In this study, Ferretti et al. provide novel insights into melanoma biology by investigating the oncogenic functions of BMI1 and melanoma progression. The findings show that BMI1 acts to determine the metastatic progression of melanoma by promoting every step of the metastatic cascade; however, BMI1 has no effect on either cell proliferation or primary tumor development.
Keywords: BMI1, melanoma, metastases, invasive signature, therapeutic resistance
Abstract
Melanoma can switch between proliferative and invasive states, which have identifying gene expression signatures that correlate with good and poor prognosis, respectively. However, the mechanisms controlling these signatures are poorly understood. In this study, we identify BMI1 as a key determinant of melanoma metastasis by which its overexpression enhanced and its deletion impaired dissemination. Remarkably, in this tumor type, BMI1 had no effect on proliferation or primary tumor growth but enhanced every step of the metastatic cascade. Consistent with the broad spectrum of effects, BMI1 activated widespread gene expression changes, which are characteristic of melanoma progression and also chemoresistance. Accordingly, we showed that up-regulation or down-regulation of BMI1 induced resistance or sensitivity to BRAF inhibitor treatment and that induction of noncanonical Wnt by BMI1 is required for this resistance. Finally, we showed that our BMI1-induced gene signature encompasses all of the hallmarks of the previously described melanoma invasive signature. Moreover, our signature is predictive of poor prognosis in human melanoma and is able to identify primary tumors that are likely to become metastatic. These data yield key insights into melanoma biology and establish BMI1 as a compelling drug target whose inhibition would suppress both metastasis and chemoresistance of melanoma.
Skin melanoma is the sixth most commonly diagnosed cancer in the United States, whose incidence has increased dramatically in the past 30 years. Localized melanoma is treated quite successfully by surgery but spreads rapidly if not caught early. Metastatic melanoma is one of the most aggressive and therapy-resistant human cancers, and, in 2011, the 5-year relative survival was just 16% (SEER Cancer Statistics Review, 1975–2011, National Cancer Institute, http://seer.cancer.gov/archive/csr/1975_2011). Activating mutations in the serine/threonine kinase BRAF are the most prevalent driving mutations for melanoma, with additional lesions being required for tumor development (Davies et al. 2002). Two selective BRAF kinase inhibitors have been approved for treatment of BRAF mutant metastatic melanomas. These drugs yield significant tumor regression, but most patients develop resistance that induces relapse (Flaherty et al. 2010; Holderfield et al. 2014). This reinforces the need to elucidate mechanisms of melanoma metastasis. Classic models postulate that cancer progression reflects acquisition of mutations that enable a more metastatic state. In contrast to this model, melanoma cells are able to switch between two distinct states, one highly proliferative and the other highly invasive, which are characterized by distinct gene expression signatures (Hoek et al. 2008; Ghislin et al. 2012). It seems likely that epigenetic alterations underlie this switch, but the regulatory mechanisms are unknown.
BMI1 is an epigenetic regulator that represses gene transcription via its participation in the Polycomb-repressive complex 1. BMI1 was originally identified as an oncogene, and subsequent studies showed that BMI1 maintains the self-renewal and proliferative capacity of adult stem cells via transcriptional silencing of the p16-INK4a, p19-ARF, and p21-Cip1 tumor suppressor loci (Park et al. 2004; Valk-Lingbeek et al. 2004). Accordingly, BMI1 loss was found to impair the development of various autochthonous tumor types at least in part via derepression of p16-INK4a, p19-ARF, and p21-Cip1 (Lessard and Sauvageau 2003; Dovey et al. 2008; Maynard et al. 2014). Notably, several of these studies reported impaired proliferative and self-renewal potential of the tumor-initiating cells (Lessard and Sauvageau 2003; Dovey et al. 2008; Maynard et al. 2014), establishing that BMI1 plays a key role in both adult stem cells and tumor-initiating cells. However, this does not rule out other mechanisms of BMI1's oncogenic action, particularly with regard to tumor progression. In support of this latter role, BMI1's expression increases with progression in many human tumors, and this is an excellent predictor of both progression and poor prognosis (Glinsky et al. 2005). Moreover, in vitro studies have implicated BMI1 in cell invasion, metastasis, and chemoresistance through a variety of activities (Berezovska et al. 2006; Song et al. 2009; Wellner et al. 2009; Yang et al. 2010; Gieni et al. 2011; Du et al. 2012; Liu et al. 2012a, 2014; Sun et al. 2012; Chou et al. 2013). However, to date, the negative impact of BMI1 depletion on cell proliferation and primary tumor development has precluded a clear evaluation of BMI1's contribution to tumor progression. In this study, we identify melanoma as a tumor in which BMI1 levels increase with progression without having an effect on proliferation or primary tumor growth. Analysis of this tumor type reveals a critical role for BMI1 in both melanoma metastasis and resistance to BRAF inhibitor treatment, which reflects activation of a widespread gene expression signature that predicts the invasive state and poor patient outcome.
Results
BMI1 controls melanoma cell metastatic dissemination without promoting cell proliferation
Prior studies have yielded conflicting results about BMI1 expression in melanoma, with one study concluding that BMI1 increases with progression (Mihic-Probst et al. 2007), and another concluding the opposite (Bachmann et al. 2008). Thus, we compared BMI1 expression levels in metastatic versus primary melanoma samples from three different human data sets (GSE8401 [Xu et al. 2008], The Cancer Genome Atlas-Skin Cutaneous Melanoma [TCGA-SKCM] [The Cancer Genome Atlas Network 2015], and GSE46517 [Kabbarah et al. 2010]) and also tumors derived from metastatic versus nonmetastatic melanoma mouse models [GSE29074 data set (Scott et al. 2011)]. In all four cases, BMI1 was significantly elevated (P = 0.000225, P = 0.0175, P < 0.0001, and P = 0.0022) in the metastatic lesions (Fig. 1A; Supplemental Fig. S1A). Accordingly, quantitative real-time PCR (qPCR) showed that BMI1 mRNA levels were typically higher in human cell lines derived from metastatic sites versus the primary tumor (Supplemental Fig. S1B). We also examined two existing melanoma cell line series in which parental cells had been used to derive more metastatic variants: human A375 and its more metastatic variant, MA2, which are the BRAF and CDKN2A mutants (Xu et al. 2008), and murine B16F0 and the increasingly metastatic variants B16F1 and B16F10, which are the Braf wild type and Cdkn2a mutant (Fidler 1973). We found that BMI1 levels did not differ significantly between the parental and derivative lines (Supplemental Fig. S1C,E). Thus, these data show that elevated BMI1 is often associated with, but is not a prerequisite for, enhanced metastatic potential of melanoma.
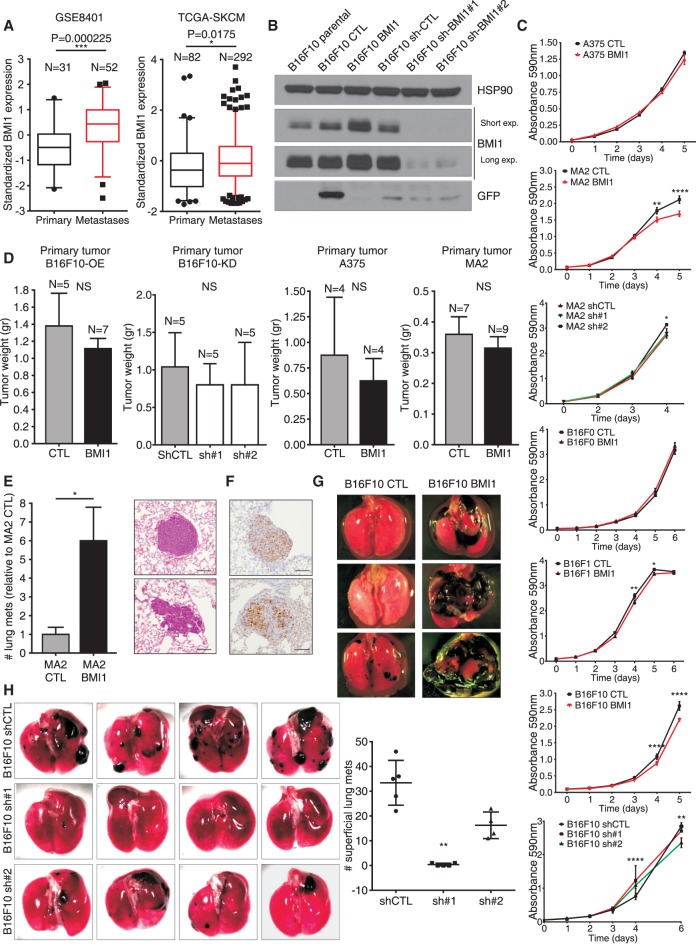
Figure 1.
BMI1 promotes metastatic potential of melanoma cells. (A) Box and whisker plots (fifth to 95th percentile) show BMI1 expression in metastatic versus primary melanoma samples (GSE8401 and TCGA-SKCM data sets). (B) Western blotting showing GFP, BMI1, and HSP90 loading control [CTL] in B16F10 variants. (C) In vitro growth curves of the indicated cell variants. P-values were calculated by two-way ANOVA with Sidak correction. (D) Primary tumor weight after subcutaneous injection of B16F10, A375, and MA2 CTL and variant cells. (E) Relative number of lung metastases (normalized to CTL) resulting from tail vein injection of CTL or BMI1 MA2 cells. (E,F) Representative images of H&E-stained (E) and anti-Ki67-stained (F) sections. Bars, 100 µm. (G,H) Representative images of lungs showing superficial metastasis after tail vein injections with CTL or BMI1 B16F10 cells (G) or sh-CTL versus sh-BMI1 (#1 or #2) B16F10 (H) cells with quantification. For H, statistical significance was assayed by one-way ANOVA and Kruskal-Wallis test. See also Supplemental Figure S1.
Given these findings, we wanted to determine whether BMI1 expression influenced the metastatic potential of melanoma cell lines. Thus, we infected A375, MA2, Cloudman S91, B16F0, B16F1, and B16F10 cells with lentiviruses expressing murine BMI1 or GFP as a control to yield stable pools of cells here called BMI1 or CTL (Fig. 1B; Supplemental Fig. S1D). BMI1 levels did not alter in CTL cells relative to parental controls but increased between 1.65-fold and eightfold in the BMI1 variants (Fig. 1B; Supplemental Fig. S1D,E). We also created knockdown variants of the more metastatic lines, MA2 and B16F10, by infection with lentiviruses expressing different shRNAs for human (MA2) or murine (B16F10) BMI1. We assessed the degree of knockdown in the resulting sh-BMI1 pools compared with sh-Ctl and selected two sh-BMI1 variants for both MA2 (sh1 = 30% and sh2 = 95% knockdown) and B16F10 (sh1 = 91% and sh2 = 85% knockdown) for further analysis (Fig. 1B; Supplemental Fig. S1D). We then compared the properties of BMI1 versus CTL and sh-BMI1 versus sh-Ctl in various assays. In all cases, the CTL and sh-Ctl cells closely resembled the parental (uninfected) cells (data not shown).
We first examined proliferation rates in vitro (Fig. 1C). In stark contrast to the proproliferative effect of BMI1 in most cell types, elevated BMI1 expression had no detectable effect on the proliferation of A375 and B16F0 cells and actually yielded a small but significant reduction in B16F1, B16F10, and MA2 proliferation (Fig. 1C). Moreover, near complete BMI1 knockdown did not reduce the proliferative capacity of either B16F10 or MA2 cells (Fig. 1C). We next examined the ability of representative cell lines to yield tumors in vivo. When injected subcutaneously into NOD/SCID (A375 and MA2 cells) or syngeneic (B16F10 cells) mice, we saw no significant increase in size or development rate of primary tumors arising from BMI1 versus CTL cells (Fig. 1D). If anything, BMI1 tumors trended toward reduced tumor growth, although not statistically significant. Moreover, the B16F10 sh-BMI1 cells formed primary tumors as well as their sh-Ctl counterparts (Fig. 1D). Thus, in melanoma cells, BMI1 levels play little or no role in proliferation or primary tumor growth.
We also analyzed mice with subcutaneous tumors from CTL or BMI1 B16F10 cells for the presence of lung metastases. As the primary tumors grow rapidly, we had to screen only 3 wk after tumor cell injection. Despite this short time frame, one of seven B16F10 BMI1-expressing tumors actually yielded lung metastases, compared with zero of five B16F10 CTL tumors. This raised the possibility that BMI1 variants have an increased ability to colonize distant organs. Thus, we performed an in vivo metastasis assay in which CTL versus BMI1 MA2 or B16F10 cells were injected into the tail vein (n = 7 mice per variant), and lung tumors were assessed (Fig. 1E,G; Supplemental Fig. S1F). Strikingly, in both MA2 and B16F10, higher BMI1 levels caused many more lung tumors (Fig. 1E,G; Supplemental Fig. S1F). Importantly, consistent with our in vitro proliferation assays, there was no difference in the proliferative index of BMI1 versus CTL lung tumors (Fig. 1F). We also conducted tail vein injections with the B16F10 sh-CTL, shBMI1#1, and shBMI#2 cells and saw a significant reduction in lung tumor numbers that was proportional to the degree of BMI1 knockdown (Fig. 1H). Thus, BMI1 acts in a dose-dependent manner to determine the metastatic potential of melanoma cells, and this is independent of altered proliferation or primary tumor growth.
BMI1 overexpression promotes melanoma cell invasion and distant site colonization
Having shown that BMI1 promotes metastases formation, we assessed BMI1's ability to modulate specific stages of the metastatic cascade. First, we examined migration using wound healing assays (for A375, B16F0, B16F1, and B16F10 BMI1 vs. CTL cells, and MA2 and B16F10 sh-Ctl vs. sh-BMI1 lines) (Fig. 2A; Supplemental Fig. S2A) and transwell migration assays (for A375, S91, and B16F10 BMI1 vs. CTL cells) (Supplemental Fig. S2B). Migration was significantly increased by BMI1 in all cases and significantly decreased with the highest degree of BMI1 knockdown (sh#2 for MA2 and sh#1 for B16F10). We then used an in vivo extravasation assay to assess BMI1's contribution to metastasis seeding. BMI1 or parental B16F10 cells were labeled with a red fluorescent dye (CMRA) and injected into the tail veins of nude mice (n = 3 for each variant and time point). Two hours later, equivalent numbers of BMI1 and parental cells were found in the lungs, indicating comparable capillary entrapment (data not shown). In contrast, 48 h after injection showed a significant increase (1.6-fold) in lung occupancy of BMI1 versus parental cells (Fig. 2B). Importantly, we confirmed that the CRMA-labeled tumor cells had exited the blood vessels (detected by CD31 immunostaining) and colonized the lung parenchyma (Supplemental Fig. S2C). We then used in vitro extravasation assays to compare the ability of BMI1 and CTL A375 cells to invade through an endothelial cell monolayer (Fig. 2C). Again, the BMI1 cells migrated significantly better than CTL cells (+35%; P = 0.001), showing that BMI1 enhances extravasation in a cell-autonomous manner.
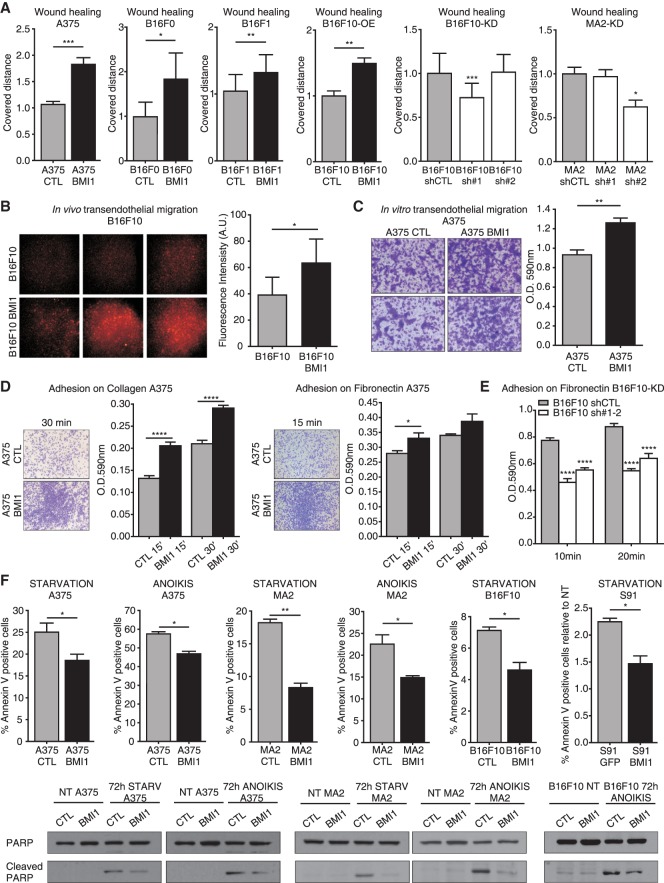
Figure 2.
BMI1 promotes melanoma cell movement, extravasation, adhesion, and survival. (A) Wound healing assays with CTL versus BMI1 A375, B16F0, B16F1, and B16F10 cells (first four bar graphs) and sh-Ctl versus sh-BMI1 B16F10, or MA2 cells (last two bar graphs). For the last two bar graphs, statistical significance was assayed by one-way ANOVA and Kruskal-Wallis test. (*) Significance versus sh-Ctl. (B–E) Representative images and/or quantification of in vivo extravasation of CMRA-labeled parental versus BMI1 B16F10 cells 48 h after tail vein injection (B), in vitro transendothelial migration analysis of CTL versus BMI1 A375 cells at 24 h (C), adhesion of CTL versus BMI1 A375 cells on collagen (left panel) or fibronectin (right panel) at 15 or 30 min (D), and adhesion assay of sh-Ctl versus sh-BMI1 B16F10 cells on fibronectin at 10 and 20 min (E). For E, statistical significance was assayed by two-way ANOVA and Kruskal-Wallis test. (*) Significance versus sh-Ctl. (F) CTL and BMI1 A375, MA2, B16F10, and S91 cells were subjected to starvation or anoikis for 72 h and analyzed for apoptosis by APC-Annexin V staining and Western blotting of total versus cleaved PARP levels. Graphs show mean ± SEM except B, which shows mean ± SD. See also Supplemental Figure S2.
Metalloproteinases (MMPs) are known to enable melanoma cell migration and invasion (Bartolome et al. 2006). Using gelatin zymography with conditioned media derived from equal cell numbers, we showed that BMI1 A375 and MA2 variants had higher MMP2 gelatinase activity than the CTL cells (Supplemental Fig. S2F). Since metastatic cells often show modulation of adhesion molecules and altered ability to bind extracellular matrix (ECM) components, we also tested BMI1's influence on cell adhesion to collagen and/or fibronectin. For all lines examined, adherence was significantly enhanced by BMI1 up-regulation (Fig. 2D; Supplemental Fig. S2D) and significantly reduced by BMI1 knockdown (Fig. 2E). Finally, BMI1 expression also caused morphological changes by which BMI1 A375 and MA2 cells had a more elongated (mesenchymal type) morphology versus the more round (amoeboid type) morphology of CTL cells (Supplemental Fig. S2E). Thus, our in vivo and in vitro data show that BMI1 increases the metastatic potential of melanoma cells, and this reflects enhancement of all stages of the metastatic cascade.
BMI1 promotes cell survival
Apoptosis resistance is a powerful enabler of metastasis because proapoptotic stimuli are encountered at many stages, including nutrient depletion, while in the circulation and anoikis resulting from ECM detachment (Mehlen and Puisieux 2006). By culturing BMI1 and CTL melanoma cells without serum (starvation) or substratum contact (anoikis), we showed that BMI1 significantly reduced starvation-induced apoptosis in A375 (−26%; P < 0.05), MA2 (−54%; P < 0.01), B16F10 (−35%; P < 0.05), and S91 (−34%; P < 0.05) cells (Fig. 2F) and also the propensity of A375 (−19%; P < 0.05) and MA2 (−34%; P < 0.05) cells to undergo anoikis (Supplemental Fig. S2G). BMI1 also reduced PARP cleavage in response to starvation and anoikis (Fig. 2F). Finally, we tested BMI1 and CTL A375, MA2, and B16F0 cells in low-seeding colony formation assays (Supplemental Fig. S2G) and found that BMI1 significantly increased single-cell colony formation (2.6-fold, P < 0.01 for A375; 2.8-fold, P < 0.001 for MA2; and 1.5-fold, P < 0.05 for B16F0). Thus, we conclude that BMI1 enables acquired resistance to apoptotic stimuli and promotes the survival and/or colony-forming potential of single cells.
BMI1 induces expression of an invasive gene signature in melanoma
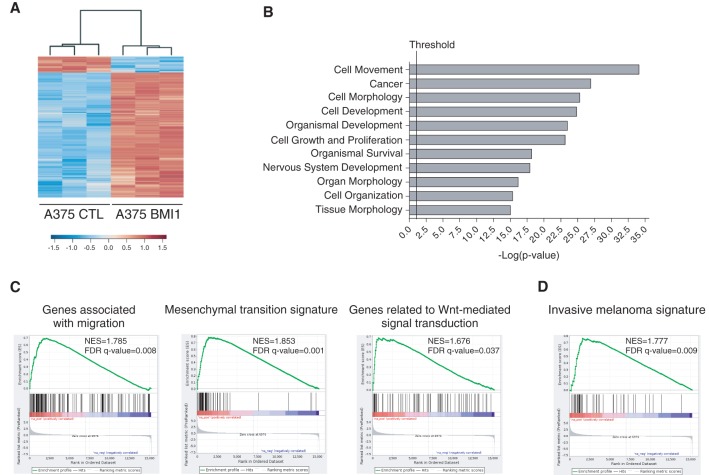
Given BMI1's role as an epigenetic regulator, it seemed likely that BMI1 promotes metastasis via gene expression changes. Previous studies have shown that BMI1 can modulate the expression of metastatic regulators PTEN and AKT (Song et al. 2009; Liu et al. 2012b), which are important in melanoma (Dankort et al. 2009; Madhunapantula et al. 2011). Thus, we examined the levels of PTEN, AKT, the active phospho-AKT isoform, and their downstream target, GSK-3β (Supplemental Fig. S3A–I). Unexpectedly, BMI1 overexpression had no effect on the levels or activity of these regulators in A375, MA2, or B16F10 cells. Having ruled out these known BMI1 targets, we performed RNA sequencing (RNA-seq) analysis on BMI1 versus CTL A375 cells using three independent samples per variant. This identified 949 genes that were differentially expressed (DE) between BMI1 and CTL cells (q-value ≤ 0.05; fold change ≥ 1.5), of which 842 were up-regulated and 107 down-regulated by BMI1 overexpression. We also identified a higher confidence signature (q-value ≤ 0.05; fold change ≥ 3) that comprised 288 up-regulated genes and 16 down-regulated genes in the BMI1 variant. The data set is available on Gene Expression Omnibus (GEO; GSE71890), and the DE genes are listed in Supplemental Table S1, with a heat map showing their hierarchical clustering in Figure 3A. We used Ingenuity Pathway Analysis (IPA) to classify the DE genes into functional groups. This revealed significant enrichment of pathways involved in cellular movement and morphology (Fig. 3B; Supplemental Fig. S4A). These included Wnt and TGFβ signaling pathways, which play key roles in migration, invasion, and tumor progression in cancer generally and melanoma especially. We also saw significant enrichment of EMT regulators (Supplemental Fig. S4A). Gene set enrichment analysis (GSEA) (Subramanian et al. 2005) revealed enrichment of Molecular Signatures Database (MSigDB) gene sets associated with activation of Wnt, TGFβ, and EMT signaling pathways and genes associated with invasion, migration, and ECM organization (Fig. 3C; Supplemental Fig. S4B). To further explore this interplay, we used custom gene sets that had been assembled based on gene ontology to include defining components of noncanonical Wnt, TGFβ, EGF/PDGF, EMT, and adhesion programs. For all of these custom gene sets, the majority of the constituent genes were differentially regulated between BMI1 and CTL A375 cells, consistent with increased pathway signaling in the BMI1 cells (Supplemental Fig. S4C).
Figure 3.
BMI1 induces the invasive signature in melanoma. (A) Heat map showing DE genes (q-value ≤ 0.05; fold change ≥ 1.5) between BMI1 and CTL A375 cells. (B) Top biological processes identified in the BMI1-induced signature by IPA. The vertical line represents the significance threshold (P-value of 0.05). (C,D) GSEA shows significant enrichment (false discovery rate [FDR] <0.05) of the following gene sets within the BMI1-induced signature: MsigDB gene sets with examples shown for migration, EMT, and Wnt signaling pathways (C) and the invasive melanoma gene signature identified by Widmer et al. (2012) (D). (NES) Normalized enrichment score. See also Supplemental Figures S3 and S4.
Previous studies have identified two distinct gene signatures in melanoma, corresponding to proliferative versus invasive transition states (Hoek and Goding 2010). The proliferative state correlates with better prognosis and is characterized by high expression of melanocytic markers, including the transcription factor MITF, while the invasive signature is characterized by TGFβ and noncanonical Wnt signaling (Hoek and Goding 2010). We used GSEA to determine whether our BMI1-induced signature correlated with either of these signatures. We found highly significant enrichment of the invasive signature (q-value = 0.009) (Fig. 3D) in our BMI1 gene set. In contrast, there was no significant enrichment, including no anti-correlation, with the proliferative state (q-value = 0.177) (data not shown). Accordingly, our BMI1 variants show no alterations in the expression of MITF or its downstream targets (e.g., TYR, CDKN1B, and SERPINF1). Thus, BMI1 is able to activate the invasive program without impacting MITF expression.
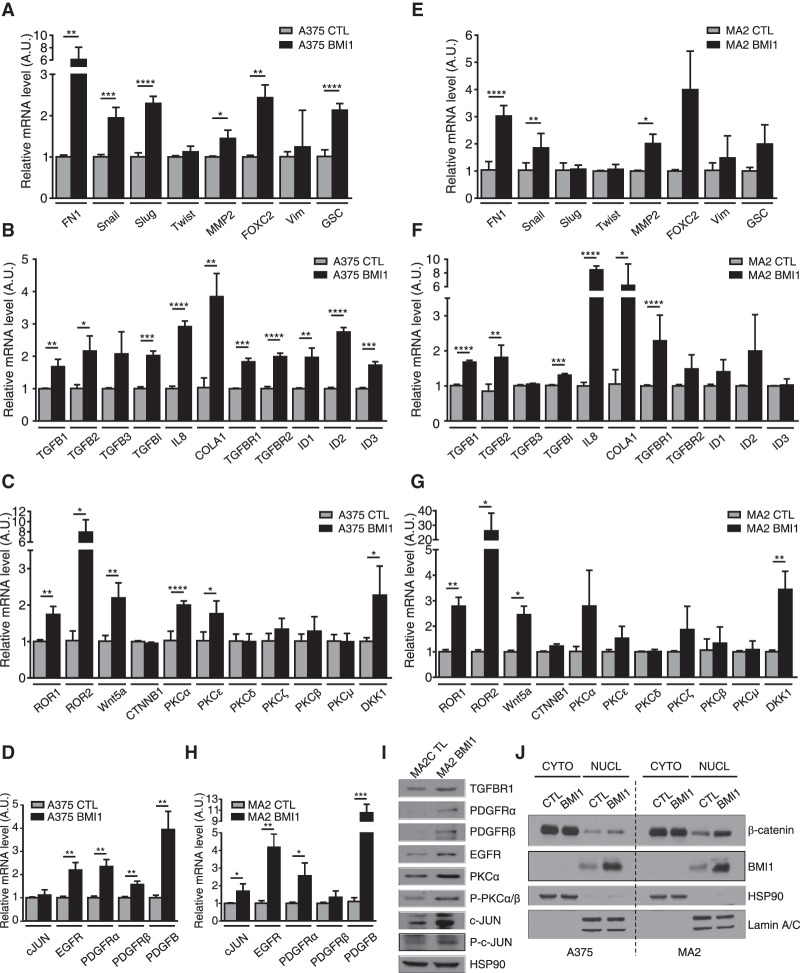
To further validate BMI1's effect on the invasive program, we used qPCR to compare the levels of key components of the EMT (Fig. 4A), TGFβ (Fig. 4B), noncanonical Wnt/PKC (protein kinase C) (Fig. 4C), and EGF/PDGF (Fig. 4D) pathways in BMI1 versus CTL A375 cells and saw significant up-regulation of many of these regulators. We also extended this qPCR analysis to the MA2 and B16F10 cell lines. Although there were some gene-to-gene differences, we found that key regulators of the TGFβ, noncanonical Wnt, EMT, and EGFR pathways were also up-regulated in these BMI1-expressing variants (Fig. 4E–H; Supplemental Fig. S5A). Moreover, regulators of EMT, noncanonical Wnt, TGFβ, and PDGF pathways were down-regulated in B16F10 cells after BMI1 knockdown (Supplemental Fig. S5B). Validation by Western blotting of selected targets and downstream regulators was carried out in A375 and MA2 cells (Fig. 4I; Supplemental Fig. S5C–F).
Figure 4.
BMI1 induces the invasive signature with concomitant nuclear accumulation of β-catenin. (A–H) qPCR analysis on total RNA from A375 (A–D) and MA2 (EH) cells confirmed the differential expression of representative genes involved in EMT (A–E), TGFβ (B–F), noncanonical Wnt/PKC (C–G), and EGF/PDGF (D–H) pathways in CTL versus BMI1 cells. Results shown are mean ± SD. (I) Expression of selected proteins evaluated by Western blot analysis of CTL and BMI1 MA2 cells. (J) Subcellular fractionation and Western blot analysis of CTL and BMI1 A375 and MA2 cells, with HSP90 and Lamin A/C as control for cytoplasmic and nuclear fractions. See also Supplemental Figure S5.
We looked more closely at the noncanonical Wnt pathway because this is one of the best-known markers of malignant melanoma. We observed significant up-regulation of the Wnt5a ligand and its coreceptor, ROR2, in BMI1 variants (Fig. 4C,G; Supplemental Fig. S5A) and down-regulation in B16F10-shBMI1 cells (Supplemental Fig. S5B). Additionally, we saw up-regulation and activation of PKCα (Fig. 4C,G,I; Supplemental Fig. S5C), which has been linked to Wnt5a-induced melanoma migration (Dissanayake et al. 2007). Accordingly, we found that the PKCα inhibitor Gö6976 suppressed the migration of BMI1 melanoma cells (data not shown). Noncanonical Wnt signaling is frequently associated with down-regulation of the canonical Wnt pathway in melanoma (Lucero et al. 2010). However, there are clear exceptions to this rule (Larue and Delmas 2006) and conflicting views on whether canonical Wnt facilitates or impedes melanoma metastasis (Lucero et al. 2010). We saw no difference in the levels of total or nonphosphorylated (active) β-catenin between the BMI1 and CTL variants in the three cell lines examined (Supplemental Fig. S4E–G). However, BMI increased the levels of nuclear β-catenin in both A375 (1.8-fold) and MA2 (twofold) cells (Fig. 4J). Moreover, many β-catenin-responsive genes were up-regulated in the BMI1-induced signature (Supplemental Table S1), and representative targets (AXIN2, NRCAM, and RHOU) were validated by qPCR (data not shown). These data show that high BMI1 levels lead to a widespread gene expression change that includes activation of key hallmarks of the invasive state of metastatic melanoma while sustaining expression of core determinants of the proliferative state (MITF and β-catenin).
BMI1 confers resistance to BRAF inhibitor treatment
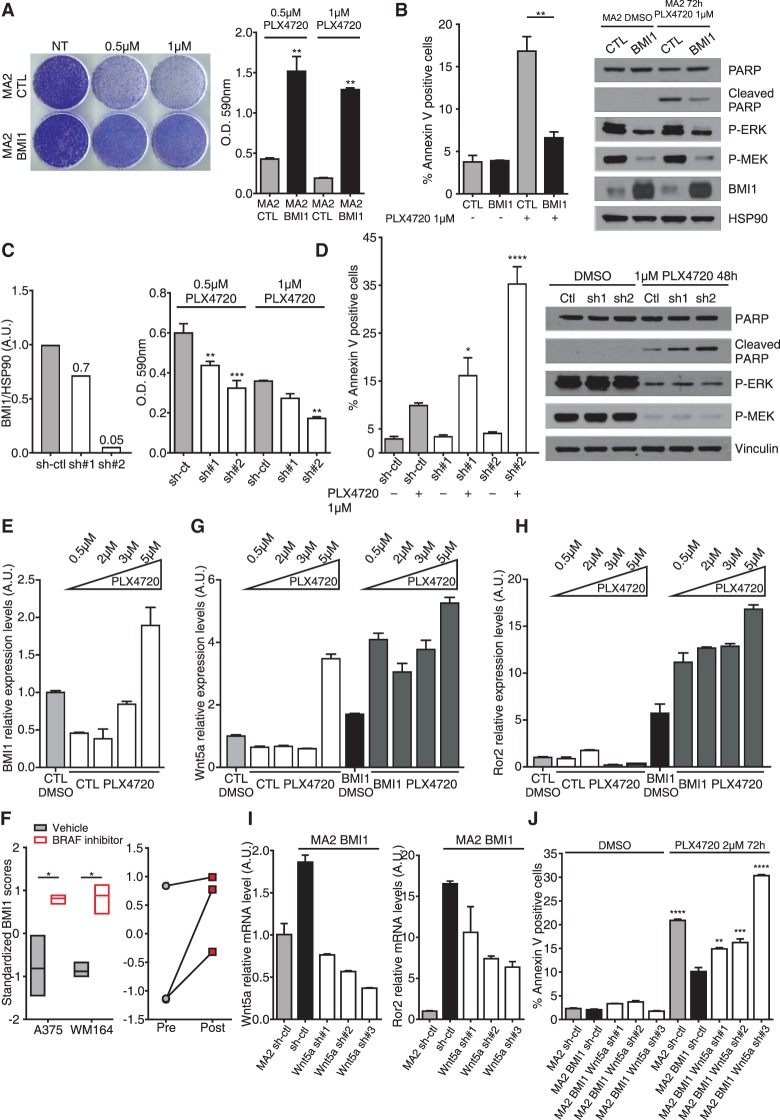
Approximately 50% of skin melanomas carry activating mutations in BRAF (Davies et al. 2002), and selective inhibitors are now used to treat patients with BRAF mutant metastatic melanoma. Having shown that BMI1 promotes apoptotic resistance to starvation and anoikis, we wanted to assess response to BRAF inhibition (BRAFi). We conducted this analysis on MA2 cells and their variants because these carry the BRAFV600E mutation. Accordingly, we found that MA2 CTL cells were sensitive to the BRAF inhibitor PLX4720 (Fig. 5). Initially, we tested the BMI1 variant and showed that this had acquired significant resistance. We assayed the cells response to long-term (18-d) drug treatment (Fig. 5A) and found that the final cell number was significantly higher for the BMI1 cells (3.5-fold with 0.5 µM PLX4720 and 6.7-fold with 1 µM PLX4720) compared with CTLs. Importantly, this reflected a significant reduction in the levels of apoptotic cells (−60%) and cleaved PARP 72 h after 1 µM PLX4720 treatment (Fig. 5B). Interestingly, the increased survival of BMI1 cells was specific to PLX4720 and not the classic chemotherapeutics cisplatin, paclitaxel, vinorelbine, and gencitabine (Supplemental Fig. S5G). By assaying the sh-Ctl and sh-BMI1 MA2 variants, we showed that BMI1 deficiency increased sensitivity to PLX4720 that was proportional to the degree of BMI1 knockdown. After long-term drug treatment, the final cell number was reduced by 27% for sh1 (P < 0.01) and 46% for sh2 (P < 0.001) with 0.5 µM PLX4720 and 24% for sh1 (N.S.) and 52% for sh2 (P < 0.01) with 1 µM PLX4720 (Fig. 5C, right bar graph; Supplemental Fig. S5H). The reduced viability of these cells correlated with increases in apoptosis (1.6-fold, P < 0.05 for sh1; 3.5-fold, P < 0.0001 for sh2) and PARP cleavage 48 h after treatment with 1 µM PLX4720 (Fig. 5D). Thus, either increases or decreases in BMI1 levels significantly impact the response of BRAF mutant melanoma cells to BRAFi.
Figure 5.
BMI1 confers resistance to the BRAF inhibitor by activation of the noncanonical Wnt pathway. (A,B) CTL and BMI1 MA2 cells were treated with the indicated levels of PLX4720 and assayed for cell number after 18 d by crystal violet staining and quantification (A) or cell death at 72 h by quantification of APC-Annexin V staining and Western blot analysis of total and cleaved PARP (B). HSP90 was used as a loading control, and P-ERK and P-MEK were used to verify drug efficacy. (C,D) sh-Ctl and sh_BMI1 MA2 cells were assessed for degree of BMI1 knockdown (by quantification of the Western blot shown in Supplemental Fig. S1D) (C, left), total accumulated cell number 18 d after culture in PLX4720 (C, right), or apoptosis in response to 48 h of culture in PLX4720 by quantification of APC-Annexin V staining and Western blot analysis of total and cleaved PARP (D). (E,G,H) CTL and BMI1 MA2 cell clones selected after long-term culture with DMSO vehicle or the indicated doses of PLX4720 were assessed for the level of BMI1 (E), Wnt5a (G), and Ror2 (H) mRNA relative to that of the CTL DMSO cells. (F) Levels of BMI1 in gene expression data sets from A375 (GSE42872 [Parmenter et al. 2014]) and WM164 (GSE54711) cells after culture in vehicle or BRAFi (left panel) or three paired biopsies from melanoma patients before and after treatment with vemurafenib (GSE50535 [Sun et al. 2014]) (right panel). (I) Levels of Wnt5a (left panel) and Ror2 (right panel) in sh-ctl and sh-Wnt5a MA2 BMI1 cells relative to sh-ctl MA2 cells. (J) Quantification of apoptosis by analysis of APC-Annexin V staining of the cell lines from I 72 h after treatment with DMSO or 2 µM PLX4720. Statistical significance was determined by one-way ANOVA with Tukey's multiple comparisons test, where the asterisk indicates significance versus the treated sh-Ctl samples (C,D), and one-way ANOVA with Dunnet's multiple comparisons test, where the asterisk indicates significance versus MA2 BMI1 sh-ctl treated with PLX4720 (J). See also Supplemental Fig. S5.
To gain additional insight into the molecular pathways responsible for the acquired resistance of BMI1 cells, we derived resistant populations (called CTL PLX4720 and BMI PLX4720) by exposing MA2 CTL and MA2-BMI cells to different concentrations of PLX4720 (0.5, 2, 3, and 5 µM) for 2 mo. The augmented invasive potential of the BMI1 variant was maintained in the post-drug selection cells (Supplemental Fig. S5I). Interestingly, we found that BMI1 levels were specifically increased in the CTL PLX4720 variant exposed to the highest drug concentration (Fig. 5E). Given this finding, we examined available data sets generated from clones of A375 (GSE42872) and WM164 (GSE54711) human cell lines before or after culture with BRAFi. In both cases, BMI1 expression was significantly increased in the post-treatment clones (Fig. 5F). Expression data are also available for three patient tumors before and after BRAFi treatment, and the two samples with low pretreatment BMI1 levels also showed BMI1 up-regulation after treatment (Fig. 5F). Thus, exposure to BRAFi can result in increased BMI1.
We also evaluated our CTL PLX4720 and BMI PLX4720 populations for the expression of DE genes that are up-regulated by BMI1 and known to be key determinants for BRAFi resistance in melanoma, such as EGFR, PDGFR, and Wnt5a (Nazarian et al. 2010; Anastas et al. 2014; Sabbatino et al. 2014; Sun et al. 2014). We found that Wnt5a was further up-regulated in all of the BMI PLX4720 populations, compared with vehicle-treated BMI1 controls, and was also specifically induced in the CTL PLX4720 line that had up-regulated its endogenous BMI1 (Fig. 5G). Notably, the BMI PLX4720 populations, but not the CTL PLX4720 line, also elevated the Wnt5a coreceptor ROR2 (Fig. 5H). Given these findings, we used three different shRNAs to knock downWnt5a in the MA2 BMI1 variant, yielding 41%, 30%, and 19% of starting Wnt5a mRNA levels (Fig. 5I). Interestingly, these yielded a correlative reduction in Ror2 mRNA (64%, 44%, and 38%) (Fig. 5I) and also down-regulation of other key components of the EMT, TGFβ, and PDGF pathways (Supplemental Fig. S5J). We assayed the apoptotic responses of the Wnt5a knockdown variants to PLX4720 (Fig. 5J) and saw increased sensitivity that was proportional to the degree of Wnt5a knockdown (1.47-fold, P ≤ 0.01; 1.6-fold, P ≤ 0.001; threefold, P ≤ 0.0001). These data show that BMI1 confers resistance to BRAF inhibitor, and the activation of the noncanonical Wnt5a–Ror2 pathway is critical for this response.
BMI1 loss in vivo impedes melanoma metastasis formation
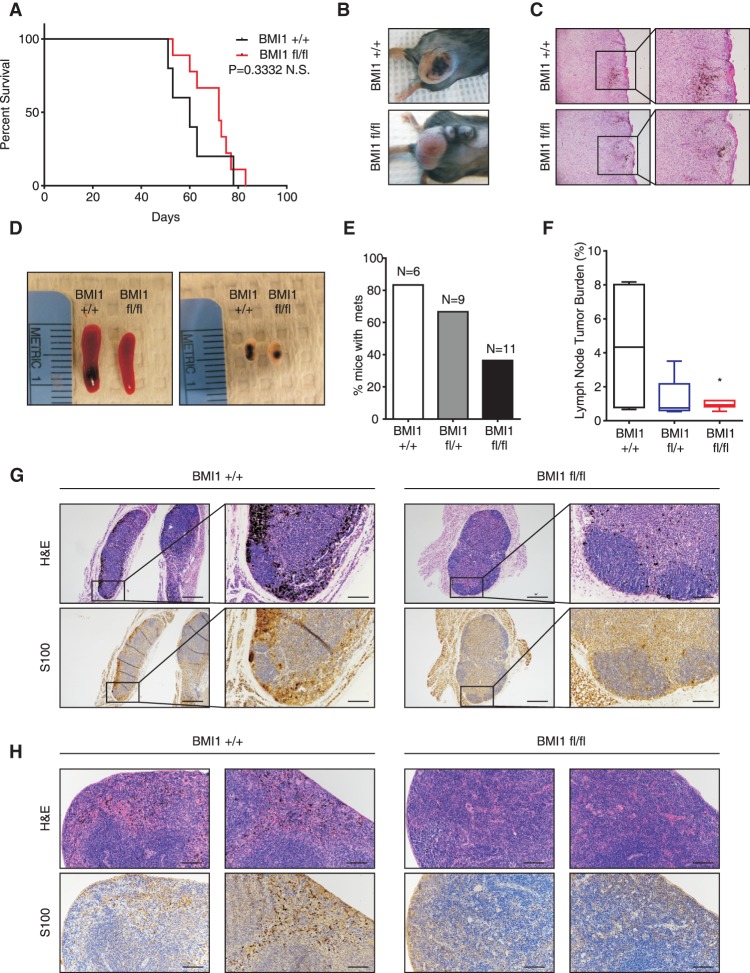
Tumor–stroma interactions are known to play a key role in cancer progression, but these are not fully modeled in transplant assays. Thus, to investigate BMI1's role in autochthonous melanoma formation, we used a well-established melanoma mouse model (Dankort et al. 2009) in which melanocyte-specific activation of heterozygous mutant Braf (BrafV600E) and loss of Pten are induced by treatment with hydroxytamoxifen (4-OHT). These treated animals (here called Braf/Pten) develop metastatic melanomas (Dankort et al. 2009; Damsky et al. 2011). We crossed a Bmi1 conditional allele (Maynard et al. 2014) into this Braf/Pten model and confirmed that topical 4-OHT application yielded efficient recombination of the Bmi1 allele in the skin (Supplemental Fig. S6A). Initially, we focused on assessing the effect of BMI1 deletion on primary melanoma formation. Previous studies showed that localized administration of 4-OHT onto the shaved backs of adult mice causes the Braf/Pten mice to develop pigmented lesions in 2–3 wk, which yield tumors that require euthanasia ∼10 wk after initiation (Damsky et al. 2011). We generated Braf/Pten mice that were Bmi1+ /+ or Bmi1fl/fl, initiated tumor development at 6–8 wk of age, and euthanized these animals once the tumors reached maximal volume (2-cm diameter) and/or displayed ulceration. Animals with more than one tumor or with tumors outside the 4-OHT-treated region (due to Tyr-Cre-ERT2 leakiness) were excluded from consideration. The remaining study animals showed no significant difference (P = 0.33) in time to euthanasia for Bmi1+/+ mice (51–78 d; n = 5) versus Bmi1fl/fl mice (53–83 d; n = 9) (Fig. 6A). Moreover, there was no difference in the histology of primary Bmi1+/+ versus Bmi1fl/fl mutant tumors (Fig. 6B,C). Thus, Bmi1 loss does not alter the initiation or development of primary autochthonous melanomas resulting from Braf/Pten mutations.
Figure 6.
BMI loss impedes metastasis formation, but not primary tumor formation, in an autochthonous mouse model of melanoma. (A–C) Primary melanoma formation was induced by local 4-OHT treatment on Braf/Pten mice that were Bmi1fl/fl or Bmi1+/+. Shown are Kaplan-Meier curves (A), representative images of the resulting tumors (B), and H&E staining of tumor sections showing both pigmented and nonpigmented regions (C). (D–H) Newborn Braf/Pten mice that were Bmi1fl/fl, Bmi1fl/+, or Bmi1+/+ were treated with 4-OHT. Shown are representative images of spleen and inguinal lymph nodes (D); the percentage of Bmi1fl/fl, Bmi1fl/+, and Bmi1+/+ mice with lymph node and/or spleen metastases (E); box plots (from minimum to maximum) of lymph node tumor burden evaluated by S100 positive area (F); and representative images of H&E and S100 immunostaining (G,H) of melanin-containing lymph nodes (G) or spleens (H). Bars, 100 µm. See also Supplemental Figure S6.
We next assessed the impact of Bmi1 status on metastases. Metastases are rare in the localized induction protocol used above because the primary tumors require euthanasia before metastases can arise. Thus, we used an alternative protocol (Damsky et al. 2011) in which perinatal 4-OHT treatment yields pigmented lesions that metastasize to lymph nodes, the lung, and the spleen with high penetrance. We performed this perinatal tumor induction and found that all of the mice, irrespective of Bmi1 genotype, developed extensive primary pigmented lesions (Supplemental Fig. S6B) and were moribund around the time of weaning (between 25 and 41 d). We then screened lymph nodes and spleens of Bmi1+/+, Bmi1fl/+, or Bmi1fl/fl Braf/Pten littermates for the presence of metastases (Fig. 6D–H). Since melanin-containing macrophages are often abundant in the lymph nodes of animals with melanoma, we used immunostaining for the melanocytic marker S100 to distinguish metastatic cells from melanin-containing macrophages (Fig. 6G,H). Strikingly, quantification revealed significantly fewer S100-positive metastatic melanoma cells in the inguinal lymph nodes of Bmi1fl/fl versus Bmi1+/+ mice (LN tumor burden of 0.9520 ± 0.08421 vs. 4.380 ± 2.014) (Fig. 6F,G), indicating that BMI1 loss inhibits metastatic lesion formation.
BMI1-driven gene signature correlates with highly metastatic phenotype in human samples
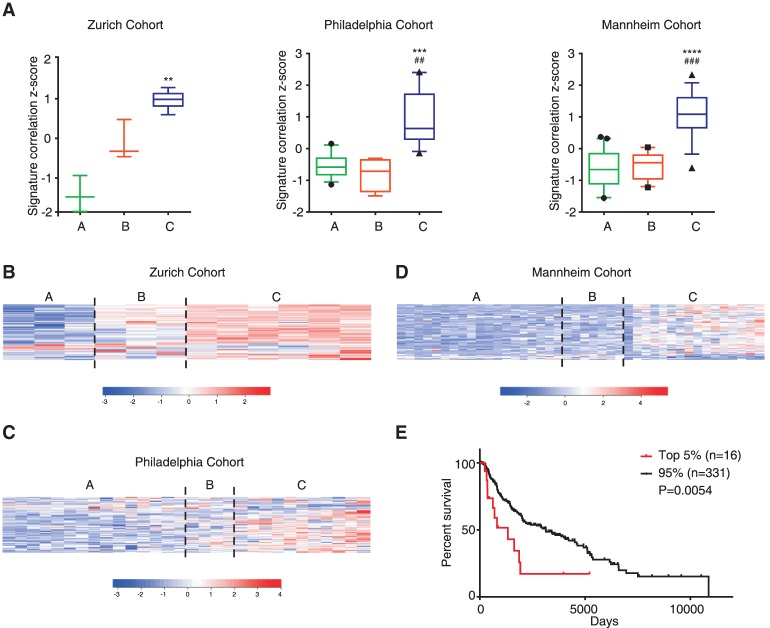
Our in vitro and in vivo studies establish BMI1 levels as a key determinant of melanoma metastasis. Given these findings, we asked whether our BMI1-induced signature has predictive value for human melanomas. For this, we took advantage of a previous study (Hoek et al. 2006) that had clustered 86 human melanoma samples from three different human data sets (the Zurich, Philadelphia, and Mannheim cohorts) and segregated them into weakly (group A), intermediate (group B), and highly (group C) invasive subclasses based on gene expression and invasive potential. We calculated the correlation score of our BMI1-induced signature for samples in the A, B, and C subclasses of the Zurich, Philadelphia, and Mannheim data sets. This revealed a significant enrichment in the highly invasive samples (group C) compared with the remaining samples (groups A and B) for all three data sets (Fig. 7A). Genes in our BMI1-induced signature with a twofold or greater difference between A+B versus C subclasses in at least two of the three cohorts are shown in the heat maps in Figure 7, B–D, and listed in Supplemental Table S2. We then assessed the BMI1-induced signature in the TCGA-SKCM data set that is comprised of both primary and metastatic melanoma samples. Notably, the BM1 signature was able to predict reduced survival (Fig. 7E) in correlated samples. Moreover, the BMI1-induced signature could also predict reduced survival in primary melanoma samples, as evidenced by analysis of the Winnepenninckx data set (Winnepenninckx et al. 2006), which is comprised of 83 primary melanomas (P = 0.03643) (data not shown). Thus, the BMI1-induced signature is associated with melanoma metastasis and drug resistance and is highly predictive of poor prognosis of melanoma patients.
Figure 7.
BMI1-induced signature correlates with invasiveness and clinical outcome of human melanoma. (A) Standardized BMI1 RNA-seq signature correlation scores in the weakly invasive A , intermediate invasive B, and highly invasive C subclasses of melanoma within the Zurich, Philadelphia, and Mannheim cohorts. Data are presented as box plots (10th to 90th percentile), with P-values calculated by Dunn's test following a significant Kruskal-Wallis test. (*) P-values for the C versus A subclasses; (#) P-values for the C versus B subclasses. (B–D) Heat maps show expression of BMI1 DE genes with a fold change greater than two between the A+B versus C subclasses and common to at least two data sets. (E) Patient outcome based on BMI1-induced signature correlation (top 5% vs. the remaining 95%) using the TCGA-SKCM data set. Log rank P-value is shown. See also Supplemental Figure S7.
Discussion
This study has revealed key insight into melanoma metastasis and the role of BMI1 in this process. Our data show that ectopic expression or loss of BMI1 has no effect on cell proliferation or primary tumor growth. This allowed us to assess BMI1's role in tumor progression independently of its usual proproliferative role. Remarkably, we found that BMI1 acts to shift melanoma cells to a more metastatic state irrespective of their original driving mutations or metastatic potential by promoting all of the steps of the metastatic cascade. Accordingly, loss of BMI1 reduces invasive properties and severely impedes metastatic dissemination, including in autochthonous tumors. Moreover, up-regulation or down-regulation of BMI1 is sufficient to render BRAF mutant melanoma cells resistant or sensitive to the BRAF inhibitor PLX4720, and the Wnt5a–Ror2 pathway plays a key role in this effect. Finally, we identify a BMI1 gene expression signature that is predictive of the invasive subclass of human melanomas and poor survival of melanoma patients.
Previous studies report that human melanomas display two gene expression signatures that correlate with more proliferative or more invasive states (Hoek and Goding 2010). Computational analyses show that our BMI1-induced signature correlates strongly with the invasive state. We found that TGFβ and noncanonical Wnt signaling, which are core components of the human invasive signature (Hoek and Goding 2010), plus an EMT program, are key components of our BMI1-induced signature. Up-regulation of the noncanonical Wnt pathway is the most consistent change observed across human malignant melanoma and also across our BMI1-induced signature. We found that BMI1 also induces the expression of ROR2, a coreceptor for Wnt5a, and increases expression and activity of PKCα. PKCα is known to contribute to the ability of Wnt5a–ROR2 to promote the invasion and migration of melanoma cells (Dissanayake et al. 2007; O'Connell et al. 2010), and, accordingly, we found that PKCα inhibition suppresses the migration of the BMI1-expressing melanoma cells (data not shown). Thus, we conclude that this Wnt5a–ROR2–PKC pathway is playing a key role in the elevated metastatic potential of our BMI1-expressing cells.
Canonical Wnt–β-catenin signaling is important for normal melanocyte development and melanoma initiation (Larue and Delmas 2006). In contrast, its role in metastasis is controversial (Chien et al. 2009; Lucero et al. 2010; Arozarena et al. 2011; Damsky et al. 2011). In concert with the noncanonical Wnt5a–ROR2–PKC signaling, our BMI1 cells show increased levels of nuclear β-catenin and expression of numerous β-catenin targets. Interestingly, it has been recently reported that Wnt5a can induce β-catenin to be released from N-Cadherin, allowing it to accumulate into the nucleus and promote melanoma cell invasion (Grossmann et al. 2013). Together, this and our findings suggest that the BMI1-mediated induction of Wnt5a activates both noncanonical and canonical Wnt signaling in a manner that enables invasion and metastasis.
We found that BMI1 up-regulates the invasive melanoma signature without a corresponding decrease in the proliferative signature. Indeed, our BMI1 variants show no alterations in MITF, the key determinant of the proliferative state, or many of its downstream targets. We speculate that the persistence of MITF expression reflects the continued presence of canonical Wnt signaling, which is known to induce MITF. This could explain why we saw little or no alteration of proliferative capacity in the BMI1 cells or the resulting xenograft tumors. However, the continued presence of MITF does create a significant quandary; high MITF is known to impede melanoma invasiveness by suppression of RHO (Carreira et al. 2006) and also promote sensitivity to treatment with BRAFi (Muller et al. 2014), and yet our BMI1 cells show the opposite properties: enhanced invasion and drug resistance. These findings yield two conclusions. First, it is possible to uncouple up-regulation of the invasive program from down-regulation of the proliferative program. Second, the invasive program, and not the proliferative program, appears to be the dominant determinant of therapeutic response, since this clearly overrides the negative effect of MITF on both invasiveness and drug response. Interestingly, MITF is known to confer sensitivity to BRAFi via inhibition of EGFR expression (Ji et al. 2015). We found that BMI1 expression leads to up-regulation of both EGFR and PDGFR, offering a simple mechanism to counteract the effect of MITF. We believe that EGFR and PDGFR do not act alone to promote resistance in the BMI1-expressing cells but work in concert with Wnt5a. WNT5a is known to modulate chemotherapeutic response (Anastas et al. 2014), and we observed a marked increase of Wnt5a and Ror2 expression in BMI1-overexpressing populations that are resistant to BRAFi. Moreover, Wnt5a knockdown was sufficient to restore BRAFi sensitivity to the MA2-BMI1 variant. Together, these observations show that the Wnt5a–Ror2 pathway makes a critical contribution to BMI1-induced resistance. Since even partial knockdown of BMI1 confers drug sensitivity in our assays, we believe that BMI1 inhibitors could have significant clinical impact, at least in combination with BRAFi or MEKi, in metastatic melanoma treatment.
Our BMI1-induced signature also has predictive value for human melanomas. First, we found that the signature scores highly in the subset of tumors previously characterized as highly invasive in the Zurich, Philadelphia, and Mannheim cohorts. Second, our analysis of the TCGA-SKCM and Winnepenninckx human data sets showed that the BMI1-induced signature correlates with poor prognosis. As BMI1 is up-regulated in metastatic melanoma, we wondered whether this correlation simply reflected the increased expression of our signature in metastatic disease. However, two observations argue against this possibility. When we considered only primary tumors (the Winnepenninckx cohort), our signature still predicts for poor prognosis and is enriched in the subset of tumors that progressed to metastatic disease during the study time course. Additionally, we re-examined the results of our TCGA data set analysis—in which we saw reduced survival for the top 5% of tumors associated with our signature versus the remaining 95%—and discovered that neither fraction (5% versus 95%) displayed overrepresentation of primary or metastatic tumors (P > 0.12, hypergeometric test) or a significant relationship with tumor grade (P = 0.47, χ2 test). Thus, the predictive value of our signature is independent of the starting tumor grade.
We are not suggesting that BMI1 is the only route to melanoma metastasis. Our analysis of the paired A375 and MA2 cell lines and the B16 series showed that BMI1 is not always up-regulated during tumor progression. Moreover, we can find human tumor samples that have activated our BMI1-induced signature but do not have high BMI1 expression. That said, our cell-based studies show that BMI1 is a major driver of invasion and metastasis. Moreover, in four different human and murine melanoma gene expression data sets, we saw a strong association between high BMI1 levels and more metastatic tumors. Thus, we conclude that BMI1 offers a major route, but not the only route, to activate our identified invasive gene expression signature and thereby promote metastasis.
BMI1 is up-regulated in a variety of tumor types, and, in several cases, including lung and breast cancer, its expression increases with progression (Glinsky et al. 2005). We characterized TCGA data from lung adenocarcinoma, breast invasive carcinoma, and colon adenocarcinoma and found that our BMI1-induced signature showed no correlation with survival in these nonmelanoma tumor types (Supplemental Fig. S7). Interestingly, previous studies in nonmelanoma cell lines have linked BMI1 to regulators of migration (PTEN) (Song et al. 2009) and EMT (Twist) (Yang et al. 2010), which differ from the BMI1-responsive genes identified in our melanoma cells. Thus, we speculate that BMI1 also regulates invasion, migration, and EMT in other tissues, but the employed pathways will be context-dependent. Notably, our ability to establish BMI1's metastatic role was entirely dependent on the unexpected finding that the proliferative capacity and the rate of primary tumor growth were unaffected by changes in BMI1 levels. For other tumor types, genetic tricks will be necessary to circumvent the known requirement of BMI1 for proliferation and thus allow assessment of BMI1's contribution to progression and drug response.
Materials and methods
Cell line culture and analyses
B16F0, B16F1, B16F10, Cloudman S91, 293T, and human umbilical vein endothelial cell lines were obtained from American Type Culture Collection and maintained as recommended. A375 and MA2 (Xu et al. 2008) were grown in DMEM with 10% heat-inactivated FBS and 5% penicillin–streptomycin. Standard procedures were used to generate stable cell lines and assay cell proliferation, migration, adhesion, and survival (see the Supplemental Material).
Gene expression and bioinformatics analyses
All computations were performed in the R statistical computing environment (http://www.R-project.org). The GSE8401 (Xu et al. 2008), TCGA-SKCM (The Cancer Genome Atlas Network 2015), GSE46517 (Kabbarah et al. 2010), and GSE29074 (Scott et al. 2011) data sets were used to compare BMI1 expression in primary and metastatic melanomas. Differential expression analysis was performed using R/limma (Smyth 2005).
Total RNA from A375 CTL and BMI1 variants was prepared and sequenced as single-end 50mers on the Illumina HiSeq 2000 (see the Supplemental Material). Reads were aligned using RSEM version 1.2.12 (Li and Dewey 2011), and differential expression analysis was performed using EBSeq version 1.4.0 (Leng et al. 2013). Heat maps of row-normalized expression values were created using the Heatplus package in R. The identified RNA-seq DE signature (false discovery rate [FDR] <0.05, fold change >1.5) was used to conduct pathway analyses by Qiagen IPA (Ingenuity Systems, http://www.ingenuity.com) and GSEA using the preranked mode (Subramanian et al. 2005). MSigDB gene sets (http://www.broadinstitute.org/gsea/msigdb) were considered enriched at a significance level of FDR ≤0.05. Additionally, proliferative and invasive gene signatures (Widmer et al. 2012) were included in GSEAs to test for enrichment. The DE signature was used to score expression profiles of individual samples in the Zurich, Philadelphia, and Mannheim data sets (Hoek et al. 2006) as well as profiles of individual TCGA tumors in skin cutaneous melanoma, breast-invasive carcinoma, lung adenocarcinoma, and colon adenocarcinoma data sets using single-sample GSEA (ssGSEA). Combined up-regulated and down-regulated signature correlation scores were calculated and standardized (z-score). TCGA tumors were stratified based on the standardized score and compared for differences in survival times.
Transplant, metastasis, and in vivo extravasation assays
All assays were performed in 7-wk-old female mice. Primary tumors were assessed by subcutaneous injections of 2 × 104 B16F10 cells (into C57/BL6) or 106 A375 and MA2 cells (into NOD/SCID; Jackson) and dissection and measurement of tumors after 3 or 6 wk, respectively. Tail vein injections used 5 × 104 B16F10-BMI1 cells, 105 B16F10-sh cells, or 106 MA2 cells, and the lungs were analyzed for metastasis at 4 or 6 wk, respectively. For in vivo extravasation assays, 106 B16F10 cells were labeled with CellTracker Orange CMRA (Molecular Probes, Invitrogen Life Technologies) and injected into the tail veins of nude mice (Jackson). After 2 or 48 h, mice were sacrificed, and 4% paraformaldehyde was injected into the trachea. The lungs were dissected, and separated lobes were photographed using fluorescence stereomicroscopy. Six-micrometer cryosections were assessed with the anti-CD31 antibody (BD Biosciences, 550274) and DAPI counterstaining.
Mouse colony and melanoma induction
All animal procedures followed protocols approved by Massachusetts Institute of Technology's Committee on Animal Care. BRafCA;Ptenex5lox and Tyr::CreER mice (Jackson) were crossed to BMI1fl/fl mice (Maynard et al. 2014). Localized tumors were induced with 1 µL of 8.3mg/mL 4-OHT (70% Z-isomer; Sigma) in ethanol vehicle. Tumor volume was measured [volume mm3= 1/2(largest diameter × smallest diameter2)] after harvesting. For perinatal treatment, 15 µL of 50 mg/mL 4-OHT in DMSO was painted on the ventral abdomens of 2- and 4-d-old pups. Ki67 and S100 immunohistochemistry was performed as described in the Supplemental Material.
Statistical analyses
Unless otherwise indicated, statistical analyses were performed using Graphpad Prism software with unpaired t-test with Welch's correction for comparison of two conditions or one-way ANOVA with Tukey's test for multiple comparisons. In figures, unless otherwise noted, data are presented as mean ± SEM, where P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***), and P ≤ 0.0001 (****) were considered statistically significant, and “NS” indicates not statistically significant.
Supplementary Material
Acknowledgments
We thank the Koch Institute Swanson Biotechnology Center for technical support; R.O. Hynes for A375 and MA2 cells; Samanta Sharma, John Lamar, Boyang Zhao, and Daniel Karl for reagents and technical support; and A. Naba, M. Sullivan, K.R. Mattaini, and Lees laboratory members, especially A. Amsterdam, for input during the study. This work was supported by a grant from the Melanoma Research Foundation to J.A.L. and a Ludwig Post-doctoral Fellowship to R.F. J.A.L. is a Ludwig Scholar at Massachusetts Institute of Technology.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.267757.115.
References
- Anastas JN, Kulikauskas RM, Tamir T, Rizos H, Long GV, von Euw EM, Yang PT, Chen HW, Haydu L, Toroni RA, et al. 2014. WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors. J Clin Invest 124: 2877–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arozarena I, Bischof H, Gilby D, Belloni B, Dummer R, Wellbrock C. 2011. In melanoma, β-catenin is a suppressor of invasion. Oncogene 30: 4531–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann IM, Puntervoll HE, Otte AP, Akslen LA. 2008. Loss of BMI-1 expression is associated with clinical progress of malignant melanoma. Mod Pathol 21: 583–590. [DOI] [PubMed] [Google Scholar]
- Bartolome RA, Molina-Ortiz I, Samaniego R, Sanchez-Mateos P, Bustelo XR, Teixido J. 2006. Activation of Vav/Rho GTPase signaling by CXCL12 controls membrane-type matrix metalloproteinase-dependent melanoma cell invasion. Cancer Res 66: 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovska OP, Glinskii AB, Yang Z, Li XM, Hoffman RM, Glinsky GV. 2006. Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle 5: 1886–1901. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2015. Genomic classification of cutaneous melanoma. Cell 161: 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. 2006. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 20: 3426–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien AJ, Moore EC, Lonsdorf AS, Kulikauskas RM, Rothberg BG, Berger AJ, Major MB, Hwang ST, Rimm DL, Moon RT. 2009. Activated Wnt/β-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci 106: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CH, Yang NK, Liu TY, Tai SK, Hsu DS, Chen YW, Chen YJ, Chang CC, Tzeng CH, Yang MH. 2013. Chromosome instability modulated by BMI1–AURKA signaling drives progression in head and neck cancer. Cancer Res 73: 953–966. [DOI] [PubMed] [Google Scholar]
- Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, Taketo MM, Dankort D, Rimm DL, McMahon M, et al. 2011. β-Catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell 20: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr, You MJ, DePinho RA, McMahon M, Bosenberg M. 2009. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 41: 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. 2002. Mutations of the BRAF gene in human cancer. Nature 417: 949–954. [DOI] [PubMed] [Google Scholar]
- Dissanayake SK, Wade M, Johnson CE, O'Connell MP, Leotlela PD, French AD, Shah KV, Hewitt KJ, Rosenthal DT, Indig FE, et al. 2007. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem 282: 17259–17271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey JS, Zacharek SJ, Kim CF, Lees JA. 2008. Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc Natl Acad Sci 105: 11857–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Li L, Ou Z, Kong C, Zhang Y, Dong Z, Zhu S, Jiang H, Shao Z, Huang B, et al. 2012. FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast Cancer Res Treat 131: 65–73. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. 1973. Selection of successive tumour lines for metastasis. Nat New Biol 242: 148–149. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. 2010. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363: 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislin S, Deshayes F, Middendorp S, Boggetto N, Alcaide-Loridan C. 2012. PHF19 and Akt control the switch between proliferative and invasive states in melanoma. Cell Cycle 11: 1634–1645. [DOI] [PubMed] [Google Scholar]
- Gieni RS, Ismail IH, Campbell S, Hendzel MJ. 2011. Polycomb group proteins in the DNA damage response: a link between radiation resistance and ‘stemness’. Cell Cycle 10: 883–894. [DOI] [PubMed] [Google Scholar]
- Glinsky GV, Berezovska O, Glinskii AB. 2005. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 115: 1503–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann AH, Yoo JH, Clancy J, Sorensen LK, Sedgwick A, Tong Z, Ostanin K, Rogers A, Grossmann KF, Tripp SR, et al. 2013. The small GTPase ARF6 stimulates β-catenin transcriptional activity during WNT5A-mediated melanoma invasion and metastasis. Sci Signal 6: ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Goding CR. 2010. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res 23: 746–759. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, et al. 2006. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res 19: 290–302. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. 2008. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res 68: 650–656. [DOI] [PubMed] [Google Scholar]
- Holderfield M, Deuker MM, McCormick F, McMahon M. 2014. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer 14: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Erin Chen Y, Kumar R, Taylor M, Jenny Njauw CN, Miao B, Frederick DT, Wargo JA, Flaherty KT, Jonsson G, et al. 2015. MITF modulates therapeutic resistance through EGFR signaling. J Invest Dermatol 135: 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, Scott KL, Kwong LN, Xiao Y, Cordon-Cardo C, et al. 2010. Integrative genome comparison of primary and metastatic melanomas. PLoS One 5: e10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Delmas V. 2006. The WNT/β-catenin pathway in melanoma. Front Biosci 11: 733–742. [DOI] [PubMed] [Google Scholar]
- Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C. 2013. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29: 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. 2003. Polycomb group genes as epigenetic regulators of normal and leukemic hemopoiesis. Exp Hematol 31: 567–585. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Tetzlaff MT, Cui R, Xu X. 2012a. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am J Pathol 181: 1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu F, Yu H, Zhao X, Sashida G, Deblasio A, Harr M, She QB, Chen Z, Lin HK, et al. 2012b. Akt phosphorylates the transcriptional repressor bmi1 to block its effects on the tumor-suppressing ink4a-arf locus. Sci Signal 5: ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sanchez-Tillo E, Lu X, Huang L, Clem B, Telang S, Jenson AB, Cuatrecasas M, Chesney J, Postigo A, et al. 2014. The ZEB1 transcription factor acts in a negative feedback loop with miR200 downstream of Ras and Rb1 to regulate Bmi1 expression. J Biol Chem 289: 4116–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero OM, Dawson DW, Moon RT, Chien AJ. 2010. A re-evaluation of the ‘oncogenic’ nature of Wnt/β-catenin signaling in melanoma and other cancers. Curr Oncol Rep 12: 314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhunapantula SV, Mosca PJ, Robertson GP. 2011. The Akt signaling pathway: an emerging therapeutic target in malignant melanoma. Cancer Biol Ther 12: 1032–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard MA, Ferretti R, Hilgendorf KI, Perret C, Whyte P, Lees JA. 2014. Bmi1 is required for tumorigenesis in a mouse model of intestinal cancer. Oncogene 33: 3742–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen P, Puisieux A. 2006. Metastasis: a question of life or death. Nat Rev Cancer 6: 449–458. [DOI] [PubMed] [Google Scholar]
- Mihic-Probst D, Kuster A, Kilgus S, Bode-Lesniewska B, Ingold-Heppner B, Leung C, Storz M, Seifert B, Marino S, Schraml P, et al. 2007. Consistent expression of the stem cell renewal factor BMI-1 in primary and metastatic melanoma. Int J Cancer 121: 1764–1770. [DOI] [PubMed] [Google Scholar]
- Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Foppen MH, et al. 2014. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun 5: 5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. 2010. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MP, Fiori JL, Xu M, Carter AD, Frank BP, Camilli TC, French AD, Dissanayake SK, Indig FE, Bernier M, et al. 2010. The orphan tyrosine kinase receptor, ROR2, mediates Wnt5A signaling in metastatic melanoma. Oncogene 29: 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Morrison SJ, Clarke MF. 2004. Bmi1, stem cells, and senescence regulation. J Clin Invest 113: 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmenter TJ, Kleinschmidt M, Kinross KM, Bond ST, Li J, Kaadige MR, Rao A, Sheppard KE, Hugo W, Pupo GM, et al. 2014. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov 4: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbatino F, Wang Y, Wang X, Flaherty KT, Yu L, Pepin D, Scognamiglio G, Pepe S, Kirkwood JM, Cooper ZA, et al. 2014. PDGFRα up-regulation mediated by sonic hedgehog pathway activation leads to BRAF inhibitor resistance in melanoma cells with BRAF mutation. Oncotarget 5: 1926–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, Min C, Jaskelioff M, Xiao Y, Wu CJ, et al. 2011. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell 20: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and Bioconductor. (ed. Gentleman R, et al. ), pp. 397–420. Springer, New York. [Google Scholar]
- Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, Kong QL, Xu LH, Zhang X, Liu WL, et al. 2009. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest 119: 3626–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yao Y, Liu B, Lin Z, Lin L, Yang M, Zhang W, Chen W, Pan C, Liu Q, et al. 2012. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene 31: 432–445. [DOI] [PubMed] [Google Scholar]
- Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, Haanen J, Blank C, Wesseling J, Willems SM, et al. 2014. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508: 118–122. [DOI] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. 2004. Stem cells and cancer; the polycomb connection. Cell 118: 409–418. [DOI] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. 2009. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 11: 1487–1495. [DOI] [PubMed] [Google Scholar]
- Widmer DS, Cheng PF, Eichhoff OM, Belloni BC, Zipser MC, Schlegel NC, Javelaud D, Mauviel A, Dummer R, Hoek KS. 2012. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res 25: 343–353. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, et al. 2006. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst 98: 472–482. [DOI] [PubMed] [Google Scholar]
- Xu L, Shen SS, Hoshida Y, Subramanian A, Ross K, Brunet JP, Wagner SN, Ramaswamy S, Mesirov JP, Hynes RO. 2008. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res 6: 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK, et al. 2010. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol 12: 982–992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.