CH, Chang CC, Zu Y. Mod Pathol. 2010;23:1553–1558. doi: 10.1038/modpathol.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han ME, Baek S, Kim HJ, Lee JH, Ryu SH, Oh SO. Nanoscale Res Lett. 2014;9:104. doi: 10.1186/1556-276X-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shigdar S, Qian C, Lv L, Pu C, Li Y, Li L, Marappan M, Lin J, Wang L, Duan W. PLoS One. 2013;8:e57613. doi: 10.1371/journal.pone.0057613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simmons SC, McKenzie EA, Harris LK, Aplin JD, Brenchley PE, Velasco-Garcia MN, Missailidis S. PLoS One. 2012;7:e37938. doi: 10.1371/journal.pone.0037938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pu Y, Liu Z, Lu Y, Yuan P, Liu J, Yu B, Wang G, Yang CJ, Liu H, Tan W. Anal Chem. 2015;87:1919–1924. doi: 10.1021/ac504175h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tammela T, Saaristo A, Holopainen T, Yla-Herttuala S, Andersson LC, Virolainen S, Immonen I, Alitalo K. Sci Transl Med. 2011;3:69ra11. doi: 10.1126/scitranslmed.3001699. [DOI] [PubMed] [Google Scholar]
- 72.Li X, An Y, Jin J, Zhu Z, Hao L, Liu L, Shi Y, Fan D, Ji T, Yang CJ. Anal Chem. 2015;87:4941–4948. doi: 10.1021/acs.analchem.5b00637. [DOI] [PubMed] [Google Scholar]
- 73.Gupta S, Thirstrup D, Jarvis TC, Schneider DJ, Wilcox SK, Carter J, Zhang C, Gelinas A, Weiss A, Janjic N, Baird GS. Appl Immunohistochem Mol Morphol. 2011;19:273–278. doi: 10.1097/PAI.0b013e3182008c29. [DOI] [PubMed] [Google Scholar]
- 74.Chen ZY, Wang YX, Lin Y, Zhang JS, Yang F, Zhou QL, Liao YY. Biomed Res Int. 2014;2014:819324. doi: 10.1155/2014/819324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu X, Zhao Z, Bai H, Fu T, Yang C, Hu X, Liu Q, Champanhac C, Teng IT, Ye M, Tan W. Theranostics. 2015;5:985–994. doi: 10.7150/thno.11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi H, Cui W, He X, Guo Q, Wang K, Ye X, Tang J. PLoS One. 2013;8:e70476. doi: 10.1371/journal.pone.0070476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma H, Gao Z, Yu P, Shen S, Liu Y, Xu B. Biochem Biophys Res Commun. 2014;449:44–48. doi: 10.1016/j.bbrc.2014.04.148. [DOI] [PubMed] [Google Scholar]
- 78.Yan L, Shi H, He X, Wang K, Tang J, Chen M, Ye X, Xu F, Lei Y. Anal Chem. 2014;86:9271–9277. doi: 10.1021/ac5024149. [DOI] [PubMed] [Google Scholar]
- 79.Shi H, He X, Wang K, Wu X, Ye X, Guo Q, Tan W, Qing Z, Yang X, Zhou B. Proc Natl Acad Sci U S A. 2011;108:3900–3905. doi: 10.1073/pnas.1016197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mashinchian O, Johari-Ahar M, Ghaemi B, Rashidi M, Barar J, Omidi Y. Bioimpacts. 2014;4:149–166. doi: 10.15171/bi.2014.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang C, Ji X, Zhang Y, Zhou G, Ke X, Wang H, Tinnefeld P, He Z. Anal Chem. 2013;85:5843–5849. doi: 10.1021/ac400606e. [DOI] [PubMed] [Google Scholar]
- 82.Kim B, Yang J, Hwang M, Choi J, Kim HO, Jang E, Lee JH, Ryu SH, Suh JS, Huh YM, Haam S. Nanoscale Res Lett. 2013;8:399. doi: 10.1186/1556-276X-8-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heo D, Lee E, Ku M, Hwang S, Kim B, Park Y, Han Lee Y, Huh YM, Haam S, Cheong JH, Yang J, Suh JS. Nanotechnology. 2014;25:275102. doi: 10.1088/0957-4484/25/27/275102. [DOI] [PubMed] [Google Scholar]
- 84.Lim EK, Kim B, Choi Y, Ro Y, Cho EJ, Lee JH, Ryu SH, Suh JS, Haam S, Huh YM. J Biomed Mater Res A. 2014;102:49–59. doi: 10.1002/jbm.a.34678. [DOI] [PubMed] [Google Scholar]
- 85.Varmira K, Hosseinimehr SJ, Noaparast Z, Abedi SM. Nucl Med Biol. 2013;40:980–986. doi: 10.1016/j.nucmedbio.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 86.Wu X, Liang H, Tan Y, Yuan C, Li S, Li X, Li G, Shi Y, Zhang X. PLoS One. 2014;9:e90752. doi: 10.1371/journal.pone.0090752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jacobson O, Yan X, Niu G, Weiss ID, Ma Y, Szajek LP, Shen B, Kiesewetter DO, Chen X. J Nucl Med. 2015;56:616–621. doi: 10.2967/jnumed.114.149484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pieve CD, Perkins AC, Missailidis S. Nucl Med Biol. 2009;36:703–710. doi: 10.1016/j.nucmedbio.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 89.Tang L, Yang X, Dobrucki LW, Chaudhury I, Yin Q, Yao C, Lezmi S, Helferich WG, Fan TM, Cheng J. Angew Chem Int Ed Engl. 2012;51:12721–12726. doi: 10.1002/anie.201205271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao Z, Fan H, Zhou G, Bai H, Liang H, Wang R, Zhang X, Tan W. J Am Chem Soc. 2014;136:11220–11223. doi: 10.1021/ja5029364. [DOI] [PubMed] [Google Scholar]
- 91.Hwang do W, Ko HY, Lee JH, Kang H, Ryu SH, Song IC, Lee DS, Kim S. J Nucl Med. 2010;51:98–105. doi: 10.2967/jnumed.109.069880. [DOI] [PubMed] [Google Scholar]
- 92.Lao YH, Phua KK, Leong KW. ACS Nano. 2015;9:2235–2254. doi: 10.1021/nn507494p. [DOI] [PubMed] [Google Scholar]
- 93.Lin H, Zhang W, Jia S, Guan Z, Yang CJ, Zhu Z. Biomicrofluidics. 2014;8:041501. doi: 10.1063/1.4890542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Biotechnol Adv. 2015 doi: 10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 95.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, Flather D, Forbes A, Foreman T, Fowler C, Gawande B, Goss M, Gunn M, Gupta S, Halladay D, Heil J, Heilig J, Hicke B, Husar G, Janjic N, Jarvis T, Jennings S, Katilius E, Keeney TR, Kim N, Koch TH, Kraemer S, Kroiss L, Le N, Levine D, Lindsey W, Lollo B, Mayfield W, Mehan M, Mehler R, Nelson SK, Nelson M, Nieuwlandt D, Nikrad M, Ochsner U, Ostroff RM, Otis M, Parker T, Pietrasiewicz S, Resnicow DI, Rohloff J, Sanders G, Sattin S, Schneider D, Singer B, Stanton M, Sterkel A, Stewart A, Stratford S, Vaught JD, Vrkljan M, Walker JJ, Watrobka M, Waugh S, Weiss A, Wilcox SK, Wolfson A, Wolk SK, Zhang C, Zichi D. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.