ABSTRACT
The life cycle of high-risk human papillomaviruses (HPVs) is dependent upon epithelial differentiation. Following infection of basal cells, HPV genomes are stably maintained at low copy numbers, and productive replication or amplification is restricted to highly differentiated suprabasal cells. In high-risk HPV infections, the ATM pathway is constitutively activated in the absence of external DNA-damaging agents and is required for productive viral replication. The ataxia telangiectasia (ATM) pathway repairs double-strand breaks in DNA, while the ataxia telangiectasia and Rad3-related (ATR) pathway targets single-strand breaks. Our studies show that the ATR pathway, like the ATM pathway, is activated in HPV-positive cells and that inhibitors of ATR or CHK1 phosphorylation block both amplification and late viral gene expression in differentiated cells while moderately reducing stable copy numbers in undifferentiated cells. TopBP1 is a critical upstream activator of the ATR pathway and is expressed at elevated levels in HPV-positive cells. This increased expression of TopBP1 is necessary for ATR/CHK1 activation in HPV-positive cells, and knockdown blocks amplification. Furthermore, TopBP1 activation is shown to be regulated at the level of transcription initiation by the innate immune regulator STAT-5, which is activated by HPV proteins. STAT-5 has also been shown to be a regulator of the ATM response, demonstrating that these two pathways are coordinately regulated in HPV-positive cells. These findings identify a novel link between the innate immune response and activation of the ATR DNA damage response in regulating the life cycle of high-risk HPVs.
IMPORTANCE
High-risk human papillomaviruses (HPVs) are the causative agents of cervical and other anogenital cancers, as well as many oral cancers. HPVs infect epithelial cells and restrict productive viral replication or amplification and virion production to differentiated cells. Our studies demonstrate that HPVs activate the ATR single-strand DNA repair pathway and this activation is necessary for HPV genome amplification. The innate immune regulator STAT-5 is shown to regulate transcription of the ATR binding factor TopBP1, and this is critical for the induction of the ATR pathway. Our study identifies important links between innate immune signaling, the ATR DNA damage pathway, and productive HPV replication that may lead to the characterization of new targets for the development of therapeutics to treat HPV-induced infections.
INTRODUCTION
High-risk human papillomaviruses (HPVs) are the causative agents of cervical as well as other anogenital and oral cancers (1–4). HPVs infect cells in the basal layer of stratified epithelia and establish their genomes as low-copy-number episomes that replicate in synchrony with cellular chromosomes. The HPV life cycle is dependent upon epithelial differentiation with productive replication or amplification restricted to differentiated suprabasal cells (5–10). Upon differentiation, HPV-positive cells remain active in the cell cycle and reenter S/G2 for amplification in suprabasal cells. Recent studies have implicated the ataxia telangiectasia (ATM) double-strand DNA damage response in HPV-positive cells as being necessary for differentiation-dependent genome amplification. This activation occurs in the absence of external DNA damage agents (11–13) in part through the action of the innate immune regulator STAT-5, but the mechanism by which this occurs is not well understood. A second DNA repair pathway, ATR (ataxia telangiectasia and Rad3 related), mediates the repair of single-strand breaks and is activated through complex formation with the topoisomerase IIβ-binding protein 1 (TopBP1) (14), the ATR-interacting protein (ATRIP) (15), and claspin (16). The formation of ATR complexes activates a different set of downstream effectors than ATM that includes CHK1, BRCA 1, and MCM proteins. The ATR pathway has been reported to be activated in undifferentiated HPV-positive cells, and treatment with ATR inhibitors can moderately reduce stable genome copy number (17). These studies suggest the ATR pathway may modulate stable HPV replication in undifferentiated cells, but it is unclear whether it plays any role in the differentiation-dependent events or what regulates its activation.
In this study, we investigated whether the ATR pathway played any role in the differentiation-dependent amplification of HPV genomes. Our studies demonstrate that both ATR and CHK1 kinases are activated in differentiating HPV-positive cells and that treatment with inhibitors blocks differentiation-dependent genome amplification. Furthermore, we show that a member of the JAK/STAT signaling pathway, STAT-5, mediates activation of ATR. STAT-5 is a transcriptional activator, and we determined that it directly regulates the transcription of TopBP1, resulting in ATR activation. These studies identify STAT-5 as an important regulator of the ATR DNA damage pathways that acts by regulating transcription of TopBP1 and link the innate immune response to induction of single-strand DNA break repair.
RESULTS
ATR and CHK1 are constitutively activated in undifferentiated and differentiated HPV-positive keratinocytes.
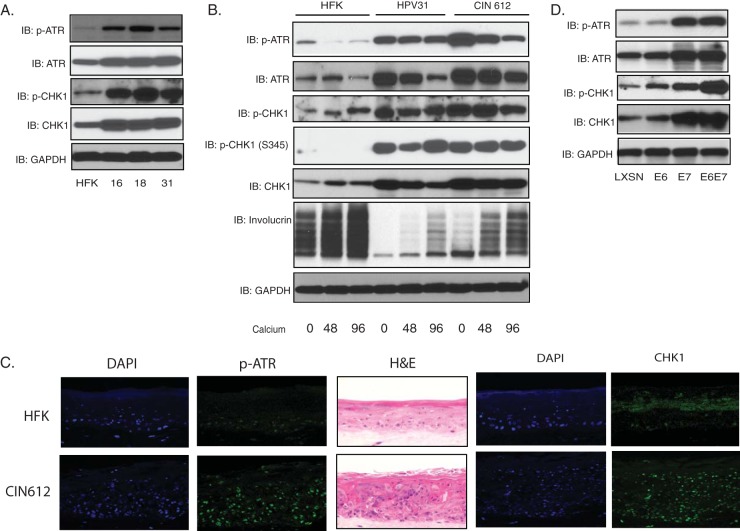
To investigate what role, if any, the ATR pathway plays in the HPV life cycle, we first investigated whether the levels of total and activated forms of the members of this pathway were altered in HPV-positive cells. For this analysis, we examined three cell lines that were generated by transfection of normal human keratinocytes with recircularized HPV16, 18, or 31 DNAs (human foreskin keratinocyte [HFK] lines HFK-16, HFK-18, and HFK-31) and stably maintain viral DNAs as episomes (18). We also examined an immortal cell line that was derived from a cervical biopsy specimen and maintains episomal copies of HPV31 (CIN612) (19). The levels of total ATR and CHK1 as well as the active, phosphorylated forms are significantly increased in undifferentiated cultures of these three HPV-positive cell lines compared to normal keratinocytes (HFKs) (Fig. 1A). Epithelial differentiation can be induced by the addition of high-calcium medium to monolayer cultures and plateaus between 72 and 96 h, which correlates with HPV genome amplification (20). The levels of total ATR and CHK1 along with p-ATR and p-CHK1 are maintained at elevated levels in these HPV-positive lines upon differentiation, which is in contrast with HFKs, where levels remain low (Fig. 1B). While there is a slight decrease in levels of p-ATR at 72 h, they are still significantly higher than levels in HFKs (Fig. 1B). Two phosphorylation sites of CHK1 were examined, and the levels of both S345 and S296 phosphorylation are greatly increased in HPV-positive cells. Similar high levels of p-ATR and CHK1 are observed by immunofluorescence in all layers of stratified organotypic raft cultures of HPV31-positive cells (Fig. 1C). These experiments indicate that components of the ATR pathway are constitutively activated in both undifferentiated and differentiated HPV-positive cells in the absence of external DNA-damaging agents.
FIG 1 .
Activation of ATR pathway in HPV-positive cells. (A) Western blot analysis of ATR, p-ATR, CHK1, p-CHK1, and GAPDH levels in HFK, HFK-16, HFK-18, and HFK-31 cells grown in monolayer cultures. (B) Western blot analysis for ATR, p-ATR, CHK1, p-CHK1 (S296) (p-CHK1), p-CHK1 (S345), involucrin, and GAPDH in HFK, HFK-31, and HPV31-positive CIN612 cells differentiated in high-calcium medium for the indicated times (in hours). (C) Sections of organotypic raft cultures generated from HFKs (top) and CIN612 (bottom) were stained with hematoxylin and eosin (H&E), p-ATR, and CHK1, as well as 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei. (D) Western blot analysis for ATR, p-ATR, CHK1, p-CHK1, and GAPDH levels in HFK cells expressing LXSN vector, HPV31E6, HPV31E7, and HPV31E6E7. All results are representative of observations from three independent experiments.
To investigate which viral proteins were responsible for the activation of ATR or CHK1, the levels of total ATR, CHK1, p-ATR, and p-CHK1 were examined by Western blot analysis using primary HFKs that had been stably infected with retroviruses expressing HPV31 E6, E7, or the combination of E6 and E7. In these experiments, E7 expression alone was sufficient to induce an increase in the levels of ATR and CHK1 as well as p-ATR and p-CHK1, while E6 had only a modest effect. The combination of E6 and E7 had the most significant effect (Fig. 1D).
Inhibition of CHK1 or ATR activation by inhibitors blocks HPV31 genome amplification.
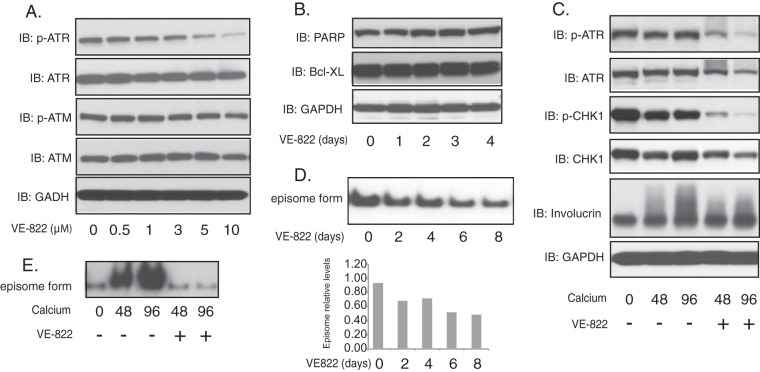
We next investigated whether the ATR pathway played any role in the differentiation-dependent HPV life cycle using inhibitors. For this analysis, CIN612 cells were first treated for 48 h with the ATR inhibitor VE822, and this resulted in a dose-dependent suppression of the levels of p-ATR, as seen by Western blot analysis. VE822 treatment significantly reduced the levels of p-ATR, but not p-ATM, while the total levels of ATR were minimally altered (Fig. 2A). In addition, VE822 had no effect on cell proliferation or induction of markers of apoptosis, such as PARP (poly ADP-ribose polymerase) or Bcl-XL, at this concentration (Fig. 2B). Treatment of HPV-positive cells induced to differentiate in high-calcium medium with VE822 substantially reduced the levels of p-ATR and p-CHK1 (Fig. 2C). To investigate the effects of inhibition of p-ATR on maintenance of stable viral genomes, undifferentiated monolayer cultures of HPV31-positive cells were treated for 8 days with VE822 and examined by Southern blot analysis. Treatment with VE822 resulted in a moderate reduction of around 40% in the levels of stable episomes that plateaus after 8 days (Fig. 2D). In contrast, treatment of HPV-positive cells with VE822 upon differentiation in high-calcium medium for 48 h completely blocked the ability to amplify viral genomes (Fig. 2E).
FIG 2 .
Suppression of ATR phosphorylation by VE822 blocks HPV genome amplification upon keratinocyte differentiation. (A) Western blot analysis of ATR, p-ATR, ATM, p-ATM, and GAPDH levels in CIN612 cells treated with VE822 at various concentrations for 48 h. (B) Western blot analysis of Bcl-XL, PARP, and GAPDH levels in CIN612 cells treated with VE822 (5 µM) for the indicated times. (C) Western blot analysis of ATR, p-ATR, CHK1, p-CHK1, and GAPDH proteins in differentiated CIN612 cells in the presence or absence of VE822 (5 µM) in high-calcium medium for the indicated times. (D) Southern blot analysis for HPV31 episomes in monolayer cultures of CIN612 cells untreated or treated with VE822 (5 µM) for the indicated times. Band densities, as determined by ImageJ software, are shown underneath. (E) Southern blot analysis for HPV31 episomes in CIN612 cells that were untreated or treated with VE822 (5 µM) following differentiation in high-calcium medium for indicated times (in hours). All results are representative of observations from three independent experiments.
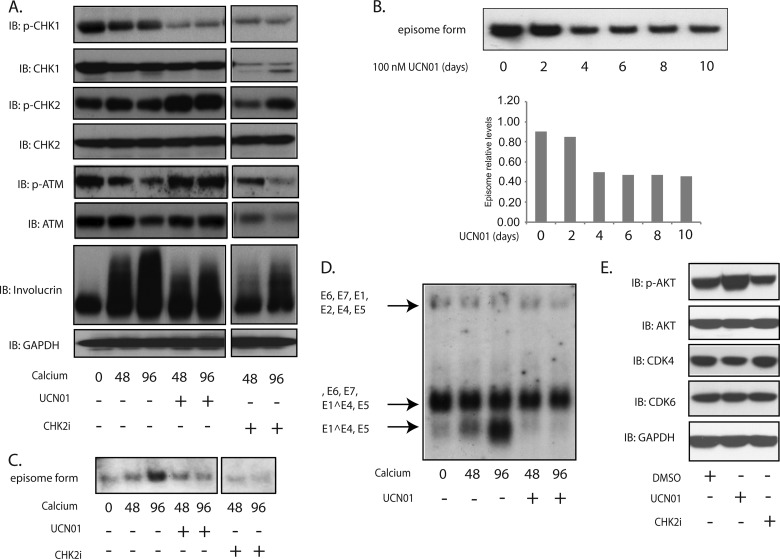
It was next important to determine if CHK1 was also important for genome amplification or whether p-ATR was the sole critical regulator. UCN01 is a CHK1-specific inhibitor, while CHK2i has been reported to inhibit CHK2 activity (21). HPV31-positive CIN612 cells were grown in monolayer cultures and switched to high-calcium medium to induce differentiation in the presence or absence of these inhibitors for 48 and 96 h. As shown in Fig. 3A, the level of p-CHK1, but not of total CHK1, was suppressed by UCN01 treatment. In addition, the levels of p-ATM as well as total ATM were not affected by UNC01 treatment (Fig. 3A). Surprisingly, CHK2i treatment decreased the levels of both p-CHK1 and total CHK1 protein along with a moderate decrease in p-CHK2. In addition, CHK2i treatment also reduced levels of p-ATM and ATM (Fig. 3A). Neither UCN01 nor CHK2i interfered with the proliferation of cells in culture at this concentration. To determine if UCN01 had an effect on stable maintenance replication of viral episomes, monolayer cultures of HPV31-positive cells were grown for 8 days in the presence or absence of UCN01, and genome copy number was assayed by Southern analysis. Our studies confirm a modest decrease in copy numbers, approximately 40% (Fig. 3B), which was similar to effects seen with VE822. To determine if activation of the ATR pathway was required for either late gene expression or differentiation-dependent genome amplification, HPV31-positive CIN612 cells were grown in high-calcium medium for 48 and 96 h in the presence of the CHK1 inhibitor UCN01 or the CHK2 inhibitor CHK2i, and total DNA was assayed by Southern blot analysis. As seen in Fig. 3C, UNC01 as well as CHK2i treatment blocks HPV genome amplification. To investigate if the ATR pathway also played a role in regulating late gene expression, total RNA was also isolated from differentiated HPV31-positive cells treated with UCN01 or not treated and examined by Northern blot analysis. Upon differentiation, late viral transcripts encoding E1^E4, E5 are initiated from a differentiation-dependent promoter located around nucleotide 742 in the middle of the E7 open reading frame. The addition of UCN01 inhibited expression of viral late transcripts (Fig. 3D), indicating that activation of the ATR pathway is necessary for both differentiation-dependent amplification and late gene expression. To exclude the possibility of possible off-target effects of these inhibitors, we also screened for the levels of AKT, p-AKT, CDK4, and CDK6 in CIN612 cells following treatment with UNC01 or CDK2i (Fig. 3E). No change in the levels of these factors was seen with CHK2i treatment, while UNC01 had no effect on total AKT, CDK4, and CDK6. Interestingly, UNC01 treatment slightly increased the levels of p-AKT, but this is unlikely to have any effect on amplification.
FIG 3 .
Suppression of CHK1 phosphorylation by UCN01 blocks HPV genome amplification upon keratinocyte differentiation. (A) Western blot analysis of p-CHK1, CHK1, p-CHK2, CHK2, p-ATM, ATM, and GAPDH levels in CIN612 cells treated with UCN01 (100 nM) or CHK2i (5 µM) in high-calcium medium for the indicated times (in hours). (B) Southern blot analysis for HPV31 episomes in monolayer cultures of CIN612 cells that were untreated or treated with UCN01 (100 nM) for the indicated times. (C) Southern blot analysis for HPV31 episomes in CIN612 cells that were untreated or treated with UCN01 (100 nM) or CHK2i (5 µM) following differentiation in high-calcium medium for the indicated times (in hours). Band densities determined by ImageJ software are shown below. (D) Northern blot analysis for HPV31 early and late gene expression in CIN612 cells in the presence or absence of UCN01 (100 nM) following differentiation in high-calcium medium for the indicated times (in hours). (E) Western blot analysis of p-AKT, AKT, CDK4, CDK6, and GAPDH levels in CIN612 cells treated with UCN (100 nM) and CHK2i (5 µM) for 48 h. All results are representative of observations from two or more independent experiments.
HPV regulates STAT5 to induce activation of the ATR/CHK1 pathway.
The results described above show that inhibition of ATR as well as CHK1 kinases blocks differentiation-dependent HPV late events, and it was important to determine how HPV proteins activate these kinases. Previous studies have identified the innate immune regulator STAT-5 as a critical activator of the ATM pathway in HPV-positive cells (22). To investigate if STAT-5 could affect activation of the ATR DNA damage pathway, CIN612 cells were grown in the presence of pimozide, an inhibitor of STAT-5 phosphorylation, and cell extracts were screened by Western blot analysis for effects on the levels of p-ATR and p-CHK1. As shown in Fig. 4A, pimozide treatment substantially reduced the levels of p-ATR and p-CHK1 but had minimal effects on total levels of these proteins. In addition, pimozide treatment had no effect on the proliferation of cells, as shown previously (22). We next investigated the effect of reducing STAT-5 levels with short-hairpin RNAs (shRNAs) to confirm the effects seen with pimozide. STAT-5 consists of two isoforms, α and β, and we previously identified shRNA-expressing lentiviruses with each of these isoforms that abrogated their expression (22). The two sets of lentiviruses were combined and used to infect CIN612 cells. Following infection for 72 h and differentiation in high-calcium medium, the suppression of phosphorylation of ATR and CHK1 was observed. The levels of activated ATR and CHK1 were further decreased upon keratinocyte differentiation in high-calcium medium (Fig. 4B). There was a slight decrease in p-ATR and p-CHK1 levels at 72 h in the scrambled control, but the levels were still significantly higher than those seen with STAT-5-specific knockdowns (Fig. 4B). We conclude that STAT-5 is an upstream activator of ATR and CHK1 phosphorylation in HPV-positive cells.
FIG 4 .
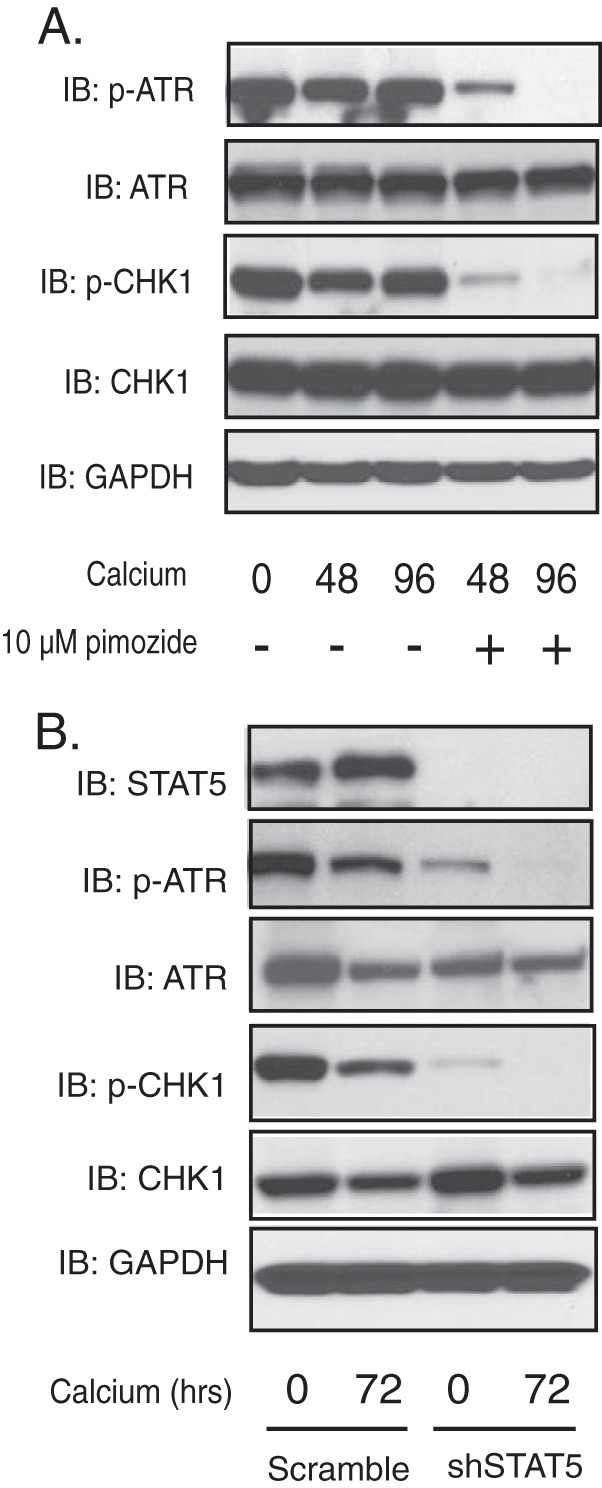
Knockdown of STAT-5 suppresses ATR DNA damage responses. (A) Western blot analysis of ATR, p-ATR, CHK1, p-CHK1, and GAPDH levels in CIN612 cells treated with pimozide (10 µM) in high-calcium medium for the indicated times (in hours). (B) Western blot analysis of STAT-5, ATR, p-ATR, CHK1, p-CHK1, and GAPDH protein levels in shRNA control and shRNA lentivirus-infected CIN612 cells upon differentiation in high-calcium medium for 72 h. All results are representative of observations from two or more independent experiments.
TopBP1 is responsible for STAT-5-dependent ATR/CHK1 activation.
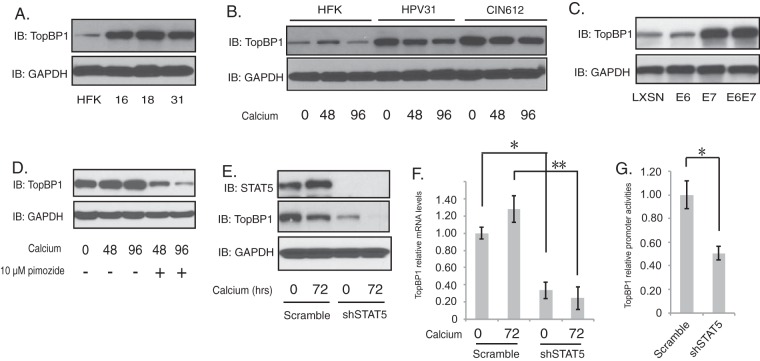
STAT-5 is a transcriptional activator, and our analyses indicate that it regulates the phosphorylation state of ATR and CHK1. This suggests that STAT-5 regulates the expression of an intermediary factor, which in turn activates the ATR pathway. Recent studies demonstrate that TopBP1 regulates the activation of ATR (14), and we investigated if the expression of TopBP1 was altered by STAT-5. The levels of TopBP1 proteins were found to be elevated in undifferentiated monolayer cultures of HPV31-positive cells along with cells with HPV16 and 18 in comparison to normal HFKs (Fig. 5A). Upon differentiation in high-calcium medium, the levels of TopBP1 are retained at high levels in HPV-positive cells (Fig. 5B), in contrast to HFKs. High levels of TopBP1 were also observed in cells that express E7 alone but not in cells expressing E6 (Fig. 5C) and correlate with STAT-5 activation (22). To examine whether TopBP1 is regulated by STAT-5, CIN612 cells were treated with pimozide and induced to differentiate in high-calcium medium. Inhibition of STAT-5 activation by pimozide resulted in decreased levels of TopBP1 after 48 and 96 h (Fig. 5D). Similarly, knockdown of STAT-5 by transduction of lentiviruses encoding STAT-5 shRNAs completely blocked TopBP1 expression (Fig. 5E). To determine if this effect was mediated at the transcriptional or posttranscriptional levels, undifferentiated and differentiated knockdown cells were assayed by RT-PCR for TopBP1 mRNA levels. Knockdown of STAT-5 significantly reduced the levels of TopBP1 transcripts in both sets of cells (Fig. 5F). We next examined if the effects of STAT-5 were mediated at the level of initiation of transcription or by mRNA stabilization through the use of TopBP1 promoter reporters. The 1,498-nucleotide upstream promoter region of TopBP1 was cloned into the pGL3-basic dual luciferase reporter. These reporter plasmids were then transfected into HPV-positive cells that had been previously infected with either lentiviruses expressing shRNAs to STAT-5 or control shRNAs. As seen in Fig. 5G, knockdown of STAT-5 inhibited TopBP1 reporter expression, confirming that regulation occurs at the initiation of transcription. These results demonstrate that STAT-5 regulates TopBP1 expression at the level of transcription (Fig. 5).
FIG 5 .
TopBP1 is the downstream target of STAT-5. (A) Western blot analysis of TopBP1 and GAPDH levels in HFK, HFK-16, HFK-18, and HFK-31 cells grown in monolayer cultures. (B) Western blot analysis for TopBP1 and GAPDH in HFK, HFK-31, and HPV31-positive CIN612 cells differentiated in high-calcium medium for the indicated times (in hours). (C) Western blot analysis for TopBP1 and GAPDH levels in HFK cells expressing LXSN vector, HPV31E6, HPV31E7, and HPV31E6E7. (D) Western blot analysis of TopBP1 and GAPDH levels in CIN612 cells treated with pimozide (10 µM) in high-calcium medium for the indicated times (in hours). (E) Western blot analysis of STAT-5, TopBP1, and GAPDH protein levels in shRNA control and shRNA lentivirus-infected CIN612 cells upon differentiation in high-calcium medium for 72 h. (F) RT-PCR analysis of TopBP1 at mRNA levels in CIN612 cells in shRNA control and shRNA lentivirus-infected CIN612 cells upon differentiation in high-calcium medium for 72 h. Data are means and standard errors. *, P < 0.05; **, P < 0.01. (G) The relative activities of TopBP1 promoter were compared in shRNA control and shRNA lentivirus-infected CIN612 cells. Data are means and standard errors. *, P < 0.01. All results are representative of observations from three independent experiments.
It was next important to confirm that high levels of TopBP1 were important for ATR/CHK1 activation as well as HPV genome amplification. For this analysis, lentiviruses encoding TopBP1-specific shRNAs were used to infect CIN612 cells, and at 72 h after transduction, the cell lysates were collected and assayed for TopBP1 protein levels by Western blot analysis. Figure 6A shows that the levels of TopBP1 were reduced by lentiviral shRNA infection compared to the mock-infected cells or cells transduced with scrambled shRNA-expressing lentiviruses and those encoding shRNAs 02 and 05 were pooled for subsequent experiments. HPV-positive cells transduced with lentiviruses were then induced to differentiation in high-calcium medium for an additional 72 h before analysis. The cells were then collected and assayed for TopBP1 protein levels by Western blot analysis as well as for genome amplification by Southern blot analysis. Our results show that the levels of TopBP1 were greatly reduced in CIN612 cells transduced with TopBP1 shRNA lentiviruses, compared to the control transduced cells (Fig. 6B). The knockdown of TopBP1 led to substantial reductions in the levels of p-ATR and p-CHK1, as well as slight reductions in the levels of total CHK1. In contrast, total ATR levels were not affected by knockdown of TopBP1. The slight decreases in p-ATR and p-CHK1 levels seen at 72 h of differentiation (Fig. 6B) are consistent with the results shown in Fig. 1, but the levels are substantially higher than the levels seen in TopBP1 knockdowns (Fig. 6B). Importantly, Southern blot analysis revealed that the loss of TopBP1 in CIN612 cells blocks HPV31 genome amplification (Fig. 6C). These results demonstrate a critical role of STAT-5 in regulating the transcription of TopBP1, which is necessary for activation of the ATR/CHK1 pathway and genome amplification in HPV-positive cells.
FIG 6 .
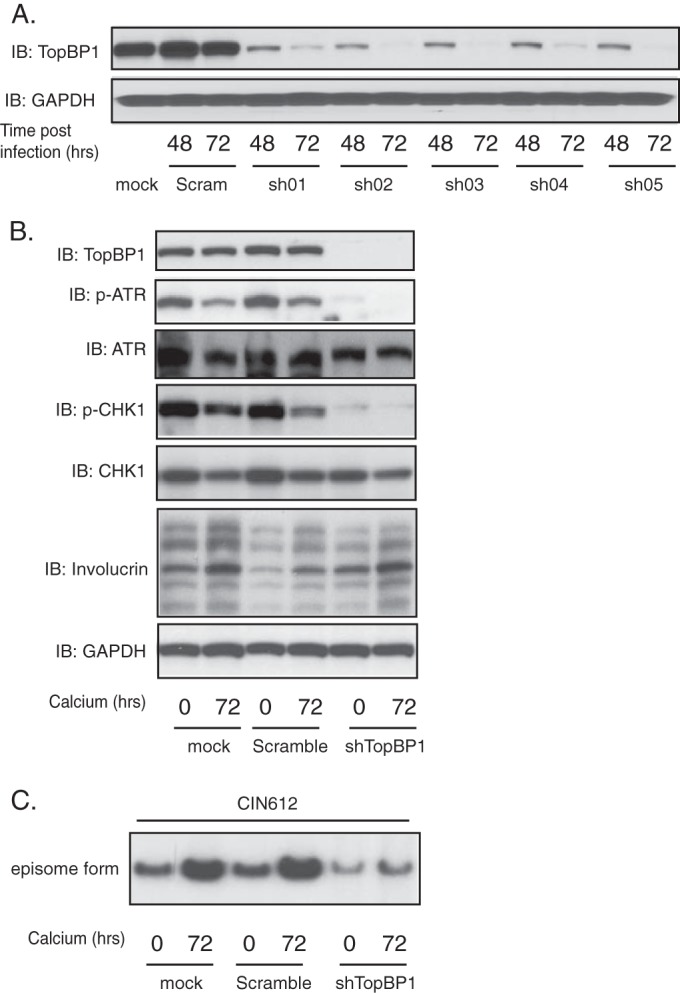
TopBP1 knockdown results in a loss of differentiation-dependent HPV genome amplification. (A) Western blot analysis of TopBP1 and GAPDH proteins in CIN612 cell monolayers infected with multiple shRNA lentiviruses targeting TopBP1 for the indicated times. “Scram” refers to the scrambled vector controls for TopBP1. (B) Western blot analysis of TopBP1, p-ATR, ATR, p-CHK1, CHK1, and GAPDH proteins following differentiation of CIN612 cells that had been infected with combined shRNA lentiviruses targeting TopBP1 for the indicated times. (C) Southern blot analysis of HPV31 genomes in CIN612 cells following infection with pooled TopBP1 shRNA lentiviruses and differentiation in high-calcium medium for indicated times. All results are representative of observations from two or more independent experiments.
DISCUSSION
The differentiation-dependent amplification of human papillomaviruses genomes is dependent upon host pathways, including those for DNA damage repair. The ATR pathway repairs single-strand breaks, and our studies show that it plays a critical role in HPV differentiation-dependent amplification. The levels of ATR kinase and its downstream effector, CHK1, are substantially increased in HPV-positive cells and are maintained at high levels upon differentiation in contrast to normal cells where levels rapidly decrease. Interestingly, the active phosphorylated forms of ATR and CHK1 are induced in the absence of any external DNA-damaging agents. Our studies demonstrate that HPV proteins activate the ATR pathway through the action of the innate immune regulator STAT-5. STAT-5 is a transcription factor that is usually activated in response to growth factors or cytokines. STAT-5 contributes to the regulation of tumorigenesis by controlling the transcription of tumor repressor genes, such as the IRF-8 gene (23), as well as prosurvival signals mediated through AKT1 (24). Our studies identify another mechanism by which STAT-5 regulates the stress response through the ATR DNA damage pathway. In HPV-positive cells, STAT-5 is constitutively activated, and we found that it regulates the transcription of TopBP1, a regulator of ATR activation. For ATR activation, our studies show by means of transcript and reporter assays that STAT-5 directly regulates transcription of the TopBP1 gene and that this is necessary for ATR activation. These observations demonstrate a link between the innate immune response and ATR activation. Interestingly, STAT-5 also regulates the ATM pathway, where it acts by modulating Tip60 phosphorylation and activation. Activated Tip60 acetylates ATM, which is necessary for ATM autophosphorylation, and Tip60 activation is mediated through the action of the GSK-3β kinase (12). Since STAT-5 is a transcription factor, it must act through an as-yet-uncharacterized intermediary factor to activate GSK-3β and its downstream target Tip60. Our studies show that HPVs act in a coordinated manner to activate both ATR and ATM through the same transcription factor, STAT-5.
ATR activation in HPV-positive cells is dependent upon p-STAT-5 regulated transcription of the TopBP1 gene. TopBP1 forms a complex with ATRIP and claspin, which binds ATR and regulates its activation through autophosphorylation. Putative STAT-5 binding sequences are located in the TopBP1 promoter, and our analyses show that STAT-5 directly activates transcription. Furthermore, our studies show that knockdown of TopBP1 blocks genome amplification. Previous studies have shown that knockdown of TopBP1 moderately increases short-term replication of transiently transfected HPV31 plasmids and has a similar modest effect on stable maintenance of HPV31 episomes (25). Our analyses have identified a critical role for ATR in the differentiation-dependent late phase of the viral life cycle as well as demonstrating that TopBP1 is an important regulator of this pathway. Interestingly, TopBP1 also forms complexes with the HPV E2 protein, resulting in its recruitment to viral genomes. E2 mutants that are no longer able to bind TopBP1 have reduced replication ability, indicating that it is a positive regulator of viral replication (26). In addition to its role in ATR activation, TopBP1 helps to load replication factors onto origins (27) as well as acting as a transcriptional regulator (28), but it is not clear if these functions are enhanced through its interactions with E2.
The E7 protein was shown to be sufficient to induce increased levels of ATR and CHK1 levels as well as activation of phosphorylation. The E1 origin recognition protein has previously been shown to induce the recruitment of ATR and ATRIP to repair foci when overexpressed by itself in short-term assays in undifferentiated cells (29). This suggests that E7 may act together with E1 for full activation of ATR. The E7 protein has also been shown to activate STAT-5 phosphorylation, but maximal effects are seen when complete viral genomes are present (22). It seems likely that the combination of E7 with E1 or some other viral protein mediates full activation of these DNA damage pathways. While high-risk viral proteins activate the ATR response, it has been reported that E6 protein of cutaneous HPV8 blocks ATR activation (30), indicating that ATM and ATR activation may be a property specific to high-risk HPV. Since no system for stably replicating HPV8 genomes types has yet been developed, it is not clear what effects on ATR activation are seen with complete genomes. ATR has also been shown to positively regulate the replication of Merkel cell polyomaviruses, BK virus, and herpes simplex virus (HSV) (31–33).
Our studies indicate that activation of both ATM and ATR is necessary for HPV genome amplification, but it is not immediately obvious why both are required. Previous studies have implicated the homologous repair (HR) arm of the ATM pathway but not those involved in nonhomologous end joining (NHEJ) as important for HPV genome amplification. Homologous recombination repair occurs in G2 and requires resection of double-strand breaks by the exonuclease activity of MRN (Mre11, Rad50, Nbs1), exposing 5′ and 3′ single-strand regions. These single-strand regions may recruit ATR factors to facilitate repair replication, and this could explain why they are important for HPV genome amplification. It is also possible that both double-strand and single-strand breaks are generated in HPV genomes prior and during amplification and that these recruit ATM and ATR factors to these sites. For simian virus 40 (SV40), ATM and ATR have been suggested to act as a part of a quality control mechanism to block rolling circle replication and concatemer formation to favor theta structure replication (34). Since both pathways are required for HPV amplification, it is interesting that they are activated through the same mechanism involving STAT-5. Overall, these studies identify TopBP1 as a critical regulator of HPV replication functions whose expression is regulated by STAT-5.
MATERIALS AND METHODS
Cell culture.
Human foreskin keratinocytes (HFKs) were isolated from neonatal foreskins as previously described (35). HPV16, 18, and 31 cell lines, which maintain viral episomes, as well as E6- and E7-expressing cells, were prepared as previously described (18). CIN612 cells are derived from a cervical intraepithelial neoplasia (CIN) II biopsy specimen that stably maintains HPV31 episomes. For the genome amplification assay, HPV-positive cells were induced to differentiate in keratinocyte basal medium without supplements and containing 1.5 mM CaCl2 for up to 96 h (35).
Inhibitors, antibodies, and Western blot analysis.
The inhibitors used in this study were as follows: 100 nM UCN01 and 5 µM VE822 (Selleckchem, Houston, TX); 10 µM pimozide (Sigma-Aldrich, St. Louis, MO); and 5 µM CHK2i (Calbiochem, San Diego, CA). The antibodies used in this study were as follows: antiinvolucrin, anti-TopBP1, and anti-GAPDH (Santa Cruz, Santa Cruz, CA) and anti-STAT-5, anti-CHK2, anti-ATM, anti-p-CHK2 (Thr68), anti-p-ATM (Ser1981), anti-CHK1, anti-ATR, anti-p-CHK1 (Ser296), anti-p-CHK1 (Ser345), and anti-p-ATR (Cell Signaling, Inc., Danvers, MA). For Western blot analysis, cell lysates were processed as previously described (36). Briefly, keratinocytes were first isolated from J2 feeders by treatment with Versene (phosphate-buffered saline [PBS] containing 0.5 mM EDTA) and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer on ice for 30 min. The protein samples were separated on SDS-PAGE gels and then transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were finally developed using Enhanced Chemiluminescence (ECL) Prime or ECL reagents (Amersham, Pittsburgh, PA). Chemiluminescence signals were detected using Eastman Kodak X-ray films.
Immunofluorescence.
HFKs as well as CIN612 HPV31-positive keratinocytes were differentiated in raft cultures as previously described (35). Sections from HFK raft cultures or CIN612 cell raft cultures were processed by the Pathology Core Facility of Northwestern University. Cross sections of rafts were stained using a 1:100 dilution of anti-p-ATR (Cell Signaling, Inc., Danvers, MA) and 1:100 of anti-CHK1 (Cell Signaling, Inc., Danvers, MA). For the secondary antibody, Alexa Fluor 488 goat anti-mouse antibody and anti-rabbit antibody were used (Life Technologies, Grand Island, NY). The sections were then mounted with Gelvatol (Sigma-Aldrich, St. Louis, MO). Images were captured using a Nikon C2+ spectral confocal microscope at Center of Advanced Microscopy of Northwestern University.
Southern and Northern blot analysis.
Total DNA was extracted with phenol-chloroform, and the DNA samples were then processed by Southern blot analysis as previously described (22). Total DNA levels were determined by multiple NanoDrop measurements and equal loading of gels was confirmed following electrophoresis by ethidium bromide staining. For Northern blot analysis, the cells were lysed in STAT-60 (Tel-Test, Friendswood, TX), and the RNA samples were then processed as previously described (22).
Lentiviral virion production and transduction.
Mission short hairpin RNA (shRNA) lentiviral vectors were purchased from Sigma-Aldrich, St. Louis, MO. Lentiviral particles expressing five TopBP1-specific shRNAs (constructs 1 to 5) were prepared as previously described (22). CIN612 cells were then incubated with 5 ml fresh E medium containing concentrated TopBP1 shRNA or scrambled shRNA lentiviral soup in the presence of 4 µg/ml hexadimethrine bromide (Polybrene; Sigma-Aldrich, St. Louis, MO) overnight at 37°C. The culture medium was changed, and the transduced cells were cultured in fresh E medium for an additional 48 h before analysis. The shRNA against STAT-5 has been tested and applied in our previous work (22).
Luciferase reporter assay.
The 1,498-nucleotide upstream promoter region of TopBP1 was cloned into the pGL3-basic dual luciferase reporter by GenScript USA, Inc. (Piscataway, NJ). CIN612 HPV-positive cells were used to seed 6-well plates. When the cell confluence reached around 50 to 60%, the cells were transduced with lentiviral soups expressing STAT-5-specific shRNA in the presence of 4 µg/ml Polybrene (Sigma-Aldrich, St. Louis, MO) overnight. The transduced cells were washed with PBS and transfected with the pGL3-TopBP1 promoter plasmid using PEI reagents. The following day, the transfected cells were transferred to fresh E medium and incubated at 37°C for 48 h. Luciferase activity was measured using the dual-luciferase reporter assay system (Promega, Madison, WI), with Renilla luciferase as an internal control, according to the manufacturer’s instructions. Significance was determined using Student’s t test.
ACKNOWLEDGMENTS
This work was supported by grants from NCI to L.A.L. (CA59655 and CA42861).
Footnotes
Citation Hong S, Cheng S, Iovane A, Laimins LA. 2015. STAT-5 regulates transcription of the topoisomerase IIβ-binding protein 1 (TopBP1) gene to activate the ATR pathway and promote human papillomavirus replication. mBio 6(6):e02006-15. doi:10.1128/mBio.02006-15.
REFERENCES
- 1.zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 2.Egawa N, Egawa K, Griffin H, Doorbar J. 2015. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses 7:3863–3890. doi: 10.3390/v7072802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ndiaye C, Mena M, Alemany L, Arbyn M, Castellsagué X, Laporte L, Bosch FX, de Sanjosé S, Trottier H. 2014. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol 15:1319–1331. doi: 10.1016/S1470-2045(14)70471-1. [DOI] [PubMed] [Google Scholar]
- 4.Whang S, Filippova M, Duerksen-Hughes P. 2015. Recent progress in therapeutic treatments and screening strategies for the prevention and treatment of HPV-associated head and neck cancer. Viruses 7:5040–5065. doi: 10.3390/v7092860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas M, Narayan N, Pim D, Tomaić V, Massimi P, Nagasaka K, Kranjec C, Gammoh N, Banks L. 2008. Human papillomaviruses, cervical cancer and cell polarity. Oncogene 27:7018–7030. doi: 10.1038/onc.2008.351. [DOI] [PubMed] [Google Scholar]
- 6.Moody CA, Laimins LA. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 7.Bodily J, Laimins LA. 2011. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol 19:33–39. doi: 10.1016/j.tim.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones DL, Münger K. 1996. Interactions of the human papillomavirus E7 protein with cell cycle regulators. Semin Cancer Biol 7:327–337. doi: 10.1006/scbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- 9.Kajitani N, Satsuka A, Kawate A, Sakai H. 2012. Productive lifecycle of human papillomaviruses that depends upon squamous epithelial differentiation. Front Microbiol 3:152. doi: 10.3389/fmicb.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conway MJ, Meyers C. 2009. Replication and assembly of human papillomaviruses. J Dent Res 88:307–317. doi: 10.1177/0022034509333446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moody CA, Laimins LA. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog 5:e1000605. doi: 10.1371/journal.ppat.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong S, Dutta A, Laimins LA. 2015. The acetyltransferase tip60 is a critical regulator of the differentiation-dependent amplification of human papillomaviruses. J Virol 89:4668–4675. doi: 10.1128/JVI.03455-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinney C, Hussmann K, McBride A. 2015. The role of the DNA damage response throughout the papillomavirus life cycle. Viruses 7:2450–2469. doi: 10.3390/v7052450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumagai A, Lee J, Yoo HY, Dunphy WG. 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 15.Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 16.Kumagai A, Dunphy WG. 2003. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat Cell Biol 5:161–165. doi: 10.1038/ncb921. [DOI] [PubMed] [Google Scholar]
- 17.Edwards TG, Helmus MJ, Koeller K, Bashkin JK, Fisher C. 2013. Human papillomavirus episome stability is reduced by aphidicolin and controlled by DNA damage response pathways. J Virol 87:3979–3989. doi: 10.1128/JVI.03473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong S, Mehta KP, Laimins LA. 2011. Suppression of STAT-1 expression by human papillomaviruses is necessary for differentiation-dependent genome amplification and plasmid maintenance. J Virol 85:9486–9494. doi: 10.1128/JVI.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedell MA, Hudson JB, Golub TR, Turyk ME, Hosken M, Wilbanks GD, Laimins LA. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol 65:2254–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moody CA, Fradet-Turcotte A, Archambault J, Laimins LA. 2007. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc Natl Acad Sci U S A 104:19541–19546. doi: 10.1073/pnas.0707947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arienti KL, Brunmark A, Axe FU, McClure K, Lee A, Blevitt J, Neff DK, Huang L, Crawford S, Pandit CR, Karlsson L, Breitenbucher JG. 2005. Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J Med Chem 48:1873–1885. doi: 10.1021/jm0495935. [DOI] [PubMed] [Google Scholar]
- 22.Hong S, Laimins LA. 2013. The JAK-STAT transcriptional regulator, STAT-5, activates the ATM DNA damage pathway to induce HPV 31 genome amplification upon epithelial differentiation. PLoS Pathog 9:e1003295. doi: 10.1371/journal.ppat.1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waight JD, Banik D, Griffiths EA, Nemeth MJ, Abrams SI. 2014. Regulation of the interferon regulatory factor-8 (IRF-8) tumor suppressor gene by the signal transducer and activator of transcription 5 (STAT5) transcription factor in chronic myeloid leukemia. J Biol Chem 289:15642–15652. doi: 10.1074/jbc.M113.544320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt JW, Wehde BL, Sakamoto K, Triplett AA, Anderson SM, Tsichlis PN, Leone G, Wagner K-. 2014. Stat5 regulates the phosphatidylinositol 3-kinase/Akt1 pathway during mammary gland development and tumorigenesis. Mol Cell Biol 34:1363–1377. doi: 10.1128/MCB.01220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanginakudru S, DeSmet M, Thomas Y, Morgan IM, Androphy EJ. 2015. Levels of the E2 interacting protein TopBP1 modulate papillomavirus maintenance stage replication. Virology 478:135–142. doi: 10.1016/j.virol.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donaldson MM, Mackintosh LJ, Bodily JM, Dornan ES, Laimins LA, Morgan IM. 2012. An interaction between human papillomavirus 16 E2 and TopBP1is required for optimum viral DNA replication and episomal genome establishment. J Virol 86:12806–12815. doi: 10.1128/JVI.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gauson EJ, Donaldson MM, Dornan ES, Wang X, Bristol M, Bodily JM, Morgan IM. 2015. Evidence supporting a role for TopBP1 and Brd4 in the initiation but not continuation of human papillomavirus 16 E1/E2-mediated DNA replication. J Virol 89:4980–4991. doi: 10.1128/JVI.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu K, Lin F-T, Ruppert JM, Lin W-C. 2003. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol 23:3287–3304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadaja M, Isok-Paas H, Laos T, Ustav E, Ustav M. 2009. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog 5:e1000397. doi: 10.1371/journal.ppat.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace NA, Robinson K, Howie HL, Galloway DA. 2012. HPV 5 and 8 E6 abrogate ATR activity resulting in increased persistence of UVB induced DNA damage. PLoS Pathog 8:e1002807. doi: 10.1371/journal.ppat.1002807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang SH, Wang X, Li J, Buck CB, You J. 2014. Host DNA damage response factors localize to Merkel cell polyomavirus DNA replication sites to support efficient viral DNA replication. J Virol 88:3285–3297. doi: 10.1128/JVI.03656-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, Lou S, Deng X, Liu Z, Li Y, Kleiboeker S, Qiu J. 2011. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J Virol 85:8046–8055. doi: 10.1128/JVI.00831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohni KN, Dee AR, Smith S, Schumacher AJ, Weller SK. 2013. Efficient herpes simplex virus 1 replication requires cellular ATR pathway proteins. J Virol 87:531–542. doi: 10.1128/JVI.02504-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sowd GA, Li NY, Fanning E. 2013. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog 9:e1003283. doi: 10.1371/journal.ppat.1003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fehrmann F, Laimins LA. 2005. Human papillomavirus type 31 life cycle: methods for study using tissue culture models. Methods Mol Biol 292:317–330. [DOI] [PubMed] [Google Scholar]
- 36.Hong S, Brass A, Seman M, Haag F, Koch-Nolte F, Dubyak GR. 2007. Lipopolysaccharide, IFN-gamma, and IFN-beta induce expression of the thiol-sensitive ART2.1 ecto-ADP-ribosyltransferase in murine macrophages. J Immunol 179:6215–6227. [DOI] [PubMed] [Google Scholar]