Abstract
The expressions and activities of hepatic drug-metabolizing enzymes and transporters (DMETs) are altered during infection and inflammation. Inflammatory responses in the liver are mediated primarily by Toll-like receptor (TLR)-signaling, which involves recruitment of Toll/interleukin (IL)-1 receptor (TIR) domain containing adaptor protein (TIRAP) and TIR domain containing adaptor inducing interferon (IFN)-β (TRIF) that eventually leads to induction of proinflammatory cytokines and mitogen-activated protein kinases (MAPKs). Lipopolysaccharide (LPS) activates the Gram-negative bacterial receptor TLR4 and polyinosinic:polycytidylic acid (polyI:C) activates the viral receptor TLR3. TLR4 signaling involves TIRAP and TRIF, whereas TRIF is the only adaptor protein involved in the TLR3 pathway. We have shown previously that LPS-mediated downregulation of DMETs is independent of TIRAP. To determine the role of TRIF, we treated TRIF+/+ and TRIF−/− mice with LPS or polyI:C. LPS downregulated (∼40%–60%) Cyp3a11, Cyp2a4, Ugt1a1, Mrp2 mRNA levels, whereas polyI:C downregulated (∼30%–60%) Cyp3a11, Cyp2a4, Cyp1a2, Cyp2b10, Ugt1a1, Mrp2, and Mrp3 mRNA levels in TRIF+/+ mice. This downregulation was not attenuated in TRIF−/− mice. Induction of cytokines by LPS was observed in both TRIF+/+ and TRIF−/− mice. Cytokine induction was delayed in polyI:C-treated TRIF−/− mice, indicating that multiple mechanisms mediating polyI:C signaling exist. To assess the role of MAPKs, primary hepatocytes were pretreated with specific inhibitors before treatment with LPS/polyI:C. We found that only the c-jun-N-terminal kinase (JNK) inhibitor attenuated the down-regulation of DMETs. These results show that TRIF-independent pathways can be involved in the downregulation of DMETs through TLR4 and 3. JNK-dependent mechanisms likely mediate this downregulation.
Introduction
Inflammation and infection have been known to downregulate the expression and activity of drug-metabolizing enzymes and transporters (DMETs) and cause changes in the clearance of drugs; however, the exact mechanism is unclear (Morgan, 1997; Renton, 2000; Renton and Nicholson, 2004). Toll-like receptors (TLRs) are the major mediators of inflammatory responses in the liver. TLRs promote initial host defense against microorganisms by recognizing distinct microbial/viral components as pathogen-associated molecular patterns (Rock et al., 1998). Upon recognition of pathogen-associated molecular patterns, TLR signaling induces potent innate immune responses that signal through adaptor molecules, including myeloid differentiation factor 88 (MyD88), Toll/interleukin (IL)-1 receptor (TIR) domain containing adaptor protein (TIRAP), and TIR domain containing adaptor inducing interferon (IFN)-β (TRIF). TLRs promote proinflammatory signaling by activating downstream signaling pathways such as the nuclear factor (NF)-κB, mitogen-activated protein kinases (MAPKs), c-Jun-N-terminal kinase (JNK), p38, and extracellular related protein kinase (ERK) pathways in the liver (Abdulla et al., 2005).
The TLRs are present on nonparenchymal cells, including Kupffer cells, as well as on hepatocytes in the liver (Akira et al., 2001; Liu et al., 2002; Matsumura et al., 2003). We have shown that activation of TLR4 by lipopolysaccharide (LPS; the Gram-negative bacterial component) leads to downregulation of gene expression and activity of DMETs (Ghose et al., 2008, 2009; Shah et al., 2014). LPS-treated rodents are well established models of inflammation, and LPS treatment induces proinflammatory cytokines, interleukin (IL)-6, IL-1β, and tumor necrosis factor (TNF)α. These cytokines act on hepatocytes to reduce the expression of DMET genes (Renton, 2004; Aitken et al., 2006). TLR3 detects viral double-stranded RNA in the endolysosome. TLR3 is involved in the recognition of polyinosinic:polycytidylic acid (polyI:C), a synthetic double-stranded RNA analog (Alexopoulou et al., 2001); however, the role of TLR3-mediated regulation of DMETs has not yet been extensively studied. Although TRIF is the only adaptor protein involved in mediating TLR3-signaling, TLR4-signaling can be mediated by TRIF or TIRAP. TRIF is important for induction of interferon (IFN)-regulating factor 3, production of IFN-β, and activation and maturation of dendritic cells (Yamamato et al., 2003). TRIF is involved in mediating nuclear factor (NF)-κB activation in response to TLR3 ligands and is involved in the MyD88-independent prolonged NF-κB activation in response to TLR4 ligands (Yamamato et al., 2003). Whereas we found no role of TIRAP in TLR4-mediated DMETs downregulation and cytokine induction in the liver, the plasma cytokine induction by LPS seems to be mediated by TIRAP (Ghose et al., 2011).
The goal of this investigation was to determine the role of TRIF in TLR4- and TLR3-mediated downregulation of DMETs (Scheme 1). We observed that LPS administration led to significant downregulation (∼40%–60%) of gene expression of key DMEs, including Cyp3a11, Cyp2a4, Ugt1a1, and Mrp2 in TRIF+/+, as well as TRIF−/− mice. Induction of proinflammatory cytokines IL-1β, IL-6, and TNFα was not attenuated in LPS-treated TRIF−/− mice. Upon treatment of TRIF+/+ mice with polyI:C, we observed significant downregulation (∼30%–60%) of gene expression of Cyp3a11, Cyp2a4, Cyp2b10, Ugt1a1, Sult1a1, Mrp2, and Mrp3. Surprisingly, the downregulation of DMETs by polyI:C was not attenuated in TRIF−/− mice. PolyI:C administration also leads to induction of gene expression of proinflammatory cytokines IL-1β, IL-6, and TNFα at 1 and 4 hours; however, this induction was not observed at 1 hour, but only at 4 hours, in TRIF−/− mice. This delayed induction indicates that alternate pathways may be involved in recognizing polyI:C-mediated responses.
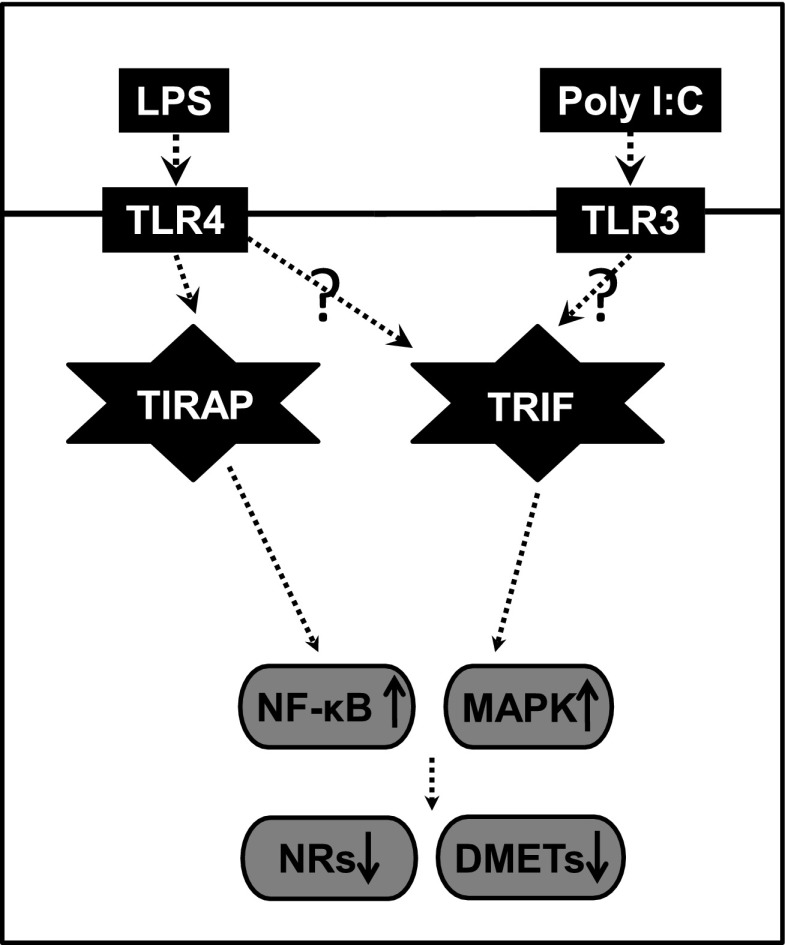
Scheme 1.
Inflammation-associated cell-signaling pathways in the liver. Activation of TLR4 and TLR3 recruits the adaptor proteins (TIRAP and TRIF) to induce downstream signaling, which eventually leads to alterations in gene expression of DMETs.
To study the role of MAPKs in regulating gene expression of DMETs, primary hepatocytes were treated with specific inhibitors of JNK, p38, and ERK before treatment with LPS or polyI:C. We found that inhibition of JNK led to complete attenuation of LPS or polyI:C-mediated downregulation of gene expression of DMETs. Our results indicate that the downregulation of DMETs during TLR4- and TLR3-induced inflammation can occur through TRIF-independent pathways and in all likelihood involves JNK-dependent signaling pathways.
Materials and Methods
Materials.
Highly purified LPS (Escherichia coli) and polyI:C (high-molecular-weight) were purchased from InvivoGen (San Diego, CA) and freshly diluted to the required concentration in 0.9% saline. The sequences of the primers and probes were obtained from the literature as reported previously (Shah et al., 2014). All oligonucleotides were purchased from Sigma Genosys (Houston, TX), and all real-time PCR reagents were purchased from Applied Biosystems (Foster City, CA). Anti-JNK and anti-phospho-JNK were purchased from Cell-Signaling (Beverly, MA). Anti-IκB-α antibody (sc-371) was purchased from Santa Cruz Biotechnology (San Diego, CA). JNK inhibitor (SP600125), ERK inhibitor (PD98059), and p38 inhibitor (SB203580) were purchased from Calbiochem (Gibbstown, NJ). Cell culture media and media supplements were purchased from Gibco BRL (Gaithersburg, MD). Unless specified, all other materials were purchased from Sigma-Aldrich (St. Louis, MO).
Animals.
Adult male C57BL/6 mice (∼6–8 weeks old) weighing 20–25 g were obtained from Harlan Laboratories (Houston, TX). Adult male TRIF+/+ and TRIF−/− mice (∼6–8 weeks old) were obtained from Jackson Laboratories (Bar Harbor, ME). We did not notice any overt phenotype in our experiments with TRIF−/− mice. The animals were maintained in a temperature- and humidity-controlled environment and were provided with water and rodent chow ad libitum. All animal experimental and surgical procedures were approved by the Institutional Animal Care and Use Committee.
Treatments.
The mice were injected i.p. with saline, polyI:C (5 mg/kg b.wt.) or LPS (2 mg/kg b.wt.) for 1, 4, 8, and 16 hours. Livers were harvested, cryopreserved in liquid nitrogen, and stored at −80°C for further studies. All experiments were performed in triplicate and repeated three times. The in vivo doses for polyI:C and LPS were found to be nontoxic as evidenced by unchanged liver toxicity markers; aspartate transaminase and alanine transaminase.
Real-Time PCR.
Total RNA was isolated from mouse liver using TRIzol reagent (Sigma-Aldrich) according to the manufacturer’s protocol. cDNA was synthesized using the High Capacity Reverse Transcription Kit from Applied Biosystems. Real-time PCR was performed using an ABI PRISM 7300 Sequence Detection System instrument and software (Applied Biosystems) as described previously (Shah et al., 2014). In short, each 25-µl reaction mixture contained 50–100 ng of cDNA, 300 nM forward primer, 300 nM reverse primer, 200 nM fluorogenic probe, and 15 μl of TaqMan Universal PCR Master Mix. We extrapolated the quantitative expression values from standard curves, and these values were normalized to cyclophilin.
Primary Hepatocyte Isolation and Treatment.
Primary mouse hepatocytes were isolated from C57BL/6 mice using the two-step collagenase perfusion technique as described previously (Li et al., 2002; Ghose et al., 2011; Shah et al., 2014). In short, after digestion, the liver was excised and the hepatocytes were purified using a Percoll gradient (33%), washed and screened for viability using trypan blue exclusion technique. Only isolations with viability greater than 90% were used for these studies. Cells were plated at a density of 500,000 cells/well in six-well Primaria plates (BD Biosciences; San Diego, CA) and allowed to attach for 4 hours. Cells were maintained for 48 hours with daily change of medium.
The cells were incubated with serum-free Williams medium E 2 hours before treatment with 50 μg/ml polyI:C (8 hours) or 1 μg/ml LPS (16 hours). To identify the role of MAPKs, the cells were pretreated with specific inhibitors 1 hour before treatment with polyI:C or LPS. RNA was then isolated for real-time PCR analysis as described already herein. The in vitro doses for polyI:C and LPS were found to be nontoxic as evidenced by unchanged liver toxicity markers aspartate transaminase and alanine transaminase and MTT assays in hepatocytes.
Immunoblotting.
Whole-cell extracts were prepared as described previously (Ghose et al., 2004), and the protein concentration was determined using the bicinchoninic acid (BCA) assay according to the manufacturer’s protocol (Pierce, Rockford, IL). Equal amounts of protein (30 μg) were analyzed by SDS-PAGE and transferred onto a nitrocellulose membrane. The membranes were probed with anti-JNK (1:500), anti-P-JNK (1:500), or IκB-α (1:1000) antibody, followed by probing with a goat anti-rabbit IgG-AP secondary antibody (1:2000). The membranes were then washed and incubated with Tropix CDP star nitroblock II TM ECL reagent per the manufacturer’s instructions (Applied Biosystems). The membranes were analyzed using FlourChem FC imaging system (Alpha Innotech, San Leandro, CA).
Statistical Analysis.
Treatment groups were compared using two-way analysis of variance, followed by a post hoc test (Bonferroni’s post hoc test) with P < 0.05. The results were presented as mean ± S.D
Results
Role of TRIF in Downregulation of Gene Expression of DMETs by LPS.
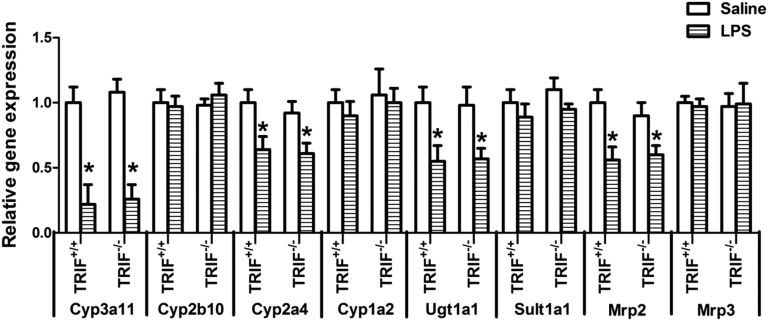
Administration pf LPS leads to downregulation of DME genes, and this downregulation is independent of TIRAP (Ghose et al., 2008). To investigate the role of TRIF in LPS-mediated downregulation of DMEs, we treated TRIF+/+ and TRIF−/− mice with LPS (Fig. 1). We found that mRNA levels of Cyp3a11 (∼80%), Cyp2a4 (∼35%), Ugt1a1 (∼45%), and Mrp2 (∼45%) were significantly downregulated in LPS-treated TRIF+/+ mice; however, this downregulation was not attenuated in TRIF−/− mice, indicating redundancy in TLR4 signaling pathways.
Fig. 1.
Regulation of DME mRNA levels in TRIF+/+ and TRIF−/− mice after LPS administration. TRIF+/+ and TRIF−/− were injected i.p. with saline or LPS (2 mg/kg), and livers were harvested at 16 hours (n = 5 or 6/group). RNA was isolated from the livers, and mRNA levels of Cyp3a11 and Ugt1a1 were determined by real-time PCR analysis as described earlier. All data are presented as ±S.D. and standardized for cyclophilin mRNA levels. Expression in saline-treated TRIF+/+ mice was set to 1, and the -fold change after LPS-treatment was compared with that of the saline-treated controls. *Significant difference (P < 0.05) between saline and LPS groups. The experiments were repeated three times.
Role of TRIF in Regulation of Hepatic Cytokine mRNA Levels by LPS.
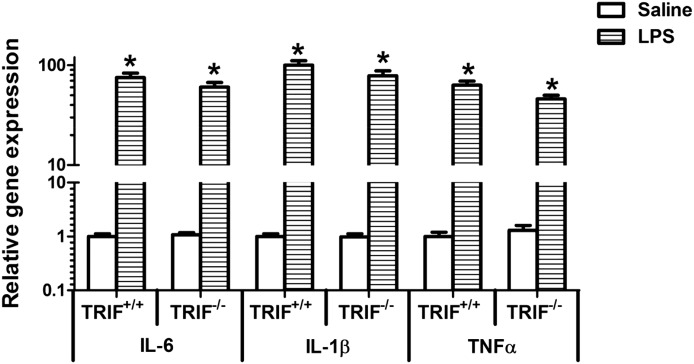
Since cytokines are involved in the downregulation of hepatic DMET genes during inflammation, we determined the changes in gene expression of proinflammatory cytokines (Fig. 2). As expected, there was significant induction of IL-1β, IL-6, and TNFα mRNA levels in the livers of LPS-treated TRIF+/+ mice; however, no attenuation of this induction was observed in LPS-treated TRIF−/− mice. We have shown previously that TLR4-mediated induction of cytokines in the liver is independent of the adaptor protein TIRAP (Ghose et al., 2008). Our results indicate that TLR4 signaling can proceed through TIRAP or TRIF, suggesting redundancy in the signaling pathway.
Fig. 2.
Regulation of cytokine mRNA levels in TRIF+/+ and TRIF−/− mice after LPS administration. TRIF+/+ and TRIF−/− mice were injected i.p. with saline or LPS (2 mg/kg), and livers were harvested at 1 hour (n = 5 or 6/group). RNA was isolated from the livers, and mRNA levels of IL-1β, IL-6, and TNFα were determined by real-time PCR analysis as described earlier. All data are presented as ±S.D. and standardized for cyclophilin mRNA levels. Expression in saline-treated TRIF+/+ mice was set to 1, and the -fold change after LPS-treatment was compared with that of the saline-treated controls. *Significant difference (P < 0.05) between saline and LPS groups. The experiments were repeated three times.
Regulation of Gene Expression of DMETs in PolyI:C-Treated C57BL/6 Mice.
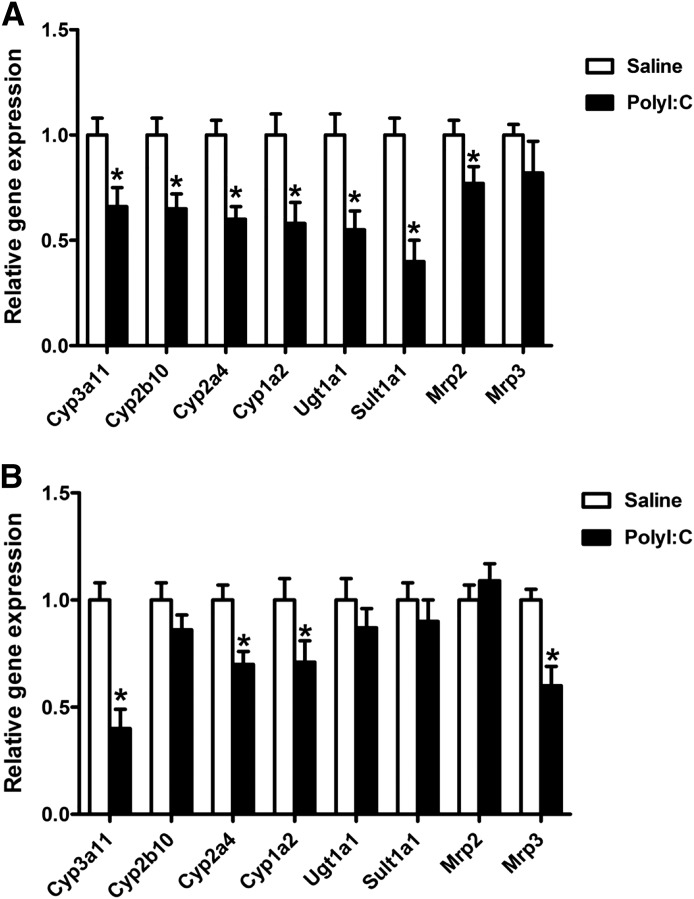
Our next goal was to determine the role of the TLR3 ligand, polyI:C, in regulating gene expression of DMETs; this would provide the basis for further investigation into the role of TRIF. Therefore, we treated C57BL/6 mice with 5 mg/kg of polyI:C for 8 and 16 hours (Fig. 3). We show for the first time that activation of TLR3 by polyI:C leads to downregulation of several DMET genes. mRNA levels of the key phase 1 enzyme Cyp3a11 were significantly downregulated at 8 hours (∼35%) and 16 hours (∼60%) in C57BL/6 mice. The mRNA levels of Cyp1a2 and Cyp2a4 (∼30%–40%) were significantly downregulated at 8 and 16 hours. Cyp2b10 mRNA levels were downregulated (∼35%) at 8 hours. The mRNA levels of key phase 2 genes Ugt1a1 and Sult1a1 were downregulated significantly (∼40% and ∼60%, respectively) at 8 hours. The mRNA levels of key transporters Mrp2 and Mrp3 were significantly (∼30%–35%) downregulated at 8 and 16 hours, respectively.
Fig. 3.
Regulation of DMET mRNA levels in C57BL/6 mice by poly I:C. C57BL/6 mice were injected i.p. with saline or polyI:C (5 mg/kg), and livers were harvested at 8 hours (A) and at 16 hours (B) (n = 5 or 6/group). RNA was isolated from the livers and mRNA levels of phase 1 and phase 2, and transporter genes were determined by real-time PCR analysis as described earlier. All data are presented as ±S.D. and standardized for cyclophilin mRNA levels. Expression in saline-treated mice was set to 1, and the -fold change after polyI:C-treatment was compared with that of the saline-treated controls. *Significant difference (P < 0.05) between saline and poly I:C groups. The experiments were repeated three times.
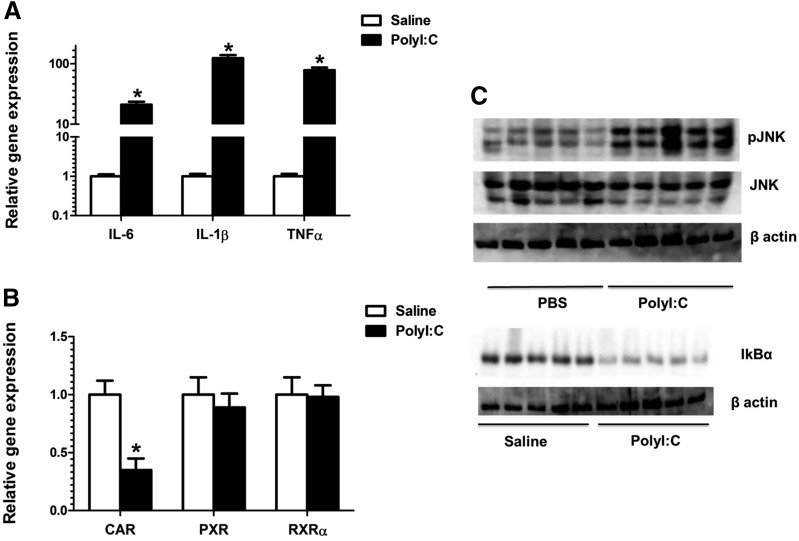
Activation of Cytokines, Nuclear Receptors (NRs) and Cell-Signaling Pathways by PolyI:C.
We have previously shown that activation of TLR2 and TLR4 leads to the induction of proinflammatory cytokines IL-1β, IL-6, and TNFα (Ghose et al., 2008, 2009). JNK and NF-κB are known to be critical components of the TLR signaling pathway (Kawai and Akira, 2005), and the alterations of hepatic DMETs in LPS-induced inflammation involves cross-talk between cell-signaling components and nuclear receptors (NRs). We have also shown that activation of TLR2 and TLR4 leads to the activation of JNK and NF-κB and alterations in the expression of NRs (Ghose et al., 2008, 2009). It is not clear, however, whether activation of polyI:C will lead to activation of these signaling components and whether they play a role in mediating the effects on DMETs. We show significant induction of mRNA levels of proinflammatory cytokines IL-1β (∼120-fold), IL-6 (∼25-fold), and TNFα (∼80-fold) in the livers of polyI:C-treated C57BL/6 mice (Fig. 4A). There was downregulation of the gene expression of CAR (∼60%) and RXRα (∼25%) at 8 and 16 hours, respectively, where as a slight upregulation of gene expression of PXR at 16 hours was seen (Fig. 4B). We also observed activation of phosphorylated JNK (P-JNK) in whole-cell extracts (Fig. 4C). Activation of NF-κB was determined by degradation of the inhibitory subunit IκBα after polyI:C treatment (Fig. 4C). We see activation of NF-κB at 1 hour, indicating that these signaling components are involved in mediating TLR3-signaling pathways.
Fig. 4.
Regulation of cell-signaling components on activation of TLR3. (A) Cytokines: C57BL/6 mice were injected i.p. with saline or polyI:C (5 mg/kg), and livers were harvested at 1 hour (n = 5 or 6/group). RNA was isolated from the livers, and mRNA levels of IL-1β, IL-6, and TNFα were determined by real-time PCR analysis as described earlier. (B) Nuclear receptors: C57BL/6 mice were injected i.p. with saline or polyI:C (5 mg/kg), and livers were harvested at 8 and 16 hours (n = 5 or 6/group). RNA was isolated from the livers, and mRNA levels of CAR, PXR, and RXRα were determined by real-time PCR analysis as described earlier. All data are presented as ±S.D. and standardized for cyclophilin mRNA levels. Expression in saline-treated mice was set to 1, and the -fold change after polyI:C treatment was compared with that of the saline-treated controls. *Significant difference (P < 0.05) between saline and polyI:C groups. The experiments were repeated three times. (C) Cell-signaling pathways: Whole-cell extracts were prepared from the livers of saline and polyI:C-treated mice at 1 hour, and the samples were analyzed by immunoblotting. The phosphorylated forms of JNK (P-JNK) and degradation of IkBα were measured as markers of JNK and NF-κB activation, respectively. The experiments were repeated three times.
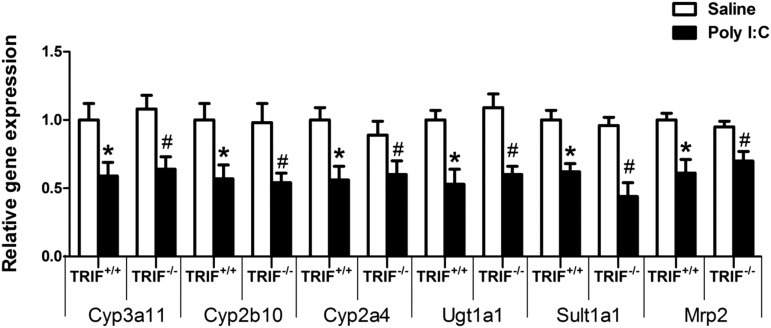
Role of TRIF in Regulation of Gene Expression of DMETs by PolyI:C.
Since we have shown that the TLR3 ligand, polyI:C downregulates DMET genes, our next objective was to determine the role of TRIF in polyI:C-mediated regulation of DMET genes. Maximal downregulation of DMETs was observed at 8 hours on treatment with polyI:C; hence, we chose the 8-hour time point for this study (Fig. 5). We found that the RNA levels of Cyp3a11 (∼40%), Cyp2a4 (∼40%), Cyp2b10 (∼45%), Ugt1a1 (∼50%), Sult1a1 (∼50%), and Mrp2 (∼30%) were significantly downregulated in TRIF+/+ as well as TRIF−/− mice at 8 hours, and the extent of downregulation was similar. These results suggest that the downregulation of DMETs through TLR3 was independent of TRIF.
Fig. 5.
Regulation of DMET mRNA levels in TRIF+/+ and TRIF−/− mice after polyI:C administration. TRIF+/+ and TRIF−/− mice were injected i.p. with saline or polyI:C (5 mg/kg), and livers were harvested at 8 hours (n = 5 or 6/group). RNA was isolated from the livers and mRNA levels of phase 1, phase 2, and transporter genes were determined by real-time PCR analysis as described earlier. All data are presented as ±S.D. and standardized for cyclophilin mRNA levels. Expression in saline-treated TRIF+/+ mice was set to 1, and the -fold change after polyI:C-treatment was compared with the saline-treated controls. *Significant difference (P < 0.05) between saline and polyI:C group in TRIF+/+ mice; #Significant difference (P < 0.05) between saline and polyI:C group in TRIF−/− mice. The experiments were repeated three times.
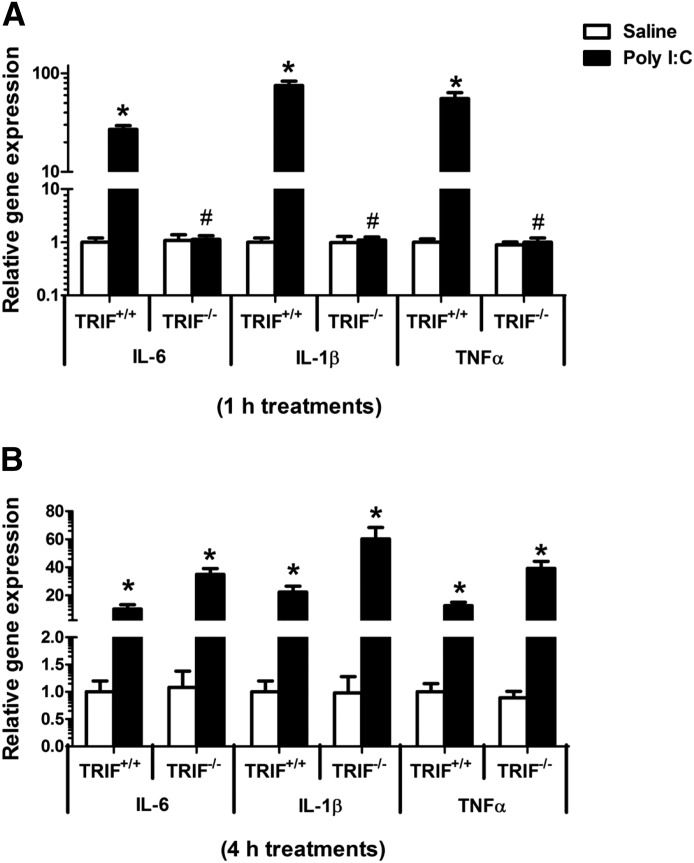
Role of TRIF in the Regulation of Hepatic Cytokine mRNA Levels by PolyI:C.
Induction of proinflammatory cytokines IL-1β, IL-6, and TNFα in TRIF+/+ mice was completely suppressed in polyI:C-treated TRIF−/− mice at 1 hour (Fig. 6A). Since cytokines have been implicated in altering the gene expression of DMETs, this finding was surprising. On further investigation, we found significant induction of cytokines at 4 hours (IL-1β, ∼60-fold; IL-6, ∼35-fold; and TNFα; ∼40-fold) in polyI:C-treated TRIF−/− mice (Fig. 6B). There is a delayed response in the induction of cytokines in TRIF−/− mice, suggesting that polyI:C can potentially mediate viral responses through multiple mechanisms.
Fig. 6.
Regulation of hepatic cytokine mRNA levels in TRIF+/+ and TRIF−/− mice after poly I:C administration. TRIF+/+ and TRIF−/− mice (n = 5/group) were injected i.p. with saline or polyI:C (5 mg/kg), and livers were harvested at 1 hour (A) and 4 hours (B). RNA was isolated from the livers, and mRNA levels of IL-1β, IL-6, and TNFα were determined by real-time PCR analysis as described earlier. All data are presented as ±S.D. and standardized for cyclophilin mRNA levels. Expression in saline-treated TRIF+/+ mice was set to 1, and the -fold change after polyI:C-treatment was compared with that of the saline-treated controls. *Significant difference (P < 0.05) between saline and polyI:C groups. #Significant difference in polyI:C-treated TRIF+/+ and TRIF−/− mice. The experiments were repeated three times.
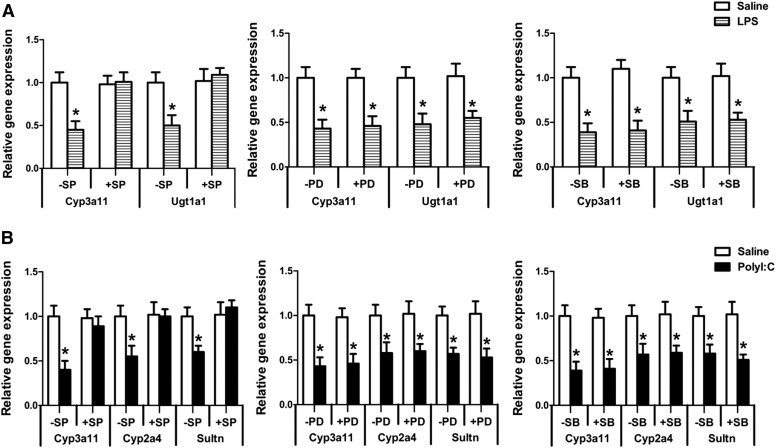
Roles of MAPKs in TLR4- or TLR3-Mediated Downregulation of DMETs.
We studied the role of NF-κB in regulating DMETs during TLR3 and 4 activation by using specific NF-kB inhibitor NAI [6-amino-4-(4-phenoxyphenylethylamino)quinazoline] in primary mouse hepatocytes (data not shown). We found that inhibiting activation of NF-κB did not attenuate downregulation of DMET genes, which is why we chose to focus on the MAPKs. MAPKs have been implicated in altering the gene expression of DMETs during inflammation because of their cross-talk with NRs (Li et al., 2002; Ghose et al., 2004). To determine their roles in TLR4- and TLR3-mediated downregulation of DMETs, we isolated primary hepatocytes from C57BL/6 mice and treated them with chemical inhibitors of JNK (SP600125, 10 µM), ERK (PD89059, 20 µM), and p38 (SB203580, 25 µM), 1 hour before treatment with LPS (1 μg/ml; 16 hours) or polyI:C (50 μg/ml; 8 hours). Treatment of primary hepatocytes with LPS led to downregulation of gene expression of Cyp3a11 and Ugt1a1 (Fig. 7A). PolyI:C treatment led to downregulation of gene expression of Cyp3a11, Cyp2a4, and Sultn (Fig. 7B). This downregulation of DMEs was completely attenuated on treatment of specific JNK inhibitor, whereas ERK and p38 inhibitor had no effect in attenuating the LPS or polyI:C mediated downregulation of DMEs.
Fig. 7.
Regulation of DMEs by MAPKs on LPS or polyI:C treatments. Primary hepatocytes from C57BL/6 mice were treated with DMSO or JNK inhibitor (SP600125, 10 μM), ERK inhibitor (PD98059, 20 μM), or p38 inhibitor (SB203580, 25 μM) for 1 hour before treatment with saline or (A) LPS (1 μg/ml) for 16 hours or (B) polyI:C (50 μg/ml) for 8 hours. RNA was isolated, and real-time PCR was performed as described earlier. n = 5 or 6/group. All data are presented as ±S.D. and standardized for cyclophilin mRNA levels. *Significant difference (P < 0.05) between saline and LPS or polyI:C groups. The experiments were repeated three times.
Discussion
Infection and inflammation can alter the expression and activities of several DMETs (Morgan, 1997) which can have potentially harmful consequences in patients; therefore, it is extremely important to understand the mechanisms behind alterations of DMET during inflammation. We have shown that expression and activities of hepatic DMETs are altered in a Gram-negative and Gram-positive bacterial-inflammatory model (Ghose et al., 2009; Gandhi et al., 2012); however, the role of viral infections in altering DMETs has not been studied. We show for the first time that treatment of mice with the TLR3 ligand polyI:C leads to downregulation of gene expression of DMET, induction of proinflammatory cytokines, alterations in nuclear receptor levels, and activation of cell signaling components. We also show that TRIF-independent signaling pathways may be involved in the downregulation of DMETs by TLR3 or TLR4; however, in vitro results show that this downregulation is likely mediated by the MAPK, JNK.
Activation of TLR4 can recruit the downstream adaptor proteins, TIRAP or TRIF. Initial studies with TIRAP−/− mice have shown that TIRAP is important for the induction of cytokines by LPS in dendritic cells and peritoneally derived macrophages (Horng et al., 2002; Yamamoto et al., 2002); however, our studies with TIRAP−/− mice indicated that TIRAP is not essential for down-regulation of hepatic DMETs through TLR4 signaling (Ghose et al., 2008). In this study, we show that LPS-mediated induction of proinflammatory cytokines and downregulation of DMETs occurs in TRIF−/− mice, indicating redundancy in TIRAP or TRIF-dependent signaling pathways. Experiments in TIRAP/TRIF double-knockout mice will most likely block the effects of LPS on DMET genes.
PolyI:C administration led to downregulation of several DMET genes such as Cyp3a11, Cyp2a4, Cyp2b10, Cyp1a2, Ugt1a1, Sult1a1, Mrp2, and Mrp3 in TRIF+/+ mice at 8 and 16 hours. We show for the first time that TLR3 activation by viral components can downregulate hepatic DMET genes. We also show that activation of TLR3 by polyI:C leads to: (1) induction of proinflammatory cytokine, IL-1β, IL-6, and TNFα; (2) alterations of nuclear receptor levels of CAR, PXR, and RXRα and; (3) activation of signaling components like JNK and NF-κB. Since TRIF is the only adaptor protein involved in TLR3 signaling, we expected attenuation of downregulation of DMET genes in TRIF−/− mice. Surprisingly, the downregulation of DMETs on polyI:C treatment was also observed in TRIF−/− mice. Since cytokines have been implicated in altering gene expression of DMETs during inflammation, we expected no changes in the induction pattern of proinflammatory cytokines in polyI:C-treated TRIF−/− mice. Surprisingly, the induction of hepatic IL-1β, IL-6, and TNFα mRNA levels was completely abolished in polyI:C-treated TRIF−/− mice at 1 hour. Further investigation showed that this induction is delayed, and there is significant induction of these cytokines in livers of TRIF−/− mice after 4 hours of polyI:C treatment. This finding indicates that in the absence of TRIF, polyI:C might be activating other signaling pathways to induce cytokine expression in the liver.
Retinoic acid-inducible gene 1 (RIG-I) and melanoma differentiation-associated protein 5 contain caspase-recruiting domains (CARDs) and can detect intracellular viral products such as genomic RNA to signal IRF3, IRF7, and NF-κB (Yoneyama et al., 2004; Onoguchi et al., 2007). These receptors are important in sensing dsRNA because dendritic cells or fibroblast cells lacking in TLR3 secrete type I IFNs on exposure to dsRNA (Matsumoto et al., 2003). RIG-I and melanoma differentiation-associated protein 5 have been implicated in mediating antiviral responses via the IFN-β promoter stimulator (IPS-1 or CARDIF) adaptor protein (Kawai et al., 2005). Previous studies have shown that induction of interferons was attenuated and delayed in TRIF−/− mice treated with polyI:C (Kumar et al., 2008). Cytokine production was totally inhibited only in IPS-1 and TRIF-double knockout mice, indicating that both TRIF and IPS-1 play important roles in mediating anti-viral responses. This evidence indicates that in our studies, the redundancy of downstream pathways (IPS-1 and TRIF) is likely to be responsible for DMET downregulation by polyI:C in TRIF−/− mice. Studies in polyI:C-treated IPS-1/TRIF double-knockout mice will likely attenuate the downregulation of DMETs during viral infections.
Inflammation and infection lead to alterations in the activity of a variety of target genes and transcription factors through several signal transduction cascades, including the three main MAPKs: JNK, ERK, and p38. To investigate the roles of JNK, ERK and p38 in regulating DMET genes during Gram-negative and viral infections, we treated primary hepatocytes with specific inhibitors 1 hour before treatment with LPS or polyI:C. We found that the TLR4- and TLR3-mediated downregulation of DMETs was completely attenuated by the JNK inhibitor, whereas ERK and p38 inhibitors had no effects on attenuating the downregulation of DMETs. JNK is found in three isoforms and the JNK inhibitor we used inhibited all three isoforms (Bennett et al., 2001). JNK1 and JNK2 are present in most tissues, and JNK3 is found almost exclusively in the brain. JNK1/JNK2 double-knockout mice are embryonically lethal, and JNK1 knockout and JNK2 knockout mice show redundancy in mediating the effects of LPS in the liver (Kosters et al., 2009). Experiments with polyI:C in JNK1 and JNK2 knockout mice will help identify which isoform plays an important role in regulating DMET genes during viral infections.
Our study is the first of its kind to show that activation of TLR3 will lead to downregulation of gene expression of several DMETs. The downregulation of DMETs and transporters during inflammation and infection is a complex process, and the molecular mechanisms are not fully understood. We show that TLR4- and TLR3-mediated downregulation of DMET genes can occur through TRIF-independent pathways, indicating redundancy in the signaling mediated by downstream adaptor proteins; however, the MAPK JNK is a major player in regulating DMET genes during infections caused by Gram-negative and viral components. Although a safe and potent human JNK inhibitor is yet to be discovered, our in vitro studies indicate that JNK-dependent phosphorylation processes could be targeted to reverse or attenuate inflammation- mediated effects on drug metabolism.
Abbreviations
- DMET
drug-metabolizing enzymes and transporters
- ERK
extracellular related protein kinase
- IFN-β
interferon β
- IL
interleukin
- JNK
c-jun-N-terminal kinase
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- NF
nuclear factor
- NRs
nuclear receptors
- PolyI:C
polyinosinic:polycytidylic acid
- RIG-I
retinoic acid-inducible gene 1
- TIR
toll/interleukin-1 receptor
- TLR
toll-like receptor
- TIRAP
toll/interleukin-1 receptor (TIR) domain containing adaptor protein
- TNF
tumor necrosis factor
- TRIF
TIR domain containing adaptor inducing IFN-β
Authorship Contributions
Participated in research design: Ghose, Shah.
Conducted experiments: Shah, Omoluabi.
Contributed new reagents and analytic tools: Ghose, Moorthy
Performed data analysis: Shah, Ghose
Wrote or contributed to the writing of the manuscript: Shah, Ghose, Moorthy
Footnotes
This work was supported in part by the National Institutes of Health [Grants R21DA035751-01A1, R01HL112516, R01ES 009132, and R01ES-019689] to B.M. and R.G.
References
- Abdulla D, Goralski KB, Garcia Del Busto Cano E, Renton KW. (2005) The signal transduction pathways involved in hepatic cytochrome P450 regulation in the rat during a lipopolysaccharide-induced model of central nervous system inflammation. Drug Metab Dispos 33:1521–1531. [DOI] [PubMed] [Google Scholar]
- Aitken AE, Richardson TA, Morgan ET. (2006) Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu Rev Pharmacol Toxicol 46:123–149. [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K, Kaisho T. (2001) Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2:675–680. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. (2001) Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738. [DOI] [PubMed] [Google Scholar]
- Bennett BL, Sasaki DT, Murray BW, O’Leary EC, Sakata ST, Xu W, Leisten JC, Motiwala A, Pierce S, Satoh Y, et al. (2001) SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA 98:13681–13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi AS, Guo T, Shah P, Moorthy B, Chow DS, Hu M, Ghose R. (2012) CYP3A-dependent drug metabolism is reduced in bacterial inflammation in mice. Br J Pharmacol 166:2176–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose R, Guo T, Haque N. (2009) Regulation of gene expression of hepatic drug metabolizing enzymes and transporters by the Toll-like receptor 2 ligand, lipoteichoic acid. Arch Biochem Biophys 481:123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose R, Guo T, Vallejo JG, Gandhi A. (2011) Differential role of Toll-interleukin 1 receptor domain-containing adaptor protein in Toll-like receptor 2-mediated regulation of gene expression of hepatic cytokines and drug-metabolizing enzymes. Drug Metab Dispos 39:874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose R, White D, Guo T, Vallejo J, Karpen SJ. (2008) Regulation of hepatic drug-metabolizing enzyme genes by Toll-like receptor 4 signaling is independent of Toll-interleukin 1 receptor domain-containing adaptor protein. Drug Metab Dispos 36:95–101. [DOI] [PubMed] [Google Scholar]
- Ghose R, Zimmerman TL, Thevananther S, Karpen SJ. (2004) Endotoxin leads to rapid subcellular re-localization of hepatic RXRalpha: a novel mechanism for reduced hepatic gene expression in inflammation. Nucl Recept 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng T, Barton GM, Flavell RA, Medzhitov R. (2002) The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature 420(6913):329-333. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. (2005) Toll-like receptor downstream signaling. Arthritis Res Ther 7:12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. (2005) IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. [DOI] [PubMed] [Google Scholar]
- Kosters A, White DD, Sun H, Thevananther S, Karpen SJ. (2009) Redundant roles for cJun-N-terminal kinase 1 and 2 in interleukin-1beta-mediated reduction and modification of murine hepatic nuclear retinoid X receptor alpha. J Hepatol 51:898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Koyama S, Ishii KJ, Kawai T, Akira S. (2008) Cutting edge: cooperation of IPS-1- and TRIF-dependent pathways in poly IC-enhanced antibody production and cytotoxic T cell responses. J Immunol 180:683–687. [DOI] [PubMed] [Google Scholar]
- Li D, Zimmerman TL, Thevananther S, Lee HY, Kurie JM, Karpen SJ. (2002) Interleukin-1 beta-mediated suppression of RXR:RAR transactivation of the Ntcp promoter is JNK-dependent. J Biol Chem 277:31416–31422. [DOI] [PubMed] [Google Scholar]
- Liu S, Gallo DJ, Green AM, Williams DL, Gong X, Shapiro RA, Gambotto AA, Humphris EL, Vodovotz Y, Billiar TR. (2002) Role of toll-like receptors in changes in gene expression and NF-kappa B activation in mouse hepatocytes stimulated with lipopolysaccharide. Infect Immun 70:3433–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, Yamamoto A, Seya T. (2003) Subcellular localization of Toll-like receptor 3 in human dendritic cells. J Immunol. 2003 171(6):3154–3162. [DOI] [PubMed] [Google Scholar]
- Matsumura T, Degawa T, Takii T, Hayashi H, Okamoto T, Inoue J, Onozaki K. (2003) TRAF6-NF-kappaB pathway is essential for interleukin-1-induced TLR2 expression and its functional response to TLR2 ligand in murine hepatocytes. Immunology 109:127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan ET. (1997) Regulation of cytochromes P450 during inflammation and infection. Drug Metab Rev 29:1129–1188. [DOI] [PubMed] [Google Scholar]
- Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. (2007) Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem 282:7576–7581. [DOI] [PubMed] [Google Scholar]
- Renton KW. (2004) Cytochrome P450 regulation and drug biotransformation during inflammation and infection. Curr Drug Metab 5:235–243. [DOI] [PubMed] [Google Scholar]
- Renton KW, Nicholson TE. (2000) Hepatic and central nervous system cytochrome P450 are down-regulated during lipopolysaccharide-evoked localized inflammation in brain. J Pharmacol Exp Ther 294:524–530. [PubMed] [Google Scholar]
- Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. (1998) A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA 95:588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Guo T, Moore DD, Ghose R. (2014). Role of constitutive androstane receptor in Toll-like receptor-mediated regulation of gene expression of hepatic drug-metabolizing enzymes and transporters. Drug Metab Dispos 42(1):172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, Hoshino K, Takeuchi O, Kobayashi M, Fujita T, et al. (2002) Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature 420:324–329. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. [DOI] [PubMed] [Google Scholar]