ABSTRACT
Marine methane seeps are globally distributed geologic features in which reduced fluids, including methane, are advected upward from the subsurface. As a result of alkalinity generation during sulfate-coupled methane oxidation, authigenic carbonates form slabs, nodules, and extensive pavements. These carbonates shape the landscape within methane seeps, persist long after methane flux is diminished, and in some cases are incorporated into the geologic record. In this study, microbial assemblages from 134 native and experimental samples across 5,500 km, representing a range of habitat substrates (carbonate nodules and slabs, sediment, bottom water, and wood) and seepage conditions (active and low activity), were analyzed to address two fundamental questions of seep microbial ecology: (i) whether carbonates host distinct microbial assemblages and (ii) how sensitive microbial assemblages are to habitat substrate type and temporal shifts in methane seepage flux. Through massively parallel 16S rRNA gene sequencing and statistical analysis, native carbonates are shown to be reservoirs of distinct and highly diverse seep microbial assemblages. Unique coupled transplantation and colonization experiments on the seafloor demonstrated that carbonate-associated microbial assemblages are resilient to seep quiescence and reactive to seep activation over 13 months. Various rates of response to simulated seep quiescence and activation are observed among similar phylogenies (e.g., Chloroflexi operational taxonomic units) and similar metabolisms (e.g., putative S oxidizers), demonstrating the wide range of microbial sensitivity to changes in seepage flux. These results imply that carbonates do not passively record a time-integrated history of seep microorganisms but rather host distinct, diverse, and dynamic microbial assemblages.
IMPORTANCE
Since their discovery in 1984, the global distribution and importance of marine methane seeps have become increasingly clear. Much of our understanding of methane seep microorganisms—from metabolisms to community ecology—has stemmed from detailed studies of seep sediments. However, it has become apparent that carbonates represent a volumetrically significant habitat substrate at methane seeps. Through combined in situ characterization and incubation experiments, this study demonstrates that carbonates host microbial assemblages distinct from and more diverse than those of other seep habitats. This emphasizes the importance of seep carbonates as biodiversity locales. Furthermore, we demonstrate that carbonate-associated microbial assemblages are well adapted to withstand fluctuations in methane seepage, and we gain novel insight into particular taxa that are responsive (or recalcitrant) to changes in seep conditions.
INTRODUCTION
Marine methane seeps serve as islands of diverse and dense deep-sea life, with food webs extending from microorganisms to varied megafauna, including clams, mussels, and tube worms (1–3). Distinct habitats associated with methane seeps include sediments, bottom water, loosely consolidated carbonate protoliths (herein called “nodules”), fully lithified carbonate blocks and pavements (herein called “carbonates”), and, occasionally, wood. Marine methane seep microbial communities and corresponding geochemistry within sediments have been intensively investigated and have been found to frequently be dominated by microbial taxa performing anaerobic oxidation of methane (AOM), notably, anaerobic methane-oxidizing archaea (ANME) and deltaproteobacterial sulfate-reducing bacteria (SRB) (4–6). More broadly, seep sediments are biologically diverse locales that host microorganisms spanning many phyla and are often rich in Epsilonproteobacteria and Gammaproteobacteria in addition to the canonical AOM-associated taxa (6–9). A distinct “seep microbiome,” rich in Deltaproteobacteria, Methanomicrobia, and candidate divisions Hyd24-12 and JS1, is apparent when seep sediment- and nodule-associated microbial assemblages are compared to other marine environments (10).
Authigenic carbonates, which are believed to form as a result of increased alkalinity associated with AOM metabolism, constitute the most pervasive solid habitat substrate at methane seeps but are historically less well sampled than sediments. Carbonates are known to host lipid (11, 12) and ribosomal DNA (9, 11, 13) biomarkers, as well as record carbon isotopic compositions reflective of microbial AOM processes (14, 15). Seep carbonates have recently been shown to host viable autoendolithic (organisms whose metabolism induces self-entombing mineral formation) Archaea and Bacteria capable of methane oxidation (16, 17), as well as metazoan communities (18). Carbonates themselves occur in a variety of sizes, morphologies, and mineralogies. These include millimeter- to centimeter-scale poorly consolidated precipitates, termed “nodules” or “concretions,” occurring within seep sediments (19–21). Seep-associated carbonates are also frequently found exposed at the seafloor in isolated blocks with sizes from centimeters to tens of meters and continuous pavements (22, 23), often extending both laterally and vertically from the site of contemporary methane seepage (24, 25). Observations of carbonates at sites lacking contemporary seepage provide evidence that carbonates can outlive seepage processes on the seafloor, supported by the recovery of demonstrably seep-associated carbonates from geologic outcrops as old as 300 million years (26). Diversity relationships between microbial assemblages associated with seep sediments, nodules, and carbonates have just recently begun to be explored (9, 19).
Seepage flux can increase and decrease, as well as shift spatially, on a scale of days (27) to weeks (28) to centuries (27, 29). Microbial assemblages presumably adapt to spatial and temporal changes in seepage flux, but the extent and rate of response in situ remain uncharacterized. Contemporary seepage activity is often defined categorically based on the presence or absence of diagnostic seafloor chemosynthetic communities within methane seeps. Active sites are defined, in this study and elsewhere (18, 27, 30, 31), as hosting sulfur-oxidizing bacterial mats, clam beds, dense snail colonies, and/or methane ebullition, while low-activity areas lack those diagnostic indicators of contemporary seepage. Notably, low-activity sites are often within <102 m of active sites, frequently host carbonates, and can still exhibit microbial activity, including AOM, at reduced rates (16). Diversity surveys using conventional cloning and sequencing have shown that seep-associated archaeal assemblages, in which only a fraction of the taxa were ANME subgroups, differed based on local seepage activity. The same trend was not apparent in bacterial assemblage composition, which instead was more influenced by habitat substrate (sediment vs. nodule vs. carbonate) (9). Lipid biomarker profiles from seep sediment and microbial mat samples have been shown to be differentiated partially by sulfate reduction rate, which is likely in turn correlated with seep activity (32). Off-seep sites host microbial assemblages that are distinct from both active and low-activity sites, further indicating the existence of a seep microbiome (6, 9, 10).
Here, a combined comparative and experimental in situ approach was applied to characterize the relationship between seep microbial assemblages, habitat substrata (carbonate vs. sediment vs. nodule vs. bottom water vs. wood), and varying seep activity (active vs. low-activity stations). By coupling a massive sampling effort of native, unperturbed seep carbonates to in situ transplantation and colonization experiments, we can leverage these compatible datasets to address two fundamental microbial ecology questions. (i) Do seep carbonates host distinct microbial assemblages? (ii) How sensitive are microbial assemblages to habitat substrate type and availability and temporal shifts in methane seepage flux?
RESULTS AND DISCUSSION
Carbonates host distinct and diverse seep microbial assemblages.
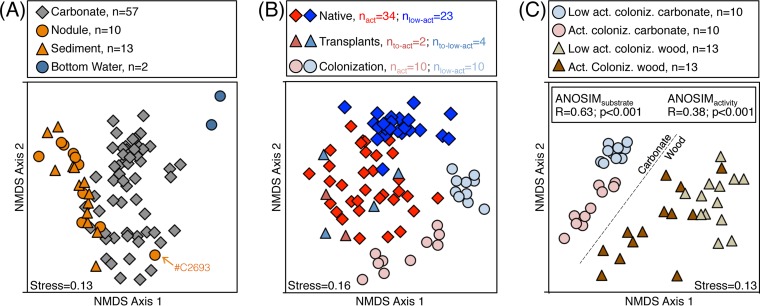
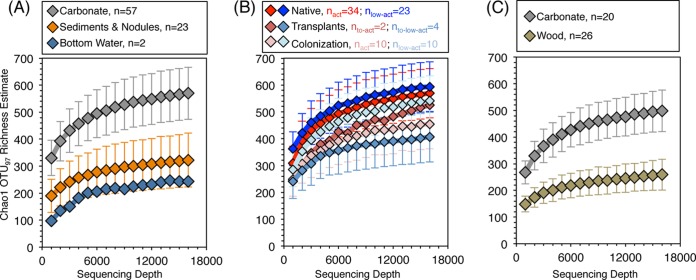
Ordination of the sample set reveals the microbial assemblages to be most strongly differentiated by habitat substrate (i.e., carbonate, sediments and nodules, bottom water) (Fig. 1A) (R = 0.49; P < 0.001; all analysis of similarity [ANOSIM] results are presented in Table S2 in the supplemental material). Habitat substrate is also the most significant factor associated with microbial assemblages as determined by distLM, accounting for 25% of the intersample variability. Furthermore, carbonates exhibit higher operational taxonomic unit (OTU) richness than the other substrates included in this study (Fig. 2A) (Chao1 estimates are given in the text; raw OTU rarefactions are given in Fig. S2 in the supplemental material). These trends are also observed in the macrofauna recovered from seep carbonates (18), confirming that carbonates host diverse benthic life across multiple trophic levels. Overall microbial assemblages of sediments and nodules are not statistically differentiable, as determined from ANOSIM tests, indicating that sediment-hosted nodules and exhumed seafloor carbonates behave as separate, distinct habitat substrates for microbial habitation (Fig. 1A; also, see Table S2 in the supplemental material). Sediments, nodules, and carbonates have recently been shown to host different bacterial, but not archaeal, assemblages in 16S clone library surveys (9), while recent examination of a subset of our iTag data demonstrated similar microbial communities inhabiting nodules and adjacent sediments, especially in active seep settings (19).
FIG 1 .
Nonmetric multidimensional scaling ordination of microbial assemblages in this study. Each point represents the entire recovered microbiological assemblage from one sample; samples plotting closer to each other are more similar in microbial composition. Lower stress values indicate better representation of the intersample (dis)similarities in two dimensions. (A) Native, unperturbed samples of sediment, nodule, bottom water, and carbonate habitat substrates. Sample C2693 (orange arrow) represents a nodule-hosted microbial assemblage recently determined to be a biological outlier among sediment and nodules (19). (B) Ordination of only carbonate samples, representing the native, transplantation, and colonization treatments. (C) Ordination of only colonization samples, representing carbonate and wood substrates at active and low-activity stations. We cannot rule out the possibility that in panel A, bottom water microbial assemblages could be different from those of sediments, nodules, and carbonates because they were extracted by a different method (see “Genomic DNA extraction and 16S rRNA gene sequencing and processing”); the same could be true for the observed difference between carbonate- and wood-hosted assemblages in panel C.
FIG 2 .
Collector’s curves of estimated Chao1 OTU97 richness. Error bars show 1σ standard deviations. (A) Native microbial assemblages associated with carbonates, sediments, and nodules plus the two bottom water samples. Sediments and nodules were binned as one group because their associated microbial assemblages were indistinguishable according to ANOSIM tests (see Table S2 in the supplemental material). (B) Carbonate samples in this study, separated by treatment category. (C) Carbonate and wood colonization samples. Standard deviations are not given for bottom water and transplant-to-active sample groups, due to the low number of analyzed samples. Raw OTU rarefaction curves are given in Fig. S2A to C in the supplemental material.
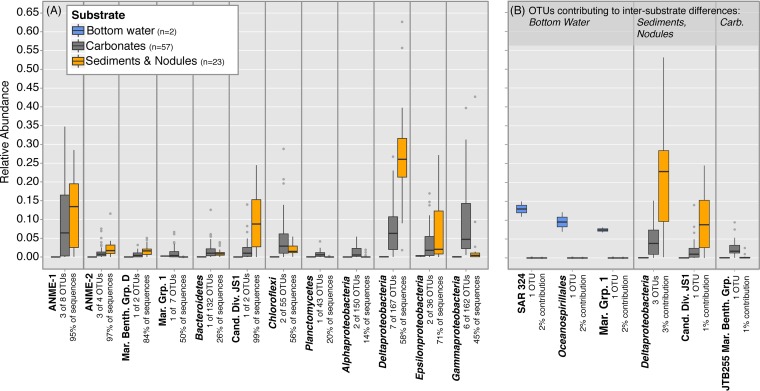
Examination the top thirty most abundant OTUs in our data set reveals a variety of Archaea and Bacteria comprising the samples (Fig. 3A), including taxonomies common to methane seep settings (e.g., ANME subgroups and Deltaproteobacteria). The higher relative abundance of ANME-1 in sediments and nodules than carbonates is in agreement with previous clone library observations at Hydrate Ridge, while the recovery of epsilonproteobacterial sequences from sediments, nodules, and carbonates is in contrast to previous findings in which they were almost exclusively recovered from sediments (Fig. 3A) (9). Data from sequencing of mock communities suggest a slight bias for ANME-1 and a stronger bias against the recovery of ANME-2 sequences by the modified Earth Microbiome Project (EMP) protocol (David H. Case and Victoria J. Orphan, unpublished data). Thus, we note the relative abundance of these groups may in reality be slightly lower (ANME-1) or higher (ANME-2) than recovered in our iTag data set. However, the intersubstrate trends, which are similar for ANME-1 and ANME-2, should be unaffected. Abundance patterns of ANME and other taxa are discussed in detail in the sections below, in the context of results from our experimental manipulations.
FIG 3 .
Boxplot of OTU relative abundances from the 82 native samples in this study. Sediments and nodules are binned as one group because ANOSIM tests revealed their associated microbial assemblages to be statistically indistinguishable. Boxplot centerline represents the median (50th percentile [Q50]). The top and bottom hinges represent Q75 and Q25 quartiles, respectively. The upper and lower whiskers correspond to the highest and lowest data points within 1.5 times the interquartile range (Q75 minus Q25) from the median. Any data points outside that range are identified by gray dots. The same plotting format is applied to Fig. 5. (A) Relative abundances of the top 30 most abundant OTUs in the data set, grouped by taxonomy. The full data set contains 1,057 OTUs, but the top 30 OTUs account for 1%, 67%, and 43% of the sequences recovered from bottom water, sediment/nodule, and carbonate substrates, respectively. (B) Relative abundances of OTUs revealed to be strongly associated with particular habitat substrates. Intersubstrate differences in microbial assemblages are a cumulative result of contributions from many OTUs; even OTUs strongly associated with a particular habitat substrate only contribute several percent to the total intersubstrate variability. Note that the JS1 OTU is the same in panels A and B—it is both highly abundant and strongly associated with sediments and nodules. The Marine Group 1 OTU in panels A and B is different—there is one Marine Group 1 OTU highly abundant in the data set in panel A and another Marine Group 1 OTU strongly associated with bottom water samples (panel B). This highlights the variable distribution of phylogenetically similar OTUs. The y axis of panel A also applies to panel B. Raw OTU data used to generate this plot are available in Table S3 in the supplemental material.
Intersubstrate differences in microbial assemblage are the cumulative result of contributions from many OTUs, with no single OTU accounting for more than 2% of the total intersubstrate differences. Nonetheless, several OTUs can be identified which are strongly associated with one habitat type (Fig. 3B). Notably, taxa previously identified as diagnostic of the “seep microbiome” (i.e., JS1 archaea and Deltaproteobacteria [10]) are observed in our data set to be characteristic of sediments and nodules but not carbonate habitats (Fig. 3B). In determining the “seep microbiome,” Ruff et al. (10) examined methane seep sediments and nodules exclusively; our data thus corroborate their results but also further demonstrate that seep carbonates host distinct microbial assemblages. Carbonates, to the exclusion of other habitat substrates, are observed to host an OTU associated with the gammaproteobacterial JTB255 Marine Benthic group (Fig. 3B). The physiology of this group remains undetermined, though uncultured members have been recovered from a variety of marine sediments (33, 34), including methane seeps (35). OTUs associated with the deltaproteobacterial SAR324 clade and thaumarchaeal Marine Group 1 are particularly abundant in the bottom water samples (Fig. 3B), although we note that a separate thaumarchaeal Marine Group 1 OTU is more abundant on carbonates than on other substrates (Fig. 3A). This exemplifies the potential for OTUs of similar phylogeny to be differentially distributed in the environment.
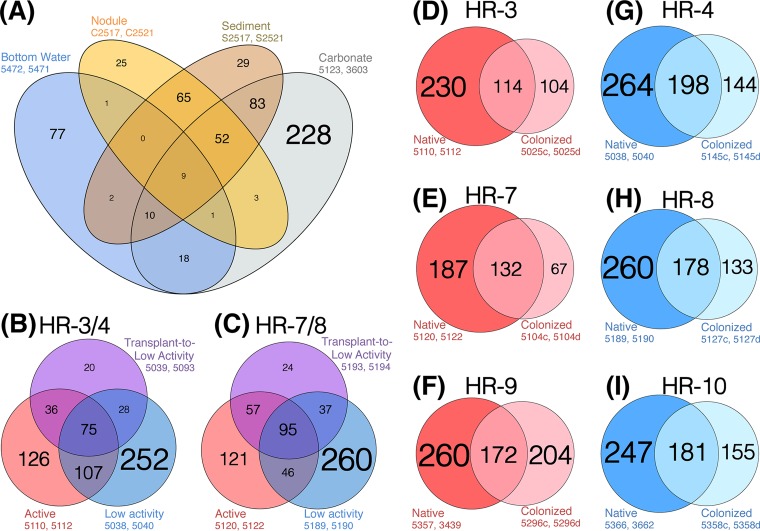
The Shannon diversity index (H′), which measures evenness in addition to richness, is higher in the carbonates than either the sediments/nodules or the bottom waters (see Fig. S2D in the supplemental material). Carbonate-associated assemblages may exhibit distinct microbial molecular signatures due to either geochemical (i.e., preferential adsorption of metabolites to the carbonate matrix [36]), physical (i.e., a site for microbial biofilm attachment), or historic (i.e., formation within or above the sediment column [37]) factors. Examination of the OTU overlap among native habitat substrates (Fig. 4A) demonstrates that carbonates share more OTUs with sediments and nodules than bottom waters, supporting the hypothesis of formation within the sediments, followed by subsequent exhumation and exposure at the seafloor (14, 37). However, bottom waters share more OTUs with carbonates than sediments or nodules, revealing that a subset of bottom water microorganisms do passively or actively inhabit carbonates exposed at the seafloor (Fig. 4A).
FIG 4 .
Comparison of OTU97 overlap among various samples and treatments. In order to ensure equal depth of sampling across each substrate type, two representative samples of each substrate were chosen randomly (sample numbers are given). The number in each region denotes the number of OTUs, and text size is proportional to OTU count (as is circle size for panels D to I). (A) OTU overlap between the four native seep habitat substrates examined in this study: sediments, nodules, carbonates, and bottom water. In order to minimize geographic bias in the analysis, samples were chosen from active stations at Hydrate Ridge south (the only exception was bottom water sample 5472, which was from an HR-South low-activity station). Note that carbonates host the richest OTU diversity (see the collector’s curve in Fig. 2A), including a large number of OTUs which are distinct to carbonates. Carbonates share more OTUs with sediments and nodules than with bottom waters, possibly indicative of an origin within the sediment column and subsequent exhumation and exposure at the seafloor. Bottom waters contribute more OTUs to carbonates than to either sediments or nodules—consistent with the recovery of our carbonates from directly on the seafloor. (B and C) OTU overlap of active and low-activity control carbonates, and transplant-to-low-activity carbonates, for the HR-3/-4 and HR-7/-8 transplant experiments. Transplant-to-active carbonates were not included due to their low sample number (n = 1 each for HR-3/-4 and HR-7/-8). (D to I) OTUs observed in native carbonate samples versus colonized carbonate samples as a function of Hydrate Ridge Station. Stations were included only if they received colonization carbonate deployments and we had recovered native carbonates from the same station (these criteria excluded HR-1, HR-2, HR-6, HR-11, and the Southeast Knoll). Left column (red [D to F]) are active stations, right column (blue [G to I]) are low-activity stations. In each case, the darker color represents the native carbonates and the lighter color represents the colonized carbonates. In most cases, the majority of recovered OTUs from colonization carbonates were also present in native carbonates.
Close overlap in assemblage composition is observed between some of the carbonates (~10 of 57, all from active seep stations) and sediments/nodules (Fig. 1A; also note the 10 carbonates highlighted in Fig. S3A in the supplemental material). It is possible that carbonate samples hosting microbial assemblages similar to sediments/nodules may have contained excess sediment entrained in the rock matrix upon recovery (37); alternatively, nodules in the overlapping region may have been sufficiently lithified to begin hosting carbonate-like microbial assemblages (e.g., nodule C2693 in Fig. 1A), though this does not necessarily explain similarity of some sediment samples. The compositional overlap between ~10 active-station carbonate assemblages and sediment/nodule assemblages is not derived from geographic proximity, as the sediments/nodules from HR do not exclusively plot in close proximity to the carbonate samples, which are dominantly from HR (see Fig. S3A in the supplemental material), nor are the overlapping carbonates unified by seafloor station, mineralogy, or collection year.
Demonstration of microbial variability within seep carbonates.
The high total OTU richness of carbonates (Fig. 2A), combined with OTU overlap between carbonates and other substrates (Fig. 4A) could indicate that carbonates represent a passive repository of preserved and extant microorganisms. We tested this possibility by first examining in detail the native carbonate samples (n = 57), which allows inference of the environmental indicators associated with differences between carbonate-hosted microbial assemblages. We then coupled these interpretations to the in situ transplantation (n = 6) and colonization (ncarbonate = 20; nwood = 26) experiments, respectively (see below).
On their own, native carbonate-associated microbial assemblages demonstrate clear differentiation according to seep activity (R = 0.45; P < 0.001) (Fig. 1B; also, see Table S2 in the supplemental material), mineralogy (R = 0.44; P < 0.001) (see Fig. S4 and Table S2 in the supplemental material), and seafloor station (Ractive stations = 0.31, Pactive stations = 0.002; Rlow-activity stations = 0.27, Plow-activity stations = 0.037) (see Table S2 in the supplemental material). The similar parsing of native carbonate-hosted assemblages by seep activity and mineralogy is partially explained by our observation of a qualitative relationship between seep activity and carbonate mineralogy, with a higher proportion of aragonite-bearing carbonates recovered from low-activity stations (see Text S1 and Fig. S4 in the supplemental material). This suggests that seep activity and carbonate mineralogy are not independent environmental factors in our data set. The biogeographic differences between stations (~102 to 104 m) are in agreement with previous observations of within-seep microbial and geochemical heterogeneity (6, 38) and recent findings that sediment-associated microorganisms in seeps exhibit “global dispersion and local diversification” (10). A distance-decay curve demonstrated that if a biogeographic effect on microbial similarity exists on a 105- to 106-m scale, it is masked by other environmental factors (see Fig. S5 in the supplemental material). We frame our further discussion in terms of seep activity because it is strongly associated with differences between carbonate-hosted assemblages, and importantly, our sample collection and in situ transplantation and colonization experiments were explicitly performed in order to test biological variability as a function of seep station activity. However, we emphasize that seep activity is a qualitative environmental indicator that may be correlated with other environmental factors, such as carbonate mineralogy.
With regard to standard ecological metrics of OTU richness and evenness (Fig. 2B; also, see Fig. S2B in the supplemental material), seep activity does not differentiate the native carbonate-associated microbial assemblages. This indicates that while carbonates at low-activity stations host distinct assemblages, they are not less diverse than microbial assemblages from carbonates at active stations. Active-station carbonates are particularly rich in OTUs associated with putative sulfur-oxidizing organisms belonging to the epsilonproteobacterial and gammaproteobacterial families, Helicobacteraceae and Thiotrichaceae, respectively, compared to carbonates from low-activity stations (Fig. 5; also, see Table S3 in the supplemental material). These organisms are likely supported by high sulfide concentrations produced by sulfate-coupled AOM at active seep stations. Among seep sediments in the Mediterranean Sea, epsilonproteobacterial Helicobacteraceae were found to be an indicator taxon for seepage (6), which our data corroborate. Data from hydrothermal vent systems also exhibit clear differences in abundance of putative sulfur-oxidizing Epsilonproteobacteria between active and low-activity (or inactive) sites, with increased abundance at active vent sites where delivery of reduced fluids is high (39). Furthermore, Epsilonproteobacteria have been observed in time-resolved experiments to rapidly respond to geochemical heterogeneity and experimental perturbations (i.e., colonization of fresh substrate) in hydrothermal vent systems (39–41). Physiologies of specific groups of the Gammaproteobacteria often include oxidation of either sulfur or methane (42, 43), both of which are common at settings with increased delivery of reduced fluids.
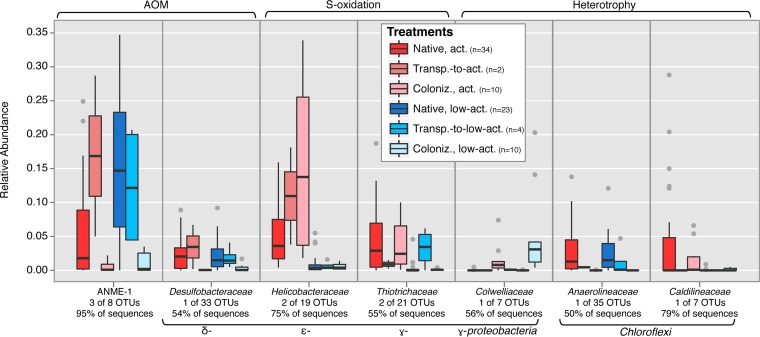
FIG 5 .
Boxplot of carbonate-associated relative abundance data of selected key OTUs identified by SIMPER, representing notable taxonomic groups. Note that data for some groups are combined from several OTUs (OTU data are reported individually in Table S3 in the supplemental material). Although in all cases a minority of the OTUs were identified for presentation (e.g., 3 of 8 for ANME-1), these generally represented the majority of the total sequences recovered from each taxonomic group (e.g., 95% of all sequences for ANME-1). When generated with all OTUs associated with each taxon, this plot does not change substantially (data not shown).
ANME-1 archaea, which are the most abundantly recovered ANME in the entire iTag data set, exhibit wide ranges of relative abundance in both active and low-activity seep stations, with higher average relative abundance at low-activity stations, in agreement with previous clone library observations (Fig. 5) (9). Similarly, the deltaproteobacterial family Desulfobacteraceae does not exhibit a clear difference in observed relative abundance as a function of seep activity (Fig. 5). It thus appears that some ANME-1 and deltaproteobacterial OTUs may be relatively insensitive to seepage level. This was unexpected, as these are key taxa involved in the AOM process and therefore hypothesized to occur at higher relative abundance in methane-replete, presumably “active” seep stations. ANME-1 may perform methanotrophy even within carbonates at low-activity stations, consistent with recent reports of AOM associated with carbonates on the periphery of active seepage (16). Alternatively, relic DNA from AOM-associated organisms may be preserved within carbonate rocks, as the carbonate precipitation process causes self-entombment, potentially sealing off inhabited pores (9, 11, 13, 17). Evidence for biomarker preservation within carbonates has been described for lipids, which are more recalcitrant to degradation than DNA (11, 12, 37).
Demonstration of successional dynamics: transplantation experiments.
The “snapshot” view of carbonate-associated microbial ecology is augmented by the seafloor transplantation experiments, which allow us to observe in situ microbial successional patterns by simulating seep quiescence and activation. In situ flux measurements at Hydrate Ridge have shown that seep activity can shift on week- to month-long timescales (27, 28), indicating our 13-month transplantation experiments are relevant to contemporary processes at Hydrate Ridge and potentially in other methane seep regions.
The OTU composition of the four active-to-low-activity transplanted microbial assemblages are statistically differentiable from both the native, active carbonate-associated microbial assemblages (R = 0.32; P = 0.008) and the native, low-activity carbonate-associated assemblages (R = 0.88; P < 0.001) (Fig. 1B; also, see Table S2 in the supplemental material). The four microbial assemblages transplanted from active to low-activity stations are more similar to the native, active carbonate assemblages (i.e., where they originated) than to the native low-activity assemblages (i.e., where they were transplanted) (Fig. 2B and ANOSIM results). The four transplanted carbonates exhibit approximately 30% lower overall OTU richness than native carbonates (Fig. 2B), but in-depth analysis of OTU overlap between transplanted and native carbonates reveals a level of fine structure to the microbial turnover and succession (Fig. 4B and C). At the paired HR-3/-4 and HR-7/-8 stations, 68% and 52%, respectively, of the OTUs associated with native, active control carbonates were not recovered upon simulated seep quiescence after 13 months (see Table S3 in the supplemental material). The “lost” OTUs are supplanted by characteristic OTUs gained from the low-activity sites (28 and 37 OTUs, representing 18% and 17% of the recovered OTUs for HR-3/-4 and HR-7/-8 transplants, respectively) as well as OTUs unique to the transplants and not recovered from native carbonates (20 and 24 OTUs for HR-3/-4 and HR-7/-8, respectively) (see Table S3 in the supplemental material). Nearly half of the OTUs recovered among the HR-3/-4 and HR-7/-8 transplants were cosmopolitan OTUs that were also observed in both the native active and the native low-activity carbonates (Fig. 4B and C; also, see Table S3 in the supplemental material). Thus, a loss of over half the initial OTUs upon seep quiescence is masked by gain of new OTUs, both unique and shared with the low-activity controls.
Combining the observation of overall similarity to native, active assemblages, diminished overall OTU richness, and specific turnover among the carbonates transplanted to low-activity stations, we can begin to paint a picture of microbial succession upon seep quiescence. Most major (i.e., highly abundant) constituent members of carbonate-associated microbial assemblages are resilient to 1 year of quiescence (or their DNA does not degrade), as evidenced by the fact that transplanted carbonates plot among the native, active controls in Fig. 1B. Indeed, of the four carbonates transplanted to low-activity sites, we observe that 49 to 90% of the recovered sequences are from resilient OTUs shared with the active-station controls (see Table S3 in the supplemental material). However, over the course of a year, low-abundance assemblage members are vulnerable to cessation of seep activity: the average relative abundance of lost OTUs in the native, active controls upon simulated quiescence was <0.5% (see Table S3 in the supplemental material).
Examining specific taxa of interest, we find the gammaproteobacterial Thiotrichaceae OTUs remain at a relative abundance similar to that of the native, active carbonates, consistent with resilience to seep quiescence (Fig. 5). In contrast, epsilonproteobacterial Helicobacteriaceae OTUs that are highly abundant in native, active carbonates had mostly disappeared after 13 months of simulated seep quiescence (Fig. 5). Thus, two putative sulfur-oxidizing groups exhibit different 16S rRNA gene distribution, highlighting the potential for variable response to environmental change, even among taxa putatively belonging to the same guild. ANME-1 OTUs were recovered at high relative abundance in the carbonates transplanted to low-activity stations, consistent with the trend observed in native, low-activity carbonates and suggesting an ability to respond over a period of time that may represent, to ANME archaea, only a few generations (44–46).
The two carbonates which experienced simulated seep activation (transplanted from low-activity to active stations) host microbial assemblages different from those in low-activity, native carbonates and somewhat similar to those in native, active assemblages (Fig. 1B), although this experimental set suffers from low sample number as a result of technical difficulties in recovering two of four originally transplanted carbonates. In juxtaposition to seep quiescence, which demonstrated resilience of the bulk microbial assemblages, our simulation of seep activation indicates that assemblages are relatively quick to respond to renewed seepage conditions. This is especially true among the epsilonproteobacterial Helicobacteraceae OTUs, which are recovered in high relative abundance in the carbonates transplanted to active stations, despite low relative abundance in the low-activity carbonates (Fig. 5). Other OTUs (for example, gammaproteobacterial Thiotrichaceae) clearly demonstrate a slower response to seep activation (Fig. 5). Examination of two OTUs of putatively heterotrophic Chloroflexi, the Anaerolineaceae and Caldilineaceae, also reveals slow response to seep activation, despite their relatively high recovery among native, active seep carbonates (Fig. 5). The Anaerolineaceae OTU also exhibits markedly higher tolerance to low-activity conditions than the Caldilineaceae OTU (Fig. 5), highlighting the potential for different ecological expression among groups of similar phylogeny.
The coupled transplant experiments provide strong evidence that many carbonate-associated seep microbial taxa are adapted to cycles of seep quiescence and activation. This may be ecologically advantageous in an environment where fluid flow has a tendency to fluctuate rapidly and frequently (27, 28). Recalcitrance to seep quiescence is consistent with low but measurable AOM from carbonates at low-activity stations (16), and the physical buffering provided by carbonate habitats has been proposed as a factor for maintenance of microbial assemblage viability during periods of diminished seepage (17). Alternatively, we note that 3 of the 4 carbonates transplanted from active to low-activity stations were composed of a mix of calcite and dolomite—mineralogies more common at active stations than low-activity stations (see Fig. S4 in the supplemental material). If mineralogy significantly drives microbial composition, the observed recalcitrance to community shift may be explained by the fact that the transplanted carbonates bore mineralogies qualitatively associated with active-seep-type microbial assemblages. In contrast, the two samples transplanted from low-activity to active stations were aragonite/calcite mixes—a mineralogical composition regularly recovered from all seep stations regardless of activity (see Fig. S4). Thus, the observed shift to an active-seep-type community is more likely a function of the seep activity shift than of mineralogy. The rapid microbial rebound upon simulated seep activation may be analogous to previous observations of microbial community activation from deep terrestrial and marine subsurface environments (44, 47). Species richness in carbonates transplanted to active stations is higher than the reciprocal transplants—though still lower than native carbonates—further indicating microbial assemblage responsiveness to simulated seep activation (Fig. 2B). Diminished OTU richness upon transplantation (in either direction) is also evidence against a “time-integrative” model of carbonate microbial assemblages: if carbonates were passive recorders of all historic seep microbial DNA, OTU richness would not be expected to decrease.
Demonstration of successional dynamics: colonization experiments.
Though our transplantation experiments best simulate the temporal variability of seepage for established microbial assemblages, they are limited in scope. To increase the interpretative power of our data set, we supplemented the transplant experiments with carbonate (calcite and dolomite) and wood (fir and pine) colonization experiments to address the successional patterns and responsiveness of seep microorganisms colonizing at the seabed under conditions of differing seep activity and colonization substrate type.
Results from these experiments follow similar trends observed in the survey of native microbial assemblages where both habitat substrate (Rcarbonate vs wood = 0.63, P < 0.001) and seep activity (R = 0.38, P < 0.001) differentiate the recovered microbial diversity (Fig. 1C; also, see Table S2 in the supplemental material). In contrast to the survey of native carbonates, mineralogy did not contribute significantly to differences in total colonizing assemblage diversity (P = 0.109) (see Table S2 in the supplemental material), further suggesting that the relationship between mineralogy and microbial diversity in the native carbonates may be due to a qualitative link between mineralogy and seep activity (see Fig. S4 in the supplemental material). Microbial assemblages colonizing carbonates exhibited higher OTU richness and evenness than those colonizing wood (Fig. 2C; also, see Fig. S2C and D in the supplemental material), substantiating the role of seep carbonates, specifically, as hosts of diverse microbial populations.
While hosting comparable OTU richness to the native carbonates (Fig. 2), the microbial assemblages colonizing the sterile carbonates at the seabed were, after 13 months, significantly different from native microbial assemblages collected in this study (R = 0.65, P < 0.001) (Fig. 1B; also, see Table S2 in the supplemental material). This supports general trends in the transplant experiments, suggesting that more than 13 months is required to achieve a mature successional phase if it is assumed that given enough time the colonizing assemblages would eventually mimic the native assemblages. Alternatively, the colonization carbonates might never host microbial assemblages completely similar to the native carbonates, considering the different history of colonization carbonates (located at the seabed) and native carbonates (believed to have formed within the sediment column and later to have been exhumed). Notably, however, sterile carbonates incubated at the seafloor share most of their observed OTUs with the native carbonates (Fig. 4D to I). The discrepancy between colonization and native carbonates hosting quite different microbial assemblages (Fig. 1B) and yet sharing many OTUs (Fig. 4D to I) implies that assemblage differences are generally a function of differential OTU relative abundance, not of the presence/absence of different OTUs themselves. Indeed, an ANOSIM test on presence/absence-normalized data reveals a diminished, though still significant, strength of difference between native and colonized carbonate microbial assemblages (R = 0.53, P < 0.001). In further support, among the six colonization/native pairings examined in detail (Fig. 4D to I), the majority (average 63%, range 46 to 84%; ncolonization samples = 12) of the recovered colonization sequences were from OTUs shared between the colonization and native carbonates.
In-depth analysis of OTU overlap at station HR-9, chosen because of the wide array of habitat types and experimental samples obtained there, reveals that of the various OTUs shared between native and colonized carbonate assemblages, many are also shared with sediment and nodule assemblages (see Fig. S6 in the supplemental material). This suggests some transference of sediment-hosted microbes onto the colonization carbonates. The mode of transfer is currently not known but may be associated with direct microbial motility (48, 49), macrofaunal grazing/bioturbation (2, 48), and/or advection from fluid flow or gas ebullition (50). At station HR-9, where 376 OTUs were reproducibly recovered from both colonization carbonates, 19% (n = 71), 3% (n = 11), and 4% (n = 14) were exclusively sourced from carbonates, sediments/nodules, and bottom waters, respectively. The bottom water samples associated with this station contained 1% to 2% relative abundance of an OTU associated with the gammaproteobacterial Colwelliaceae, which were also recovered at moderate relative abundances from the colonization carbonates (<1% to 20%) (Fig. 5; also, see Table S3 in the supplemental material) despite a lack of detection on either native or transplanted carbonates. This further indicates some transfer of bottom water microorganisms onto carbonates during early-phase succession and is consistent with common ecophysiology of Colwellia as generally marine, psychrophilic, motile, chemoorganotrophic microorganisms (42). Thus, OTU recovery from multiple nearby substrates, coupled to the observed difference between colonized carbonate and wood microbial assemblages after 13 months (R = 0.63, P < 0.001) (Fig. 1C), could be explained by two hypotheses: either (i) OTUs are recruited from all surrounding habitats, followed by assemblage differentiation according to habitat substrate (i.e., carbonates diverge from woods), or (ii) carbonate colonization is a substrate-specific process from the very first microbial succession, and then over time occasional passive capture of OTUs from other habitat substrates occurs. In either case, the colonization data support the observation from native samples that carbonates host distinct microbial assemblages. Furthermore, carbonate distinctiveness is not simply a product of time-integrated passive capture of sediment-hosted microorganisms, nor does it depend on a history of burial in sediment.
Microbial diversity within the carbonate colonization experiments is almost wholly explained by seep activity differences, in further support of observations from native carbonates (Fig. 1C) (R = 0.81, P < 0.001). Indeed, OTUs associated with the epsilonproteobacterial Helicobacteraceae exhibit a wide range of relative abundances in the colonization carbonates at active stations but only a very minor amount of colonization at low-activity stations (Fig. 5; also, see Table S3 in the supplemental material). The Thiotrichaceae OTUs also demonstrate colonization patterns reminiscent of distributions observed in the native carbonates, again indicating that putative sulfur-oxidizing OTUs are dynamic responders to carbonate substrate availability in regions of seep activity at the seabed. However, the specific Thiotrichaceae OTU observed to most strongly colonize experimental carbonates was different than the Thiotrichaceae OTU more frequently observed in the native carbonates (see Table S3 in the supplemental material)—demonstrating the potential for within-group variability in ecological expression. The recovery of Helicobacteraceae or Thiotrichaceae OTUs was not obviously tied to qualitative observations of bacterial mats upon recovery of colonized carbonates from the seafloor. Previous studies of microbial colonization in shallow marine sediments and near hydrothermal vents have observed a dominance of early-stage colonization by Epsilonproteobacteria (40, 41, 48, 51), and similar ecological behavior appears to be occurring in methane seeps. The rapid colonization by Epsilonproteobacteria in various marine settings has been attributed to both a tolerance for rapidly changing physicochemical conditions and motility within many members of the class (40, 48). We observe that our key Helicobacteraceae OTUs were recovered in high relative abundance in methane seep sediments and low relative abundance in bottom water samples (see Table S3 in the supplemental material); therefore, it appears likely the Helicobacteraceae recovered in the colonization experiments were inoculated from underlying sediments, in contrast to Colwelliaceae OTUs derived from overlying bottom waters.
Colonization by the ANME-1-associated OTUs (the same OTUs as recovered from native carbonates) on the sterile carbonates was observed at low levels at both active and low-activity stations (Fig. 5). Any level of colonization by ANME-1 is intriguing for two reasons. First, ANME-1 are believed to have doubling times on the order of several months, so the 13-month course of the colonization experiments could reasonably be expected not to have provided enough time for ANME-1 archaea to colonize and become established on the fresh carbonate substrates (44–47). Second, ANME-1 are obligate anaerobes typically associated with highly reducing conditions located deeper within the sediment column at seeps and near the sulfate-methane transition zone, not at the sediment/water interface, where the colonization experiments were located (52). An exception to this are the Black Sea “reefs,” composed partly of ANME-1; however, these grow into permanently stratified bottom water of the euxinic Black Sea (53). That ANME-1 OTUs are observed at significant levels in the sediment samples but at negligible levels in the aerobic bottom water samples (Fig. 3) indicates that ANME-1 almost certainly colonize the carbonates seeded by the underlying sediments. This highlights the complexity of potential mechanisms driving regional and global between-seep dispersion of ANME-1 archaea and perhaps other ANME subclades, as previously observed (10) and perhaps accomplished through periodic sediment disturbance. In contrast to our observations, Archaea were not observed as early colonizers in hydrothermal vent colonization experiments, despite their presence within in situ vent communities (40). Our experiments suggest that ANME-1 archaea may exhibit phenotypes thus far undiscovered in seep settings or may be distributed by hydrological flow or macrofaunal movements (pumping, filtering, burrowing, defecation, etc.).
Wood-colonizing microbial assemblages at methane seeps in the Mediterranean Sea have been observed to be different than surrounding, off-seep sediment-hosted microbial assemblages (54). Our data further demonstrate that even among active and low-activity seep stations, wood-colonizing microbial assemblages differ after 13 months (Fig. 1C). The stark difference between carbonate- and wood-colonizing assemblages in our data set highlights the importance of habitat substrate to deep-sea microbial assemblages. The mere presence of putative sulfur-oxidizing Epsilonproteobacteria and Gammaproteobacteria in the wood colonization experiments suggests that wood falls may act as ephemeral sulfide-rich reducing habitats, possibly representing stepping stones between seeps and vents for chemosynthetic communities, as has been hypothesized for metazoans and Bacteria (54, 55). Our results are consistent with previous characterizations of native wood fall samples, as well as deep-sea benthic wood colonization experiments, which yielded observations of phylogenetically diverse microbial assemblages, including, but not limited to, the Bacteroidetes, Firmicutes, Spirochaetes, Epsilonproteobacteria, and Gammaproteobacteria (54, 56, 57), but very limited recovery of methanogenic and methanotrophic archaeal taxa (57). The lack of significant ANME colonization in the wood experiments (see Table S3 in the supplemental material) indicates that AOM-related archaeal taxa may have more difficulty spreading geographically via wood substrates than many Bacteria. That AOM-related archaeal taxa appear to be able to colonize carbonate substrates, even on relatively short timescales, indicates a possible mode of wide geographic dispersion. Other hypotheses have included transportation in the guts of deep-sea metazoans or distribution during ocean anoxic events (10), both of which may complement the apparent suitability of carbonate habitats for ANME.
In summary, the deployment of in situ manipulation experiments, coupled to an extensive characterization of native microbial assemblages in association with various seep habitat substrates, has enabled unique insights into the ecology of seep microorganisms. Microbial assemblages associated with carbonates at methane seeps are distinct from, and more diverse than, other habitat substrates examined in this study: sediments, nodules, and bottom waters. Further, bulk carbonate-associated microbial assemblages are adapted to resist seep quiescence and poised to respond to seep activation over 13 months. OTUs associated with the epsilonproteobacterial Helicobacteraceae are particularly sensitive to seep activity. Colonization experiments corroborate the idea that carbonates host distinct and diverse microbial assemblages, and recovery of ANME-1 OTUs associated with the carbonates suggests more dynamic physiologies and/or distribution processes for these organisms than previously hypothesized.
The difference in the microbial assemblages associated with native active and low-activity carbonates, coupled to the dynamics and decreased OTU richness observed in the transplant experiments, suggests that upon the final quiescence of a historic methane seep, the genomic microbial signatures recorded in carbonates could differ from those microbes which were present during active seepage. Investigation of our same research questions should be applied to lipid profiles, to investigate whether trends observed at the genomic level are likely to be preserved in the rock record and, in particular, whether microbial signatures in the rock record merely reflect the final, low-activity period of seep activity rather than the biological assemblage present during the most active phases of seepage and AOM.
MATERIALS AND METHODS
Sample collection and deployment of experiments.
The majority of samples in this study (114 out of 134) (see Table S1 in the supplemental material), including all transplantation and colonization treatments (see below), are from an extensively studied natural laboratory of methane seepage, namely, the northern and southern promontories of Hydrate Ridge (HR), on the Cascadia margin in Oregon, USA (HR-North: 44°40′N, 125°6′W, ~600 m below sea level [MBSL]; HR-South; 44°34′N, 125°9′W, ~800 MBSL) (see Fig. S1A, B, and D to H in the supplemental material) (31, 58–62). Active and low-activity stations were identified by the presence (or absence) of benthic chemosynthetic communities throughout HR and given sequential names for experimental purposes (stations spaced 101 to 104 m apart on the seafloor) (see Fig. S1D and E and Table S1 in the supplemental material). Our active and low-activity station designations were confirmed by pore water sulfide concentrations from 0- to 3-cm-below-seafloor horizons of sediment cores collected within active stations (1 to 14 mM range, 6 mM average; n = 9) and low-activity stations (0 to 0.9 mM range, 0.2 mM average; n = 5) (for more details, see Text S1 in the supplemental material). Of 114 HR samples, 110, including carbonates, sediments, nodules, bottom water, and woods, were collected from these stations, with four additional carbonate samples obtained from a seep promontory approximately 20 km SSE of HR (Southeast Knoll; 44°27.0′N, 125°7.8′W, ~620 MBSL). Of the remaining 20 samples in this study, 10 carbonates were collected from seeps off the Costa Rica coast: Mound 11, Mound 12, Quepos Mound, and Jaco Scarp (see Fig. S1C in the supplemental material) (25, 63, 64). As a point of comparison to sediments and nodules collected at HR, we also included ten sediment and nodule samples from Eel River Basin (ERB; 40°48.7′N, 124°36.7′W, 517 MBSL) (30, 65). Recently published sequencing data from 18 sediment and nodule samples (13% of our 134-sample data set) provide valuable context for this study regarding habitat substrate and are indicated in Table S1 in the supplemental material (19).
Among all the collected samples, 82 out of 134 represent native, unperturbed microbial assemblages associated with a variety of habitat substrates (ncarbonate = 57; nnodule = 10; nbottom water = 2; nsediment = 13) and seep activity levels (nactive = 52, nlow-activity = 28, noff seep = 2). Samples were collected in 2006, 2009, 2010, and 2011 during R/V Atlantis cruises AT15-11, AT15-44, AT15-68, and AT18-10, respectively. Upon shipboard retrieval, subsamples were immediately frozen at −80°C and transferred to an onshore lab for downstream processing. Mineralogy of carbonate samples was examined by powder X-ray diffraction (see Text S1 in the supplemental material).
Six transplanted carbonate and 46 introduced carbonate and wood (ncarbonate = 20, nwood = 26) samples represent microbial assemblages after 13 months of incubation on the seafloor. Transplantation experiments were conducted using the DSV Alvin in August 2010 by moving seafloor carbonates at HR-North from active to low-activity stations (n = 4) and vice versa (n = 2), followed by collection and freezing in September 2011 using the ROV Jason II. Colonization experiments were conducted with fir and pine woods (n = 26) and autoclaved, aseptically stored calcite and dolomite seep carbonates (n = 20) deposited at selected seafloor stations, including those of the transplantation experiments, in August 2010 (AT15-68) and recovered in September 2011 (AT18-10). More methodological details regarding the transplantation and colonization experiments can be found in Text S1 in the supplemental material.
Genomic DNA extraction and 16S rRNA gene sequencing and processing.
Onshore, the carbonates, sediments, and nodules were separately ground into powder with a sterile porcelain mortar and pestle. The nodules, which were only loosely consolidated and thus could have contained sediment-phase contamination, were preprocessed in order to thoroughly remove sediment as previously described (19), with the exception of nodule 5118N (see Table S1 in the supplemental material). Genomic DNA was extracted following the general procedure of the MoBio PowerSoil kit (MoBio, St. Louis, MO) (see reference 5 for variations from default protocol), using ~400 mg powder. For wood samples, a sterile razor blade was used to collect shavings from the exterior, avoiding the bark and any observed animals (e.g., shipworms) whenever possible. DNA from wood samples was extracted using the MoBio PowerPlantPro kit’s recommended protocol, with 40 µl of phenolic separation solution and ~70 mg wood shavings. Bottom water samples from nearby station HR-9 (see Fig. S1 in the supplemental material) were collected on a 0.2-µm filter and extracted by phenol-chloroform followed by CsCl density gradient centrifugation (66).
Preparation for sequencing of the V4 region of the 16S rRNA gene was performed with universal primers according to the protocol recommended by the EMP (iTag sequencing [67]) (http://www.earthmicrobiome.org/emp-standard-protocols/16s/) (68, 69), with minor modifications as previously described (19). Raw sequences were generated on an Illumina MiSeq platform at Laragen, Inc. (Los Angeles, CA). In-house data processing was completed in QIIME1.8.0 and included joining paired ends, quality trimming, chimera checking, 97% OTU clustering, singleton removal, PCR contaminant removal, 0.01% relative abundance threshold removal, and rarefaction to 16,051 sequences per sample (see Text S1 in the supplemental material). Taxonomic assignments were generated according to an appended version of the Silva 115 database (for details, see reference 19).
Diversity analyses.
Alpha diversity calculations (Shannon diversity index [H′], observed OTUs, and Chao1) were carried out in QIIME1.8.0 (alpha_diversity.py). Nonmetric multidimensional scaling (NMDS) analyses were carried out in the R environment (70) after application of a square root transformation to the relative abundance data. For all analysis of similarity (ANOSIM) tests, P values of <0.05 were considered significant. R values are reported only for tests which yielded significant results. Examples of the R commands, including options used, are given in Text S1 in the supplemental material. Distance-based linear modeling (distLM) was applied with Primer-E software to complement the ANOSIM testing (71). The similarity percentage (SIMPER) test was applied in R to identify specific OTUs which demonstrate different relative abundances between sample groups; key OTUs were selected for presentation and usually represented the majority of sequences associated with each taxonomy (Fig. 3 and 5; also, see Table S3 in the supplemental material).
Sequence accession numbers.
Raw sequences are available in the Sequence Read Archive under accession numbers SRP055767 and SRP049675.
SUPPLEMENTAL MATERIAL
Overview map of sampling locations in this study. For seep descriptions, see “Sample collection and deployment of experiments” in the text. (A) Overview of the three seep locations sampled in this study; (B) map of Hydrate Ridge, including sampling locations at HR-North and HR-South; (C) map of Costa Rica, including the four mounds sampled for native carbonates; (D) map of HR-North showing the spatial relationship between stations HR-3, -4, -5, -6, -7, and -8; (E) map of HR-South showing spatial relationship between stations HR-1, -2, -9, -10, -11, -12, and -V1; (F and G) seafloor images of active stations HR-3 and HR-7 exhibiting orange bacterial mats (HR-3), clam beds (HR-3 and HR-7), and methane ebullition (HR-7); (H) seafloor image of low-activity station HR-4 lacking the diagnostic indicators seen in panels F and G. Panels A to C were generated in GeoMapApp (http://www.geomapapp.org) (72). For panel B, contour lines represent 100-m bathymetric relief, with HR-North being the shallowest, at 700 MBSL. For panel C, contour lines represent 250-m bathymetric relief, with the shallow continental shelf at 250 MBSL in the NE corner. Red and blue points plotted in panels B to E represent active and low-activity stations, respectively. Download
Additional alpha diversity metrics for the samples in this study. (A to C) Collector’s curves of raw OTU97 for the native samples (A), carbonate samples (B), and colonization samples (C). (D) Shannon diversity indices (H′) for the major sample groups analyzed in this study. Error bars represent standard deviations among the sample groups and are not included for the bottom water and transplant-to-active samples due to the low number of replicates. Download
Additional nonmetric multidimensional scaling analyses. (A) Ordination of the sample data from native carbonates, sediments, nodules, and bottom water. Ordination is identical to that in Fig. 1A, but the carbonates, sediments, and nodules are colored by geographic origin. The overlap between Costa Rica and Hydrate Ridge carbonates, in the same vicinity on the plot as Hydrate Ridge and Eel River Basin sediments and nodules, demonstrates that the region where habitat substrates overlap (roughly indicated by the dashed circle) is not a geographic effect. (B) All 134 samples in this study. The same intrasample relationships are visible as in Fig. 1. The wood colonization samples plot in a distinct region away from all other samples, though they are most similar to the colonization carbonates. Sample C2693 is highlighted as a biological outlier among the nodule-hosted samples. Sample 5471 is highlighted because in this ordination plot of all samples, it appears to be biologically similar to some of the carbonate colonization samples. Sample 5471 was bottom water from an active seep site (see Table S1), which may be related to its similarity to some colonization samples; more bottom water data points would be necessary in order to draw a stronger conclusion. Download
Mineralogical analysis. (A and B) Distribution of recovered carbonate mineralogies at low-activity (A) and active seep stations (B). Note that aragonite-bearing mineralogies are dominant at low-activity stations, while active seep stations host more varied mineralogies, including a greater proportion of dolomite-bearing carbonates. In order to emphasize the distributions, text size is proportional to value. From this data, it appears that carbonate mineralogy may have a loose correlation with seep activity. From our data set we are unable to determine whether mineralogy and seep activity are truly dependent or independent environmental variables (see Text S1). (C) Nonmetric multidimensional scaling analysis of microbial assemblages from native carbonate samples according to seep activity and mineralogy. Samples are separated according to seep activity (Fig. 1B), but such a difference is also qualitatively correlated with mineralogical differences. Download
Distance-decay plot of the 57 native carbonates collected from Hydrate Ridge and Costa Rica. Geographical distance is calculated in km along a Great Circle between each sample pair (R package geosphere v1.3-13, function distHaversine, assuming a spherical Earth with a radius of 6,371 km). Bray-Curtis similarity was calculated as described in the text, with a value of 1 representing identical similarity and 0 representing complete dissimilarity. Note that the y axis is linear and the x axis is logarithmic. No correlation is observed between Bray-Curtis similarity and geographic distance. Although this does not preclude the existence of such a correlation, it means that such a correlation may be masked by influences of other environmental factors (e.g., seep activity). Download
OTU overlap between colonized substrates at active site HR-9, including representative native substrates. Site HR-9 was chosen because it was the associated site for both bottom water samples, it hosted both carbonate and wood colonization experiments, it hosted multiple native carbonates which were recovered and analyzed, and it was an active seep site. Since sediment and nodule samples were not collected from HR-9, representatives of those substrates were chosen from another HR-South location (HR-V1); shallow (0 to 3 cm below the sea floor [CMBSF]) sediments and nodules were chosen because they were most likely to be relevant for colonization experiments which were performed on the seafloor. In order to keep the Venn diagram relatively simple, and because ANOSIM tests revealed sediments and nodules to be indistinguishable from one another, sediments and nodules were binned together as one habitat type for this analysis. In order to ensure equal depth of sampling across each substrate type, two representative samples of each substrate were chosen randomly (sample numbers are provided). In order to emphasize the distributions, text size is proportional to OTU occurrence. For all OTU overlap diagrams, the specific samples analyzed are given in small text next to the diagram bubbles. In order for an OTU to be counted as present for a category (e.g., for an OTU to be associated with bottom water), it had to be present in both replicates. This approach ensured that the OTUs under examination were reproducibly recovered from every category, rather than spurious observations. Download
All samples in this study are listed, with their accompanying metadata. Bold boxes denote the three different experimental treatments in the study: native samples, transplantation samples, and colonization samples.
List of ANOSIM tests. Significance values (P values) of >0.05 are in green, and those that are <0.05 are in red.
Relative abundance information is listed for OTUs presented in Fig. 3 to 5 of the main text. Data are provided in four tabs: tab 1, raw data associated with Fig. 3; tab 2, raw data associated with Fig. 4B; tab 3, raw data associated with Fig. 4C; tab 4, raw data associated with Fig. 5. For the data used to generate Fig. 5, in all cases a minority of the OTUs were identified for presentation (e.g., 3 of 8 for ANME-1). However, these represented the majority of the total sequences recovered within each taxonomic group (e.g., 95% of all sequences for ANME-1). Bold boxes outline the sets of OTUs which were binned for presentation in Fig. 5. Color scales are meant to aid the reader in assessing OTU distribution. OTUs of the same phylogeny were combined for presentation in Fig. 5
Supplemental methods. Download
ACKNOWLEDGMENTS
We thank Connor Skennerton and Elizabeth Trembath-Reichert for helpful discussions on data processing. We acknowledge Stephanie Connon, Patricia Tavormina, Josh Steele, Heather Grotzinger, and shipboard teams from the Orphan, Levin, Rathburn, and Rouse labs for assistance with sample collection and processing. We are indebted to the captain, crew, and pilots of the DSV Alvin and ROV Jason II from cruises AT15-11, AT15-44, AT15-68, who made this work possible. We thank the two anonymous reviewers, whose comments strengthened the study.
This research was supported by a grant to V.O. from the NASA Astrobiology Institute (award NNA13AA92A). This work was also support by a National Science Foundation (NSF) grant (OCE-0825791) and a Gordon and Betty Moore Foundation Marine Microbiology Initiative grant (3780) to V.O. D.C. was supported by an NSF Graduate Research Fellowship. Levin lab research was supported by NSF grants OCE-0826254 and OCE-0939557.
Footnotes
Citation Case DH, Pasulka AL, Marlow JJ, Grupe BM, Levin LA, Orphan VJ. 2015. Methane seep carbonates host distinct, diverse, and dynamic microbial assemblages. mBio 6(6):e01348-15. doi:10.1128/mBio.01348-15.
REFERENCES
- 1.Niemann H, Linke P, Knittel K, MacPherson E, Boetius A, Brückmann W, Larvik G, Wallmann K, Schacht U, Omoregie E, Hilton D, Brown K, Rehder G. 2013. Methane-carbon flow into the benthic food web at cold seeps—a case study from the Costa Rica subduction zone. PLoS One 8:e74894. doi: 10.1371/journal.pone.0074894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thurber AR, Levin LA, Orphan VJ, Marlow JJ. 2012. Archaea in metazoan diets: implications for food webs and biogeochemical cycling. ISME J 6:1602–1612. doi: 10.1038/ismej.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin L. 2005. Ecology of cold seep sediments: interactions of fauna with flow, chemistry, and microbes. Oceanogr Mar Biol Annu Rev 43:1–46. doi: 10.1201/9781420037449.ch1. [DOI] [Google Scholar]
- 4.Hinrichs KU, Hayes JM, Sylva SP, Brewer PG, DeLong EF. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 5.Orphan VJ, Hinrichs K-, Ussler W, Paull CK, Taylor LT, Sylva SP, Hayes JM, Delong EF. 2001. Comparative analysis of methane-oxidizing Archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl Environ Microbiol 67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pop Ristova P, Wenzhöfer F, Ramette A, Felden J, Boetius A. 2015. Spatial scales of bacterial community diversity at cold seeps (Eastern Mediterranean Sea). ISME J 9:1306–1318. doi: 10.1038/ismej.2014.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nunoura T, Takaki Y, Kazama H, Hirai M, Ashi J, Imachi H, Takai K. 2012. Microbial diversity in deep-sea methane seep sediments presented by SSU rRNA gene tag sequencing. Microbes Environ 27:382–390. doi: 10.1264/jsme2.ME12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruff SE, Arnds J, Knittel K, Amann R, Wegener G, Ramette A, Boetius A. 2013. Microbial communities of deep-sea methane seeps at Hikurangi Continental margin (New Zealand). PLoS One 8:e72627. doi: 10.1371/journal.pone.0072627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marlow JJ, Steele JA, Case DH, Connon SA, Levin LA, Orphan VJ. 2014. Microbial abundance and diversity patterns associated with sediments and carbonates from the methane seep environments of Hydrate Ridge, OR. Front Mar Sci 1:1–16. doi: 10.3389/fmars.2014.00044. [DOI] [Google Scholar]
- 10.Ruff SE, Biddle JF, Teske AP, Knittel K, Boetius A, Ramette A. 2015. Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci U S A 112:4015–4020. doi: 10.1073/pnas.1421865112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadnitskaia A, Muyzer G, Abbas B, Coolen MJL, Hopmans EC, Baas M, van Weering TCE, Ivanov MK, Poludetkina E, Sinninghe Damsté JS. 2005. Biomarker and 16S rDNA evidence for anaerobic oxidation of methane and related carbonate precipitation in deep-sea mud volcanoes of the Sorokin Trough, Black Sea. Mar Geol 217:67–96. doi: 10.1016/j.margeo.2005.02.023. [DOI] [Google Scholar]
- 12.Thiel V, Peckmann J, Richnow HH, Luth U, Reitner J, Michaelis W. 2001. Molecular signals for anaerobic methane oxidation in Black Sea seep carbonates and a microbial mat. Mar Chem 73:97–112. doi: 10.1016/S0304-4203(00)00099-2. [DOI] [Google Scholar]
- 13.Heijs SK, Aloisi G, Bouloubassi I, Pancost RD, Pierre C, Sinninghe Damsté JS, Gottschal JC, van Elsas JD, Forney LJ. 2006. Microbial community structure in three deep-sea carbonate crusts. Microb Ecol 52:451–462. doi: 10.1007/s00248-006-9099-8. [DOI] [PubMed] [Google Scholar]
- 14.Gieskes J, Mahn C, Day S, Martin JB, Greinert J, Rathburn T, McAdoo B. 2005. A study of the chemistry of pore fluids and authigenic carbonates in methane seep environments: Kodiak Trench, Hydrate Ridge, Monterey Bay, and Eel River Basin. Chem Geol 220:329–345. doi: 10.1016/j.chemgeo.2005.04.002. [DOI] [Google Scholar]
- 15.Greinert J, Bohrmann G, Suess E. 2001. Gas hydrate-associated carbonates and methane-venting at Hydrate Ridge: classification, distribution, and origin of authigenic lithologies, p 99–113. In Paull CK, Dillon WP (ed), Natural gas hydrates. American Geophysical Union, Washington, DC. [Google Scholar]
- 16.Marlow JJ, Steele JA, Ziebis W, Thurber AR, Levin LA, Orphan VJ. 2014. Carbonate-hosted methanotrophy represents an unrecognized methane sink in the deep sea. Nat J Commun 5:5094. [DOI] [PubMed] [Google Scholar]
- 17.Marlow J, Peckmann J, Orphan V. 2015. Autoendoliths: a distinct type of rock-hosted microbial life. Geobiology 13:303–307. doi: 10.1111/gbi.12131. [DOI] [PubMed] [Google Scholar]
- 18.Levin LA, Mendoza GF, Grupe BM, Gonzalez JP, Jellison B, Rouse G, Thurber AR, Waren A. 2015. Biodiversity on the rocks: macrofauna inhabiting authigenic carbonate at Costa Rica methane seeps. PLoS One 10:1–31. doi: 10.1371/journal.pone.0131080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason OU, Case DH, Naehr TH, Lee RW, Thomas RB, Bailey JV, Orphan VJ. 2015. Comparison of archaeal and bacterial diversity in methane seep carbonate nodules and host sediments, Eel River Basin and Hydrate Ridge, USA. Microb Ecol 70:766–784. doi: 10.1007/s00248-015-0615-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Yan W, Chen M, Wang S, Lu J, Zhang F, Xiang R, Xiao S, Yan P, Gu S. 2006. Discovery of seep carbonate nodules as new evidence for gas venting on the northern continental slope of South China sea. Chin Sci Bull 51:1228–1237. doi: 10.1007/s11434-006-1228-8. [DOI] [Google Scholar]
- 21.Watanabe Y, Nakai S, Hiruta A, Matsumoto R, Yoshida K. 2008. U–Th dating of carbonate nodules from methane seeps off Joetsu, Eastern margin of Japan Sea. Earth Planet Sci Lett 272:89–96. doi: 10.1016/j.epsl.2008.04.012. [DOI] [Google Scholar]
- 22.Hovland M, Talbot MR, Qvale H, Olaussen S, Aasberg L. 1987. Methane-related carbonate cements in pockmarks of the North Sea. J Sedimentary Petrology 57:881–892. [Google Scholar]
- 23.Naehr TH, Eichhubl P, Orphan VJ, Hovland M, Paull CK, Ussler W, Lorenson TD, Greene HG. 2007. Authigenic carbonate formation at hydrocarbon seeps in continental margin sediments: a comparative study. Deep Sea Res II 54:1268–1291. doi: 10.1016/j.dsr2.2007.04.010. [DOI] [Google Scholar]
- 24.Teichert BMA, Bohrmann G, Suess E. 2005. Chemoherms on Hydrate Ridge—unique microbially-mediated carbonate build-ups growing into the water column. Palaeogeogr Palaeoclimatol Palaeoecol 227:67–85. doi: 10.1016/j.palaeo.2005.04.029. [DOI] [Google Scholar]
- 25.Sahling H, Masson DG, Ranero CR, Hühnerbach V, Weinrebe W, Klaucke I, Bürk D, Brückmann W, Suess E. 2008. Fluid seepage at the continental margin offshore Costa Rica and southern Nicaragua. Geochem Geophys Geosyst 9:Q05S05. doi: 10.1029/2008GC001978. [DOI] [Google Scholar]
- 26.Birgel D, Himmler T, Freiwald A, Peckmann J. 2008. A new constraint on the antiquity of anaerobic oxidation of methane: late Pennsylvanian seep limestones from southern Namibia. Geology 36:543–546. doi: 10.1130/G24690A.1. [DOI] [Google Scholar]
- 27.Tryon MD, Brown KM, Torres ME. 2002. Fluid and chemical flux in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR, II: hydrological processes. Earth Planet Sci Lett 201:541–557. doi: 10.1016/S0012-821X(02)00732-X. [DOI] [Google Scholar]
- 28.Tryon MD, Brown KM, Torres ME, Tréhu AM, McManus J, Collier RW. 1999. Measurements of transience and downward fluid flow near episodic methane gas vents, Hydrate Ridge, Cascadia. Geology 27:1075–1078. doi:. [DOI] [Google Scholar]
- 29.Bekins BA, Dreiss SJ. 1992. A simplified analysis of parameters controlling dewatering in accretionary prisms. Earth Planet Science Lett 109:275–287. doi: 10.1016/0012-821X(92)90092-A. [DOI] [Google Scholar]
- 30.Orphan VJ, Ussler W, Naehr TH, House CH, Hinrichs K-, Paull CK. 2004. Geological, geochemical, and microbiological heterogeneity of the seafloor around methane vents in the Eel River Basin, offshore California. Chem Geol 205:265–289. doi: 10.1016/j.chemgeo.2003.12.035. [DOI] [Google Scholar]
- 31.Boetius A, Suess E. 2004. Hydrate Ridge: a natural laboratory for the study of microbial life fueled by methane from near-surface gas hydrates. Chem Geol 205:291–310. doi: 10.1016/j.chemgeo.2003.12.034. [DOI] [Google Scholar]
- 32.Rossel PE, Elvert M, Ramette A, Boetius A, Hinrichs K. 2011. Factors controlling the distribution of anaerobic methanotrophic communities in marine environments: evidence from intact polar membrane lipids. Geochim Cosmochim Acta 75:164–184. doi: 10.1016/j.gca.2010.09.031. [DOI] [Google Scholar]
- 33.Bowman JP, McCuaig RD. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl Environ Microbiol 69:2463–2483. doi: 10.1128/AEM.69.5.2463-2483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schauer R, Bienhold C, Ramette A, Harder J. 2010. Bacterial diversity and biogeography in deep-sea surface sediments of the south Atlantic Ocean. ISME J 4:159–170. doi: 10.1038/ismej.2009.106. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Kato C, Horikoshi K. 1999. Microbial diversity in sediments collected from the deepest cold-seep area, the Japan Trench. Mar Biotechnol 1:391–400. doi: 10.1007/PL00011793. [DOI] [PubMed] [Google Scholar]
- 36.Ijiri A, Tsunogai U, Gamo T, Nakagawa F, Sakamoto T, Saito S. 2009. Enrichment of adsorbed methane in authigenic carbonate concretions of the Japan Trench. Geo-Marine Lett 29:301–308. doi: 10.1007/s00367-009-0143-9. [DOI] [Google Scholar]
- 37.Blumenberg M, Walliser E, Taviani M, Seifert R, Reitner J. 2015. Authigenic carbonate formation and its impact on the biomarker inventory at hydrocarbon seeps—a case study from the Holocene Black sea and the plio-Pleistocene Northern Apennines (Italy). Mar Petrol Geol 66:532–541. doi: 10.1016/j.marpetgeo.2015.05.013. [DOI] [Google Scholar]
- 38.Treude T, Boetius A, Knittel K, Wallmann K, Jørgensen B. 2003. Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar Ecol Prog Ser 264:1–14. doi: 10.3354/meps264001. [DOI] [Google Scholar]
- 39.Sylvan JB, Toner BM, Edwards KJ. 2012. Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. mBio 3:e00279-11. doi: 10.1128/mBio.00279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alain K, Zbinden M, Le Bris N, Lesongeur F, Quérellou J, Gaill F, Cambon-Bonavita. 2004. Early steps in microbial colonization processes at deep-sea hydrothermal vents. Environ Microbiol 6:227–241. doi: 10.1111/j.1462-2920.2003.00557.x. [DOI] [PubMed] [Google Scholar]
- 41.Sylvan JB, Pyenson BC, Rouxel O, German CR, Edwards KJ. 2012. Time-series analysis of two hydrothermal plumes at 9°50′N East Pacific Rise reveals distinct, heterogeneous bacterial populations. Geobiology 10:178–192. doi: 10.1111/j.1472-4669.2011.00315.x. [DOI] [PubMed] [Google Scholar]
- 42.Garrity GM, et al.(ed). 2005. The Proteobacteria. Bergey’s manual of systematic bacteriology, vol 2 Springer Verlag, New York, NY. [Google Scholar]
- 43.Sorokin DY, Tourova TP, Bezsoudnova EY, Pol A, Muyzer G. 2007. Denitrification in a binary culture and thiocyanate metabolism in Thiohalophilus thiocyanoxidans gen. nov. sp. nov.—a moderately halophilic chemolithoautotrophic sulfur-oxidizing Gammaproteobacterium from hypersaline lakes. Arch Microbiol 187:441–450. doi: 10.1007/s00203-006-0208-3. [DOI] [PubMed] [Google Scholar]
- 44.Morono Y, Terada T, Nishizawa M, Ito M, Hillion F, Takahata N, Sano Y, Inagaki F. 2011. Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc Natl Acad Sci U S A 108:18295–18300. doi: 10.1073/pnas.1107763108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orphan VJ, Turk KA, Green AM, House CH. 2009. Patterns of 15N assimilation and growth of methanotrophic ANME-2 archaea and sulfate-reducing bacteria within structured syntrophic consortia revealed by FISH-SIMS. Environ Microbiol 11:1777–1791. doi: 10.1111/j.1462-2920.2009.01903.x. [DOI] [PubMed] [Google Scholar]
- 46.Girguis PR, Orphan VJ, Hallam SJ, DeLong EF. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl Environ Microbiol 69:5472–5482. doi: 10.1128/AEM.69.9.5472-5482.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajala P, Bomberg M, Kietäväinen R, Kukkonen I, Ahonen L, Nyyssönen M, Itävaara M. 2015. Rapid reactivation of deep subsurface microbes in the presence of C-1 compounds. Microorganisms 3:17–33. doi: 10.3390/microorganisms3010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernard C, Fenchel T. 1995. Mats of colourless sulphur bacteria. II. Structure, composition of biota and successional patterns. Mar Ecol Prog Ser 128:171–179. doi: 10.3354/meps128171. [DOI] [Google Scholar]
- 49.Sievert SM, Wieringa EBA, Wirsen CO, Taylor CD. 2007. Growth and mechanism of filamentous-sulfur formation by Candidatus Arcobacter sulfidicus in opposing oxygen-sulfide gradients. Environ Microbiol 9:271–276. doi: 10.1111/j.1462-2920.2006.01156.x. [DOI] [PubMed] [Google Scholar]
- 50.Schmale O, Leifer I, Schneider von Deimling J, Stolle C, Krause S, Kießlich K, Frahm A, Treude T. 2015. Bubble transport mechanism: indications for a gas bubble-mediated inoculation of benthic methanotrophs into the water column. Contin Shelf Res 103:70–78. doi: 10.1016/j.csr.2015.04.022. [DOI] [Google Scholar]
- 51.Taylor CD, Wirsen CO, Gaill F. 1999. Rapid microbial production of filamentous sulfur mats at hydrothermal vents. Appl Environ Microbiol 65:2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol 71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reitner J, Peckmann J, Reimer A, Schumann G, Thiel V. 2005. Methane-derived carbonate build-ups and associated microbial communities at cold seeps on the lower Crimean shelf (Black Sea). Facies 51:66–79. doi: 10.1007/s10347-005-0059-4. [DOI] [Google Scholar]
- 54.Bienhold C, Pop Ristova P, Wenzhöfer F, Dittmar T, Boetius A. 2013. How deep-sea wood falls sustain chemosynthetic life. PLoS One 8:e53590. doi: 10.1371/journal.pone.0053590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Distel DL, Baco AR, Chuang E, Morrill W, Cavanaugh C, Smith CR. 2000. Marine ecology: do mussels take wooden steps to deep-sea vents? Nature 403:725–726. doi: 10.1038/35001667. [DOI] [PubMed] [Google Scholar]
- 56.Fagervold SK, Romano C, Kalenitchenko D, Borowski C, Nunes-Jorge A, Martin D, Galand PE. 2014. Microbial communities in sunken wood are structured by wood-boring bivalves and location in a submarine canyon. PLoS One 9:e96248. doi: 10.1371/journal.pone.0096248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fagervold SK, Galand PE, Zbinden M, Gaill F, Lebaron P, Palacios C. 2012. Sunken woods on the ocean floor provide diverse specialized habitats for microorganisms. FEMS Microbiol Ecol 82:616–628. doi: 10.1111/j.1574-6941.2012.01432.x. [DOI] [PubMed] [Google Scholar]
- 58.Suess E, Carson B, Ritger SD, Moore JC, Jones ML, Kulm LD, Cochrane GR. 1985. Biological communities at vent sites along the subduction zone off Oregon. Bull Biol Soc Wash 6:475–484. [Google Scholar]
- 59.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 60.Sahling H, Rickert D, Lee R, Linke P, Suess E. 2002. Macrofaunal community structure and sulfide flux at gas hydrate deposits from the Cascadia convergent margin, NE Pacific. Mar Ecol Prog Ser 231:121–138. doi: 10.3354/meps231121. [DOI] [Google Scholar]
- 61.Levin LA, Mendoza GF, Gonzalez JP, Thurber AR, Cordes EE. 2010. Diversity of bathyal macrofauna on the northeastern Pacific margin: the influence of methane seeps and oxygen minimum zones. Mar Ecol 31:94–110. doi: 10.1111/j.1439-0485.2009.00335.x. [DOI] [Google Scholar]
- 62.Guilini K, Levin LA, Vanreusel A. 2012. Cold seep and oxygen minimum zone associated sources of margin heterogeneity affect benthic assemblages, diversity and nutrition at the Cascadian margin (NE Pacific Ocean). Prog Oceanogr 96:77–92. doi: 10.1016/j.pocean.2011.10.003. [DOI] [Google Scholar]
- 63.Levin LA, Orphan VJ, Rouse GW, Rathburn AE, Ussler W, Cook GS, Goffredi SK, Perez EM, Waren A, Grupe BM, Chadwick G, Strickrott B. 2012. A hydrothermal seep on the Costa Rica margin: middle ground in a continuum of reducing ecosystems. Proc R Soc Lond 279:2580–2588. doi: 10.1098/rspb.2012.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dekas AE, Chadwick GL, Bowles MW, Joye SB, Orphan VJ. 2014. Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME archaea. Environ Microbiol 16:3012–3029. doi: 10.1111/1462-2920.12247. [DOI] [PubMed] [Google Scholar]
- 65.Levin L, Ziebis W, Mendoza G, Growney V, Tryon M, Brown K, Mahn C, Gieskes J, Rathburn A. 2003. Spatial heterogeneity of macrofauna at northern California methane seeps: influence of sulfide concentration and fluid flow. Mar Ecol 265:123–139. doi: 10.3354/meps265123. [DOI] [Google Scholar]
- 66.Tavormina PL, Ussler W, Joye SB, Harrison BK, Orphan VJ. 2010. Distributions of putative aerobic methanotrophs in diverse pelagic marine environments. ISME J 4:700–710. doi: 10.1038/ismej.2009.155. [DOI] [PubMed] [Google Scholar]
- 67.Gilbert JA, Meyer F, Jansson J, Gordon J, Pace NR, Tiedje JM, Ley RE, Fierer N, Field D, Kyrpides NC, Gloeckner FO, Klenk HP, Wommack KE, Glass E, Docherty K, Gallery R, Stevens R, Knight R. 2011. The Earth Microbiome Project: meeting report of the “1st EMP meeting on sample selection and acquisition” at Argonne National Laboratory, October 6th, 2010, p 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Nalt Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Core Team 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 71.Clarke KR, Warwick RM. 2001. Change in marine communities, 2nd ed. Primer-E Ltd., Plymouth, United Kingdom. [Google Scholar]
- 72.Ryan WBF, Carbotte SM, Coplan JO, O’Hara S, Melkonian A, Arko R, Weissel RA, Ferrini V, Goodwillie A, Nitsche F, Bonczkowski J, Zemsky R. 2009. Global multi-resolution topography synthesis. Geochem Geophys Geosyst 10:1–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview map of sampling locations in this study. For seep descriptions, see “Sample collection and deployment of experiments” in the text. (A) Overview of the three seep locations sampled in this study; (B) map of Hydrate Ridge, including sampling locations at HR-North and HR-South; (C) map of Costa Rica, including the four mounds sampled for native carbonates; (D) map of HR-North showing the spatial relationship between stations HR-3, -4, -5, -6, -7, and -8; (E) map of HR-South showing spatial relationship between stations HR-1, -2, -9, -10, -11, -12, and -V1; (F and G) seafloor images of active stations HR-3 and HR-7 exhibiting orange bacterial mats (HR-3), clam beds (HR-3 and HR-7), and methane ebullition (HR-7); (H) seafloor image of low-activity station HR-4 lacking the diagnostic indicators seen in panels F and G. Panels A to C were generated in GeoMapApp (http://www.geomapapp.org) (72). For panel B, contour lines represent 100-m bathymetric relief, with HR-North being the shallowest, at 700 MBSL. For panel C, contour lines represent 250-m bathymetric relief, with the shallow continental shelf at 250 MBSL in the NE corner. Red and blue points plotted in panels B to E represent active and low-activity stations, respectively. Download
Additional alpha diversity metrics for the samples in this study. (A to C) Collector’s curves of raw OTU97 for the native samples (A), carbonate samples (B), and colonization samples (C). (D) Shannon diversity indices (H′) for the major sample groups analyzed in this study. Error bars represent standard deviations among the sample groups and are not included for the bottom water and transplant-to-active samples due to the low number of replicates. Download
Additional nonmetric multidimensional scaling analyses. (A) Ordination of the sample data from native carbonates, sediments, nodules, and bottom water. Ordination is identical to that in Fig. 1A, but the carbonates, sediments, and nodules are colored by geographic origin. The overlap between Costa Rica and Hydrate Ridge carbonates, in the same vicinity on the plot as Hydrate Ridge and Eel River Basin sediments and nodules, demonstrates that the region where habitat substrates overlap (roughly indicated by the dashed circle) is not a geographic effect. (B) All 134 samples in this study. The same intrasample relationships are visible as in Fig. 1. The wood colonization samples plot in a distinct region away from all other samples, though they are most similar to the colonization carbonates. Sample C2693 is highlighted as a biological outlier among the nodule-hosted samples. Sample 5471 is highlighted because in this ordination plot of all samples, it appears to be biologically similar to some of the carbonate colonization samples. Sample 5471 was bottom water from an active seep site (see Table S1), which may be related to its similarity to some colonization samples; more bottom water data points would be necessary in order to draw a stronger conclusion. Download
Mineralogical analysis. (A and B) Distribution of recovered carbonate mineralogies at low-activity (A) and active seep stations (B). Note that aragonite-bearing mineralogies are dominant at low-activity stations, while active seep stations host more varied mineralogies, including a greater proportion of dolomite-bearing carbonates. In order to emphasize the distributions, text size is proportional to value. From this data, it appears that carbonate mineralogy may have a loose correlation with seep activity. From our data set we are unable to determine whether mineralogy and seep activity are truly dependent or independent environmental variables (see Text S1). (C) Nonmetric multidimensional scaling analysis of microbial assemblages from native carbonate samples according to seep activity and mineralogy. Samples are separated according to seep activity (Fig. 1B), but such a difference is also qualitatively correlated with mineralogical differences. Download
Distance-decay plot of the 57 native carbonates collected from Hydrate Ridge and Costa Rica. Geographical distance is calculated in km along a Great Circle between each sample pair (R package geosphere v1.3-13, function distHaversine, assuming a spherical Earth with a radius of 6,371 km). Bray-Curtis similarity was calculated as described in the text, with a value of 1 representing identical similarity and 0 representing complete dissimilarity. Note that the y axis is linear and the x axis is logarithmic. No correlation is observed between Bray-Curtis similarity and geographic distance. Although this does not preclude the existence of such a correlation, it means that such a correlation may be masked by influences of other environmental factors (e.g., seep activity). Download
OTU overlap between colonized substrates at active site HR-9, including representative native substrates. Site HR-9 was chosen because it was the associated site for both bottom water samples, it hosted both carbonate and wood colonization experiments, it hosted multiple native carbonates which were recovered and analyzed, and it was an active seep site. Since sediment and nodule samples were not collected from HR-9, representatives of those substrates were chosen from another HR-South location (HR-V1); shallow (0 to 3 cm below the sea floor [CMBSF]) sediments and nodules were chosen because they were most likely to be relevant for colonization experiments which were performed on the seafloor. In order to keep the Venn diagram relatively simple, and because ANOSIM tests revealed sediments and nodules to be indistinguishable from one another, sediments and nodules were binned together as one habitat type for this analysis. In order to ensure equal depth of sampling across each substrate type, two representative samples of each substrate were chosen randomly (sample numbers are provided). In order to emphasize the distributions, text size is proportional to OTU occurrence. For all OTU overlap diagrams, the specific samples analyzed are given in small text next to the diagram bubbles. In order for an OTU to be counted as present for a category (e.g., for an OTU to be associated with bottom water), it had to be present in both replicates. This approach ensured that the OTUs under examination were reproducibly recovered from every category, rather than spurious observations. Download
All samples in this study are listed, with their accompanying metadata. Bold boxes denote the three different experimental treatments in the study: native samples, transplantation samples, and colonization samples.
List of ANOSIM tests. Significance values (P values) of >0.05 are in green, and those that are <0.05 are in red.
Relative abundance information is listed for OTUs presented in Fig. 3 to 5 of the main text. Data are provided in four tabs: tab 1, raw data associated with Fig. 3; tab 2, raw data associated with Fig. 4B; tab 3, raw data associated with Fig. 4C; tab 4, raw data associated with Fig. 5. For the data used to generate Fig. 5, in all cases a minority of the OTUs were identified for presentation (e.g., 3 of 8 for ANME-1). However, these represented the majority of the total sequences recovered within each taxonomic group (e.g., 95% of all sequences for ANME-1). Bold boxes outline the sets of OTUs which were binned for presentation in Fig. 5. Color scales are meant to aid the reader in assessing OTU distribution. OTUs of the same phylogeny were combined for presentation in Fig. 5
Supplemental methods. Download